Significance
Eastern equine encephalitis virus (EEEV) is a neuroinvasive alphavirus that causes severe neurological diseases. Entry of alphaviruses requires host–virus membrane fusion in endosomes to release the RNA genome into the cytosol for virus replication. How alphaviruses utilize their structural proteins to facilitate membrane fusion remains unclear at a molecular level. This study presents cryoelectron microscopy structures of EEEV under acidic conditions that demonstrate conformational rearrangements of the E2 glycoproteins for the exposure of the E1 fusion loops, essential for membrane fusion. A protective antibody that stabilizes E2 blocks this “prefusion” structural rearrangement. These findings suggest that development of antibodies or agents that stabilize E2 may serve as strategies for treatments of diseases caused by EEEV and other alphaviruses.
Keywords: alphavirus, EEEV, cryoelectron microscopy, infection, endosome
Abstract
Alphaviruses can cause severe human arthritis and encephalitis. During virus infection, structural changes of viral glycoproteins in the acidified endosome trigger virus–host membrane fusion for delivery of the capsid core and RNA genome into the cytosol to initiate virus translation and replication. However, mechanisms by which E1 and E2 glycoproteins rearrange in this process remain unknown. Here, we investigate prefusion cryoelectron microscopy (cryo-EM) structures of eastern equine encephalitis virus (EEEV) under acidic conditions. With models fitted into the low-pH cryo-EM maps, we suggest that E2 dissociates from E1, accompanied by a rotation (∼60°) of the E2-B domain (E2-B) to expose E1 fusion loops. Cryo-EM reconstructions of EEEV bound to a protective antibody at acidic and neutral pH suggest that stabilization of E2-B prevents dissociation of E2 from E1. These findings reveal conformational changes of the glycoprotein spikes in the acidified host endosome. Stabilization of E2-B may provide a strategy for antiviral agent development.
Eastern equine encephalitis virus (EEEV) belongs to the Alphavirus genus in the family Togaviridae. Alphaviruses are broadly classified into two groups: Old World alphaviruses (such as Sindbis virus [SINV] and Semiliki Forest virus [SFV]) and New World alphaviruses (such as EEEV, Western equine encephalitis virus, and Venezuelan equine encephalitis virus) (1). In general, Old World alphaviruses cause rashes and musculoskeletal disease, whereas New World alphaviruses can cause encephalitis in humans or domestic animals (1). EEEV is normally maintained in an enzootic cycle between Culiseta mosquitos and avian hosts in freshwater swamps. However, EEEV also can be transmitted by Aedes, Coquillettidia, and Culex mosquitoes to dead-end hosts, such as humans and horses. Although there are relatively few reported human cases of EEEV infection in the United States annually, EEEV infections can cause severe neurological disease with high mortality rates above 50% (2, 3), such as in a 2019 emergence that resulted in 38 cases of infection and 19 deaths (3).
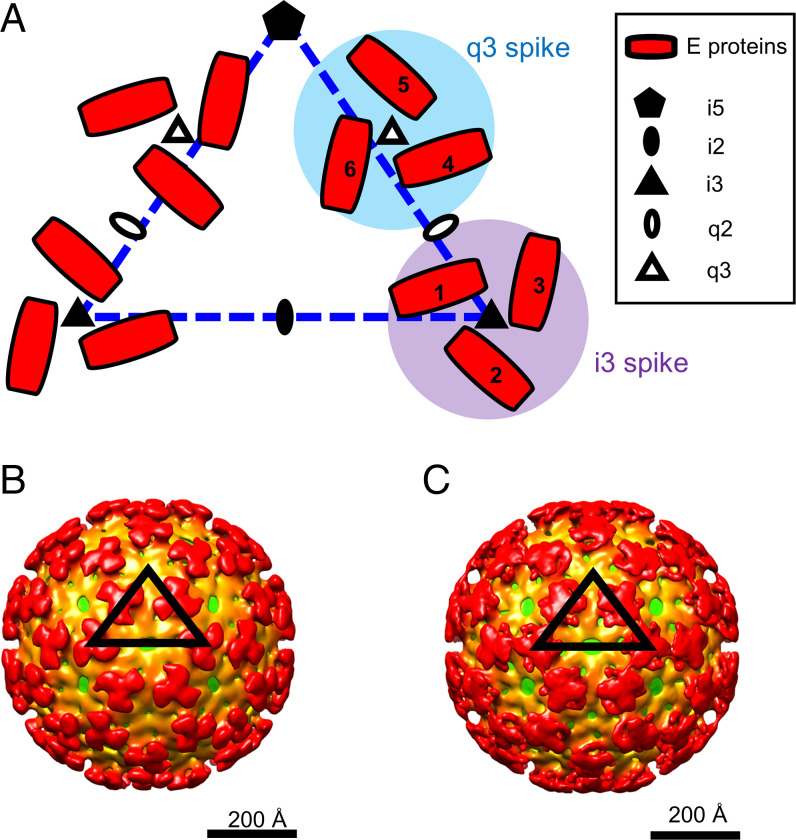
EEEV is an enveloped virus of ∼650 Å in diameter, a characteristic shared with other alphaviruses (4–7). The host-derived viral membrane contains 80 surface glycoprotein spikes (60 spikes residing at the quasithreefold [q3] positions and 20 spikes at the icosahedral threefold [i3] positions) arranged with T = 4 icosahedral symmetry (Fig. 1A) (8). Each q3 or i3 glycoprotein spike contains three threefold related E1–E2 heterodimers. Within the viral envelope, 240 copies of the capsid proteins form a core enclosing the ∼12-kb positive-sense viral RNA genome, which encodes four nonstructural proteins (nsP1, nsP2, nsP3, and nsP4) and six structural proteins (E1, E2, E3, 6K, TF, and capsid) (1, 9, 10). Each capsid monomer utilizes its N-terminal domain to bind the viral RNA genome and its C-terminal domain to form contacts with the C-terminal cytoplasmic domain of each E2 glycoprotein (11), resulting in T = 4 icosahedral symmetry of the capsid core.
Fig. 1.
Low-pH EEEV cryo-EM reconstructions with icosahedral symmetry imposed. (A) Diagram of alphavirus E1–E2 glycoproteins arranged with T = 4 icosahedral symmetry. The dashed triangle (blue) defines an asymmetric unit in an alphavirus structure with T = 4 icosahedral symmetry. Quasitwofold (q2), q3, icosahedral twofold (i2), i3, and icosahedral fivefold (i5) symmetry elements are labeled. The envelope glycoproteins (E proteins) containing E1 and E2 are shown in red. One asymmetric unit contains one i3 E2–E1 protein (labeled 1) and three q3 E2–E1 proteins (labeled 4, 5, and 6). (B) Cryo-EM reconstruction of prefusion state 1 of EEEV. (C) Cryo-EM reconstruction of prefusion state 2 of EEEV. The black triangles indicate an asymmetric unit of the EEEV cryo-EM reconstruction with T = 4 icosahedral symmetry. The cryo-EM maps are radially colored. The virus membrane is shown in green (∼240 Å from the virus center), and the glycoprotein shell is depicted in orange (∼250 Å from the virus center) and red (∼350 Å from the virus center).
