Significance
Genome engineering in Bifidobacterium remains challenging, despite successful CRISPR deployment in other bacteria, hindering the comprehensive understanding of the molecular mechanisms of health-promoting effects. In this study, we adapted existing CRISPR effectors and editing strategies to generate a variety of editing outcomes in B. animalis subsp. lactis. We also showed that the editing efficiency and genome accessibility are shaped by the genomic and epigenetic landscapes, despite their monomorphic genomes. Together, these findings emphasize that strain-to-strain variation can impact the deployment of genome editing and lead to phenotypic and functional differences. This study expands the genome editing platform of Bifidobacterium to enhance their probiotic efficacy and opens opportunities for the engineering of bacterial biotherapeutics.
Keywords: CRISPR-Cas, bifidobacterium, genomics, epigenomics, probiotics
Abstract
Bifidobacterium is a commensal bacterial genus ubiquitous in the human gastrointestinal tract, which is associated with a range of health benefits. The advent of CRISPR-based genome editing technologies provides opportunities to investigate the genetics of important bacteria and transcend the lack of genetic tools in bifidobacteria to study the basis for their health-promoting attributes. Here, we repurpose the endogenous type I-G CRISPR-Cas system and adopt an exogenous CRISPR base editor for genome engineering in B. animalis subsp. lactis, demonstrating that both genomic and epigenetic contexts drive editing outcomes across strains. We reprogrammed the endogenous type I-G system to screen for naturally occurring large deletions up to 27 kb and to generate a 500-bp deletion in tetW to abolish tetracycline resistance. A CRISPR-cytosine base editor was optimized to install C•G-to-T•A amber mutations to resensitize multiple B. lactis strains to tetracycline. Remarkably, we uncovered epigenetic patterns that are distributed unevenly among B. lactis strains, despite their genomic homogeneity, that may contribute to editing efficiency variability. Insights were also expanded to Bifidobacterium longum subsp. infantis to emphasize the broad relevance of these findings. This study highlights the need to develop individualized CRISPR-based genome engineering approaches for distinct bacterial strains and opens avenues for engineering of next generation probiotics.
As an important bacterial genus, Bifidobacterium typically colonizes the human gastrointestinal tract early in life and remains a key component of the gut microbiome throughout the lifespan (1). The presence of Bifidobacterium has been associated with a variety of potential health benefits including the prevention of enteropathogenic infection via the production of short chain fatty acids (2), alleviating irritable bowel syndrome (3), enhancing the host immune system (4), particularly in infants (5), and antitumor effects that can modulate cancer immunotherapy (6). Given the numerous health-promoting effects and long-term history of safe usage of Bifidobacterium, some strains of B. animalis subsp. lactis and B. longum subsp. infantis have been commercialized as probiotics (7, 8). Consequently, Bifidobacterium genomes have been studied extensively to characterize the genetic basis for important functional attributes, such as the ability to metabolize human milk oligosaccharides (9), which some bifidobacteria leverage to colonize the relatively complex human gastrointestinal tract. Bifidobacterium genomes are also enriched with diverse CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR-associated proteins) systems (10), presenting tremendous opportunities to repurpose these systems for next-generation genome editing, as demonstrated previously in Lactobacillus (11).
Functioning as the prokaryotic adaptive immune system, CRISPR-Cas typically provides DNA-encoded (12), RNA-mediated (13), and DNA-targeting (13–15) resistance against invasive nucleic acids (12, 16). Promptly following the characterization of the Cas9 endonuclease (14) as a programmable dual-nickase (17), the advent of the single-guide RNA (sgRNA) technology (18) enabled and revolutionized genome editing (19, 20). Despite its rapid adoption in eukaryotic genome editing, the relatively slow pace of CRISPR technology deployment in prokaryotic genome engineering (21) is attributed to technical barriers such as Cas effectors cytotoxicity and limited DNA repair mechanisms typically present in bacterial genomes (22). Nonetheless, several studies have reported successful application of CRISPR-based genome editing in bacteria, especially hinging on the combination of homologous recombination of DNA templates and DNA targeting by a programmable CRISPR nuclease (23–26).
For bacteria such as Lactobacillus and Clostridium that are enriched with functional CRISPR-Cas systems, the endogenous system can be repurposed for genome engineering by delivering a repair template alongside a self-targeting CRISPR array (11, 27). Despite the characterization of endogenous CRISPR-Cas systems (10, 28), CRISPR-based genome editing has yet to be achieved in Bifidobacterium. Unlike other genetically tractable bacteria, Bifidobacterium manipulation is hindered by a relatively limited transformation and molecular biology toolbox (29), exacerbated by a refractory cell wall structure, an abundance of restriction and modification (R-M) systems, and lack of a universal replicon that replicates in a wide range of Bifidobacterium species (30, 31). Altogether, these limiting factors hamper our ability to investigate and manipulate this important genus, and novel molecular tools will enable the engineering of Bifidobacterium strains with enhanced probiotic efficacy and the development of biotherapeutic applications.
Here, we illustrate how B. animalis subsp. lactis can be engineered via both an endogenous type I-G system and an exogenous base editor, in a programmable and strain-dependent manner, and characterize the endogenous type I-E CRISPR-Cas system in B. longum subsp. infantis. We highlight the need to individualize the deployment of molecular tools for this recalcitrant species given the variable methylation patterns across select strains that possess otherwise monomorphic genomes. Collectively, these findings show that CRISPR-based genome editing strategies should be customized for individual strains, taking both genomic and epigenomic contexts into consideration, which presumably alter genome accessibility and editing outcomes. These insights provide a framework for future studies of bifidobacteria, the deciphering of their probiotic efficacy, and their genomic enhancement to manipulate the gut microbiome composition and function for enhanced human health.
Results
Characterization of Endogenous Bifidobacterium CRISPR-Cas Systems.
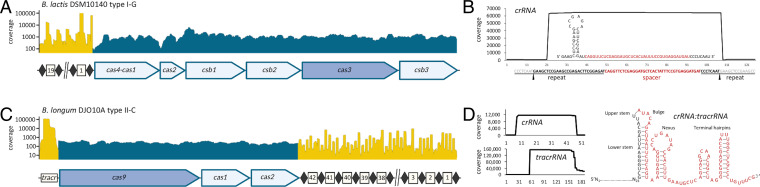
To verify that the endogenous CRISPR-Cas systems previously characterized (10, 28) are actively expressed, we studied the transcriptional profile of B. animalis subsp. lactis DSM 10140, carrying a type I-G system, and B. longum DJO10A, carrying a type II-C system, through RNA-seq analyses. In the type I-G system, the cas4-cas1 fusion was expressed in a monocistronic transcript whereas the cascade (cas2-csb1-csb2-csb3) and cas3 genes were expressed in a polycistronic transcript (Fig. 1A). Furthermore, small RNA analysis of the type I-G revealed a mature crRNA structure encompassing a 5′ handle consisting of 8 nucleotides (Fig. 1B), similar to the canonical crRNA processed by endoribonuclease Cas6 (32). Regarding the type II-C system in B. longum DJO10A, a polycistronic transcript was observed for the cas operon (Fig. 1C). The crRNA:tracrRNA duplex structure was bioinformatically predicted based on the cleavage sites (Fig. 1D). The RNA-seq results confirmed that these systems are actively transcribed during the exponential growth phase.
Fig. 1.
CRISPR-Cas systems transcription profiles based on RNA-seq data. (A) Transcriptional profile of the type I-G CRISPR locus in B. lactis DSM 10140, with mRNA in blue and small RNA (smRNA) in yellow. (B) Mature type I-G crRNA determined by smRNA sequencing (smRNA-seq). The pre-crRNA processing sites are indicated by black arrows. The spacer sequence is highlighted in red and the repeat sequence is underlined, with the cleaved sequence in gray and retained sequence in black. (C) Transcriptional profile of the type II-C CRISPR locus in B. longum DJO10A. (D) Mature type II-C crRNA and tracrRNA determined by smRNA-seq, along with the predicted crRNA:tracrRNA duplex.
