Significance
Hypothiocyanite and hypothiocyanous acid (OSCN−/HOSCN) have long been considered highly specific antimicrobials which rapidly damage proteins of invading bacteria while leaving mammalian cells unharmed. In this study, we have described a specific bacterial enzyme capable of reducing HOSCN to provide substantial protection against the damaging effects of this compound.
Keywords: oxidative stress resistance, bacteria, enzymology
Abstract
Hypothiocyanite and hypothiocyanous acid (OSCN−/HOSCN) are pseudohypohalous acids released by the innate immune system which are capable of rapidly oxidizing sulfur-containing amino acids, causing significant protein aggregation and damage to invading bacteria. HOSCN is abundant in saliva and airway secretions and has long been considered a highly specific antimicrobial that is nearly harmless to mammalian cells. However, certain bacteria, commensal and pathogenic, are able to escape damage by HOSCN and other harmful antimicrobials during inflammation, which allows them to continue to grow and, in some cases, cause severe disease. The exact genes or mechanisms by which bacteria respond to HOSCN have not yet been elucidated. We have found, in Escherichia coli, that the flavoprotein RclA, previously implicated in reactive chlorine resistance, reduces HOSCN to thiocyanate with near-perfect catalytic efficiency and strongly protects E. coli against HOSCN toxicity. This is notable in E. coli because this species thrives in the chronically inflamed environment found in patients with inflammatory bowel disease and is able to compete with and outgrow other important commensal organisms, suggesting that HOSCN may be a relevant antimicrobial in the gut, which has not previously been explored. RclA is conserved in a variety of epithelium-colonizing bacteria, implicating its HOSCN reductase activity in a variety of host–microbe interactions. We show that an rclA mutant of the probiotic Limosilactobacillus reuteri is sensitive to HOSCN and that RclA homologs from Staphylococcus aureus, Streptococcus pneumoniae, and Bacteroides thetaiotaomicron all have potent protective activity against HOSCN when expressed in E. coli.
Hypothiocyanite (OSCN−) is a potent antimicrobial oxidant and is widely considered a highly specific immune defense mechanism used by the human body that selectively damages microbes and not host tissue. OSCN−, which exists in equilibrium in aqueous solution with protonated hypothiocyanous acid (HOSCN), is a pseudohypohalous acid closely related to hypohalous acids such as hypochlorous acid (HOCl) and hypobromous acid, which are known for being aggressive oxidants that cause damage to nearly every type of macromolecule found in cells, with HOCl being the most damaging and reactive of the three (1–4).
During inflammatory response by the innate immune system, heme peroxidase enzymes convert (pseudo)halide ions into (pseudo)hypohalous acids, which are then released into the phagosome and the immediate environment to kill pathogenic bacteria (5, 6). Myeloperoxidase (MPO), which is released by degranulation of leukocytes during inflammation (7), and lactoperoxidase (LPO), which is secreted in the lungs, breastmilk, and saliva (8, 9), are primarily responsible for the production of HOCl and HOSCN, respectively (10–12). These enzymes catalyze the formation of hypohalous and pseudohypohalous acids through a two-electron reaction with H2O2 and the corresponding halide or pseudohalide ions (Cl−, Br−, and thiocyanate [SCN−]). Hypohalous acids are capable of oxidizing nearly all biomolecules (1) but react most quickly with sulfur-containing amino acid residues, and this is thought to be the basis of their antimicrobial activity (1). In the case of HOCl, both Cys and Met residues are rapidly oxidized, forming irreversibly oxidized sulfinic and sulfonic acids and methionine sulfoxide, respectively (2). HOSCN more specifically oxidizes Cys residues, forming sulfenic acids and disulfide bonds, which are oxidation products that can be reversed by cellular reductants (2).
SCN−, the precursor of HOSCN, is found in many human secretions, with the highest reported concentrations (from 0.01 to 2 mM) in the lungs and oral cavity (13). The levels of SCN− in human fluids are dependent on diet and can be increased by consuming brassica vegetables such as broccoli or Brussels sprouts (14), as well as being present in much higher concentrations in the plasma of smokers (up to 3 mM) (13, 15). In mammalian tissues, accumulation of SCN− acts as a chemical shield against damaging hypohalous acids (16). Not only is SCN− the preferred substrate of heme peroxidases (including MPO and LPO), leading to production of higher concentrations of HOSCN than of HOCl when SCN− is present, but when HOCl reacts with SCN−, it forms HOSCN, which is less reactive than HOCl and is also readily reduced by mammalian selenocysteine-containing thioredoxin reductase (10, 17). This cycle limits damage to the host during inflammation. Bacterial thioredoxin reductase, lacking selenocysteine, is potently inhibited by HOSCN (18)
While work from many laboratories over the last decade has identified multiple mechanisms by which bacteria defend themselves against HOCl and other reactive chlorine species (19–25), to date, no specific bacterial defense system against HOSCN has been identified. This contributes to the consensus that HOSCN is a highly specific antimicrobial which is nearly harmless to mammalian cells (26). Some bacterial species, both pathogenic and commensal, are able to survive in areas of high inflammation where HOSCN is found in abundance (4, 9, 27), but the mechanism allowing their survival is unknown. In this study, we have identified the flavoprotein RclA, previously implicated in HOCl stress response (6, 28, 29), as a highly active, broadly conserved HOSCN reductase which strongly protects bacteria against HOSCN stress.
In Escherichia coli, RclA is a flavin-dependent oxidoreductase transcribed as part of the rcl operon, which consists of the transcriptional activator rclR and three genes: rclA, rclB, and rclC (29). Only RclR and RclA have been studied in depth, but overall, this operon is known to play a role in oxidative stress resistance, particularly against HOCl. Following exposure to HOCl and other reactive chlorine compounds, RclR rapidly up-regulates transcription of the rclABC operon more than 500-fold (29, 30). Recently, Derke et al. (28) and Baek et al. (6) reported that RclA has Cu(II) reductase activity. When E. coli is exposed to a combination of intracellular Cu(II) and HOCl, RclA provides modest protection (28), although the mechanism by which this occurs remains unclear (6). RclA is also involved in colonizing the intestine of fruit flies (28) and, in Salmonella, resisting killing by macrophages (6). However, the in vitro Cu(II) reductase activity of RclA is extremely slow, which is odd for a flavin-dependent enzyme like RclA. Perplexingly, in vitro Cu(II) reductase activity was also reported to require O2, and mutation of the conserved active-site cysteine 43 in RclA enhanced the Cu(II) reductase activity of the enzyme (6). These points prompted us to continue probing the function of RclA and its role in oxidative stress response, leading us to the identification of HOSCN as a physiologically relevant RclA substrate.
