Significance
The microbial decomposition of organic matter is a critical component of the global biogeochemical cycle that emerges from cellular interactions on microscale resource patches. The ecological processes controlling community dynamics in these microenvironments remain poorly understood. We show, through metagenomics, metabolomics, and modeling, that complex marine microbial communities growing on microscale resource particles diverge both taxonomically and functionally due to stochastic colonization, historical contingencies, and growth processes affected by community contexts. Prophage induction, especially among keystone organisms, is one significant contributor to covariation in community composition, species diversity, and biomass. Our results suggest that variability in microscale community assembly promotes coexistence among diverse metapopulations and that divergence in microscale functional states may translate to large-scale variation in particle degradation rates.
Keywords: community assembly, marine particles, prophages, historical contingencies, microscale
Abstract
In many natural environments, microorganisms decompose microscale resource patches made of complex organic matter. The growth and collapse of populations on these resource patches unfold within spatial ranges of a few hundred micrometers or less, making such microscale ecosystems hotspots of heterotrophic metabolism. Despite the potential importance of patch-level dynamics for the large-scale functioning of heterotrophic microbial communities, we have not yet been able to delineate the ecological processes that control natural populations at the microscale. Here, we address this challenge by characterizing the natural marine communities that assembled on over 1,000 individual microscale particles of chitin, the most abundant marine polysaccharide. Using low-template shotgun metagenomics and imaging, we find significant variation in microscale community composition despite the similarity in initial species pools across replicates. Chitin-degrading taxa that were rare in seawater established large populations on a subset of particles, resulting in a wide range of predicted chitinolytic abilities and biomass at the level of individual particles. We show, through a mathematical model, that this variability can be attributed to stochastic colonization and historical contingencies affecting the tempo of growth on particles. We find evidence that one biological process leading to such noisy growth across particles is differential predation by temperate bacteriophages of chitin-degrading strains, the keystone members of the community. Thus, initial stochasticity in assembly states on individual particles, amplified through ecological interactions, may have significant consequences for the diversity and functionality of systems of microscale patches.
A central challenge in microbial ecology is to connect the microscale world experienced by microbial cells to observations of large-scale community functions (1, 2). In many environments—ranging from soils (3) and sediments (4) to bioreactors (5) and hosts (6)—microbes live not in homogeneous, well-mixed cultures but rather in diverse, spatially structured assemblages, attached to surfaces and other cells in nutrient-dense patches on the order of 100 µm in size. Patches often exist in otherwise nutrient-limiting environments, creating hotspots of ecological interactions and nutrient fluxes (7, 8).
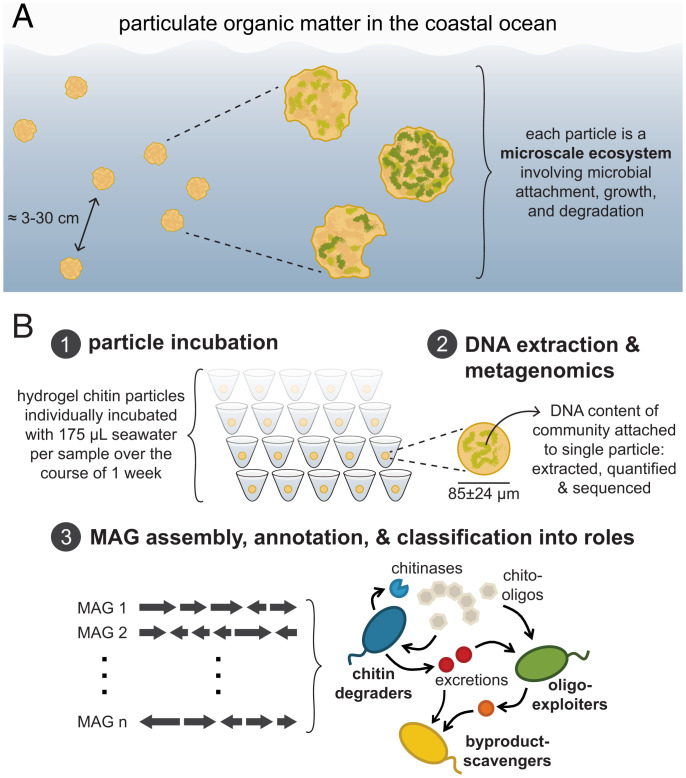
A well-known example of a system of micrometer-scale ecological hotspots is marine particulate organic matter (POM). Individual organic particles (Fig. 1A) serve as both nutrients and scaffolds around which complex communities of bacteria, archaea, viruses, and eukaryotes assemble (9), with global biogeochemical consequences (10). These interacting community members can be broadly classified as primary degraders (that produce extracellular enzymes to hydrolyze particle biopolymers), exploiters and scavengers (that are facilitated by primary degraders) (11–14), and predators [such as bacteriophages (15) and grazers (16)]. Although these assemblages are often ephemeral, cells can undergo multiple generations of growth while residing on a single patch of nutrient-rich POM, implying that the ecological functions of POM-associated microbes are heavily influenced by their dynamics and interactions on microscale particles. However, little is known about the processes governing community assembly at these scales.
Fig. 1.
Modeling POM degradation with a laboratory system of enriching of marine microbes on chitin particles. (A) Microscale marine particles are discrete, spatially separated nutrient-rich habitats dynamically populated and degraded by complex communities of heterotrophic bacteria. The interparticle distance range is estimated from data reported in Simon et al. (16). (B) Schematic depicting experimental design and analysis. Microscale chitin particles were individually incubated in seawater, and the DNA content of particle-attached communities was quantified and submitted for shotgun metagenomic sequencing. Communities were characterized using MAGs, which were classified into three predicted ecological roles for this ecosystem: chitin degraders, chitooligosaccharide exploiters, and metabolic byproduct scavengers.
A major obstacle to understanding the factors that control populations at the patch level is the difficulty of characterizing natural microscale communities with high replication. Microbial communities are usually sampled at spatial scales orders of magnitude larger than those relevant for microbial life (2), which homogenizes their inherent patchiness and results in inconsistent inferences about ecological interactions (1, 17). Recent technological advances now permit the sequencing of only thousands of cells (18, 19), presenting an opportunity to systematically characterize microbial populations in units more closely approximating in scale the ecological contexts experienced by microbes.
Here, we leveraged high-replicate sequencing of individual microscale communities to evaluate the outcomes of assembly processes without the homogenizing effects of standard bulk sampling procedures. We employed a hybrid natural-laboratory approach that paired the complexity of environmental microbial species pools with the controllability of synthetic microparticles as discrete resource patches (11, 12). In order to have sufficient statistical power to assess the variability in microscale assembly outcomes, we immersed 1,222 individual model resource particles (85.0 ± 24.0 µm in diameter) in samples of seawater containing microbes in their native states. The hydrogel particles were made of chitin, a highly abundant biopolymer in marine POM (9), enabling the enrichment of chitin-degrading communities on the particle surface. By incubating single particles separately under identical abiotic conditions, each one became a microenvironment harboring a replicate community assembled from initially similar species pools. We performed a comparative analysis across these microscale ecosystems to investigate the natural variability in community composition and function among particles and to identify biological processes that contribute to particle-level variability.
Results
Bacterial Community Composition Varies Significantly across Individual Particles.
