Significance
Hoogsteen base pairs, which are thermodynamically less stable than the canonical Watson-Crick base pairs, often feature in complexes of DNA with proteins and small molecules, where they play key roles in DNA recognition, repair, and replication. Distinguishing between Hoogsteen and Watson-Crick base pairs in large DNA complexes and assemblies presents significant challenges for traditional structural biology techniques. Here, we demonstrate that Hoogsteen base pairs can be detected if present and distinguished from Watson-Crick base pairs in a wide range of DNA systems based on characteristic NMR chemical shifts and dipolar couplings by using multidimensional dynamic nuclear polarization-enhanced solid-state NMR spectroscopy.
Keywords: Hoogsteen base pairing, DNA, nucleosome, solid-state NMR, dynamic nuclear polarization
Abstract
The majority of base pairs in double-stranded DNA exist in the canonical Watson-Crick geometry. However, they can also adopt alternate Hoogsteen conformations in various complexes of DNA with proteins and small molecules, which are key for biological function and mechanism. While detection of Hoogsteen base pairs in large DNA complexes and assemblies poses considerable challenges for traditional structural biology techniques, we show here that multidimensional dynamic nuclear polarization–enhanced solid-state NMR can serve as a unique spectroscopic tool for observing and distinguishing Watson-Crick and Hoogsteen base pairs in a broad range of DNA systems based on characteristic NMR chemical shifts and internuclear dipolar couplings. We illustrate this approach using a model 12-mer DNA duplex, free and in complex with the antibiotic echinomycin, which features two central adenine-thymine base pairs with Watson-Crick and Hoogsteen geometry, respectively, and subsequently extend it to the ∼200 kDa Widom 601 DNA nucleosome core particle.
In the canonical DNA double helix, Watson-Crick base pairs (bps) exist in a dynamic equilibrium with short-lived (<1 ms) low-abundance (<1%) Hoogsteen conformations (1). Starting from a Watson-Crick guanine-cytosine (G-C) or adenine-thymine (A-T) bp, the corresponding Hoogsteen conformation can be obtained by flipping the purine base 180° around its glycosidic bond and then bringing the two bases into close proximity to create a new set of hydrogen bonds (Fig. 1B). While Hoogsteen bps exist as minor conformations in naked DNA duplexes, they have been observed as the dominant conformation in crystal structures of B-DNA in complex with proteins (2, 3) and drug molecules (4), where they play unique roles in DNA recognition. Hoogsteen bps have also been observed in structures of B-DNA duplexes containing damaged nucleotide bases, where they are believed to play roles in damage accommodation (5, 6), recognition, and repair (7), as well as in the active sites of Y-family low-fidelity polymerases, which replicate DNA using Hoogsteen base pairing as a means of bypassing mutagenic lesions on the Watson-Crick face of nucleotide bases (8, 9). In addition, Hoogsteen bps have been shown to increase the susceptibility of double-stranded DNA to damage (10, 11).
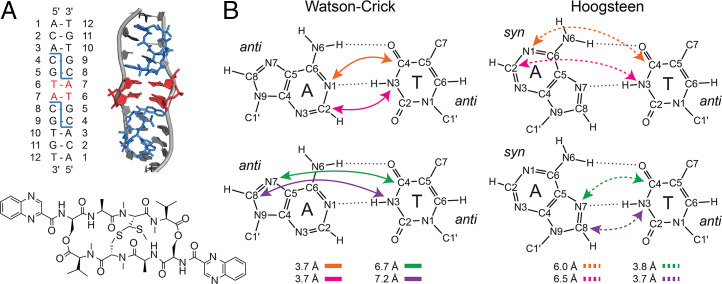
Fig. 1.
Model DNA duplex and Watson-Crick and Hoogsteen A-T base pairs (bps). (A) The double helix structure of 13C,15N-6T-7A-labeled nucleotide, which has two echinomycin C-G binding sites (4C-5G and 8C-9G). The bound echinomycin molecules are highlighted in blue and the trapped Hoogsteen bps are highlighted in red. The chemical structure of echinomycin is also shown. (B) The structures of Watson-Crick and Hoogsteen A-T bps with key internuclear distances are highlighted in colored arrows with solid lines (Watson-Crick) or dashed lines (Hoogsteen).
Hoogsteen bps are often observed in stressed regions of DNA structure, in which they appear to lubricate the DNA by providing an alternative conformation when the canonical Watson-Crick conformation comes under stress (12, 13). Considering that many proteins induce large distortions in the DNA upon complex formation, it is surprising that Hoogsteen bps have not been more widely observed in structures of DNA–protein complexes. A number of studies have documented that, depending on the quality of the electron density, it can be difficult to resolve Hoogsteen from Watson-Crick bps using X-ray crystallography (3, 14, 15). Such ambiguity can arise even in high-resolution structures due to poor electron density at specific local sites (14). Thus, it remains conceivable that many Hoogsteen bps in crystal structures of DNA–protein complexes have been potentially mismodeled as Watson-Crick bps. Indeed, a recent study identified several X-ray crystal structures of DNA–protein complexes in which one or two Hoogsteen base pairs were mismodeled as Watson-Crick (16). These Hoogsteen bps were observed in stressed regions of the DNA, including near-mismatches, nicks, and lesions.
In this regard, the nucleosome core particle (NCP), the basic repeating unit of chromatin, provides an interesting test case and a starting point for exploring the occurrence of Hoogsteen bps within the genome. Nucleosomes govern the accessibility of the genetic code to transcriptional machinery (17). They consist of a spool-like, highly conserved histone octamer protein core, around which a 145- to 147-bp DNA duplex is wrapped around 1.65 superhelical turns. Approximately 75% of eukaryotic DNA in the nucleus is wrapped around the histone core proteins. In the NCP, B-DNA experiences significant curvature (∼45° per helical turn), underwinding, and stretching (18, 19). These forces stress B-DNA and potentially promote alternative base pairing conformations, such as Hoogsteen bps. There are ∼180 NCP X-ray structures deposited in the Protein Data Bank (PDB) (20), and in all cases, the bps are modeled exclusively as Watson-Crick rather than Hoogsteen. These studies have contributed to the present view that bps in chromatin are purely Watson-Crick.
It has previously been noted that the DNA electron density in existing high-resolution crystal structures of NCPs is, in general, much lower than the protein electron density (21). For example, in one of the NCP structures containing a Widom 601 DNA sequence (22) (PDB entry 3LZ1, with overall resolution of 2.5 Å), while the DNA electron density is unambiguously modeled as Watson-Crick, up to 28% of bps have ambiguous electron density (15) (SI Appendix, Fig. S1). Therefore, discriminating between Watson-Crick and Hoogsteen bps in NCPs can be challenging based solely on X-ray electron density. The weak electron density at certain DNA bps could reflect DNA dynamics possibly related to increased propensities to form Hoogsteen bps. Compounding these limitations is the fact that high-resolution crystal structures are only available for a small number of DNA sequences that satisfy the crystallization requirement. Given that Hoogsteen bps are strongly dependent on sequence (23), a broader exploration of bps over many DNA sequence contexts is needed to assess the presence or absence of Hoogsteen bps in NCPs and chromatin.
Given the aforementioned challenges, there is an urgent need for complimentary experimental techniques that enable the identification of Hoogsteen bps in large DNA–protein complexes such as nucleosomes, without the limitations posed by crystallization. Cryoelectron microscopy (cryo-EM), while obviating the need for crystallization, suffers from the same problem as X-ray crystallography of potentially having ambiguous electron density for DNA bps. Although solution-state NMR can visualize the conformations of DNA bps and their dynamics (1), nucleosomes and other high-molecular-weight DNA–protein complexes exceed the size limit of solution NMR by roughly an order of magnitude. However, magic angle spinning (MAS) solid-state NMR does not suffer from inherent limitations related to molecular size and allows high-resolution spectra to be collected for large assemblies of biological macromolecules (24–26). For example, high-resolution MAS NMR spectra of condensed nucleosomes and nucleosome arrays containing 13C,15N-labeled histone proteins and unlabeled DNA have been reported in several recent studies focused on the analysis of histone protein structure, dynamics, and interactions in chromatin (27–31). Moreover, while the vast majority of applications of biomolecular MAS solid-state NMR to date have been to peptides and proteins, the utility of this methodology toward detailed atomic-level characterization of nucleic acids and nucleic acid–protein complexes has also been demonstrated (32–37).
