Abstract
A high yield of lactic acid per gram of glucose consumed and the absence of additional metabolites in the fermentation broth are two important goals of lactic acid production by microrganisms. Both purposes have been previously approached by using a Kluyveromyces lactis yeast strain lacking the single pyruvate decarboxylase gene (KlPDC1) and transformed with the heterologous lactate dehydrogenase gene (LDH). The LDH gene was placed under the control the KlPDC1 promoter, which has allowed very high levels of lactate dehydrogenase (LDH) activity, due to the absence of autoregulation by KlPdc1p. The maximal yield obtained was 0.58 g g−1, suggesting that a large fraction of the glucose consumed was not converted into pyruvate. In a different attempt to redirect pyruvate flux toward homolactic fermentation, we used K. lactis LDH transformant strains deleted of the pyruvate dehydrogenase (PDH) E1α subunit gene. A great process improvement was obtained by the use of producing strains lacking both PDH and pyruvate decarboxylase activities, which showed yield levels of as high as 0.85 g g−1 (maximum theoretical yield, 1 g g−1), and with high LDH activity.
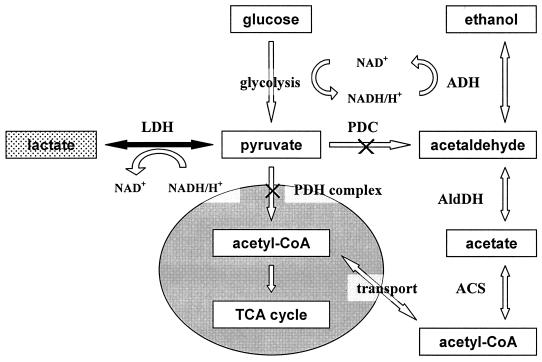
Lactic acid is widely used in industry. Pure lactate can be obtained from bacteria: fermentation processes are carried out in buffered conditions at neutral pH in order to avoid metabolic and growth inhibition caused by the accumulation of the acidic product (2, 4, 10). Genetically engineered fermentative yeasts can be used for the production of lactic acid from glucose by transformation with heterologous lactate dehydrogenase (LDH) genes. Conversion of pyruvate to lactic acid by LDH requires cytosolic NADH/H+. A general scheme of pyruvate catabolism in yeast is shown in Fig. 1.
FIG. 1.
Scheme of pyruvate metabolism and pyruvate bypass in K. lactis. Enzymes: ACS, acetyl-CoA synthetase; ADH, alcohol dehydrogenase; AldDH, aldehyde dehydrogenase. The gray oval represents the mitochondrial compartment. The black arrow and the dotted box indicate the metabolic step added by transformation with heterologous LDH gene and the new metabolite, respectively.
The yeast Saccharomyces cerevisiae, which has a predominant fermentative metabolism, has been used for lactic acid production (5, 14). However, the bioprocess yield (grams of lactic acid produced per gram of glucose consumed) was low because of the simultaneous production of ethanol due to the competition for pyruvate by the heterologous LDH and the homologous pyruvate decarboxylase (PDC) activities. S. cerevisiae has two active structural PDC genes: PDC1 and PDC5 (16, 17). A third gene, PDC6, is inactive (9). Increased production and yield of lactic acid were obtained by the use of single pdc1 or pdc5 mutant strains, but the amount of ethanol could only be slightly decreased (1). On the other hand, S. cerevisiae mutant strains with both PDC1 and PDC5 genes inactivated are strongly impaired for growth on glucose medium (9) and thus are not useful for production purposes.
The yeast Kluyveromyces lactis has a single gene, KlPDC1, expressing PDC activity (3). The deletion of KlPDC1 leads to strains without PDC activity and which do not produce ethanol. In contrast to S. cerevisiae pdc1 pdc5 double mutant strains, K. lactis Klpdc1Δ strains grow at the same rate as the wild-type strains on glucose medium (3). The difference between the two yeasts might be ascribed to active acetyl-coenzyme A (CoA) transport from mitochondria to cytosol in K. lactis (Fig. 1). KlPDC1 is subjected to autoregulation by its own gene product, and it is induced by glucose and repressed by ethanol at the transcriptional level (6). In the presence of both carbon sources, KlPDC1 promoter-driven expression shows intermediate levels (6). In previous works, we reported the use of wild-type and Klpdc1Δ K. lactis strains transformed with the bovine LDH gene (15), as well as with bacterial LDH genes (patent application WO1998EP0005758), for the production of lactic acid.
