Abstract
Background
Glioblastoma is the most frequent and the most aggressive primary malignant brain tumor in adults. Standard treatment includes surgical removal of the tumor followed by concomitant chemotherapy and radiotherapy. Temozolomide, an oral alkylating agent, is currently the most commonly used chemotherapy. However, the median survival of glioblastoma multiforme (GBM) patients remains very low. Epidermal growth factor receptor variant III (EGFRvIII) is a novel marker for GBM patients of Indian origin as very few studies have been done on this molecular marker in our country. This is the first study utilizing this molecular marker among GBM patients in Rajasthan, India. This was a single institutional study that aimed to estimate the proportion of EGFRvIII mutation in GBM patients of Indian origin.
Methodology
This was a non-randomized, ambispective, single institutional observational study done on 35 brain tissue biopsies of histopathologically diagnosed and confirmed cases of GBM based on the World Health Organization 2007 Classification received in the pathology department of Dr. Sampurnanand Medical College, Jodhpur from 2015 to 2020 after applying inclusion and exclusion criteria. Molecular study of the EGFRvIII marker was conducted in all cases of GBM in the same institution on the RNA extracted from selected biopsy samples. Statistical analysis was performed using the SPSS version 22.0 software package (IBM Corp., Armonk, NY USA). The correlation between age and gender with EGFR-positive cases was analyzed, and EGFR positivity compared with previous studies.
Results
The occurrence of the EGFRvIII mutation was found to be 17.4% (6/35 cases). The mean age of presentation of a tumor with this mutation was estimated to be 54.3 years. Males were more commonly found to be affected (66.6%, 4/6 cases).
Conclusions
Thus, the identification of this mutation would segregate patients who may benefit from newer therapeutic approaches. In the future, personalized treatment may be advised for GBM patients depending on the presence of the EGFRvIII mutation.
Keywords: world health organization classification 2007, genetic pathways, rt-pcr, targeted therapy, egfr mutation variant iii, molecular markers
Introduction
The epidermal growth factor receptor (EGFR), also known as HER1 or ERBB1, is an important regulator of cellular growth in tissues of epithelial origin. The EGFR gene is the cellular homolog of the v-erbB oncogene originally identified in avian erythroblastosis viruses [1]. In glioblastoma, a particular group of EGFR deletions and point mutations are frequently found. These include EGFRvI (N-terminal deletion), vII (deletions of exons 14-15), vIII (deletions of exons 2-7), vIV (deletions of exons 25-27), and vV (deletions of exons 25-28), among which vII and vIII are oncogenic [1].
Among these mutations, EGFRvIII occurs most frequently with a global incidence of 10-50% [2]. EGFRvIII is 145 KDa in molecular weight [3]. Compared with the EGFR, EGFRvIII has an in-frame deletion of exons 2-7, resulting in a shorter extracellular domain. Compared to the wild-type EGFR, EGFRvIII lacks amino acids 6-273, and deletion of those 267 amino acids creates a junction site with a new glycine residue between amino acids 5 and 274 [4-6]. It mimics ligand binding effects and initiates conformational changes in receptors, leading to the activation of signaling pathways.
Being a constitutively active mutant of EGFR, EGFRvIII transduces signals via traditional EGFR pathways, namely, RAS/mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K)/Akt, and Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathways. Although EGFRvIII activates several downstream pathways, evidence shows that it preferentially activates the PI3K/Akt signal transduction pathway [6-9]. Recently, EGFRvIII has been found to activate the mammalian target of rapamycin complex 2 (mTORC2) (CREB-regulated transcription co-activator 2) via the PI3K/Akt pathway, which, in turn, leads to stimulation of the nuclear factor-kappa B (NF-κB) pathway, thus leading to resistance to chemotherapy [10]. Another consequence of EGFRvIII activation of PI3K/Akt is increased proliferation and cell cycle progression, which is mediated by a decrease in the level of p27KIP1, a cyclin-dependent kinase (CDK) inhibitor that binds and inactivates cyclin-dependent kinase 2 (CDK2)-cyclin E complexes, thus inhibiting the transition of cells from the G1 to the S phase [11]. The significance of PI3K/Akt activation by EGFRvIII has been confirmed by Klingler-Hoffmann et al., who showed in their study that treatment of U87MG.D2-7 cells with the PI3K inhibitor wortmannin, or by reconstitution of the physiological levels of phosphatase and tensin homolog (PTEN) (a negative regulator of PI3K), resulted in the discontinuation of the EGFRvIII-conferred growth advantage [12]. Selective activation of the PI3K/Akt pathway by EGFRvIII is thought to mediate the resistance to radiation that is observed in EGFRvIII-positive tumors [13-17].