RclA is a broadly conserved enzyme (28), with homologs in many bacteria that colonize or infect epithelial surfaces, including Limosilactobacillus reuteri, Bacteroides thetaiotaomicron, Streptococcus pneumoniae, and Staphylococcus aureus (31), and we show that the RclA homologs from these distantly related bacteria also potently protect bacteria against HOSCN. The identification of RclA as a HOSCN-detoxifying enzyme therefore has important implications for understanding how a wide variety of bacteria, both pathogenic and commensal, survive interactions with antimicrobials that are released by the mammalian immune system.
Results
RclA Is a Highly Active Bacterial HOSCN Reductase.
RclA is a member of the pyridine nucleotide-disulfide oxidoreductase family, and these enzymes (including RclA) have a pair of cysteine residues in their active site that are used to reduce their disulfide-containing substrates using NAD(P)H (32). During catalysis, the cysteine pair cycles between disulfide and dithiol states as reducing equivalents are transferred from NAD(P)H to the disulfide-containing substrate, and a flavin adenine dinucleotide prosthetic group mediates electron transfer between NAD(P)H and the enzymatic cysteine pair. We found it noteworthy that RclA’s active site contains these thiols, which are presumably important for the enzyme’s function, as they are the biological functional group most susceptible to oxidation by the antimicrobial oxidants that RclA provides resistance against in vivo (28, 29). However, in RclA, the intramolecular disulfide that would result upon oxidation of its cysteine pair can be rapidly reduced back to the dithiol state by NAD(P)H via the enzyme’s flavin. We therefore hypothesized that RclA may provide resistance against the antimicrobial oxidants produced by the immune system by rapidly reducing one of those oxidants using NAD(P)H, thereby detoxifying the oxidant before it has a chance to react with other cellular targets. We investigated this hypothesis in this study.
HOCl, N-chlorotaurine (NCT), and HOSCN are among the most abundant antimicrobial oxidants produced by the innate immune system (4, 33, 34), and we evaluated them as potential substrates for RclA in vitro using NAD(P)H oxidation as a readout. HOCl was too reactive to evaluate, as it spontaneously reacts with NAD(P)H too fast to determine if RclA enhances the rate of NAD(P)H oxidation (35). NCT and HOSCN spontaneously reacted with NAD(P)H slowly enough that we could measure the enhancement in NAD(P)H oxidation rate upon adding RclA (SI Appendix, Fig. S1), and we therefore evaluated these two oxidants as potential substrates for RclA. RclA exhibited slow NAD(P)H oxidase activity under aerobic conditions due to the intrinsic ability of flavins to be oxidized by O2. Both NCT and HOSCN significantly enhanced the NAD(P)H oxidase activity of RclA at 200 µM oxidants, though the rate enhancement was much more dramatic with HOSCN than with NCT (Fig. 1A). Notably, the NAD(P)H oxidation rate was more than 3 times and more than 100 times greater with NCT and HOSCN, respectively, than the NAD(P)H oxidation rate in the presence of Cu(II), indicating that both of these oxidants react much more quickly with RclA than with Cu(II). The enzyme activity with NADH was greater than the activity with NADPH for both NCT and HOSCN, so kinetic parameters with these oxidants were determined using NADH as the pyridine nucleotide in the following in vitro experiments.
Fig. 1.
RclA is a potent HOSCN reductase. (A) NAD(P)H oxidation rates with various potential substrates. Note the logarithmic y axis. The reaction was initiated by adding RclA into a buffered solution containing 200 µM NAD(P)H and 200 µM potential substrate. 10 nM RclA was used in assays with HOSCN, and 2 µM RclA was used with all other substrates. The rates indicate the apparent activity per enzyme active site with each substrate. The control indicates the NAD(P)H oxidation rate in the absence of added substrate (error bars of one standard deviation). (B) Michaelis-Menten plot of the NADH oxidation reaction velocity with 180 µM NADH, 10 nM RclA, and various concentrations of HOSCN. (C) 13C-NMR spectra of SCN− and HOSCN. Upon addition of RclA and NADH, the signal for HOSCN decreased and the signal for SCN− increased, indicating that HOSCN was converted to SCN−. Addition of NADH or RclA alone did not convert HOSCN to SCN−.
We next measured the NADH oxidation rate at several NCT and HOSCN concentrations in order to determine steady-state kinetic parameters for RclA with these two oxidants. With NCT, RclA had an apparent kcat (turnover number) of 33 s−1 and an apparent KM (Michaelis constant) of 12.7 mM for NCT, giving a relatively low kcat/KM value of 2.6 × 103 M−1s−1 (SI Appendix, Fig. S2). The related enzyme glutathione reductase (GR) displays similar activity with NCT, suggesting that the ability to react with NCT is a general property of pyridine nucleotide-disulfide oxidoreductases and is not specific to RclA (SI Appendix, Fig. S3). We also measured RclA’s activity with a panel of amino acid chloramines to determine if RclA can generally reduce all chloramines, but only N-chloroglycine (NCG) enhanced the NADH oxidation rate above background, suggesting that only smaller chloramines can fit within RclA’s active site (Fig. 1A and SI Appendix, Fig. S4). With HOSCN, RclA had an apparent kcat of 180 s−1 and an apparent KM of 2 µM, giving a kcat/KM of 9 × 107 M−1s−1 at 180 µM NADH (Fig. 1B). This value is near the diffusion limit, indicating that RclA has near-perfect catalytic efficiency with HOSCN. Notably, the KM for HOSCN is well below the HOSCN concentrations that bacteria are predicted to encounter in vivo (estimates of steady-state concentrations in saliva, for example, range between 10 and 60 µM) (36, 37). In contrast, HOSCN reacts more poorly with GR, with GR having an apparent kcat of 6.1 s−1, an apparent KM of 81 µM, and kcat/KM of 7.5 × 104 M−1s−1 for HOSCN, indicating that the ability to react with HOSCN is specific to RclA and is not a general property of pyridine nucleotide-disulfide oxidoreductases (SI Appendix, Fig. S5). Measuring RclA’s activity at a variety of NADH and HOSCN concentrations and generating a double-reciprocal plot revealed a series of parallel lines, which demonstrates that RclA follows the ping-pong kinetic mechanism with respect to HOSCN and NADH (SI Appendix, Fig. S6).
We expected that SCN− would be the product resulting from the RclA-catalyzed reaction of HOSCN with NADH. 13C NMR was used to verify this. 13C-labeled HOSCN was first generated from 13C SCN− using LPO, and the 13C resonances for SCN− and HOSCN were assigned based on prior reports (33, 38). Note that SCN− is always present in the sample containing HOSCN due to an inability to achieve complete conversion using LPO. Addition of NADH and a catalytic amount of RclA to 13C HOSCN caused the signal for HOSCN to disappear and the intensity of the peak for SCN− to increase, indicating that HOSCN was reduced to SCN− by RclA (Fig. 1C). Addition of NADH or RclA alone did not perturb the signal for 13C HOSCN.