Here, we illustrate the potential of using solid-state NMR coupled with dynamic nuclear polarization (DNP) (38), which has been applied toward the structural analysis of numerous proteins and protein assemblies (38, 39), to obtain atomic-level information regarding base pairing in DNA on the basis of 13C and 15N chemical shifts and interbase 13C-15N dipolar couplings (Fig. 1B). We focus specifically on A-T bps, given that A-T Hoogsteen bps are more energetically favored (by ∼1 kcal/mol) relative to G-C+ Hoogsteen bps, under physiological conditions (1). DNP solid-state NMR is ideally suited for this purpose, and this methodology has recently been successfully used to investigate several nucleic acid and DNA/RNA–protein systems (40–44). In addition to offering NMR sensitivity enhancements of ∼1 to 2 orders of magnitude, which are crucial for studies of DNA complexes in amount-limited samples potentially containing different bp conformers, the fact that DNP solid-state NMR measurements are typically conducted at low temperatures (∼100 K or below) makes this approach particularly well positioned to generally address this problem since, in principle, it enables direct observation of “frozen out” noncanonical Hoogsteen DNA bp conformations that at ambient temperatures exist in dynamic equilibrium with Watson-Crick bps. We initially apply this DNP solid-state NMR-based approach to a model DNA duplex containing two central 13C,15N-labeled A-T bps, which convert from Watson-Crick to Hoogsteen conformation upon the binding of an antibiotic molecule echinomycin (45) and subsequently extend it to probe DNA bp conformation in nucleosomes (∼200 kDa) reconstituted with 13C,15N-labeled DNA and natural abundance histone proteins.
Results
Model DNA Duplex with Watson-Crick and Hoogsteen Base Pairing.
To establish the DNP solid-state NMR approach, we used a 12-mer DNA duplex (5′-ACACGTACGTGT-3′) to model the Watson-Crick and Hoogsteen conformations (Fig. 1A). Prior studies (45–47) have shown that two echinomycin molecules bind to the CpG steps in this Watson-Crick duplex to induce two A-T Hoogsteen bps in the central TpA step. Echinomycin shows limited direct interactions with the Hoogsteen bps, inducing relatively small chemical shift perturbations (<2 ppm) around the binding sites (45), thus providing a means of measuring the NMR spectroscopic signatures of A-T Hoogsteen conformation in the context of double-stranded DNA. The 12-mer DNA duplex in the absence of echinomycin serves as the corresponding A-T Watson-Crick control sample.
The DNA duplex was prepared using chemical synthesis to uniformly enrich the central A and T residues involved in the two A-T Hoogsteen bps with 13C and 15N isotopes, and the DNA–echinomycin complex was formed as described previously (45). Solution-state NMR was used to confirm that the duplex adopts a Watson-Crick conformation in the absence of echinomycin and that the two central A-T bps adopt Hoogsteen conformations when bound to echinomycin, identified by the ∼2.5 ppm downfield shift of the adenine C8 resonance (SI Appendix, Table S1), in line with our previous study (45). Samples for the DNP solid-state NMR measurements were generated to closely mimic those used for the solution-state NMR studies and consisted of the DNA duplex with and without bound echinomycin at concentrations of ∼3–4 mM and the biradical polarizing agent AMUPol (48) in a phosphate-buffered glycerol–water mixture, as described in the Materials and Methods section.
DNP Enhanced Solid-State NMR Spectra of DNA Duplex Samples.
To establish the feasibility of using DNP solid-state NMR to monitor bp conformations in DNA, we commenced our study with experiments on the model duplex at natural abundance isotope concentrations. DNP solid-state NMR experiments on biological samples typically use a solvent matrix consisting of deuterated glycerol, D2O, and H2O in a 60:30:10 vol/vol/vol ratio (38). Given that imino protons can readily exchange with water (49), we investigated whether the use of a solvent matrix that does not contain any D2O would permit reasonable DNP NMR signal enhancements to be obtained for these DNA samples. A comparison of the 13C cross-polarization MAS (CP-MAS) NMR spectra of the DNA duplex recorded in the D2O-containing solvent matrix and those in the corresponding solvent matrix containing H2O in place of D2O (SI Appendix, Fig. S2) shows significant 13C NMR signal enhancements for samples in both solvent matrices in spectra recorded with the microwaves (MWs) turned on versus corresponding spectra with the MWs turned off. The DNP enhancements, εDNP = Ion/Ioff – 1, where Ion and Ioff are the NMR signal intensities integrated over the entire spectral window with MWs turned on and off, respectively, were found to be on the order of ∼100. Importantly, for the deuterated glycerol/H2O solvent matrix, the resonance intensities for most 13C sites, including those located in the vicinity of the imino protons, are comparable to or exceed the corresponding 13C resonance intensities for the deuterated glycerol/D2O/H2O matrix, indicating that the absence of D2O in the solvent matrix does not preclude the acquisition of high-sensitivity DNP-enhanced solid-state NMR spectra for the DNA samples.
Next, we explored the spectral changes induced by the binding of echinomycin to the DNA duplex at natural abundance isotope concentrations. Comparison of the DNP-enhanced 13C CP-MAS NMR spectra of the DNA duplex with and without bound echinomycin (SI Appendix, Fig. S3A) reveals changes in several 13C resonance frequencies in the base and sugar spectral regions, presumably indicative of the conversion from Watson-Crick to Hoogsteen conformation for the two central A-T bps. However, spectral overlap with the remaining Watson-Crick bps prevents the verification of Hoogsteen bps in this sample. Moreover, a number of additional 13C resonances appear in the ∼20- to 50-ppm and the ∼120- to 180-ppm spectral regions for the DNA–echinomycin complex, corresponding to bound echinomycin molecules, which are absent in the free DNA duplex.
Finally, relative to natural abundance DNA, the 13C,15N isotope enrichment of central A and T residues in the DNA duplex results in major increases in 13C spectral sensitivity for these residues for both free DNA (Fig. 2A) and the DNA–echinomycin complex (Fig. 2C), with associated εDNP values of 156 and 102, respectively. Remarkably, 13C CP-MAS NMR spectra of the 13C,15N-labeled DNA samples with and without bound echinomycin exhibit clear and unambiguous differences, which not only confirms a conformational change due to the binding of echinomycin to the 12-mer DNA duplex but also reveals that A-T Watson-Crick and Hoogsteen bps have distinct spectral signatures.
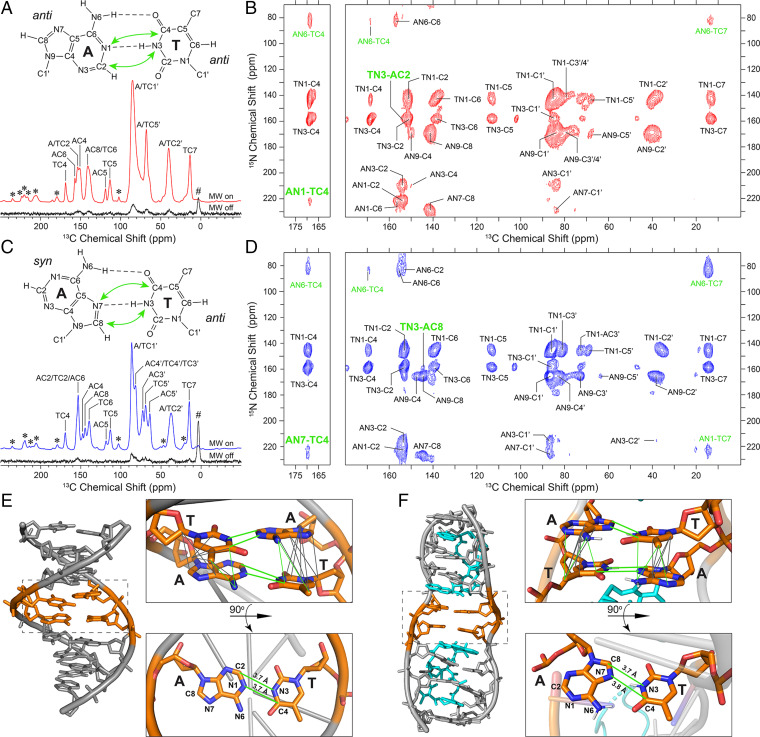
Fig. 2.