The life cycle of alphaviruses involves virus attachment, low pH–mediated membrane fusion, release of the RNA genome from the capsid into the cytoplasm, virus translation, replication, and finally, assembly and budding of mature virions at the plasma membrane. Alphaviruses can utilize host–cell surface glycosaminoglycans (e.g., heparan sulfate and chondroitin sulfate) (12, 13), dendritic cell-specific intercellular adhesion molecule (ICAM) 3-grabbing non-integrin (DC-SIGN) (14), liver/lymph node-specific ICAM 3-grabbing non-integrin (L-SIGN) (14), or membrane proteins [e.g., matrix remodeling-associated 8 (Mxra8) (15), low density lipoprotein receptor class A domain containing 3 (LDLRAD3) (16), or natural resistance-associated macrophage protein 2 (NRAMP2) (17)] to attach to the cell surface. Virus particles are then internalized into host cells through clathrin-dependent endocytosis (18–20). Within the increasingly acidic pH environment of the endosome, alphavirus glycoproteins undergo conformational changes, resulting in dissociation of the E1–E2 heterodimers (SINV E1–E2 ectodomains; Protein Data Bank [PDB] ID code 3MUU) (21) and the formation of fusogenic E1 trimers (SFV E1 trimer; PDB ID code 1RER) (22) that mediate virus–host membrane fusion, followed by capsid core release and virus replication.
Previous studies proposed that the viral–host membrane fusion process occurring in the acidified endosomes involves conformational rearrangements of the E1 and E2 glycoproteins (21, 22). The E1 glycoprotein contains a transmembrane helix and an ectodomain, which can be divided into three subdomains, domains I, II, and III (E1-I, E1-II, and E1-III, respectively). The E2 glycoprotein contains a short cytosolic helix, a transmembrane helix, and an ectodomain consisting of domains A, B, and C and a β-ribbon connector (E2-A, E2-B, E2-C, and E2-β, respectively) (23). The E1 and E2 glycoproteins form heterodimers through hydrophobic and electrostatic interactions (24). It has been shown that the E1 glycoproteins form homotrimers under acidic conditions, and the “fusion loop” at the tip of E1-II can insert itself into the host endosomal membrane to trigger virus–host membrane fusion (25). However, little is known about conformational intermediates of the glycoprotein spikes in this process. In addition, the role of the E2 domains in the fusion process remains unclear. The E2-B domain likely functions to protect the E1 fusion loop from premature exposure. At low pH, the E2-B domain is destabilized, is unable to interact with the E1 fusion loop, and thus, leads to exposure of the E1 fusion loop and dissociation of E1–E2 heterodimers (21). In this study, we determined cryoelectron microscopy (cryo-EM) structures of EEEV at acidic pH mimicking the environment of the host cell endosome and identified two prefusion states of EEEV before the formation of fusogenic E1 trimers. The two cryo-EM reconstructions of EEEV suggest a possible mechanism as to how the E2 glycoproteins dissociate from E1 by rotation of the domains around the q3 and i3 axes, resulting in exposure of the E1 fusion loops. We also demonstrate that stabilization of the E2-B domain by Fab22, an antigen-binding fragment (Fab) derived from the protective antibody EEEV-22 (26), prevents conformational changes of the glycoprotein spikes, suggesting that the E2-B domain is important in maintaining the structural integrity of the E1–E2 glycoprotein spikes. Our results provide insight into the fusion mechanism of alphaviruses and suggest that stabilization of the E2-B domain by Fab22 may serve as a strategy for future antiviral agent development.
Results
Cryo-EM Structures of EEEV Prefusion States under Low-pH Conditions.
We examined prefusion-state intermediates of alphaviruses using cryo-EM. To prevent virus aggregation at low pH, chimeric SINV/EEEV virus particles (13), which encode for the nonstructural proteins of SINV and structural proteins of EEEV, were applied onto glow-discharged lacey carbon grids. Buffer was exchanged (50 mM sodium citrate/phosphate-buffered saline [PBS], pH 5.5) with the sample on the grid after manual blotting to produce a low-pH environment, followed by standard plunge-freezing procedures (Materials and Methods). We successfully obtained micrographs showing well-distributed virus particles on the grids with minimal aggregation (SI Appendix, Figs. S1A and S2A). The Thon rings in the Fourier transform of the micrographs show reasonable data quality, and the contrast transfer function (CTF) fitting in both cases shows maximum resolution greater than 5 Å (SI Appendix, Figs. S1 B and C and S2 B and C).
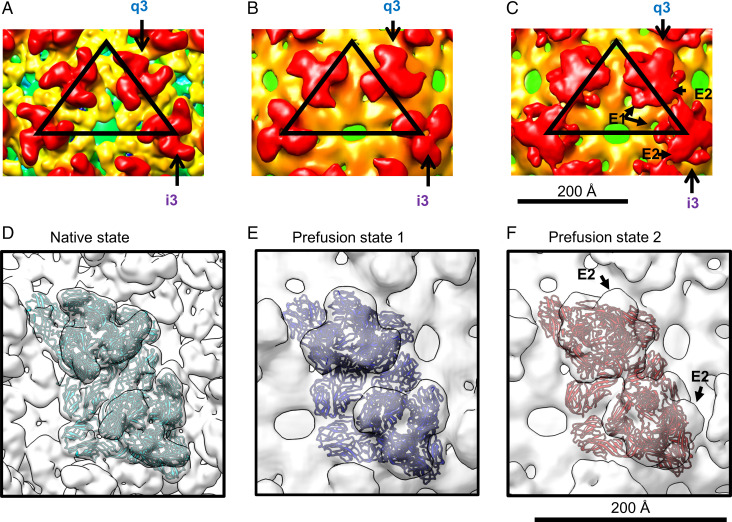
Two cryo-EM reconstructions representing prefusion states of EEEV under low-pH conditions are shown (Fig. 1B and C), with overall resolutions of ∼20 Å (prefusion state 1) and 16.4 Å (prefusion state 2) (SI Appendix, Fig. S3). Similar to the native EEEV reconstruction (Fig. 2A), reconstructions of the prefusion states of EEEV (Fig. 2B and C) have the E1–E2 glycoprotein spikes consistently arranged with T = 4 icosahedral symmetry. However, the glycoprotein spikes in the two low-pH EEEV structures differ from those in native EEEV, indicating structural rearrangements of E1–E2 glycoproteins under acidic conditions. We defined one structure as “prefusion state 1” of EEEV (Fig. 2B), in which the q3 and i3 spikes are slightly twisted compared with the native EEEV structure and have distinguishable glycoprotein densities similar to those in native EEEV (Fig. 2A). The structure of “prefusion state 2” of EEEV (Fig. 2C) showed disparately shaped spikes with dissociated E1 and E2 densities in each of the q3 and i3 spikes (Figs. 1C and 2C). By comparing the cryo-EM map of prefusion state 2 of EEEV with the crystal structure of low-pH SINV E1–E2 (PDB ID code 3MUU) (21), we found that the E1 density of prefusion state 2 of EEEV was roughly similar in position to the E1 structure of low-pH SINV. However, the EEEV E2 density did not align to the SINV E2 structural model. Thus, the individual domains of E2 may undergo rotation about each q3 and i3 axis to dissociate from E1, resulting in exposure of the E1 fusion loops in a low-pH environment.
Fig. 2.
Interpretation of cryo-EM reconstructions of native, prefusion state 1, and prefusion state 2 of EEEV. (A) Native EEEV. The EEEV map (EMD-20025) (12) was low-pass filtered to 14.3 Å for comparison with structures shown in B and C. (B) Prefusion state 1 of EEEV. (C) Prefusion state 2 of EEEV. E1 and E2 densities from q3 and i3 spikes are labeled and highlighted by arrows (a description is in Fig. 1A). The black triangles indicate an asymmetric unit in each of the cryo-EM reconstructions. (D) Rigid body–fitted E1–E2 model (cyan) in the localized-reconstruction native EEEV map (white). (E) MDFF-fitted E1–E2 model (blue) in the localized-reconstruction prefusion state 1 of the EEEV map (white). (F) MDFF-fitted E1–E2 model (red) in the localized-reconstruction prefusion state 2 of the EEEV map (white). Procedures for the model fitting in D–F are also described in Materials and Methods and SI Appendix, Fig. S5.