RclA Protects E. coli against HOSCN.
After determining that RclA possesses HOSCN reductase activity in vitro, we investigated the effect of this enzyme on the E. coli oxidative stress response in vivo. Wild-type (WT) E. coli and an isogenic rclA knockout strain were exposed to varying concentrations of HOSCN over 24 h (Fig. 2A). In the presence of HOSCN, WT E. coli recovered from the stress and entered log-phase growth more quickly than the ΔrclA knockout. Chromosomal complementation of the ΔrclA mutation in a single copy restored WT HOSCN resistance, and expression of rclA from a multicopy plasmid completely protected E. coli from the tested HOSCN concentrations. Mutants lacking rclB and rclC were also more sensitive to HOSCN than WT (SI Appendix, Fig. S7).
Fig. 2.
rclA protects E. coli against HOSCN. (A) WT, ΔrclA, ΔrclA attλ::pRCLA15(PrclA-rclA, cat+), and ΔrclA/pRCLA1 (rclA+, bla+) strains of E. coli MG1655 were exposed to the indicated HOSCN concentrations and incubated 24 h at 37 °C with shaking (n = 8 technical replicates with error bars of 1 SD, representative of three independent experiments). (B) qRT-PCR was performed using rclA-specific primers on RNA isolated from WT E. coli after exposure to HOSCN in biological quadruplicate with error bars representing SD. A one-way ANOVA was performed to test significance between groups (*P < 0.05, **P < 0.01).
Expression of rclA is regulated by RclR, which responds to reactive chlorine species (29), and the RclR homolog of Pseudomonas aeruginosa also responds to HOSCN (39). We therefore examined how different concentrations of HOSCN affect E. coli rclA expression using qRT-PCR (Fig. 2B). At 20 and 200 µM HOSCN, expression of rclA increased more than 256-fold, indicating that RclR is strongly activated by HOSCN in E. coli. At 2 mM HOSCN, rclA expression was variable, probably because the bacteria are damaged or dead at that concentration (Fig. 2B).
Active-Site Cysteines C43 and C48 Are Required for HOSCN Reductase Activity of RclA.
There are two cysteine residues within the active site of RclA (C43 and C48) which are characteristic of flavoprotein-disulfide oxidoreductases (40). Typically, in flavoprotein-disulfide reductases, the active-site cysteines in the dithiol form (EH2) transfer electrons to the substrate, with the cysteines becoming oxidized to an intramolecular disulfide (Eox) that can subsequently be reduced back to EH2 by NAD(P)H (Fig. 3A). The EH2 state in flavoprotein-disulfide oxidoreductases is known to produce a charge-transfer band in the flavin-visible absorbance spectrum above ∼520 nm, and addition of NADH to oxidized RclA results in the formation of this charge-transfer absorbance (Fig. 3B). Subsequent addition of HOSCN converted the flavin absorbance spectrum back to that of Eox, consistent with HOSCN oxidizing RclA’s cysteines into the disulfide. We confirmed that C43 and C48 in RclA undergo reversible disulfide bond formation, as depicted in Fig. 3A using a mass spectrometry (MS)–based thiol-labeling approach (Fig. 3C and SI Appendix, Table S1). Analysis of purified RclA treated with dithiothreitol (DTT) (Fig. 3C, red) shows that C43 and C48 are primarily reduced. After complete removal of the reductant and exposure to HOSCN (Fig. 3C, ox), C43 and C48 were primarily in the oxidized form, indicating that HOSCN had oxidized the active-site cysteines to the disulfide. Subsequent treatment with an excess of NADH to rereduce the protein (Fig. 3C, rered) caused C43 and C48 to become reduced again. C396, another cysteine that is not in the active site of RclA, did not show major changes in oxidation state upon the various treatments. SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) analysis and reverse thiol trapping with methoxypolyethylene glycol (mPEG) maleimide further demonstrated that two cysteines become oxidized upon HOSCN treatment and are rereduced by NADH and showed that HOSCN treatment does not induce the formation of intermolecular disulfides between different monomers (SI Appendix, Fig. S8). Combined, these results show that like other flavoprotein-disulfide oxidoreductases during catalysis, RclA’s active-site cysteines cycle between a disulfide and a dithiol form upon reacting with HOSCN and NADH.
Fig. 3.
Active-site cysteines C43 and C48 are required for HOSCN reduction by RclA. (A) Schematic representation of RclA-mediated reduction of HOSCN using NAD(P)H. (B) UV-vis absorbance spectrum of RclA after treatment with NADH (EH2) and subsequent reoxidation with HOSCN (Eox). (C) MS–based analysis of the oxidation status of RclA’s cysteines after different treatments; red, after reducing enzyme with DTT; ox, after removing the DTT and oxidizing with HOSCN; rered, subsequent addition of an excess of NADH. (D) Activity of C43A and C48A mutants of RclA compared with WT. Reactions were initiated by adding 10 nM RclA into a buffered solution containing 200 µM HOSCN and 180 µM NADH. (E) E. coli ΔrclA complemented with pBAD30 plasmids expressing the indicated mutant forms of rclA. Strains were exposed to HOSCN, with A600 measured for 24 h. Experiments were performed in biological triplicate with error bars representing SD.
Based on the in vitro behavior of C43 and C48 upon HOSCN treatment, we hypothesized that, if HOSCN is the physiological substrate of RclA, mutating either of these residues would drastically impair HOSCN reduction by the enzyme. We mutated each active-site cysteine residue to alanine and measured the in vitro HOSCN reductase activity for each mutant. While WT RclA is capable of rapidly reducing HOSCN (concomitant with NADH oxidation), the C43A (RclAC43A) and C48A (RclAC48A) mutants were catalytically inactive (Fig. 3D), confirming that these two cysteine residues are critical for RclA’s HOSCN reductase activity. To verify that these two cysteines are important for RclA’s function in vivo, we individually expressed RclAC43A and RclAC48A in the rclA knockout strain and measured growth in the presence of HOSCN (Fig. 3E). The rclA knockout complemented with WT RclA on a plasmid was resistant to HOSCN, but complementation with either RclAC43A or RclAC48A provided no protection. These results taken together show that HOSCN is a more typical and more physiologically relevant substrate of RclA than Cu(II).
RclA Homologs Also Protect against HOSCN.
