Significance
Heme availability in the cell enables the proper folding and function of enzymes, which carry heme as a cofactor. Using genome-scale modeling, we identified metabolic fluxes and genes that limit heme production. Our study experimentally validates ecYeast8 model predictions. Moreover, we developed an approach to predict gene combinations, which provides an in silico design of a viable strain able to overproduce the metabolite of interest. Using our approach, we constructed a yeast strain that produces 70-fold-higher levels of intracellular heme. With its high-capacity metabolic subnetwork, our engineered strain is a suitable platform for the production of additional heme enzymes. The heme ligand-binding biosensor (Heme-LBB) detects the cotranslational incorporation of heme into the heme-protein hemoglobin.
Keywords: genome-scale modeling, heme, Saccharomyces cerevisiae, metabolic engineering, heme ligand-binding biosensor
Abstract
Heme is an oxygen carrier and a cofactor of both industrial enzymes and food additives. The intracellular level of free heme is low, which limits the synthesis of heme proteins. Therefore, increasing heme synthesis allows an increased production of heme proteins. Using the genome-scale metabolic model (GEM) Yeast8 for the yeast Saccharomyces cerevisiae, we identified fluxes potentially important to heme synthesis. With this model, in silico simulations highlighted 84 gene targets for balancing biomass and increasing heme production. Of those identified, 76 genes were individually deleted or overexpressed in experiments. Empirically, 40 genes individually increased heme production (up to threefold). Heme was increased by modifying target genes, which not only included the genes involved in heme biosynthesis, but also those involved in glycolysis, pyruvate, Fe-S clusters, glycine, and succinyl-coenzyme A (CoA) metabolism. Next, we developed an algorithmic method for predicting an optimal combination of these genes by using the enzyme-constrained extension of the Yeast8 model, ecYeast8. The computationally identified combination for enhanced heme production was evaluated using the heme ligand-binding biosensor (Heme-LBB). The positive targets were combined using CRISPR-Cas9 in the yeast strain (IMX581-HEM15-HEM14-HEM3-Δshm1-HEM2-Δhmx1-FET4-Δgcv2-HEM1-Δgcv1-HEM13), which produces 70-fold-higher levels of intracellular heme.
Heme is a cofactor of essential enzymes for aerobic life within the three domains of life (archaea, bacteria, and eukarya). The heme molecule consists of a porphyrin ring that surrounds an iron atom, which alternates between its ferric and ferrous states in the oxidation and reduction reactions. Heme-containing proteins (HCPs) have several functions. For example, HCPs transport electrons in the respiratory chain in mitochondria and are crucial for energy production, transport molecular oxygen in globin proteins (e.g., hemoglobin in humans), and protect cells from oxidative damage (1–4). The heme biosynthetic pathway is conserved and tightly regulated to supply heme at levels to meet cellular demands. The cotranslational incorporation of heme into heme proteins governs their folding process (5, 6). The intracellular availability of heme is crucial for the production of heme proteins, which denature and lose their function without heme.
Because of their biological importance, heme and HCPs are a central topic in molecular cell biology, with basic research occurring together with applications in medicine and technology. The production of heme and heme proteins has been a focus of research in microbial metabolic engineering. For example, research on blood substitutes focuses on human hemoglobin (7, 8), and plant-derived hemoglobin provides vegetarian protein (artificial meat with a lower carbon footprint) (9). Heme was used to improve charging of lithium batteries (10) and in the bioremediation of sulfite waste (11). Cytochromes and their new mutant forms catalyze novel chemical reactions with silicon (12) and were evolved to perform novel chemical reactions (13). The heterologous production of heme proteins is, however, challenging due to the limited amount of free heme and the complexity of the metabolic network in the cell.
While a heme-biosynthesis pathway is conserved in nature, the precursor 5-aminolevulinic acid (5-ALA) is synthesized distinctly in different organisms. In the C4 pathway, the precursor 5-ALA is produced from glycine and from succinyl-coenzyme A (CoA) (the C4 intermediate of the tricarboxylic acid [TCA] cycle) in yeast, birds, mammals, and purple nonsulfur photosynthetic bacteria. In contrast, in the C5 pathway, the precursor 5-ALA is produced from alpha-ketoglutarate (the C5 intermediate of the TCA cycle) in algae, plants, and bacteria such as Escherichia coli (14).
In E. coli, heme production has been increased by metabolic engineering of the pathways for 5-ALA synthesis, both native (C5) and heterologous (C4). The metabolic engineering studies using the C4 pathway increased heme production by overexpression of the Rhodobacter sphaeroides hemA gene (encoding ALA synthase), which produces the 5-ALA precursor; by overexpressing the native coaA gene (encoding pantothenate kinase), which produces CoA; and by overexpression of genes for heme biosynthesis. This engineering strategy yielded 3.3 μmol/L (15) and 9.1 μmol/gcell (16) of heme. By overexpressing genes for heme production via the C5 pathway and by deleting genes of competing pathways, 51.5 mg/L total heme was produced (17). In the same strain, metabolic engineering of a heme-secretory pathway and feed-control optimization of substrates in fed-batch cultivation increased the production of total heme to 239 mg/L (17).
In the unicellular eukaryote and established yeast cell-factory Saccharomyces cerevisiae, heme is synthesized through the C4 pathway (14). To improve the production of heme and heme proteins in S. cerevisiae, metabolic engineering studies have overexpressed genes encoding the known rate-limiting enzymes for heme biosynthesis (18–21) and have engineered oxygen sensing involved in heme biosynthesis regulation (22). To increase the production of the first intermediate of the heme pathway, 5-ALA, the HEM1 and ACO2 genes were overexpressed (23). However, the contribution of overall metabolism to heme production has not been analyzed.
The impressive development of heme production in E. coli, however, has had some limitations, such as weak tolerance to acidic pH and phage sensitivity. As E. coli produces endotoxins, it is difficult to use E. coli directly in food production. In contrast, the S. cerevisiae yeast has greater tolerance for acidic pH and has been used for food production for millennia.
The S. cerevisiae has been analyzed with genome-scale metabolic models (GEMs) (24). For S. cerevisiae, GEM analysis has guided the construction of strains with optimized yields of industrial molecules (e.g., bioethanol, sesquiterpenes, vanillin, 2,3-butanediol, fumaric acid, succinate, amorphadiene, 3-hydroxypropionate, β-farnesene, and dihydroxyacetone phosphate [DHAP]) (24, 25). The measurement of metabolic compounds in screening has facilitated the development of new biosensors that can be used for novel applications in other organisms (26).
For S. cerevisiae, the consensus GEM (version 7.6) informed the engineering of strains with increased production of acetyl-CoA and malonyl-CoA in 2019 (27). The updated consensus Yeast8 model was followed by ecYeast8, which has additional constraints on the metabolic fluxes, representing enzymatic abundances. Enzyme-constrained GEMs improved the prediction of specific phenotypes (28, 29).