DNP solid-state NMR analysis of Watson-Crick and Hoogsteen DNA duplexes. (A) 13C CP-MAS solid-state NMR spectra of the 13C,15N-TA DNA duplex with Watson-Crick base pairing recorded with microwaves (MWs) on (red) and off (black; the MW off spectrum is shown at 10× intensity). The use of DNP yields an NMR signal enhancement (εDNP) of 156. Resonance assignments determined based on 15N-13C TEDOR and 13C-13C DARR spectra are indicated (for details, see text and SI Appendix, Figs. S4 and S5). Spinning sidebands and background signal arising from a silicone rotor insert are denoted by (*) and (#), respectively. (B) DNP enhanced 15N-13C ZF-TEDOR (Right) and band-selective TEDOR (Left) solid-state NMR spectra of the 13C,15N-TA DNA duplex recorded with dipolar mixing times of 10 ms and total measurement time of 96 h per spectrum. For band-selective TEDOR, the frequency selective 13C 180° refocusing pulses were applied at 169 ppm, corresponding to TC4 resonance frequency. The A-T 15N-13C correlations are denoted in green in the spectra, with correlations that provide direct spectroscopic signatures of the bp conformation highlighted in large bold font and indicated with green arrows in the bp structure in (A). (C and D) Same as (A and B), but for the 13C,15N-TA DNA duplex bound to echinomycin with Hoogsteen conformation for the two central A-T bps (εDNP = 102 for the 13C CP-MAS spectrum). (E and F) DNA duplex structures with Watson-Crick (E) and Hoogsteen (F) conformation for the central A-T bps highlighted in orange. The structure in (E) was generated using 3DNA, assuming an idealized B-form geometry (54), and the structure in (F) corresponds to the experimental crystal structure (PDB entry 1XVN) (4). Mapped onto the structures are the dipolar coupling-based A-T correlations observed in 15N-13C TEDOR (green) and 13C-13C DARR (gray) solid-state NMR spectra, corresponding to the shortest interatomic distance in the structure.
Chemical Shift Assignments of A-T Watson-Crick and Hoogsteen bps in DNA Duplex Samples.
Two-dimensional (2D) z-filtered transferred echo double-resonance (ZF-TEDOR) DNP solid-state NMR experiments (51), which generate 13C-15N correlations via polarization transfers based on corresponding through-space 13C-15N dipolar couplings, were performed to establish the 13C and 15N chemical shift assignments for A-T Watson-Crick and Hoogsteen bps. ZF-TEDOR spectra recorded with short dipolar mixing times of 1.33 ms (SI Appendix, Fig. S4) primarily contain correlations corresponding to directly bonded 13C-15N pairs and those separated by two bonds, allowing the nearly complete chemical shift assignments of A-T Watson-Crick and Hoogsteen base 13C and 15N resonances as well as sugar C1′ and C2′ resonances. Additional chemical shifts, such as TC7 and sugar C3′-C5′, are extracted from longer mixing time 15N-13C ZF-TEDOR (Fig. 2B and D) and 13C-13C dipolar assisted rotational resonance (DARR) spectra (SI Appendix, Fig. S5). Relative to solution NMR experiments, which typically only detect the protonated carbons and nitrogens, the DNP-enhanced ZF-TEDOR solid-state NMR spectra readily yield the chemical shifts for all resolved base and sugar 13C and 15N sites, thereby dramatically increasing the amount of data that may be used to discriminate Watson-Crick and Hoogsteen bp conformations. In summary, all A and T base and sugar carbons and nitrogens were unambiguously assigned for the model Watson-Crick and Hoogsteen DNA duplex samples (SI Appendix, Table S1), with the exception of AC3′-C5′ and TC3′-C5′ for the Watson-Crick form due to spectral overlap. The latter is to be expected given that the Watson-Crick double helix features similar sugar backbone conformations, leading to indistinguishable chemical shifts for these A and T sugar carbons. Note that the majority of resonances in the Watson-Crick and Hoogsteen DNA duplex samples were found to have uncertainties of less than 0.5 ppm in the 13C and 15N chemical shift values and linewidths in the ∼1.5 to 3 ppm and ∼4 to 6 ppm range for 13C and 15N, respectively (SI Appendix, Table S1). The observed resonance linewidths are consistent with those reported in prior low-temperature DNP solid-state NMR studies of nucleic acids and other biomolecules (39, 41), and, as expected (39), exceed those that can be obtained in solid-state NMR spectra recorded at ambient temperature for microcrystalline nucleic acid samples (36, 41).
Prior solution state NMR studies of model DNA duplexes have established several 13C chemical shift signatures that can reliably distinguish A and T Watson-Crick versus Hoogsteen bp conformations (5, 45), including ∼2 to 3 ppm downfield shifted AC1′ and AC8 for Hoogsteen bps relative to Watson-Crick bps. Chemical shift differences (Δδ) of this magnitude between Watson-Crick and Hoogsteen bps are robustly observed in a wide variety of sequence contexts for both A-T and G-C+ Hoogsteen bps (1, 23) stabilized by chemical modification (1, 5, 6) or through the binding of echinomycin (45, 46), as well as transient Hoogsteen bps in naked unmodified DNA (1, 5). These shift differences could be rationalized using density functional theory (DFT) calculations as arising due to changes in the local conformation at the bp, including changes in χ angle at the glycosidic bond, base pairing hydrogen bonding, and protonation of cytosine (1, 52). Altogether, these findings imply that these chemical shift signatures report primarily on the bp conformation and do not simply result from changes in local electronic structure at the base and sugar sites due to direct interactions with the bound echinomycin ligand. In addition, echinomycin does not appear to engage in any specific interactions with the A-T Hoogsteen bps in the 12-mer DNA duplex used for this study (45). These unique chemical shift signatures were also observed in the present study in DNP solid-state NMR spectra of the model DNA duplex samples, with Δδ values for Hoogsteen versus Watson-Crick bps of 2.74 ppm for AC1′ and 3.28 ppm for AC8 (SI Appendix, Table S1 and Fig. S6).
Importantly, the solid-state NMR experiments provide a number of additional Δδ signatures, beyond those accessible by solution NMR, that may be used to discriminate between Watson-Crick and Hoogsteen bps with high confidence in a wide range of DNA systems. For example, we observed the following substantial 13C and 15N chemical shift differences for Hoogsteen versus Watson-Crick bps: ∼7 ppm downfield for AN3, ∼3 ppm upfield for AN7, ∼3.5 ppm upfield for AN9, ∼3 ppm downfield for TN1, and ∼4 ppm upfield for TC2′ (SI Appendix, Table S1). The sizeable bp conformation-dependent chemical shift changes for adenine likely stem from the change from an anti to a syn conformer due to the rotation of the purine about the glycosidic bond, whereas thymine generally experiences smaller chemical shift changes, particularly within the base, which remains anti in both Watson-Crick and Hoogsteen conformations (5). While a direct comparison of these additional solid-state NMR Hoogsteen versus Watson-Crick bp Δδ signatures to solution NMR data are not feasible, many of them are found to be in remarkably good agreement with the corresponding Δδ values predicted by DFT calculations for Watson-Crick and Hoogsteen duplex DNA structures carried out in the absence of echinomycin (SI Appendix, Fig. S6), despite the inherent limitations on the accuracy of DFT chemical shift predictions when modeling biomolecules as single structures rather than dynamic ensembles (5, 53). The latter further underscores the notion that the A-T bp conformation, rather than a direct interaction with the bound echinomycin ligand, is primarily responsible for the base and sugar 13C and 15N chemical shift differences observed in the model Watson-Crick and Hoogsteen DNA duplexes. Notably, several of these signature chemical shift changes correspond to directly bonded 13C and 15N nuclei, such as AN3-C4, AN7-C8, AN9-C4, and AN9-C8, and are strongly correlated with one another. This leads to significant shifts in 15N-13C resonance frequencies, by amounts that considerably exceed the uncertainties in experimental 13C and 15N chemical shifts (SI Appendix, Table S1), and migration of cross-peaks to empty regions of the 2D ZF-TEDOR spectrum (SI Appendix, Fig. S4), which can generally serve as spectral fingerprints of A-T Hoogsteen versus Watson-Crick bp conformation and even facilitate the observation of a minor population of A-T Hoogsteen bps in the presence of a predominant Watson-Crick conformer (or vice versa). Indeed, this possibility of concurrent detection of Watson-Crick and Hoogsteen bp conformers in the same sample by the low-temperature DNP solid-state NMR approach described in the present study is supported by our data for the duplex DNA–echinomycin complex. Specifically, close inspection of the 2D ZF-TEDOR spectra in SI Appendix, Fig. S4C reveals that the spectrum for the Hoogsteen DNA duplex (blue contours) contains several minor “residual” cross-peaks at 15N-13C frequencies associated with signature Watson-Crick bp correlations (e.g., AN7-C8, AN9-C8), in addition to the major Hoogsteen bp cross-peaks. These residual cross-peaks most likely stem from populations of Hoogsteen and Watson-Crick bp conformers that are engaged in dynamic exchange at ambient temperature but frozen out under low-temperature conditions used for the DNP solid-state NMR measurements. Prior solution NMR studies (46) have found the Hoogsteen bps in this duplex 12-mer DNA–echinomycin complex to be dynamic, and a more recent quantitative relaxation dispersion solution NMR study of this system (45) indicates that for the central A-T bps the major Hoogsteen conformer undergoes exchange on the millisecond time scale with a minor Watson-Crick state with a population of ∼3 to 4%.