Glioblastoma multiforme (GBM) is a highly proliferative, vascular, and locally invasive tumor [18-20]. Evidence suggests that EGFRvIII-transfected GBM cell lines have higher rates of proliferation, angiogenesis, increased ability to form tumor xenografts, reduced apoptosis, and higher invasiveness when compared to matched parental cell lines [11,21-26]. The fact that this mutated EGFR variant has been found in tumors and not in normal tissues has made it an interesting target for antitumor therapies [27]. In this study, we aimed to estimate the proportion of EGFRvIII mutations in GBM patients in the Indian population. This study will contribute to the framing of targeted therapies in the future for these patients, which may improve their survival rate.
Materials and methods
This study is an ambispective, single institutional observational study done on brain tissue biopsies of histopathologically diagnosed and confirmed cases of GBM based on the World Health Organization (WHO) 2007 Classification received in the Pathology Department of Dr. Sampurnanand Medical College, Jodhpur, from 2015 to 2020. The proposal for the study was reviewed thoroughly and approved by the Institutional Ethical Committee of Dr. Sampurnanand Medical College, Jodhpur (EC/MC/JU/2018/319) before the commencement of the study.
After applying the inclusion and exclusion criteria, as depicted in Table 1, paraffin-embedded tissue blocks of 35 cases of histopathologically confirmed GBM were retrieved. The epidemiological data of the selected cases were procured from the patient records maintained in the Pathology Department.
Table 1. Inclusion and exclusion criteria.
| Inclusion criteria | Exclusion criteria |
| All brain biopsies with histopathological diagnosis of glioblastoma multiforme from 2015 to 2020 | Extremely small biopsies (<5 mm) |
| Glioblastoma multiforme cases with single tissue block | |
| Biopsies of the brain with histopathological diagnoses other than glioblastoma multiforme |
Reverse transcription-polymerase chain reaction
Initially, excess paraffin was trimmed off the tissue block. Subsequently, 10-15 sections of 10 µ thickness were taken from each case, and the excess paraffin was trimmed using a sterile blade and placed in labeled Eppendorf tubes of 2 mL volume. RNA from these tissue sections was extracted using the ReliaPrepTM FFPE Total RNA Miniprep System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Initially, samples were deparaffinized using mineral oil, followed by sample lysis by adding lysis buffer and proteinase K. Then, the DNAase treatment mix was added to the sample. Further, isopropanol and buffer were incorporated for nucleic acid binding. A wash solution containing 95-100% ethanol was then used as a washing solution to obtain eluted RNA. It was stored at -30°C to -10°C in an elution tube. Extracted RNA concentration was analyzed on DS-11 from DeNovix (Wilmington, DE, USA). RNA samples with an adequate concentration were then processed for EGFRvIII detection using the TRUPCR EGFRvIII detection kit (3B Black Bio, Biotech India Pvt. Ltd.) according to the manufacturer’s protocol. An ABL1 primer-probe mix was taken as an internal control. The sample mixtures were then tested in the Microbiology Department (Dr. Sampurnanand Medical College) using reverse transcription-polymerase chain reaction (RT-PCR) (Bio-Rad, Hercules, CA, USA). The cycle threshold value (Ct value) was 33 for EGFRvIII mutation detection.