In aerobic conditions the yeast K. lactis has a predominantly respiratory metabolism on glucose media (8, 11, 13) and produces a limited amount of ethanol. In this yeast, pyruvate is largely channeled into the tricarboxylic acid cycle by the pyruvate dehydrogenase (PDH) complex. On the other hand, K. lactis strains with no PDH activity have a vigorous fermentative metabolism on glucose in aerobic batch cultures, and only in glucose-limited aerobic chemostat conditions can the Klpda1Δ strains metabolize glucose exclusively through the cytoplasmic acetyl-CoA pathway (20).
In this work, we present the results of new metabolic host configurations for the conversion of glucose into lactate, based on engineered rerouting of pyruvate. We used K. lactis strains either lacking PDH activity or lacking both PDC and PDH activities (Fig. 1), transformed with the bovine LDH gene placed under the transcriptional control of the inducible promoter of KlPDC1 gene and cloned into a stable multicopy vector. The heterologous LDH enzyme could efficiently compete for available cytoplasmic NADH/H+ and pyruvate in these strains when they were grown in glucose excess conditions.
MATERIALS AND METHODS
Strains and media.
The Escherichia coli strain used in the molecular cloning procedures was DH5αF′ [ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deo rec1 end1 sup44 λ THI-1 gyrA96 relA1]. The K. lactis strains used in this work are listed in Table 1. The deleted strains PMI and MW341-5/Klpdc1Δ were obtained from wild-type strains PM6-7A and MW341-5, respectively, by disruption of KlPDC1 with a deletion cassette containing the marker gene URA3 from S. cerevisiae (3, 6). The deleted strains GG1993 (20) and PM6-7A/Klpda1Δ were obtained from wild-type strains CBS2359 and PM6-7A, respectively, by disruption of KlPDA1 with the bacterial Tn5BLE gene from plasmid pUT322 (7). Double-deleted strains were constructed as follows. In strain PM6-7A/DD, the KlPDC1 gene was deleted and replaced with the URA3 marker gene by integrative transformation of strain PM6-7A/Klpda1Δ with vector pBSU7, as described in Bianchi et al. (3). The deletion of KlPDC1 was verified by PCR and growth on glucose plus the mitochondrial inhibitor antimycin A. The double-deleted strain BM1-3C was selected as a phleomycin-resistant and antimycin A-sensitive segregant strain issued from a diploid strain obtained by crossing strain MW341–5/Klpdc1Δ with the Klpda1Δ strain GG1993.
TABLE 1.
Yeast strains
| Straina | Genotype | Source or reference |
|---|---|---|
| PM6-7A | MATaadeT-600, uraA1-1 | 18 |
| MW109-8C | MATα lysA1-1 | 18 |
| PMI | MATaadeT-600 uraA1-1 Klpdc1::URA3 | 3 |
| MW341-5/Klpdc1Δ | MATα lac4-8 leu2 lysA1-1 uraA1-1 Klpdc1::URA3 | 6 |
| 7C(pLAZ10) | MATα lysA1-1 adeT-600 Klpdc1::URA3, pLAZ10+ | This work |
| GG1993 | MATaura3-49 Klpda1::Tn5BLE | 20 |
| PM6-7A/Klpda1Δ | MATaadeT-600 uraA1-1 Klpda1::Tn5BLE | H. Y. Steensma |
| PM6-7A/DD | MATaadeT-600 uraA1-1 Klpdc1::URA3 Klpda1::Tn5BLE | This work |
| BM1-3C | MATaleu2 Klpdc1::URA3 Klpda1::Tn5BLE | This work |
| BM3-12D (pLAZ10) | MATaKlpdc1::URA3 Klpda1::Tn5BLE, pLAZ10+ | This work |
All of the K. lactis strains used in this work originate from the standard NRRL strains Y-1140 (CBS2359; MATa genotype) and Y-1205 (CBS2360; MATα genotype) (19). Auxotrophic markers were introduced by selection of spontaneous or induced mutations or transferred by genetic crosses.