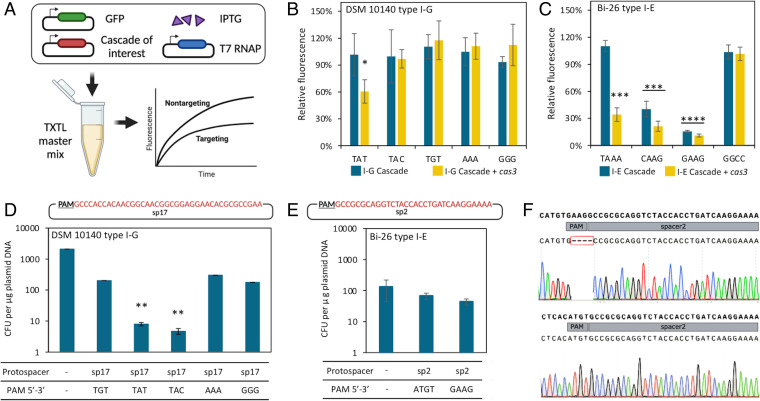
Next, we validated the functionality of the type I-G system in vitro using an Escherichia coli (E. coli) cell-free transcription-translation (TXTL) system by measuring reductions in fluorescence caused by Cascade targeting and subsequent transcriptional repression of gfp (Fig. 2A) (33). Unexpectedly, none of type I-G Cascade plasmids demonstrated GFP repression using any of the predicted PAMs (5′-TAN-3′) (Fig. 2B). However, type I-G Cascade targeting combined with Cas3-mediated cleavage significantly repressed the GFP fluorescence by 60.6% compared to the nontargeting control when the 5′-TAT-3′ PAM was used while other PAMs remained ineffective (Fig. 2B). This result indicated that Cas3 might be required to form a functional Cascade in type I-G systems, which contradicts with the common understanding that type I Cascade recruits Cas3 protein upon recognition and hybridization of the target DNA with the crRNA (34).
Fig. 2.
Functionality of endogenous CRISPR-Cas systems in Bifidobacterium. (A) A schematic diagram of CRISPR-Cas system characterization using a cell-free transcription-translation (TXTL) system. (B, C) Characterization of type I-G and type I-E functionality in TXTL, respectively. The relative fluorescence reported in the bar graphs is calculated by dividing the background-corrected fluorescence of targeting spacer by the nontargeting spacer at the 16 h end point. (D, E) Plasmid interference assay of type I-G and type I-E system in Bifidobacterium, respectively. (F) Sanger sequencing revealed that the surviving colonies in Bi-26 (type I-E) transformation with 5′-GAAG-3′ PAM carried mutations in the PAM sequence but not in the negative control associated with a 5′-ATGT-3′ PAM. Data shown in the bar graphs represents the average of three independent biological replicates (except for E where two biological replicates were performed), with the SD displayed as error bars. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 based on Welch’s t test to compare the sample average with the negative control.
To verify Cas3 is also required for Cascade formation specific to type I-G, we then tested the type I-E system of B. longum subsp. infantis Bi-26 in TXTL. Significant fluorescence repression was observed with type I-E Cascade targeting, using either 5′-CAAG-3′ or 5′-GAAG-3′ PAMs (Fig. 2C). The addition of cas3 lead to increased GFP repression using adenine-rich PAMs, particularly in the case of a 5′-TAAA-3′ PAM, for which GFP expression was down to 34.2% compared to the nontargeting control. Further research is required to investigate whether the augmented GFP repression using a 5′-TAAA-3′ PAM was via the Cas3-mediated Cascade stabilization and improved tolerance for less ideal PAMs, or simply a result of GFP cleavage from the Cas3 nuclease.
We then performed plasmid interference assays to verify the predicted PAMs and functionality in vivo. In contrast to the TXTL results, plasmid interference results showed that the endogenous type I-G system can recognize and cleave foreign plasmids carrying a protospacer flanked by either a 5′-TAT-3′ or a 5′-TAC-3′ PAM, decreasing the transformation efficiency by over two log units compared to the empty (nontargeted) vector (Fig. 2D). This discrepancy further suggested that, although TXTL can provide a rapid preliminary characterization and read out, in vivo testing is essential for systems isolated from distantly related bacteria such as Bifidobacterium.
Intriguingly, no significant reduction in transformation efficiency was observed in the type I-E plasmid interference assay using a 5′-GAAG-3′ PAM, the strongest PAM predicted using TXTL. Upon further analysis of the surviving transformants, we discovered that all screened colonies (8/8) contained a mutated 5′-GAAG-3′ PAM sequence in the plasmid, while the control plasmid did not possess any mutations (Fig. 2E), suggesting a high level of selective pressure from the endogenous type I-E system recognizing the 5′-GAAG-3′ PAM. Mutation in PAM sequences can be an escape mechanism to avoid CRISPR cleavage (35) and still enable survival under antibiotic pressure. Notably, B. infantis Bi-26 transformed less efficiently with the shuttle vector pTRK1278 (∼1 × 102 colony-forming unit [cfu]/μg), compared to B. lactis DSM 10140 (∼5 × 103 cfu/μg), as a reminder of the necessity for transformation optimization across different species. Overall, these results showed that the endogenous CRISPR-Cas systems in Bifidobacterium are active and functional despite the lack of spacer diversity across strains (10), providing an opportunity to repurpose these systems for genome editing.
Generating Large Deletions Using Endogenous CRISPR-Cas Systems.
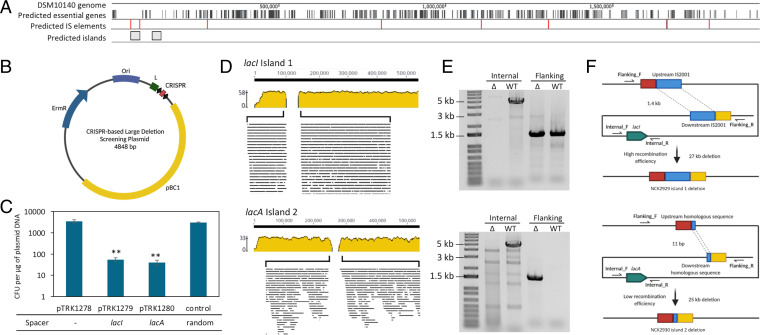
Next, we investigated whether the type I-G system can be repurposed to generate large, naturally occurring deletion events, as previously demonstrated in Streptococcus (36). The aim of the experiment was to target regions devoid of essential genes and flanked by homologous sequences to select for rare variants in which the expendable island is removed, by killing cells carrying the target sequence using a self-targeting CRISPR array. By mapping important features on the DSM 10140 genome, encompassing mobile genetic elements, defense islands, essential genes, known epigenetic modifications, and transcriptional level, we tested the feasibility of selecting for expendable genomic island removal using CRISPR self-targeting (Fig. 3A). Genomic regions lacking essential genes and/or flanked by potential transposases were identified as potential islands to be targeted. Accordingly, a CRISPR array targeting either lacI (island 1) or lacA (island 2) was cloned into pTRK1278, generating CRISPR targeting plasmids (Fig. 3B). Self-targeting showed a high level of cytotoxicity, lowering the transformation efficiency by over 2 orders of magnitude (Fig. 3C).
Fig. 3.