Our low-resolution cryo-EM reconstructions of EEEV under low-pH conditions suggest structural instability of the glycoprotein spikes in each of the asymmetric units. To improve the resolution of the two prefusion EEEV cryo-EM reconstructions, localized reconstructions were performed using the localrec software package (27) to extract 60 asymmetric units from each of the particle micrographs and cisTEM (28) to reconstruct volumes from the extracted particles. The localized reconstructions of native EEEV and prefusion state 1 and prefusion state 2 of EEEV are shown in Fig. 2D–F, respectively. After this analysis, the cryo-EM reconstruction resolution of native EEEV was estimated at 5.1 Å (SI Appendix, Fig. S4A), similar to the resolution of EEEV with icosahedral symmetry imposed (12). However, the resolution of prefusion state 1 of EEEV improved from 18.2 to 14.3 Å (SI Appendix, Fig. S4B) and in prefusion state 2, from 16.4 to 14.2 Å (SI Appendix, Fig. S4C). In each case, an improved resolution with the localized reconstruction suggested that under low-pH conditions, EEEV particles undergo conformational changes resulting in spatially displaced E1–E2 glycoproteins in each of the asymmetric units and imperfect icosahedral symmetry on the viral surface. The relatively low cryo-EM map resolution implies that the E1 and E2 glycoproteins in each of the q3 and i3 spikes become heterogeneous and have imperfect local threefold symmetry, resulting in lower-resolution reconstructions of EEEV under low-pH conditions.
Molecular Dynamics Flexible Fitting of E1–E2 Glycoproteins into the Localized-Reconstruction Cryo-EM Maps.
The localized-reconstruction map of native EEEV (SI Appendix, Fig. S5A) was low-pass filtered to a resolution of ∼14.0 Å (SI Appendix, Fig. S5B), followed by rigid-body fitting of a previously determined cryo-EM structure of EEEV (PDB ID code 6MX4) (24) into the localized-reconstruction EEEV map (Fig. 2D and SI Appendix, Fig. S5C) using Chimera (29). Prefusion state 1 of the EEEV cryo-EM map (SI Appendix, Fig. S5D) showed a spike shape similar to that in the native EEEV map (SI Appendix, Fig. S5A). However, the E1–E2 structure of native EEEV did not fit well into prefusion state 1 of the EEEV map, suggesting a different conformation of E1–E2 glycoproteins in prefusion state 1 of EEEV. To obtain the glycoprotein structures of EEEV in prefusion state 1, we used molecular dynamics flexible fitting (MDFF) (Materials and Methods) to place glycoprotein models into the glycoprotein densities and simulate their conformations in the q3 and i3 spikes. To facilitate this process, a segmented map was generated from prefusion state 1 of the EEEV map (SI Appendix, Fig. S5D), and densities around the q3 and i3 spikes were extracted (SI Appendix, Fig. S5E) and used for MDFF (Fig. 2E and SI Appendix, Fig. S5F). The simulation was performed until convergence by monitoring the rmsd with respect to the original structure (SI Appendix, Fig. S6A) and the cross-correlation coefficient (CCC) between the atomic structure and the cryo-EM map (Table 1 and SI Appendix, Fig. S6B) from 0 to 300 ps. These results suggest that the glycoprotein model after MDFF is more representative of the low-pH prefusion state 1 of EEEV.
Table 1.
Cryo-EM data statistics
| Data collection and processing | Prefusion state 1 | Prefusion state 2 | EEEV–Fab22 (pH 5.5) | EEEV–Fab22 (pH 7.4) | Native EEEV (localrec) | Prefusion state 1 (localrec) | Prefusion state 2 (localrec) |
|---|---|---|---|---|---|---|---|
| Nominal magnification (×) | 64,000 | 81,000 | 81,000 | 64,000 | 38,000 | 64,000 | 81,000 |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 | 300 | 300 |
| Defocus range (mm) | 0.5–3.5 | 0.5–3.5 | 0.5–3.5 | 0.5–3.5 | 0.5–3.5 | 0.5–3.5 | 0.5–3.5 |
| Pixel size (Å) | 1.328 | 1.748 | 1.748 | 1.328 | 1.62 | 1.328 | 1.748 |
| Symmetry imposed | Icosahedral | Icosahedral | Icosahedral | Icosahedral | C1 | C1 | C1 |
| Initial particle images | 7,573 | 12,277 | 4,234 | 4,453 | — | — | — |
| Final particle images | 2,261 | 3,201 | 3,974 | 4,302 | 848,700 | 135,660 | 192,060 |
| Map resolution (Å) | |||||||
| Unmasked (jspr) (31) | 18.2 | 16.4 | 8.1 | 6.7 | 5.1 | 14.3 | 14.1 |
| Masked (jspr) (31) | 15.3 | 15.8 | 7.8 | 6.1 | — | — | — |
| Sharpened B factor | −1,491 | −1,343 | −566 | −274 | — | — | — |
| FSC threshold* | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Refinement | |||||||
| Initial model used | — | — | — | — | — | 6mx4 | Homology model from 3muu |
| Model composition | — | — | — | — | — | Ectodomain of E1–E2 | Ectodomain of E1–E2 (without E2-B) |
| Nonhydrogen atoms | — | — | — | — | — | 34,974 | 28,704 |
| Protein residues | — | — | — | — | — | 4,506 | 3,704 |
| rmsds | |||||||
| Bond lengths (Å) | — | — | — | — | — | 0.019 | 0.019 |
| Bond angles (°) | — | — | — | — | — | 2.398 | 2.331 |
| Ramachandran plot | |||||||
| Favored (%) | — | — | — | — | — | 79.56 | 85.26 |
| Allowed (%) | — | — | — | — | — | 13.34 | 10.22 |
| Disallowed (%) | — | — | — | — | — | 7.1 | 4.51 |
| CCC | 0.96 | 0.94 | |||||
| CCmask/CCbox | — | — | — | — | — | 0.73/0.95 | 0.75/0.95 |
*The threshold of the Fourier Shell Correlation (FSC) of two independently determined half maps was set at 0.143 for map resolution determination.
In the same way, the localized-reconstruction cryo-EM map of EEEV prefusion state 2 (SI Appendix, Fig. S5G) was segmented in Chimera (29), and densities around the q3 and i3 spikes were extracted (SI Appendix, Fig. S5H) followed by model fitting using MDFF. It has been suggested that the E2-B domain becomes destabilized in an acidic environment to facilitate E1–E2 dissociation (21). Therefore, we generated an MDFF starting model of E1–E2 from the crystal structure of low-pH SINV E1–E2 (21) (PDB ID code 3MUU) using the SWISS-MODEL homology-modeling server (30). MDFF fitting (Fig. 2F and SI Appendix, Fig. S5I) was performed as previously described for prefusion state 1 of EEEV and compared with the starting model until convergence of the rmsd (SI Appendix, Fig. S6C) and the CCC (Table 1 and SI Appendix, Fig. S6D). These results suggest that the model after MDFF better represents the density map and structure in prefusion state 2 of EEEV under low-pH conditions.
Movement of the E2-B Domain Exposes the E1 Fusion Loops under Acidic Conditions.
