Abstract
Folate metabolism plays an essential role in tumor development. Various cancers display therapeutic response to reagents targeting key enzymes of the folate cycle, but obtain chemoresistance later. Therefore, novel targets in folate metabolism are highly demanded. Methylenetetrahydrofolate dehydrogenase/methylenetetrahydrofolate cyclohydrolase 2 (MTHFD2) is one of the key enzymes in folate metabolism and its expression is highly increased in multiple human cancers. However, the underlying mechanism that regulates MTHFD2 expression remains unknown. Here, we elucidate that SIRT4 deacetylates the conserved lysine 50 (K50) residue in MTHFD2. K50 deacetylation destabilizes MTHFD2 by elevating cullin 3 E3 ligase-mediated proteasomal degradation in response to stressful stimuli of folate deprivation, leading to suppression of nicotinamide adenine dinucleotide phosphate production in tumor cells and accumulation of intracellular reactive oxygen species, which in turn inhibits the growth of breast cancer cells. Collectively, our study reveals that SIRT4 senses folate availability to control MTHFD2 K50 acetylation and its protein stability, bridging nutrient/folate stress and cellular redox to act on cancer cell growth.
Keywords: folate metabolism, MTHFD2, CUL3, SIRT4, acetylation, breast cancer
Introduction
The one-carbon (1C) cycle is the essential metabolic network integrating the folate cycle and methionine cycle. In 1C metabolism, folate accepts, carries, and transfers 1C units. Intermediate metabolites produced in 1C metabolism are precursors of various essential macromolecules, including thymidylate, purine, S-adenosyl methionine, and the antioxidant nicotinamide adenine dinucleotide phosphate (NADPH) (Zhao et al., 2021). Folate fortification is recommended to prevent fetal neural tube defects, congenital eye defects, and anemia in adults (Sijilmassi et al., 2021), but its role in cancer remains controversial. While some studies suggest that adequate folic acid intake reduces the risk of breast cancer (Chen et al., 2014; Di Maso et al., 2020), recent evidence has shown that folic acid supplementation fosters the malignant procedure of tumors and avoiding excessive folate supplementation benefits the patients by impeding tumor progression. For example, a folate diet significantly increases the weight and volume of breast tumors in mice (Ly et al., 2011; Hansen et al., 2017). In addition, clinical trials also provide evidence that daily supplementation of 1 mg folic acid increases the risk of prostate cancer and promotes cell proliferation and invasion in prostate cancer (Figueiredo et al., 2009). Collectively, these observations underscore the necessity for further investigation into folate metabolism reprogramming in cancer development.
Methylenetetrahydrofolate dehydrogenase/methylenetetrahydrofolate cyclohydrolase 2 (MTHFD2) is an integral bifunctional enzyme in mitochondrial folate metabolism, sequentially converting 5,10-methylenetetrahydrofolate (5,10-meTHF) to 5,10-methenyltetrahydrofolate (5,10-me+THF) and 10-formyl-tetrahydrofolate (10-formyl-THF). In the reaction, adenine dinucleotide phosphate (NADP+) or nicotinamide adenine dinucleotide (NAD+) acts as a cofactor to generate NADPH or NADH. MTHFD2 is expressed at a relatively low level in normal tissues (Di Pietro et al., 2002). In contrast, upregulation of MTHFD2 has been found in different cancers, including breast, colorectal, and hepatocellular cancers, and plays a key role in remodeling folate metabolism in tumor cells (Nilsson et al., 2014). However, the molecular mechanism that regulates MTHFD2 expression remains elusive. Previous studies have shown that the KRAS or MYC oncogene regulates MTHFD2 transcriptionally (Moran et al., 2014; Pikman et al., 2016), but there is little knowledge on posttranslational regulation of MTHFD2 and how this regulation functions on tumor progression, especially under the stressful conditions that frequently occur in the tumor microenvironment.
Protein lysine acetylation is a conserved posttranslational modification (PTM), which is catalyzed reversibly by acetyltransferases and deacetylases. Metabolic enzyme acetylation regulates multiple cellular processes, involving protein stability, subcellular location, protein interaction, and signal transduction, by removing the positive charge from lysine or changing steric hindrance (Choudhary et al., 2014). A proteomic acetylome survey of mouse liver in the status of nutrient enrichment or deficiency indicates that >20% of mitochondrial proteins are acetylated, including many metabolic enzymes (Kim et al., 2006). Nevertheless, few have been studied in depth till now (Baeza et al., 2016).
SIRT4 is one of the most elusively investigated mitochondrial sirtuins, which functions as ADP-ribosyltransferase and lipoamidase, but its deacetylase activity was previously considered to be weak (Laurent et al., 2013; Mathias et al., 2014). Notably, SIRT4 is closely related to lipid and amino acid metabolic processes, but little is known about its role in folate metabolism.
Here, we report that SIRT4 deacetylates MTHFD2 at lysine 50 (K50) residue in response to folate deprivation. The deacetylation enhances proteasomal degradation of MTHFD2 protein by strengthening the binding of MTHFD2 to cullin 3 (CUL3) E3 ligase and MTHFD2 ubiquitylation, resulting in the disruption of folate metabolism and cellular redox homeostasis and the impairment of breast cancer cell growth.
Results
MTHFD2 is acetylated at K50
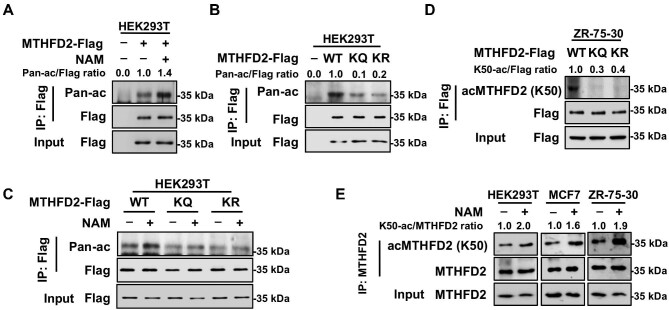
Protein acetylome studies indicated that MTHFD2 is potentially modified by acetylation at K50 according to mass spectrometry analysis (Choudhary et al., 2009; Weinert et al., 2013). Cross-species sequence alignment revealed that the sequence constituted of K50 residue and the adjacent amino acids are highly conserved from zebrafish to humans, suggesting that K50 residue may be important for the function of MTHFD2 (Supplementary Figure S1A). To test whether MTHFD2 is acetylated at K50 residue, Flag-tagged MTHFD2 was overexpressed in HEK293T cells and the sirtuin inhibitor nicotinamide (NAM) was added into cells. Using the antibody against pan-acetylated lysine (pan-ac), western blotting result revealed that immunopurified MTHFD2 was acetylated and NAM treatment significantly increased the acetylation level of MTHFD2, indicating that MTHFD2 is dynamically modified by lysine acetylation (Figure 1A). Next, we mutated K50 into glutamine (K50Q, referred to as KQ afterward) or arginine (K50R, referred to as KR afterward), respectively. Either Flag-tagged wild-type (WT) MTHFD2 or its mutants were overexpressed in HEK293T cells and both substitutions resulted in a markedly reduced acetylation level of MTHFD2 (Figure 1B). Of note, NAM increased the pan-acetylation level of WT MTHFD2 but not that of KQ and KR mutants, indicating that K50 is the major acetylated site of MTHFD2 under our tested conditions (Figure 1C). To further investigate the acetylation of MTHFD2 at K50 residue, we generated the site-specific antibody against acetylated K50 [acMTHFD2 (K50)]. Dot blot assay showed that the antibody specifically recognized the modified rather than unmodified peptide (Supplementary Figure S1B). And peptide competition tests verified that the modified peptide could counteract the recognition of the antibody but the unmodified peptide had no interfering effect (Supplementary Figure S1C). Besides, the antibody efficiently reacted with WT MTHFD2, but not with its KQ or KR mutant (Figure 1D). These results suggest that the antibody is specifically against K50 acetylation of MTHFD2. Moreover, the acetylation level of K50 residue in endogenous MTHFD2 was significantly increased when treated with NAM in HEK293T or breast cancer MCF7 and ZR-75-30 cells (Figure 1E). Taken together, MTHFD2 is acetylated at K50 residue.