Studies of heme production have explored the modification of genes and their expression, improving our knowledge of particular pathways. Using metabolic GEMs to maximize the production of heme is the focus of this study. We used the 2019 enzyme-constrained ecYeast8 (29) to identify metabolic fluxes that are important for heme biosynthesis. Our systems-biology analysis and modification of the gene expression guided the optimization of a heme strain with 58 genes in silico. The sequential strain engineering increased intracellular heme production 70-fold. In optimization of sequentially accumulated gene modifications, we developed a heme biosensor, which detects heme availability and the incorporation of heme into hemoglobin protein. This heme ligand-binding biosensor (Heme-LBB), like earlier genetically encoded ratiometric heme sensors (30), is likely useful for heme detection in other organisms.
Our results are striking in terms of the dramatic increase in heme production and as a showcase of model-assisted synthetic biology. More importantly, our case study is one of the most rigorous in terms of evaluation of model-predicted targets for the widely used cell factory S. cerevisiae. As several of the model-predicted targets resulted in improved production, our paper represents a significant milestone in terms of a wider use of model-based design of yeast cell factories.
Results
Yeast8 Simulations of Metabolic Fluxes Impacting Heme Production.
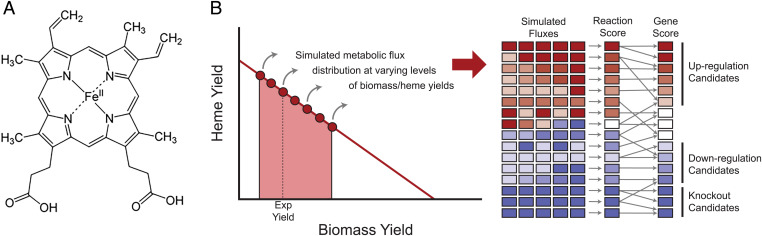
As an initial screening, we quantified the fluxes impacting heme production using flux balance analysis (FBA) tools available for S. cerevisiae at the start of our study. Using Yeast8 (29), we computed the theoretical biomass yield on glucose to be 0.1168 gDW/g for batch cultures, which is very close to the experimentally validated value of our strain (0.122 gDW/g). We followed a published approach (27, 31), which is the adaptation of the flux scanning based on enforced objective flux (FSEOF) method (32). To simulate physiologically relevant conditions and analyze heme production at suboptimal growth yields, we ran several simulations on glucose as the single carbon source, varying the biomass yield from half of the experimental yield to twice the value (Fig. 1). In each simulation, the objective function was to maximize heme production, computing for each biomass-yield condition an optimum solution. In these simulation-generated optimal solutions, the number of active fluxes was reduced by parsimonious FBA (33). From these simulations, scores were computed for each metabolic reaction in the network to detect which fluxes were consistently either increasing or decreasing as the biomass requirements decreased, an established strategy (27, 31, 32). Finally, using known reaction-gene associations, we converted those flux scores to gene scores, which indicate whether a gene has a monotonic behavior—that is, the flux scores selected genes that are consistently upregulated (score > 1), downregulated (0 < score < 1), or completely silenced (score = 0) (Fig. 1). This scoring predicted that 84 genes had a monotonic effect, including 62 genes being overexpressed and 8 genes deleted. Additionally, 14 genes were downregulated (among them, 6 were essential or required additional growth supplements when deleted: OLE1, FAS1, FAS2, RNR2, CDS1, and CHO1) (Dataset S1).
Fig. 1.
The Yeast8 genome-scale model was used to find fluxes important for heme production to enable the construction of a heme yeast cell factory. (A) The structure of heme b, which is protoporphyrin IX with ferrous iron. (B) Simulations of heme production using S. cerevisiae Yeast8.0.1 model.
Validation of Individual Gene Targets Predicted by Yeast8.
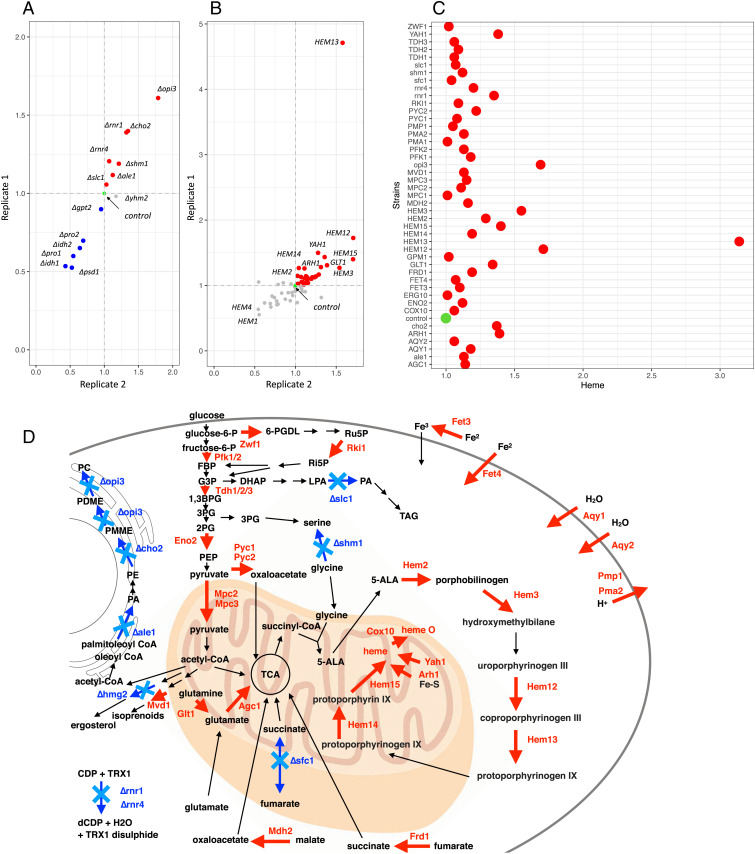
The gene targets predicted by Yeast8 were experimentally tested for their impact on heme production by modifying genes one at a time and by measuring intracellular heme concentration. As gene downregulation requires more fine tuning (e.g., by promoter modifications or gene silencing approaches), we tested the effect of gene deletions first by using the deletion strains from the yeast knockout (YKO) collection (34). We analyzed 16 S. cerevisiae BY4741 strains carrying single gene deletions: 8 strains from the downregulation group (Δrnr1, Δrnr3, Δrnr4, Δcho2, Δopi3, Δpsd1, Δgpt2, Δale1) and 8 deletion strains from the deletion group (Δshm1, Δslc1, Δpro1, Δpro2, Δsfc1, Δyhm2, Δidh1, Δidh2) (Fig. 2A and Dataset S1). The Δrnr3 strain grew poorly and was excluded from further experimental analysis. The heme production of 15 strains was measured in two biological replicates after 24 h of cultivation in yeast extract-peptone-dextrose (YPD) medium (Fig. 2A).
Fig. 2.