Probing 15N-13C Dipolar Couplings in A-T Watson-Crick and Hoogsteen bps in DNA Duplex Samples.
Inspection of A-T Watson-Crick and Hoogsteen bp structures indicates that several distances between 13C and 15N atoms for the two bases are highly characteristic for each conformation (Fig. 1B). Namely, the AN1-TC4 and TN3-AC2 distances are both ∼3.7 Å for Watson-Crick bps (both distances increase to ∼6 to 6.5 Å for Hoogsteen bps), and, similarly, the AN7-TC4 and TN3-AC8 distances are ∼3.7 to 3.8 Å for Hoogsteen bps (both distances increase to ∼7 Å for Watson-Crick bps). In principle, 15N-13C through-space dipolar couplings corresponding to the ∼3.5- to 4-Å distances should be detectable by using TEDOR solid-state NMR experiments (51) recorded with longer (∼5 to 10 ms) mixing times, while dipolar couplings associated with the ∼6- to 7-Å distances are too weak to generate observable 15N-13C cross-peaks. In addition, the unambiguous chemical shift assignments of the aforementioned 15N and 13C sites, in the Watson-Crick and Hoogsteen DNA duplex samples, are expected to facilitate the observation of the A-T 15N-13C correlations of interest.
The rightmost panels of Fig. 2B and D show 15N-13C ZF-TEDOR spectra recorded with a dipolar mixing time of 10 ms for the model Watson-Crick and Hoogsteen DNA duplex samples, respectively. The spectrum for the Watson-Crick DNA duplex contains three A-T 15N-13C correlations, TN3-AC2 (appearing as a downfield shoulder adjacent to the TN3-C2 cross-peak), AN6-TC4, and AN6-TC7. The spectrum for the Hoogsteen DNA duplex contains four A-T 15N-13C cross-peaks, TN3-AC8, AN1-TC7, AN6-TC4, and AN6-TC7. Notably absent from these spectra, however, are the expected cross-peaks corresponding to distances in the ∼3.7- to 3.8-Å regime involving the TC4 site, AN1-TC4 (Watson-Crick) and AN7-TC4 (Hoogsteen). Given that the 15N-13C cross-peak intensities in ZF-TEDOR experiments are modulated by homonuclear 13C-13C J-couplings and consequently are significantly attenuated, we recorded analogous band-selective TEDOR spectra (51) for the DNA duplex samples with a 10-ms dipolar mixing time and frequency-selective 13C 180° pulses applied at the resonance frequency of TC4 to refocus the ∼65-Hz TC4-C5 J-coupling (54) (Fig. 2B and D, leftmost panels). These band-selective TEDOR spectra were found to contain the correlations characteristic of Watson-Crick and Hoogsteen bps, AN1-TC4 and AN7-TC4, respectively, as well as AN6-TC4 cross-peaks that are more intense relative to those observed in the ZF-TEDOR spectra.
Interpretation of 15N-13C dipolar contacts in terms of bp topology must consider the fact that both central A-T pairs are 13C,15N-labeled for the model DNA duplexes used in this study. This means that, in general, the correlations observed in TEDOR spectra recorded for these samples will contain contributions from both A and T bases located in different DNA strands and constituting the bp as well as from successive A and T bases of the same strand. Table 1 lists the inter- and intrastrand A-T 15N-13C distances for the model Watson-Crick and Hoogsteen DNA duplexes, associated with correlations observed in the TEDOR solid-state NMR spectra. These data reveal that the AN1-TC4 and TN3-AC2 (AN7-TC4 and TN3-AC8) correlations for the model Watson-Crick (Hoogsteen) DNA duplex are primarily interstrand in nature and therefore provide direct spectroscopic signatures that report on the bp conformation; note that the ∼3.7- to 3.8-Å interstrand A-T distances are associated with 15N-13C dipolar coupling constants of ∼60 Hz, which exceed by a factor of ∼1.5 to 2 the magnitudes of corresponding intrastrand couplings, given the inverse cube dependence of the dipolar coupling constant magnitude on the internuclear distance. However, the AN6-TC4 correlations observed for the two model DNA duplex samples arise largely from ∼3.3- to 3.5-Å intrastrand contacts and do not report on the bp conformation. Irrespective of this, the AN6-TC4 cross-peaks could not be used to reliably distinguish between Watson-Crick and Hoogsteen bps since these two conformers are associated with nearly identical interstrand AN6-TC4 distances in the ∼3.7- to 3.9-Å range. Similarly, the AN1-TC7 and AN6-TC7 correlations for both DNA duplex samples arise from ∼3.5- to 4.5-Å intrastrand contacts, with the corresponding interstrand contacts being associated with 15N-13C distances in the ∼6- to 8-Å regime and beyond the detection limit of TEDOR experiments. Finally, in addition to the 15N-13C TEDOR spectra, we recorded long (500 ms) mixing time 13C-13C DARR spectra for both DNA duplex samples (SI Appendix, Fig. S5). While these spectra contain a multitude of cross-peaks, including a considerable number of A-T correlations involving base and sugar 13C nuclei (9 and 10 for the Watson-Crick and Hoogsteen DNA duplex samples, respectively), all of these A-T correlations are associated with intrastrand distances, which are significantly shorter than the corresponding interstrand distances between the same 13C sites (SI Appendix, Table S2) and hence do not report directly on the base pair conformation. Fig. 2E and F, respectively, show the A-T contacts observed in 15N-13C TEDOR and 13C-13C DARR spectra, mapped onto the Watson-Crick and Hoogsteen DNA duplex structures.
Table 1.
Inter- and intrastrand A-T distances for model Watson-Crick and Hoogsteen DNA duplexes
| Atoms | Watson-Crick | Hoogsteen | |||
|---|---|---|---|---|---|
| Interstrand distance (Å) | Intrastrand distance (Å) | Interstrand distance (Å) | Intrastrand distance (Å) | ||
| AN1 | TC4 | 3.7 | 4.4 | 6.0 | 4.1 |
| AN1 | TC7 | 6.2 | 6.3 | 8.0 | 3.8 |
| AN6 | TC4 | 3.7 | 3.5 | 3.9 | 3.3 |
| AN6 | TC7 | 6.1 | 4.6 | 5.7 | 3.7 |
| AN7 | TC4 | 6.7 | 4.5 | 3.8 | 4.1 |
| TN3 | AC2 | 3.7 | 4.3 | 6.5 | 5.1 |
| TN3 | AC8 | 7.2 | 4.8 | 3.7 | 4.1 |
Distances corresponding to A-T 15N-13C correlations observed in TEDOR solid-state NMR spectra of model Watson-Crick or Hoogsteen DNA duplexes are highlighted in bold font. Note that while, in general, both inter- and intrastrand 15N-13C dipolar couplings contribute to the cross-peaks, the primary contribution arises from the largest coupling associated with the shortest interatomic distance (see Results section for details). Distances for the Watson-Crick DNA duplex were determined from a structure generated using 3DNA, assuming an idealized B-form geometry (54). Distances for the Hoogsteen DNA duplex were determined from the crystal structure (4).
Probing DNA Base Pairing in Nucleosomes.
To assess the potential of DNP solid-state NMR to probe base pairing in chromatin and other large DNA–protein complexes, we applied this methodology to an NCP. The NCP sample was prepared by reconstituting uniformly 13C,15N-labeled 147-bp Widom 601 DNA (22) with natural abundance histone octamer, composed of two copies each of histone H2A, H2B, H3, and H4 proteins (Fig. 3A and SI Appendix, Figs. S7 and S8), as described in the Materials and Methods section. The sucrose gradient–purified NCP shows the expected shift in DNA electrophoretic mobility on an acrylamide gel upon binding histone octamer (SI Appendix, Fig. S9). For the DNP solid-state NMR measurements, the NCP sample was pelleted by ultracentrifugation and resuspended in a mixture of 60:40 d8,12C-glycerol:H2O (vol/vol) containing the AMUPol polarizing agent. DNP enhanced 13C and 15N CP-MAS solid-state NMR spectra for the NCP sample are shown in Fig. 3B and C, respectively; DNP enhancement of 122 was found for the 13C spectrum, consistent with εDNP values in the ∼100 to 150 regime obtained for the model DNA duplex samples (cf., Fig. 2). Fig. 3D shows a DNP enhanced 15N-13C ZF-TEDOR solid-state NMR spectrum of NCPs reconstituted with 13C,15N-labeled Widom 601 DNA recorded with a dipolar mixing time of 1.33 ms. The quality of this spectrum is remarkable given the large molecular size of the NCP (∼200 kDa) and the fact that the sample used for the solid-state NMR measurements contains only ∼1 to 1.5 mg (∼10 to 15 nmol) of DNA, and, to the best of our knowledge, these data correspond to the initial observation of any unmodified DNA signals in NCPs by NMR spectroscopy.