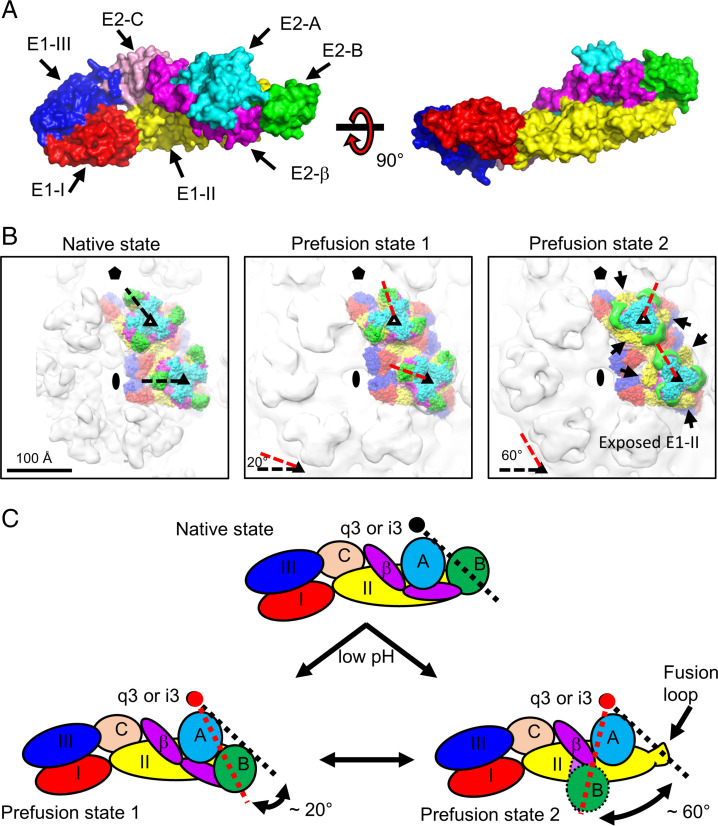
The domains of E1 and E2 glycoproteins of EEEV are shown in Fig. 3A. Compared with the native E1–E2 glycoproteins of EEEV (Fig. 3B, Left and C, Upper), the structural model of prefusion state 1 of EEEV (Fig. 3B, Center) shows that the E2-B domains (density shown in green) are rotated ∼20° clockwise around the center of either the q3 or i3 axis (Fig. 3B, Center [red dashed lines] and C, Lower Left and Table 2). The structural model of prefusion state 2 of EEEV (Fig. 3B, Right) shows comparable movement of a larger magnitude, and the E2-B domains (green) are rotated by ∼60° around the center of either the q3 or i3 axis (Fig. 3B, Right and C, Lower Right and Table 2) when compared with native E1–E2 glycoproteins of EEEV (Fig. 3B, Left and C, Upper). Conformational changes of the E2 glycoproteins, especially in the E2-B domains, expose the fusion loops on the E1-II domain, which may facilitate the fusion process upon contact of the host endosomal membrane.
Fig. 3.
Domain mapping of E1–E2 glycoproteins in the native and two low-pH EEEV structures. (A) Surface representation of the E1–E2 ectodomains. Domains from E1 and E2 glycoproteins are colored separately and labeled accordingly. Residue ranges were used for defining the domain boundaries. E1-I (red): 1 to 38, 130 to 169, and 273 to 293; E1-II (yellow): 39 to 129 and 170 to 272; E1-III (dark blue): 294 to 381; E2-A (cyan): 1 to 134; E2-B (green): 170 to 228; E2-C (light pink): 266 to 340; E2-β (magenta): 135 to 169 and 229 to 265. (B) Domain mapping in the structures of native (Left), prefusion state 1 (Center), and prefusion state 2 (Right) of EEEV. Domains are colored coded as described for A. Symmetry elements are marked as described in Fig. 1A. In Right, arrows indicate the exposure of E1-B (yellow). (C) Cartoon diagrams of E1–E2 ectodomains under native (Upper) and acidic (Lower Left and Lower Right) conditions for demonstrating the rotation of E2-B. The black circle indicates the center of the q3 or i3 spike. In prefusion state 2, unresolved regions of E2-B (circle with dashed outline) and a part of β-ribbon (oval with dashed outline) are highlighted. In B and C, black and red dashed lines indicate the E2-B positions at native and low-pH states, respectively.
Table 2.
Rotation of the E2-B domain
| Native EEEV | Prefusion state 1 | Prefusion state 2 | |
|---|---|---|---|
| Coordinate (x, y, z) | |||
| i3-1* | (73.8, –3.13, 311.42) | (74.13, 6.95, 303.37) | (97.63, 28.72, 296.84) |
| i3-2* | (135.42, –35.5, 286.69) | (122.6, –41.09, 283.2) | (98.88, –34.04, 295.22) |
| i3-3* | (130.77, 38.19, 288.96) | (138.75, 28.23, 278.12) | (149.61, –4.51, 288.66) |
| i3-center | (113.33, –0.15, 295.69) | ||
| q3-4* | (99.3, 96.99, 288.52) | (94.36, 79.86, 287.59) | (80.37, 58.23, 300.08) |
| q3-5* | (31.72, 126.39, 293.6) | (47.48, 128.34, 284.35) | (73.12, 122.4, 286.33) |
| q3-6* | (42.91, 55.97, 312.91) | (25.3, 66.6, 305.92) | (18.87, 85.27, 302.5) |
| q3-center | (57.98, 93.12, 298.34) | ||
| Vector (a, b, c)† | |||
| i3-1* | (–39.53, –2.98, 15.73) | (–39.2, 7.1, 7.68) | (–15.7, 28.87, 1.15) |
| i3-2* | (22.09, –35.35, –9) | (9.27, –40.94, –12.49) | (–14.45, –33.89, –0.47) |
| i3-3* | (17.44, 38.34, –6.73) | (25.42, 28.38, –17.57) | (36.28, –4.36, –7.03) |
| q3-4* | (41.32, 3.87, –9.82) | (36.38, –13.26, –10.75) | (22.39, –34.89, 1.74) |
| q3-5* | (–26.26, 33.27, –4.74) | (–10.5, 35.22, –13.99) | (15.14, 29.28, –12.01) |
| q3-6* | (–15.07, –37.15, 14.57) | (–32.68, –26.52, 7.58) | (–39.11, –7.85, 4.16) |
| Rotation angle (°) | |||
| i3-1* | 17.61 | 66.79 | |
| i3-2* | 19.14 | 55.80 | |
| i3-3* | 22.81 | 71.15 | |
| q3-4* | 24.66 | 64.09 | |
| q3-5* | 25.50 | 64.95 | |
| q3-6* | 29.47 | 56.61 | |
| Mean± SD (q3 + i3) | 23.20 ± 4.35 | 63.23 ± 5.97 |
*The q3 and i3 numbers from one to six represent the positions of E glycoproteins at q3 and i3 spikes as indicated in Fig. 1A.
†Each of the vectors was defined from the center of the q3 or i3 spike to the center of mass of each of the E2-B domains. The center of the spike was defined by the three E2-B domains in either q3 or i3 position.
Cryo-EM Structure of EEEV Bound with Fab22 at pH 5.5.