Experimental validation of Yeast8 gene targets. (A) Heme production (fold-change) of 15 gene KO strains from the YKO collection (BY4741 strain background). BY4741 strain served as a control to normalize data (shown in green). Two replicates were used in the analysis. Heme was extracted from eight OD600 of cells. The gene targets where heme production was higher than the control are highlighted in red. The gene targets where heme production was lower than the control are highlighted in blue. (B) Heme production (fold-change) of strains carrying 61 model genes overexpressed under control of the TEF1 promoter using a centromeric plasmid in CEN.PK113-11C strain background. Heme was extracted from eight OD600 of cells. CEN.PK113-11C carrying empty vector served as a control to normalize data (shown in green). Two replicates were used in the analysis. (C) Heme production of strains with gene modifications that improved heme production the most. Average value of two replicates was used. Heme was extracted from eight OD600 of cells. (D) Schematic overview of metabolism with Yeast8 targets, which experimentally improved heme production.
Deletion of five out of the seven tested genes in the downregulation group (OPI3 [encoding methylene-fatty-acyl-phospholipid synthase], CHO2 [encoding phosphatidylethanolamine methyltransferase], RNR1 [encoding major isoform of large subunit of ribonucleotide-diphosphate reductase], RNR4 [encoding ribonucleotide-diphosphate reductase small subunit], and ALE1) validated the model predictions and increased heme production up to 70% compared to the BY4741 control strain (Fig. 2A). The deletion of two genes, GPT2 (encoding glycerol-3-phosphate/DHAP sn-1 acyltransferase) and PSD1 (encoding phosphatidylserine decarboxylase of the mitochondrial inner membrane), decreased heme production ∼10 to 50% compared to the BY4741 control strain (Fig. 2A).
Deletion of three out of the eight genes (identified to be deleted by Yeast8) increased heme production (Fig. 2A). The deletion of SHM1 (encoding mitochondrial serine hydroxylmethyltransferase) resulted in a ∼11.5% increase in heme production, the deletion of the ALE1 gene (encoding broad-specificity lysophospholipid acyltransferase) resulted in a ∼13% increase, and the deletion of SFC1 (encoding mitochondrial succinate-fumarate transporter) resulted only in a ∼4% increase (Fig. 2A). On the other hand, the deletion of SLC1 (encoding 1-acyl-sn-glycerol-3-phosphate acyltransferase) and YHM2 (encoding citrate and oxoglutarate carrier protein), did not result in a significant increase in heme production compared with BY4741 (Fig. 2A and Dataset S1). Deletions of PRO1 (encoding gamma-glutamyl kinase), PRO2 (gamma-glutamyl phosphate reductase), and IDH1 and IDH2 (encoding subunits of mitochondrial NAD [+]-dependent isocitrate dehydrogenase) genes decreased heme production, contrary to the model predictions (Fig. 2A and Dataset S1). Both PRO1 and PRO2 gene deletions resulted in proline auxotrophy, and the resulting strains grew poorly in YPD. In summary, among the 15 tested gene candidates identified to be deleted or downregulated, 8 genes increased heme production.
We evaluated the overexpression of 61 of the 62 model gene targets (we could not amplify the HMG2 gene) in the S. cerevisiae CEN.PK.113–11c strain background (Fig. 2B). For this purpose, we cloned the open reading frames (ORFs) of the 61 genes into the centromeric expression plasmid pRS316+prTEF1-terADH1 under control of strong constitutive promoter TEF1. Two transformants with expression cassettes for each of the 61 model target genes (predicted to be overexpressed) were used to evaluate heme production (Fig. 2B and Dataset S1). The highest heme production (∼300% average increase) was observed upon the overexpression of the HEM13 (encoding coproporphyrinogen III oxidase) heme biosynthetic gene (Fig. 2B). Under normal conditions, the HEM13 is transcriptionally repressed by Rox1 (22, 35), and expressing it under the promoter TEF1 will increase the protein abundance independent of the oxygen and heme levels. Overexpressing other heme biosynthetic genes—such as HEM14 (encoding protoporphyrinogen oxidase), HEM2 (encoding aminolevulinate dehydratase), HEM15 (encoding ferrochelatase), HEM3 (encoding porphobilinogen deaminase), and HEM12 (encoding uroporphyrinogen decarboxylase)—also increased heme production from ∼20 to 70%, respectively (Fig. 2 B and C). The HEM2, HEM3, and HEM12 genes have been reported to be rate-limiting steps in heme biosynthesis (18–20). Overexpression of HEM1 (encoding 5-aminolevulinate synthase) did not improve heme production at 48 h of fermentation (Fig. 2B), and the overexpression of HEM4 (encoding uroporphyrinogen III synthase) resulted in substantially reduced yeast growth. We speculate this was caused by the accumulation of uroporphyrinogen III, which is toxic when oxidized (36). In addition to heme-biosynthetic genes, the overexpression of single genes involved in iron homeostasis and Fe-S cluster proteins (YAH1 and ARH1), glutamate biosynthesis (GLT1), pyruvate metabolism and its transport (PYC1, PYC2, MPC1, MPC2, MPC3), fumarate reductase (FRD1), malate dehydrogenase (MDH2), glycolysis (PFK1, PFK2, TDH1, TDH2, TDH3), amino acids, iron, protons, and water transport (AGC1, FET4, FET3, PMA1, PMA2, AQY1, and AQY2) increased heme production up to 40% compared to the control strain carrying the empty vector pRS316 (Fig. 2 B and C). In summary, among the 61 overexpression targets tested, 32 increased heme production (Fig. 2C), which is a 52% success rate of model predictions.
Refining the Simulations of Heme Production Fluxes Using ecYeast8.