Statistical analysis was performed using the SPSS version 22.0 software package (IBM Corp., Armonk, NY, USA).
Results
Out of the total 35 cases, an EGFRvIII mutation was noted in 17.1% (6/35) cases (Figure 1). The mean age of presentation of EGFRvIII-positive cases was 54.3 years. Among positive cases, most were beyond 40 years of age (83.3%). Further, out of eight cases with an age less than 40 years, only one case was EGFRvIII positive (12.5%), while positivity was seen in five out of 27 cases beyond the 40-year age group (18.5%). As shown in Table 2, the majority of EGFRvIII-negative cases (37.9%) were 41 to 50 years old. However, the association between age and EGFRvIII positivity was not statistically significant (Fisher exact test, p-value = 1.000).
Table 2. Age distribution.
P-value was 1.0000 for the association between age and EGFRvIII positivity.
EGFRvIII: epidermal growth factor receptor variant III
| Age (years) | EGFRvIII | Total | ||||
| Positive | Negative | |||||
| N | % | N | % | N | % | |
| 17–30 | 0 | 0.00 | 4 | 100.00 | 4 | 11.43 |
| 31–40 | 1 | 25.00 | 3 | 75.00 | 4 | 11.43 |
| 41–50 | 1 | 8.33 | 11 | 91.67 | 12 | 34.29 |
| 51–60 | 2 | 40.00 | 3 | 60.00 | 5 | 14.29 |
| 61–70 | 2 | 20.00 | 8 | 80.00 | 10 | 28.57 |
| Total | 6 | 17.14 | 29 | 82.86 | 35 | 100.00 |
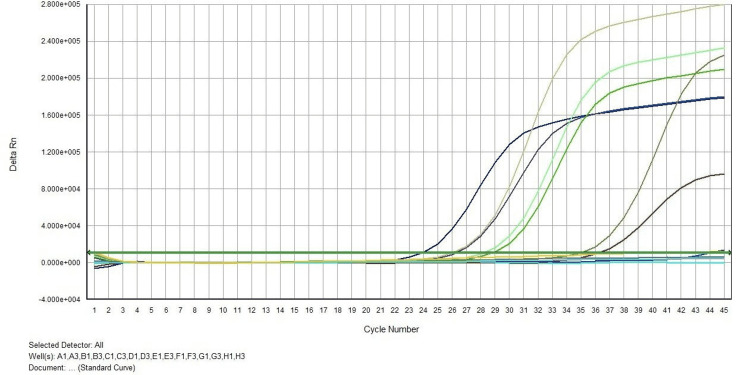
Figure 1. RT-PCR graph of tissue samples showing the delta run versus cycle number.
RT-PCR: reverse transcription-polymerase chain reaction
In EGFRvIII-positive cases, males were affected twice as commonly as females, while the male-to-female ratio was 1.4:1 in EGFRvIII-negative cases. EGFRvIII positivity was more in males (19.05%) compared to females (14.29%); however, the difference was not statistically significant (p = 1.000) (Table 3).
Table 3. Gender distribution.
EGFRvIII: epidermal growth factor receptor variant III
| Gender | EGFRvIII | Total | ||||
| Positive | Negative | |||||
| N | % | N | % | N | % | |
| Male | 4 | 19.05 | 17 | 80.95 | 21 | 60.00 |
| Female | 2 | 14.29 | 12 | 85.71 | 14 | 40.00 |
| Total | 6 | 17.14 | 29 | 82.86 | 35 | 100.00 |
The association between gender and EGFRvIII-positive cases was not significant statistically (Fisher exact test, p-value = 1.000).
Most of the EGFRvIII-positive cases presented unilaterally (66.67%). The tumor was located in the right frontoparietal region in two cases, the left frontal region in one case, the left temporal region in one case, the right frontal and left parietal regions in one case, and the corpus callosal region in one case.