To quantify the variation in community states across replicate microscale ecosystems, we separately incubated single chitin particles in coastal seawater sampled from a common reservoir (Fig. 1B and Materials and Methods). Assembly outcomes were assessed by removing particles from the seawater at 13 time points over the course of 167 h, a duration that aligns with previous measurements (12, 20) of particle lifetimes in bulk incubations (Materials and Methods). Of the 1,222 chitin particle microcosms harvested for metabolomics, shotgun metagenomes of 495 particle-attached communities from throughout the incubation period were used to construct a set of metagenome-assembled genomes (MAGs). These MAGs were annotated to infer strains’ potential ecological roles in a chitin-degrading community as primary degraders, chitooligosaccharide exploiters (21), or metabolic byproduct scavengers (Dataset S1 and Materials and Methods), and they served as the references for characterizing the taxonomic and functional composition of each particle (Fig. 1B, Dataset S1, and Materials and Methods).
We found a remarkable degree of compositional variability across individual particle communities at the end of the time course (n = 149, after 154 h to 167 h of incubation). The distributions of taxon (MAG) relative abundances across these endpoint particles spanned more than three orders of magnitude (Fig. 2 and SI Appendix, Fig. S1A) and were approximately lognormal, with a skew toward high abundances (SI Appendix, Fig. S2). As a result, the community compositions observed at the single-particle level diverged so significantly that the rank abundances of taxa on individual particles were poorly predicted by their average, global abundances (SI Appendix, Fig. S1B and Materials and Methods). To assess whether nonecological factors, such as sampling bias in initial species pools, could have contributed to this compositional divergence, we compared the variability in communities across endpoint particles to that across unincubated aliquots of the seawater used as the inoculum (SI Appendix, Extended Methods). Intersample variation was significantly higher across particles than across seawater samples (Mann–Whitney U test on Aitchison distances: P = 1.3 × 10−13; SI Appendix, Fig. S3), indicating that the observed variability stemmed more from the community assembly process than from differences across inocula. Because other technical sources of noise (Materials and Methods) also did not significantly impact the measured particle compositions (SI Appendix, Fig. S4) and all particles were chemically identical, we concluded that the variation in taxon relative abundances across particles was due to biological and ecological factors that amplified stochasticity in the initial assembly states of these communities.
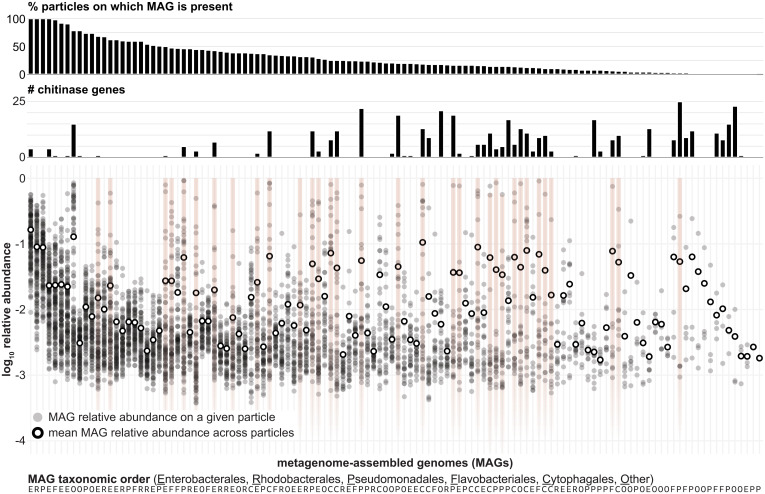
Fig. 2.
High compositional variability across replicate endpoint particles is driven by conditionally rare degrader taxa. Smaller black dots indicate the relative abundance of each MAG per endpoint particle (n = 149), with larger white dots indicating the log10[mean relative abundance] across the particles on which the MAG was found. Light red highlights indicate which MAGs display the “jackpot” phenomenon. MAGs are sorted from left to right by their prevalence across particles (i.e., the percent of particles on which they are detected; top bar graph), with the number of chitinases encoded in each MAG indicated in the bar graph beneath. The annotations at the bottom of the plot show each MAG’s taxonomic order (E, Enterobacterales; R, Rhodobacterales; P, Pseudomonadales; F, Flavobacteriales; C, Cytophagales; O, other). See SI Appendix, Fig. S1 for additional details.
Rare Chitin Degraders Dominate Individual Particles, Striking the “Jackpot.”
The skew toward high frequencies in the relative abundance distributions implied that taxa that were rare on average became dominant on a small number of particles (Fig. 2, red highlights). As a result, those particles harbored low-complexity communities (SI Appendix, Fig. S5 A and B) that diverged highly from the average particle taxonomic composition (SI Appendix, Fig. S5C). We termed the species that displayed this phenomenon “jackpot taxa” for their simultaneous local success and global rarity (Materials and Methods). The strains in this phylogenetically broad group of organisms included members of the Enterobacterales, Cytophagales, Pseudomonadales, Flavobacteriales, Rhodobacterales, Fibrobacterales, and Chitinophagales orders and were mostly (87.9%) classified as chitin degraders (SI Appendix, Fig. S1 and Dataset S1). Jackpot taxa were more prevalent across endpoint particles than other taxa that were equally rare in the initial seawater inocula (Mann–Whitney U test: P = 7.1 × 10−3; SI Appendix, Fig. S5D), indicating that the probability of their success on particles, while influenced by their scarcity in seawater, was also determined by ecological factors during community assembly. Notably, while taxon-specific interactions did not explain the abundance patterns observed across particles (SI Appendix, Fig. S6 and Materials and Methods), the most variable strains were likely to be degraders enriched in genes encoding chitinases (Fig. 2 and SI Appendix, Fig. S7; coefficient of variation vs. chitinase copy number, Spearman’s ρ = 0.44, P = 8.5 × 10−7). These observations indicated that the conditional success of specialized degraders from a diverse initial species pool contributed to the differentiation of the many rare community states found at the single-particle level.
Taxonomic Variability Translates to Divergent Community-Level Productivity.
We found that the communities that formed on particles in our individual microscale ecosystems were highly functionally divergent (Fig. 3A and SI Appendix, Fig. S8). By the end of the time course, most particles (63.8%)—and especially particles dominated by jackpot taxa—harbored majority-degrader communities (Fig. 3A), highlighting the importance of degraders for establishing and maintaining chitin-associated communities. However, the percentage of putative degraders on each endpoint particle was as low as 13.1% and as high as 97.3%, indicating that chitin-degrading communities did not self-assemble to “optimized” or conserved ratios of ecological roles after a fixed incubation period. Read mapping to chitinase protein sequences rather than MAGs supported our interpretation that variability in the estimated proportion of degraders was not due to the use of MAGs as reference genomes (SI Appendix, Fig. S9 and Materials and Methods). We hypothesized that this extensive variability in community composition, primed by stochasticity in assembly processes, could have had significant consequences for overall community function.
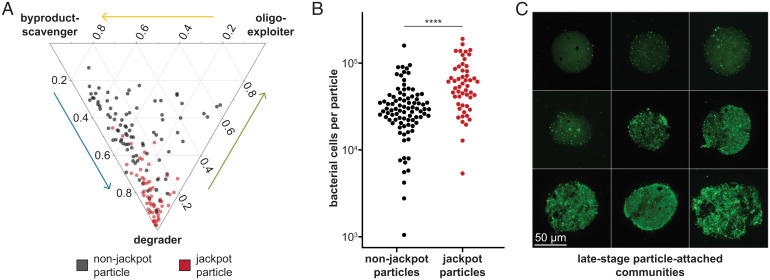
Fig. 3.