A different approach was followed for the construction of the prototrophic double-deleted strain bearing the heterologous LDH gene. The Klpdc1Δ strain PMI was transformed with pLAZ10 (see below) and crossed with the wild-type strain MW109-8C. After sporulation of the resulting diploid strain and tetrad dissection, the strain called 7C(pLAZ10) was selected as a MATα, Geneticin-resistant, and antimycin A-sensitive segregant strain. The double-deleted BM1-3C strain and 7C strain carrying pLAZ10 were then crossed, and haploid segregant strains were isolated after sporulation of the diploid strain. All of them were sensitive to antimycin A (Klpdc1Δ) and resistant to Geneticin (pLAZ10+), while the phleomycin-resistance phenotype (Klpda1Δ) and auxotrophic markers correctly segregated 2:2. Among these strains, we selected for fermentation processes a prototrophic Klpda1Δ segregant strain, BM3-12D(pLAZ10).
Yeast-rich medium (YP) contained 1% (wt/vol) yeast extract and 2% (wt/vol) peptone. Synthetic medium (SM) contained 0.67% (wt/vol) yeast nitrogen base without amino acids. Media were supplemented with 2% (wt/vol) or 5% (wt/vol) glucose (D) and/or 2% (vol/vol) ethanol (E). The YP media used for the selection and identification of the Klpda1Δ and Klpdc1Δ mutant strains contained 8 mg of phleomycin liter−1 and 2% (wt/vol) glucose or 5 μM antimycin A and 5% (wt/vol) glucose, respectively. Geneticin was added to a final concentration of 200 mg liter−1, when needed. Solid media contained 2% (wt/vol) agar.
Construction of the LDH expression vector
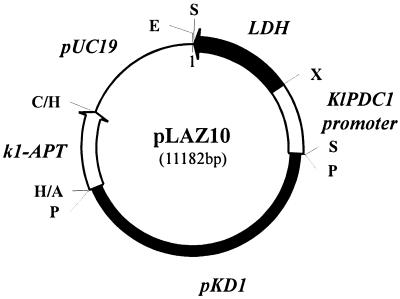
Vector pLAZ10 (Fig. 2) was obtained by cloning the 2.8-kbp SalI fragment of vector pEPL2 (15), bearing the KlPDC1 promoter fused in front of the cDNA of the bovine LDH-A gene (14), into the unique SalI site of vector p3K31. Common cloning procedures have been followed. Vector p3K31 is composed of pUC19 DNA and of the kanamycin resistance cassette of vector pKan707 inserted in the unique SphI site of the natural multicopy plasmid pKD1 (12). The kanamycin resistance gene is the aminoglycoside phosphotransferase gene (APT) of the bacterial transposon Tn903, which confers Geneticin resistance on yeast, fused downstream from the K. lactis killer promoter k1. Yeast strains were transformed with pLAZ10 by the electroporation procedure (3), and the transformants were selected on YPD medium containing Geneticin.
FIG. 2.
Map of vector pLAZ10. The elements of pLAZ10 are marked in the figure as follows: thin line, pUC19 bacterial plasmid; black box, entire pKD1 genome; empty arrow, k1-APT cassette; black arrow, bovine LDH-A cDNA; empty box, KlPDC1 promoter sequence. Relevant cloning sites: A, ScaI; C, HincII; E, EcoRI; H, HindIII; P, SphI; S, SalI; X, XbaI.
Stirred-tank cultures.
Growth on glucose medium of strains PM6-7A, PMI, and PM6-7A/Klpda1Δ was monitored in a 5-liter Biostat-B stirred-tank bioreactor (B-Braun). Cells were inoculated in 4 liters of SM, containing 2% (wt/vol) glucose and 200 mg of adenine liter−1and supplemented, when required, with 100 mg of uracil liter−1. The bioreactor was kept at 30°C and pH 5 and was aerated at 2 liters min−1. A pO2 level higher than 40% was maintained throughout the process by controlling the stirring rate. The PM6-7A/DD strain was grown on mixed glucose-ethanol substrate and compared to the wild-type strain PM6-7A grown on the same carbon sources. The process conditions were the same as described above. The medium contained 2% (wt/vol) glucose and 2% (vol/vol) ethanol.
Fermentative processes for lactate production from strain BM3-12D(pLAZ10) were performed in a Biostat-Q 1-liter stirred-tank fermentor (B-Braun) containing 0.8 liter of SM supplemented with 5% (wt/vol) glucose, 2% (vol/vol) ethanol, and 200 mg of Geneticin liter−1. Temperature and stirring were kept at 30°C and 400 rpm, respectively. During the fermentation processes, air was fed at 0.8 liter min−1, and glucose was added to a concentration of 4.5% ± 5% (wt/vol). In the fermentation tests, the pH was maintained at 4.5 by the automatic addition of 2 M KOH.