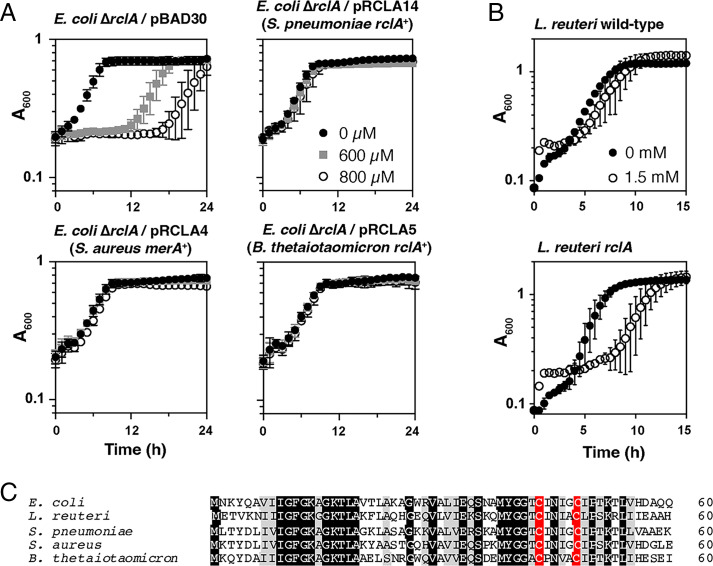
Because RclA was so effective at protecting against HOSCN stress in E. coli (Figs. 2A and 3E), we hypothesized that HOSCN reduction might be a conserved function of this enzyme. RclA homologs are found in diverse bacteria (SI Appendix, Fig. S9) (28). We therefore complemented the E. coli ΔrclA mutant with plasmids encoding homologs of RclA from the gram-positive pathogens S. pneumoniae and S. aureus and the gram-negative gut commensal B. thetaiotaomicron (these enzymes range from 47 to 49% amino acid sequence identity to E. coli RclA). Both S. pneumoniae and S. aureus were selected because of their role in colonizing tissues during chronic inflammation (41, 42), especially in the lungs, where they would be expected to come into contact with high concentrations of HOSCN (27, 43), as well as previous studies investigating the role of the S. aureus RclA homolog MerA, which was found to protect S. aureus against HOCl (44). B. thetaiotaomicron was chosen because it is an important commensal organism found, like E. coli, in the human gut (45). Fig. 4A shows that all three rclA homologs provided substantial resistance to all concentrations of HOSCN tested, similar to the resistance provided by the complementation with rclA from E. coli. Additionally, we tested the sensitivity of an rclA mutant of L. reuteri (46), a gut-dwelling probiotic bacterium (47), to HOSCN (Fig. 4B). While L. reuteri was generally more resistant to HOSCN than E. coli, the rclA knockout was substantially more sensitive than WT. L. reuteri mutants lacking other predicted redox response genes (46) were tested, but none were as sensitive to HOSCN as the rclA mutant (SI Appendix, Fig. S10). Fig. 4C shows a comparison of the amino acid sequences of the N termini of the four RclA homologs in comparison to the E. coli RclA, with the active-site Cys residues in red. The full-length alignment is shown in SI Appendix, Fig. S11. The conserved region N terminus of the active site contains two lysine residues previously reported to affect RclA metalloreductase activity (6) and is predicted to make up one side of the cleft leading into the active site in the RclA structure (SI Appendix, Fig. S12).
Fig. 4.
Homologs of rclA protect bacteria against HOSCN. (A) Growth of E. coli ΔrclA expressing homologs of rclA from S. pneumoniae and B. thetaiotaomicron and merA from S. aureus from pBAD30-derived plasmids. Strains were exposed to the indicated concentrations of HOSCN and incubated for 24 h at 37 °C with shaking. Experiments were performed in biological triplicate with error bars representing SD. (B) L. reuteri WT and rclA mutant cultures were exposed to 1.5 mM HOSCN for 15 h at 37 °C, with A600 measured every 15 min. Averages of biological triplicate are shown with error bars representing SD. Additional L. reuteri mutants tested can be found in SI Appendix, Fig. S10. (C) Alignment of the N-terminal 60 residues of RclA from E. coli and homologs from the indicated species (Clustal Omega). Fully and partially conserved residues are indicated in black and gray, respectively, and active-site cysteines are indicated in red. Full-length alignment is shown in SI Appendix, Fig. S12.
Discussion
During inflammation, the human immune system releases a variety of reactive and damaging antimicrobials meant to fight off invading pathogens. Understanding how bacteria can evade these powerful oxidants, including the hypohalous acids, is crucial to human health. We have discovered that the pseudohypohalous acid HOSCN is the physiologically relevant substrate of the widely conserved bacterial flavin-dependent oxidoreductase RclA, an enzyme that plays a role in the survival of oxidative stress for E. coli (28). While we have not yet directly addressed the effect of this enzyme on host colonization in vivo, we have laid an important foundation for future studies with the data we have gathered here.
When comparing the rate of reduction between HOSCN and other potential substrates, including Cu(II), which was previously thought to be the most relevant substrate of RclA (6, 28), HOSCN reduction was substantially faster than any other compound tested. This reduction rate was shown to be fast in vitro for RclA. Furthermore, when the two Cys residues of the RclA active site were mutated, all HOSCN reductase activity by the enzyme was eliminated. This is also in contrast with previously published results on Cu(II) reductase activity (6), and it provides more evidence that RclA is primarily a HOSCN reductase. At this time, it is unclear what role the Cu(II) reduction plays in this system, although the fact that rclA mutants have a Cu-dependent HOCl sensitivity phenotype (28) suggests there might be a physiological connection. Additionally, a recent study from Hajj et al. (48) demonstrated that E. coli two-component system HprSR responds to HOCl and regulates some copper response genes, reinforcing the idea that there is a link between the two during stress. In light of our current results, the mechanism by which RclA (and its S. aureus homolog MerA) (44) protects bacteria against reactive chlorine compounds remains unclear (28, 29). RclA has modest NCT and NCG reductase activity (Fig. 1A), which could contribute to detoxification of chloramines formed in vivo during HOCl exposure (5). We were also unable to determine if HOCl is a substrate for RclA because HOCl spontaneously oxidizes NAD(P)H faster than we can measure using our instrumentation, leaving open the possibility that RclA may also be capable of reducing HOCl. However, the rapid, spontaneous reaction of HOCl with biomolecules like NAD(P)H raises doubts that an RclA-like reductase would be capable of detoxifying HOCl before it reacts with other cellular targets. MerA has also been reported to have allicin reductase activity and to contribute to resistance to this thiol-targeting compound in S. aureus (31). It seems likely that RclA homologs provide protection against a variety of oxidative stresses, but the exceptional speed and catalytic efficiency of RclA’s HOSCN reductase activity (Fig. 1), especially compared with the lack of spontaneous reaction between HOSCN and NAD(P)H (SI Appendix, Fig. S1), argue for this being the primary physiological role of this enzyme.