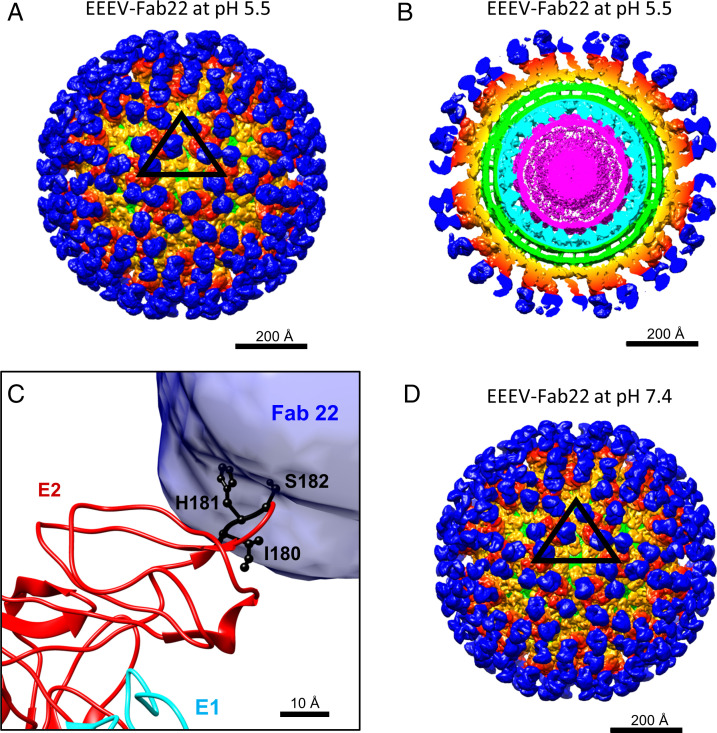
EEEV-22 is a protective, neutralizing antibody that prevents EEEV infection by blocking viral fusion (26). Based on prior alanine-scanning mutagenesis studies and neutralization escape mapping, this antibody recognizes an epitope in E2-B centered at residues I180, H181, and S182. To examine how this antibody prevents fusion between host and viral membranes under acidic conditions, we prepared Fab fragments of EEEV-22 (Fab22) and incubated purified EEEV virus particles with Fab22, followed by incubation with pH 5.5 buffer prior to cryo-EM sample preparation. The cryo-EM reconstruction of the EEEV–Fab22 complex at pH 5.5 (Fig. 4A and B) reached a resolution of 8.1 Å (Table 1 and SI Appendix, Fig. S7A). The density corresponding to the E1–E2 glycoproteins was nearly identical to that of wild-type EEEV at neutral pH. Although the density belonging to Fab22 was easily identifiable, a difference map containing only the Fab22 densities was generated by subtracting the density map of the native EEEV from the low-pH EEEV–Fab22 map to aid interpretation (31). To determine the epitope of Fab22 on EEEV, the E1–E2 glycoprotein structure (PDB ID code 6MX4) was fitted into the low-pH EEEV–Fab22 cryo-EM map as a rigid body, followed by superimposition with the difference map (Fig. 4C and SI Appendix, Fig. S8A and B). The previously determined epitope (I180-H181-S182) was located within the binding interface between E2-B and Fab22 (Fig. 4C and SI Appendix, Fig. S8C). The low-pH EEEV–Fab22 reconstruction showed little difference compared with the neutral pH EEEV–Fab22 (Fig. 4D), suggesting that the EEEV–Fab22 complex is not affected by low pH. Our findings further suggest that the EEEV-22 antibody recognizes the E2-B epitope and prevents fusion by stabilizing the interactions between E1-II and E2-B under low-pH conditions.
Fig. 4.
Cryo-EM reconstructions of EEEV bound to Fab22 under low-pH and neutral pH conditions. In A, B, and D, densities belonging to Fab22 (blue), E1–E2 glycoproteins (orange and red), the viral membrane (green), capsid proteins (cyan), and the viral RNA genome (magenta) are highlighted. (A) Cryo-EM reconstruction of EEEV–Fab22 at low pH. The black triangle indicates an asymmetric unit as described in Fig. 1A. (B) Central section of the cryo-EM reconstruction of low-pH EEEV–Fab22 as presented in A. (C) Superimposition of the EEEV E1–E2 glycoprotein model with the difference map generated by subtracting the native EEEV densities from the low-pH EEEV–Fab22 map. E1 (cyan) and E2 (red) glycoproteins are labeled, and the Fab22 density is shown in blue. Key binding residues for EEEV-22 (I180, H181, and S182) are colored in black. (D) Cryo-EM reconstruction of EEEV–Fab22 at neutral pH. The map resolution was estimated to be ∼6.7 Å (Table 1 and SI Appendix, Fig. S7B).
Discussion
The dissociation of E1–E2 glycoproteins in alphaviruses has been suggested as a prerequisite for the formation of E1 fusogenic trimers primed for membrane fusion. One mechanism involves destabilization of the E2-B domain. It is known that binding of E2-B to E1-II prevents the premature formation of E1 homotrimers during alphavirus assembly in the trans-Golgi network (21). Therefore, disruption of this interaction is presumed to occur during the process of virus–host membrane fusion. The crystal structure of SINV E1–E2 ectodomains at acidic pH (21) lacked electron density of the E2-B domain, implying that this region is disordered and possibly flexible. Our results suggest that the conformational changes of the glycoprotein spikes are initiated by destabilization of E2-B under low-pH conditions. The E2 glycoproteins undergo a rotation around each of the q3 and i3 spikes, resulting in exposure of E1-II and the fusion loops, while the capsid cores in both cases are similar to those in native EEEV (SI Appendix, Fig. S9). In both low-pH cryo-EM maps of EEEV, E1 glycoproteins do not form E1 trimers or transition toward E1 trimers.
The low-pH cryo-EM samples of EEEV were prepared from the same virus purification and with similar procedures prior to blotting. Since conformational changes take place in milliseconds (32), performing manual-blotting and buffer exchange procedures with differences of a few seconds may not be sufficient to trap virus particles in different states. One possible reason that we were, nevertheless, able to obtain two prefusion EEEV structures may be due to different final pH values in different grids. In the manual-blotting procedure, the grids were not completely drained of buffer. Therefore, in the buffer exchange step, each grid may have a different amount of original neutral pH buffer with a roughly equal amount of low-pH buffer, resulting in slightly different pH values on the grids. Currently, there is no method to determine the pH value on a grid. In the cryo-EM reconstructions of EEEV at low pH, E1 trimer formation was not observed, and the E1 and E2 glycoproteins were not fully dissociated from each other. Therefore, both cryo-EM structures belong to prefusion states in an early stage of the fusion process. To interpret the cryo-EM maps of the two prefusion states of EEEV, rigid-body fitting (rigid fit) and MDFF were performed to place the E1–E2 glycoprotein models into each of the density maps. With either rigid-fit or MDFF method, we observed the rotation of the E2-B domain in the q3 and i3 spikes (Table 1 and SI Appendix, Table S1). However, the results show that MDFF allows better model fitting into the density with less steric clash (SI Appendix, Table S2). With MDFF fitting, the domains in the E1–E2 glycoproteins can move based upon density, while the secondary structures are preserved in both prefusion states (SI Appendix, Figs. S10 and S11 and Table S3).
The crystal structure of SINV glycoprotein spikes under acidic conditions proposed conformational changes in which the E2-B domain becomes unfolded and the E2 glycoprotein then can dissociate from the E1 glycoprotein (21). However, the E1-II domain in the crystal structure of SINV E1–E2 at low pH (PDB ID code 3MUU) has interactions with neighboring E2-C, E1-I, and E1-III domains (SI Appendix, Fig. S12A–C), and a part of the E1-II fusion loop formed contacts with neighboring E1-I (SI Appendix, Fig. S11D). In this case, dissociation of E2-B from E1-II could be due to the low-pH condition or crystal packing. Therefore, cryo-EM structures of EEEV under low-pH conditions were needed to address these questions. The cryo-EM reconstructions presented here confirm that conformational changes of the glycoprotein spikes are initiated by destabilization of E2-B under low-pH conditions. Based on these observations, we suggest that the two low-pH cryo-EM EEEV reconstructions represent two prefusion states of EEEV, mimicking the structures that would be present in acidified host endosomes. Interestingly, the cryo-EM study of SFV under low-pH conditions (32) showed a virus structure with a spike rotation, similar to what we observed in prefusion state 1 of the EEEV structure. These results may suggest a consensus mechanism for initiating fusion in alphaviruses.
