Summary
Impaired protein stability/trafficking underlies diverse ion channelopathies and represents an unexploited unifying principle to develop common treatments for otherwise dissimilar diseases. Ubiquitination limits ion channel surface density, but targeting this pathway for basic study or therapy is challenging because of its prevalent role in proteostasis. We developed engineered deubiquitinases (enDUBs) that enable ubiquitin chain removal selectively from target proteins to rescue functional expression of disparate mutant ion channels underlying Long QT syndrome (LQT1) and cystic fibrosis (CF). In a LQT1 cardiomyocyte model, enDUB treatment restored delayed rectifier K+ currents and normalized action potential duration. CF-targeted enDUBs synergistically rescued common (F508del) and pharmacotherapy-resistant (N1303K) CF mutations when combined with the FDA-approved drugs, Orkambi and Trikafta. Altogether, targeted deubiquitination via enDUBs provides a powerful protein stabilization method that not only corrects diverse diseases caused by impaired ion channel trafficking, but also introduces a new tool for deconstructing the ubiquitin code in situ.
Inherited or de novo mutations in ion channels underlie diseases spanning the nervous (epilepsy)1, cardiovascular (long QT syndrome)2, respiratory (cystic fibrosis)3, endocrine (diabetes)4, and urinary (Bartter syndrome) systems5. Hundreds of disease-causing mutations are typically found in individual ion channels, presenting an extraordinary challenge for study and treatment. Mechanism-based approaches to correct underlying abnormalities that can be generally applied across different ion channels would be advantageous, but are lacking5,6.
Long QT syndrome type 1 (LQT1) and cystic fibrosis (CF) arise from loss-of-function mutations in KCNQ12,7 and cystic fibrosis transmembrane regulator (CFTR)3 channels, respectively. LQT1 increases risk of sudden cardiac death, while CF patients display impaired airway mucus clearance leading to recurrent bacterial infection, uncontrolled inflammation, lung damage, and low life expectancy. For both channels, a prominent mechanism underlying loss-of-function mutations is impaired channel trafficking to the cell surface8–12. We sought to exploit this shared mechanism to develop a general cell biological approach that is amenable to therapeutic development and applicable to diverse ion channels.
Results
enDUBs selectively reverse NEDD4L ubiquitination of KCNQ1
Because ubiquitination/deubiquitination is a primary determinant of ion channel surface density (Fig. 1a)13, we hypothesized that ubiquitin removal would rescue trafficking-deficient ion channels. Since ubiquitination is widespread, we developed a targeted deubiquitination approach that circumvents problematic off-target effects associated with targeting the ubiquitin/proteasomal system14,15. For initial proof-of-concept, we utilized YFP-tagged KCNQ1, which is known to be down-regulated by NEDD4L16. We developed a YFP-targeted engineered deubiquitinase (enDUB-O1) by fusing the catalytic unit of ovarian tumor deubiquitinase 1 (OTUD1)17 to a nanobody specific for GFP/YFP18 (Fig. 1a, inset). We tested efficacy and selectivity of enDUB-O1 using biochemical and functional assays in transiently transfected HEK293 cells (Fig. 1b-h).
Figure 1. enDUBs reverse NEDD4L-mediated ubiquitination of KCNQ1.
a, Schematic of targeted deubiquitination via enDUBs (nano, PDB: 3K1K). Inset, Modular domains of OTUD1 and enDUB-O1. b, Left, KCNQ1 pulldowns probed with anti-KCNQ1 antibody from HEK293 cells expressing KCNQ1-YFP ± NEDD4L with nano alone or enDUB-O1. The four bands represent KCNQ1 monomer, dimer, trimer, and tetrameric species, respectively. Right, Anti-ubiquitin labeling of KCNQ1 pulldowns after stripping previous blot. c, Relative KCNQ1 ubiquitination computed by ratio of anti-ubiquitin to anti-KCNQ1 signal intensity (n = 4 independent experiments; mean). *p=0.0012, **p=0.0005, one-way ANOVA with Tukey’s multiple comparison test. d, Flow cytometry contour plots showing surface (BTX647 fluorescence) and total (YFP fluorescence) KCNQ1 expression in cells expressing BBS-KCNQ1-YFP. Vertical and horizontal lines represent thresholds for YFP and BTX647-positive cells, respectively, based on analyses of single color controls. e,f, Quantification of flow cytometry experiments for e, surface and f, total KCNQ1 expression, analyzed from YFP- and CFP-positive cells (n ≥ 5000 cells per experiment; N = 4; mean ± s.e.m). Data are normalized to values from the control group, KCNQ1 without NEDD4L (dotted line). e, *p=0.002; f, *p=0.0097; unpaired two-tailed Student’s t test. g, Exemplar family of KCNQ1 currents from whole-cell patch clamp measurements in CHO cells. h, Population I-V curves for nano (black circles, n = 9), nano + NEDD4L (red squares, n = 9), and enDUB-O1 + NEDD4L (blue triangles, n = 12) (mean ± s.e.m). *p<0.01 versus nano + NEDD4L, two-way ANOVA with Tukey’s multiple comparison test. Here and throughout, population electrophysiology data are pooled from n cells across three or more independent experiments conducted on different days.
Immunoprecipitation experiments in control cells expressing KCNQ1-YFP and anti-GFP nanobody (nano) yielded a characteristic KCNQ1 banding pattern of monomer, dimer, trimer, and tetrameric species19, and showed robust expression of ubiquitinated KCNQ1 channels, reflecting endogenous E3 ligase activity (Fig. 1b,c; Extended Data Fig. 1). Co-expressing NEDD4L with KCNQ1-YFP and nano resulted in decreased KCNQ1 levels (Fig. 1b, left), and increased ubiquitination (Fig. 1b,c; Extended Data Fig. 1). In the presence of NEDD4L, enDUB-O1 rescued KCNQ1 expression, and prevented the increase in channel ubiquitination (Fig. 1b,c; Extended Data Fig. 1).
We utilized flow cytometry to simultaneously measure KCNQ1-YFP total expression and surface density, and to assess the ability of enDUB-O1 (expressed in a 1:1 ratio with CFP using a P2A self-cleaving peptide plasmid) to antagonize the impact of NEDD4L (Extended Data Fig. 2). NEDD4L significantly decreased KCNQ1 surface density (assessed by fluorescent bungarotoxin binding to an extracellular epitope tag) and total expression (YFP fluorescence), and both effects were reversed by enDUB-O1 (Fig. 1d-f). Catalytically dead enDUB-O1* did not rescue KCNQ1-YFP surface density, demonstrating DUB enzymatic activity is necessary for this effect (Extended Data Fig. 3b). Importantly, enDUB-O1 did not rescue surface density of KCNQ1 channels lacking YFP, indicating specificity of the targeted enDUB approach (Extended Data Fig. 3c).
We used whole-cell electrophysiology to determine whether enDUB-O1-rescued KCNQ1 channels are functional. Control cells expressing KCNQ1/nano displayed robust KCNQ1 currents that were abolished with NEDD4L expression (Fig. 1g,h); co-expressing enDUB-O1 fully rescued KCNQ1 currents (Fig. 1g,h), confirming the efficacy of targeted deubiquitination with enDUBs to specifically deubiquitinate and stabilize functional channels at the cell surface.
enDUBs rescue trafficking-deficient mutant KCNQ1 channels that cause LQT1
We wondered whether enDUBs would rescue trafficking-deficient mutant KCNQ1 channels that underlie LQT1. We used flow cytometry to determine the impact of 14 distinct LQT1 mutations in KCNQ1 C-terminus7,20, and a previously described endoplasmic reticulum-associated degradation (ERAD)-dependent mutation in the N-terminus (L114P)21, on channel surface density (Fig. 2a). In addition to L114P, 9 of the 14 C-terminus mutations showed significantly reduced surface trafficking compared to WT KCNQ1 (Fig. 2a, red bars). Remarkably, the surface density of 6 mutant channels was either partially or fully rescued with enDUB-O1 co-expression (Fig. 2a, blue bars and inset). The responsive mutant channels were clustered along the KCNQ1 coiled-coil tetramerization domain (helix D), defining a spatial ‘hotspot’ amenable to enDUB-mediated rescue of trafficking (Fig. 2a, purple text).