Measurement of cell concentration, metabolites, and enzymatic activities.
Fermentation processes were monitored at regular time intervals. Cell concentrations were determined by measuring the optical density at 660 nm (OD660). Glucose, ethanol, acetate, l-(+)-lactate, and LDH activities were determined by using diagnostic kits (Boehringer Mannheim 716251, 176290, 148261, and 139084 and Sigma DG1340-K, respectively) according to instructions. The concentration of pyruvate was assayed by high-pressure liquid chromatography (Jasco Corporation). Separations were achieved on HPX-87H 300-by-7.8-mm column (Bio-Rad), and peaks were detected at 210 nm with UV-VIS detector (Jasco Corporation). Diluted sulfuric acid (4 mM in water) was used as solvent at 35°C and at a 0.6-ml min−1 flow rate. Yields of lactate were calculated by linear regressions obtained by plotting the grams of glucose consumed in the course of fermentation processes versus the grams of lactate produced.
RESULTS AND DISCUSSION
Growth and metabolite productions of wild-type, Klpdc1Δ, and Klpda1Δ deleted strains
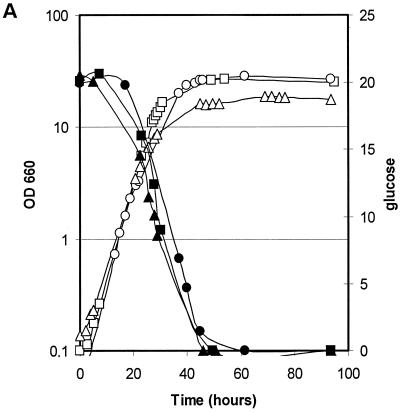
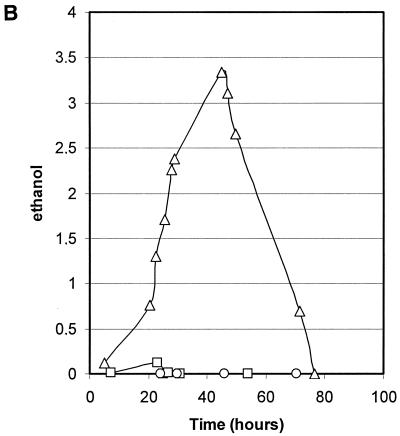
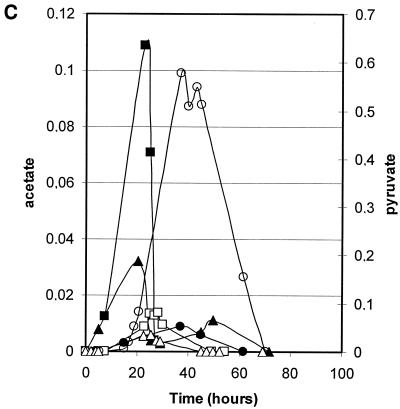
Growth and consumption or production of carbon compounds of strains harboring defects in pyruvate metabolism were studied in stirred-tank fermentor supplemented with defined minimal medium containing glucose as carbon sources. In the first experiment, the parental strain PM6-7A, the Klpdc1Δ strain PMI, and the PM6-7A/Klpda1Δ strain were compared. These strains were isogenic except for the deletion of the single structural gene encoding for PDC activity in strain PMI and the deletion of KlPDA1, which is the gene encoding for the E1α subunit of the PDH complex (Fig. 1). Time courses of the fermentation processes are shown in Fig. 3. Specific growth and glucose consumption rates (Fig. 3A) were similar for the three strains, while the overall biomass production at the stationary phase was lower for the PM6-7A/Klpda1Δ strain. Ethanol was produced and accumulated—up to 3.3 g liter−1—during the log growth phase only by strain PM6-7A/Klpda1Δ (Fig. 3B). The production of other carbon metabolites (Fig. 3C) showed modest peaks of acetate or pyruvate excretion during the log phase of the wild-type and PM6-7A/Klpda1Δ strains and the late log phase of the PMI strain, respectively.
FIG. 3.
Growth and metabolites composition of wild-type (squares), Klpda1Δ (circles), and Klpdc1Δ (triangles) isogenic strain cultures. (A) Cell growth (OD660 [open symbols]) and residual glucose (grams liter−1 [solid symbols]). (B) Ethanol (grams liter−1). (C) Pyruvate (grams liter−1 [open symbols]) and acetate (grams liter−1 [solid symbols]).