Figure 1.
MTHFD2 is acetylated at K50. (A) Pan-acetylation of MTHFD2 is increased by NAM treatment. HEK293T cells with Flag-tagged MTHFD2 overexpression were treated with NAM for 6 h. Lysine acetylation of immunopurified MTHFD2 was detected with anti-acetyl lysine (pan-ac) antibody. Relative ratios of MTHFD2 acetylation were normalized by MTHFD2 protein level. (B) The pan-acetylation level of WT MTHFD2 is much higher than that of the KQ or KR mutant. Flag-tagged WT MTHFD2 and KQ and KR mutants were ectopically expressed into HEK293T cells and purified by Flag beads. Pan-acetylation of immunopurified MTHFD2 was analyzed by western blotting. (C) Pan-acetylation of WT MTHFD2 rather than that of the KQ or KR mutant is increased by NAM treatment. Flag-tagged WT MTHFD2 and KQ and KR mutants were ectopically expressed in HEK293T cells treated with or without NAM and purified by Flag beads. Pan-acetylation of immunopurified MTHFD2 was analyzed by western blotting. (D) K50 site-specific acetylation antibody [acMTHFD2 (K50)] recognizes WT MTHFD2, but not its KQ and KR mutants. Flag-tagged WT MTHFD2 and its KQ and KR mutants were immunopurified with Flag beads and blotted with acMTHFD2 (K50) antibody. (E) Endogenous MTHFD2 is acetylated at K50 in multiple cell lines. HEK293T, MCF-7, and ZR-75-30 cells were treated with NAM for 6 h. K50 acetylation of immunopurified endogenous MTHFD2 was detected by western blotting.
SIRT4 deacetylates MTHFD2 at K50
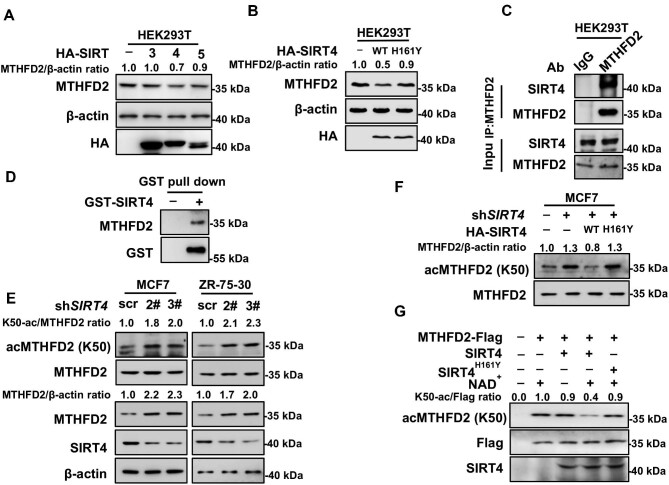
As shown in Figure 1E, NAM treatment increased K50 acetylation of MTHFD2, indicating that some member of the SIRT deacetylase family, like SIRT3, SIRT4, or SIRT5, is probably involved in MTHFD2 K50 deacetylation. Due to the mitochondrial localization of MTHFD2, we co-transfected mitochondrial-located SIRT3–SIRT5, respectively, with MTHFD2 into HEK293T cells to identify the enzyme that catalyzes MTHFD2 K50 deacetylation. The MTHFD2 protein level decreased when SIRT4 was overexpressed; however, other sirtuins had little effect on it (Figure 2A). H161Y is the enzyme-dead mutant of SIRT4 (Ahuja et al., 2007). Overexpression of SIRT4WT but not SIRT4H161Y decreased the MTHFD2 protein level as well (Figure 2B). To investigate how SIRT4 regulates the deacetylation of MTHFD2, we performed an immunoprecipitation (IP) assay of endogenous MTHFD2 in HEK293T cells and the presence of SIRT4 in precipatates turned out the endogenous interaction of MTHFD2 and SIRT4 (Figure 2C). Next, we purified glutathione-S-transferase (GST)-tagged SIRT4WT and SIRT4H161Y, respectively, from the prokaryotic system (Anderson et al., 2017). The GST pull-down experiment result proved that MTHFD2 and SIRT4 interacted directly (Figure 2D). To further validate that SIRT4 is the deacetylase of MTHFD2, we constructed MCF7 and ZR-75-30 cells with stable knockdown of SIRT4 and found that both acetylation and protein levels of MTHFD2 increased (Figure 2E). Additionally, the K50 acetylation level of MTHFD2 declined when SIRT4WT but not SIRT4H161Y was restored into SIRT4-knockdown cells (Figure 2F). More importantly, in vitro deacetylation assay was carried out by incubating purified SIRT4 and MTHFD2-Flag immunoprecipitated from HEK293T cells. SIRT4 indeed deacetylated MTHFD2 at K50 residue and subsequently decreased the enzymatic activity of MTHFD2. In contrast, the SIRT4H161Y mutant was not able to deacetylate MTHFD2 (Figure 2G). Above all, these results reveal that SIRT4 is the deacetylase of MTHFD2. Particularly, SIRT4 knockdown increases MTHFD2 K50 acetylation, simultaneously with the upregulation of the protein level.
Figure 2.
SIRT4 deacetylates MTHFD2 at K50. (A) Overexpression of SIRT4, but not other mitochondrial sirtuins, decreases the protein level of MTHFD2. HA-tagged SIRT3–SIRT5 were individually coexpressed with Flag-tagged MTHFD2 into HEK293T cells and protein levels of MTHFD2 were detected by western blotting. (B) WT SIRT4 but not its catalytic-dead H161Y mutant decreases the endogenous MTHFD2 protein level. WT SIRT4 and H161Y mutants were transfected into HEK293T cells. The endogenous MTHFD2 protein level was detected by western blotting. (C) MTHFD2 interacts with SIRT4. The interaction between endogenous MTHFD2 and SIRT4 in HEK293T cells was determined by co-IP and western blotting. (D) Verification of direct interaction between SIRT4 and MTHFD2 using GST pull-down assay. GST-tagged SIRT4 protein was purified with GST beads and incubated with HEK293T cell lysates. The beads were collected and subjected to western blotting. (E) SIRT4 knockdown increases both MTHFD2 acetylation and protein levels. MCF7 and ZR-75-30 cells stably expressing scramble shRNA (scr) or two independent shRNAs (2# and 3#) against SIRT4, respectively, were harvested for western blotting analysis. (F) WT SIRT4, but not the H161Y mutant, decreases K50 acetylation. SIRT4 or its H161Y mutant was re-expressed into stable SIRT4-knockdown MCF7 cells. Cell extracts were analyzed by western blotting. (G) WT SIRT4 but not its H161Y mutant deacetylates MTHFD2 in vitro. Recombinant SIRT4 (with GST tag cleaved off) and MTHFD2 immunoprecipitated from HEK293T cells were incubated with or without NAD+. After in vitro deacetylation, the reaction mixture was subjected to K50 acetylation detection by western blotting.