Figure 2. enDUBs rescue trafficking-deficient mutant LQT1 channels in HEK293 cells.
a, Left, Schematic of LQT1 mutations along C-terminus of KCNQ1. Right, Quantification of flow cytometry experiments for surface expression (BTX647) of LQT1 mutant channels in presence of nano alone (red) or enDUB-O1 (blue), analyzed from YFP- and CFP-positive cells (n ≥ 5000 cells per experiment; N = 3; mean ± s.e.m). Data are normalized to values from the WT KCNQ1 control group (dotted line). †p=0.051, *p<0.03, unpaired two-tailed Student’s t test. Right inset, Confocal image of live cells expressing BBS-tagged WT KCNQ1-YFP (top) or G589D-YFP + nano (middle) or enDUB-O1 (bottom), stained with BTX647 (magenta). Scale bar, 5 µm. b, Exemplar families of WT and mutant KCNQ1 currents reconsitituted in CHO cells. c, Population I-V curves for WT + nano (black squares, n = 10), R591H + nano (pink triangles, n = 8), and R591H + nano + ML277 (red triangles, n = 13); n cells examined over 3 independent experiments (mean ± s.e.m). d, Population I-V curves for R591H + enDUB-O1 (cyan circles, n = 9), and R591H + enDUB-O1 + ML277 (blue circles, n = 9); n cells examined over 3 independent experiments (mean ± s.e.m). Data for WT KCNQ1 and R591H + nano are reproduced from c (black and pink lines). *p<0.01, **p<0.001, two-way ANOVA with Tukey’s multiple comparison test. e, Flow cytometry contour plots showing surface and total KCNQ1 expression in cells expressing L114P (top) or G589D (bottom) mutants with cumulative distribution histograms generated from population of YFP- and CFP-positive cells (n ≥ 5000 cells per experiment; N = 3). f, KCNQ1 pulldowns probed with anti-KCNQ1 antibody from HEK293 cells expressing WT, G589D, and V524G KCNQ1-YFP channels (representative of two independent experiments). g, Anti-ubiquitin labeling of KCNQ1 pulldowns after stripping Western blot from f (representative of two independent experiments).
Homotetrameric R591H displayed dramatically reduced currents compared to WT KCNQ1, consistent with the impaired surface trafficking (Fig. 2b,c). The KCNQ1 activator, ML27722 (1 µM), modestly increased R591H currents (Fig. 2b,c). Co-expressing enDUB-O1 significantly rescued R591H currents to approximately 50% of WT KCNQ1, which in light of the full rescue in surface density (Fig. 2a), suggests the mutation causes an additional impairment in gating or conductance (Fig. 2b,d). Strikingly, 1 μM ML277 increased enDUB-O1-rescued R591H currents to beyond WT KCNQ1 levels (Fig. 2b,d).
Could the differential functional rescue of LQT1 mutations be predicted a priori based on relative ubiquitination status? One possibility was that rescuable mutations would display enhanced basal ubiquitination compared to WT KCNQ1 using standard biochemical assays. We investigated this hypothesis by measuring ubiquitination levels of two representative trafficking-deficient KCNQ1 C-terminus mutants— G589D which is amenable to enDUB-O1-mediated surface rescue, and V524G which is not. Interestingly, while both G589D and V524G mutations were deubiquitinated by enDUB-O1, the baseline ubiquitin signal of G589D was actually less than that observed for V524G (Fig. 2f,g). For the ERAD-sensitive L114P mutation, which displayed deficits in both total protein expression and surface trafficking, enDUB-O1 restored total protein levels but not surface density (Fig. 2e), suggesting an additional layer of regulation for misfolded proteins regardless of ubiquitin status.
enDUBs normalize electrophysiological properties in LQT1 cardiomyocyte model
LQT1 is typically autosomal dominant wherein patients possess one WT and one mutant allele2. We sought to recapitulate the heterotetrameric essence of LQT1 in cardiomyocytes from a species expressing endogenous KCNQ1 and KCNE1 subunits that result in IKs critical for cardiac action potential repolarization. We used adenovirus to express YFP-tagged WT or LQT1 mutant KCNQ1 in adult guinea pig cardiomyocytes (Fig. 3a). Compared to cardiomyocytes expressing WT KCNQ1, those with G589D displayed reduced late outward current measured by slow voltage ramps to +100 mV (Ipeak at 100 mV = 1.12±0.12 nA/pF, n=17 for WT; and 0.71±0.06 nA/pF, n=16 for G589D; P=0.03, one-way ANOVA) (Fig. 3b,c), consistent with a dominant negative impact of this mutation on WT KCNQ1 trafficking. Co-expressing enDUB-O1 with G589D completely rescued the decrement in late outward current (Ipeak at 100 mV = 1.32±0.15 nA/pF, n=14 for G589D+enDUB-01, P=0.0018 compared to G589D) (Fig. 3b,c). Control cardiomyocytes expressing WT KCNQ1 displayed action potentials with duration after 90% repolarization (APD90) of 263±40 ms, n=13 (Fig. 3,d,e). Cardiomyocytes expressing G589D exhibited markedly prolonged APD (APD90 = 753±93 ms, n=13, P<0.0001 compared to WT) (Fig. 3d,e). Remarkably, co-expressing enDUB-O1 with G589D normalized APD90 (APD90 = 324±36 ms, n=11, P=0.0002 compared to G589D, and P=0.88 compared to WT) (Fig. 3d,e). These results reveal efficacy of targeted deubiquitination as a stand-alone therapeutic approach in reversing pathological electrophysiological signatures underlying LQT1.
Figure 3. enDUBs restore action potential duration in LQT1 cardiomyocyte model.
a, Representative confocal images of adult guinea pig cardiomyocytes expressing WT KCNQ1-YFP (top) or G589D-YFP + nano (middle) or enDUB-O1 (bottom). Scale bar, 20 µm. b, Average current response of slow voltage ramp to +100 mV from cardiomyocytes expressing WT KCNQ1-YFP (left; n = 17, N = 3) or G589D-YFP (right) + nano alone (red; n = 16, N = 3) or enDUB-O1 (blue; n = 14, N = 3); n cells from N independent experiments / animals (mean ± s.e.m). c, Quantification of Ipeak at +100 mV of individual cells from data shown in b (mean ± s.d.). Same number of cells and independent experiments as in b. *p=0.03, **p=0.0018, one-way ANOVA with Tukey’s multiple comparison test. d, Representative action potential recordings from cardiomyocytes expressing WT KCNQ1-YFP (left) or G589D-YFP (right) + nano alone (red) or enDUB-O1 (blue). e, Quantification of action potential duration at 90% repolarization (APD90). WT KCNQ1-YFP (grey; n = 13, N = 3) or G589D-YFP + nano alone (red; n = 13, N = 3) or enDUB-O1 (blue; n = 11, N = 3); n cells from N independent experiments / animals (mean ± s.d.). **p=0.0002, ***p<0.0001, one-way ANOVA with Tukey’s multiple comparison test.
enDUB rescue of recombinant mutant CFTR channels that cause cystic fibrosis
Could enDUBs similarly rescue functional channels in an entirely different ion channelopathy for which impaired trafficking is an underlying cause? We turned to cystic fibrosis (CF) a devastating monogenic disease arising from loss-of-function mutations in CFTR. Over 2000 distinct CF mutations have been mapped to CFTR, many of which reduce channel surface density due to impaired folding/trafficking (class II) or decreased plasma membrane stability (class VI)23,24. Discovery of pharmacologic chaperones (correctors) and gating modifiers (potentiators) from high throughput screening has led to an FDA-approved combination therapy, Orkambi, consisting of lumacaftor (VX809; corrector) and ivacaftor (VX770; potentiator), for treating homozygous F508del mutations25–27. However, the clinical efficacy of Orkambi is often sub-optimal and a substantial number of CFTR mutations are refractory to current modulators, emphasizing an urgent need to develop complementary therapies24,28.
For initial screening, we engineered BBS-tagged YFP-CFTR to enable simultaneous assessment of total channel expression and surface density using flow cytometry, and probed the impact of six distinct mutations previously categorized as Class II (F508del, R560T, N1303K) or Class VI (Q1412X, 4279insA, 4326delTC) mutations, respectively (Fig. 4a). All six mutations impaired channel surface density compared to WT CFTR (Fig. 4b, black bars). Pre-incubation of cells with VX809 for 24 hrs did not increase F508del and R560T surface expression, but improved trafficking of the remaining 4 mutations (Fig. 4b, red bars). We utilized a second enDUB (enDUB-U21) that comprises the catalytic component of ubiquitin-specific protease USP2129 (Fig. 4a). In pilot experiments, enDUB-U21 was more efficacious for rescuing CFTR trafficking compared to enDUB-O1 (Extended Data Fig. 4), leading us to adopt the former for CFTR experiments. Similar to VX809, enDUB-U21 did not significantly rescue F508del and R560T surface density; however, it was either equal to or more effective in correcting the other four mutations, two of which (N1303K and 4279insA) were rescued to WT CFTR levels (Fig. 4b, blue bars). Both DUB activity and CFTR targeting were required for reversing trafficking deficits as catalytically inactive enDUB-U21 and mCh-targeted enDUB-U21 did not improve surface expression of YFP-tagged N1303K (Extended Data Fig. 5). Most importantly, co-applying VX809 and enDUB-U21 yielded synergistic rescue of mutant CFTR surface density (Fig. 4b; green bars).