Discussion
In this study, a total of six cases were found to be positive for the EGFRvIII mutation (17.1%), which is similar to the findings of another study that investigated 75 GBM cases and found eight cases positive for the EGFRvIII mutation (10.7%) [28]. All of these cases were aged over 40 years. They also used RT-PCR for the identification of this mutation. These results corroborate our findings. In our study, only one case was under 40 years of age. Das et al. also found EGFRvIII positivity in 12.2% of cases [29]. A mutation was found in one out of 13 cases (7.7%) in the 45-year age group, while five out of 37 cases were found positive in the >45-year age group. These findings are similar to our study findings. In both studies, no significant correlation was noted between age or gender with EGFRvIII mutation, which is in line with our study.
In an Indian study done by Jose et al. in 2020, mutated EGFRvIII was found in 57.5% of GBM cases, which is higher than our study finding [28]. The use of different molecular techniques may be the prime reason for such variation. They performed multiplex ligation-dependent probe amplification (MLPA) to identify this mutation. In a study by Perne et al. on a comparison between MLPA and RT-PCR, it was concluded that MLPA is a superior technique compared to RT-PCR in copy number quantification [30].
Rourke et al. performed a clinical trial on GBM patients with EGFRvIII-directed chimeric antigen receptor T cells on 369 patients with histologically confirmed GBM and tested for EGFRvIII using the next-generation sequencing (NGS) assay over the course of two years, of which 79 (21%) cases tested positive for EGFRvIII [31].
Sugawa et al. conducted a similar study using Southern blot analysis and discovered an EGFRvIII mutation in 17% of GBM cases [4]. Some researchers used more than one technique for estimating EGFRvIII positivity. Feldkamp et al. performed three techniques, namely, immunohistochemical testing, RT-PCR, and Western blot, and reported 25% EGFRvIII mutation positivity, which is in line with our results [32]. Thus, EGFRvIII mutation has been found in 10-50% of GBM cases worldwide in most studies.
The mean age of EGFRvIII-positive cases in our study was 54.3 years, while it was 56.1 ± 13.8 years in newly diagnosed GBM cases in the study by Shinojima et al. [33]. The mean age of GBM cases with EGFRvIII mutation was 47.13 years in the study by Jose et al. [28].
In our study, the male-to-female ratio was estimated to be 2:1 and 1.4:1 in EGFR-vIII positive cases aged ≤40 years and >40 years, respectively, while in the study of Jose et al., it was found to be 1:1 in ≤40 years and male predominance in >40 years of age, similar to the present study [28].
Studies done in the past on this molecular marker in GBM are listed in Table 4.
Table 4. Previous studies on epidermal growth factor receptor in glioblastoma multiforme patients.
| Year | Authors | Study region | Technique used | Positivity |
| 2020 | Jose et al. [28] | India | Multiplex ligation-dependent probe amplification | 57.5% |
| 2017 | Felsberg et al. [34] | Germany | Immunohistohemistry | 50.5% |
| Reverse transcription-polymerase chain reaction | 56.8% | |||
| 2017 | O’Rourke et al. [31] | Philadelphia, United States | Next-generation sequencing | 21% |
| 2011 | Jha et al. [35] | India | Reverse transcription-polymerase chain reaction | 10.7% |
| 2011 | Montano et al. [36] | Italy | Reverse transcription-polymerase chain reaction | 43.8% |
| 2008 | Yashimoto et al. [37] | California, United States | Reverse transcription-polymerase chain reaction) | 27% |
| 2008 | Stutz et al. [38] | United States | Southern blot | 17% |
| 2007 | Saikali et al. [39] | France | Immunohistochemistry and reverse transcription-polymerase chain reaction) | 64% |
| 2004 | Aldape et al. [40] | Immunohistochemistry) | 43.2% | |
| Reverse transcription-polymerase chain reaction | 40.9% | |||
| 2003 | Shinojima et al. [33] | Japan | Immunohistochemistry) | 25.3% |
| 2000 | Frederick et al. [41] | Minnesota | Sequence analysis | 67% |
| 1999 | Feldkamp et al. [32] | United States | Reverse transcription-polymerase chain reaction, immunohistochemistry, and Western blot | 25% |
| 1995 | Moscatello et al. [27] | Philadelphia | Western blot | 56% |
| 1990 | Sugawa et al. [23] | Sweden | Southern blot | 17% |
The mean age of presentation in EGFR-negative cases in our study was 49.3 years, while it was 52 years in the study by Jose et al. [28]. In their study, Shinojima et al. reported the mean age of EGFRvIII-negative cases to be 51.0 ± 13.8 years, which is similar to our study. Most of the EGFRvIII-negative cases occurred beyond 40 years of age (75.9%), with the maximum cases between 41 and 50 years (91.7%) in our study.