K50 acetylation blocks MTHFD2 degradation mediated by CUL3
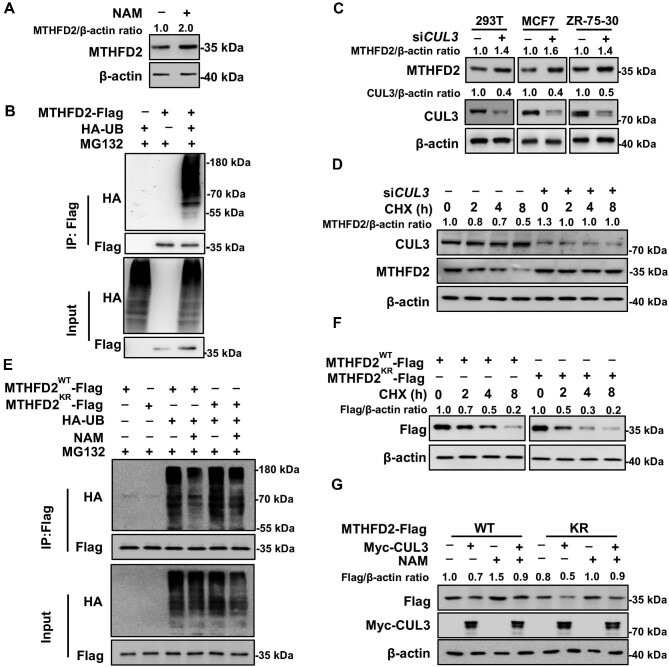
This study initially found that NAM treatment upregulates the intracellular MTHFD2 protein level (Figure 3A). Most acetylated lysine residues are also subject to other PTMs, such as ubiquitylation. It is estimated that one-third of the acetylation sites in human cells are ubiquitylated as well (Wagner et al., 2011). As MTHFD2 is predicted to have seven potential ubiquitylation sites by the Phosphosite database, we speculate that MTHFD2 might be degraded through the proteasome pathway. In order to confirm the ubiquitylation of MTHFD2, Flag-tagged MTHFD2 and/or HA-tagged ubiquitin were expressed individually or coexpressed in 293T cells. The typical ubiquitylation ladder was present when MTHFD2 and ubiquin were coexpressed in cells (Figure 3B). According to the Biogrid database (https://thebiogrid.org/116011/summary/homo-sapiens/mthfd2.html), E3 ligase CUL3 binds to MTHFD2 potentially. To verify whether CUL3 is the E3 ligase for MTHFD2, short interfering RNA (siRNA) targeting CUL3 was transfected in HEK293T, MCF7, and ZR-75-30 cells. After silencing CUL3, the endogenous MTHFD2 protein level increased ∼1.5-fold (Figure 3C). Moreover, the protein synthesis inhibitor cycloheximide (CHX) chase experiment indicated that the half-life of endogenous MTHFD2 was ∼4 h, and CUL3 knockdown prolonged the half-life of MTHFD2 (Figure 3D). These data demonstrate that MTHFD2 is degraded by CUL3. Acetylation often occurs at highly conserved domains, changing the charge and steric hindrance of the protein, and thus may alter the stability of the modified proteins. To determine the effect of acetylation on MTHFD2 stability, MTHFD2WT or MTHFD2KR was ectopically expressed with HA-Ub in cells in the presence or absence of NAM. Results showed that the ubiquitylation signal of MTHFD2WT was less than that of MTHFD2KR, and NAM treatment decreased the ubiquitylation of MTHFD2WT (Figure 3E). Moreover, the half-life of MTHFD2WT was significantly longer than that of MTHFD2KR (Figure 3F). Furthermore, the NAM-accumulated acetylation level of MTHFD2 blocks CUL3-induced degradation of MTHFD2WT but not that of MTHFD2KR (Figure 3G). In conclusion, MTHFD2 acetylation mitigates its ubiquitylation to enhance protein stabilization.
Figure 3.
K50 acetylation blocks MTHFD2 degradation by CUL3. (A) NAM treatment increases the MTHFD2 protein level. HEK293T cells were cultured with or without NAM for 6 h before harvest. (B) MTHFD2 is ubiquitylated. Ubiquitylation of MTHFD2 was detected by IP of exogenous Flag-tagged MTHFD2 and analyzed by western blotting. (C) CUL3 knockdown increases the MTHFD2 protein level. HEK293T, MCF7, and ZR-75-30 cells expressing control siRNA or siRNA targeting CUL3 were collected for western blotting analysis as indicated. (D) CUL3 knockdown stabilizes MTHFD2. Control siRNA or siRNA targeting CUL3 was expressed into HEK293T cells. Cells were treated with CHX (10 mg/ml) as indicated. The endogenous MTHFD2 protein level was detected by western blotting. (E) The KR mutant increases MTHFD2 ubiquitylation. Indicated plasmids were transfected into HEK293T cells, and the ubiquitylation of immunoprecipitated MTHFD2-Flag proteins was determined after NAM treatment. (F) The half-life of WT MTHFD2 is longer than that of the KR mutant. ZR-75-30 cells stably expressing WT MTHFD2 or the KR mutant were treated with CHX for the indicated time points before harvest. (G) Acetylation blocks CUL3-induced MTHFD2 degradation. ZR-75-30 cells stably expressing WT MTHFD2 or the KR mutant were transfected with or without Myc-CUL3, followed by treatment with or without NAM for 6 h. Flag-tagged MTHFD2 was determined.
Folate deprivation induces MTHFD2 hypoacetylation and destabilization
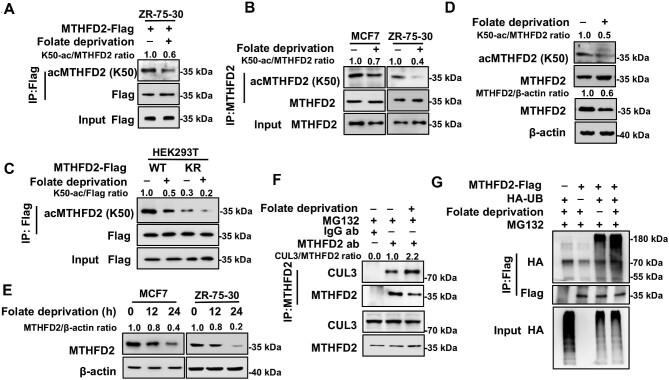
To figure out the physiological signal that regulates the K50 acetylation level, we tested different culture conditions and found that folate dynamically regulated the K50 acetylation level. In ZR-75-30 cells, the K50 acetylation level of exogenous or endogenous MTHFD2 was reduced by 2-fold in response to folate deprivation (Figure 4A and B). In addition, a similar reduction of MTHFD2 K50 acetylation was obtained in purified mitochondria from ZR-75-30 cells upon folate deprivation (Supplementary Figure S2A). Furthermore, folate deprivation decreased the K50 acetylation level of MTHFD2WT but not that of MTHFD2KR (Figure 4C). Notably, folate deprivation-induced reduction of the K50 acetylation level was accompanied by a decrease of the MTHFD2 protein level either in HEK293T cells or in purified mitochondria from ZR-75-30 cells, whereas the MTHFD2 mRNA level remained unchanged (Figure 4D; Supplementary Figure S2B and C). Complementarily, the significant decline of the MTHFD2 protein level caused by folate deprivation was in a time-dependent manner (Figure 4E). Thus, the folate metabolic signal has pivotal effects on MTHFD2 acetylation as well as the protein level. As MTHFD2 acetylation prevents CUL3-mediated ubiquitylation, we then performed an endogenous IP assay and found that, indeed, CUL3 interacted with MTHFD2. Particularly, the interaction between CUL3 and MTHFD2 was fortified in response to folate deprivation (Figure 4F). Consequently, folate deprivation increased MTHFD2 ubiquitylation (Figure 4G). Besides, we determined the binding between MTHFD2 and SIRT4 in the absence of folate and found that folate deprivation substantially promoted the binding between SIRT4 and MTHFD2 (Supplementary Figure S2D). Furthermore, we explored the regulation of MTHFD2 ubiquitylation upon folate deprivation. SIRT4 overexpressing enhanced the MTHFD2 ubiquitylation level. By contrast, the ubiquitylation signal of MTHFD2 could not be increased by SIRT4 expression after silencing CUL3 (Supplementary Figure S2E). Our data demonstrate that Sirt4-mediated degradation of MTHFD2 is dependent on CUL3. These data suggest that the extrinsic folate metabolic signal is essential for the maintenance of MTHFD2 protein by acetylating modification.