Endpoint particles diverge in community-level functional potential and biomass. (A) Ternary plot of the relative abundances of organisms occupying the three ecological roles (degrader, exploiter, scavenger) on each endpoint particle (n = 149), calculated by summing the relative abundances of MAGs classified into each role. Red dots represent jackpot particles, and black ones represent nonjackpot particles. Jackpot particles harbored significantly higher degrader populations than nonjackpot particles (79.8% vs. 47.4% on average; Mann–Whitney U test: P < 2.2 × 10−16). (B) Estimates of absolute bacterial cell counts on endpoint particles through qPCR of the 16S rRNA gene in DNA extracted from particle-attached communities. Jackpot particles (red dots) harbored significantly higher numbers of cells (Mann–Whitney U test: P = 2.3 × 10−7) than nonjackpot particles (black dots). (C) Representative images of endpoint particles that were harvested after 167 h of incubation in seawater and stained with the DNA-intercalating dye SYTO 9. (Scale bar, 50 µm.) Particle-attached communities spanned a range of growth states, from sparsely to densely populated.
Consistent with this hypothesis, individual particles sustained highly variable particle-attached biomass levels that were correlated with their community compositions. The number of bacterial cells in each endpoint community, estimated using qPCR of the 16S ribosomal RNA (rRNA) region (Materials and Methods), ranged from ∼1,000 to nearly 200,000 cells (Fig. 3B) and was strongly correlated with the overall frequency of degraders (Spearman’s ρ = 0.45, P = 1.6 × 10−8; SI Appendix, Fig. S10). Accordingly, particles that displayed the jackpot phenomenon had significantly higher cell counts (Fig. 3B; Mann–Whitney U test: P = 2.3 × 10−7), revealing that jackpot taxa were dominant in terms of not only relative abundances but also absolute abundances. The distribution of cells per particle was approximately lognormal, with a skew toward low cell numbers, indicating that some particles were highly productive while others harbored small populations even by the end of the incubation. These measurements were supported by direct observations of particle-attached cells using a DNA stain (Fig. 3C and Materials and Methods), which revealed a similar distribution in bacterial density per particle.
Importantly, the initial colonization of single particles incubated together in the same volume of seawater, rather than individually, resulted in particle-associated cell biomass that also spanned several orders of magnitude (SI Appendix, Fig. S11 and Materials and Methods). This variability in initial particle colonization was observed across a range of particle densities 15 to 140 times more concentrated than the conditions of the individual particle incubations, indicating that phenomena such as jackpot colonization are not specific to the environmental regime established in our separate microscale ecosystems. Collectively, these results suggested that variation in taxonomic composition and total biomass were correlated at the individual particle level, raising the question of which biological or ecological factors could explain this covariance.
Predation by Bacteriophages Contributes to Variability in Particle-Associated Community Composition and Biomass.
Our observation that most (63.7%) of our MAGs contained sequences homologous to those of bacteriophages led us to examine the potential importance of lysogeny in the development of particle-associated communities. High viral densities have been measured on marine particles relative to ambient seawater (15), but it is unknown to what extent this is the consequence of passive adsorption of free virions as opposed to active proliferation due to infection. Furthermore, the prevalence and roles of temperate phages (i.e., those that conditionally employ both lytic and lysogenic cycles) remain open questions in marine microbial ecology (22, 23). Therefore, we sought to identify populations of induced prophages within our single-particle communities to determine whether heterogeneous lysogeny–lysis switching could explain the observed variability in community composition and yield.
To detect actively replicating phages, we first classified contigs in our MAGs as phage-derived or bacteria-derived using tools (24, 25) that annotate phages from mixed metagenomes (Materials and Methods). We reasoned that contigs classified as phage-derived, especially those belonging to the genomes of temperate phages, were likely to be binned into the MAGs of their bacterial hosts. Phage k-mer signatures tend to be more similar to those of their specific hosts than to those of random bacteria (25, 26), and phages in a lysogenic cycle will have the same sequencing read coverage patterns as their hosts across samples. Therefore, phages that were lysogenic in most single-particle communities would tend to be binned with their hosts and have similar coverage levels, reflected in an inferred virus-to-microbial cell ratio (VMR) close to one (Fig. 4 A, Top Left). In contrast, phages in a productive cycle (lytic or chronic) would have higher coverage than their hosts because of the multiple virion copies produced per bacterial cell (27, 28) (Fig. 4 A, Top Right). Therefore, we considered a phage-derived contig to be productive in a sample if it was one of the most highly covered elements of its MAG (Materials and Methods). Because our approach relied on comparisons between co-binned phages and MAGs, phages that exclusively employ a lytic cycle were unlikely to be detected. The VMRs of three representative examples of lysogenic and productive phage contigs are shown for each particle in Fig. 4B (Dataset S2). Comparing the coverage patterns of phage- and bacteria-derived contigs provided evidence that variable phage coverage was not due to sequencing noise, lending confidence to our estimates of VMRs for specific phages (SI Appendix, Fig. S12 and Materials and Methods). Using the VMRs of individual productive phages, we calculated the total productive VMR per particle as a measure of overall phage replication in each community (Materials and Methods).
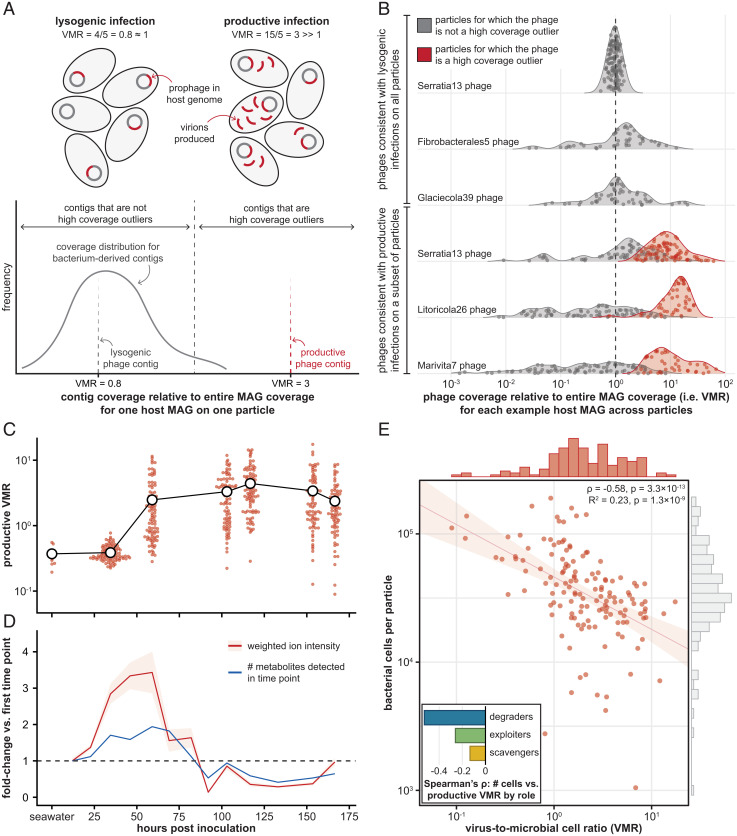
Fig. 4.