Figure 4. enDUBs facilitate novel rescue of mutant CFTR channels in combination with Orkambi.
a, Schematic of six CF patient mutations (Class II, VI) across BBS-CFTR-YFP channel. Inset, Modular components of USP21 and enDUB-U21. b, Quantification of flow cytometry experiments for surface expression (BTX647) of CFTR mutant channels in presence of nano alone (black) or + VX809 (3µM) (red), and enDUB-O1 alone (blue) or + VX809 (3µM) (green), analyzed from YFP- and CFP-positive cells (n ≥ 5000 cells per experiment; N = 3; mean ± s.e.m). Data are normalized to values from the WT CFTR control group (dotted line). *p<0.02, **p<0.0001, two-way ANOVA followed by Dunnett’s test. c, Exemplar family of basal, forskolin-activated (10 µM), and CFTRinh-172-treated (10 µM) WT CFTR currents from whole-cell patch clamp measurements in HEK293 cells. d, Population I-V curves for basal (black squares, n = 16), forskolin-activated (red squares, n = 16) WT CFTR currents; n cells examined over ≥3 independent experiments (mean ± s.e.m). e-g, Exemplar family of basal and forskolin-activated currents from e, untransfected; and f, 4326delTC; g, N1303K CFTR mutant expressing cells. h, Exemplar family of forskolin-activated, VX770-potentiated (5µM) currents for 4236delTC mutant channels after 24hr VX809 treatment (3µM) and co-expression with nano (left) or enDUB-U21 (right). i, Population I-V curves for forskolin-activated, VX770-potentiated currents from 4326delTC mutants expressing nano (black circles, n = 17), versus VX809-treated 4326delTC cells expressing nano (red squares, n =15) or enDUB-U21 (green triangles, n = 14); n cells examined over ≥3 independent experiments (mean ± s.e.m). j,k, Same format as h,i for N1303K mutants expressing nano (black; n = 8), versus VX809-treated N1303K cells expressing nano (red; n = 9) or enDUB-U21 (green; n = 11); n cells examined over ≥3 independent experiments (mean ± s.e.m). ns (p=0.0861), **p<0.0001, two-way ANOVA with Tukey’s multiple comparison test.
We next determined whether enDUB-U21-rescued mutant CFTR channels are functional, focusing on N1303K and 4326delTC to represent Class II and Class VI mutations, respectively. HEK293 cells expressing WT CFTR display robust forskolin-activated chloride currents that are blocked by CFTR inhibitor (Fig. 4c,d), and not observed in untransfected cells (Fig. 4e). By contrast, cells expressing either N1303K or 4326delTC alone yielded no forskolin-induced currents (Fig. 4f,g). In nano-expressing cells, pre-incubation with VX809 yielded relatively small forskolin-induced 4326delTC and N1303K currents (Extended Data Fig. 6), which were modestly improved by VX770 (Fig. 4h-k). Excitingly, under the same conditions, cells co-expressing enDUB-U21 yielded substantially larger forskolin-stimulated (Extended Data Fig. 6) and VX770-potentiated 4326delTC or N1303K currents (Fig. 4h-k).
CFTR-targeted enDUB rescues modulator-unresponsive mutation N1303K
While GFP-targeted enDUBs provided critical proof-of-concept for efficacy of the targeted deubiquitination approach, a key next step was to develop nanobodies towards CFTR itself to enable targeting of endogenous channels. We used purified CFTR NBD1 domain (Fig. 5a, PDB: 5UAK30) as bait to identify binders using a yeast nanobody library surface display approach31 (Fig. 5b). After several rounds of magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS) selection, we isolated 14 unique nanobody binders with a range of affinities (from 20 nM to ∼1 uM) for NBD1 as reported by an on-yeast binding assay (Extended Data Fig. 7a). Reassuringly, when co-expressed with WT CFTR, these nanobody binders did not intrinsically interfere with surface trafficking of the channel (Extended Data Fig. 7b).
Figure 5. CF-targeted enDUB combination therapy functionally rescues rare trafficking-deficient CFTR mutations.
a, Structure of full-length CFTR channel30 (PDB: 5UAK). NBD1 highlighted in red. b, Top, Schematic for nanobody selection via yeast surface display library. Bottom, Example flow cytometry plots after MACS/FACS enrichment of yeast library with target binders (red). c, Top, Schematic for FRET binding assay in HEK293 cells co-expressing Cerulean-nb.E3h (donor) and Venus-CFTR (acceptor). Bottom, Flow cytometric FRET binding curves with FRET donor efficiency as function of free acceptor, with Cerulean-nb.E3h (blue) and Cerulean alone control (black) (n ≥ 10,000 cells per experiment; N = 2). d, Left, Exemplar traces showing YFP quenching in HEK293 cells expressing mCh (grey) or mCh-tagged N1303K mutants alone (red), and N1303K mutants treated with VX809 (green) or VX809 + enDUB-U21CF.E3h (blue) after addition of forskolin and VX770. Right, Summary of iodide influx rates (n = 8); n samples from 4 independent experiments (mean ± s.d.). e, Same formats as d for 4326delTC mutant channels (n = 9); n samples from 4 independent experiments (mean ± s.d.). **p<0.0001 versus mutant alone, one-way ANOVA with Tukey’s multiple comparison test. f, Population I-V curves for forskolin-activated currents from mCh-tagged WT CFTR channels (black circles, n = 41) or forskolin-activated, VX770-potentiated currents from 4326delTC mutants treated with VX809 and co-expressing CFP alone (red squares, n = 29), nb.E3h (green triangles, n = 9) or enDUB-U21CF.E3h (blue triangles, n = 12); n cells examined over ≥3 independent experiments (mean ± s.e.m.). *p=0.013, **p<0.0001, two-way ANOVA with Tukey’s multiple comparison test. g, Exemplar family of forskolin-activated, VX770-potentiated currents in FRT cells stably expressing WT CFTR (left) or N1303K after 24hr VX809 treatment and co-expressing either CFP alone (middle) or enDUB-U21CF.E3h (right). h, Population I-V curves for forskolin-activated WT (black circles, n = 7) and N1303K (red squares, n = 8) cells, compared to VX809-treated, forskolin-activated, and VX770-potentiated N1303K cells expressing CFP alone (green triangles, n = 12) or enDUB-U21CF.E3h (blue triangles, n = 10); n cells examined over ≥3 independent experiments (mean ± s.e.m.). **p=0.0008, ***p<0.0001, two-way ANOVA with Tukey’s multiple comparison test.
We used a flow cytometry fluorescence resonance energy transfer (flow-FRET) assay to determine whether nanobodies raised against NBD1 bound full-length CFTR in live cells. HEK293 cells were transiently transfected with Cerulean-tagged nanobodies and Venus-CFTR. Fig. 5c shows robust FRET between Cerulean-nb.E3h (our lead nanobody chosen for its efficacy in functional experiments described below) and Venus-CFTR compared to negative control cells expressing Cerulean + Venus-CFTR (Fig. 5c), confirming binding of this nanobody to full-length CFTR in situ. We utilized a halide-sensitive YFP quenching assay32 to initially screen different nanobodies for enDUB-mediated functional rescue of CFTR iodide currents. HEK293 cells transiently co-expressing either N1303K or 4326delTC and halide-sensitive YFP displayed YFP fluorescence that slowly declined monotonically over a 200-s recording period in response to forskolin + VX770 (Fig. 5d,e). For both these mutants, VX809 alone had marginal impact on the rate of quenching of YFP fluorescence (Fig. 5d,e). By contrast, in cells co-expressing enDUB-U21CF.E3h in the presence of VX809, the rate of forskolin-induced quenching of YFP fluorescence was accelerated, consistent with a rescue of functional CFTR channels (Fig. 5d,e). Patch clamp experiments directly confirmed the marked salutary effect of enDUB-U21CF.E3h on VX809 + VX770 in rescuing 4326delTC reconstituted in HEK293 cells (Fig. 5f). Further, we confirmed in pull-down experiments that enDUB-U21CF.E3h resulted in decreased ubiquitination of both co-expressed mCherry-tagged WT and N1303K CFTR channels (Extended Data Fig. 8).