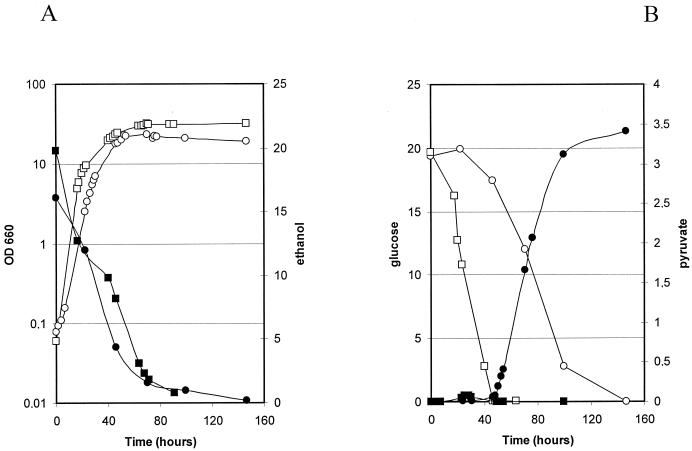
In a second series of experiments, we compared growth and metabolite production or consumption of the wild-type strain PM6-7A and of the double-deleted strain PM6-7A/DD. These processes were carried out on medium containing both glucose and ethanol as carbon sources, because double-deleted strains cannot grow on minimal medium containing only C6 sugars or C3 compounds as the sole carbon sources (data not shown). Results are reported in Fig. 4. Growth profiles (Fig. 4A) were similar for both strains, with only a slightly reduced specific growth rate and a lower cell concentration at the stationary phase for PM6-7A/DD strain. Ethanol and glucose (Fig. 4A and B, respectively) were simultaneously consumed by the wild-type strain PM6-7A. In contrast, the double-deleted strain consumed exclusively ethanol in the first phase of the fermentation process, during which the bulk of biomass was produced. Glucose assimilation was observed only when ethanol was completely consumed. Interestingly, glucose consumption was associated with the accumulation of pyruvate at up to 3.5 g liter−1.
FIG. 4.
Growth and metabolite composition of wild-type (squares) and Klpda1Δ Klpdc1Δ (circles) isogenic strain cultures. (A) Cell growth (OD660 [open symbols]) and residual ethanol (grams liter−1 [closed symbols]). (B) Residual glucose (grams liter−1 [open symbols]) and pyruvate (grams liter−1 [closed symbols]).
Stirred-tank fermentation of a Klpda1Δ Klpdc1Δ(pLAZ10) strain
Production of lactic acid by single-deleted Klpda1Δ strains transformed with pLAZ10 was tested. The maximum yield obtained (0.35 g g−1) was unsatisfactory because of competition for pyruvate by the PDC enzyme, which channeled carbon flux into the ethanologenic pathway and/or into the pyruvate bypass (not shown). In order to increase the product yield by completely channeling pyruvate flux toward lactate formation, we planned to assay lactic fermentation in strains defective in both PDC and PDH activities. Since the double-deleted strains PM6-7A/DD and BM1-3C were recalcitrant to direct transformation, we genetically transduced pLAZ10 and selected double-deleted transformants as described in Materials and Methods. This procedure also allowed us to select a prototrophic host, BM3-12D, that was more suitable for fermentation processes.
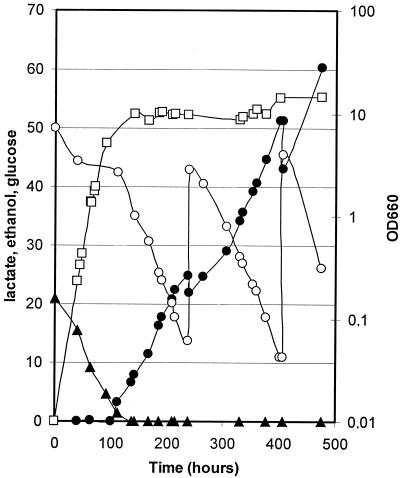
Lactic acid production of BM3-12D(pLAZ10) transformant strain was tested by cultivation in a 1-liter stirred-tank bioreactor at constant pH by KOH addition. The time course of the process is shown in Fig. 5. During the first few hours of the process, cells consumed ethanol for biomass production. Glucose assimilation was observed only when ethanol was completely consumed, as for the nontransformed double-deleted strain (see Fig. 4). However, because of the heterologous LDH activity, in this case the glucose assimilation was associated with lactate production. Further additions of glucose to the bioreactor (after 238 and 405 h of fermentation) resulted in a continuous trend of glucose consumption and lactate accumulation, up to a concentration of 60 g of lactate liter−1 at 500 h of incubation, when the process was stopped. Lactate was not produced in control fermentation processes without glucose added in the culture medium (not shown). The yield of product formation (0.85 g g−1), not far from the maximum theoretical value (1 g g−1), was much higher than yields obtained with single Klpda1Δ strains (0.35 g g−1) or the single Klpdc1Δ strain (0.58 g g−1 [15]).