Stabilization of the alphavirus E2-B domain by protective antibodies, such as EEEV-22 against EEEV in this study and 5F10 against chikungunya virus (CHIKV) (26, 33), can block structural rearrangement of the glycoprotein spikes. Notably, these two Fab fragments recognize different epitopes on the E2-B domains of EEEV (Ile180-His181-Ser182) and CHIKV (Gly205-Asn219), and the binding orientations of the two Fab fragments are also different. Fab fragments have been used in cryo-EM studies for improving particle alignment and reconstructions (34) as well as for trapping macromolecules at different conformational states (35). In this study, the cryo-EM structure of EEEV–Fab22 at low pH is similar to the one at neutral pH, and no other conformational states were identified. This suggests that Fab22 not only recognizes the epitope but also, stabilizes E2-B in its the native conformation and prevents virus fusion. The CHIKV Fab (5F10) and other EEEV fusion-blocking antibodies may function similarly to prevent alphavirus conformational changes at low pH (26, 33).
During alphavirus maturation, furin protease processes the precursor of E2 (P62) to generate E2 glycoprotein and E3 peptide. For many alphaviruses, E3 dissociates from the virus particle following cleavage under suitable temperature and buffer conditions. However, it has also been observed that E3 peptides may remain attached to E2 glycoproteins via hydrophobic interactions after cleavage (36), and the attached or uncleaved E3 fragments may affect the alphavirus fusion process (37). In the case of the furin-resistant SFVSQL mutant virus, virus particles were treated with protease K, resulting in ∼50% uncleaved P62 and ∼50% E3-attached E2 glycoproteins on the virus particles. The E3-bound virus particles tolerate a pH drop and show a delay of E1 trimer formation (37). Interestingly, this effect was not observed in the wild-type SFV, which contains ∼25% P62 precursor proteins and no E3 attached to E2, suggesting that E3 can influence the pH-dependent alphavirus fusion process when significant amounts of E3 (cleaved or uncleaved) are present on the virus particles.
The cryo-EM density of attached or uncleaved E3 on alphaviruses can be identified across different resolution ranges (from 3.5 to 17 Å) (36, 38). In previous and current EEEV cryo-EM structures (12, 24), we did not observe E3 density in either the whole virus or the localized reconstructions, indicating that E3 is either at a low abundance or absent and may not serve as a factor for trapping either of the observed prefusion states under low-pH conditions. However, in CHIKV E1–E2 bound with E3 (PDB ID code 3N41), E3 binds to E1 and the β-ribbon. If the CHIKV E1–E2–E3 map is superimposed over the cryo-EM maps of the prefusion states (Fig. 5), CHIKV E3 is in a position that would block the movement observed in the E2-B domain in both EEEV prefusion states 1 and 2. This suggests that E3 bound to the glycoproteins may interfere with the movement of E2-B and prevent subsequent exposure of the fusion loops under acidic conditions.
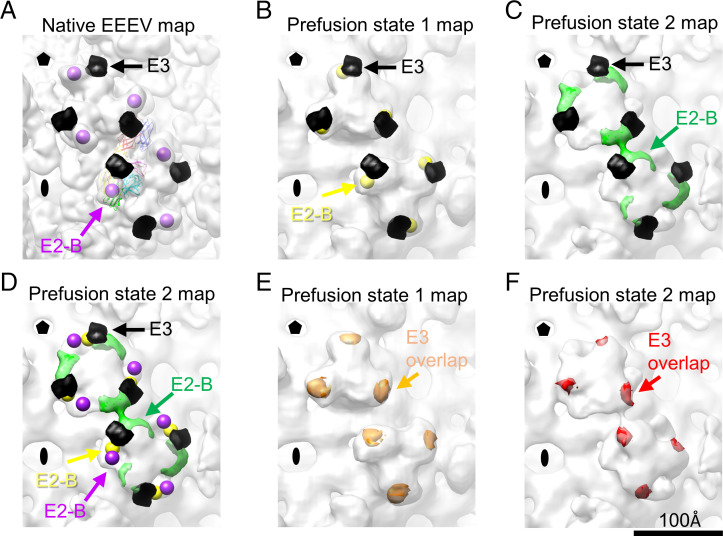
Fig. 5.
Inhibitory role of E3 in the pH-dependent conformational changes of glycoprotein spikes. (A) Cryo-EM reconstruction of native EEEV (low-pass filtered to 14 Å). The centers of E2-B domains are labeled and indicated by purple spheres. The crystal structure of CHIKV containing E3 (PDB ID code 3N41) was aligned to the native EEEV structure (PDB ID code 6MX4). Simulated CHIKV E3 density at 14-Å resolution was generated in Chimera (29) as a black surface and is presented in A–D. (B) Cryo-EM reconstruction of prefusion state 1 of EEEV. The centers of E2-B domains are labeled and indicated by yellow spheres. (C) Cryo-EM reconstruction of prefusion state 2 of EEEV. The density of E2-B is indicated by green density. (D) Comparison of the centers or density of E2 domains at native, prefusion state 1, and prefusion state 2. The presentation of centers and density is described in A–C. The distance between the E2-B domains in native EEEV and prefusion state 1 of EEEV is about 18 Å (SI Appendix, Table S4). The distance between the E2-B domains in native EEEV and prefusion state 2 of EEEV is about 42 Å (SI Appendix, Table S4). (E) Overlapped density of CHIKV E3 (orange) in prefusion state 1 of EEEV. E3 density outside prefusion state 1 of EEEV was manually erased in Chimera (29). The density volume values were measured, and an average of overlapped density (red) is about 35% of the simulated E3 density (SI Appendix, Table S5). (F) Overlapped density of CHIKV E3 (red) in prefusion state 2 of EEEV. E3 density outside prefusion state 2 of EEEV was manually erased in Chimera. The density volume values were measured, and an average of overlapped density (red) is about 20% of the simulated E3 density (SI Appendix, Table S5).
In conclusion, we demonstrate here conformational changes of the EEEV glycoprotein spikes under low-pH conditions and that E2-B structural integrity serves an essential role in controlling the fusion state. The E2-B domain can move upon reduction of pH to expose the fusion loop. Antibodies that preserve E2-B in its native conformation can prevent conformational changes of the spike and thus, block the fusion process. The presence of E3 also would prevent movement of the E2-B domain to block fusion. Electron tomography, which provides opportunities to examine samples with a high degree of heterogeneity at a molecular level, may be necessary for studying the formation of E1 homotrimers primed for fusion.
Materials and Methods
Cryo-EM Sample Preparation of SINV–EEEV Chimeric Virus Particles.