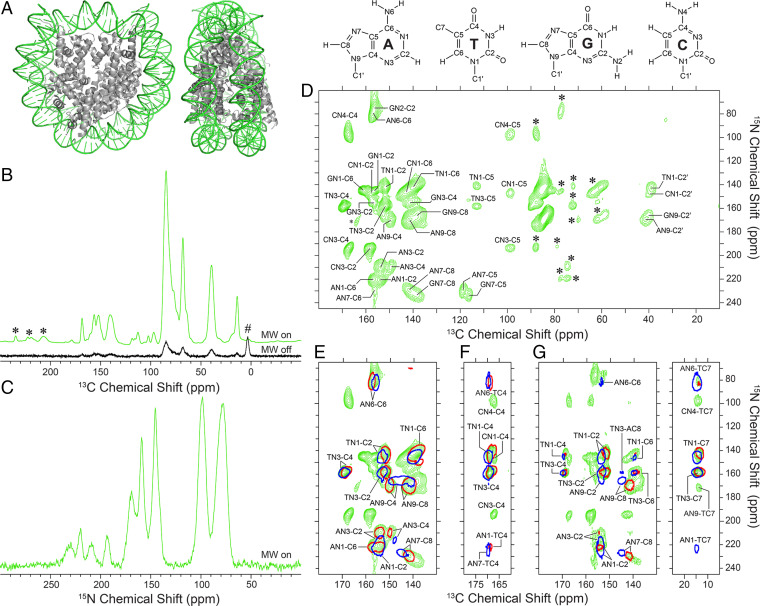
Fig. 3.
DNP solid-state NMR analysis of Widom 601 DNA nucleosomes. (A) X-ray crystal structure of the nucleosome core particle (NCP) with Widom 601 DNA (PDB entry 3LZ1) (56). The DNA is colored in green and histones H2A, H2B, H3, and H4 are gray. (B) 13C and (C) 15N DNP enhanced CP-MAS solid-state NMR spectra of the NCP sample reconstituted with U-13C,15N-DNA (green). For the 13C spectrum, spinning sidebands and background signal arising from a silicone rotor insert are denoted by (*) and (#), respectively, and comparison of the MWs on spectrum with a spectrum recorded with MWs off (black; shown at 10× intensity) yields a εDNP value of 122. The 15N MW off spectrum had negligible intensity and is not shown. (D) DNP enhanced 15N-13C ZF-TEDOR solid-state NMR spectrum of the NCP sample recorded with a dipolar mixing time of 1.33 ms and total measurement time of 40 h, with resonance assignments for A, T, G, and C base and sugar sites indicated. (E–G) Small regions of NCP 15N-13C spectra (green contours) including (E) ZF-TEDOR (1.33 ms dipolar mixing time), (F) band-selective TEDOR (10 ms dipolar mixing time) with frequency-selective 13C 180° refocusing pulses applied at the TC4 resonance frequency (169 ppm) and (G) ZF-TEDOR (10 ms dipolar mixing time) recorded with total measurement times of 40, 53, and 53 h, respectively. Overlaid on the NCP spectra are the corresponding spectra of model DNA duplex samples with Watson-Crick (single red contour) and Hoogsteen (single blue contour) conformation for the central A-T bps.
Analogous to the model DNA duplex samples, the NCP 15N-13C ZF-TEDOR spectrum enabled complete chemical shift assignments to be established for all of the base 15N and 13C sites for each of the four nucleotide types in Widom 601 nucleosomal DNA. In addition, C2′ sites were unambiguously assigned for each type of nucleotide from the ZF-TEDOR dataset, and the TC7 methyl resonance at ∼14 ppm was readily identified in a 1D 13C CP-MAS spectrum and confirmed in a 2D 13C-13C DARR spectrum (SI Appendix, Fig. S10 and Table S3). While the DNA nucleosome positioning sequence used in the current study contains 31, 33, 36, and 47 A, T, G, and C nucleotides per strand, respectively, no residue-specific resonance assignments could be determined from the 15N-13C correlation spectrum given the similarity of the chemical shifts for each type of nucleotide. Interestingly, a comparison of the NCP 15N-13C ZF-TEDOR spectrum with that of free 13C,15N-labeled Widom 601 DNA in glycerol/water/AMUPol matrix (SI Appendix, Fig. S11) shows a small degree of broadening for multiple correlations upon NCP formation, indicative of minor chemical shift perturbations at different sites likely induced by DNA structural changes caused by wrapping around the histone octamer. These perturbations are in line with small base and sugar 13C chemical shift changes (up to ∼1 to 2 ppm) observed upon the formation of other DNA–protein complexes with DNA curvature comparable to that found in nucleosomes (56).
The finding that DNA 13C and 15N chemical shift perturbations resulting from NCP assembly are relatively minor immediately suggests that most bps in Widom 601 DNA NCPs, for which roughly one-third were found to have ambiguous electron density in a high-resolution crystal structure (cf., SI Appendix, Fig. S1) (15), are Watson-Crick (assuming that free Widom 601 DNA in the absence of histone octamer is fully Watson-Crick). In addition, this finding allows us to explore the possible presence of A-T Hoogsteen bps by examining in detail the spectral regions identified for the model DNA duplex samples to contain 15N-13C cross-peaks that can serve as unique signatures of Watson-Crick versus Hoogsteen bp conformation. In Fig. 3E we show a small region of the short mixing time 15N-13C ZF-TEDOR spectrum recorded for the NCP sample (cf., Fig. 3D), overlaid with the corresponding spectra for the Watson-Crick and Hoogsteen DNA duplexes (SI Appendix, Fig. S4). Longer mixing time (10 ms) ZF-TEDOR (Fig. 3G) and band-selective TEDOR (Fig. 3F) spectra were also recorded for the NCP sample, assigned based on the established NCP DNA chemical shifts (SI Appendix, Table S3) and compared with corresponding datasets for the model DNA duplex samples (cf., Fig. 2). Inspection of the short mixing time ZF-TEDOR spectra (cf., Fig. 3E) reveals a high degree of correspondence between the 15N-13C correlations for A and T bases in the model Watson-Crick DNA duplex and the corresponding cross-peaks for the NCP sample, further confirming that the A-T bps in nucleosomal Widom 601 DNA are Watson-Crick. Notably absent is evidence of any appreciable intensity in the relatively isolated regions of the spectrum corresponding to the signature A-T Hoogsteen bp correlations AN3-C4, AN7-C8, AN9-C4, and AN9-C8. While the spectral sensitivity attainable in our experiments does not absolutely exclude a scenario in which one or more A-T bps in Widom 601 DNA within nucleosomes can access low-populated minor states with Hoogsteen conformation, given that the NCP sample exhibits normalized per base signal-to-noise ratios in the ∼2 to 10 regime for the different A-T 15N-13C cross-peaks, our data indicate that it is unlikely that Widom 601 DNA NCPs contain one or more stable Hoogsteen A-T bps.
Interestingly, the long mixing time TEDOR spectra do contain several A-T and C-T correlations, including AN6-TC4, AN6-TC7, AN9-TC7, and CN4-TC7. Based on the data described above for the model DNA duplex samples (cf., Table 1), these correlations likely report on A-T and C-T base stacking involving adjacent nucleotides within the same DNA strand rather than on interstrand contacts within a hydrogen-bonded bp. For example, the average AN9-TC7 interstrand distance in the NCP is 10.1 Å (55), compared to average intrastrand distances of 4.1 and 7.5 Å between these sites for ApT and TpA steps, respectively, for which the ApT step can be expected to generate an observable 15N-13C cross-peak in a TEDOR spectrum. This result is also consistent with data for the model Watson-Crick DNA duplex, in which the AN9-TC7 correlation was not detected due to the fact that the sequence contains a TpA step with an intrastrand AN9-TC7 distance of 7.0 Å. Finally, it is worth noting that neither of the characteristic interstrand Watson-Crick bp dipolar contacts observed for the model DNA duplex, AN1-TC4 and TN3-AC2, could be detected for the NCP sample. This is due to insufficient spectral sensitivity and spectral crowding in the NCP sample, as these correlations were of relatively low intensity when compared to most of the remaining 15N-13C cross-peaks in the 12-mer Watson-Crick DNA duplex sample and the location of a TN3-AC2 cross-peak falling in a largely overlaid region of the NCP ZF-TEDOR spectrum.