The presence of a strong HOSCN reductase in gut-dwelling bacteria like E. coli, L. reuteri, and B. thetaiotaomicron suggests a previously underappreciated role for HOSCN during gut inflammation. In the lungs, the pneumonia-causing pathogen S. pneumoniae possesses RclA and is known to take advantage of host inflammation to cause severe infection (49), but its relationship with HOSCN has only recently been studied (50). Because SCN− outcompetes Cl− and Br− ions for oxidation by peroxidase enzymes (36), it is likely that wherever SCN− is found during inflammation, there will be a considerable amount of HOSCN produced, although the in vivo concentration of HOSCN has not been directly measured in any mammalian tissue or fluid due to its reactivity. In the lungs and oral cavity, SCN− concentrations are as high as 3 mM, while the concentration of SCN− has not, to our knowledge, been measured in the gut (9, 14, 15, 27, 38, 43, 51, 52). Notably, we exposed bacteria to a bolus addition of HOSCN, rather than attempting to simulate the steady-state production of HOSCN that cells would likely encounter in a host. The concentrations of HOSCN we found to inhibit E. coli and L. reuteri (Figs. 2–4) with this method are consistent with those reported by other groups for other bacterial species (39, 53, 54).
HOSCN reductase activity in crude lysates of some oral Streptococcus species was described more than 50 y ago, and the presence of this activity is correlated with the ability of streptococci to survive exposure to HOSCN or salivary LPO (55, 56). However, no gene or enzyme responsible for this activity has been identified. With complete genome sequences now available, we were able to determine that oral Streptococcus species such as Streptococcus sanguinis (formerly Streptococcus sanguis), Streptococcus mitis, and Streptococcus salivarius, which possess HOSCN reductase activity also possess homologs of RclA, while Streptococcus mutans has neither HOSCN reductase activity nor an RclA homolog (55, 57). In 1996, Courtois and Pourtois (58) reported the partial purification of a 21-kDa protein from S. sanguinis that they believed to be an HOSCN reductase, but the protein was never identified and was only partially purified, leading us to speculate that these authors were observing the activity of RclA contamination in their protein preps. Future experiments will be needed to establish definitively whether RclA is responsible for HOSCN reductase activity in oral streptococci.
Of course, it would be unlikely for RclA to be the sole protector against an antimicrobial such as HOSCN. The rcl operon in E. coli consists of the transcriptional regulator RclR and three additional genes: rclA, rclB, and rclC (29). A homolog of RclR (but not of RclA) is found in P. aeruginosa and responds to HOSCN to up-regulate the transcription of the peroxiredoxin RclX (39). While RclA is very broadly conserved, the complete set of RclA, RclB, and RclC is found only in Enterobacteriaceae, including E. coli, Salmonella, and other related organisms (28). RclB is a small periplasmic protein, and RclC is an inner membrane protein. We found that E. coli knockouts of either rclB or rclC (29) are sensitive to HOSCN, suggesting that those genes are also involved in HOSCN defense by currently unknown mechanisms (SI Appendix, Fig. S7). A homolog of RclC has recently been reported to play an important role in HOCl resistance in uropathogenic E. coli, but the mechanism by which it does so is not yet known (59). In L. reuteri, perR, msrB, hslO, and sigH mutants (46) were sensitive to HOSCN stress, albeit not as sensitive as the rclA mutant (SI Appendix, Fig. S10). These results show that RclA, while important for HOSCN resistance, is not the sole determinant of bacterial survival under HOSCN stress.
Perhaps our most exciting finding is that homologs of RclA, including from the gut commensal species B. thetaiotaomicron and L. reuteri and from species implicated in serious lung disease such as S. pneumoniae and S. aureus (31, 41, 42, 45), protect against HOSCN damage to the same degree as E. coli RclA. This indicates that a wide range of bacteria, both commensal and pathogenic, may possess specific defenses against HOSCN stress (28). Learning more about the scope of protection provided by this enzyme to pathogenic species will gain us better knowledge on potentially a wide range of diseases, including cystic fibrosis, inflammatory bowel disease, and oral diseases. Future in vivo experiments will be needed to provide insight into how bacteria evade the host immune response and chronic inflammation in any tissue where HOSCN is found. By identifying the function of RclA in the model organism E. coli, which notably is able to compete with commensal organisms and thrive in an inflamed gut (60), we have laid the foundation for understanding bacterial survival and the relationship to the human immune system in ways that were previously not understood.
Materials and Methods
Additional details of materials and methods are available in SI Appendix, Materials and Methods.
Enzyme Assays.
All in vitro experiments with Cu(II), NCT, and chlorinated amino acids were done in 20 mM HEPES and 100 mM NaCl buffer, pH 7. All in vitro experiments with HOSCN were done in 100 mM sodium phosphate buffer, pH 7.4. Relative NAD(P)H oxidation rates by various substrates without and with WT or mutant RclA were measured under aerobic conditions, using the change absorbance of NAD(P)H at 340 nm as a readout. A Shimadzu UV-1900 ultraviolet-visible (UV-vis) spectrophotometer (UV Probe software) was used to monitor the absorbance change. Reactions were initiated by first injecting 200 μM substrate into the solution containing 200 μM NAD(P)H, followed by addition of the enzyme. The concentration of the enzyme for the experiments with Cu(II), NCT, and other chlorinated amino acids was 2 µM. For the experiments with HOSCN, the concentration of RclA or mutant enzyme was 10 nM due to the much higher activity of RclA with HOSCN. All experiments were repeated in triplicate.
Stopped-Flow Steady-State Kinetic Assays.