We next tested the functional efficacy of CF-targeted enDUBs in a more predictive in vitro model of efficacy of CF-targeted therapeutics. We took advantage of Fischer Rat Thyroid (FRT) epithelial cells stably expressing mutant CFTR channels that have been used to generate preclinical data preceding clinical trials26,27 and to promote FDA drug label expansion of Kalydeco (VX770)33. Consistent with previous findings34,35, FRT cells stably expressing N1303K channels demonstrated little current compared to WT control cells, and were unresponsive to VX809 + VX770 treatment (Fig. 5g,h). Remarkably, enDUB-U21CF.E3h in combination with the same CFTR modulators rescued N1303K currents to ∼40% of WT cells (Fig. 5g,h). More recently, the FDA has approved Trikafta, a small molecule drug combination comprised of the potentiator VX770 and two correctors– VX661 (tezacaftor) and VX445 (elexacaftor)– for treatment of patients with at least one F508del allele36. We found that enDUB-U21CF.E3h synergized with Trikafta to rescue N1303K currents to 80% of WT CFTR levels (Extended Data Fig. 9a).
Dual-acting enDUB rescues the most common mutation F508del
F508del represents the most common CF mutation, with a phenylalanine deletion in NBD1 that leads to deficits in thermostability of CFTR folding, assembly, and trafficking37,38. In HEK293 cells, enDUB-U21CF.E3h resulted in only a modest improvement in F508del surface expression in the presence of VX809 (Fig. 6b). We hypothesized that an alternate enDUB with a dual capability to; 1) enhance NBD1 thermostability upon binding, and 2) tune CFTR ubiquitin status via catalytic action, would lead to improved F508del rescue. Notably, a recent study developed a nanobody (nb.T2a) that bound isolated WT and F508del NBD1 with thermostabilizing properties in cell-free preparations39. Flow-FRET experiments confirmed the interaction of Cerulean-nb.T2a with full-length Venus-CFTR in live cells (Fig. 6a). We tested the potential synergy of thermostabilizing enDUBs by adapting nb.T2a to our enDUB-U21 system (enDUB-U21CF.T2a). Although nb.T2a expression in combination with VX809 led to a modest increase in F508del surface trafficking, the functionalized enDUB-U21CF.T2a + VX809 demonstrated a significantly enhanced surface rescue in HEK293 cells (Fig. 6b). This superior functional rescue was corroborated in HEK293 patch-clamp studies, with enDUB-U21CF.T2a markedly improving F508del functional currents compared to VX809 ± nb.T2a alone (Fig. 6c). Finally, in FRT cells stably expressing F508del, combination treatment with enDUB-U21CF.T2a + VX809 + VX770 rescued F508del to ∼45% of WT levels, a substantial improvement compared with VX809 + VX770 ± nb.T2a treatment alone (Fig. 6d,e). When combined with Trikafta, enDUB-U21CF.T2a rescued F508del current to beyond WT CFTR levels (Extended Data Fig. 9b).
Figure 6. Dual-acting enDUBs synergize the functional rescue and apical localization of the most common F508del mutation.
a, Flow-FRET binding curves in cells expressing Venus-CFTR with either Cerulean-nb.T2a or Cerulean alone (n ≥ 10,000 cells per experiment; N = 2). b, Surface expression (BTX647) of F508del in different conditions (n ≥ 5000 cells per experiment; N = 4; mean ± s.e.m). Data are normalized to WT CFTR control group; dotted line represents F508del+VX809. †p=0.0417 versus CFP+VX809; **p<0.0002 versus all, one-way ANOVA with Tukey’s multiple comparison test. c, I-V curves for forskolin-activated currents from mCh-tagged WT CFTR (black circles, n = 41) or forskolin-activated, VX770-potentiated currents from F508del+VX809 and co-expressing CFP (red squares, n = 8), nb.T2a (green circles, n = 10) or enDUB-U21CF.T2a (blue circles, n = 9); n cells examined over ≥3 independent experiments (mean ± s.e.m.). *p=0.0401, two-way ANOVA with Tukey’s multiple comparison test. d, Exemplar forskolin-activated, VX770-potentiated currents in FRT cells stably expressing WT CFTR (left) or F508del after VX809 treatment and co-expressing either nb.T2a alone (middle) or enDUB-U21CF.T2a (right). e, I-V curves for forskolin-activated WT (black circles, n = 7) and F508del (red squares, n = 7) cells vs. VX809-treated, forskolin-activated, and VX770-potentiated F508del cells expressing CFP (orange diamonds, n = 6), nb.T2a (green triangles, n = 11), or enDUB-U21CF.T2a (blue triangles, n = 12); n cells examined over ≥3 independent experiments (mean ± s.e.m.). *p=0.0035, **p<0.0001, two-way ANOVA with Tukey’s multiple comparison test. f, Anti-CFTR immunofluorescence staining of transverse cryosections from WT or F508del homozygous patient hBECs cultured at ALI. Scale bar, 20 µm. Inset, CFTR expression at apical membrane. Scale bar, 5 µm. g, Quantification of apical CFTR density. WT + CFP (grey; n=25, N=3), F508del + CFP (red; n=24, N=3), F508del + CFP + VX809 (green; n=23, N=3), F508del + enDUB-U21CF.T2a + VX809 (blue; n=23, N=3); n transverse sections examined over N independent samples (mean ± s.e.m.). **p<0.0001, one-way ANOVA with Tukey’s multiple comparison test.
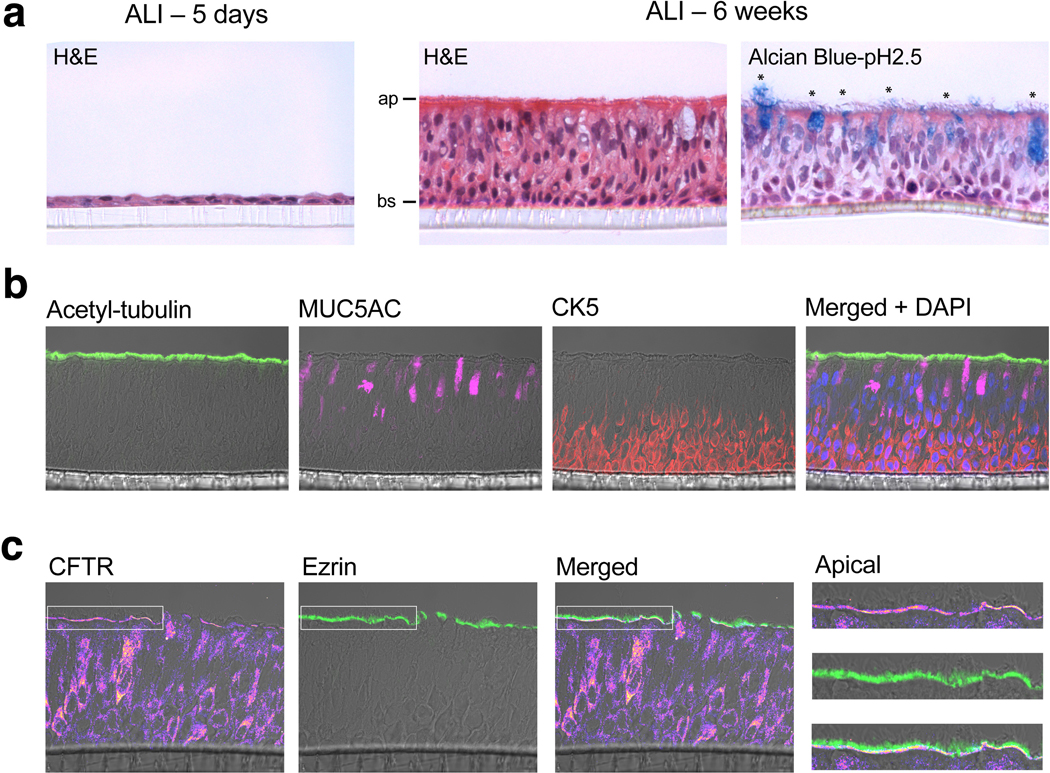
F508del disrupts apical localization of CFTR in airway epithelial cells, representing the basic archetype of CF pathology28,40,41. To assess enDUB-mediated rescue of endogenous CFTR expression in a patient-derived CF model, we cultured primary human bronchial epithelial cells (hBECs) from normal or F508del homozygous donors at air-liquid interface (ALI). After 4–6 weeks of ALI growth, hBECS formed a polarized pseudostratified epithelium and mature mucociliary phenotype, embedded with ciliated cells (acetylated-tubulin), mucus-producing goblet cells (MUC5AC), and a basal cell layer (CK5) (Extended Data Fig. 10a,b). Frozen transverse sections from mature ALI cultures were co-immunostained with antibodies against CFTR and the apical membrane marker, ezrin (Extended Data Fig. 10c). F508del mutant epithelia demonstrated a reduced apical expression of CFTR compared to WT (Fig. 6f,g). VX809 alone led to a modest upward trend in apical CFTR density, while enDUB-U21CF.T2a combination treatment resulted in a rescue of apical CFTR expression to WT levels (Fig. 6f,g).