FIG. 5.
l-(+)-Lactic acid production in a stirred-tank fermentation process of the Klpdc1Δ Klpda1Δ strain BM3-12D(pLAZ10). The time courses of product, biomass, and carbon source concentrations are reported. The fermentation was carried out on SM as described in Materials and Methods. The pH was maintained at 4.5 by KOH addition. Symbols: ●, lactate (grams liter−1); ▴, ethanol (grams liter−1); □, cells (OD660); ○, glucose (grams liter−1).
As previously described (15), LDH activity is higher in the Klpdc1Δ genetic background (55 to 60 U mg−1) than in wild-type strain (4 to 5 U mg−1), when the LDH gene is fused to the KlPDC1 promoter. This effect is a consequence of the absence of transcriptional autoregulation by KlPdc1p in Klpdc1Δ strains (6). We measured values of heterologous LDH activity that were as high as 110 to 150 U mg−1 in the Klpdc1Δ Klpda1Δ BM3-12D(pLAZ10) strain, which contained the same LDH expression cassette as in the above-mentioned transformant strains. This finding suggests that the factors involved in the induction or derepression of KlPDC1 promoter were fully operating also in the double-deleted genetic background. As a whole, the results presented here indicate that the metabolic constraints of carbon flux, growth-phase dependence of substrate assimilation, and upregulation of specific promoters might provide synergetic cooperation in metabolic engineered K. lactis strains for highly efficient transformation of glucose into lactic acid.
ACKNOWLEDGMENTS
We thank L. Alberghina, L. Frontali, and B. M. Ranzi for helpful discussions. Vector pLAZ10 was constructed by R. Menghini.
F.P. was the recipient of a fellowship from Biopolo s.c.r.l. This work was partially supported by MURST-Cofin 40% to D.P. (MM05C63814).
Footnotes
This work is dedicated to Franco Tato.
REFERENCES
- 1.Adachi E, Torigoe M, Sugiyama M, Nikawa J J, Shimizu K. Modification of metabolic pathways of Saccharomyces cerevisiae by the expression of lactate dehydrogenase and deletion of pyruvate decarboxylase genes for the lactic acid fermentation at low pH value. J Ferment Bioeng. 1998;86:284–289. [Google Scholar]
- 2.Benninga H A. A history of lactic acid making. Dordrecht, The Netherlands: Kluyver Academic Publishers; 1990. [Google Scholar]
- 3.Bianchi M M, Tizzani L, Destruelle M, Frontali L, Wésolowski-Louvel M. The “petite-negative” yeast Kluyveromyces lactis has a single gene expressing pyruvate decarboxylase activity. Mol Microbiol. 1996;19:27–36. doi: 10.1046/j.1365-2958.1996.346875.x. [DOI] [PubMed] [Google Scholar]
- 4.Buchta K. Lactic acid. In: Delleweg H, editor. Bio/technology. Vol. 3. Weinheim, Germany: Verlag Chemie; 1983. pp. 409–417. [Google Scholar]
- 5.Dequin S, Barre P. Mixed lactic acid-alcoholic fermentation by Saccharomyces cerevisiae expressing the Lactobacillus caseil-(+)-LDH. Bio/Technology. 1994;12:173–177. doi: 10.1038/nbt0294-173. [DOI] [PubMed] [Google Scholar]
- 6.Destruelle M, Menghini R, Frontali L, Bianchi M M. Regulation of the expression of the Kluyveromyces lactis PDC1 gene: carbon source responsive elements and autoregulation. Yeast. 1999;15:361–370. doi: 10.1002/(SICI)1097-0061(19990330)15:5<361::AID-YEA378>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 7.Gatignol A, Dassain M, Tirabi G. Cloning of Saccharomyces cerevisiae promoters using a probe vector based on the phleomycin resistance. Gene. 1990;9:35–41. doi: 10.1016/0378-1119(90)90159-o. [DOI] [PubMed] [Google Scholar]