Figure 4.
Folate deprivation induces MTHFD2 hypoacetylation and destabilization. (A) Folate deprivation decreases K50 acetylation of MTHFD2. ZR-75-30 cells stably expressing WT MTHFD2 were cultured in medium with or without folate for 24 h. Acetylation of MTHFD2 was determined by IP and western blotting. (B) Folate deprivation decreases K50 acetylation of endogenous MTHFD2 in breast cancer cell lines. MCF7 and ZR-75-30 cells were cultured under folate deprivation. K50 acetylation was determined by western blotting. (C) Folate deprivation decreases acetylation of WT MTHFD2 but not its KR mutant. ZR-75-30 cells stably expressing WT MTHFD2 or the KR mutant were cultured with or without folate. Acetylation of MTHFD2 was determined by IP and western blotting. (D) Folate deprivation decreases both K50 acetylation and protein levels of MTHFD2. HEK293T cells were cultured with normal or folate-deprived medium, respectively. The protein expression level of MTHFD2 was analyzed with β-actin normalization and K50 acetylation was analyzed with MTHFD2 normalization by western blotting. (E) Folate deprivation decreases the protein level of MTHFD2 in breast cancer cell lines. MCF7 and ZR-75-30 cells were cultured under folate deprivation for different time courses as indicated. Protein expression of MTHFD2 was determined by western blotting. (F) Folate deprivation modulates the interaction between MTHFD2 and CUL3. HEK293T cells were cultured with or without folate, endogenous IP against MTHFD2 was carried out, and the interaction was detected by western blotting. (G) Folate deprivation enhances the ubiquitylation of MTHFD2. Flag-tagged MTHFD2 was coexpressed with HA-tagged ubiquitin in HEK293T cells. Cells were cultured with or without folate and treated with MG132 for 6 h. Ubiquitylation of MTHFD2 was determined by western blotting.
K50 acetylation maintains cellular redox balance and promotes tumor growth
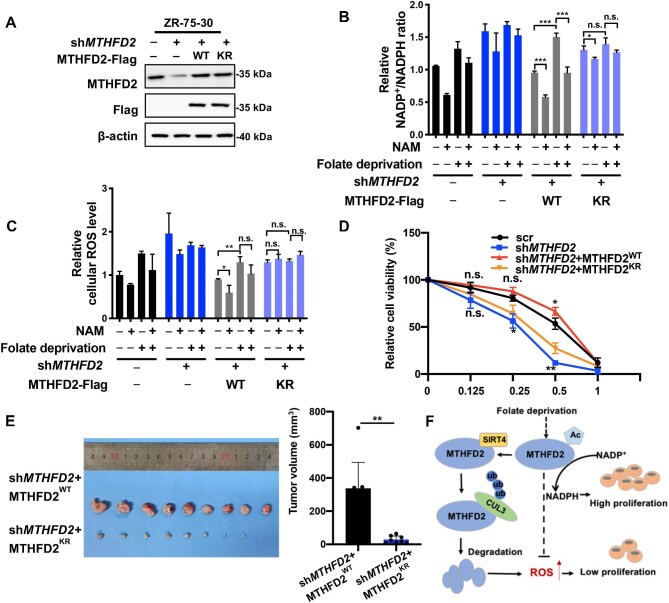
To explore the effects of MTHFD2 acetylation, we generated ZR-75-30 cells with stable knockdown of endogenous MTHFD2 using short hairpin RNA (shRNA) and the derivative cells with putback of MTHFD2WT or MTHFD2KR. MTHFD2 knockdown efficiency and restored expression of MTHFD2WT or MTHFD2KR were shown by western blotting analysis. Actually, the restored protein level of MTHFD2WT or MTHFD2KR was similar to the endogenous expression level of MTHFD2 (Figure 5A).
Figure 5.
MTHFD2 deacetylation inhibits NADPH production and breast cancer growth. (A) Characterization of stable re-expression of MTHFD2 in ZR-75-30 cells. WT MTHFD2 or the KR mutant was reintroduced into MTHFD2-knockdown cells. The knockdown efficiency and re-expression of Flag-tagged MTHFD2 were determined by western blotting. (B and C) K50 acetylation of MTHFD2 modulates the NADP+/NADPH ratio and ROS level. MTHFD2-knockdown ZR-75-30 cells re-expressing WT MTHFD2 (shMTHFD2 + MTHFD2WT) or its KR mutant (shMTHFD2 + MTHFD2KR) were cultured under folate deprivation followed by NAM incubation for 6 h. The NADP+/NADPH ratios (B) and ROS levels (C) were determined by a spectrofluorometer. (D) Re-expression of WT MTHFD2, but not the KR mutant, protects cells from oxidative injury. MTHFD2-knockdown ZR-75-30 cells re-expressing WT MTHFD2 (shMTHFD2 + MTHFD2WT) or the KR mutant (shMTHFD2 + MTHFD2KR) were treated with indicated concentrations of H2O2 for 12 h. Cell viability was assayed. (E) The KR mutant significantly reduces tumor growth by xenograft assay. ZR-75-30 breast cancer cells were transplanted to nude mice. After tumor harvest, tumor weight and size were measured. (F) Working model. SIRT4 deacetylates MTHFD2 at K50 residue in response to folate deprivation, which promotes proteasomal degradation of MTHFD2 via CUL3-triggered ubiquitylation, leading to an inhibition of NADPH production and cancer cell proliferation. Therefore, stressful signal of inadequate folate is sensed by the SIRT4–MTHFD2 axis to cause disorder of cellular redox by reducing MTHFD2-catalyzed NADPH production and hamper proliferation of breast cancer cells. Data shown represent mean ± SD obtained from triplicate independent experiments. n.s. indicates no significance, *P < 0.05, **P < 0.01, and ***P < 0.001.