Bacteriophages become increasingly activated during community development and contribute to variability in bacterial abundances on endpoint particles. (A) Schematic of approach to detect productive phage infections from metagenomic data. (Left) During lysogenic infections, prophages replicate with their bacterial hosts (VMR ≈ 1; Top); lysogenic phage contigs have read coverage values similar to those of most bacterial contigs of their host MAG (Bottom). (Right) During productive infections, prophages replicate much more than their hosts (VMR ≫ 1; Top); productive phage contigs have read coverage values much higher than those of most bacterial contigs of their host MAG (Bottom). (B) Representative examples of phages with lysogenic coverage patterns on all endpoint particles (top three rows), and of phages with productive coverage patterns on a subset of particles (bottom three rows). For each phage contig, VMR is shown across endpoint particles on which each MAG is present; gray dots, particles on which the phage contig is not a coverage outlier; red dots, particles on which the phage is a high coverage outlier; dashed line, VMR = 1. (C) Total VMRs for productive phages over time. The first time point shows productive VMRs of initial seawater samples; subsequent time points show productive VMRs for chitin particle-attached communities incubated in seawater; smaller red dots, values for individual samples; larger white dots, mean VMR for each time point. (D) Metabolomic profiles of the seawater surrounding chitin particles as a function of incubation duration. Values are depicted in terms of fold-change at each time point relative to the first time point (dashed line, no change); red line (and shading), mean (±1 SD) weighted ion intensity (Materials and Methods); blue line, number of unique metabolites. (E) Absolute bacterial cell counts on endpoint particles (n = 142), estimated through qPCR, vs. each particle’s total VMR for productive phages. Cell counts were negatively correlated with productive VMRs (Spearman’s ρ = −0.56, P = 3.3 × 10−13; red line and shading, log–log linear regression and 95% CI; R2 = 0.23, P = 1.3 × 10−9). Marginal histograms are distributions of productive VMRs (red) and bacterial cell counts (light gray). (Inset) Bar plot of values of Spearman’s ρ between cell counts and productive VMRs of bacterial populations by ecological role (blue, degraders; green, exploiters; yellow, scavengers; see SI Appendix, Fig. S14B for details).
We analyzed earlier time points in our experiment to assess the dynamics of replicating phages. The mean productive VMR was lowest for the initial seawater inocula and rose sharply until the middle of the incubation period (59 h), suggesting that phages became induced as their particle-associated hosts began to grow (Fig. 4C). Concomitant with this increase in productive VMRs, we observed the accumulation of diverse metabolites in the seawater surrounding each particle until 59 h of incubation, followed by a decrease in metabolite concentrations (Fig. 4D, SI Appendix, Fig. S13, Dataset S3, and Materials and Methods). These observations could be explained by metabolite release upon the initiation of bacterial growth (29) or lysis by phages (30) and by subsequent metabolite consumption by the remaining viable bacteria (31). The coinciding timescales of metabolite liberation and rising VMRs are consistent with our hypothesis that a particle-associated lifestyle among bacteria promotes phage proliferation; therefore, we sought to assess the impact of variable phage induction on each community’s composition and consequent yield.
There was a striking negative relationship between cell counts and productive VMRs on endpoint particles (Fig. 4E, red data: Spearman’s ρ = −0.56, P = 3.3 × 10−13), suggesting that phage predation impacted bacterial growth success on particles upon induction. The degrader populations contributed the most to this signal, indicating that strains among this trophic level may have been especially prone to phage activation (Fig. 4 E, Inset, SI Appendix, Fig. S14, and Dataset S4). Importantly, jackpot degrader taxa had lower productive VMRs than nonjackpot degraders (SI Appendix, Fig. S15A; Mann–Whitney U test: P = 1.3 × 10−49). This translated to jackpot particles having significantly lower productive VMRs than nonjackpot particles (Mann–Whitney U test: P = 4.3 × 10−8), even controlling for differences in cell counts between these groups of communities (analysis of covariance: F(1,139) = 16.92, P = 4.1 × 10−4, partial η2 = 0.09; SI Appendix, Fig. S15B). Collectively, these data point to increased activation of prophages in degrader taxa that exist as part of taxonomically diverse communities, more so than activation in degrader taxa that dominate particles.
A Stochastic Model Recapitulates the Observed Variability in Taxonomic Compositions and Biomass on Particles.
While cell counts were significantly correlated with both phage abundances and community compositions, these features explained, respectively, 23% (Fig. 4E) and 34% (SI Appendix, Fig. S10) of the observed variation in yields, indicating that other factors also contributed to variable growth returns. Therefore, we sought a more general framework in which to understand the key quantitative features of the data—namely, the lognormal-like distributions of relative taxon abundances (with right skews consistent with jackpot taxa) and of absolute cell abundances (with a left skew corresponding to low-biomass communities). We considered four mathematical models of increasing complexity describing bacterial population dynamics on individual particles, all of which assume that cells arrive on particles stochastically and then grow exponentially with different mean growth rates across taxa (SI Appendix, SI Text). Models in which individual taxa have identical growth rates (SI Appendix, Fig. S16) or randomly fluctuating growth rates on different particles (SI Appendix, Figs. S17 and S18) do not recapitulate the observed trends in taxon abundances. Importantly, the key features in our data could be reproduced as a result of the interplay between multiple stochastic processes (SI Appendix, Fig. S19): 1) noisy growth rates and 2) the requirement that a degrader must first colonize a particle by stochastic arrival before a community can develop. Taken together with our experimental data, this model indicates that the biological processes which contribute to the stochasticity of particle colonization and growth rates—and especially those processes that affect degraders—will result in variable growth returns for strains across particles.
Discussion
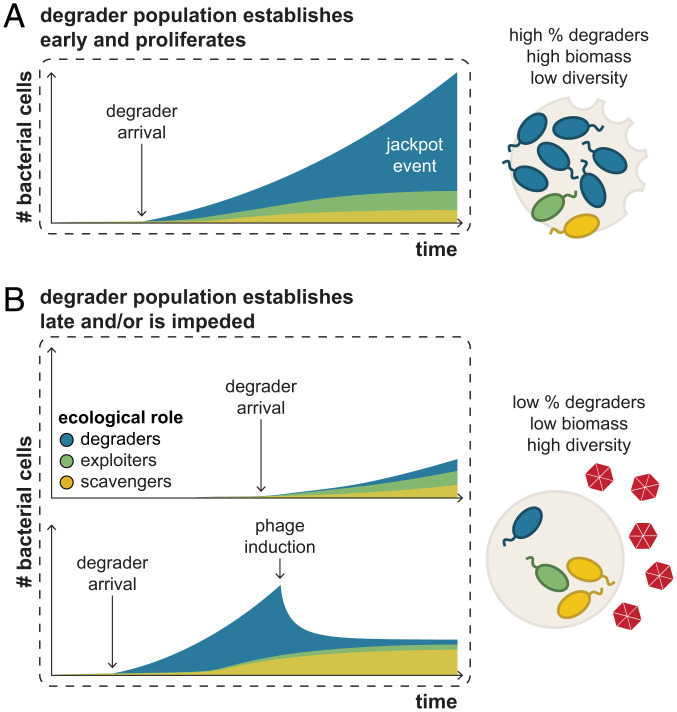
Together, a synthesis of our mathematical model with our observations of bacterial and phage abundances across individual chitin particles suggests a conceptual framework for key processes promoting variability in microscale community composition and function (Fig. 5). Consistent with prior work demonstrating small-scale heterogeneity on aquatic resource particles (32), we found that key community features—namely, taxonomic compositions, functional gene content, and absolute abundances—were not conserved at the single particle scale. We posit that 1) stochastic arrival on particles diversifies initial assembly states; 2) phage induction and subsequent host lysis, primarily among degraders, contribute to variable growth returns; and 3) the timescale and magnitude of degrader colonization determine the extent to which scavengers and exploiters are supported. Therefore, in this conceptual framework, the high-biomass jackpot particles are those in which degraders arrive early and resist phage induction, leading to high relative and absolute degrader abundances (Fig. 5A). By contrast, low-biomass particles are those in which degraders are not able to proliferate, either because they become established on a particle relatively late due to stochastic arrival (Fig. 5 B, Top) or because phage induction impedes the establishment of a robust population (Fig. 5 B, Bottom).