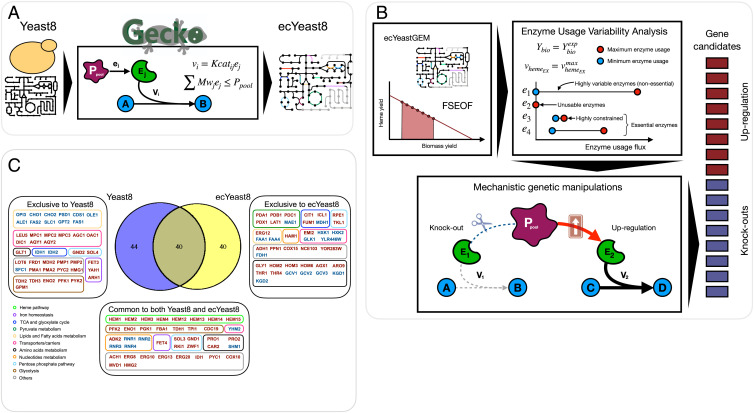
We used the enzyme-constrained version of the Yeast8 model (ecYeast8) to refine model simulations and to evaluate the combinatorial effects of the gene targets (Fig. 3A and SI Appendix). The ecYeast8 model accounts for the activity of metabolic enzymes as constraints on the reactions in the network. These constraints are limited by the total amount of available protein mass, yielding a drastic reduction of the variability of the metabolic fluxes and notable improvements on phenotype predictions for S. cerevisiae’s metabolism (28, 29). Simulations for optimization of heme production using ecYeast8 were performed following the same procedure as with the Yeast8 model; in this case, candidate gene targets for downregulation (0 < gene score ≤ 1) were discarded. Additionally, as enzyme-constrained models enable a direct assessment of the effects of enzyme activity perturbations, the enzyme usage variability analysis and mechanistic simulations for the individual gene modifications were implemented for heme production (Fig. 3B and SI Appendix). This allowed the prediction of 80 gene targets (Dataset S2) by ecYeast8. Comparing the target lists of both models (Datasets S1 and S2), 40 genes were found to overlap between Yeast8 and ecYeast8, 44 genes were detected exclusively by Yeast8, and 40 genes were detected exclusively by ecYeast8 (Fig. 3C). The genes exclusive to ecYeast8 were heme A synthase (COX15), nucleoside triphosphate pyrophosphohydrolase (HAM1), pentose phosphate pathway (TKL1, RPE1), alcohol dehydrogenase (ADH1), glucose uptake (YRL446W, HXK1, HXK2, GLK1, EMI2), isoprenoids and sterol biosynthesis (ERG12), pyruvate metabolism (PDA1, PDB1, PDC1, PDX1, LAT1, MAE1), TCA cycle (CIT1, MDH1, FUM1), glyoxylate cycle (ICL1), glycine biosynthesis (GLY1, AGX1), glycine cleavage system (GCV1, GCV2, GCV3), fatty acids synthesis (FAA1, FAA4), L-lysine degradation (KGD1, KGD2), L-threonine metabolism (HOM2, HOM3, HOM6, THR1, THR4), phosphatase (YOR283W), polyphosphate metabolism (PPN1), formate dehydrogenase (FDH1), carbonic anhydrase (NCE103), and aromatic amino acids synthesis (ARO9) (Fig. 3C). Interestingly, among genes common to both Yeast8 and ecYeast8, PRO1 was predicted to be downregulated by Yeast8, whereas it was predicted to be overexpressed by the ecYeast8 simulations. Experimental validation showed that deletion of this gene reduced the heme production drastically (Fig. 2A).
Fig. 3.
The ecYeast8 model was used to find new targets for improved heme production. (A) Following use of the Yeast8, simulation using the enzyme-constrained model ecYeast8 was performed for increased heme production. (B) Following the adapted FSEOF approach (19, 22, 23), the enzyme usage variability analysis and mechanistic genetic manipulations for the individual gene modifications were used to refine the heme target list. (C) In simulations, the Yeast8 model identified 84 targets, and the ecYeast8 model identified 80 targets. Of the gene targets identified by the two models, 40 genes overlapped between Yeast8 and ecYeast8; 44 genes were identified by only the Yeast8 model, and 40 genes were identified by only the ecYeast8 model.
The small portion of positive Yeast8 genes (including OPI3, CHO2, SLC1, PMA2, MPC3, MDH2, GLT1, FRD1, AQY1, AQY2, ALE1, SFC1, and AGC1) were not detected by ecYeast8. However, these genes proved to improve heme production by the engineering genes one at a time (Fig. 2). These data can also be used for further improvement of the ecYeast8 model predictions.
Predicting Compatible Gene Combinations for Improved Heme Production.
The list of genetic targets in Dataset S2 represents individual strategies for enhancing heme production, and we next used ecYeast8 to assess the viability of combining these strategies in silico. First, metabolic function redundancy was assessed by identification of identical genes in a genes-metabolites network (i.e., a bipartite graph that connects a metabolite with a gene if they are both involved in the same reaction). This allowed classification of gene targets in groups, where each gene group contains genes that are linked to the same metabolites according to the model. This grouping allowed a further reduction of the total number of candidates by discarding all genes that did not fit into any of the following categories: 1) gene target candidates with a unique metabolic function; 2) genes encoding for enzymes with the highest specific activity in a given group of redundant candidates for overexpression, due to their lower impact on the total protein burden for the cell; and 3) all gene candidates for deletion whose enzymes did not carry any flux in a reference flux distribution for optimized heme production (SI Appendix). Overall, this filtering procedure reduced the number of candidates from 80 to 71 genes.
We ranked the remaining modification targets according to the categories described above. Within each category, the fold-change in heme production was predicted for each individual target. An optimal mutant strain was then constructed in silico by implementing the remaining modifications in a sequential and cumulative way. Gene modifications that decreased the optimal production yield when compared to the previous iteration were discarded. This allowed us to obtain a list of “compatible” 58 gene target modifications that, according to the ecYeast8 model simulations, should yield a viable strain with enhanced heme production capabilities if they are combined (Dataset S3).
Constructing Compatible Gene Combinations for Improved Heme Production.
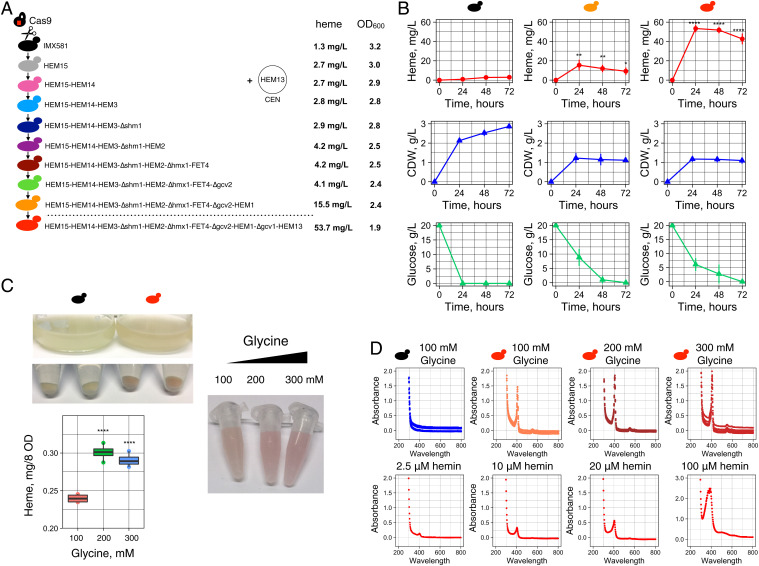
We used the CRISPR-Cas9 toolbox developed for S. cerevisiae (37) to combine positive gene targets, which were predicted by the ecYeast8 model to yield higher production of intracellular heme, resulting in a viable strain. From our list of 58 compatible genes (Dataset S3), we overexpressed the HEM13 gene first, as this gene had the maximum experimental effect (Fig. 2). The choice of the sequential targets to be combined with HEM13 gene was evaluated experimentally. If the individual gene modification did not increase the output, then it was declared a failure at that time (but kept as a possible modification for later experimentation with a new gene combination). Plasmids expressing single guide RNAs (sgRNAs) targeting different genomic loci were constructed (using the pMEL10 plasmid vector as a base; ref. 37) and used to integrate the expression cassettes of gene targets (as described in SI Appendix). The sequential gene modifications, which lead to sequential increases in heme production, are presented in Fig. 4A. The HEM13 gene was expressed from centromeric plasmid under promoter TEF1 of S. cerevisiae (Fig. 4A and SI Appendix, Fig. S1). The effects of introduced strain modifications were verified both by heme production measurement and by using Heme-LBB (Fig. 5 and SI Appendix, Figs. S9–S12). The Heme-LBB is a fusion protein of green fluorescent protein (GFP) and hemoglobin alpha-gamma subunits (SI Appendix) and was expressed under the copper-inducible promoter CUP1 of S. cerevisiae. The biosensor fluorescence was designed to reflect the intracellular heme amount. Hemoglobin is a HCP, and heme incorporation during its translation determines correct folding (5, 6). Thus, we inferred that greater intracellular heme is associated with an increase in correctly folded GFP-hemoglobin protein that can be measured by the biosensor’s activity (Fig. 5A). As the Heme-LBB is a new biosensor, it was used in parallel with direct heme measurement to study its response.