As a major source of NADPH production, the 1C cycle pathway produces ∼40% of cellular NADPH, which powers reductive biosynthesis and protects against redox (Fan et al., 2014; Wei et al., 2018; Ju et al., 2019). Therefore, we measured the NADP+/NADPH ratios in stable cells treated with NAM or folate deprivation to detect the effect of K50 acetylation on NADPH production. The results showed that NADPH production in MTHFD2-knockdown or MTHFD2KR cells was lower than that in MTHFD2WT cells. Furthermore, NAM addition enhanced NADPH production in both control and MTHFD2WT cells but was unable to enhance NADPH production in MTHFD2-knockdown or MTHFD2KR cells (Figure 5B). Thus, K50 acetylation of MTHFD2 sustained by NAM addition supports NADPH production by counteracting against folate deprivation. It is known that persistent elevation of the reactive oxygen species (ROS) level in response to oxidative stress is harmful to cell viability. Cell damage could be prevented by NADPH as a reducing agent to quench cellular ROS. Therefore, we next detected cellular ROS levels in established ZR-75-30 cells. Indeed, expression of MTHFD2WT led to a reduction of the ROS level in cells, whereas the MTHFD2KR mutant could not fully simulate the scavenging effect of MTHFD2WT on ROS. Moreover, folate deprivation-induced accumulation of cellular ROS in MTHFD2WT or MTHFD2KR cells could be partially lessened by NAM treatment (Figure 5C). According to the observations, we speculated that SIRT4-mediated MTHFD2 acetylation is critical to connecting the folate metabolic signal and cellular redox maintenance by controlling MTHFD2 stability and sequential NADPH production. To define the speculation, we determined the effect of SIRT4 knockdown on NADPH production and redox balance. SIRT4 knockdown resulted in a dramatic increase of NADPH production and decrease of ROS levels in control but not MTHFD2-knockdown cells (Supplementary Figure S3A and B). We further measured ZR-75-30 cell viability after challenge with hydrogen peroxide. MTHFD2-knockdown cells were highly sensitive to oxidative stress and induced a remarkable increase of cell death. Re-expression of MTHFD2WT, but not the MTHFD2KR mutant, protected cells from oxidative damage and the most protective effect was observed in cells introduced with MTHFD2WT (Figure 5D).
Previous studies reported that MTHFD2 overexpression accelerates cancer cell proliferation and cancer progression, especially in breast cancer. Inhibition of key enzymes of the 1C metabolism pathway impairs tumor cell growth (Selcuklu et al., 2012; Nilsson et al., 2014; Pikman et al., 2016). Next, we examined the effect of MTHFD2 acetylation on the tumor burden and cell proliferation in ZR-75-30 cells. Overexpression of MTHFD2WT increased cell proliferation and knockdown of endogenous MTHFD2 significantly inhibited cell growth, whereas cells expressing the MTHFD2K50R mutant could not completely replenish the cell proliferation suppressed by MTHFD2 knockdown both in vivo and in vitro (Figure 5E; Supplementary Figure S3C–E). Our data showed that the tumor volume and weight were significantly reduced in mice receiving cells expressing KR mutation compared with mice bearing WT MTHFD2 cells (Figure 5D; Supplementary Figure S3D). Consistently, reintroduction of the KR mutant into cells significantly attenuated the staining intensity of the cell proliferation marker Ki-67 (Supplementary Figure S3E). In addition, we examined enzymatic activities of WT and mutant (KR, KQ) MTHFD2 and found that the KR mutant significantly decreased whereas the KQ mutant significantly increased the enzymatic activity (Supplementary Figure S3F). Moreover, results of the cell proliferation assay and enzyme activity assay showed that the MTHFD2 KR mutant had a dominant inhibition on WT MTHFD2 (Supplementary Figure S3G–I). Co-IP experiments revealed the interaction between the Flag-tagged KR mutant and HA-tagged WT MTHFD2 when they were simultaneously expressed in cells, suggesting that the KR mutant and WT MTHFD2 could form a heterodimer (Supplementary Figure S3G and H). Notably, their interaction significantly inhibited the enzymatic activity of WT MTHFD2 (Supplementary Figure S3H). As expected, overexpression of the KR mutant significantly impeded cell viability and growth in ZR-75-30 cells with endogenous MTHFD2 in either the absence or the presence of folate (Supplementary Figure S3I). These results prove that acetylation-stabilized MTHFD2 substantially potentiates breast cancer cell growth.
Discussion
Previous research of acetyl proteomics of mouse revealed that 63% of mitochondria proteins, >700 proteins, contain acetylation sites (Baeza et al., 2016). Nevertheless, few acetylated mitochondrial proteins are functionally defined (Baeza et al., 2016).
The mitochondrial MTHFD2 plays a crucial role in the 1C unit cycle and is postulated as an oncoprotein. MTHFD2 overexpression occurs in multiple types of cancers and is associated with poor survival of patients (Nilsson et al., 2014). The underlying mechanism of MTHFD2-driven cancer development has been comprehensively investigated. On the one hand, cancer progression is dependent on MTHFD2-enhanced biosynthesis of nucleotide, such as lung cancer and MYCN-amplified neuroblastoma (Cheung et al., 2019; Nishimura et al., 2019). Specifically, MTHFD2-sustained nucleotide synthesis profoundly contributes to the stemness and chemoresistance of lung cancer cells as well as the adaptation of the tumor immune microenvironment (Nishimura et al., 2019; Wang et al., 2019; Sugiura et al., 2022). On the other hand, serine catabolism becomes a major NADPH source through MTHFD2 to balance the cellular oxidative condition and facilitate cell growth by ROS detoxification (Fan et al., 2014). Studies revealed that MTHFD2 modulates serine–glycine and mitochondrial 1C metabolism in response to oxidative stress and oxPAPC and the suppression of MTHFD2 disturbs NADPH and redox homeostasis and accelerates cell death (Hitzel et al., 2018; Ju et al., 2019). In this research, we find that K50 residue in MTHFD2 is acetylated. Deacetylation of MTHFD2 K50 residue by SIRT4 triggers CUL3 E3 ubiquitin ligase-mediated MTHFD2 degradation, particularly under folate deprivation. Notably, MTHFD2 knockdown or its KR mutant putback decreases the enzymatic activity of MTHFD2 and upregulates intracellular ROS, resulting in the suppression of breast cancer cell proliferation and suggesting the essential role of MTHFD2 in oxidative homeostasis (Figure 5F). Furthermore, it would be interesting to perform stable isotope tracing, like utilizing U-[13C]-serine, to explore the effect of MTHFD2 on nucleotide biosynthesis. Altogether, this finding reveals that SIRT4-induced deacetylation of MTHFD2 is indispensable for folate sensing, metabolism remodeling, and tumor cell proliferation.
Combination or crosstalk of different PTMs makes the regulation of protein function or turnover more flexible and precise in response to stimulating signals of different nutrients and/or growth factors (Cai et al., 2012; Yang et al., 2015; Li et al., 2020). Here, we find that acetylation and ubiquitylation of MTHFD2 are coupled together. When a folate metabolic signal is present, persistent K50 residue acetylating hampers the binding of MTHFD2 to CUL3 E3 ubiquitin ligases and prevents degradation of MTHFD2 via the proteasome pathway, potentiating the 1C unit cycle to generate a methyl group for epigenetic modulation, precursors for nucleotide synthesis, and reductive power for cell redox maintenance in breast cancer cells. On the other hand, when folate is deficient, the stressful signal initiates SIRT4-mediated deacetylation of MTHFD2 K50. Sequentially, ubiquitylation is triggered to promote MTHFD2 degradation, resulting in the obstruction of the 1C unit cycle and impairment of cell growth by elevating cellular ROS. Further research is planned to define the sensing mechanism of the folate signal in cells and the deubiquitylase and precise ubiquitylation sites of MTHFD2 to fully explain the mechanism underlying MTHFD2 ubiquitylation.