Fig. 5.
Conceptual model of key processes contributing to the diversification of communities on microscale particles. Schematics of community development over time and the resultant features measured on particles are shown for different scenarios highlighting how historical contingencies and phage induction, particularly affecting keystone degraders, can contribute to variability in microscale community assembly. Bacterial populations are indicated by their ecological role (blue, degraders; green, exploiters; yellow, scavengers), and phages are depicted in red. (A) Jackpot particles are those on which degraders arrive early and proliferate, leading to communities with high relative and absolute degrader abundances and low species diversity. (B) Low-biomass particles are those on which degraders are not able to proliferate, either because they become established on a particle relatively late due to stochastic arrival (Top) or because phage induction impedes the establishment of a robust population (Bottom).
Our results highlight that the first step in community assembly at the microscale—the arrival of cells to a particle—is an intrinsically random process dependent on encounter probabilities. Chitin degraders are taxonomically diverse and exist at low abundance in the bacterioplankton, two factors that favor stochasticity and strong priority effects (33) at the single-particle level. Our model demonstrates that historical contingencies, in our case created by stochastic arrival times and the growth dependency of non-degraders on degraders, can be magnified through variable growth rates to reproduce the wide distributions of bacterial abundances experimentally observed across particles. Because of this high compositional variability, we hypothesize that interspecific interactions on individual particles are based on organisms’ functional roles within a community as degraders, exploiters, or scavengers, rather than on their specific taxonomic identities (34).
Our study suggests the interesting possibility that the differential induction of prophages may be one factor contributing to the variable growth rate of degraders on particles. We hypothesize that the relatively low levels of biomass observed on particles with elevated VMRs at the end of our experiment are partially the consequence of phage-mediated lysis and community impairment. Furthermore, our observation that phages became increasingly activated as communities developed may indicate that particles are hotspots for lysogeny–lysis transitions in temperate phages. We speculate that heterotrophs navigating a patchy nutrient landscape may be especially prone to prophage induction in particle-associated contexts, where a combination of host metabolic activity, abundant cellular resources, and proximity to potentially susceptible cells and other bacterial competitors may favor phage proliferation (35–39). However, the mechanisms underlying the regulation of lysogeny–lysis switches are complex, diverse, and largely unexplored in the contexts of complex communities. Open questions remain around which environmental and ecological conditions promote prophage induction (23), with host density and activity either promoting (40) or inhibiting (41, 42) prophage induction in different biological systems. Future studies leveraging model marine particles present an opportunity to learn more about the ecological triggers of phage activation.
Our results reveal an interesting connection between the diversity of a community and the magnitude of phage predation of chitin degraders, precisely because this functional class of organisms initiates the trophic cascade that governs community assembly around chitin particles (11, 43). Because some degraders display density-dependent growth on chitin (44), the collapse of a degrader population may be influenced by phage predation yet not contingent on induction in all degrader cells. Instead, even partial induction of a lysogenic degrader population could decrease cell densities below a critical threshold required to maintain a robust particle-associated community, amplifying the apparent effects of phage induction on degrader colonization. Phage induction of degraders may not only impair degrader proliferation but also may liberate internal metabolites for utilization by scavenging strains, leading to the observed association between lower biomass particles and more-diverse communities with higher proportions of non-degraders. In contrast, because phage induction was significantly less associated with degraders that displayed the jackpot phenomenon, we hypothesize that jackpot degraders are locally successful on a minority of particles in part because they experience less predation, resulting in communities characterized by high biomass and low species diversity. Alternatively, jackpot colonization may represent an earlier community state than particles that support more-complex communities. Indeed, an exciting future avenue of investigation will be to tease apart the link between degrader growth dynamics, prophage induction, and community assembly, using particle-derived isolates representative of the different ecological “strategies” present on chitin particles.
While we did capture a fraction of the natural viral and bacterial diversity involved in POM degradation, several experimental simplifications may have impacted the assembly outcomes we observed. For example, our initial filtration and concentration of the seawater, intended to decrease the variability across inocula, may have resulted in the partial removal of aggregate-attached protozoa (16). Protozoan predation of bacteria has significant impacts on the structure of particle-attached communities, both through selective killing of certain taxa and through the facilitation of others through the release of nutrients (45); therefore, these interactions were likely perturbed in our experimental system and were not assessed.
Our observations of wild marine communities, although made in a laboratory setting, may provide insights on the ecosystem-level consequences of microscale stochastic assembly dynamics. First, the stochasticity in bacterial growth, amplified across a patchy landscape of microscale marine particles, may promote the maintenance of a diverse regional species pool at the macroscale. This is because the variability in growth yield can effectively offset differences in competitive fitness between degraders (46). Second, the properties of a system of diverse particle-attached communities—for example, its resistance to perturbations (47)—differ from those of a system in which all individual community compositions are identical to a bulk measurement, underscoring the significance of microscale processes. Third, the variability in microscale community states could be reflected in larger-scale biogeochemical patterns since the cumulative process of POM degradation can be approximated as the sum of degradation events on individual particles. Although degraders were progressively enriched on particles, we found that endpoint communities did not converge to a fixed proportion of chitin degraders or to a fixed amount of biomass per particle within the timescale of our experiment. Both of these measures are positively correlated with the rate of particle degradation (12), suggesting that historical contingencies in community assembly promote functional divergence (33, 48). These results contrast with those of previous studies on the replicability of microbial community assembly at the functional level (49, 50), likely because of the homogenizing effect of macroscale sampling. Nevertheless, our study suggests that the diversity measured in microscale community states at any given time may be represented among a system of particles. Indeed, the lognormal-like distribution of biomass on individual particles intriguingly aligns with observations and predictions of lognormally-distributed global marine organic matter export and remineralization rates; these distributions may repeatedly emerge as a reflection of the multiplicative effects of stochastic variables in ecological settings (51–53). Although our experimental system significantly simplified the process of POM degradation in the ocean, our approach provides a quantitative link between the microscale and larger-scale processes, highlighting the importance of considering local variability when investigating the mechanisms behind microbial community development in a spatially structured environment.
Materials and Methods
Abridged methods are provided below; details and additional information are provided in SI Appendix, Extended Methods.
Seawater Collection and Individual Chitin Particle Incubation.
Nearshore coastal seawater was collected from Nahant, MA; filtered (63 µm) to remove large particulate matter; gently concentrated via centrifugation at 4,000 × g for 5 min; and aliquoted for incubations and sequencing. Hydrogel chitin particles (New England Biolabs, #E8036L) were washed in sterile artificial seawater (Sigma-Aldrich, #S9883) and individually selected beneath a dissecting microscope in a laminar flow hood. Single chitin particles (85.0 ± 24.0 µm in diameter) were transferred to sterile 96-well plates (Thermo Fisher, #AB0600L), with one chitin particle per well. Plates were inoculated consecutively with 175 µL of filtered, centrifuged seawater per well; sealed (VWR, #89092-056); and rotated end over end (7.5 rpm) at room temperature. The particles in an entire plate were harvested at each time point (after 12, 22.75, 34.5, 46, 59, 69, 82, 92, 103, 116.75, 113, 153.5, and 166.5 h of incubation) by inspection and pipetting under a dissecting microscope in a laminar flow hood. Each particle was transferred into sterile 96-well plates (Thermo Fisher, #AB0600L) containing Tris-EDTA buffer and stored at −20 °C. The seawater surrounding each harvested particle was also saved in 96-well plates and stored at −20 °C.