Repurposing the endogenous type I-G CRISPR-Cas system to generate large deletion events in Bifidobacterium. (A) The DSM 10140 genome was mapped with predicted essential genes (black), insertion sequence (IS) (red), and predicted islands (gray). (B) The CRISPR-based large deletion screening plasmid, a pBC1-based shuttle vector expressing a CRISPR array driven by the native leader. (C) Plasmid transformation efficiencies. **P < 0.01, based on Welch’s t test to compare the self-targeting average transformation efficiency with the random control. (D) Genome sequencing revealed that by targeting lacI and lacA using the endogenous type I-G system, large deletion events (27 kb and 25 kb, respectively) were observed. (E) PCR amplicons generated from combinations of internal and flanking primers. (F) Schematic overview of the recombinogenic deletion events.
We screened the survived transformants and found no single-nucleotide polymorphism (SNP) or genomic mutations that may circumvent CRISPR targeting. Rather, colonies appeared to lack the spacer target gene (lacI or lacA), and were subsequently subjected to genomic DNA sequencing, revealing a 27-kb island 1 deletion (strain NCK2929) and a 25-kb island 2 deletion (strain NCK2930) (Fig. 3D). The deletion events were confirmed through polymerase chain reaction (PCR), using primers flanking the expendable island, generating a 1.5-kb deletion amplicon (Fig. 3E). Predictably, such deletion amplicons were also observed from the wild-type population, suggesting that these islands can naturally, albeit rarely, occur. Indeed, island 1 encoding lacI naturally excises from the genome, mediated by two homologous copies of IS2001 (99.9% sequence identity over a 1.4-kb fragment) flanking the island 1 (Fig. 3F). On the contrary, natural deletion of island 2 encoding lacA was not observed in the wild type (Fig. 3E), suggesting this deletion either did no preexist in the population or may occur at an extremely low rate that cannot be detected via PCR. The only homologous sequences found flanking island 2 were two copies of 100% identical 11-bp sequence, the short length of which likely contributed to the low recombination efficiency. Yet, this illustrates how microhomologies may yield such large deletion events, expanding the possible realm of inducing island removal by homologous recombination between homologous sequences. Both expendable islands encompassed genes related to carbohydrate metabolism (SI Appendix, Fig. S1), and the two large deletion mutants possessed different carbohydrate metabolism profiles compared to the wild type, predictably (SI Appendix, Fig. S2). While NCK2929 was not capable of metabolizing xylose, amygdalin and gentiobiose, NCK2930 only lost the ability to ferment gentiobiose. This result underlined that genome decay and genomic content loss via recombination can lead to minimization of genomes and loss of metabolic pathways. We also tested three more targets (a prophage remnant gene, malQ, lacZ) in other potential islands but observed no deletion events under the experimental conditions we tested. Altogether, these data suggested that CRISPR-based screening provides a feasible route to select for rare natural variants within the bacterial population, opening avenues to select for preexisting natural events that may not fall under some of the strict regulatory frameworks that apply to genetically modified organisms in various applications encompassing food.
Repurposing the Endogenous Type I-G System for Genome Editing.
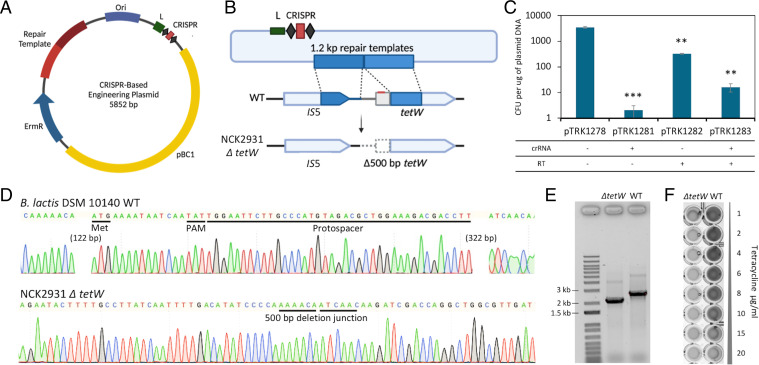
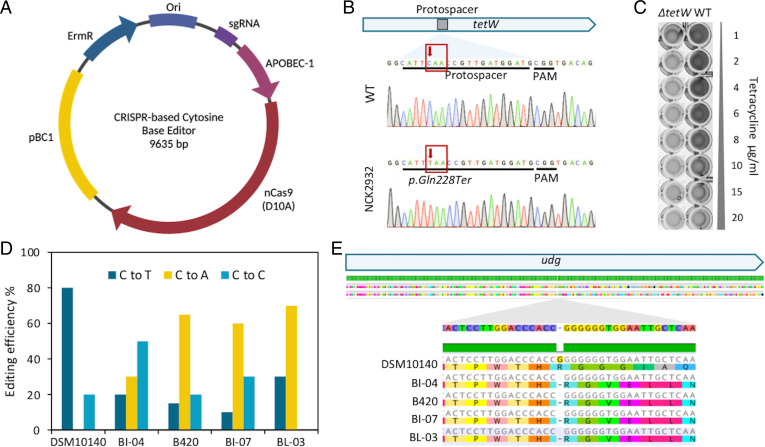
Some Bifidobacterium strains are naturally resistant to tetracycline, conferred by the tet genes encoding for ribosomal protection proteins (37, 38), posing a potential risk for horizontal transfer of antibiotic resistance cassettes. We unsuccessfully attempted to screen for a natural tetW deletion variant, presumably due to the essential genes predicted in the neighboring region. This is consistent with recent findings indicating that tetW and an adjacent IS5-like element are stable within the B. animalis subsp. lactis genome (39). We thus repurposed the endogenous type I-G system to delete the tetW gene in B. lactis, to resensitize the strain for safer probiotic usage. Initial attempts to engineer B. lactis proved challenging, even using large homologous arms (up to 3 kb each), testing various spacer designs, using replicons with different copy number, and utilizing various plasmids and plasmid designs. These challenges were compounded by historical reports of low transformation efficiency, attributed to the active R-M systems that cleave foreign nucleic acids upon recognition of restriction sites (40). Indeed, the shuttle vector pTRK1278 was predicted to contain several recognition sites including the two main R-M systems (BanLI recognizing 5′-GGW5mCC-3′ and BanLII recognizing 5′-RTC6mAGG-3′), as previously reported (41). We thus strategically designed a spacer targeting the 5′end of tetW gene, in combination with 600-bp flanking homologous arms, avoiding the predicted R-M sites to ensure a relatively high transformation efficiency (Fig. 4A and B). Transformation with tetW targeting CRISPR array was lethal with less than five colonies recovered in each transformation (Fig. 4C). The addition of repair templates overcame the Cas3-based cleavage and triggered genome editing to generate a 500-bp-deletion in the ΔtetW mutant NCK2931 (Fig. 4D and E). We were able to repeat this deletion in three independent experiments, with various efficiencies (1/49, 1/22, 1/25). The 500-bp-deletion included the promoter sequence, the start codon and a portion of the 5′ end of the tetW gene, restoring sensitivity to tetracycline at concentration as low as 1 μg × mL−1 (Fig. 4F). This result showed that in order to achieve genome editing in recalcitrant bacteria such as Bifidobacterium, a combined approach considering both genomic and epigenomic context is necessary.
Fig. 4.