The most common location in the brain involved by this tumor was the temporal region in the study by Jose et al., while the frontal lobe was found to be most commonly involved by EGFRvIII-mutated tumors in our study [28].
Study limitations
The principal limitations of this study were that patients were from a single center and the small sample size. Therefore, the findings cannot be readily generalized or considered conclusive unless confirmed by subsequent studies in our population. Moreover, limited clinical data restricted the subdivision of tumors into primary and secondary glioblastoma, which could have provided information regarding the correlation of EGFRvIII to subtypes.
Conclusions
EGFRvIII detection is a major breakthrough in the improved management of GBM tumors through targeted gene therapies. Several clinical trials are underway globally to find a specific therapy against this mutation. However, research related to this mutation is way behind in India. Although extensive literature in Western countries is available on this marker, knowledge about this marker in the Indian population, particularly regarding brain tumors, is limited. Identification of this mutation may segregate patients who would be more responsive to newer therapeutic approaches. Previous studies also found that tyrosine kinase inhibitors only worked in cases of EGFRvIII mutations with a retained PTEN gene. The simultaneous presence of molecular markers in GBM cases would also need to be considered before initiating any gene therapy for its improved effectiveness.
Acknowledgments
The authors would like to acknowledge the support and efforts of the Indian Council of Medical Research (ICMR)-Department of Health Research (DHR) and GC Life Sciences.
The content published in Cureus is the result of clinical experience and/or research by independent individuals or organizations. Cureus is not responsible for the scientific accuracy or reliability of data or conclusions published herein. All content published within Cureus is intended only for educational, research and reference purposes. Additionally, articles published within Cureus should not be deemed a suitable substitute for the advice of a qualified health care professional. Do not disregard or avoid professional medical advice due to content published within Cureus.
Funding Statement
This study was funded by the Indian Council of Medical Research (Department of Health Research, Government of India) through the Multidisciplinary Research Unit, Dr. Sampurnanand Medical College, Jodhpur, Rajasthan, India.
The authors have declared that no competing interests exist.
Human Ethics
Consent was obtained or waived by all participants in this study. Institutional Ethics Committee, Dr. Sampurnanand Medical College, Jodhpur issued approval EC/MC/JU/2018/319
Animal Ethics
Animal subjects: All authors have confirmed that this study did not involve animal subjects or tissue.