Mock Communities and Negative Controls.
To quantify the technical error associated with creating metagenomic libraries from low DNA inputs, mock communities were simulated by combining the DNA of two strains previously isolated from a chitin particle enrichment (11). Libraries from three technical replicates of mock communities totaling 50 pg or 5 pg of DNA (SI Appendix, Extended Methods), as well as from six negative controls (containing only nuclease-free water), were prepared and analyzed with the same protocols used for individual chitin particle-attached communities.
DNA Extraction and Metagenomic Sequencing.
DNA extractions were performed for twelve 175-µL-volume aliquots of the initial, unincubated seawater and for particles harvested after 34.5, 59, 103, 116.75, 113, 153.5, and 166.5 h of incubation (see Dataset S5 for sample metadata; in total, metagenomes from 495 particle-attached communities were analyzed in this study). DNA was extracted from all samples with the Agencourt DNAdvance Genomic DNA Isolation Kit (Beckman Coulter; modifications noted in SI Appendix, Extended Methods). Metagenomic libraries were prepared with the Nextera XT DNA Library Prep Kit and index primers (Illumina) using the protocol developed by Rinke et al. (18) for low DNA inputs (SI Appendix, Extended Methods). Libraries were quantified on an Agilent 4200 TapeStation system with High Sensitivity D5000 ScreenTapes (Agilent Technologies) and pooled by time point in equimolar amounts. Sequencing was performed on an Illumina HiSeq 2500 machine (250-bp paired-end reads) at the Whitehead Institute for Biomedical Research.
MAG Generation, Taxonomic Assignment, and Role Classification.
Raw sequencing reads were quality trimmed with Trimmomatic v0.36 (54). Reads mapping to the PhiX and human genomes were filtered out using BBDuk v38.16 (55) and BBMap v38.16, respectively (SI Appendix, Extended Methods). Trimmed, filtered reads that were error corrected using BayesHammer (56) were pooled within each time point and coassembled using MEGAHIT v1.2.9 (57). Bins were generated with MaxBin v2.2.7 (58) and CONCOCT v1.1.0 (59), consolidated and filtered using DAS Tool v1.1.1 (60), and evaluated for completeness and contamination using CheckM v1.1.2 (61). The resulting 251 bins were used as reference metagenome-assembled genomes, i.e. MAGs (≥50% complete, ≤10% contaminated; median completeness 93.7%, median contamination 3.9%; Dataset S1). Highly similar MAGs obtained from separate coassemblies were grouped into 132 clusters (SI Appendix, Extended Methods). MAG taxonomic classifications were made using GTDB-Tk v1.1.1 (62). MAGs were functionally annotated using a custom database of profile hidden Markov models of proteins involved in growth on chitin (SI Appendix, Extended Methods and Dataset S6). Ecological roles for MAGs (as degraders, chitooligosaccharide exploiters, or metabolic byproduct scavengers) were defined based on the gene content patterns observed for sequenced and phenotyped (14) strains previously isolated (11, 13) from particle enrichments (SI Appendix, Extended Methods).
Read Mapping to MAGs for Relative Abundance Estimation.
Trimmed, filtered reads were mapped competitively against the MAGs generated from sequencing particle-attached communities, initial seawater samples, and negative controls. Read mapping was performed using the approach described in Leventhal et al. (63) (SI Appendix, Extended Methods). Reads that best mapped to predicted contaminant MAGs (SI Appendix, Extended Methods) were removed from consideration. MAG relative abundances were calculated for each sample by 1) tallying the hits to all MAGs in each MAG cluster, 2) normalizing the tally by the average genome length of all MAGs in each MAG cluster, and 3) dividing the normalized tallies for each MAG cluster by their sum for each sample. Therefore, for MAGs clustered together based on similarity, their relative abundances are represented in that of the entire MAG cluster to which they belong; this calculation circumvents the artificial underestimation of MAG relative abundances that would otherwise be obtained with a nondereplicated reference set. The relative abundances of organisms occupying the three ecological roles (degrader, exploiter, scavenger) on each particle were calculated by summing the relative abundances of MAGs classified into each role.
Definitions of Jackpot MAGs and Jackpot Particles.
A jackpot score was calculated for each MAG cluster to quantitatively reflect the properties of rarity across most particles and dominance on a few particles (SI Appendix, Extended Methods) such that MAGs with high scores strongly displayed the jackpot phenomenon. Each particle’s jackpot score was calculated as the weighted average of MAG jackpot scores (i.e., the sum of the relative abundance of each MAG cluster multiplied by its jackpot score). Particles with high jackpot scores and low Pielou’s evenness were categorized as “jackpot particles” (SI Appendix, Extended Methods).
Bacteriophage Analyses.
Binned contigs were classified as phage-derived or bacteria-derived using VirSorter v1.0.3 with its RefSeqABVir database (24) and VirFinder v1.1 (25), two tools designed to detect phage sequences among mixed metagenomes (SI Appendix, Extended Methods). We used a read coverage-based approach to categorize phage-derived contigs as productive or lysogenic in particle-attached communities (Dataset S2; see SI Appendix, Extended Methods for analysis controls). Based on read mapping to MAGs, per-base coverage values for all binned contigs were computed with BEDTools v2.27.0 (64) and were used to calculate contig-wide average coverage values. For each MAG and for each sample, a phage-derived contig was considered to be productive if its coverage was greater than the coverage of the 95th percentile bacteria-derived contig in the same MAG; otherwise, it was considered to be lysogenic in that sample. The virus-to-microbial cell ratio (VMR) of an individual phage contig in one sample is defined as the phage contig coverage divided by average coverage of the MAG with which it is binned (which was calculated using only the bacteria-derived contigs). Total VMRs—that is, the total number of phage copies relative to the total number of bacterial MAG copies in an entire sample—were calculated separately for productive and lysogenic phage contigs. The total productive VMR for a sample was defined as
where n is the number of MAGs found in a sample. This calculation is equivalent to
where n is the number of MAGs found in a sample. Total lysogenic VMRs were calculated using the same formula while considering only lysogenic-annotated contigs. VMRs for each ecological role (i.e., for the subpopulation in a community that belongs to one of the three roles of degrader, exploiter, or scavenger) were calculated using the same formula considering only the MAGs of each role and their associated phages.
Cell Count Estimation.
Bacterial DNA extracted from individual particle-attached communities was quantified through qPCR of the 16S rRNA gene using the Femto Bacterial DNA Quantification Kit (Zymo Research), which has a lower limit of detection of 20 fg. Two sets of standards and negative controls were included in each qPCR run. The number of bacterial cells for each particle was estimated from the absolute DNA amounts based on measurements indicating a mean of 2.5 fg DNA per bacterial cell in seawater samples (65).
Imaging of Chitin Particle Colonization.
Subsets of chitin particles incubated individually in seawater were stained at time points by adding the DNA stain SYTO9 (Invitrogen, #S34854) at a final concentration of 500 nM directly to the particle incubations. Particles were incubated in the dark at room temperature for 15 min before being mounted separately on microscope slides and imaged with a Zeiss epifluorescence microscope at 100× magnification.