Repurposing the endogenous type I-G CRISPR-Cas system for genome editing in Bifidobacterium. (A) Plasmid pTRK1278, a pBC1-based E. coli–Bifidobacterium shuttle vector, was used to generate CRISPR-based genome editing in Bifidobacterium. (B) Schematic overview of a 500-bp-deletion containing the promoter region, start codon and the 5′ portion of tetW. The CRISPR array expresses a spacer matching the 5′ end of tetW (within the deletion region, indicated in red), flanked by a 5′-TAT-3′ PAM. (C) Plasmid transformation efficiencies. **P < 0.01, ***P < 0.001 based on Welch’s t test to compare each plasmid transformation efficiency with the positive control pTRK1278. (D) Chromatogram profiles demonstrating the 500-bp-deletion was achieved in B. lactis DSM 10140 by repurposing its native type I-G CRISPR-Cas system. (E) PCR amplicons generated from flanking primers revealed a 500-bp-deletion in the ΔtetW mutant NCK2931 compared to the wild type. (F) The minimal inhibitory concentration (MIC) test demonstrated that the 500-bp-deletion rendered the mutant sensitive to tetracycline.
A CRISPR Cytosine Base Editor Can Introduce SNPs in Bifidobacterium.
Repurposing the endogenous CRISPR-Cas system to genome editing requires extensive characterization and validation of the system and is not scalable or broadly applicable to bacteria, since approximately half of all bacteria lack a CRISPR-Cas system (42), and it is unclear what portion of endogenous systems is actually active. Therefore, we applied a CRISPR-based cytosine base editor (CBE), a fusion system of a cytidine deaminase APOBEC-1 and a SpCas9(D10A) nickase previously developed by the Ji group (Fig. 5A) (43), for portable base editing in Bifidobacterium.
Fig. 5.
Application of a portable CRISPR-Cas cytosine base editor for genome editing in Bifidobacterium. (A) The nCas9-APOBEC-1 fusion along with a sgRNA was cloned into pTRK1278, generating the base editing plasmid pTRK1284. (B) Schematic overview of the cytosine base editor introducing C•G-to-T•A mutation in tetW in B. lactis DSM 10140. The sgRNA targets a protospacer flanked by a 5′-NGG-3′ PAM at the 5′ end of tetW, targeting the fifth nucleotide within the protospacer. (C) MIC testing revealed that the p.Gln228Ter nonsense mutation rendered the mutant NCK2932 sensitive to tetracycline. (D) The portable base editor generated C•G-to-T•A mutation in B. lactis strains with various efficiencies. (E) Alignment of the uracil-DNA glycosylase coding sequence (udg) from all five B. lactis strains revealed a c.351_delinsC insertion in DSM 10140.
A protospacer at the 5′ end of the tetW was targeted, in which a cytosine was in the editing window (the first 3–8 nucleotide at the 5′ end of the protospacer) (Fig. 5B). We screened 20 recovered colonies and over half (12 out of 20) showed the intended C•G-to-T•A mutation, or a coexisting mixed population of mutant and wild type (Fig. 5B). The g.682C > T mutation lead to a p.Gln228Ter nonsense mutation, rendering the mutant strain NCK2932 sensitive to tetracycline (Fig. 5C). We did not observe any significant difference in tetracycline sensitivity between the ΔtetW deletion mutant (NCK2931) and the p.Gln228Ter mutant (NCK2932), indicating the base-editing strategy can efficiently remove the antibiotic resistance and restore tetracycline sensitivity.
To demonstrate the portability of the CBE, we then generated base editing in four other B. lactis probiotic strains, albeit with different editing efficiencies across strains (Fig. 5D). Surprisingly, a high level of unintended editing (C•G-to-A•T) was observed for over half of the screened colonies, which was not observed in DSM 10140 (Fig. 5D). Alignment of the native uracil-DNA glycosylase (udg) gene from the five B. lactis strains revealed a cytosine insertion at the g.351 position of the udg gene in DSM 10140, generating a frameshift mutation (Fig. 5E). This SNP in DSM 10140 was also confirmed by the RNA sequencing (RNA-seq) reads. This resulted in lower efficiency of base editing in other B. lactis strains, in which a functional UDG can initiate base-excision repair (BER), generating the conversion of U•G to the wild-type C•G or incorrect A•G. New generations of CBE fused with an uracil glycosylase inhibitor (44) should be implemented in Bifidobacterium to improve editing efficiencies. This result also showed that a single SNP could lead to significant differences in genome editing efficiencies, emphasizing the necessity of strain-resolution approach for genetic modification in recalcitrant bacteria. This is noteworthy given the documented homogeneity of B. lactis genomes and their near identity at the genome-wide level in numerous polished genomes.
Epigenetic Analysis Revealed Strain-Level Variation in B. lactis Methylation Pattern.
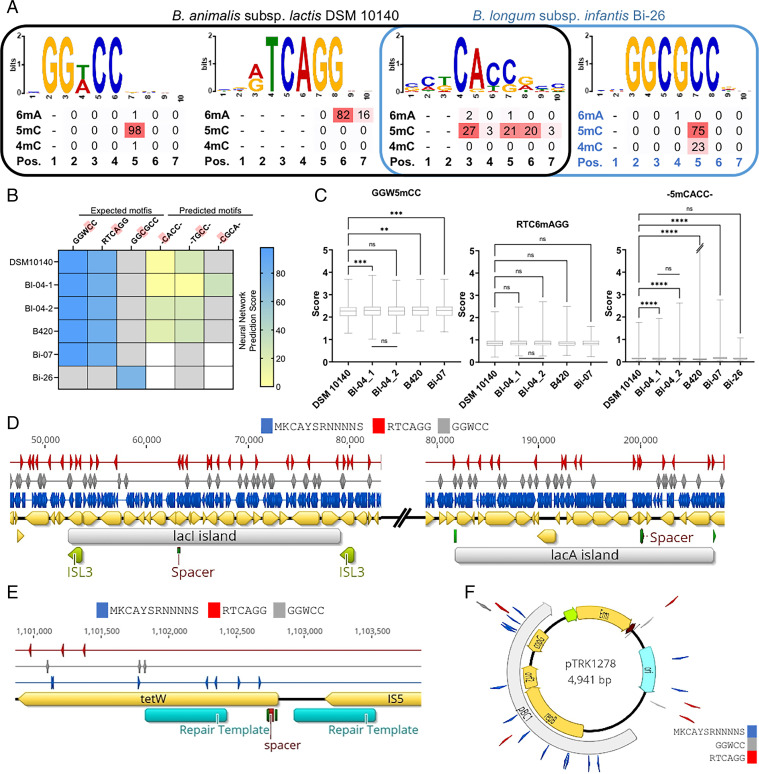
In order to investigate potential factors involved in the observed differences in base editing efficiencies, as well as the substantial variations in transformation efficiencies among B. lactis strains that are otherwise genomically nearly identical (SI Appendix, Fig. S3), we next investigated their epigenomes. Comparative genomic analysis revealed that although all five B. lactis strains harbor the same methylation cassettes, consisting of 6mA and 5mC methyltransferases, multiple SNPs were found in genes or regulatory elements related to the R-M systems in DSM 10140. We hypothesized that these SNPs might affect the overall epigenomes and the cognate restriction profile of strains, resulting in higher genetic accessibility for DSM 10140 compared to other B. lactis strains. To reveal methylation patterns in B. lactis, particularly 5mC modification that can be overlooked using the traditional approach, we sequenced native and PCR-amplified genomic DNA from each strain using Oxford Nanopore Technologies and analyzed the nanopore current differences using Nanodisco (45). In accordance with motifs described in previous work (41), de novo motif characterization showed that 5′-GGW5mCC-3′ and 5′-RTC6mAGG-3′ motifs are present in all B. lactis strains indicated by strong machine learning prediction scores against the training dataset (Fig. 6A). Remarkably, and unexpectedly, we also observed a group of 5mC motifs that are shared by B. lactis strains and B. longum subsp. infantis Bi-26, indicating cross-species methylation motifs (Fig. 6A and SI Appendix, Fig. S4). These newly discovered motifs had various neural network prediction scores across strains with many prediction scores consistent with the motif training set (Fig. 6A and B), albeit lower than scores of the previously established motifs 5′-GGW5mCC-3′ and 5′-RTC6mAGG-3′.