Steady-state kinetic assays used to determine kcat and KM for the reactions of RclA or GR with NCT or HOSCN were carried out using a TgK Scientific SF-61DX2 KinetAsyst stopped-flow spectrophotometer (with Kinetic Studio software). Stopped-flow experiments with NCT were carried out under anaerobic conditions to eliminate the NAD(P)H oxidase activity of the enzymes. The reaction of both RclA and GR with NCT is slow enough that the NAD(P)H oxidase activity of the enzymes contributes significantly to the apparent reaction velocity under aerobic conditions. Accordingly, for experiments with NCT, the enzyme solution was made anaerobic in a glass tonometer by cycling with vacuum and anaerobic argon (61), with NAD(P)H in a side arm separated from the enzyme solution. After the solution was made anaerobic, NAD(P)H from the side arm was added to the enzyme solution. The 10 µM enzyme + 100 µM NAD(P)H solution was then loaded onto the instrument and mixed with buffer solutions containing varying concentrations of NCT (0.1 to 49.8 mM) that had been made anaerobic by sparging with argon (all concentrations listed are after mixing in the stopped-flow instrument), and the change in absorbance at 340 nm was used as a readout. With HOSCN, the NAD(P)H oxidase activity of RclA and GR is insignificant compared with the NAD(P)H oxidation rate in the presence of HOSCN such that steady-state assays could be performed under aerobic conditions. However, the enzyme solution, NAD(P)H solution, and HOSCN solution were all kept in separate syringes on the stopped-flow instrument to prevent the slow oxidation of NAD(P)H that would otherwise have occurred if NAD(P)H were premixed with either aerobic RclA or HOSCN prior to initiating the experiment. Accordingly, the stopped-flow instrument’s double-mixing mode was utilized, with NAD(P)H and RclA/GR combined in the instrument’s first mix, which was then mixed with the HOSCN solution in the second mix, using the shortest delay time possible between the two mixes for the instrument (50 ms), and the change in absorbance of NAD(P)H at 340 nm was used as a readout. Stopped-flow experiments with HOSCN were done using 10 nM enzyme, 180 µM NAD(P)H, and 1 to 100 µM HOSCN for RclA or 50 to 250 µM HOSCN for GR (all concentrations after mixing). Initial velocities were plotted against the substrate concentration and fit to the Michaelis-Menten equation using KaleidaGraph to determine kcat and Km. For determining the kinetic mechanism of NADH and HOSCN binding with RclA, stopped-flow steady-state assays were repeated using several fixed concentrations of NADH (25, 50, 90, and 180 µM), and double-reciprocal (Lineweaver-Burk) plots of the reaction velocities were made.
Oxidative Half-Reaction Kinetics.
The direct reaction between RclA having reduced active-site cysteines and HOSCN in Fig. 3B was monitored anaerobically in a stopped-flow spectrophotometer. The RclA solution was made anaerobic in a glass tonometer, with 0.9 equivalents of NADH in a side arm separated from the enzyme solution. After the solution was made anaerobic, NADH from the side arm was added to the enzyme solution to prereduce the active-site cysteines. The 18 µM RclA-NADH solution was loaded onto the stopped-flow instrument and mixed with 18 µM or 50 µM HOSCN (all concentrations after mixing), and the reaction was monitored using the instrument’s multiwavelength charge-coupled device detector. The absorbance spectrum at 1.6 ms after initiating the reaction for both HOSCN concentrations indicated that RclA’s cysteines had been oxidized to the disulfide state at that timepoint and no further changes in signal occurred after that.
13C-NMR Spectroscopy.
All 13C-NMR spectra were acquired on a JEOL ECZ-400S, 400-MHz digital FT-NMR (Fourier Transform Nuclear Magnetic Resonance) spectrometer equipped with a 400 MHz, 5 mM field gradient ROYAL digital autotune probe, ZNM-03811RO5S-4S, and controlled by Delta 5.3 software. The samples were prepared in 50 mM phosphate buffer, pH 7; 0.1% 1,4-dioxane; and 10% D2O, at 25 °C using 5-mm NMR tubes. 1,4-dioxane was used as a chemical shift reference (66.6 ppm). The concentration of HOSCN in the samples was 1.6 mM, with 5 mM NADH and 1 µM RclA. Reactions were allowed to proceed for 1 min before adding NaOH to the samples. Due to instability of HOSCN, 100 mM NaOH was added to diminish spontaneous decomposition of HOSCN during the ∼1 h required for data collection. 1,012 scans were taken for each sample over the course of ∼1 h at 20 °C using 5-mm NMR tubes.
Measuring Growth of Bacteria under HOSCN Stress.
Single colonies of E. coli were inoculated into 5 mL of M9 minimal media with 100 µM FeCl3 and grown overnight at 37 °C with shaking. The next day, E. coli was subcultured into fresh M9 and grown to early log phase [absorbance at 600 nm (A600) = 0.3 to 0.4] before harvesting. Cultures were normalized to A600 = 0.05 in M9 medium containing the indicated concentrations of HOSCN and, for strains containing pBAD30-derived plasmids, 0.2% arabinose. The plate was then covered with a Breathe-Easy plate-sealing film (Andwin Scientific) and placed in a Tecan M1000 Infinite plate reader. A600 was measured every 30 min for 24 h at 37 °C, with shaking in between each measurement.
L. reuteri strains were grown overnight at 37 °C in malic enzyme induction broth without cysteine (MEI-C) (46) without shaking and then diluted to A600 = 0.01 in MEI-C broth containing the indicated concentration of HOSCN and aliquoted (200 μL) to clear 96-well plates. The plates were sealed with a transparent, gas-impermeable membrane and incubated for 15 h at 37 °C in a Tecan Sunrise plate reader, measuring A600 every 30 min with shaking (2 s) before each measurement.
qRT-PCR.
Analysis of rclA gene expression was measured as previously described (29). The purified RNA from HOSCN-treated E. coli was used to generate complementary DNA (cDNA) using the SuperScript IV VILO reverse-transcription kit (Thermo Fisher Scientific), and qRT-PCR was performed using a Bio-Rad CFX96 thermocycler. Expression of rclA was normalized against 16S (rrsD) gene expression, and changes were calculated using the 2−ΔΔCt method (62). SsoAdvanced Universal SYBR Green system dye (Bio-Rad) was used with rclA primers 5′ CAA AAC TTT AAG GAT AAC GGG GTT C 3′ and 5′ CCC GTT TTT AGC GAC CTT AAT ATC T 3′ and rrsD primers 5′ GAG CAA GCG GAC CTC ATA AA 3′ and 5′ TCC CGA AGG TTA AGC TAC CTA 3′.
MS Analysis of Differential Thiol-Labeled RclA.
Thiol labeling and MS analysis were performed as described by Bazopoulou et al. (63). Briefly, reduced cysteines of recombinant RclA (50 µM) were blocked with 20 mM N-ethylmaleimide (NEM) under denaturing conditions (4 M urea, 0.25% SDS, and 10 mM ethylenediaminetetraacetic acid in 100 mM sodium phosphate, pH 7.5) for 30 min at 25 °C. After trichloroacetic acid precipitation, the pellet was redissolved in denaturing buffer containing 2 mM DTT and incubated for 30 min 37 °C. Newly reduced cysteines were labeled with 10 mM iodoacetamide for 45 min at 25 °C in the dark and run on an SDS gel. MS and data analyses were performed by MS Bioworks. After in-gel digestion with trypsin at 37 °C for 4 h, peptides were analyzed by nano-liquid chromatography with tandem mass spectrometry with a Waters M-class high-pressure liquid chromatography system interfaced to a Thermo Fisher Fusion Lumos mass spectrometer. Peptides were loaded on a trapping column and eluted over a 75-μm analytical column at 350 nL/min using Luna C18 resin (Phenomenex). The mass spectrometer was operated in data-dependent mode, with the Orbitrap operating at 60,000 and 15,000 full width at half maximum for MS and tandem MS, respectively. Advanced Peak Determination was enabled, and the instrument was run with a 3-s cycle for MS and MS/MS. The target protein was identified by matching spectra, with a sequence coverage of <99% for each sample. The number of spectral counts was compared for cysteines labeled with either NEM or iodoacetamide.