Metabolomics.
We performed untargeted metabolomics of the seawater that surrounded 1,222 harvested chitin particles and of the initial, unincubated seawater (SI Appendix, Extended Methods). We used a binary LC pump (Agilent Technologies) and an MPS2 Autosampler (Gerstel) coupled to an Agilent 6520 time-of-flight mass spectrometer (Agilent Technologies) operated in negative mode, at 2 GHz, extended dynamic range, with an m/z (mass/charge) range of 50 to 1,000. Ions (Dataset S3) were annotated against a curated library of metabolites present in marine microbes, based on the BioCyc database (66). For metabolites that exceeded the limit of detection (SI Appendix, Extended Methods), the intensities of each ion were normalized between zero (the limit of detection) and one (the highest measured intensity of a given ion). Weighted ion intensities for each time point were calculated by taking the sum of all normalized intensities of ions in all samples for each time point.
Supplementary Material
Acknowledgments
We extend our gratitude to all past and present members of the O.X.C. lab for their support and critical feedback, as well as members of the Simons Collaboration on Principles of Microbial Ecosystems, for stimulating discussions. In particular, we would like to thank Manoshi S. Datta, for contributing to the genesis of this project; José T. Saavedra, for developing the DNA extraction and metagenomic library preparation pipeline for single particle-attached communities; Gabriel E. Leventhal and Jakob Russel, for bioinformatic mentorship and assistance; Gonzalo Gonzalez Del Pino and Matti Gralka, for feedback on analyses and this manuscript; Elise Ledieu-Dherbécourt, for quantifying chitin particle sizes; Anthony Gaca, for advice on metagenomic library preparation; Fatima Aysha Hussain, for feedback on the bacteriophage analysis; Emily Zakem, for insights on global particle remineralization rates; Akshit Goyal, for comments on this manuscript; and Sara Szabo and William Mandella, for assistance with seawater sampling. This material is based upon work supported by the NSF Graduate Research Fellowship under Grant 174530. This project was supported by the Simons Collaboration: Principles of Microbial Ecosystems (PriME) Award 542395. S. Pontrelli was supported by a grant from the Simons Foundation (Grant ID608247) as part of PriME. S. Pollak was supported by the European Molecular Biology Organization Postdoctoral Fellowship (ALTF Grant 800-2017).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117748119/-/DCSupplemental.
Data Availability
Supplementary Datasets include: (i) metadata for MAGs (Dataset S1); (ii) metadata for phage-annotated sequences (Dataset S2); (iii) metadata for detected metabolites (Dataset S3); (iv) summary statistics for phage-annotated sequences (Dataset S4); (v) metadata for metagenomic samples (Dataset S5); (vi) information on hidden Markov models used in this study (Dataset S6); and (vii) MAG relative abundances in each metagenome (Dataset S7). Metagenomic reads from individual particle-attached communities and from initial seawater samples are accessible through NCBI BioProject (accession number PRJNA847179) (67) and BioSample (accession numbers SAMN29046748 through SAMN29047344) (68, 69). Data available through the repository at doi: 10.17632/7t5mscdtwb.1 (70) include: (i) MAGs generated and used in this study; (ii) images of microbial communities attached to individually-incubated chitin particles; (iii) images of microbial communities attached to bulk-incubated chitin particles; and (iv) code for the mathematical models described in the text. Code for analyzing particle image data is accessible at https://github.com/jaschwartzman/seawater_colonize (71). All mass spectra files from the metabolomics are accessible through MassIVE (ftp://MSV000087936@massive.ucsd.edu). All custom scripts for processing the data listed above are available from the authors upon request.
References
- 1.Cordero O. X., Datta M. S., Microbial interactions and community assembly at microscales. Curr. Opin. Microbiol. 31, 227–234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nemergut D. R., et al. , Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Donnell A. G., Young I. M., Rushton S. P., Shirley M. D., Crawford J. W., Visualization, modelling and prediction in soil microbiology. Nat. Rev. Microbiol. 5, 689–699 (2007). [DOI] [PubMed] [Google Scholar]
- 4.McGlynn S. E., Chadwick G. L., Kempes C. P., Orphan V. J., Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Gil G., Holliger C., Aerobic granules: Microbial landscape and architecture, stages, and practical implications. Appl. Environ. Microbiol. 80, 3433–3441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mark Welch J. L., Rossetti B. J., Rieken C. W., Dewhirst F. E., Borisy G. G., Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U.S.A. 113, E791–E800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stocker R., Marine microbes see a sea of gradients. Science 338, 628–633 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Dann L. M., et al. , Microbial micropatches within microbial hotspots. PLoS One 13, e0197224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang H., Lovell C. R., Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiao N., et al. , Microbial production of recalcitrant dissolved organic matter: Long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 8, 593–599 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Datta M. S., Sliwerska E., Gore J., Polz M. F., Cordero O. X., Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat. Commun. 7, 11965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enke T. N., Leventhal G. E., Metzger M., Saavedra J. T., Cordero O. X., Microscale ecology regulates particulate organic matter turnover in model marine microbial communities. Nat. Commun. 9, 2743 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enke T. N., et al. , Modular assembly of polysaccharide-degrading marine microbial communities. Curr. Biol. 29, 1528–1535.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Pontrelli S., et al. , Metabolic crossfeeding structures the assembly of polysaccharide degrading communities. Sci. Adv. 8, eabk3076 (2022). [DOI] [PMC free article] [PubMed]
- 15.Weinbauer M. G., et al. , Viral ecology of organic and inorganic particles in aquatic systems: Avenues for further research. Aquat. Microb. Ecol. 57, 321–341 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon M., Grossart H. P., Schweitzer B., Ploug H., Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 28, 175–211 (2002). [Google Scholar]
- 17.Armitage D. W., Jones S. E., How sample heterogeneity can obscure the signal of microbial interactions. ISME J. 13, 2639–2646 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rinke C., et al. , Validation of picogram- and femtogram-input DNA libraries for microscale metagenomics. PeerJ 4, e2486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheth R. U., et al. , Spatial metagenomic characterization of microbial biogeography in the gut. Nat. Biotechnol. 37, 877–883 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iversen M. H., Ploug H., Temperature effects on carbon-specific respiration rate and sinking velocity of diatom aggregates – Potential implications for deep ocean export processes. Biogeosciences 10, 4073–4085 (2013). [Google Scholar]
- 21.Pollak S., et al. , Public good exploitation in natural bacterioplankton communities. Sci. Adv. 7, eabi4717 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breitbart M., Bonnain C., Malki K., Sawaya N. A., Phage puppet masters of the marine microbial realm. Nat. Microbiol. 3, 754–766 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Tuttle M. J., Buchan A., Lysogeny in the oceans: Lessons from cultivated model systems and a reanalysis of its prevalence. Environ. Microbiol. 22, 4919–4933 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Roux S., Enault F., Hurwitz B. L., Sullivan M. B., VirSorter: Mining viral signal from microbial genomic data. PeerJ 3, e985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren J., Ahlgren N. A., Lu Y. Y., Fuhrman J. A., Sun F., VirFinder: A novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome 5, 69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahlgren N. A., Ren J., Lu Y. Y., Fuhrman J. A., Sun F., Alignment-free d2* oligonucleotide frequency dissimilarity measure improves prediction of hosts from metagenomically-derived viral sequences. Nucleic Acids Res. 45, 39–53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kieft K., Anantharaman K., Deciphering active prophages from metagenomes. mSystems 7, e00084-22 (2022). [DOI] [PMC free article] [PubMed]