Fig. 6.
Epigenomic mapping of Bifidobacterium to assess genomic context variability. (A) The expected motifs of B. lactis and B. infantis, as well as a de novo 5mC motif that is shared between the species, are shown with neural network (NN) scoring using Nanodisco. (B) The heatmap shows the NN scoring prediction for each motif in the various strains. Red boxes indicate the fine mapping locations in the motifs. White boxes indicate the motif was detected de novo with MEME, but was below NN prediction scoring, while gray boxes show the motif was not detected de novo with MEME. (C) The metagenomic binning scores across the genomes of all strains are compared using one-way ANOVA with DSM 10140 as the control (**P < 0.05, ***P < 0.001, ****P < 0.0001). (D) Three methylation motifs 5′-GGW5mCC-3′ (gray), 5′-RTC6mAGG-3′ (red), and 5′-5mCACC-3′ (blue) were mapped across (D) the lac islands in B. lactis DSM 10140, (E) the tetW and IS5-like element region in DSM 10140, and (F) plasmid pTRK1278 using Geneious Prime. Green regions show transposase sequence for lacI island, and homologous sequences for lacA island. The deletion repair templates for the tetW region are shown below the gold gene annotations as blue arrows for base editing and green arrows for endogenous CRISPR-Cas targets.
To confirm methylation patterns are strain-specific, we used several tools within Nanodisco to measure the strength of the methylation signals of the various motifs: meme level, neural network prediction against the training dataset, and metagenomic binning scoring. Some motifs only were strong enough to be detected by meme input, but not through machine learning prediction (Fig. 6B). Specifically, Bl-04 and B420 had occurrences where the motifs were modified in combination that affected overall smooth values of current differences, whereas DSM 10140 and Bi-07 did not. This is shown by multiple motifs in a single meme level (SI Appendix, Fig. S4) and could indicate stronger overall methylation in Bl-04 and B420. However, the Bl-04 replicates showed different meme motifs and neural network prediction scores, indicating testing with more replicates is needed. Since the 5′-GGW5mCC-3′, 5′-RTC6mAGG-3′, and 5′-MK5mCAYSRNNNNS-3′ motifs were detected in the meme analysis for all B. lactis strains, and the Bl-04 replicates (Fig. 6B), binning scores were calculated for each strain and Bl-04 replicates. The 5′-GGW5mCC-3′ and 5′-MK5mCAYSRNNNNS-3′ motifs revealed significant differences between binning scores of DSM 10140 and the other strains (Fig. 6C). Interestingly, the 5′-MK5mCAYSRNNNNS-3′ motif was not significantly different between DSM 10140 and Bi-26, which are different species. No significant differences in binning scores were observed for 5′-RTC6mAGG-3′ motif, nor were there significant differences in the binning scores of the Bl-04 replicates in all three motifs. This indicates that binning score is a useful metric to distinguish strain-specific methylation patterns, which are different between highly genomically similar strains of B. lactis.
The methylation motif patterns were then assessed for their effects on genome editing. The crRNAs used for editing the tetW region and the lac islands were designed to avoid the 5′-GGWCC-3′ and 5′-RTC6mAGG -3′ motifs (Fig. 6D and E), however, after epigenomic screening, we noted that the lacA protospacer overlapped with the 5′-MK5mCAYSRNNNNS-3′ motif (Fig. 6D). Nonetheless, the lacA deletion was successful, which may indicate partial methylation or that the motif was from an orphan methyltransferase. To investigate if the overall epigenome contributes to the low genetic accessibility in nonmodel B. lactis strains, we mapped the 5′-GGW5mCC-3′, 5′-TC6mAGG-3′, and 5′-MK5mCAYSRNNNNS-3′ degenerative motifs to the shuttle vector pTRK1287 sequence (Fig. 6F). When mapping all motifs discovered by Meme for each strain (SI Appendix, Fig. S4), we observed that due to the degenerative nature of the motifs, over 40 recognition sites belonging to Bl-04 and B420 were mapped to pTRK1287 whereas less than 20 motifs belonging to DSM 10140 and Bi-07 were mapped (SI Appendix, Fig. S5). This may explain the different transformation efficiencies observed among the strains, although the methyltransferases responsible for these motifs and their cognate endonucleases remain to be discovered and will require further investigation.
Discussion
The molecular mechanisms underpinning Bifidobacterium probiotic efficacy, especially host colonization, metabolic repertoire, immunogenicity and safety, have been increasingly studied over the past decade (1). This progress has mainly been driven by advances in next generation sequencing technologies along with a burst of omics-based research encompassing genomics, transcriptomics, proteomics, and metabolomics (46, 47). However, the development of robust genome modification systems for Bifidobacterium has been lagging with a paucity of reports in select species (30, 31), hindering the functional analysis of specific genes and the development of next-generation probiotics with enhanced efficacy. Although CRISPR-based editing tools have been established for a range of beneficial bacteria including Lactobacillus (23) and Clostridium (27), successful application of CRISPR-mediated editing in Bifidobacterium has yet to be reported. To date, knockout studies have only been performed in a relatively limited number of Bifidobacterium strains, such as B. breve UCC2003 and B. longum 105-A, to study the molecular mechanism of bacteria-host interactions including gut colonization (48) and human milk oligosaccharides (HMOs) metabolism (49). The CRISPR-based genome editing platform established in this study can enable the genesis of specific knockout mutants broadly in other genetically refractory Bifidobacterium species such as B. bifidum (50). In the future, this could be useful to determine and enhance probiotic efficacy in this diverse and important probiotic genus.
Here, we characterized multiple CRISPR-Cas systems previously identified in the Bifidobacterium genomes (10). The type I-G CRISPR-Cas system, recently renamed from the type I-U system (42), has been relatively understudied, although it is generally a more compact system with potential for portability. DNA targeting by a type I-G CRISPR-Cas system was previously described in Pyrococcus furious (51). It was suggested that the type I-G ribonucleoprotein (crRNP) complexes form and bind to target DNA without Cas3 nuclease (52). This contradicts with the TXTL data in our study, suggesting that the Cas3 protein was in complex with the type I-G Cascade in B. lactis. Although Cas3 is usually a stand-alone nuclease in most type I systems, Cas3 fusion proteins have also been described in the literature (53–56). In particular, it was previously documented that a Cas2-Cas3 fusion protein interacts with Cas1, suggesting Cas3 is involved in spacer acquisition besides interference (53). Additionally, a Cas2-Cas3 fusion may lead to a more stable Cas3-Cascade complex in the type I-F system (54). The variation in effector interaction may result in different underlying mechanism among systems. Further study will be needed to determine the exact structure and the occurrence of Cas3 fusion proteins in the type I-G system in Bifidobacterium.
Harnessing the proper DNA repair mechanisms is crucial to obtain the desirable mutations. Besides the broadly discussed nonhomologous end joining (NHEJ) repair and homology-directed repair (HDR) mechanisms, there are other pathways that may contribute to genome editing outcomes. Of note, microhomology-mediated end joining (MMEJ) repair, hinging on the local target site sequence, can deliver precise template-free deletions using short stretches of homologous sequences that flank the cleavage site. Indeed, it was previously reported that over half of the Cas9-mediated double-stranded breaks (DSB) are repaired through MMEJ in human cell line models (57). Recently, this error-prone DSB repair mechanism was utilized to enable genome editing in Zymomonas mobilis (58), although there is a need to better characterize the factors driving MMEJ-based editing in bacteria. Previous studies have established that CRISPR-based targeting can induce or screen for large chromosomal deletion (59), as well as select for preexisting natural mutants lacking protospacer or PAM sequences (36). Our results show that Cas3-based targeting can both screen for preexisting natural mutants and induce large deletion via a recombinogenic mechanism. These large deletion events may contribute to the genome decay in B. lactis genomes, leading to loss of metabolic pathways (60). Alternatively, regions that are not subject to recombinogenic events may indicate stable genomic infrastructure, as shown with the tetW and IS5-like element discussed in this study. The nature of CRISPR-based self-targeting can be exploited to develop a high-throughput genome-wide screening platform. A CRISPR array library can be designed to interrogate the bacterial genomes to study genotype-phenotype association and select for natural variants with valuable attributes, opening new avenues to generate products not subject to genetically modified organism regulations.