Supplementary Material
Acknowledgments
This work was funded by NIH Grants R35 GM124590 (to M.J.G.) and R35 GM122506 (to K.U.) and Western Michigan University research startup funds (to F.S.). We thank Ursula Jakob (University of Michigan) for advice and technical support, Emily Schwessinger (University of Michigan) for construction of plasmids pRCLA4 and pRCLA5, and Rhea Derke (University of Alabama at Birmingham) for construction of plasmids pRCLA11, pRCLA11a, and pRCLA11b.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. A.B. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2119368119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Ulfig A., Leichert L. I., The effects of neutrophil-generated hypochlorous acid and other hypohalous acids on host and pathogens. Cell. Mol. Life Sci. 78, 385–414 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hawkins C. L., Hypochlorous acid-mediated modification of proteins and its consequences. Essays Biochem. 64, 75–86 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Hawkins C. L., Pattison D. I., Davies M. J., Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids 25, 259–274 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Pattison D. I., Davies M. J., Hawkins C. L., Reactions and reactivity of myeloperoxidase-derived oxidants: Differential biological effects of hypochlorous and hypothiocyanous acids. Free Radic. Res. 46, 975–995 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Winterbourn C. C., Kettle A. J., Redox reactions and microbial killing in the neutrophil phagosome. Antioxid. Redox Signal. 18, 642–660 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Baek Y., et al. , Structure and function of the hypochlorous acid-induced flavoprotein RclA from Escherichia coli. J. Biol. Chem. 295, 3202–3212 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies M. J., Hawkins C. L., The role of myeloperoxidase in biomolecule modification, chronic inflammation, and disease. Antioxid. Redox Signal. 32, 957–981 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Reiter B., Härnulv G., Lactoperoxidase antibacterial system: Natural occurrence, biological functions and practical applications. J. Food Prot. 47, 724–732 (1984). [DOI] [PubMed] [Google Scholar]
- 9.Day B. J., The science of licking your wounds: Function of oxidants in the innate immune system. Biochem. Pharmacol. 163, 451–457 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Aune T. M., Thomas E. L., Accumulation of hypothiocyanite ion during peroxidase-catalyzed oxidation of thiocyanate ion. Eur. J. Biochem. 80, 209–214 (1977). [DOI] [PubMed] [Google Scholar]
- 11.Pruitt K. M., Tenovuo J., Kinetics of hypothiocyanite production during peroxidase-catalyzed oxidation of thiocyanate. Biochim. Biophys. Acta 704, 204–214 (1982). [DOI] [PubMed] [Google Scholar]
- 12.Cupp-Sutton K., Ashby M. T., Reverse ordered sequential mechanism for lactoperoxidase with inhibition by hydrogen peroxide. Antioxidants 10, 1646 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.San Gabriel P. T., Liu Y., Schroder A. L., Zoellner H., Chami B., The role of thiocyanate in modulating myeloperoxidase activity during disease. Int. J. Mol. Sci. 21, 6450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felker P., Bunch R., Leung A. M., Concentrations of thiocyanate and goitrin in human plasma, their precursor concentrations in brassica vegetables, and associated potential risk for hypothyroidism. Nutr. Rev. 74, 248–258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madiyal A., et al. , Status of thiocyanate levels in the serum and saliva of non-smokers, ex-smokers and smokers. Afr. Health Sci. 18, 727–736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandler J. D., Day B. J., Biochemical mechanisms and therapeutic potential of pseudohalide thiocyanate in human health. Free Radic. Res. 49, 695–710 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandler J. D., Nichols D. P., Nick J. A., Hondal R. J., Day B. J., Selective metabolism of hypothiocyanous acid by mammalian thioredoxin reductase promotes lung innate immunity and antioxidant defense. J. Biol. Chem. 288, 18421–18428 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snider G. W., Ruggles E., Khan N., Hondal R. J., Selenocysteine confers resistance to inactivation by oxidation in thioredoxin reductase: Comparison of selenium and sulfur enzymes. Biochemistry 52, 5472–5481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goemans C. V., Collet J. F., Stress-induced chaperones: A first line of defense against the powerful oxidant hypochlorous acid. F1000 Res. 8, F1000 Faculty Rev-1678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sultana S., Foti A., Dahl J. U., Bacterial defense systems against the neutrophilic oxidant hypochlorous acid. Infect. Immun. 88, e00964-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray M. J., Wholey W. Y., Jakob U., Bacterial responses to reactive chlorine species. Annu. Rev. Microbiol. 67, 141–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.da Cruz Nizer W. S., Inkovskiy V., Overhage J., Surviving reactive chlorine stress: Responses of Gram-negative bacteria to hypochlorous acid. Microorganisms 8, 1220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goemans C. V., Vertommen D., Agrebi R., Collet J. F., Cno X., CnoX Is a chaperedoxin: A holdase that protects its substrates from irreversible oxidation. Mol. Cell 70, 614–627.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Gray M. J., Jakob U., Oxidative stress protection by polyphosphate—New roles for an old player. Curr. Opin. Microbiol. 24, 1–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang S. M., Schellhorn H. E., Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch. Biochem. Biophys. 525, 161–169 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Barrett T. J., Hawkins C. L., Hypothiocyanous acid: Benign or deadly? Chem. Res. Toxicol. 25, 263–273 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Lorentzen D., et al. , Concentration of the antibacterial precursor thiocyanate in cystic fibrosis airway secretions. Free Radic. Biol. Med. 50, 1144–1150 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derke R. M., et al. , The Cu(II) reductase RclA protects Escherichia coli against the combination of hypochlorous acid and intracellular copper. MBio 11, e01905-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker B. W., Schwessinger E. A., Jakob U., Gray M. J., The RclR protein is a reactive chlorine-specific transcription factor in Escherichia coli. J. Biol. Chem. 288, 32574–32584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Königstorfer A., et al. , Induction of the reactive chlorine-responsive transcription factor RclR in Escherichia coli following ingestion by neutrophils. Pathog. Dis. 79, ftaa079 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loi V. V., et al. , Staphylococcus aureus responds to allicin by global S-thioallylation—Role of the Brx/BSH/YpdA pathway and the disulfide reductase MerA to overcome allicin stress. Free Radic. Biol. Med. 139, 55–69 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Miller S. M., “Volume 2 complex flavoproteins, dehydrogenases and physical methods” in 8 Flavoprotein Disulfide Reductases and Structurally Related Flavoprotein Thiol/Disulfide-Linked Oxidoreductases, Russ H., Susan M., Bruce P., Eds. (De Gruyter, 2013). pp. 165–202. [Google Scholar]