- 28.von Boeselager R. F., Pfeifer E., Frunzke J., Cytometry meets next-generation sequencing – RNA-Seq of sorted subpopulations reveals regional replication and iron-triggered prophage induction in Corynebacterium glutamicum. Sci. Rep. 8, 14856 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noriega-Ortega B. E., et al. , Does the chemodiversity of bacterial exometabolomes sustain the chemodiversity of marine dissolved organic matter? Front. Microbiol. 10, 215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ankrah N. Y. D., et al. , Phage infection of an environmentally relevant marine bacterium alters host metabolism and lysate composition. ISME J. 8, 1089–1100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blasche S., et al. , Metabolic cooperation and spatiotemporal niche partitioning in a kefir microbial community. Nat. Microbiol. 6, 196–208 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bižić-Ionescu M., Ionescu D., Grossart H.-P., Organic particles: Heterogeneous hubs for microbial interactions in aquatic ecosystems. Front. Microbiol. 9, 2569 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukami T., Historical contingency in community assembly: Integrating niches, species pools, and priority effects. Annu. Rev. Ecol. Evol. Syst. 46, 1–23 (2015). [Google Scholar]
- 34.Louca S., et al. , Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2, 936–943 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Brum J. R., Hurwitz B. L., Schofield O., Ducklow H. W., Sullivan M. B., Seasonal time bombs: Dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J. 10, 437–449 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Touchon M., Bernheim A., Rocha E. P. C., Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 10, 2744–2754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silpe J. E., Wong J. W. H., Owen S. V., Baym M., Balskus E. P., The bacterial toxin colibactin triggers prophage induction. Nature 603, 315–320 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laganenka L., et al. , Quorum sensing and metabolic state of the host control lysogeny-lysis switch of bacteriophage T1. MBio 10, 3–8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jancheva M., Böttcher T., A metabolite of Pseudomonas triggers prophage-selective lysogenic to lytic conversion in Staphylococcus aureus. J. Am. Chem. Soc. 143, 8344–8351 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silpe J. E., Bassler B. L., A host-produced quorum-sensing autoinducer controls a phage lysis-lysogeny decision. Cell 176, 268–280.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knowles B., et al. , Lytic to temperate switching of viral communities. Nature 531, 466–470 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Tan D., et al. , High cell densities favor lysogeny: Induction of an H20 prophage is repressed by quorum sensing and enhances biofilm formation in Vibrio anguillarum. ISME J. 14, 1731–1742 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pontrelli S., et al. , Metabolic cross-feeding structures the assembly of polysaccharide degrading communities. Sci. Adv. 8, eabk3076 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ebrahimi A., Schwartzman J., Cordero O. X., Cooperation and spatial self-organization determine rate and efficiency of particulate organic matter degradation in marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 116, 23309–23316 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corno G., Salka I., Pohlmann K., Hall A. R., Grossart H. P., Interspecific interactions drive chitin and cellulose degradation by aquatic microorganisms. Aquat. Microb. Ecol. 76, 27–37 (2015). [Google Scholar]
- 46.Melbinger A., Vergassola M., The impact of environmental fluctuations on evolutionary fitness functions. Sci. Rep. 5, 15211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shade A., et al. , Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3, 417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bittleston L. S., Gralka M., Leventhal G. E., Mizrahi I., Cordero O. X., Context-dependent dynamics lead to the assembly of functionally distinct microbial communities. Nat. Commun. 11, 1440 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Louca S., et al. , High taxonomic variability despite stable functional structure across microbial communities. Nat. Ecol. Evol. 1, 0015 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Human Microbiome Project Consortium, Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cael B. B., Bisson K., Follett C. L., Can rates of ocean primary production and biological carbon export be related through their probability distributions? Global Biogeochem. Cycles 32, 954–970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zakem E. J., Cael B. B., Levine N. M., A unified theory for organic matter accumulation. Proc. Natl. Acad. Sci. U.S.A. 118, e2016896118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Limpert E., Stahel W. A., Abbt M., Log-normal distributions across the sciences: Keys and clues. Bioscience 51, 341–352 (2001). [Google Scholar]
- 54.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bushnell B., “BBMap: A fast, accurate, splice-aware aligner” (LBNL Rep. LBNL-7065E, Lawrence Berkeley National Laboratory, 2014).
- 56.Nikolenko S. I., Korobeynikov A. I., Alekseyev M. A., BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genomics 14, S7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li D., et al. , MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102, 3–11 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Wu Y. W., Simmons B. A., Singer S. W., MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Alneberg J., et al. , Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Sieber C. M. K., et al. , Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W., CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chaumeil P. A., Mussig A. J., Hugenholtz P., Parks D. H., GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leventhal G. E., et al. , Strain-level diversity drives alternative community types in millimetre-scale granular biofilms. Nat. Microbiol. 3, 1295–1303 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Button D. K., Robertson B. R., Determination of DNA content of aquatic bacteria by flow cytometry. Appl. Environ. Microbiol. 67, 1636–1645 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karp P. D., et al. , The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 20, 1085–1093 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.R. E. Szabo et al., Metagenomes of (i) microbial communities enriched on individual synthetic hydrogel chitin micro-particles from coastal seawater collected near Nahant, MA, USA; and (ii) samples of coastal seawater collected near Nahant, MA, USA. NCBI BioProject. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA847179. Deposited 8 June 2022. [Google Scholar]
- 68.R. E. Szabo et al., Metagenome or environmental sample from marine metagenome. NCBI BioSample. https://www.ncbi.nlm.nih.gov/biosample/SAMN29046748. Deposited 14 June 2022. [Google Scholar]
- 69.R. E. Szabo et al., Metagenome or environmental sample from marine metagenome. NCBI BioSample. https://www.ncbi.nlm.nih.gov/biosample/SAMN29047344. Deposited 14 June 2022. [Google Scholar]
- 70.R. E. Szabo et al., Historical contingencies and phage induction diversify bacterioplankton communities at the microscale: Datasets. Mendeley Data. https://data.mendeley.com/datasets/7t5mscdtwb/1. Deposited 4 July 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.J. A. Schwartzman, seawater_colonize. GitHub. https://github.com/jaschwartzman/seawater_colonize. Deposited 4 January 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary Datasets include: (i) metadata for MAGs (Dataset S1); (ii) metadata for phage-annotated sequences (Dataset S2); (iii) metadata for detected metabolites (Dataset S3); (iv) summary statistics for phage-annotated sequences (Dataset S4); (v) metadata for metagenomic samples (Dataset S5); (vi) information on hidden Markov models used in this study (Dataset S6); and (vii) MAG relative abundances in each metagenome (Dataset S7). Metagenomic reads from individual particle-attached communities and from initial seawater samples are accessible through NCBI BioProject (accession number PRJNA847179) (67) and BioSample (accession numbers SAMN29046748 through SAMN29047344) (68, 69). Data available through the repository at doi: 10.17632/7t5mscdtwb.1 (70) include: (i) MAGs generated and used in this study; (ii) images of microbial communities attached to individually-incubated chitin particles; (iii) images of microbial communities attached to bulk-incubated chitin particles; and (iv) code for the mathematical models described in the text. Code for analyzing particle image data is accessible at https://github.com/jaschwartzman/seawater_colonize (71). All mass spectra files from the metabolomics are accessible through MassIVE (ftp://MSV000087936@massive.ucsd.edu). All custom scripts for processing the data listed above are available from the authors upon request.