SIRT4 is a guardian for cellular energy metabolism. It inhibits fatty acid oxidation and promotes lipid anabolism by inhibiting malonyl CoA decarboxylase (MCD). There is also a report that SIRT4 downregulates glutamate dehydrogenase to suppress insulin secretion. More importantly, SIRT4 shows tumor-suppressive activity by disrupting glutamine anabolism. The SIRT4 expression level is reduced in several types of cancers (Jeong et al., 2013). When regulating these essential metabolic processes, SIRT4 displays multiple enzymatic activities, including lysine deacetylase, lipoamidase, and ADP-ribosyltransferase. Among the studies, the deacetylation of MCD by SIRT4 was only determined by in vitro assay using MCD and SIRT4 proteins isolated from cells. There is a lack of solid evidence to show the direct interaction between MCD and SIRT4. A few other substrates have been discovered for SIRT4 acting as a deacetylase (Laurent et al., 2013). In this study, the in vitro reaction of prokaryotic recombinant SIRT4 protein and isolated intracellular MTHFD2 was performed in a more direct way to confirm the deacetylase activity of SIRT4 on MTHFD2.
Folic acid supplementation has been widely applied to protect from neural tube defects of births in the world since the 1990s. However, studies in the recent two decades have shown that excess intake of folic acid promotes the incidence and/or progression of gastrointestinal, breast, pancreatic, and other tumors (Shang et al., 2021). Consistently, it has been reported that low plasma folate level is associated with decreased colorectal cancer risk (Lee et al., 2012; Gylling et al., 2014). Thus, folate deprivation for tumor treatment is feasible. Folate restriction impairs malignant phenotypes of cancer cells in vivo and in vitro (Yang et al., 2015; Coleman et al., 2021). One preclinical study revealed that relative long-term (8 weeks), but not short-period (1 week), folate restriction effectively counteracts tumorigenesis. As expected, mechanistically, folate restriction disturbs epigenetic modification, redox maintenance, as well as DNA damage repair due to the linkage of folate metabolism to methionine metabolism, reducing power production, and nucleotide syntheses (Fardous et al., 2021). In addition, folate metabolism antagonists, such as methotrexate, pemetrexed, and 5-fluorouracil, have been widely used in the treatment of many types of tumors, although being challenged by chemoresistance (Assaraf, 2007; Liu et al., 2021). However, functional folate metabolism facilitates therapeutic efficiency by targeting MYC in leukemia (Su et al., 2020). Therefore, a folate deprivation strategy in cancer treatment should be precisely defined based on personal genetic and/or epigenetic characteristics. In our study, the SIRT4–MTHFD2 axis can respond to folate deficiency signals and inhibit the growth of breast cancer. Accordingly, one recent study also showed that folate deprivation inhibits the growth of triple-negative breast cancer xenograft (Coleman et al., 2021). Intriguingly, in addition to the overexpression in various tumor cells (Pallmann et al., 2021), MTHFD2 is also involved in the expression of PD-L1 stimulated by IFN-γ, promoting immune escape (Shang et al., 2021). As targeting MTHFD2 can inhibit a variety of tumors, our research suggests that folate deprivation is a potentially auxiliary approach for tumor treatment by targeting MTHFD2, especially in breast cancer.
In summary, our discoveries demonstrate the critical role of the SIRT4–MTHFD2 axis in folate metabolism. In particular, under folate-lacking conditions, the stressful signal of inadequate nutrients is sensed by hijacking the SIRT4–MTHFD2 axis to fine-tune cellular folate metabolism, slowing down the folate cycle and curbing cell growth. Thus, our finding suggests one rational strategy of cancer therapy by targeting the SIRT4–MTHFD2 axis.
Materials and methods
Antibodies
Antibodies against Flag (Cell Signaling Technology, Cat#2368), pan-acetylated lysine (Cell Signaling Technology, Cat#9441), MTHFD2 (Cell Signaling Technology, Cat#41377), SIRT4 (Abclonal, Cat#A7585), CUL3 (Abcam, Cat#ab75851), β-actin (AOGMA, Cat#9601), and HA (SAB, Cat#T501) were purchased. To generate a site-specific antibody against acetyl K50 of MTHFD2 [acMTHFD2 (K50)], the synthesized peptide (GL Biochem) was coupled to keyhole limpet hemocyanin to immunize rabbits and the post-immunization serum was collected.
Cell culture and treatment
Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Cat#12100046/12800017) supplemented with 10% fetal bovine serum (FBS) (Gibco, Cat#0082147) and 1% penicillin/streptomycin (Hyclone, Cat#SV30010). For sirtuin inhibitor treatment, cells were treated with 5 mM NAM (Sigma, Cat#98920). For proteasome inhibitor treatment, cells were treated with 10 μM carbobenzoxy-Leu-Leu-leucinal (MG132) (Sigma, Cat#1211877-36-9) for 6 h.
IP
Cells were lysed on ice in NP-40 lysis buffer (150 mM NaCl, 50 mM Tris, 0.5% NP-40, pH 7.5) with protease inhibitors (Biotool, Cat#B14001). After centrifugation, the collected supernatant was incubated with Flag beads (Sigma, Cat#A2220) at 4°C overnight. Beads were then washed for further analysis. For endogenous IP, antibody against MTHFD2 was added to cell lysate supernatant, followed by incubation with protein G agarose (Sigma, Cat#11243233001) at 4°C for another 4 h.
Recombinant protein expression and purification
According to the protocol described previously (Guo et al., 2020), transformed BL21 Escherichia coli with the plasmid encoding GST-tagged SIRT4 was cultured in LB medium. Then, 0.2 mM isopropyl-β-D-1-thiogalactopyranoside was added to induce protein expression at 16°C when the optical density (OD) at 600 nm (OD600nm) reached 0.6. Recombinant SIRT4 was harvested using GST agarose beads (AOGMA, Cat#AGM90049) for purification. Then, GST tag was removed by protease.
In vitro deacetylation assay
MTHFD2-Flag was overexpressed in HEK293T cells and purified by IP with Flag beads (MCE, 98849-88-8). MTHFD2-Flag and recombinant SIRT4 were added to the deacetylase buffer (20 mM Tris–HCl, pH 7.8, 150 mM NaCl, 1 mM NAD+, and 2 mM dithiothreitol). After incubation at room temperature for 3 h, the mixture was boiled with loading buffer and the acetylation level of K50 was detected by western blotting.
Real-time quantitative PCR analysis
Total RNA was extracted from cells using TRNzol Universal (Tiangen, Cat#DP424) and transcribed reversely. Quantitative PCR was performed with SYBR Green premix reagent (TaKaRa, Cat#RR820A) using β-actin as a control. The primers used are as follows: hACTB forward, 5′-GCACAGAGCCTCGCCTT-3′; hACTB reverse, 5′-GTTGTCGACGACGAGCG-3′; hMTHFD2 forward, 5′-GCGGCAGGAGGTAGAAGA-3′; and hMTHFD2 reverse, 5′-TGTTGAGGACATAGGAGTGACT-3′.
Construction of stable cell pools
To generate stable MTHFD2-knockdown cells, shRNA targeting MTHFD2 was constructed in pMKO plasmid and delivered into cells with a two-plasmid packaging retrovirus system. Infected cells were selected in 2 μg/ml puromycin (Ameresco, Cat#J593). The targeting sequence is shMTHFD2: 5′-GGTGGGTGTTTCTGCACATAC-3′.
To generate MTHFD2-knockdown and rescue stable cell pools, pQCXIH vector containing Flag-tagged human MTHFD2WT or MTHFD2KR was co-transfected with the gag and vsvg vectors into HEK293T cells to produce retroviruses to infect MTHFD2-knockdown cells, followed by selection in 50 μg/ml hygromycin B (Ameresco, Cat#K547).