Fig. 4.
CRISPR-Cas9 genome engineering for increased heme production. (A) The IMX581 strain carrying CRISPR-Cas9 gene integrated in the genome was used to carry the combinatorial engineering of heme gene targets deduced by Yeast8 and ecYeast8 genome-scale model. The gene integrations and deletions were performed using the gRNA constructs targeting different genome loci. The gene HEM13 was overexpressed from the centromeric plasmid. The HEM13 expression cassette was integrated into the genome in the final strain. Absolute heme (mg/L) was extracted from the entire biomass of the strains. (B) Heme production, CDW, and glucose consumption in different strains at 24, 48, and 72 h of cultivation in buffered SD ura- or SD with 2% glucose, 100 mM glycine supplemented with 0.1 mM Fe3+. Four biological replicates (transformants) were used in the experiment. Error bars represent the SD. Commercial hemin was used to calibrate data. Strains: IMX581 carrying an empty vector; IMX581/HEM15 HEM14 HEM3 Δshm1 HEM2 Δhmx1 FET4 Δgcv2 HEM1 carrying the HEM13 centromeric plasmid; IMX581/HEM15 HEM14 HEM3 Δshm1 HEM2 Δhmx1 FET4 Δgcv2 HEM1 Δgcv1 carrying HEM13 expression cassette integrated into genome. Statistical analysis was performed using one-way ANOVA (*P ≤ 0.02741, **P ≤ 0.00594, ****P ≤ 0). (C) The culture, cells, and cell extracts (obtained with oxalic acid treatment) of engineered strain IMX581/HEM15 HEM14 HEM3 Δshm1 HEM2 Δhmx1 FET4 Δgcv2 HEM1 Δgcv1 HEM13 had a red color. Increasing the glycine amount from 100 to 300 mM resulted in a further increase in heme production. Statistical analysis was performed using one-way ANOVA (****P ≤ 0.00007). (D) Spectral analysis of yeast extracts (obtained with oxalic acid treatment) shows the presence of the Soret peak (at 400 nm) characteristic to heme in IMX581/HEM15 HEM14 HEM3 Δshm1 HEM2 Δhmx1 FET4 Δgcv2 HEM1 Δgcv1 HEM13 strain. Hemin (2.5, 10, 20, and 100 μM) spectra were used in comparison.
Fig. 5.
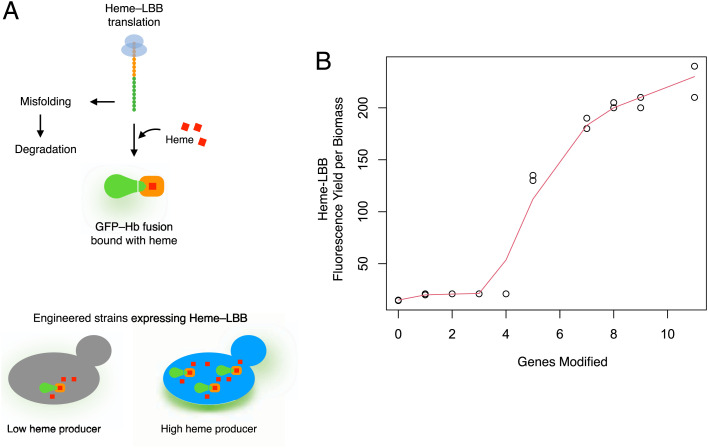
Heme biosensor in engineered strains. (A) Heme-LBB is a fusion construct of GFP (highlighted in green) and hemoglobin (Hb, highlighted in orange). Heme (highlighted in red) is cotranslationally incorporated into the hemoglobin part of the biosensor polypeptide and promotes its correct folding. Heme-less biosensor molecules are misfolded and subjected to degradation. GFP-Hb fusion bound with heme is active and fluorescent. An increase in heme supply by the strain engineering will subsequently increase the number of correctly folded Heme-LBB molecules and, therefore, increase the strain’s fluorescence. (B) Yield of Heme-LBB fluorescence per biomass with sequential heme-modeling targets engineered. Genes modified: 1: HEM15; 2: HEM15, HEM14; 3: HEM15, HEM14, HEM3; 4: HEM15, HEM14, HEM3, Δshm1; 5: HEM15, HEM14, HEM3, Δshm1, HEM2; 7: HEM15, HEM14, HEM3, Δshm1, HEM2, Δhmx1, FET4; 8: IMX581, HEM15, HEM14, HEM3, Δshm1, HEM2, Δhmx1, FET4, Δgcv2; 9: HEM15, HEM14, HEM3, Δshm1, HEM2, Δhmx1, FET4, Δgcv2, HEM1; 11: HEM15, HEM14, HEM3, Δshm1, HEM2, Δhmx1, FET4, Δgcv2, HEM1, Δgcv1, HEM13. Quantile regression with nondecreasing shape constraint (49) was used to estimate the biosensor response. To calculate the yield, the fluorescence of the Heme-LBB and the growth of each strain was monitored using a BioLector.