The chimeric SINV–EEEV with nonstructural proteins from SINV (TR339) and structural proteins from EEEV (FL91-4679) was described previously (13). Virus preparation of EEEV virions was also carried out as described previously (12, 24). Briefly, baby hamster kidney cells were infected with an SINV–EEEV virus stock at a multiplicity of infection of one. Twenty hours postinfection, virions were harvested, precipitated by polyethylene glycol 8000 (PEG-8000) (12 to 14%), and subjected to a density gradient centrifugation (Opti-prep; 247,000 × g for 1.5 h at 4 °C; SW-41 swinging-bucket rotor) for further purification. Finally, the viral sample was buffer exchanged and stored in PBS. The cryo-EM sample preparations for obtaining the two prefusion states of EEEV were performed using the same batch of virus and following the same procedures. Ahead of cryo-EM studies, we tested a few conditions for grid preparation with different batches of virus samples and found the following method reproducible for preparing samples at low pH. Purified EEEV virions (∼3.3 μL) were applied onto a lacey carbon grid (glow discharged for 1 min before use). After manually blotting the solution, a small amount of the sample (in PBS buffer) was intentionally left on the grids to prevent sample denaturation due to interactions at the air–water interface. A drop of 3.5 μL PBS containing 50 mM citrate at pH 5.5 was added onto the grid for the low-pH treatment. The low pH–treated grids were mounted onto the Gatan CP3 instrument, blotted, and plunged into liquid ethane as described previously (12, 24).
Cryo-EM Data Collection and Three-Dimensional Reconstruction of Low-pH EEEV.
Two separate cryo-EM datasets of EEEV treated with low pH were imaged using a Titan Krios cryoelectron microscope, and micrographs were recorded as movies on either the Gatan K3 detector (nominal magnification: 64,000×, magnified pixel size: 1.328 Å) or the Gatan K2 summit detector (nominal magnification: 81,000×, magnified pixel size: 1.73 Å). The total electron dose for data collection was ∼32 e-/A2. Motion correction to align the movie frames for each of the datasets was performed using the motioncor2 software (39). The CTF of each aligned micrograph was estimated using CTFFind4 (40). Particle picking, two-dimensional (2D) classification, initial model generation, and three-dimensional (3D) classification were performed in Relion (41). After 3D classification, selected particles were used for cryo-EM 3D refinement using the jspr program (31), in which the gold standard 3D reconstruction procedures with independent subdatasets were performed in both cases. The resolution was determined based on the Fourier shell correlation cutoff of 0.143 (42). Statistics for all datasets are summarized in Table 1.
Localized Reconstruction of EEEV q3–i3 Spikes in an Asymmetric Unit under Normal and Low-pH Conditions.
For the dataset of native EEEV, 60 asymmetric units were extracted from each of the particle images (box size: 256 pixels, pixel size: 1.62 Å) based on predetermined Euler angles and centers using the localrec program (27). The center of the extracted image was set to the center of the ectodomains of q3 and i3 glycoprotein spikes in order to include the two spikes in an asymmetric unit. After subparticle extraction, each of the images was then aligned against a model generated by cisTEM (28), and the reconstruction of the subvolume was performed using cisTEM (28). Similarly, localized reconstructions for the two datasets of EEEV at low pH were performed using the same procedure but with the box size at 108 pixels and the pixel size at 5.312 Å (binning eight times) for prefusion state 1 of the EEEV dataset and 144 pixels and 3.496 Å (binning four times) for prefusion state 2 of the EEEV dataset.
MDFF.
The ectodomains of mature E1 (residues 1 to 400) and E2 (residues 1 to 351) glycoproteins were obtained from PDB coordinates 6MX4 (24) and rigid body fitted into the density of q3 and i3 spikes in the map from the localized reconstruction of native EEEV as a starting point. The maps of the two prefusion states of EEEV were segmented in Chimera (29) to define the map volumes for further MDFF procedures (43–45). For map segmentation in Chimera, segments at the q3 and i3 spike positions were grouped and extracted from each of the maps. The extracted maps containing density of q3 and i3 spikes were used in MDFF. The fitted E1–E2 domains with updated coordinates were flexible fitted into the segmented low-pH map, again using MDFF, and the grid-scaling (gscale) value was set to 0.5. Suggested restraints regarding secondary structures, peptide bonds, and chirality also were applied upon MDFF fitting (44, 45). Each MDFF simulation was performed for 300 ps until convergence of the model’s rmsd (45), followed by 3,000 steps (3 ps) of energy minimization with a gscale value of 0.5. Quality of the MDFF fittings was estimated using the CCC generated in VMD (45) and cross-correlation values (CCmask, CCbox) generated using phenix.real_space_refine (only apply Group B-factor refinement) (46). The results are summarized in Table 1.
E2-B Domain Rotation Calculation.
The central coordinates of the E2-B domain (residue numbers 170 to 228) in q3 and i3 spikes were obtained using Chimera (29) for both native EEEV and prefusion state 1 of the EEEV models. For visualization of the E2-B domain in the q3 and i3 spikes of prefusion state 2 of EEEV, we subtracted a simulated map of the flexible-fitted model without the E2 domains from prefusion state 2 of the EEEV map, which highlights the densities belonging to E2-B (Fig. 3B, Right). We placed point markers at these densities and obtained the coordinates representing the center of all entities of the E2-B domain. With the coordinates of E2-B of EEEV in the native, prefusion state 1, and prefusion state 2, using the native EEEV as reference, the vectors and the angles between vectors for the q3 and i3 locations were calculated and are summarized in Table 2.
Preparation of EEEV in Complex with Fab22 Derived from the Protective EEEV-22 Antibody.
The production and purification of EEEV-22 immunoglobulin G were carried out as described previously (26). EEEV-22 Fab fragments (Fab22) were generated using the Pierce Fab preparation kit following the manufacturer’s recommended protocol (ThermoFisher; catalog no. 44985). Purified Fab22 fragments were analyzed by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and gel filtration chromatography. Purified EEEV was mixed with the purified Fab22 for 24 h at 4 °C at an E2 glycoprotein/Fab22 molar ratio of 1:4. After incubation, the mixture was subjected to low-pH treatment with the addition of 50 mM citrate buffer at pH 5.0 for 30 min before cryo-EM sample preparation. The final pH value was at pH ∼ 5.5. Cryo-EM samples were prepared with the virus–Fab mixture at either neutral pH (pH 7.0) or acidic pH (pH 5.5).
Supplementary Material
Acknowledgments
We thank Valorie D. Bowman (Purdue University) for technical help in cryo-EM data collection. Cryo-EM data were collected at the Purdue University Cryo-EM Facility. This work was supported by NIH Grants R01 AI095366 (to M.G.R. and R.J.K), U19 AI142790 (to M.S.D. and R.J.K.), and R01 AI095436 (to W.B.K.) and Defense Threat Reduction Agency Awards HDTRA1-15-1-0013 (to M.S.D. and W.B.K.) and HDTRA1-15-1-0047 (to W.B.K.).
Footnotes
Competing interest statement: M.S.D. is a consultant for Inbios, Vir Biotechnology, Fortress Biotech, and Carnival Corporation and on the scientific advisory boards of Moderna and Immunome. The laboratories of M.S.D. at Washington University School of Medicine received unrelated support under sponsored research agreements from Emergent BioSolutions, Moderna, and Vir Biotechnology. The laboratories of W.B.K. at the University of Pittsburgh receive unrelated support under sponsored research agreements from SAb Biotherapeutics and ICON gps.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114119119/-/DCSupplemental.
Data Availability
The atomic coordinates of E1–E2 glycoproteins of prefusion state 1 and prefusion state 2 of EEEV have been deposited in PDB (ID codes 7N6A and 7N69, respectively) (47, 48). Whole-virus cryo-EM density maps of prefusion state 1 and prefusion state 2 of EEEV; the localized-reconstruction maps of the native, prefusion state 1, and prefusion state 2 of EEEV; and the cryo-EM reconstructions of EEEV–Fab22 under neutral pH and acidic pH conditions have been deposited in the Electron Microscopy Data Bank (accession nos. EMD-24204, EMD-24200, EMD-24199, EMD-24205, EMD-24201, EMD-24203, and EMD-24202, respectively) (49–55).