References
- 1.Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Downward J, Yarden Y, Mayes E, et al. Nature. 1984;307:521–527. doi: 10.1038/307521a0. [DOI] [PubMed] [Google Scholar]
- 2.The somatic genomic landscape of glioblastoma. Brennan CW, Verhaak RG, McKenna A, et al. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epidermal growth factor ligand-independent, unregulated, cell-transforming potential of a naturally occurring human mutant EGFRvIII gene. Batra SK, Castelino-Prabhu S, Wikstrand CJ, Zhu X, Humphrey PA, Friedman HS, Bigner DD. https://pubmed.ncbi.nlm.nih.gov/8845302/ Cell Growth Differ. 1995;6:1251–1259. [PubMed] [Google Scholar]
- 4.Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Sugawa N, Ekstrand AJ, James CD, Collins VP. Proc Natl Acad Sci U S A. 1990;87:8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.A deletion mutation within the ligand binding domain is responsible for activation of epidermal growth factor receptor gene in human brain tumors. Yamazaki H, Ohba Y, Tamaoki N, Shibuya M. Jpn J Cancer Res. 1990;81:773–779. doi: 10.1111/j.1349-7006.1990.tb02644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genes for epidermal growth factor receptor, transforming growth factor alpha, and epidermal growth factor and their expression in human gliomas in vivo. Ekstrand AJ, James CD, Cavenee WK, Seliger B, Pettersson RF, Collins VP. https://pubmed.ncbi.nlm.nih.gov/2009534/ Cancer Res. 1991;51:2164–2172. [PubMed] [Google Scholar]
- 7.Monoclonal antibodies against EGFRvIII are tumor specific and react with breast and lung carcinomas and malignant gliomas. Wikstrand CJ, Hale LP, Batra SK, et al. https://pubmed.ncbi.nlm.nih.gov/7606735/ Cancer Res. 1995;55:3140–3148. [PubMed] [Google Scholar]
- 8.Mutant epidermal growth factor receptor displays increased signaling through the phosphatidylinositol-3 kinase/AKT pathway and promotes radioresistance in cells of astrocytic origin. Li B, Yuan M, Kim IA, Chang CM, Bernhard EJ, Shu HK. Oncogene. 2004;23:4594–4602. doi: 10.1038/sj.onc.1207602. [DOI] [PubMed] [Google Scholar]
- 9.The EGF receptor family as targets for cancer therapy. Mendelsohn J, Baselga J. Oncogene. 2000;19:6550–6565. doi: 10.1038/sj.onc.1204082. [DOI] [PubMed] [Google Scholar]
- 10.Oncogenic EGFR signaling activates an mTORC2-NF-κB pathway that promotes chemotherapy resistance. Tanaka K, Babic I, Nathanson D, et al. Cancer Discov. 2011;1:524–538. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mutant epidermal growth factor receptor signaling down-regulates p27 through activation of the phosphatidylinositol 3-kinase/Akt pathway in glioblastomas. Narita Y, Nagane M, Mishima K, Huang HJ, Furnari FB, Cavenee WK. https://pubmed.ncbi.nlm.nih.gov/12438278/ Cancer Res. 2002;62:6764–6769. [PubMed] [Google Scholar]
- 12.Inhibition of phosphatidylinositol 3-kinase signaling negates the growth advantage imparted by a mutant epidermal growth factor receptor on human glioblastoma cells. Klingler-Hoffmann M, Bukczynska P, Tiganis T. Int J Cancer. 2003;105:331–339. doi: 10.1002/ijc.11085. [DOI] [PubMed] [Google Scholar]
- 13.Constitutive activation of phosphatidylinositol 3-kinase by a naturally occurring mutant epidermal growth factor receptor. Moscatello DK, Holgado-Madruga M, Emlet DR, Montgomery RB, Wong AJ. J Biol Chem. 1998;273:200–206. doi: 10.1074/jbc.273.1.200. [DOI] [PubMed] [Google Scholar]
- 14.Resistance to tyrosine kinase inhibition by mutant epidermal growth factor receptor variant III contributes to the neoplastic phenotype of glioblastoma multiforme. Learn CA, Hartzell TL, Wikstrand CJ, et al. Clin Cancer Res. 2004;10:3216–3224. doi: 10.1158/1078-0432.ccr-03-0521. [DOI] [PubMed] [Google Scholar]