- 33.Arlandson M., et al. , Eosinophil peroxidase oxidation of thiocyanate. Characterization of major reaction products and a potential sulfhydryl-targeted cytotoxicity system. J. Biol. Chem. 276, 215–224 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Gottardi W., Nagl M., N-chlorotaurine, a natural antiseptic with outstanding tolerability. J. Antimicrob. Chemother. 65, 399–409 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Prütz W. A., Measurement of copper-dependent oxidative DNA damage by HOCl and H2O2 with the ethidium-binding assay. J. Biochem. Biophys. Methods 32, 125–135 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Chandler J. D., Day B. J., Thiocyanate: A potentially useful therapeutic agent with host defense and antioxidant properties. Biochem. Pharmacol. 84, 1381–1387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tenovuo J., Pruitt K. M., Thomas E. L., Peroxidase antimicrobial system of human saliva: Hypothiocyanite levels in resting and stimulated saliva. J. Dent. Res. 61, 982–985 (1982). [DOI] [PubMed] [Google Scholar]
- 38.Nagy P., Alguindigue S. S., Ashby M. T., Lactoperoxidase-catalyzed oxidation of thiocyanate by hydrogen peroxide: A reinvestigation of hypothiocyanite by nuclear magnetic resonance and optical spectroscopy. Biochemistry 45, 12610–12616 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Farrant K. V., Spiga L., Davies J. C., Williams H. D., Response of Pseudomonas aeruginosa to the innate immune system-derived oxidants hypochlorous acid and hypothiocyanous acid. J. Bacteriol. 203, e00300-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammerstad M., Hersleth H. P., Overview of structurally homologous flavoprotein oxidoreductases containing the low Mr thioredoxin reductase-like fold—A functionally diverse group. Arch. Biochem. Biophys. 702, 108826 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Tigabu A., Getaneh A., Staphylococcus aureus, ESKAPE bacteria challenging current health care and community settings: A literature review. Clin. Lab. 67, 1539–1549 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Subramanian K., Henriques-Normark B., Normark S., Emerging concepts in the pathogenesis of the Streptococcus pneumoniae: From nasopharyngeal colonizer to intracellular pathogen. Cell. Microbiol. 21, e13077 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moskwa P., et al. , A novel host defense system of airways is defective in cystic fibrosis. Am. J. Respir. Crit. Care Med. 175, 174–183 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loi V. V., et al. , Redox-sensing under hypochlorite stress and infection conditions by the Rrf2-family repressor HypR in Staphylococcus aureus. Antioxid. Redox Signal. 29, 615–636 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porter N. T., Luis A. S., Martens E. C., Bacteroides thetaiotaomicron. Trends Microbiol. 26, 966–967 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Basu Thakur P., et al. , Complex responses to hydrogen peroxide and hypochlorous acid by the probiotic bacterium Lactobacillus reuteri. mSystems 4, e00453-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mu Q., Tavella V. J., Luo X. M., Role of Lactobacillus reuteri in human health and diseases. Front. Microbiol. 9, 757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El Hajj S., et al. , HprSR is a reactive chlorine species-sensing, two-component system in Escherichia coli. J. Bacteriol. 204, e00449-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiser J. N., Ferreira D. M., Paton J. C., Streptococcus pneumoniae: Transmission, colonization and invasion. Nat. Rev. Microbiol. 16, 355–367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gingerich A. D., et al. , Oxidative killing of encapsulated and nonencapsulated Streptococcus pneumoniae by lactoperoxidase-generated hypothiocyanite. PLoS One 15, e0236389 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson I. T., Glucosinolates: Bioavailability and importance to health. Int. J. Vitam. Nutr. Res. 72, 26–31 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Narbad A., Rossiter J. T., Gut glucosinolate metabolism and isothiocyanate production. Mol. Nutr. Food Res. 62, e1700991 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Groitl B., Dahl J. U., Schroeder J. W., Jakob U., Pseudomonas aeruginosa defense systems against microbicidal oxidants. Mol. Microbiol. 106, 335–350 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shearer H. L., Paton J. C., Hampton M. B., Dickerhof N., Glutathione utilization protects Streptococcus pneumoniae against lactoperoxidase-derived hypothiocyanous acid. Free Radic. Biol. Med. 179, 24–33 (2022). [DOI] [PubMed] [Google Scholar]
- 55.Carlsson J., Iwami Y., Yamada T., Hydrogen peroxide excretion by oral streptococci and effect of lactoperoxidase-thiocyanate-hydrogen peroxide. Infect. Immun. 40, 70–80 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oram J. D., Reiter B., The inhibition of streptococci by lactoperoxidase, thiocyanate and hydrogen peroxide. The effect of the inhibitory system on susceptible and resistant strains of group N streptococci. Biochem. J. 100, 373–381 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen I. A., et al. , The IMG/M data management and analysis system v.6.0: New tools and advanced capabilities. Nucleic Acids Res. 49 (D1), D751–D763 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Courtois P. H., Pourtois M., Purification of NADH: Hypothiocyanite oxidoreductase in Streptococcus sanguis. Biochem. Mol. Med. 57, 134–138 (1996). [DOI] [PubMed] [Google Scholar]
- 59.Sultana S., et al. , Redox-mediated inactivation of the transcriptional repressor C3600 makes uropathogenic Escherichia coli exquisitely resistant to reactive chlorine species. bioRxiv 10.1101/2021.08.31.458474, 2021.2008.2031.458474 (2021). [DOI] [PMC free article] [PubMed]
- 60.Mirsepasi-Lauridsen H. C., Vallance B. A., Krogfelt K. A., Petersen A. M., Escherichia coli pathobionts associated with inflammatory bowel disease. Clin. Microbiol. Rev. 32, e00060-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moran G. R., Anaerobic methods for the transient-state study of flavoproteins: The use of specialized glassware to define the concentration of dioxygen. Methods Enzymol. 620, 27–49 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Bazopoulou D., et al. , Developmental ROS individualizes organismal stress resistance and lifespan. Nature 576, 301–305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.