References
- 1.Strauss J. H., Strauss E. G., The alphaviruses: Gene expression, replication, and evolution. Microbiol. Rev. 58, 491–562 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindsey N. P., Staples J. E., Fischer M., Eastern equine encephalitis virus in the United States, 2003-2016. Am. J. Trop. Med. Hyg. 98, 1472–1477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindsey N. P., Martin S. W., Staples J. E., Fischer M., Notes from the field: Multistate outbreak of Eastern equine encephalitis virus - United States, 2019. MMWR Morb. Mortal. Wkly. Rep. 69, 50–51 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang R., et al. , 4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J. 30, 3854–3863 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W., et al. , Placement of the structural proteins in Sindbis virus. J. Virol. 76, 11645–11658 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukhopadhyay S., et al. , Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 14, 63–73 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun S., et al. , Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. eLife 2, e00435 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caspar D. L., Klug A., Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 27, 1–24 (1962). [DOI] [PubMed] [Google Scholar]
- 9.Gaedigk-Nitschko K., Schlesinger M. J., The Sindbis virus 6K protein can be detected in virions and is acylated with fatty acids. Virology 175, 274–281 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Snyder J. E., et al. , Functional characterization of the alphavirus TF protein. J. Virol. 87, 8511–8523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S., et al. , Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure 4, 531–541 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Chen C. L., et al. , Cryo-EM structure of eastern equine encephalitis virus in complex with heparan sulfate analogues. Proc. Natl. Acad. Sci. U.S.A. 117, 8890–8899 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner C. L., Ebel G. D., Ryman K. D., Klimstra W. B., Heparan sulfate binding by natural eastern equine encephalitis viruses promotes neurovirulence. Proc. Natl. Acad. Sci. U.S.A. 108, 16026–16031 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klimstra W. B., Nangle E. M., Smith M. S., Yurochko A. D., Ryman K. D., DC-SIGN and L-SIGN can act as attachment receptors for alphaviruses and distinguish between mosquito cell- and mammalian cell-derived viruses. J. Virol. 77, 12022–12032 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R., et al. , Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 557, 570–574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma H., et al. , LDLRAD3 is a receptor for Venezuelan equine encephalitis virus. Nature 588, 308–314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose P. P., et al. , Natural resistance-associated macrophage protein is a cellular receptor for sindbis virus in both insect and mammalian hosts. Cell Host Microbe 10, 97–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernard E., et al. , Endocytosis of chikungunya virus into mammalian cells: Role of clathrin and early endosomal compartments. PLoS One 5, e11479 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vonderheit A., Helenius A., Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol. 3, e233 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeTulleo L., Kirchhausen T., The clathrin endocytic pathway in viral infection. EMBO J. 17, 4585–4593 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L., Jose J., Xiang Y., Kuhn R. J., Rossmann M. G., Structural changes of envelope proteins during alphavirus fusion. Nature 468, 705–708 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibbons D. L., et al. , Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427, 320–325 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Voss J. E., et al. , Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468, 709–712 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Hasan S. S., et al. , Cryo-EM structures of Eastern equine encephalitis virus reveal mechanisms of virus disassembly and antibody neutralization. Cell Rep. 25, 3136–3147.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lescar J., et al. , The Fusion glycoprotein shell of Semliki Forest virus: An icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105, 137–148 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Kim A. S., et al. , Protective antibodies against Eastern equine encephalitis virus bind to epitopes in domains A and B of the E2 glycoprotein. Nat. Microbiol. 4, 187–197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ilca S. L., et al. , Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun. 6, 8843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grant T., Rohou A., Grigorieff N., cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettersen E. F., et al. , UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Waterhouse A., et al. , SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46 (W1), W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo F., Jiang W., Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol. Biol. 1117, 401–443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuller S. D., Berriman J. A., Butcher S. J., Gowen B. E., Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell 81, 715–725 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Porta J., et al. , Structural studies of chikungunya virus-like particles complexed with human antibodies: Neutralization and cell-to-cell transmission. J. Virol. 90, 1169–1177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Q., et al. , Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1. Nature 595, 600–605 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao J., Sidhu S. S., Wells J. A., Two-state selection of conformation-specific antibodies. Proc. Natl. Acad. Sci. U.S.A. 106, 3071–3076 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L., et al. , Implication for alphavirus host-cell entry and assembly indicated by a 3.5Å resolution cryo-EM structure. Nat. Commun. 9, 5326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sjöberg M., Lindqvist B., Garoff H., Activation of the alphavirus spike protein is suppressed by bound E3. J. Virol. 85, 5644–5650 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu S. R., Haag L., Sjöberg M., Garoff H., Hammar L., The dynamic envelope of a fusion class II virus. E3 domain of glycoprotein E2 precursor in Semliki Forest virus provides a unique contact with the fusion protein E1. J. Biol. Chem. 283, 26452–26460 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheres S. H., Chen S., Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trabuco L. G., Villa E., Mitra K., Frank J., Schulten K., Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 (1996). [DOI] [PubMed] [Google Scholar]
- 46.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen C.-L., et al., Pre-fusion state 1 of EEEV with localized reconstruction. PDB. https://www.rcsb.org/structure/7N6A. Deposited 7 June 2021.
- 48.Chen C.-L., et al., Pre-fusion state 2 of EEEV with localized reconstruction. PDB. https://www.rcsb.org/structure/7N69. Deposited 7 June 2021.
- 49.Chen C.-L., et al., Pre-fusion state 1 of EEEV. EMDB. https://www.ebi.ac.uk/emdb/EMD-24204. Deposited 7 June 2021.
- 50.Chen C.-L., et al., Pre-fusion state 2 of EEEV. EMDB. https://www.ebi.ac.uk/emdb/EMD-24200. Deposited 7 June 2021.
- 51.Chen C.-L., et al., Localized reconstruction of EEEV. EMDB. https://www.ebi.ac.uk/emdb/EMD-24199. Deposited 7 June 2021.
- 52.Chen C.-L., et al., Pre-fusion state 1 of EEEV with localized reconstruction. EMDB. https://www.ebi.ac.uk/emdb/EMD-24205. Deposited 7 June 2021.
- 53.Chen C.-L., et al., Pre-fusion state 2 of EEEV with localized reconstruction. EMDB. https://www.ebi.ac.uk/emdb/EMD-24201. Deposited 7 June 2021.
- 54.Chen C.-L., et al., EEEV in complex with Fab22 at neutral pH (pH 7.4). EMDB. https://www.ebi.ac.uk/emdb/EMD-24203. Deposited 7 June 2021.
- 55.Chen C.-L., et al., EEEV in complex with Fab22 at acidic pH (pH 5.5). EMDB. https://www.ebi.ac.uk/emdb/EMD-24202. Deposited 7 June 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates of E1–E2 glycoproteins of prefusion state 1 and prefusion state 2 of EEEV have been deposited in PDB (ID codes 7N6A and 7N69, respectively) (47, 48). Whole-virus cryo-EM density maps of prefusion state 1 and prefusion state 2 of EEEV; the localized-reconstruction maps of the native, prefusion state 1, and prefusion state 2 of EEEV; and the cryo-EM reconstructions of EEEV–Fab22 under neutral pH and acidic pH conditions have been deposited in the Electron Microscopy Data Bank (accession nos. EMD-24204, EMD-24200, EMD-24199, EMD-24205, EMD-24201, EMD-24203, and EMD-24202, respectively) (49–55).