- 15.Analysis of the phosphatidylinositol 3'-kinase signaling pathway in glioblastoma patients in vivo. Choe G, Horvath S, Cloughesy TF, et al. https://pubmed.ncbi.nlm.nih.gov/12782577/ Cancer Res. 2003;63:2742–2746. [PubMed] [Google Scholar]
- 16.Loss of PTEN/MMAC1/TEP in EGF receptor-expressing tumor cells counteracts the antitumor action of EGFR tyrosine kinase inhibitors. Bianco R, Shin I, Ritter CA, et al. Oncogene. 2003;22:2812–2822. doi: 10.1038/sj.onc.1206388. [DOI] [PubMed] [Google Scholar]
- 17.The epidermal growth factor receptor pathway mediates resistance to sequential administration of radiation and chemotherapy in primary human glioblastoma cells in a RAS-dependent manner. Chakravarti A, Chakladar A, Delaney MA, Latham DE, Loeffler JS. https://pubmed.ncbi.nlm.nih.gov/12154034/ Cancer Res. 2002;62:4307–4315. [PubMed] [Google Scholar]
- 18.Immunolocalization of basic fibroblast growth factor to the microvasculature of human brain tumors. Brem S, Tsanaclis AM, Gately S, Gross JL, Herblin WF. Cancer. 1992;70:2673–2680. doi: 10.1002/1097-0142(19921201)70:11<2673::aid-cncr2820701118>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 19.Cost of migration: invasion of malignant gliomas and implications for treatment. Giese A, Bjerkvig R, Berens ME, Westphal M. J Clin Oncol. 2003;21:1624–1636. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- 20.Malignant glioma biology: role for TGF-beta in growth, motility, angiogenesis, and immune escape. Platten M, Wick W, Weller M. Microsc Res Tech. 2001;52:401–410. doi: 10.1002/1097-0029(20010215)52:4<401::AID-JEMT1025>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 21.A common mutant epidermal growth factor receptor confers enhanced tumorigenicity on human glioblastoma cells by increasing proliferation and reducing apoptosis. Nagane M, Coufal F, Lin H, Bögler O, Cavenee WK, Huang HJ. https://pubmed.ncbi.nlm.nih.gov/8895767/ Cancer Res. 1996;56:5079–5086. [PubMed] [Google Scholar]
- 22.IkappaBalphaM suppresses angiogenesis and tumorigenesis promoted by a constitutively active mutant EGFR in human glioma cells. Wu JL, Abe T, Inoue R, Fujiki M, Kobayashi H. Neurol Res. 2004;26:785–791. doi: 10.1179/016164104225014139. [DOI] [PubMed] [Google Scholar]
- 23.Function of aberrant EGFR in malignant gliomas. Sugawa N, Yamamoto K, Ueda S, et al. Brain Tumor Pathol. 1998;15:53–57. doi: 10.1007/BF02482101. [DOI] [PubMed] [Google Scholar]
- 24.Mutant epidermal growth factor receptor up-regulates molecular effectors of tumor invasion. Lal A, Glazer CA, Martinson HM, Friedman HS, Archer GE, Sampson JH, Riggins GJ. https://pubmed.ncbi.nlm.nih.gov/12067969/ Cancer Res. 2002;62:3335–3339. [PubMed] [Google Scholar]
- 25.Expression of collagenase-3 (MMP-13) enhances invasion of human fibrosarcoma HT-1080 cells. Ala-Aho R, Johansson N, Baker AH, Kähäri VM. Int J Cancer. 2002;97:283–289. doi: 10.1002/ijc.1619. [DOI] [PubMed] [Google Scholar]
- 26.Cumulative influence of matrix metalloproteinase-1 and -2 in the migration of melanoma cells within three-dimensional type I collagen lattices. Ntayi C, Lorimier S, Berthier-Vergnes O, Hornebeck W, Bernard P. Exp Cell Res. 2001;270:110–118. doi: 10.1006/excr.2001.5306. [DOI] [PubMed] [Google Scholar]
- 27.Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors. Moscatello DK, Holgado-Madruga M, Godwin AK, et al. https://pubmed.ncbi.nlm.nih.gov/7585629/ Cancer Res. 1995;55:5536–5539. [PubMed] [Google Scholar]