In the CEN.PK.113–11c strain background, the overexpression of the HEM13 gene resulted in an average ∼threefold increase in heme production (Fig. 2C). In contrast, the HEM13 overexpression in IMX581 strain resulted in only 1.5-fold-higher heme production (SI Appendix, Fig. S1). Next, we integrated five heme biosynthetic genes (HEM15, HEM14, HEM3, HEM2, HEM1) into different genome loci step by step using CRISPR-Cas9, and this resulted in increased heme production (SI Appendix, Figs. S1 and S2). Our initial test using Heme-LBB with 5-ALA (which is the product of Hem1) in the medium showed a response in the IMX581 strain carrying the HEM15 gene expression cassette but no response in the control strain (SI Appendix, Fig. S14A). On the other hand, the biosensor activity increased with the engineered model targets that increase heme production (SI Appendix, Figs. S9, S11, S12, and S14B). The deletion of the SHM1 gene combined with overexpression of heme genes (HEM15, HEM14, HEM3) resulted in a strain producing ∼fivefold more heme than the IMX581 control (SI Appendix, Fig. S2). The overexpression of ACH1 did not result in improvement of heme production (SI Appendix, Fig. S3). Additional deletion of FAA4, FDH1, and YLR446w resulted only in a small improvement of heme production (SI Appendix, Figs. S4 and S5). The deletion of GCV2 improved heme production in combination with only some genes (SI Appendix, Figs. S2, S4, and S7). The gene encoding the heme oxygenase (HMX1), which is responsible for heme degradation (38), was the integration locus we used for expression cassettes of the FET4, ADH1, and ARH1 genes. The overexpression of FET4 and deletion of HMX1 was a better combination for heme improvement than overexpression of either ADH1 or ARH1 and deletion of HMX1 (SI Appendix, Figs. S6 and S11). Further deletions of the GCV2 and GCV1 genes and integration of the HEM1 and HEM13 genes substantially improved heme production, resulting in the strain turning red (Fig. 4C) and the highest GFP fluorescence of the heme biosensor (Fig. 5 and SI Appendix, Fig. S12). Further evaluation of this production strain (IMX581 HEM15 HEM14 HEM3 Δshm1 HEM2 Δhmx1 FET4 Δgcv2 HEM1 Δgcv1 HEM13) using direct heme extraction and fluorescence measurement showed that it produced 53.5 mg heme per liter of the culture at 24 h of cultivation, which was 35.6 times higher than that of the initial strain, IMX581 (Fig. 4B). When normalized by the cell dry weight (CDW) (Fig. 4B), the constructed strain produced 70-fold more heme when compared to the initial strain. When the heme was extracted from the same amount of biomass, the production strain (IMX581 HEM15 HEM14 HEM3 Δshm1 HEM2 Δhmx1 FET4 Δgcv2 HEM1 Δgcv1 HEM13) contained 74.4 times more intracellular heme at 72 h of cultivation compared to the control strain, IMX581 (SI Appendix, Fig. S15). The fluorescence of the biosensor protein in the constructed strain was also the highest and was ∼20-fold higher than that of initial strain IMX581 (Fig. 5 and SI Appendix, Fig. S12). The best-performing strain also accumulated less biomass and consumed less glucose (Fig. 4B). Its growth rate was reduced by 40% (Fig. 4B and SI Appendix, Fig. S13), and its heme titer was 35-fold greater (Fig. 4B).
To test the possibility of a further increase in heme production, we studied heme produced in the IMX581 strain with genotype HEM15 HEM14 HEM3 Δshm1 HEM2 Δhmx1 FET4 Δgcv2 HEM1 Δgcv1 HEM13 when cultured with elevated amounts of the glycine, the substate of Hem1 (Fig. 4C). As shown in Fig. 4C, the cell extracts of cultures grown on the medium supplemented with 200 or 300 mM glycine had a 25 or 20% greater heme, respectively. This was also accompanied with a darker red color of the yeast extracts (Fig. 4C). While both media with 200 or 300 mM glycine resulted in significantly higher heme production than the medium with 100 mM glycine (ANOVA, ****P ≤ 0.00007), the difference in heme production between cultures grown in media with 200 or 300 mM glycine was not significant (ANOVA, P ≤ 0.19484) (Fig. 4C). Unlike the control strain, the extracts of the production strain displayed a characteristic of heme Soret peak (at 400 nm) similarly to hemin, which was used as standard (Fig. 4D). Future studies should optimize heme production using glycine in fed-batch bioreactors and introduce the remaining gene modifications deduced by the model to improve heme production further.
Discussion
Heme is a cofactor of heme proteins and enzymes crucial for aerobic cell physiology (1). Free heme, heme proteins, and heme enzymes have been used in emerging technologies, such as flavoring agents for artificial meat (39), blood substitutes (40), lithium-air batteries (10), and recently discovered chemical reactions (12, 13). High levels of intracellular heme are toxic to cells. Cytosolic heme is between 20 and 40 nM. Mitochondria tolerate higher concentrations of heme, roughly 30 μM (30). Higher levels of heme increase the production of hemoglobin and of P450 enzymes, which is apparently because the heme group insertion is essential for the proper folding and conformational stability of heme proteins (5, 6, 19, 41, 42). On the other hand, the overexpression of HCPs depletes the cellular heme pool and stresses the cell (18).
Recent advances in the GEM of S. cerevisiae (27–29) facilitate genome-scale identification of metabolic fluxes active in heme production, which can then be optimized to increase heme production. Linear and quadratic programming are needed for computing optimal, basic, feasible solutions and optimal interior solutions. Solving such problems is usually beyond the scope of humans.
Like E. coli, the yeast S. cerevisiae has GEMs of proven research supported by international communities of researchers (29, 43–45). In our study, to increase the cellular heme pool in the yeast S. cerevisiae, we used a metabolic modeling approach on the genome scale to maximize heme production by genetic modifications with in silico predictions and in vivo confirmation. Using FBA with Yeast8 and then enzymatically constrained ecYeast8 models (27, 29), we identified 84 gene as candidates to increase heme production. Our modeling suggested overexpressing 62 genes, downregulating 14 genes, and deleting 8 genes.
In the experimental phase of the study, several strategies were used. The strong constitutive promoter TEF1 was used for overexpression of candidate genes. For the deletion and downregulation of genes, we used mutants from a collection of YKOs. Our one-gene-at-a-time experiments increased heme production by many interventions: strengthening glycolysis; improving the transport of pyruvate into mitochondria; improving the flow of acetyl-CoA into TCA cycle; overexpressing genes of the TCA cycle; modifying glycine-serine metabolism; and improving the transport of iron, water, and amino acids.
Then, additional modeling was performed to optimize combinations of gene modifications. Building on previous approaches to the prediction of gene overexpression targets (32, 46, 47), our study developed a procedure to identify some gene combinations that are compatible with the specified set of growth rates. With optimization approaches, increasing the flux for a reaction (or fluxes for reactions) allows other fluxes to change unless additional constraints are introduced to fix their values. FBA approaches have had difficulty accounting for, for example, protein burden, potential inhibitions by reaction products, and regulatory feedback loops.