Discussion
The ubiquitin-proteasome system (UPS) is a sought-after pathway for biological inquiry and therapeutic development, due to its critical role in governing protein fate. Yet the tools available to selectively interrogate ubiquitin signaling in living cells remains limited. Efforts to manipulate the UPS to promote protein stabilization have focused on three strategies: 1) inhibiting E1, E2 or E3 ligases, 2) activating endogenous DUBs, and 3) inhibiting the proteasome. A significant limitation of all three approaches stems from the prevalent and promiscuous roles these enzymes play in maintaining global proteostasis; targeting them often leads to confounding off-target effects that limits their scientific and therapeutic utility. Here, we introduce enDUBs as a novel strategy for stabilizing proteins that utilizes removal of ubiquitin from target proteins. The exceptional mechanistic insight and therapeutic potential of enDUBs is demonstrated by the ability to elucidate and rescue ubiquitin-responsive mutations across two disparate ion channelopathies, namely LQT1 and CF. To our knowledge, this is the first demonstration that trafficking-deficient KCNQ1 channels can be functionally corrected by deubiquitination, offering a new approach to develop drugs that prevent the sudden cardiac death associated with LQT1. Previous studies have established the importance of ubiquitination in the decreased surface density of CFTR in CF38,42,43. The potential of targeting CFTR ubiquitination to stabilize CF-causing mutant channels has been validated in knockdown studies of E3 ligases including CHIP and Nedd4L that are involved in the CFTR ERAD and peripheral quality control pathways42. However, inhibiting these E3 ligases to treat CF is problematic given their widespread involvement in cellular protein homeostasis. Our results show that enDUBs offer a new more selective path to deubiquitinate CFTR mutants for CF therapy. The synergistic rescue of the common F508del mutant by enDUB-U21CF.T2a + Orkambi aligns with the combination therapy strategy to provide new, improved treatments for CF. The impressive ability of enDUB-U21CF.E3h to switch N1303K from being modulator-insensitive to a rescuable phenotype provides a new rational approach to develop therapeutics for other rare CF mutations that are pharmaco-resistant.
In addition to LQT1 and CF, there are many other serious ion channelopathies underlain by trafficking-deficient ion channel mutations. Investigating the efficacy of enDUBs in rescuing other disease phenotypes is of significant interest and will likely expand the broad impact of this approach. Notably, interventions such as low temperature and nonspecific chemical chaperones (e.g. glycerol)44 can rescue distinct subsets of trafficking-impaired ion channels; however, these approaches are nonspecific and not amenable to therapeutic development. By contrast, enDUBs themselves are potential gene therapy reagents or can serve to inspire development of bivalent small molecules that direct endogenous deubiquitinases to target proteins. Finally, it is worth placing this work in context of the active field of targeted protein degradation, achieved through either small molecule proteolysis targeting chimeras (PROTACS)45 or with antibody-mimetics fused to ubiquitin ligase components46,47. Several biotechnology companies have emerged in the targeted protein degradation space, with efforts culminating in the first clinical trials using this therapeutic strategy in patients48. While methods for post-translational knockdown continue to develop49, the ability to enhance expression/stabilize endogenous targets at the protein level to interrogate biology has been lacking. In this context, our work represents the first technology and report of targeted protein stabilization, and demonstrates the general mechanistic utility and therapeutic potential for such an approach. Beyond ion channel pathology, the prevalence of ubiquitin-dependent mechanisms in the etiology of cancers50, viral infections51, and many other diseases offers a target rich environment for both basic and translational applications of enDUBs.
Beyond therapeutic applications, enDUBs have tremendous promise as enabling technology to probe ubiquitin signaling mechanisms in cells. Although great strides have been made in illuminating the structural and molecular basis for ubiquitin modifications of proteins in cell-free systems, there is less clarity in elucidating the complex ubiquitin code regulation of individual proteins in living cells. A technical barrier has been the inability to precisely manipulate ubiquitin modifications of particular proteins in situ. enDUBs provide a transformative tool that enables both superior control and adaptability in manipulating ubiquitin status of target proteins in the complex cellular environment, a development that facilitates a new approach to decipher ubiquitin code regulation of diverse proteins inside cells.
Online Methods
Molecular biology and cloning of plasmid vectors.
A customized bicistronic CMV mammalian expression vector (nano-xx-P2A-CFP) was generated as described previously46; we PCR amplified the coding sequence for GFP nanobody (vhhGFP4)18 and cloned it into xx-P2A-CFP using NheI/AflII sites. To generate the enDUB-O1 construct, we PCR amplified the OTU domain + UIM (residues 287–481) from OTUD1 (Addgene #61405) using AscI/AflII sites separated by a flexible GSG linker. To create the catalytically inactive enDUB-O1*, we introduced a point mutation at the catalytic cysteine residue [C320S] by site-directed mutagenesis. A second custom bicistronic vector (CFP-P2a-nano-xx) was generated as described previously46. To generate enDUB-U21, we PCR amplified the USP domain (residues 196–565) from USP21 (Addgene #22574) and cloned this fragment into CFP-P2a-nano-xx using AscI/NotI sites. To create the catalytically inactive enDUB-U21*, we introduced a point mutation at the catalytic cysteine residue [C221S] by site-directed mutagenesis. An mCh-targeted enDUB-U21 was generated with the mCh nanobody, LaM-452, using a similar cloning strategy as above.
KCNQ1 constructs were made as described previously20. Briefly, overlap extension PCR was used to fuse enhanced yellow fluorescent proteins (EYFP) in frame to the C-terminus of KCNQ1. A 13-residue bungarotoxin-binding site (BBS; TGGCGGTACTACGAGAGCAGCCTGGAGCCCTACCCCGAC)20,53 was introduced between residues 148–149 in the extracellular S1–S2 loop of KCNQ1 using the Quik-Change Lightning Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. LQT1 mutations were introduced in the N- and C-termini of KCNQ1 via site-directed mutagenesis. NEDD4L (PCI_NEDD4L; Addgene #27000) was a gift from Joan Massague54.
CFTR constructs were derived from pAd.CB-CFTR (ATCC® 75468). To create CFTR-YFP, PCR amplification was used to fuse EYFP to the N-terminus of CFTR. To create BBS-CFTR-YFP, overlap extension PCR was used to introduce the BBS site between residues 901–902 in the fourth extracellular loop (ECL4) of CFTR55. CF patient-specific mutations were introduced in NBD1, NBD2, and C-terminus of CFTR via site-directed mutagenesis. YFP halide sensor (EYFP H148Q/I152L) was a gift from Dr. Peter Haggie56 (Addgene #25872). To generate CF-targeted enDUBs, we created a modular CFP-P2a-xx-U21 vector with an extended (GGGGS)x5(GGGTG) linker upstream the USP domain. Select NBD1 nanobody binders were then cloned using BglII/AscI sites.
Generation of adenoviral vectors.