Yet, the lack of genetic tools for Bifidobacterium remains a bottleneck for comprehensive studies of the molecular mechanisms underpinning health-promoting effects, hindering the development of next generation probiotics with enhanced efficacy. Several strategies including homologous recombination systems utilizing either nonreplicative plasmids or temperature sensitive plasmids (61), random transposon-based mutagenesis systems (62), as well as an inducible plasmid self-destruction (IPSD)-assisted systems (63) have been reported. However, these methods encompass various drawbacks such as random nontargeted mutations, extensive screening requirements, and limitation to select host species. The challenges of adapting CRISPR-based genome editing in Bifidobacterium range from CRISPR nuclease-associated cytotoxicity to insufficient transformation efficiency (22). Harnessing the endogenous CRISPR-Cas systems effectively circumvents these issues by delivering a short CRISPR array in combination with the desired repair templates (11, 59). Our results show that by strategically designing the repair templates to avoid predicted restriction sites, deletion can be achieved with 600-bp homologous arms, much shorter than the 3-kb arms from the literature (64). Cas3-induced DNA cleavage initiated the editing event as well as removed wild-type genotypes carrying the protospacer. However, this can be lethal, and bacteria often lack the adequate DNA repair mechanism to survive such damage, even in the presence of repair templates. To compensate, we applied a cytosine base editor to introduce SNPs independent of recombination efficiency. Amazingly, a single SNP mutation in udg resulted in a more efficient base editing for DSM 10140 compared to other strains, which highlighted how impactful single nucleotide differences can be, when positioned in certain genetic elements. In addition to the variation in CRISPR spacers content, only a handful of nucleotide polymorphisms have been identified among B. lactis strains (60), consisting predominantly of nonsynonymous mutations. While some SNPs have been linked to interstrain phenotypic differences (65), their relevance to genomic modification efficiency is reported here for what we believe to be the first time. Our results suggest that despite genome homogeneity in B. lactis, engineering strategies should be customized for each strain since the strain-to-strain variations can result in drastically different genome accessibilities and genome editing efficiencies, contrary to the common belief that universal editing tools can be applied broadly, even in mixed bacterial populations encompassing diverse genera and phylogenetic units.
Although target DNA sequences can significantly influence genome editing outcomes, increasing evidence suggests that methylation and other local features (e.g., chromatin) also determine editing efficiency in eukaryotes (66, 67). However, methylation presumably impacts delivery efficiency (40, 68), although there is no systematic study of the impact of DNA methylation on CRISPR-based genome editing efficiency in bacteria. We speculate that the methylation pattern may play an indirect role on editing efficiency by influencing the delivery of CRISPR payloads into bacteria, perhaps rather than the targeting efficiency per se. Other factors may be in play such as nucleoid-associated proteins affecting bacterial chromosome structure (69) and the impact of epigenetic patterns on expression level (70). Epigenetics is gaining increasing attention in the field of probiotic bacteria (70). Over 200 R-M systems have been identified in Bifidobacterium, recognizing the base modifications N6-methyladenine (m6A), N4-methylcytosine (m4C), and 5-methylcytosine (m5C) (71). Plasmid methylation to circumvent the R-M systems and improve transformation efficiency was reported previously in multiple Bifidobacterium species including B. breve (72) and B. lactis (41). Two type II R-M systems have been previously characterized in B. lactis (41). The de novo methylation characterization in this study not only confirmed the previous findings but also discovered 5mC motifs distributed unevenly across the five B. lactis strains tested (Fig. 6), presumably contributing to the observed differences in transformation efficiencies. Further research is necessary to link these motifs with associated methyltransferases and cognate restriction enzymes (45). Remarkably, we also observed genus-level methylation motif conservation in different Bifidobacterium species. This approach could be applied to metagenomic datasets to monitor mobile elements and track genes of interest such as those responsible for antibiotic resistance (45). The conservation of DNA methylation in bacteria is being increasingly discussed but the functional relevance has yet to be revealed (73). We wonder whether the conserved methylation patterns observed in this study between B. lactis and B. infantis, two distantly related species (74), may provide some fundamental cell regulatory function beyond just defense systems. Nonetheless, the different methylation profiles across the strains contributed to the different genome accessibilities among strains. Moving forward, the development and adaptation of CRISPR tools should be customized in a strain-specific manner, to enable future studies of Bifidobacterium genetics and their health-promoting effects in human health. Altogether, our data reveal a unique dynamic in bacteria recalcitrant to genome engineering, in which both genomic and epigenomic context can influence editing efficiency and outcomes, highlighting the need to individualize strategies and designs.
Materials and Methods
Bacterial Strains and Growth Conditions.
All bacterial strains used in this study are listed in SI Appendix, Table S1. The detailed bacterial culture condition is described in SI Appendix, Supplementary Materials and Methods.
Genome Sequencing and Assembly.
The bacterial genomic DNA was isolated using the DNeasy PowerLyzer Microbial Kit (Qiageny) and was sequenced in the Flongle Flow Cell R9.4.1 (Oxford Nanopore Technologies) using the Ligation Sequencing Kit (SQK-LSK109). The detailed sequencing and assembly method is described in SI Appendix, Supplementary Materials and Methods.
RNA Extraction and RNA Sequencing Analysis.
The total RNA of Bifidobacteria was isolated using the Zymo Direct-Zol RNA Miniprep Kit (Zymo Research) and sequenced at the Roy J. Carver Biotechnology Centre following the protocol as described previously ((11)). The detailed RNA-seq analysis method is described in SI Appendix, Supplementary Materials and Methods.
Methylome Analysis (Nanopore and Nanodisco).
The strategy of epigenomic analysis followed the Nanodisco method as previously described (45). The detailed epigenomics methods are described in SI Appendix, Supplementary Materials and Methods.
Construction of pTRK1278, a pBC1-Based E. coli–Bifidobacterium Shuttle Vector.
All plasmids, duplexes, and oligonucleotides used in this study are listed in SI Appendix, Tables S1 and S2. Plasmid pTRK1278 was constructed following a similar strategy of pAM1 construction as previously described (75). The detailed cloning method is described in SI Appendix, Supplementary Materials and Methods.
DNA Manipulation and Transformation.
Chemically competent E. coli cells and heat shock transformation were prepared as previously described (11). Bifidobacterial transformation was performed as previously described (64). The detailed DNA manipulation and transformation method is described in SI Appendix, Supplementary Materials and Methods.
Characterization of Endogenous CRISPR-Cas Systems In Vitro Using TXTL.
The endogenous CRISPR-Cas systems in Bifidobacteria were characterized using TXTL (Daicel Arbor Biosciences) as previously described (33). The detailed method is described in SI Appendix, Supplementary Materials and Methods.
Plasmid Interference Assay.
The plasmid interference assay for then endogenous CRISPR-Cas systems was performed as previously described (11). The detailed plasmid interference assay is described in SI Appendix, Supplementary Materials and Methods.
Repurposing the Endogenous Type I-G System to Generate Large-Deletion Events.