- 28.Frequency and prognosis of epidermal growth factor receptor variant III mutations in glioblastoma multiforme among Indian patients: a single-institution study. Jose WM, Munirathnam V, Narendranath V, Philip A, Keechilat P. South Asian J Cancer. 2020;9:126–129. doi: 10.1055/s-0041-1723078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glioblastoma multiforme in an Asian population: evidence for a distinct genetic pathway. Das A, Tan WL, Teo J, Smith DR. J Neurooncol. 2002;60:117–125. doi: 10.1023/a:1020622415786. [DOI] [PubMed] [Google Scholar]
- 30.Comparison of multiplex ligation-dependent probe amplification and real-time PCR accuracy for gene copy number quantification using the beta-defensin locus. Perne A, Zhang X, Lehmann L, Groth M, Stuber F, Book M. Biotechniques. 2009;47:1023–1028. doi: 10.2144/000113300. [DOI] [PubMed] [Google Scholar]
- 31.A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. O'Rourke DM, Nasrallah MP, Desai A, et al. Sci Transl Med. 2017;9:0. doi: 10.1126/scitranslmed.aaa0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Expression of activated epidermal growth factor receptors, Ras-guanosine triphosphate, and mitogen-activated protein kinase in human glioblastoma multiforme specimens. Feldkamp MM, Lala P, Lau N, Roncari L, Guha A. Neurosurgery. 1999;45:1442–1453. doi: 10.1097/00006123-199912000-00034. [DOI] [PubMed] [Google Scholar]
- 33.Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Shinojima N, Tada K, Shiraishi S, et al. https://pubmed.ncbi.nlm.nih.gov/14583498/ Cancer Res. 2003;63:6962–6970. [PubMed] [Google Scholar]
- 34.Epidermal growth factor receptor variant III (EGFRvIII) positivity in EGFR-amplified glioblastomas: prognostic role and comparison between primary and recurrent tumors. Felsberg J, Hentschel B, Kaulich K, et al. Clin Cancer Res. 2017;23:6846–6855. doi: 10.1158/1078-0432.CCR-17-0890. [DOI] [PubMed] [Google Scholar]
- 35.IDH1 mutations in gliomas: first series from a tertiary care centre in India with comprehensive review of literature. Jha P, Suri V, Sharma V, et al. Exp Mol Pathol. 2011;91:385–393. doi: 10.1016/j.yexmp.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 36.Expression of EGFRvIII in glioblastoma: prognostic significance revisited. Montano N, Cenci T, Martini M, et al. Neoplasia. 2011;13:1113–1121. doi: 10.1593/neo.111338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Development of a real-time RT-PCR assay for detecting EGFRvIII in glioblastoma samples. Yoshimoto K, Dang J, Zhu S, et al. Clin Cancer Res. 2008;14:488–493. doi: 10.1158/1078-0432.CCR-07-1966. [DOI] [PubMed] [Google Scholar]
- 38.LRIG1 negatively regulates the oncogenic EGF receptor mutant EGFRvIII. Stutz MA, Shattuck DL, Laederich MB, Carraway KL 3rd, Sweeney C. Oncogene. 2008;27:5741–5752. doi: 10.1038/onc.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tumor-specific anti-epidermal growth factor receptor variant III monoclonal antibodies: use of the tyramine-cellobiose radioiodination method enhances cellular retention and uptake in tumor xenografts. Reist CJ, Archer GE, Kurpad SN, et al. https://pubmed.ncbi.nlm.nih.gov/7671250/ Cancer Res. 1995;55:4375–4382. [PubMed] [Google Scholar]
- 40.Immunohistochemical detection of EGFRvIII in high malignancy grade astrocytomas and evaluation of prognostic significance. Aldape KD, Ballman K, Furth A, et al. J Neuropathol Exp Neurol. 2004;63:700–707. doi: 10.1093/jnen/63.7.700. [DOI] [PubMed] [Google Scholar]
- 41.Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Frederick L, Wang XY, Eley G, James CD. https://pubmed.ncbi.nlm.nih.gov/10728703/ Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]