Measurement of NADP+/NADPH ratios
The intracellular levels of NADP+ and NADPH were measured by NADP+/NADPH assay kit (Beyotime, Cat#S0179). Briefly, samples isolated from 1×106 cells with 200 μl NADP+/NADPH extraction buffer were heated at 60°C for 30 min to decompose NADP+, followed by sequentially adding G6PDH working solution for conversion of NADP+ to NADPH and NADPH developer. Finally, the absorbance was read at OD450nm.
Human breast cancer xenograft model
ZR-75-30 breast cancer cell line was used in tumor growth analysis. A total of 5 × 106 cells suspended in 50 µl DMEM without FBS and penicillin/streptomycin were mixed with 50 µl Matrigel matrix (Corning, Cat#354248) at a ratio of 1:1, followed by subcutaneous injection into both flanks of the mouse. Once the tumor diameter of the WT group reached 8–10 mm, both the WT and KR groups of mice were sacrificed and the tumor was fetched for weighing. The length and width of the tumor were also measured. The tumor volume was calculated by the following formula: 1/2 × length × width2. These tumors were fixed in 4% paraformaldehyde for embedding, followed by immunohistochemistry (IHC) and hematoxylin and eosin (HE) staining.
Measurement of MTHFD2 enzyme activity
The enzyme activity assay was based on the method published previously (Fu et al., 2017). Briefly, substrate (6R,S)-5,10-methylene-5,6,7,8-tetrahydrofolic acid, calcium salt (Schircks, Cat#16.226) dissolved in 50 mM Tris–HCl (pH 8.0) and 100 mM β-mercaptoethanol was stored in aliquots under nitrogen.
MTHFD2 protein immunoprecipitated from cells and substrate (400 µM as final concentration) were added to detection buffer (50 mM Tris–HCl, 30 mM β-mercaptoethanol, 5 mM MgCl2, 1 mM NAD+, pH 8.0). The mixture was incubated at 30°C for 2 min and stopped by 1 M HCl. The absorbance was measured at 350 nm and the final result was obtained by normalization with the quantified protein concentration.
IHC and HE staining
Slides were deparaffinized in xylene, hydrated in graded alcohol solutions, and blocked in peroxidase. Heat-induced antigen retrieval was performed with 0.01 M citrate buffer (pH 6.0) for 25 min. Sections were stained in hematoxylin and eosin for HE staining. For IHC, sections were immersed in goat serum for 30 min to block non-specific staining and then incubated with Ki-67 antibody (Abcam, Cat#ab15580) overnight at 4°C. Secondary anti-rabbit antibody was incubated with sections for 40 min at 37°C, followed by diaminobenzidine development and hematoxylin counterstaining.
Statistical analysis
Statistical analyses were performed with a two-tailed unpaired Student's t-test. Data shown represent the results obtained from triplicate independent experiments with standard errors of the mean (mean ± SD). n.s. indicates no significance, *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Acknowledgements
We thank members of the Lei Laboratory for discussion throughout this study. We also appreciate the Biomedical Core Facility of Fudan University for technical support.
Contributor Information
Fan Zhang, Fudan University Shanghai Cancer Center & Institutes of Biomedical Sciences & School of Basic Medical Sciences, Cancer Institutes, Key Laboratory of Breast Cancer in Shanghai, Shanghai Key Laboratory of Radiation Oncology, The Shanghai Key Laboratory of Medical Epigenetics, Shanghai Medical College, Fudan University, Shanghai 200032, China; Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, China.
Di Wang, Fudan University Shanghai Cancer Center & Institutes of Biomedical Sciences & School of Basic Medical Sciences, Cancer Institutes, Key Laboratory of Breast Cancer in Shanghai, Shanghai Key Laboratory of Radiation Oncology, The Shanghai Key Laboratory of Medical Epigenetics, Shanghai Medical College, Fudan University, Shanghai 200032, China; Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, China.
Jintao Li, Fudan University Shanghai Cancer Center & Institutes of Biomedical Sciences & School of Basic Medical Sciences, Cancer Institutes, Key Laboratory of Breast Cancer in Shanghai, Shanghai Key Laboratory of Radiation Oncology, The Shanghai Key Laboratory of Medical Epigenetics, Shanghai Medical College, Fudan University, Shanghai 200032, China; Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, China.
Ying Su, Fudan University Shanghai Cancer Center & Institutes of Biomedical Sciences & School of Basic Medical Sciences, Cancer Institutes, Key Laboratory of Breast Cancer in Shanghai, Shanghai Key Laboratory of Radiation Oncology, The Shanghai Key Laboratory of Medical Epigenetics, Shanghai Medical College, Fudan University, Shanghai 200032, China; Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, China.
Suling Liu, Fudan University Shanghai Cancer Center & Institutes of Biomedical Sciences & School of Basic Medical Sciences, Cancer Institutes, Key Laboratory of Breast Cancer in Shanghai, Shanghai Key Laboratory of Radiation Oncology, The Shanghai Key Laboratory of Medical Epigenetics, Shanghai Medical College, Fudan University, Shanghai 200032, China; Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, China.
Qun-Ying Lei, Fudan University Shanghai Cancer Center & Institutes of Biomedical Sciences & School of Basic Medical Sciences, Cancer Institutes, Key Laboratory of Breast Cancer in Shanghai, Shanghai Key Laboratory of Radiation Oncology, The Shanghai Key Laboratory of Medical Epigenetics, Shanghai Medical College, Fudan University, Shanghai 200032, China; Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, China; State Key Laboratory of Medical Neurobiology, Fudan University, Shanghai 200032, China.
Miao Yin, Fudan University Shanghai Cancer Center & Institutes of Biomedical Sciences & School of Basic Medical Sciences, Cancer Institutes, Key Laboratory of Breast Cancer in Shanghai, Shanghai Key Laboratory of Radiation Oncology, The Shanghai Key Laboratory of Medical Epigenetics, Shanghai Medical College, Fudan University, Shanghai 200032, China; Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, China.
Funding
This work was supported by the National Key R&D Program of China (2020YFA0803400/2020YFA0803402 and 2019YFA0801703 to Q.-Y.L.), the National Natural Science Foundation of China (81872240 to M.Y., 82002951 to J.L., and 81790250/81790253, 91959202, and 82121004 to Q.-Y.L.), and the Innovation Program of Shanghai Municipal Education Commission (N173606 to Q.-Y.L.).
Conflict of interest: none declared.
Author contributions: M.Y. and Q.-Y.L. conceived and designed the experiments and wrote the manuscript. F.Z. and D.W. performed most of the experiments and analyzed the data. J.L. and Y.S. helped to identify the specificity of the acMTHFD2 (K50) antibody, and S.L gave constructive advice.