The potential expendable islands in B. lactis DSM 10140 were predicted as previously described in Streptococcus (36). Within the predicted islands, protospacers flanked by a 5′-TAT-3′ PAM on the 5′end was selected for the endogenous type I-G CRISPR targeting. The detailed method is described in SI Appendix, Supplementary Materials and Methods.
Repurposing the Endogenous Type I-G System for Genome Editing.
A pTRK1278-based plasmid carrying a type I-G CRISPR array targeting the 5′ end of tetW ORF and a 1.2-kb repair template was constructed to generate a 500-bp deletion in B. lactis DSM 10140. The detailed construction method is described in SI Appendix, Supplementary Materials and Methods.
Constructing the CRISPR Base Editor Plasmid for Base Editing.
The CRISPR cytosine base editor along with the sgRNA, amplified from plasmid pnCasPA-BEC (Addgene plasmid #113349) (43), was cloned into pTRK1278, generating the pTRK1284, which served as the base plasmid for all subsequent base editing experiments. The detailed construction method is described in SI Appendix, Supplementary Materials and Methods.
Statistical Analyses.
Three independent biological replicates were performed and the average was used to represent the data in the bar graphs along with the error bars representing the SD. To test the null hypothesis that the two groups had equal means, a Welch’s t test was performed to analyze the data distribution and determine the difference between the group means were significant or not. A P value <0.05 was considered statistically significant. The statistical analyses were performed in R studio, v1.2.5. For the epigenomic analysis, one-way ANOVA with Tukey’s multiple comparison test was performed in Prism v. 9.3.1 (GraphPad).
Supplementary Material
Acknowledgments
We thank Dr. Sarah O’Flaherty and Dr. Matthew Nethery for insightful discussions. We also acknowledge funding by North Carolina State University and the North Carolina Agricultural Foundation. We also thank Chris Fields of the HPCBio team as well as Alvaro Hernandez of the DNA Services team at the University of Illinois Urbana-Champaign for sequencing and raw processing for the epigenomics analysis. Matthew Poore is acknowledged for performing growth experiments at IFF.
Footnotes
Reviewers: M.V., Universita degli Studi di Parma; and J.v.d.O., Wageningen University & Research
Competing interest statement: R.B., C.H.-C. and Y.J.G are inventors on several patents related to CRISPR-Cas systems and their uses. R.B. is a cofounder of Intellia Therapeutics, Locus Biosciences, TreeCo, CRISPR Biotechnologies and Ancilia Biosciences, and a shareholder of Caribou Biosciences, Inari Ag, Felix Biotechnologies, and Provaxus. C.H.-C. is a cofounder of Microviable Therapeutics and shareholder of CRISPR Biotechnologies. W.M. is employed by IFF Health & Biosciences, International Flavors and Fragrances, Inc., which commercializes probiotic products. R.B. and J.v.d.O. are co-authors on a 2018 news and views article and a 2020 review article.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2205068119/-/DCSupplemental.
Data Availability
The RNA-seq and DNA-seq data of Bifidobacteria studied in this paper was deposited in the NCBI Short Read Archive database, under the BioProject ID PRJNA811526 (76). The SRA accessions number are SAMN26342519–SAMN26342528 (77, 78). The raw Nanopore sequencing native and amplified current difference files and metagenomic binning score outputs are available upon request.
References
- 1.Wong C. B., Odamaki T., Xiao J. Z., Insights into the reason of Human-Residential Bifidobacteria (HRB) being the natural inhabitants of the human gut and their potential health-promoting benefits. FEMS Microbiol. Rev. 44, 369–385 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fukuda S., et al. , Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547 (2011). [DOI] [PubMed] [Google Scholar]
- 3.O’Mahony L., et al. , Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology 128, 541–551 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Fanning S., et al. , Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc. Natl. Acad. Sci. U.S.A. 109, 2108–2113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henrick B. M., et al. , Bifidobacteria-mediated immune system imprinting early in life. Cell 184, 3884–3898.e11 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Sivan A., et al. , Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science 350, 1084–1089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng J., Laitila A., Ouwehand A. C., Bifidobacterium animalis subsp. lactis HN019 effects on gut health: A review. Front. Nutr. 8, 790561 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chichlowski M., Shah N., Wampler J. L., Wu S. S., Vanderhoof J. A., Bifidobacterium longum subspecies infantis (B. infantis) in pediatric nutrition: Current state of knowledge. Nutrients 12, 1581 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zivkovic A. M., German J. B., Lebrilla C. B., Mills D. A., Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Natl. Acad. Sci. U.S.A. 108 (suppl. 1), 4653–4658 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan M., Nethery M. A., Hidalgo-Cantabrana C., Barrangou R., Comprehensive mining and characterization of CRISPR-cas systems in Bifidobacterium. Microorganisms 8, 720 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hidalgo-Cantabrana C., Goh Y. J., Pan M., Sanozky-Dawes R., Barrangou R., Genome editing using the endogenous type I CRISPR-Cas system in Lactobacillus crispatus. Proc. Natl. Acad. Sci. U.S.A. 116, 15774–15783 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrangou R., et al. , CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Brouns S. J. J., et al. , Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garneau J. E., et al. , The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Sinkunas T., et al. , Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 30, 1335–1342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horvath P., Barrangou R., CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Gasiunas G., Barrangou R., Horvath P., Siksnys V., Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 109, E2579–E2586 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinek M., et al. , A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cong L., et al. , Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrangou R., Doudna J. A., Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 34, 933–941 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Barrangou R., Horvath P., A decade of discovery: CRISPR functions and applications. Nat. Microbiol. 2, 17092 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Arroyo-Olarte R. D., Bravo Rodríguez R., Morales-Ríos E., Genome editing in bacteria: CRISPR-Cas and beyond. Microorganisms 9, 844 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goh Y. J., Barrangou R., Portable CRISPR-Cas9N system for flexible genome engineering in Lactobacillus acidophilus, Lactobacillus gasseri, and Lactobacillus paracasei. Appl. Environ.Microbiol. 87, e02669-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang W., Bikard D., Cox D., Zhang F., Marraffini L. A., RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang H., Song X., Yang S., Development of a RecE/T-assisted CRISPR-Cas9 toolbox for Lactobacillus. Biotechnol. J. 14, e1800690 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Roberts A., Barrangou R., Applications of CRISPR-Cas systems in lactic acid bacteria. FEMS Microbiol. Rev. 44, 523–537 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Maikova A., Kreis V., Boutserin A., Severinov K., Soutourina O., Using an endogenous CRISPR-Cas system for genome editing in the human pathogen Clostridium difficile. Appl. Environ. Microbiol. 85, e01416–e01419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hidalgo-Cantabrana C., Crawley A. B., Sanchez B., Barrangou R., Characterization and exploitation of CRISPR loci in Bifidobacterium longum. Front. Microbiol. 8, 1851 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Z., Baur A., Zhurina D., Yuan J., Riedel C. U., Accessing the inaccessible: molecular tools for bifidobacteria. Appl. Environ. Microbiol. 78, 5035–5042 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukiya S., Sakanaka M., Yokota A., “Chapter 15 - Genetic manipulation and gene modification technologies in Bifidobacteria” in The Bifidobacteria and Related Organisms, Mattarelli P., Biavati B., Holzapfel W. H., Wood B. J. B., Eds. (Academic Press, 2018), pp. 243–259. [Google Scholar]