In the first round of simulations with Yeast8, we identified genes that individually influence heme production. In the second round with ecYeast8 (with enzyme constraints), we developed in silico a viable mutant strain with improved heme production that had accumulated many positive modifications, successively added after having increased heme production above the previous maximum. In our model, we blocked the enzyme usage reactions for deletion targets, and for the overexpression targets, we doubled the enzyme usage. In laboratory experiments, the identified target combination was then engineered using CRISPR-Cas9. Our constraint-based model and our algorithm produced a list of 58 compatible genes. Thereafter, our implementation of changes sequentially chose the largest increase predicted by the model. If the individual gene modification did not increase the output, then it was declared a failure at that time (but kept as a possible modification for later experimentation). The first deletion of the GCV2 gene did not increase output; at a later stage (after having introduced successful modifications), GCV2 reappeared as a gene with maximum predicted increase, and it was (per our method) reintroduced, this time successfully. Increased heme production was positively and strongly associated with an increased activity of the newly developed Heme-LBB, as expected; the predicted increase in heme availability improves the cotranslational incorporation into hemoglobin. The Heme-LBB response to increased heme productivity was found to be dose dependent and sigmoidal, which is typical for biosensors. The developed biosensor provided the opportunity to measure heme in vivo without the need to extract heme for measurements. The biosensor activity in the constructed strains also assessed the expression of heme protein, which is useful for future work on the production of heme proteins using these strains.
With linear programming algorithms, our approach generated very interesting findings, which were not noticed previously in the literature. For example, our model found that heme biosynthesis is tightly coupled to central carbon metabolism with 80 genes, whose expression affects the heme production. Also, unexpectedly, the model implied that improved heme production could be achieved by reducing the lipid and deoxyribonucleotide triphosphates (dNTPs) biosynthesis and by increasing the activity of pentose phosphate pathway.
Our enzyme-constrained GEMs enabled us to develop a yeast strain with 70-fold more intracellular heme compared to the control strain when normalized per biomass. Our engineered strain produced 53.5 mg/L heme. Zhao et al. (17) achieved the intracellular production of 51.5 mg/L in E. coli, which is comparable with our yeast strain producing 53.5 mg/L total heme. Improving heme output by an order of magnitude in our strain required the simultaneous modification of 11 genes, which were selected through GEM simulation and laboratory experimentation. Our strain overexpressed the heme-biosynthetic genes HEM15, HEM14, HEM3, HEM2, HEM1, and HEM13, and it also overexpressed the low-affinity Fe (II) transporter of the plasma-membrane gene (FET4). In addition, we deleted the mitochondrial serine hydroxymethyltransferase gene (SHM1), the heme oxygenase gene (HMX1), and the two genes encoding subunits of the mitochondrial glycine decarboxylase complex (GCV1 and GCV2). The constructed strain with 11 genetic modifications can be further engineered with the 58 genetic modifications predicted to be beneficial. However, the introduction of numerous genetic modifications in one strain risks off-target mutations, and the 11 implemented modifications already increased heme production by 70 times.
Materials and Methods
All the materials and methods are detailed in SI Appendix. These include preliminary target selection using Yeast8; reference flux distribution using ecYeast8; gene target selection using ecYeast8; identification of an optimal combination of targets using ecYeast8; media and growth conditions; genome engineering; determination of glucose concentration; CDW analysis; determination of heme concentration; and heme biosensor. Briefly, the Yeast8 metabolic model of S. cerevisiae was used to identify preliminary gene targets using FBA. Then, the ecYeast8 allowed the incorporation of enzyme constraints and informed the selection of gene targets. Intracellular heme was extracted with oxalic acid (18). For the cell dry weight (CDW), cells were collected on 0.45 μm cellulose-acetate filter paper (Satorius Biolabs). We developed the Heme-HBB as a synthetic fusion protein (consisting of α-globin, γ-globin, and GFP) to detect heme in vivo, validating its increasing response experimentally. The Heme-HBB construct was expressed under the control of the copper-inducible promoter CUP1.
Statistical Analysis.
The statistical programs R (48) and Minitab 18.1 were used to analyze the data. The biosensor response was studied with quantile regression with a nondecreasing shape constraint (49, 50).
Supplementary Material
Acknowledgments
O.P.I. and D.P. were funded by a grant from Swedish Foundation for Strategic Research (RBP14-0055). I.D. has received funding from the European Union’s Horizon 2020 Framework Programme for Research and Innovation Grant Agreement 720824. I.D., B.J.S., and J.N. were funded by the EU project DD-DeCaF (grant 686070) and the Novo Nordisk Foundation (grant NNF10CC1016517). Authors thank Professor Thomas Nyström for the BY4741 deletion strains, Honzhong Lu and Mario Beck for providing insight on the predictive method implemented with ecYeast8, Eduard Kerkhoven and Feiran Li for the numerous discussions on ecYeast8, and Jim E. Blevins and Louis H. Scott for scientific editing.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. C.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108245119/-/DCSupplemental.
Data Availability
All the necessary scripts for model prediction and analysis used in this study have been deposited to GitHub and are available at https://github.com/SysBioChalmers/heme_production_ecYeastGEM/releases/tag/v1.0 (51) or through Zenodo at https://doi.org/10.5281/zenodo.6792435 (52).
References
- 1.Bernhardt R., Cytochromes P450 as versatile biocatalysts. J. Biotechnol. 124, 128–145 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Patton K. T., Anatomy & Physiology (Elsevier - Health Sciences, 2015). [Google Scholar]
- 3.Heinemann I. U., Jahn M., Jahn D., The biochemistry of heme biosynthesis. Arch. Biochem. Biophys. 474, 238–251 (2008). [DOI] [PubMed] [Google Scholar]
- 4.He L., et al. , Antioxidants maintain cellular redox homeostasis by elimination of reactive oxygen species. Cell. Physiol. Biochem. 44, 532–553 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Komar A. A., Kommer A., Krasheninnikov I. A., Spirin A. S., Cotranslational heme binding to nascent globin chains. FEBS Lett. 326, 261–263 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Komar A. A., Kommer A., Krasheninnikov I. A., Spirin A. S., Cotranslational folding of globin. J. Biol. Chem. 272, 10646–10651 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Palmer A. F., Intaglietta M., Blood substitutes. Annu. Rev. Biomed. Eng. 16, 77–101 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Moradi S., Jahanian-Najafabadi A., Roudkenar M. H., Artificial blood substitutes: First steps on the long route to clinical utility. Clin. Med. Insights Blood Disord. 9, 33–41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser R. Z., Shitut M., Agrawal P., Mendes O., Klapholz S., Safety evaluation of soy leghemoglobin protein preparation derived from Pichia pastoris, intended for use as a flavor catalyst in plant-based meat. Int. J. Toxicol. 37, 241–262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryu W. H., et al. , Heme biomolecule as redox mediator and oxygen shuttle for efficient charging of lithium-oxygen batteries. Nat. Commun. 