Discussion
While the majority of bps in duplex DNA adopt the canonical Watson-Crick conformation, the thermodynamically less stable Hoogsteen bps can feature prominently, particularly for A-T pairs, in complexes of DNA with proteins and small molecules, where they play key roles in DNA recognition, repair, and replication (2–5, 7–11). However, observation of Hoogsteen bps poses a significant challenge to traditional structural biology techniques because, unlike epigenetic modifications, Hoogsteen bps lack unique chemical groups that may aid in their detection. As such, unambiguous experimental identification of Hoogsteen bps in DNA-protein complexes by X-ray crystallography and cryo-EM methods, even for relatively high-resolution structures, is hindered by insufficiently clear electron density at specific DNA sites (14, 15, 57). Solution-state NMR spectroscopy has been successful in characterizing bp topologies and conformational dynamics in various DNA sequences. Specifically, multiple previous studies have unequivocally established in a wide variety of contexts and sample conditions that, relative to Watson-Crick bps, Hoogsteen bps are characterized by unique chemical shift fingerprints in solution NMR spectra. This includes the observation of Hoogsteen bps in DNA–protein (56) and DNA–drug (45) complexes, in 5mC epigenetically modified G-5mC bps, and in damaged A-T and G-C bps (6, 58), across a wide range of temperature (1), pH (52), and sequence contexts (23), and even in the context of RNA (59). However, since solution NMR spectroscopy relies on rapid molecular tumbling, these experiments aimed at probing bp topologies are severely restricted by molecular size to relatively small DNA oligonucleotides and generally not applicable to high-molecular-weight DNA–protein systems such as nucleosomes, chromatin, or large complexes of DNA with transcription factors or damage repair proteins.
The present study establishes DNP-enhanced MAS solid-state NMR spectroscopy as a viable tool for exploring bp conformations in nucleic acids and observing Hoogsteen bps in a broad range of complexes and assemblies of DNA with small molecules and proteins, without any fundamental molecular weight limitations on the system of interest. It is also noteworthy that in comparison to analogous solution NMR experiments, DNP solid-state NMR permits virtually all nucleotide 13C and 15N chemical shifts to be readily visualized, thereby increasing by nearly an order of magnitude the number of available spectroscopic signatures that can be accessed for DNA systems. Specifically, we identify several key signatures of Watson-Crick and Hoogsteen bp conformations using a selectively 13C,15N isotope-labeled model 12-mer DNA duplex, free and in a complex with the antibiotic echinomycin, that enable these bp conformers to be clearly distinguished from one another in solid-state NMR spectra. These conformational fingerprints include (1) multiple adenine 13C and 15N chemical shifts, which differ on the order of ∼3 to 7 ppm between Watson-Crick and Hoogsteen bps due to 180° rotation of the base about the glycosidic bond, in particular those associated with directly bonded AN3-C4, AN7-C8, AN9-C4, and AN9-C8 pairs that yield strong cross-peaks in unique regions of 15N-13C solid-state NMR chemical shift correlation spectra, and (2) characteristic 15N-13C dipolar coupling-based correlations between the A and T nucleotides making up the bp, AN1-TC4/TN3-AC2 and AN7-TC4/TN3-AC8 for Watson-Crick and Hoogsteen bps, respectively, which may be detectable for DNA systems exhibiting sufficient sensitivity in DNP solid-state NMR spectra.
Subsequently, we extended this DNP solid-state NMR approach to evaluate A-T base pairing in the ∼200-kDa NCP, reconstituted with 13C,15N-labeled Widom 601 DNA and unlabeled histone protein octamer complex. In light of the ambiguous electron densities observed for a significant fraction of bps in crystallographic studies of Widom 601 DNA nucleosomes (15), the DNP solid-state NMR data indicate that A-T bps adopt predominantly Watson-Crick conformations and effectively exclude the possibility of stable Hoogsteen A-T bps, which provides valuable insights about the nature of DNA base pairing in this system. Finally, it is worth noting that these experiments were carried out for an NCP sample containing only ∼10 to 15 nanomoles of 13C,15N-labeled DNA and thus do not display sufficiently high spectral sensitivity to permit the detection of isolated A-T Hoogsteen bps present at low abundance (<10%) relative to the major Watson-Crick conformer.
When applied to large, uniformly 13C,15N-labeled DNA molecules, as exemplified by the NCP experiments in the present study, the DNP solid-state NMR approach described herein enables rapid sequence-wide screening for Hoogsteen bps, but is generally not expected to yield residue specific resolution. Nevertheless, our results bode well for future applications of this methodology toward the detection of Hoogsteen bps in a wide range of DNA systems, potentially with site-specific resolution when combined with targeted nucleotide isotope-labeling approaches facilitated by the ready availability of segmentally and/or selectively 13C,15N-labeled large synthetic DNA sequences and DNA ligation methods. Immediate future applications of this methodology that can be readily envisioned include analogous studies of NCPs and oligonucleosome arrays reconstituted with various nucleosome positioning DNA sequences other than Widom 601, aimed at investigating the possible presence of Hoogsteen bps in eukaryotic genomes and understanding whether Hoogsteen bps impart unique functionalities to DNA, as well as studies of other challenging DNA–protein complexes, such as DNA-p53 tumor suppressor protein and DNA–TATA box binding protein complexes, that play important roles in biological function and mechanism. Finally, we anticipate that ongoing developments of high and ultrahigh magnetic field DNP solid-state NMR spectroscopy, combined with continuous improvements in methods for selective isotope labeling of DNA and preparation of optimal samples for DNP solid-state NMR studies, may ultimately enable detection of functionally relevant, low-populated Hoogsteen bps that exist in dynamic equilibrium with Watson-Crick bps at ambient temperatures.
Materials and Methods
DNA Duplex Preparation.
The 12-mer DNA with sequence 5′-ACACGTACGTGT-3′, containing two echinomycin binding sites at C-G residues (positions 4 to 5 and 8 to 9), was selectively isotope labeled with 13C,15N-thymine and 13C,15N-adenine at positions 6 and 7, respectively. In the absence of the antibiotic echinomycin, this sequence forms a DNA duplex with canonical Watson-Crick base pairing. In the presence of echinomycin in a 2:1 molar ratio of echinomycin to DNA duplex, the two central A-T bps adopt a Hoogsteen conformation. For the model Watson-Crick DNA duplex, the 13C,15N-TA DNA was dissolved in aqueous solution containing 15 mM sodium phosphate, 150 mM NaCl, and 0.1 mM ethylenediaminetetraacetate (EDTA) at pH 6.8 and lyophilized. The lyophilized pellet was redissolved in a solution of d8,12C-glycerol, and H2O in a 60:40 vol/vol ratio containing 12 mM AMUPol, to a 13C,15N-TA DNA duplex concentration of 4.1 mM, and the sample was freeze-thawed 10 times to remove excess dissolved oxygen. For DNP solid-state NMR measurements, 23.5 μL of the sample solution was packed into a 3.2-mm Bruker sapphire rotor and sealed with a silicone plug and ceramic cap. For the model Hoogsteen DNA duplex, the 13C,15N-TA DNA echinomycin complex was generated as described previously (10), with the addition of echinomycin before air drying, and the sample for DNP solid-state NMR measurements containing the 13C,15N-TA DNA duplex at a 2.7-mM concentration was prepared as described above for the model Watson-Crick DNA duplex sample.
NCP Preparation.