Adenoviral vectors were generated using the pAdEasy system (Stratagene) according to manufacturer’s instructions as previously described20. Plasmid shuttle vectors (pShuttle CMV) containing cDNA for nano-P2A-CFP, WT KCNQ1-YFP, and G589D KCNQ1-YFP were linearized with PmeI and electroporated into BJ5183-AD-1 electrocompetent cells pre-transformed with the pAdEasy-1 viral plasmid (Stratagene). PacI restriction digestion was used to identify transformants with successful recombination. Positive recombinants were amplified using XL-10-Gold bacteria, and the recombinant adenoviral plasmid DNA linearized with PacI digestion. HEK cells were cultured in 60 mm diameter dishes at 70–80% confluency and transfected with PacI-digested linearized adenoviral DNA. Transfected plates were monitored for cytopathic effects (CPEs) and adenoviral plaques. Cells were harvested and subjected to three consecutive freeze-thaw cycles, followed by centrifugation (2,500 × g) to remove cellular debris. The supernatant (2 mL) was used to infect a 10 cm dish of 90% confluent HEK293 cells. Following observation of CPEs after 2–3 d, cell supernatants were used to re-infect a new plate of HEK293 cells. Viral expansion and purification was carried out as previously described20. Briefly, confluent HEK293 cells grown on 15 cm culture dishes (x8) were infected with viral supernatant (1 mL) obtained as described above. After 48 h, cells from all of the plates were harvested, pelleted by centrifugation, and resuspended in 8 mL of buffer containing (in mM) Tris·HCl 20, CaCl2 1, and MgCl2 1 (pH 8.0). Cells were lysed by four consecutive freeze-thaw cycles and cellular debris pelleted by centrifugation. The virus-laden supernatant was purified on a cesium chloride (CsCl) discontinuous gradient by layering three densities of CsCl (1.25, 1.33, and 1.45 g/mL). After centrifugation (50,000 rpm; SW41Ti Rotor, Beckman-Coulter Optima L-100K ultracentrifuge; 1 h, 4 °C), a band of virus at the interface between the 1.33 and 1.45 g/mL layers was removed and dialyzed against PBS (12 h, 4 °C). Adenoviral vector aliquots were frozen in 10% glycerol at −80ºC until use. Generation of enDUB-O1-P2A-CFP, CFP-P2a-enDUB-U21CF.T2a, and CFP was performed by Vector Biolabs (Malvern, PA).
Cell culture and transfections.
Human embryonic kidney (HEK293) cells were a kind gift from the laboratory of Dr. Robert Kass (Columbia University). Cells were mycoplasma free, as determined by the MycoFluor Mycoplasma Detection Kit (Invitrogen). Low passage HEK293 cells were cultured at 37ºC in DMEM supplemented with 8% fetal bovine serum (FBS) and 100 mg/mL of penicillin–streptomycin. HEK293 cell transfection was accomplished using the calcium phosphate precipitation method. Briefly, plasmid DNA was mixed with 62 μL of 2.5M CaCl2 and sterile deionized water (to a final volume of 500 μL). The mixture was added dropwise, with constant tapping to 500 μL of 2x Hepes buffered saline containing (in mM): Hepes 50, NaCl 280, Na2HPO4 1.5, pH 7.09. The resulting DNA–calcium phosphate mixture was incubated for 20 min at room temperature and then added dropwise to HEK293 cells (60 – 80% confluent). Cells were washed with Ca2+-free phosphate buffered saline after 4–6 h and maintained in supplemented DMEM.
Chinese hamster ovary (CHO) cells were obtained from ATCC and cultured at 37ºC in Kaighn’s Modified Ham’s F-12K (ATCC) supplemented with 8% FBS and 100 mg/mL of penicillin–streptomycin. CHO cells were transiently transfected with desired constructs in 35 mm tissue culture dishes—KCNQ1 (0.5 µg) and nano-P2A-CFP (0.5 µg) or enDUB-O1-P2A-CFP (0.5µg) using X-tremeGENE HP (1:2 DNA/reagent ratio) according to the manufacturers’ instructions (Roche).
FRT epithelial cells stably-expressing WT and mutant CFTR channels34 were kindly provided by Dr. Eric Sorscher (Emory University). FRT cells were maintained at 37ºC in Ham’s F-12 Coon’s modification (Sigma) supplemented with 5% FBS, 100 mg/mL of penicillin–streptomycin, 7.5% w/v sodium bicarbonate, and 100 µg/mL Hygromycin (Invitrogen). FRT cell transient transfection was accomplished using Lipofectamine 3000 according to the manufacturer’s instructions (Thermo).
Cardiomyocyte isolation and transduction
Isolation of adult guinea pig cardiomyocytes was performed in accordance with the guidelines of Columbia University Animal Care and Use Committee. Prior to isolation, plating dishes were pre-coated with 15 µg/mL laminin (Gibco). Adult Hartley guinea pigs (Charles River) were euthanized with 5% isoflurane, hearts were excised and ventricular myocytes isolated by first perfusing in KH solution (mM): 118 NaCl, 4.8 KCl, 1 CaCl2 25 HEPES, 1.25 K2HPO4, 1.25 MgSO4, 11 glucose, 0.02 EGTA, pH 7.4, followed by KH solution without calcium using a Langendorff perfusion apparatus. Enzymatic digestion with 0.3 mg/mL Collagenase Type 4 (Worthington) with 0.08 mg/mL protease and 0.05% BSA was performed in KH buffer without calcium for six minutes. After digestion, 40 mL of a high K+ solution was perfused through the heart (mM): 120 potassium glutamate, 25 KCl, 10 HEPES, 1 MgCl2, and 0.02 EGTA, pH 7.4. Cells were subsequently dispersed in high K+ solution. Healthy rod-shaped myocytes were cultured in Medium 199 (Life Technologies) supplemented with (mM): 10 HEPES (Gibco), 1x MEM non-essential amino acids (Gibco), 2 L-glutamine (Gibco), 20 D-glucose (Sigma Aldrich), 1% vol vol−1 penicillin-streptomycin-glutamine (Fisher Scientific), 0.02 mg/mL Vitamin B-12 (Sigma Aldrich) and 5% (vol/vol) FBS (Life Technologies) to promote attachment to dishes. After 5 hrs, the culture medium was switched to Medium 199 with 1% (vol/vol) serum, but otherwise supplemented as described above. Cultures were maintained in humidified incubators at 37ºC and 5% CO2. Adenoviral vectors were added to the media overnight, followed by a wash with fresh media the next day. Experiments with cardiomyocytes were performed 1–2 days later.
Culturing of human bronchial epithelial cells (hBECs) at air-liquid interface (ALI)
Primary hBECs from CF (F508del/F508del) and normal control patients were obtained from a commercially available source (Lonza). Cryovials were expanded using conditional reprogramming culture (CRC) approach57–59. A feeder-free expansion media (PneumaCult-Ex Plus, STEMCELL Technologies), containing Rho-associated protein kinase (ROCK) inhibitor and TGF-B inhibitor, was used to expand cells in a basal-like state. Cells were seeded onto collagen IV coated Transwell inserts (Corning) at a density of 1.25 × 105 per insert and maintained in expansion media. Upon obtaining a confluent cell monolayer, the apical cell surface was exposed to air and the basolateral chamber was exchanged with ALI differentiation media (PneumaCult-ALI, STEMCELL Technologies). Basal media was exchanged every 2–3 days and the apical surface was washed weekly with Ca2+-free DPBS to remove mucus. Cells grown for approximately 4–6 weeks at ALI demonstrated markers of a mature mucociliary epithelium, including ciliated and mucus-containing (goblet) cells. Cells were transduced by overnight addition of adenovirus in the basolateral compartment, followed by a wash with fresh media the next day. Experiments were performed 2–3 days later.
Flow cytometry assay of total and surface channels.
Cell surface and total ion channel pools were assayed by flow cytometry in live, transfected HEK293 cells as previously described46,60. Briefly, 48 hrs post-transfection, cells cultured in 12-well plates were gently washed with ice cold PBS containing Ca2+ and Mg2+ (in mM: 0.9 CaCl2, 0.49 MgCl2, pH 7.4), and then incubated for 30 min in blocking medium (DMEM with 3% BSA) at 4ºC. HEK293 cells were then incubated with 1 μM Alexa Fluor 647 conjugated α-bungarotoxin (BTX647; Life Technologies) in DMEM/3% BSA on a rocker at 4ºC for 1 hr, followed by washing three times with PBS (containing Ca2+ and Mg2+). Cells were gently harvested in Ca2+-free PBS, and assayed by flow cytometry using a BD LSRII Cell Analyzer (BD Biosciences, San Jose, CA, USA). CFP- and YFP-tagged proteins were excited at 405 and 488 nm, respectively, and Alexa Fluor 647 was excited at 633 nm.
FRET flow cytometric assay.
FRET binding assays were performed via flow cytometry in live, transfected HEK293 cells as previously described61. Briefly, cells were cultured for 24 hours post-transfection and incubated for 2–4 hrs with cycloheximide (100 µM) and 30 min with H89 (30 µM) prior to analysis to reduce cell variation in fluorescent protein maturity and basal kinase activity. Cells were gently washed with ice cold PBS (containing Ca2+ and Mg2+), harvested in Ca2+-free PBS, and assayed by flow cytometry using a BD LSRII Cell Analyzer (BD Biosciences, San Jose, CA, USA). Cerulean (Cer), Venus (Ven), and FRET signals were analyzed using the following laser / filter set configurations: BV421 (Ex: 405 nm, Em: 450/50), FITC (Ex: 488 nm, Em: 525/50), and BV520 (Ex: 405 nm, Em: 525/50), respectively. Several controls were prepared for each experiment, including untransfected blanks for background subtraction, single color Ven and Cer for spectral unmixing, Cer + Ven co-expressed together for concentration-dependent spurious FRET estimation, as well as a series of Cer-Ven dimers for FRET calibration. Custom Matlab software was used to analyze FRET donor / acceptor efficiency and generate FRET binding curves as a function of [acceptor]free and [donor]free.