7, 12925 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mirts E. N., Petrik I. D., Hosseinzadeh P., Nilges M. J., Lu Y., A designed heme-[4Fe-4S] metalloenzyme catalyzes sulfite reduction like the native enzyme. Science 361, 1098–1101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kan S. B., Lewis R. D., Chen K., Arnold F. H., Directed evolution of cytochrome c for carbon-silicon bond formation: Bringing silicon to life. Science 354, 1048–1051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnold F. H., Directed evolution: Bringing new chemistry to life. Angew. Chem. Int. Ed. Engl. 57, 4143–4148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki K., Watanabe M., Tanaka T., Tanaka T., Biosynthesis, biotechnological production and applications of 5-aminolevulinic acid. Appl. Microbiol. Biotechnol. 58, 23–29 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Kwon S. J., de Boer A. L., Petri R., Schmidt-Dannert C., High-level production of porphyrins in metabolically engineered Escherichia coli: Systematic extension of a pathway assembled from overexpressed genes involved in heme biosynthesis. Appl. Environ. Microbiol. 69, 4875–4883 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pranawidjaja S., Choi S. I., Lay B. W., Kim P., Analysis of heme biosynthetic pathways in a recombinant Escherichia coli. J. Microbiol. Biotechnol. 25, 880–886 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Zhao X. R., Choi K. R., Lee S. Y., Metabolic engineering of Escherichia coli for secretory production of free haem. Nat. Catal. 1, 720–728 (2018). [Google Scholar]
- 18.Michener J. K., Nielsen J., Smolke C. D., Identification and treatment of heme depletion attributed to overexpression of a lineage of evolved P450 monooxygenases. Proc. Natl. Acad. Sci. U.S.A. 109, 19504–19509 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L., Martínez J. L., Liu Z., Petranovic D., Nielsen J., Balanced globin protein expression and heme biosynthesis improve production of human hemoglobin in Saccharomyces cerevisiae. Metab. Eng. 21, 9–16 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Hoffman M., Góra M., Rytka J., Identification of rate-limiting steps in yeast heme biosynthesis. Biochem. Biophys. Res. Commun. 310, 1247–1253 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Ishchuk O. P., et al. , Improved production of human hemoglobin in yeast by engineering hemoglobin degradation. Metab. Eng. 66, 259–267 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Martínez J. L., Liu L., Petranovic D., Nielsen J., Engineering the oxygen sensing regulation results in an enhanced recombinant human hemoglobin production by Saccharomyces cerevisiae. Biotechnol. Bioeng. 112, 181–188 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Hara K. Y., et al. , 5-Aminolevulinic acid fermentation using engineered Saccharomyces cerevisiae. Microb. Cell Fact. 18, 194 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopes H., Rocha I., Genome-scale modeling of yeast: Chronology, applications and critical perspectives. FEMS Yeast Res. 17, fox050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerkhoven E. J., Lahtvee P. J., Nielsen J., Applications of computational modeling in metabolic engineering of yeast. FEMS Yeast Res. 15, 1–13 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Zhang F., Keasling J., Biosensors and their applications in microbial metabolic engineering. Trends Microbiol. 19, 323–329 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Ferreira R., et al. , Model-assisted fine-tuning of central carbon metabolism in yeast through dCas9-based regulation. ACS Synth. Biol. 8, 2457–2463 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Sánchez B. J., et al. , Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol. Syst. Biol. 13, 935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu H., et al. , A consensus S. cerevisiae metabolic model Yeast8 and its ecosystem for comprehensively probing cellular metabolism. Nat. Commun. 10, 3586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanna D. A., et al. , Heme dynamics and trafficking factors revealed by genetically encoded fluorescent heme sensors. Proc. Natl. Acad. Sci. U.S.A. 113, 7539–7544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J., et al. , Combining mechanistic and machine learning models for predictive engineering and optimization of tryptophan metabolism. Nat. Commun. 11, 4880 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi H. S., Lee S. Y., Kim T. Y., Woo H. M., In silico identification of gene amplification targets for improvement of lycopene production. Appl. Environ. Microbiol. 76, 3097–3105 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis N. E., et al. , Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol. Syst. Biol. 6, 390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giaever G., Nislow C., The yeast deletion collection: A decade of functional genomics. Genetics 197, 451–465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keng T., HAP1 and ROX1 form a regulatory pathway in the repression of HEM13 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 2616–2623 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiabrando D., Vinchi F., Fiorito V., Mercurio S., Tolosano E., Heme in pathophysiology: A matter of scavenging, metabolism and trafficking across cell membranes. Front. Pharmacol. 5, 61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mans R., et al. , CRISPR/Cas9: A molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 15, fov004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Protchenko O., Philpott C. C., Regulation of intracellular heme levels by HMX1, a homologue of heme oxygenase, in Saccharomyces cerevisiae. J. Biol. Chem. 278, 36582–36587 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Fraser R., O’Reilly Brown P., Karr J., Holz-Schietiger C., Cohn E., “Methods and compositions for affecting the flavor and aroma profile of consumables.” US Patent 9700067B2 (2017).
- 40.Khan F., Singh K., Friedman M. T., Artificial blood: The history and current perspectives of blood substitutes. Discoveries (Craiova) 8, e104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reedy C. J., Gibney B. R., Heme protein assemblies. Chem. Rev. 104, 617–649 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Ji H. F., Shen L., Grandori R., Müller N., The effect of heme on the conformational stability of micro-myoglobin. FEBS J. 275, 89–96 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Fang X., Lloyd C. J., Palsson B. O., Reconstructing organisms in silico: Genome-scale models and their emerging applications. Nat. Rev. Microbiol. 18, 731–743 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orth J. D., et al. , A comprehensive genome-scale reconstruction of Escherichia coli metabolism--2011. Mol. Syst. Biol. 7, 535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mienda B. S., Dräger A., “Genome-scale metabolic modeling of escherichia coli and its chassis design for synthetic biology applications” in Computational Methods in Synthetic Biology. Methods in Molecular Biology, Marchisio M. A., Ed. (Humana, New York, NY, 2021), vol. 2189. [DOI] [PubMed] [Google Scholar]
- 46.Ranganathan S., Suthers P. F., Maranas C. D., OptForce: An optimization procedure for identifying all genetic manipulations leading to targeted overproductions. PLOS Comput. Biol. 6, e1000744 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chowdhury A., Zomorrodi A. R., Maranas C. D., k-OptForce: Integrating kinetics with flux balance analysis for strain design. PLOS Comput. Biol. 10, e1003487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.R Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2016). https://www.R-project.org/ [Google Scholar]
- 49.Koenker R., Quantile Regression (Cambridge University Press, 2005), pp. 1–366. [Google Scholar]
- 50.Koenker R., quantreg: Quantile regression. R package version 5.86 (2021). https://CRAN.R-project.org/package=quantreg.
- 51.Domenzain I., Sánchez B. J., Heme Production ecYeastGEM. https://github.com/SysBioChalmers/heme_production_ecYeastGEM/releases/tag/v1.0. Deposited 6 March 2022.
- 52.Domenzain I., Sánchez B. J., SysBioChalmers/heme_production_ecYeastGEM: Source code for publication. 10.5281/zenodo.6792435. Deposited 3 July 2022. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the necessary scripts for model prediction and analysis used in this study have been deposited to GitHub and are available at https://github.com/SysBioChalmers/heme_production_ecYeastGEM/releases/tag/v1.0 (51) or through Zenodo at https://doi.org/10.5281/zenodo.6792435 (52).