The NCP sample for DNP solid-state NMR studies was prepared as follows based on established protocols (60, 61). Histones H2A, H2B, H3, and H4 were each expressed in Escherichia coli BL21 (DE3) pLysS cells using Luria-Bertani medium, harvested and purified using gel filtration and ion-exchange chromatography in 7 M urea, followed by dialysis against a solution of 2 mM β-mercaptoethanol (BME) in ultrapure water and lyophilized. The histone octamer was prepared by dissolving the four histones at concentrations of ≤10 mg/mL and H2A:H2B:H3:H4 molar ratio of 1.2:1.2:1:1 in unfolding buffer (20 mM Tris, 7 M guanidine hydrochloride, 10 mM dithiothreitol, pH 7.5), followed by double dialysis into refolding buffer consisting of 1× TE (10 mM Tris, 1 mM EDTA, pH 8.0), 2 M NaCl, and 5 mM BME, and purification by gel filtration chromatography in 1× TE, 2 M NaCl buffer. 13C,15N-labeled DNA was prepared by amplifying the pJ201 plasmid containing 32 repeats of the 147-bp Widom 601 nucleosome positioning sequence in E. coli DH5a using minimal media containing 13C-glucose and 15NH4Cl as the sole carbon and nitrogen sources. DNA was purified using Qiagen Giga kits and digested with EcoRV (New England Biolabs). The fragments corresponding to 147-bp Widom 601 DNA were separated from the parent plasmid by polyethylene glycol precipitation, followed by collection of the supernatant and further purification by ethanol precipitation, and the DNA purity was confirmed using a 1% agarose gel. NCPs were reconstituted by preparing an aqueous solution in a 0.5× TE, 2 M NaCl, 1 mM benzamidine (BZA) buffer containing histone octamer and ∼25% molar excess of DNA, followed by the removal of NaCl by double dialysis at 4 °C against a 0.5×-TE, 1-mM BZA buffer. The resulting solution was then concentrated to ∼2 mg/mL DNA using Amicon 30 kDa cutoff centrifugal filter devices and purified using a 5 to 30% sucrose gradient in 0.5× TE. The sucrose gradient fractions were analyzed on a 5% native acrylamide gel, and those containing pure NCPs were collected and combined. To generate the DNP solid-state NMR sample, a 3.5-mg/mL NCP solution in 0.5× TE, 100 mM NaCl, 0.1 mM MgCl2 at pH 7.0 was adjusted to a Mg2+ concentration of 46 mM, and ultra-centrifuged at 500,000 × g and 4 °C for 16 h to generate a ∼3.25-mg NCP pellet. The pellet was washed with two 30-µL aliquots of d8,12C-glycerol, and H2O in a 60:40 vol/vol ratio containing 12 mM AMUPol and then resuspended and packed into a 3.2-mm Bruker sapphire rotor and sealed with a silicone plug and ceramic cap, with the final sample for DNP solid-state NMR containing ∼2.5 mg NCP.
DNP Solid-State NMR Spectroscopy.
Experiments were performed on a 600-MHz/395 GHz Bruker Avance III HD wide-bore DNP solid-state NMR spectrometer equipped with a gyrotron and a 3.2-mm HXY low-temperature MAS (LT-MAS) probe. Samples were spun with cooled N2 gas at 10 or 12 kHz (±3 Hz), with temperatures ranging from 99 to 111 K. MW irradiation was applied continuously for the duration of NMR experiments at the optimal power for AMUPol, 130 mA applied current. Standard 13C and 15N CP-MAS solid-state NMR experiments were used to characterize each sample and measure the DNP enhancement, εDNP = Ion/Ioff – 1, where Ion and Ioff are the NMR signal intensities integrated over the entire spectral window with MWs turned on and off. The DNP buildup times, τDNP, for each sample were measured from saturation-recovery experiments; recycle delays were set to 1.256τDNP and were 4.6 s and 4.1 s for the DNA duplex and NCP samples, respectively. 2D z-filtered ZF-TEDOR experiments were recorded, with dipolar mixing times ranging from 1.33 to 10 ms, z-filter delays of 250 μs, evolution times of 7 and 30 ms in the indirect (15N) and direct (13C) dimensions, respectively, and SPINAL-64 1H decoupling (62) of 90 and 100 kHz during chemical shift evolution and dipolar transfer periods, respectively. 2D band-selective TEDOR spectra were recorded with parameters similar to those used to record the ZF-TEDOR spectra, with 1,167 μs rSNOB (63) 13C 180° refocusing pulses applied at the resonant frequency of TC4 (169 ppm). 2D 13C-13C DARR spectra were recorded with a mixing time of 500 ms, evolution times of 8.5 and 30 ms in indirect and direct dimensions, respectively, and SPINAL-64 1H decoupling of 90 kHz. NMR data were processed using NMRPipe (64) and analyzed using NMRDraw and Sparky (65). 13C and 15N chemical shifts were referenced to adamantane and ammonium chloride external standards, respectively (66, 67).
Automated Fragmentation Quantum Mechanics/Molecular Mechanics (QM/MM) calculations.
The structural model of the DNA–echinomycin complex with DNA sequence 5′-ACGTACGT-3′ was downloaded from the PDB (https://www.rcsb.org), with PDB accession code 1XVN, in which the tandem A-T bps at the central 5′-TA step are Hoogsteen bps. The structural model of corresponding naked DNA with the same DNA sequence was generated by 3DNA, assuming an idealized B-form geometry (50) in which the central tandem A-T bps are Watson-Crick bps. The automated fragmentation QM/MM approach (68) was then used to calculate the chemical shifts of all four nucleotides in the central tandem A-T bps for both the Watson-Crick and Hoogsteen models. The starting structural models were subjected to an initial geometry optimization (53), and the echinomycin ligands were removed from the coordinates at this stage for the echinomycin-bound DNA. For both the Watson-Crick and Hoogsteen models, four quantum fragments centered at each nucleotide of the central tandem A-T bps were generated, with 3 to 5 additional neighboring nucleotides (68). For atoms outside this quantum region, including solvent and ions, we used point charges distributed on the surface of the quantum region, assuming a Poisson-Boltzmann distribution, to represent their effect by using the solinprot module of MEAD (69). The dielectric constant, ε, was set to 1, 4, and 80 for quantum fragments, the remaining atoms outside the quantum regions, and the solvent, respectively. Gauge invariant atomic orbitals chemical shift calculations with the OLYP functional with the TZVP basis set and the GEN-A2* fitting set using the demon-2k program (70) were carried out as described previously (5).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01GM118664, to C.P.J.; R01GM123743, to C.P.J.; and R01GM089846, to H.M.A.-H.), the National Science Foundation (MCB-1715174, to C.P.J.), and the G. Harold and Leila Y. Mathers Foundation (to H.M.A.-H. and C.P.J.). We thank Prof. Catherine Musselman for providing the pJ201 plasmid.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200681119/-/DCSupplemental.
Data Availability
All of the study data are included in the article and/or SI Appendix.
References
- 1.Nikolova E. N., et al. , Transient Hoogsteen base pairs in canonical duplex DNA. Nature 470, 498–502 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patikoglou G. A., et al. , TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 13, 3217–3230 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitayner M., et al. , Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat. Struct. Mol. Biol. 17, 423–429 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuesta-Seijo J. A., Sheldrick G. M., Structures of complexes between echinomycin and duplex DNA. Acta Crystallogr. D Biol. Crystallogr. 61, 442–448 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Shi H., et al. , Atomic structures of excited state A-T Hoogsteen base pairs in duplex DNA by combining NMR relaxation dispersion, mutagenesis, and chemical shift calculations. J. Biomol. NMR 70, 229–244 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sathyamoorthy B., et al. , Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A. Nucleic Acids Res. 45, 5586–5601 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu L., Yi C., Jian X., Zheng G., He C., Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked protein-DNA system. Nucleic Acids Res. 38, 4415–4425 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling H., Boudsocq F., Plosky B. S., Woodgate R., Yang W., Replication of a cis-syn thymine dimer at atomic resolution. Nature 424, 1083–1087 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Nair D. T., Johnson R. E., Prakash S., Prakash L., Aggarwal A. K., Replication by human DNA polymerase-ι occurs by Hoogsteen base-pairing. Nature 430, 377–380 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Xu Y., et al. , Hoogsteen base pairs increase the susceptibility of double-stranded DNA to cytotoxic damage. J. Biol. Chem. 295, 15933–15947 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bohnuud T., et al. , Computational mapping reveals dramatic effect of Hoogsteen breathing on duplex DNA reactivity with formaldehyde. Nucleic Acids Res. 40, 7644–7652 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikolova E. N., et al. , A historical account of Hoogsteen base-pairs in duplex DNA. Biopolymers 99, 955–968 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H., et al. , New insights into Hoogsteen base pairs in DNA duplexes from a structure-based survey. Nucleic Acids Res. 43, 3420–3433 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J., DNA polymerases: Hoogsteen base-pairing in DNA replication? Nature 437, E6–E7, discussion E7 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Hintze B. J., Richardson J. S., Richardson D. C., Mismodeled purines: Implicit alternates and hidden Hoogsteens. Acta Crystallogr. D Struct. Biol. 73, 852–859 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi H., et al. , Revealing A-T and G-C Hoogsteen base pairs in stressed protein-bound duplex DNA. Nucleic Acids Res. 49, 12540–12555 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrews A. J., Luger K., Nucleosome structure(s) and stability: Variations on a theme. Annu. Rev. Biophys. 