Electrophysiology.
For potassium channel measurements, whole-cell membrane currents were recorded at room temperature in CHO cells using an EPC-10 patch-clamp amplifier (HEKA Electronics) controlled by the Pulse software (HEKA). A coverslip with adherent CHO cells was placed on the glass bottom of a recording chamber (0.7–1 mL in volume) mounted on the stage of an inverted Nikon Eclipse Ti-U microscope. Micropipettes were fashioned from 1.5 mm thin-walled glass and fire-polished. Internal solution contained (mM): 133 KCl, 0.4 GTP, 10 EGTA, 1 MgSO4, 5 K2ATP, 0.5 CaCl2, and 10 HEPES (pH 7.2). External solution contained (in mM): 147 NaCl, 4 KCl, 2 CaCl2, and 10 HEPES (pH 7.4). Pipette resistance was typically 1.5 MΩ when filled with internal solution. I–V curves were generated from a family of step depolarizations (−40 to +100 mV in 10 mV steps from a holding potential of −80 mV). Currents were sampled at 20 kHz and filtered at 5 kHz. Traces were acquired at a repetition interval of 10 s.
Whole-cell recordings of cardiomyocytes were performed 48–72 hrs after infection. Internal and external solutions were used as above. Slow voltage ramp protocol (from −80 mv to +100 mV over 2 s) was used to evoke whole-cell currents. Action potential recordings under current clamp were obtained via 0.25 Hz stimulation with short current pulses (150 pA. 10 ms).
For CFTR channel measurements, whole-cell recordings were carried out in HEK293 and FRT cells at room temperature. Internal solution contained (mM): 113 L-aspartic acid, 113 CsOH, 27 CsCl, 1 NaCl, 1 MgCl2, 1 EGTA, 10 TES, 3 MgATP (pH 7.2). External contained (in mM): 145 NaCl, 4 CsCl, 1 CaCl2, 1 MgCl2, 10 glucose, and 10 TES (pH 7.4). I-V curves were generated from a family of step depolarizations (−80 to +80 mV in 20 mV steps from a holding potential of −40 mV). CFTR currents were activated by perfusion with 10 µM forskolin. In experiments utilizing VX809 (3 µM), the drug was added for 24 hrs post-transfection and incubated at 37ºC. VX770 was used acutely at 5 µM concentration. Currents were sampled at 20 kHz and filtered at 7 kHz. Traces were acquired at a repetition interval of 10 s.
Immunoprecipitation and Western blotting.
HEK293 cells were washed once with PBS without Ca2+, harvested, and resuspended in RIPA lysis buffer containing (in mM) Tris (20, pH 7.4), EDTA (1), NaCl (150), 0.1% (wt/vol) SDS, 1% Triton X-100, 1% sodium deoxycholate and supplemented with protease inhibitor mixture (10 μL/ mL, Sigma-Aldrich), PMSF (1 mM, Sigma-Aldrich), N-ethylmaleimide (2 mM, Sigma-Aldrich) and PR-619 deubiquitinase inhibitor (50 µM, LifeSensors). Lysates were prepared by incubation at 4ºC for 1 hr, with occasional vortex, and cleared by centrifugation (10,000 × g, 10 min, 4ºC). Supernatants were transferred to new tubes, with aliquots removed for quantification of total protein concentration determined by the bis-cinchonic acid protein estimation kit (Pierce Technologies). Lysates were pre-cleared by incubation with 10 µL Protein A/G Sepharose beads (Rockland) for 40 min at 4ºC and then incubated with 0.75 µg anti-Q1 (Alomone) for 1 hr at 4ºC. Equivalent total protein amounts were added to spin-columns containing 25 µL Protein A/G Sepharose beads, tumbling overnight at 4ºC. Equivalent total protein amounts of pre-cleared lysates for mCh-CFTR pulldowns were added directly to 20 µL RFP-Trap conjugated agarose beads (Chromotek, rta-20), tumbling overnight at 4ºC. Immunoprecipitates were washed twice with RIPA buffer, 3 times with high salt RIPA (500 mM NaCl), spun down at 500 × g, eluted with 40µL of warmed sample buffer [50 mM Tris, 10% (vol/vol) glycerol, 2% SDS, 100 mM DTT, and 0.2 mg/mL bromophenol blue], and boiled (55 °C, 15 min). Proteins were resolved on a 4–12% Bis·Tris gradient precast gel (Life Technologies) in Mops-SDS running buffer (Life Technologies) at 200 V constant for ∼1 h. We loaded 10 μL of the PageRuler Plus Prestained Protein Ladder (10–250 kDa, Thermo Fisher) alongside the samples. Protein bands were transferred by tank transfer onto a nitrocellulose membrane in transfer buffer (25 mM Tris pH 8.3, 192 mM glycine, 15% (vol/vol) methanol, and 0.1% SDS). The membranes were blocked with a solution of 5% nonfat milk (BioRad) in tris-buffered saline-tween (TBS-T) (25 mM Tris pH 7.4, 150 mM NaCl, and 0.1% Tween-20) for 1 hr at RT and then incubated overnight at 4 °C with primary antibodies (anti-Q1, Alomone APC-022, 1:1000; anti-CFTR, Millipore MM13–4, 1:400) in blocking solution. The blots were washed with TBS-T three times for 10 min each and then incubated with secondary horseradish peroxidase-conjugated antibody for 1 hr at RT. After washing in TBS-T, the blots were developed with a chemiluminiscent detection kit (Pierce Technologies) and then visualized on a gel imager. Membranes were then stripped with harsh stripping buffer (2% SDS, 62 mM Tris pH 6.8, 0.8% ß-mercaptoethanol) at 50ºC for 30 min, rinsed under running water for 2 min, and washed with TBST (3x, 10 min). Membranes were pre-treated with 0.5% glutaraldehyde and re-blotted with anti-ubiquitin (LifeSensors VU1, 1:500) as per the manufacturers’ instructions.
Yeast surface display for nanobody binders.
Isolation of nanobody binders were performed using a yeast surface display library approach previously described31. Human NBD1 (residues 387–646, Δ405–436) construct with an N-terminal Hisx6-Smt3 fusion was obtained from Arizona State University Plasmid Repository (clone: HsCD00287374). A FLAG tag was inserted immediately downstream the Hisx6-Smt3 tag using Gibson assembly. Proteins were expressed and His-purified via custom order (Genscript). The Hisx6-Smt3 tag was removed using SUMO protease kit (Invitrogen), with Ulp1 protease incubation overnight at 4ºC and subsequent affinity chromatography purification (HisPur spin columns; Thermo). A naïve yeast library (6 × 109 yeast) was incubated at 25ºC in galactose-containing tryptophan drop-out (Trp-) media for 2–3 days to induce nanobody expression. Induced cells were washed and resuspended in selection buffer (PBS, 0.1% BSA, 5mM maltose). First round of magnetic-activated cell sorting (MACS) selection began with a preclearing step, incubating yeast with anti-FLAG M2-FITC conjugated antibodies (Sigma, 1:100) and anti-FITC microbeads (Miltenyi) for 30 min at 4ºC and passing them through an LD column (Miltenyi) to remove antibody/microbead binders. NBD1-binding nanobodies were then MACS-enriched by incubating precleared yeast with 500 nM Hisx6-Smt3-FLAG-NBD1 and anti-FLAG M2-FITC for 1 hr at 4ºC, followed by a wash in selection buffer, and incubation with anti-FITC microbeads for 20 min at 4ºC. Labeled yeast were passed through an LS column (Miltenyi), washed three times with selection buffer, and eluted by removing the MACS magnetic stand (Miltenyi). Enriched NBD1 binders were grown up in glucose-containing Trp- media overnight at 30ºC. Induction of nanobody expression was repeated with enriched NBD1 libraries (∼1 × 108 yeast) by incubation in galactose Trp- media as outlined above. Subsequently, two rounds of positive selection were performed via fluorescence-activated cell sorting (FACS), first by incubating induced cells (∼5 × 106 yeast here, and thereafter) with 500 nM Hisx6-Smt3-FLAG-NBD1, and next with 500 nM FLAG-NBD1 to remove any Smt3 binders. Nonspecific FITC conjugated antibody binders were removed with a third round, negative selection FACS, incubating cells with anti-FLAG FITC alone. Finally, high affinity NBD1 binders were selected by incubation with 100 nM FLAG-NBD1, FACS sorted as single cells into 96-well plates, and grown up as monoclonal colonies for binding validation studies and plasmid isolation. Unique, validated NBD1 binders were subjected to on-yeast Kd measurements by labeling cells (∼105 yeast) with serial dilutions of FLAG-NBD1 (in nM): 5000, 1000, 500, 100, 50, 10, and 1.