- 8.Gonzales-Siso M I, Ramil E, Cerdan M E, Freire-Picos M A. Respirofermentative metabolism in Kluyveromyces lactis: ethanol production and the Crabtree effect. Enzyme Microbiol Technol. 1996;18:585–591. doi: 10.1016/s0141-0229(00)00161-7. [DOI] [PubMed] [Google Scholar]
- 9.Hohmann S. Characterization of PDC6, a third structural gene for pyruvate decarboxylase in Saccharomyces cerevisiae. J Bacteriol. 1991;173:7963–7969. doi: 10.1128/jb.173.24.7963-7969.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hongo M, Nomura Y, Iwahara M. Novel methods of lactic acid production by electrodialysis fermentation. Appl Environ Microbiol. 1986;32:227–234. doi: 10.1128/aem.52.2.314-319.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiers J, Zeeman A-M, Luttik M, Thiele C, Castrillo J I, Steensma Y H, Van Dijken J P, Pronk J T. Regulation of alcoholic fermentation in batch and chemostat cultures of Kluyveromyces lactis CBS2359. Yeast. 1997;14:459–469. doi: 10.1002/(SICI)1097-0061(19980330)14:5<459::AID-YEA248>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 12.Morlino G B, Tizzani L, Fleer R, Frontali L, Bianchi M M. Inducible amplification of gene copy number and heterologous protein production in the yeast Kluyveromyces lactis. Appl Environ Microbiol. 1999;65:4808–4813. doi: 10.1128/aem.65.11.4808-4813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulder W, Scholten I H J M, Grivell L A. Carbon catabolite regulation of transcription of nuclear genes coding for mitochondrial proteins in Kluyveromyces lactis. Curr Genet. 1995;28:267–273. doi: 10.1007/BF00309786. [DOI] [PubMed] [Google Scholar]
- 14.Porro D, Brambilla L, Ranzi B M, Martegani E, Alberghina L. Development of metabolically engineered Saccharomyces cerevisiae cells for the production of lactic acid. Biotechnol Prog. 1995;11:294–298. doi: 10.1021/bp00033a009. [DOI] [PubMed] [Google Scholar]
- 15.Porro D, Bianchi M M, Brambilla L, Menghini R, Bolzani D, Carrera V, Lievense J, Liu C-L, Ranzi B M, Frontali L, Alberghina L. Replacement of a metabolic pathway for large-scale production of lactic acid from engineered yeasts. Appl Environ Microbiol. 1999;65:4211–4215. doi: 10.1128/aem.65.9.4211-4215.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaaff I, Green J B A, Gozalbo D, Hohmann S. A deletion of the PDC1 gene coding for pyruvate decarboxylase of yeast causes a different phenotype than previously isolated point mutations. Curr Genet. 1989;15:75–81. doi: 10.1007/BF00435452. [DOI] [PubMed] [Google Scholar]
- 17.Schmitt H D, Ciriacy M, Zimmermann F K. The synthesis of yeast pyruvate decarboxylase is regulated by large variations in the messenger RNA level. Mol Gen Genet. 1983;192:247–252. doi: 10.1007/BF00327674. [DOI] [PubMed] [Google Scholar]
- 18.Wésolowski-Louvel M, Prior C, Bornecque D, Fukuhara H. Rag− mutations involved in glucose metabolism in yeast: isolation and genetic characterization. Yeast. 1992;8:711–719. [Google Scholar]
- 19.Wésolowski-Louvel M, Fukuhara H. A map of the Kluyveromyces lactis genome. Yeast. 1995;11:211–218. doi: 10.1002/yea.320110303. [DOI] [PubMed] [Google Scholar]
- 20.Zeeman A-M, Luttik M A H, Thiele C, Van Dijken J P, Pronk J T, Steensma Y H. Inactivation of the Kluyveromyces lactis KlPDA1 gene leads to loss of pyruvate dehydrogenase activity, impairs growth on glucose and triggers alcoholic fermentation. Microbiology. 1998;144:3437–3446. doi: 10.1099/00221287-144-12-3437. [DOI] [PubMed] [Google Scholar]