40, 99–117 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J., Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Luger K., Richmond T. J., DNA binding within the nucleosome core. Curr. Opin. Struct. Biol. 8, 33–40 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Berman H. M., et al. , The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davey C. A., Sargent D. F., Luger K., Maeder A. W., Richmond T. J., Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 319, 1097–1113 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Lowary P. T., Widom J., New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Alvey H. S., Gottardo F. L., Nikolova E. N., Al-Hashimi H. M., Widespread transient Hoogsteen base pairs in canonical duplex DNA with variable energetics. Nat. Commun. 5, 4786 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loquet A., Habenstein B., Lange A., Structural investigations of molecular machines by solid-state NMR. Acc. Chem. Res. 46, 2070–2079 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Quinn C. M., Polenova T., Structural biology of supramolecular assemblies by magic-angle spinning NMR spectroscopy. Q. Rev. Biophys. 50, e1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lecoq L., Fogeron M. L., Meier B. H., Nassal M., Böckmann A., Solid-state NMR for studying the structure and dynamics of viral assemblies. Viruses 12, 1069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao M., et al. , Histone H3 and H4 N-terminal tails in nucleosome arrays at cellular concentrations probed by magic angle spinning NMR spectroscopy. J. Am. Chem. Soc. 135, 15278–15281 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang S., et al. , Site-specific studies of nucleosome interactions by solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 57, 4571–4575 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi X., et al. , Structure and dynamics in the nucleosome revealed by solid-state NMR. Angew. Chem. Int. Ed. Engl. 57, 9734–9738 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Shi X., Prasanna C., Soman A., Pervushin K., Nordenskiöld L., Dynamic networks observed in the nucleosome core particles couple the histone globular domains with DNA. Commun. Biol. 3, 639 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zandian M., et al. , Conformational dynamics of histone H3 tails in chromatin. J. Phys. Chem. Lett. 12, 6174–6181 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riedel K., Leppert J., Ohlenschläger O., Görlach M., Ramachandran R., Characterisation of hydrogen bonding networks in RNAs via magic angle spinning solid state NMR spectroscopy. J. Biomol. NMR 31, 331–336 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Huang W., Varani G., Drobny G. P., 13C/15N-19F intermolecular REDOR NMR study of the interaction of TAR RNA with Tat peptides. J. Am. Chem. Soc. 132, 17643–17645 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cherepanov A. V., Glaubitz C., Schwalbe H., High-resolution studies of uniformly 13C,15N-labeled RNA by solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 49, 4747–4750 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Asami S., Rakwalska-Bange M., Carlomagno T., Reif B., Protein-RNA interfaces probed by 1H-detected MAS solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 52, 2345–2349 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Marchanka A., Simon B., Althoff-Ospelt G., Carlomagno T., RNA structure determination by solid-state NMR spectroscopy. Nat. Commun. 6, 7024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchanka A., Carlomagno T., “Solid-state NMR spectroscopy of RNA” in Methods in Enzymology, A. J. Wand, Ed. (Academic Press., 2019), vol. 615, pp. 333–371. [DOI] [PubMed] [Google Scholar]
- 38.Barnes A. B., et al. , High-field dynamic nuclear polarization for solid and solution biological NMR. Appl. Magn. Reson. 34, 237–263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaudzems K., Polenova T., Pintacuda G., Oschkinat H., Lesage A., DNP NMR of biomolecular assemblies. J. Struct. Biol. 206, 90–98 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Jaudzems K., et al. , Dynamic nuclear polarization-enhanced biomolecular NMR spectroscopy at high magnetic field with fast magic-angle spinning. Angew. Chem. Int. Ed. Engl. 57, 7458–7462 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Sergeyev I. V., Day L. A., Goldbourt A., McDermott A. E., Chemical shifts for the unusual DNA structure in Pf1 bacteriophage from dynamic-nuclear-polarization-enhanced solid-state NMR spectroscopy. J. Am. Chem. Soc. 133, 20208–20217 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Daube D., Vogel M., Suess B., Corzilius B., Dynamic nuclear polarization on a hybridized hammerhead ribozyme: An explorative study of RNA folding and direct DNP with a paramagnetic metal ion cofactor. Solid State Nucl. Magn. Reson. 101, 21–30 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Wenk P., et al. , Dynamic nuclear polarization of nucleic acid with endogenously bound manganese. J. Biomol. NMR 63, 97–109 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Wiegand T., et al. , Protein-nucleotide contacts in motor proteins detected by DNP-enhanced solid-state NMR. J. Biomol. NMR 69, 157–164 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Xu Y., McSally J., Andricioaei I., Al-Hashimi H. M., Modulation of Hoogsteen dynamics on DNA recognition. Nat. Commun. 9, 1473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert D. E., van der Marel G. A., van Boom J. H., Feigon J., Unstable Hoogsteen base pairs adjacent to echinomycin binding sites within a DNA duplex. Proc. Natl. Acad. Sci. U.S.A. 86, 3006–3010 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilbert D. E., Feigon J., The DNA sequence at echinomycin binding sites determines the structural changes induced by drug binding: NMR studies of echinomycin binding to [d(ACGTACGT)]2 and [d(TCGATCGA)]2. Biochemistry 30, 2483–2494 (1991). [DOI] [PubMed] [Google Scholar]
- 48.Sauvée C., et al. , Highly efficient, water-soluble polarizing agents for dynamic nuclear polarization at high frequency. Angew. Chem. Int. Ed. Engl. 52, 10858–10861 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Guéron M., Leroy J. L., Studies of base pair kinetics by NMR measurement of proton exchange. Methods Enzymol. 261, 383–413 (1995). [DOI] [PubMed] [Google Scholar]
- 50.Lu X. J., Olson W. K., 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 31, 5108–5121 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaroniec C. P., Filip C., Griffin R. G., 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly (13)C,(15)N-labeled solids. J. Am. Chem. Soc. 124, 10728–10742 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Nikolova E. N., Goh G. B., Brooks C. L. III, Al-Hashimi H. M., Characterizing the protonation state of cytosine in transient G·C Hoogsteen base pairs in duplex DNA. J. Am. Chem. Soc. 135, 6766–6769 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi H., et al. , Rapid and accurate determination of atomistic RNA dynamic ensemble models using NMR and structure prediction. Nat. Commun. 11, 5531 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wijmenga S. S., Van Buuren B. N. M., The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 32, 287–387 (1998). [Google Scholar]
- 55.Vasudevan D., Chua E. Y. D., Davey C. A., Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J. Mol. Biol. 403, 1–10 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Zhou H., et al. , Characterizing Watson-Crick versus Hoogsteen base pairing in a DNA-protein complex using nuclear magnetic resonance and site-specifically 13C- and 15N-labeled DNA. Biochemistry 58, 1963–1974 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kastrup J. S., et al. , Structure of the C-type lectin carbohydrate recognition domain of human tetranectin. Acta Crystallogr. D Biol. Crystallogr. 54, 757–766 (1998). [DOI] [PubMed] [Google Scholar]
- 58.Zhou H., et al. , m(1)A and m(1)G disrupt A-RNA structure through the intrinsic instability of Hoogsteen base pairs. Nat. Struct. Mol. Biol. 23, 803–810 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rangadurai A., et al. , Why are Hoogsteen base pairs energetically disfavored in A-RNA compared to B-DNA? Nucleic Acids Res. 46, 11099–11114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rabdano S. O., et al. , Histone H4 tails in nucleosomes: A fuzzy interaction with DNA. Angew. Chem. Int. Ed. Engl. 60, 6480–6487 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morrison E. A., et al. , DNA binding drives the association of BRG1/hBRM bromodomains with nucleosomes. Nat. Commun. 8, 16080 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fung B. M., Khitrin A. K., Ermolaev K., An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 142, 97–101 (2000). [DOI] [PubMed] [Google Scholar]
- 63.Kupče E., Boyd J., Campbell I. D., Short selective pulses for biochemical applications. J. Magn. Reson. B. 106, 300–303 (1995). [DOI] [PubMed] [Google Scholar]
- 64.Delaglio F., et al. , NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 65.Goddard T. D., Kneller D. G., SPARKY 3, (University of California, San Francisco, CA, 2006).
- 66.Morcombe C. R., Zilm K. W., Chemical shift referencing in MAS solid state NMR. J. Magn. Reson. 162, 479–486 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Bertani P., Raya J., Bechinger B., 15N chemical shift referencing in solid state NMR. Solid State Nucl. Magn. Reson. 61–62, 15–18 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Swails J., Zhu T., He X., Case D. A., AFNMR: Automated fragmentation quantum mechanical calculation of NMR chemical shifts for biomolecules. J. Biomol. NMR 63, 125–139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Richardson W. H., Peng C., Bashford D., Noodleman L., Case D. A., Incorporating solvation effects into density functional theory: Calculation of absolute acidities. Int. J. Quantum Chem. 61, 207–217 (1998). [Google Scholar]
- 70.Koster A. M., et al. , deMon2k (Version 5, The deMon Developers, Cinvestav, Mexico City, Mexico, 2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the study data are included in the article and/or SI Appendix.