References
- Ahuja N., Schwer B., Carobbio S.et al. (2007). Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J. Biol. Chem. 282, 33583–33592. [DOI] [PubMed] [Google Scholar]
- Anderson K.A., Huynh F.K., Fisher-Wellman K.et al. (2017). SIRT4 is a lysine deacylase that controls leucine metabolism and insulin secretion. Cell Metab. 25, 838–855.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaraf Y.G. (2007). Molecular basis of antifolate resistance. Cancer Metastasis Rev. 26, 153–181. [DOI] [PubMed] [Google Scholar]
- Baeza J., Smallegan M.J., Denu J.M. (2016). Mechanisms and dynamics of protein acetylation in mitochondria. Trends Biochem. Sci. 41, 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai N., Li M., Qu J.et al. (2012). Post-translational modulation of pluripotency. J. Mol. Cell Biol. 4, 262–265. [DOI] [PubMed] [Google Scholar]
- Chen P., Li C., Li X.et al. (2014). Higher dietary folate intake reduces the breast cancer risk: a systematic review and meta-analysis. Br. J. Cancer 110, 2327–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C.H.Y., Hsu C.L., Tsuei C.Y.et al. (2019). Combinatorial targeting of MTHFD2 and PAICS in purine synthesis as a novel therapeutic strategy. Cell Death Dis. 10, 786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C., Kumar C., Gnad F.et al. (2009). Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840. [DOI] [PubMed] [Google Scholar]
- Choudhary C., Weinert B.T., Nishida Y.et al. (2014). The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 15, 536–550. [DOI] [PubMed] [Google Scholar]
- Coleman M.F., O'Flanagan C.H., Pfeil A.J.et al. (2021). Metabolic response of triple-negative breast cancer to folate restriction. Nutrients 13, 1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Maso M., Dal Maso L., Augustin L.S.A.et al. (2020). Adherence to the Mediterranean diet and mortality after breast cancer. Nutrients 12, 3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro E., Sirois J., Tremblay M.L.et al. (2002). Mitochondrial NAD-dependent methylenetetrahydrofolate dehydrogenase–methenyltetrahydrofolate cyclohydrolase is essential for embryonic development. Mol. Cell. Biol. 22, 4158–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Ye J., Kamphorst J.J.et al. (2014). Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardous A.M., Beydoun S., James A.A.et al. (2021). The timing and duration of folate restriction differentially impacts colon carcinogenesis. Nutrients 14, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo J.C., Grau M.V., Haile R.W.et al. (2009). Folic acid and risk of prostate cancer: results from a randomized clinical trial. J. Natl Cancer Inst. 101, 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C., Sikandar A., Donner J.et al. (2017). The natural product carolacton inhibits folate-dependent C1 metabolism by targeting FolD/MTHFD. Nat. Commun. 8, 1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Zhang Q., Su Y.et al. (2020). Arginine methylation of ribose-5-phosphate isomerase A senses glucose to promote human colorectal cancer cell survival. Sci. China Life Sci. 63, 1394–1405. [DOI] [PubMed] [Google Scholar]
- Gylling B., Van Guelpen B., Schneede J.et al. (2014). Low folate levels are associated with reduced risk of colorectal cancer in a population with low folate status. Cancer Epidemiol. Biomarkers Prev. 23, 2136–2144. [DOI] [PubMed] [Google Scholar]
- Hansen M.F., Jensen S.O., Fuchtbauer E.M.et al. (2017). High folic acid diet enhances tumour growth in PyMT-induced breast cancer. Br. J. Cancer 116, 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzel J., Lee E., Zhang Y.et al. (2018). Oxidized phospholipids regulate amino acid metabolism through MTHFD2 to facilitate nucleotide release in endothelial cells. Nat. Commun. 9, 2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S.M., Xiao C., Finley L.W.et al. (2013). SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell 23, 450–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju H.Q., Lu Y.X., Chen D.L.et al. (2019). Modulation of Redox homeostasis by inhibition of MTHFD2 in colorectal cancer: mechanisms and therapeutic implications. J. Natl Cancer Inst. 111, 584–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.C., Sprung R., Chen Y.et al. (2006). Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618. [DOI] [PubMed] [Google Scholar]
- Laurent G., German N.J., Saha A.K.et al. (2013). SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol. Cell 50, 686–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Wei E.K., Fuchs C.S.et al. (2012). Plasma folate, methylenetetrahydrofolate reductase (MTHFR), and colorectal cancer risk in three large nested case-control studies. Cancer Causes Control 23, 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.T., Yin M., Wang D.et al. (2020). BCAT2-mediated BCAA catabolism is critical for development of pancreatic ductal adenocarcinoma. Nat. Cell Biol. 22, 167–174. [DOI] [PubMed] [Google Scholar]
- Liu Y.P., Zheng C.C., Huang Y.N.et al. (2021). Molecular mechanisms of chemo- and radiotherapy resistance and the potential implications for cancer treatment. MedComm 2, 315–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly A., Lee H., Chen J.M.et al. (2011). Effect of maternal and postweaning folic acid supplementation on mammary tumor risk in the offspring. Cancer Res. 71, 988–997. [DOI] [PubMed] [Google Scholar]
- Mathias R.A., Greco T.M., Oberstein A.et al. (2014). Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell 159, 1615–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran D.M., Trusk P.B., Pry K.et al. (2014). KRAS mutation status is associated with enhanced dependency on folate metabolism pathways in non-small cell lung cancer cells. Mol. Cancer Ther. 13, 1611–1624. [DOI] [PubMed] [Google Scholar]
- Nilsson R., Jain M., Madhusudhan N.et al. (2014). Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 5, 3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Nakata A., Chen X.et al. (2019). Cancer stem-like properties and gefitinib resistance are dependent on purine synthetic metabolism mediated by the mitochondrial enzyme MTHFD2. Oncogene 38, 2464–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallmann N., Deng K., Livgard M.et al. (2021). Stress-mediated reprogramming of prostate cancer one-carbon cycle drives disease progression. Cancer Res. 81, 4066–4078. [DOI] [PubMed] [Google Scholar]
- Pikman Y., Puissant A., Alexe G.et al. (2016). Targeting MTHFD2 in acute myeloid leukemia. J. Exp. Med. 213, 1285–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcuklu S.D., Donoghue M.T.A., Rehmet K.et al. (2012). MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. J. Biol. Chem. 287, 29516–29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang M., Yang H., Yang R.et al. (2021). The folate cycle enzyme MTHFD2 induces cancer immune evasion through PD-L1 up-regulation. Nat. Commun. 12, 1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijilmassi O., Del Rio Sevilla A., Maldonado Bautista E.et al. (2021). Gestational folic acid deficiency alters embryonic eye development: possible role of basement membrane proteins in eye malformations. Nutrition 90, 111250. [DOI] [PubMed] [Google Scholar]
- Su A., Ling F., Vaganay C.et al. (2020). The folate cycle enzyme MTHFR is a critical regulator of cell response to MYC-targeting therapies. Cancer Discov. 10, 1894–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura A., Andrejeva G., Voss K.et al. (2022). MTHFD2 is a metabolic checkpoint controlling effector and regulatory T cell fate and function. Immunity 55, 65–81.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S.A., Beli P., Weinert B.T.et al. (2011). A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.W., Shen H., Nobre L.et al. (2019). Epstein–Barr-virus-induced one-carbon metabolism drives B cell transformation. Cell Metab. 30, 539–555.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert B.T., Scholz C., Wagner S.A.et al. (2013). Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 4, 842–851. [DOI] [PubMed] [Google Scholar]
- Wei Z., Song J., Wang G.et al. (2018). Deacetylation of serine hydroxymethyl-transferase 2 by SIRT3 promotes colorectal carcinogenesis. Nat. Commun. 9, 4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.B., Xu Y.Y., Zhao X.N.et al. (2015). Acetylation of MAT IIalpha represses tumour cell growth and is decreased in human hepatocellular cancer. Nat. Commun. 6, 6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L.N., Bjorklund M., Caldez M.J.et al. (2021). Therapeutic targeting of the mitochondrial one-carbon pathway: perspectives, pitfalls, and potential. Oncogene 40, 2339–2354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.