- 31.Zuo F., Chen S., Marcotte H., Engineer probiotic bifidobacteria for food and biomedical applications - Current status and future prospective. Biotechnol. Adv. 45, 107654 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Carte J., Wang R., Li H., Terns R. M., Terns M. P., Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 22, 3489–3496 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall R., et al. , Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system. Mol. Cell 69, 146–157.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao Y., Luo M., Dolan A. E., Liao M., Ke A., Structure basis for RNA-guided DNA degradation by Cascade and Cas3. Science 61, eaat0839 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paez-Espino D., et al. , Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat. Commun. 4, 1430 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Selle K., Klaenhammer T. R., Barrangou R., CRISPR-based screening of genomic island excision events in bacteria. Proc. Natl. Acad. Sci. U.S.A. 112, 8076–8081 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aires J., Doucet-Populaire F., Butel M. J., Tetracycline resistance mediated by tet(W), tet(M), and tet(O) genes of bifidobacterium isolates from humans. Appl. Environ. Microbiol. 73, 2751–2754 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ammor M. S., Flórez A. B., Alvarez-Martín P., Margolles A., Mayo B., Analysis of tetracycline resistance tet(W) genes and their flanking sequences in intestinal Bifidobacterium species. J. Antimicrob. Chemother. 62, 688–693 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Nøhr-Meldgaard K., Struve C., Ingmer H., Agersø Y., The tetracycline resistance gene, tet(W) in Bifidobacterium animalis subsp. lactis follows phylogeny and differs from tet(W) in other species. Front. Microbiol. 12, 658943 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnston C. D., et al. , Systematic evasion of the restriction-modification barrier in bacteria. Proc. Natl. Acad. Sci. U.S.A. 116, 11454–11459 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’ Connell Motherway M., et al. , Identification of restriction-modification systems of Bifidobacterium animalis subsp. lactis CNCM I-2494 by SMRT sequencing and associated methylome analysis. PLoS One 9, e94875 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makarova K. S., et al. , Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen W., et al. , CRISPR/Cas9-based genome editing in Pseudomonas aeruginosa and cytidine deaminase-mediated base editing in Pseudomonas species. iScience 6, 222–231 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L., et al. , Enhanced base editing by co-expression of free uracil DNA glycosylase inhibitor. Cell Res. 27, 1289–1292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tourancheau A., Mead E. A., Zhang X.-S., Fang G., Discovering multiple types of DNA methylation from bacteria and microbiome using nanopore sequencing. Nat. Methods 18, 491–498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bottacini F., van Sinderen D., Ventura M., Omics of bifidobacteria: research and insights into their health-promoting activities. Biochem. J. 474, 4137–4152 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Turroni F., et al. , Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J. 10, 1656–1668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Connell Motherway M., et al. , Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U.S.A. 108, 11217–11222 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakurama H., et al. , Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression. J. Biol. Chem. 288, 25194–25206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turroni F., et al. , Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. U.S.A. 107, 19514–19519 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elmore J., Deighan T., Westpheling J., Terns R. M., Terns M. P., DNA targeting by the type I-G and type I-A CRISPR-Cas systems of Pyrococcus furiosus. Nucleic Acids Res. 43, 10353–10363 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Majumdar S., Ligon M., Skinner W. C., Terns R. M., Terns M. P., Target DNA recognition and cleavage by a reconstituted Type I-G CRISPR-Cas immune effector complex. Extremophiles 21, 95–107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richter C., Gristwood T., Clulow J. S., Fineran P. C., In vivo protein interactions and complex formation in the Pectobacterium atrosepticum subtype I-F CRISPR/Cas System. PLoS One 7, e49549 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S., et al. , A split protease-E. coli ClpXP system quantifies protein-protein interactions in Escherichia coli cells. Commun. Biol. 4, 841 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Westra E. R., et al. , CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell 46, 595–605 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Oost J., Westra E. R., Jackson R. N., Wiedenheft B., Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 12, 479–492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen M. W., et al. , Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563, 646–651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X., et al. , CRISPR-mediated host genomic DNA damage is efficiently repaired through microhomology-mediated end joining in Zymomonas mobilis. J. Genet. Genomics 48, 115–122 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Csörgő B., et al. , A compact Cascade-Cas3 system for targeted genome engineering. Nat. Methods 17, 1183–1190 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barrangou R., et al. , Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04. J. Bacteriol. 191, 4144–4151 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Motherway M. O., et al. , Characterization of ApuB, an extracellular Type II amylopullulanase from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 74, 6271–6279 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakanaka M., et al. , A transposon mutagenesis system for Bifidobacterium longum subsp. longum based on an IS3 family insertion sequence, ISBlo11. Appl. Environ. Microbiol. 84, e00824-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zuo F., Zeng Z., Hammarström L., Marcotte H., Inducible plasmid self-destruction (IPSD) assisted genome engineering in Lactobacilli and Bifidobacteria. ACS Synth. Biol. 8, 1723–1729 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Hidalgo-Cantabrana C., et al. , A single mutation in the gene responsible for the mucoid phenotype of Bifidobacterium animalis subsp. lactis confers surface and functional characteristics. Appl. Environ. Microbiol. 81, 7960–7968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Briczinski E. P., Phillips A. T., Roberts R. F., Transport of glucose by Bifidobacterium animalis subsp. lactis occurs via facilitated diffusion. Appl. Environ. Microbiol. 74, 6941–6948 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen X., et al. , Probing the impact of chromatin conformation on genome editing tools. Nucleic Acids Res. 44, 6482–6492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Daer R. M., Cutts J. P., Brafman D. A., Haynes K. A., The impact of chromatin dynamics on Cas9-mediated genome editing in human cells. ACS Synth. Biol. 6, 428–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Connell Motherway M., O’Driscoll J., Fitzgerald G. F., Van Sinderen D., Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb. Biotechnol. 2, 321–332 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Browning D. F., Grainger D. C., Busby S. J., Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 13, 773–780 (2010). [DOI] [PubMed] [Google Scholar]
- 70.Morovic W., Budinoff C. R., Epigenetics: A new frontier in probiotic research. Trends Microbiol. 29, 117–126 (2021). [DOI] [PubMed] [Google Scholar]
- 71.Roberts R. J., Vincze T., Posfai J., Macelis D., REBASE—A database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 43, D298–D299 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bottacini F., et al. , Comparative genome and methylome analysis reveals restriction/modification system diversity in the gut commensal Bifidobacterium breve. Nucleic Acids Res. 46, 1860–1877 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oliveira P. H., Fang G., Conserved D. N. A., Conserved DNA methyltransferases: A window into fundamental mechanisms of epigenetic regulation in bacteria. Trends Microbiol. 29, 28–40 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brandt K., Barrangou R., Phylogenetic analysis of the Bifidobacterium genus using glycolysis enzyme sequences. Front. Microbiol. 7, 657 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alvarez-Martín P., O’Connell-Motherway M., van Sinderen D., Mayo B., Functional analysis of the pBC1 replicon from Bifidobacterium catenulatum L48. Appl. Microbiol. Biotechnol. 76, 1395–1402 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Pan M., et al., CRISPR-based Engineering in Bifidobacterium. NCBI BioProject. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA811526. Deposited 1 Mar 2022.
- 77.Pan M., et al., Genomic and epigenetic landscapes drive CRISPR-based genome editing in Bifidobacterium. NCBI Short Read Archive. https://www.ncbi.nlm.nih.gov/sra/?term=SAMN26342519. Deposited 7 June 2022. [DOI] [PMC free article] [PubMed]
- 78.Pan M., et al., Genomic and epigenetic landscapes drive CRISPR-based genome editing in Bifidobacterium. NCBI Short Read Archive. https://www.ncbi.nlm.nih.gov/sra/?term=SAMN26342528. Deposited 7 June 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and DNA-seq data of Bifidobacteria studied in this paper was deposited in the NCBI Short Read Archive database, under the BioProject ID PRJNA811526 (76). The SRA accessions number are SAMN26342519–SAMN26342528 (77, 78). The raw Nanopore sequencing native and amplified current difference files and metagenomic binning score outputs are available upon request.