YFP halide quenching assay.
A YFP halide quenching plate-reader assay was adapted from previous work32. Briefly, HEK293 cells were split onto 24-well black-wall plates (VisiPlate; PerkinElmer) and co-transfected with halide-sensitive eYFP (H148Q/I152L), mCh alone or mCh-tagged mutant CFTR channels, and CFP alone or CFP-P2a CF-targeted enDUB-U21 constructs. After 2–3 days, cells were washed once with PBS (containing Ca2+ and Mg2+) and incubated at 37ºC for 30 min in 200 µL PBS (containing 145 mM NaCl). Baseline YFP readings (Ex: 510 nm, Em: 538 nm) were taken using a SpectraMax M5 plate-reader (Molecular Devices). An equal amount of 2x activation solution, containing iodide, was added to obtain a final concentration (70mM NaI, 10 µM forskolin, 5 µM VX770), and a time series recording YFP fluorescence was taken every 2 s. Assays were performed at 37ºC.
Histology and immunostaining of hBEC-ALI cultures
HBECs grown on Transwell inserts at air-liquid interface (ALI) were washed twice with Ca2+-free DPBS on both apical and basal compartments and fixed with 4% formaldehyde for 20 min at room temperature (RT). Inserts were washed twice with Ca2+-free DPBS, incubated in 30% sucrose at 4ºC overnight, and embedded in OCT compound. Cryosections were cut on edge at 6 µm and prepared for Hematoxylin and Eosin (H&E) or Periodic acid–Schiff (PAS) (HICCC Molecular Pathology Core).
Cryosections prepared for immunofluorescent staining were rehydrated in PBS for 10min, fixed in chilled methanol at −20ºC for 5 min and permeabilized in 0.2% Triton X-100 in PBS for 5 min at RT. Sections were subsequently blocked with 10% NGS / 1% BSA / 0.1% Triton X-100 / PBS for 1–2 hrs at RT and incubated with primary antibodies for CFTR (R&D, 24–1, 1:500) and ezrin (Sigma, clone 3C12, 1:1000) in blocking solution overnight at 4ºC. Sections were then washed with 0.2% Triton X-100 / PBS for 5min (4x), incubated with secondary antibodies (goat anti-mouse IgG1 CF647, Sigma SAB4600353, 1:1000; goat anti-mouse IgG2a CF568, Sigma SAB4600315, 1:1000) for 1 hr at RT, and washed in 0.2% Triton X-100 / PBS for 5min (4x). Coverslips were mounted with Fluoromount-G (SouthernBiotech). Other primary antibodies used to characterize ALI cultures included anti-cytokeratin 5 (Novus, EP1601Y, 1:100), anti-MUC5AC (Novus, clone 45M1, 1:100), and anti-acetylated tubulin (Sigma, clone 6–11B-1, 1:1000), as well as secondary antibodies (goat anti-mouse IgG2b CF488A, Sigma SAB4600241, 1:1000; goat anti-Rabbit IgG AF568, Thermo Cat# A-11011, 1:1000).
Confocal microscopy.
Cardiomyocytes were plated onto 35 mm MatTek dishes (MatTek Corporation) and fixed with 4% formaldehyde for 10 min at room temperature (RT). Live HEK293 cells were stained with BTX647 as above. Images were captured on a Nikon A1RMP confocal microscope with a 40x oil immersion objective. Immunostained HBEC-ALI sections were captured using a 60x oil immersion objective
Data and statistical analyses
Data were acquired using BD FACSDiva (flow cytometry), Softmax Pro (plate reader), and NIS-Elements (confocal imaging). Data were analyzed off-line using FlowJo, FitMaster (HEKA), PyMOL, Microsoft Excel, Origin and GraphPad Prism software. Statistical analyses were performed in Origin or GraphPad Prism using built-in functions. Statistically significant differences between means (p< 0.05) were determined using one-way ANOVA with Tukey’s multiple comparison test or two-tailed unpaired t test for comparisons between two groups. Data are presented as means ± s.e.m unless otherwise noted.
Data availability statement
All data generated or analyzed during this study are included in this published article (and its Extended Data files). All supporting data are available from the corresponding author upon reasonable request. CFTR structure was reproduced from PDB (PDB: 5UAK). Nanobody structure was reproduced from PDB (PDB: 3K1K).
Extended Data
Extended Data Fig. 1. KCNQ1 pulldown and ubiquitin analyses after cell lysis with modified RIPA buffer containing 1% SDS.
a, Left, KCNQ1 pulldowns probed with anti-KCNQ1 antibody from HEK293 cells expressing KCNQ1-YFP ± NEDD4L with nano alone or enDUB-O1. The four bands represent KCNQ1 monomer, dimer, trimer, and tetrameric species, respectively. Right, Anti-ubiquitin labeling of KCNQ1 pulldowns after stripping previous blot. b, Relative KCNQ1 ubiquitination computed by ratio of anti-ubiquitin to anti-KCNQ1 signal intensity (n = 3 independent experiments; mean). **p < 0.003, one-way ANOVA with Tukey’s multiple comparison test.
Extended Data Fig. 2. Flow cytometry gating strategy for BTX-647 surface labeling experiments.
a, Flow cytometry pseudocolor dot plots displaying an initial selection gate (left) for cells (vs debris); and a second selection gate (right) for singlets (vs doublets). b, Single color fluorescent controls after applying the gating strategy in a. c, Analysis gate established with single color controls in b to quantify channel surface density (BTX-647) in YFP- and CFP-positive cells (left) with cumulative distribution histograms (right). Sample 1 is exemplified by BBS-Q1-YFP + CFP-P2a-nano. Sample 2 is exemplified by BBS-Q1-YFP + CFP-P2a-nano + NEDD4L.
Extended Data Fig. 3. enDUB-O1 requires catalytic activity and target specificity for ubiquitin-dependent rescue of KCNQ1 channels.
a, (Left) Schematic of experimental strategy; BBS-Q1-YFP was co-transfected with either nanobody alone (grey line), NEDD4L + nano (red line), or NEDD4L + enDUB-O1 (blue line). (Right) Cumulative distribution histograms of Alexa647 fluorescence from flow cytometry analyses (data adapted from Fig. 1). Plot generated from population of YFP- and CFP-positive cells (n ≥ 5000 cells per experiment; N = 4). b, Same experiment as in a, but using catalytically inactive enDUB-O1* with C320S (N = 2). c, Same experiment as in a, but with untagged BBS-Q1 co-expressed with enDUB-O1 as a control for target specificity (N = 2).
Extended Data Fig. 4. enDUB-U21 has greater efficacy than enDUB-O1 in surface rescue of N1303K CFTR mutant channels.
a, Cumulative distribution histograms of Alexa647 fluorescence from flow cytometry analyses for cells expressing WT BBS-CFTR-YFP + nano (dotted line) and N1303K mutation co-expressing nano alone (red line), enDUB-O1 (cyan line), enDUB-U21 (blue line). Plot generated from population of YFP- and CFP-positive cells (n ≥ 5000 cells per experiment; N = 2). b, Same experimental design as above but with 24 hour incubation of VX809 with nano (green line), enDUB-O1 (cyan line) and enDUB-U21 (blue line) (N = 2).
Extended Data Fig. 5. enDUB-U21 requires catalytic activity and target specificity for ubiquitin-dependent rescue of CFTR mutants.
a, (Left) Schematic of experimental strategy; WT BBS-CFTR-YFP + nano (dashed line) or N1303K mutants co-transfected with nano (red line) or enDUB-U21 (blue line). Cumulative distribution histograms (middle) and quantification (right) of Alexa647 fluorescence from flow cytometry analyses (data adapted from Fig. 4). Plots generated from population of YFP- and CFP-positive cells (n ≥ 5000 cells per experiment; N = 3; mean ± s.e.m). Data are normalized to values from the WT CFTR control group (dotted line). b, Same experiment as in a, but using catalytically inactive enDUB-U21* with C221S (N = 3). c, Same experiment as in a, but with an mCherry-targeted nanobody, m-enDUB-U21, as a control for target specificity (N = 4).
Extended Data Fig. 6. enDUB-U21 increases functional rescue of 4326delTC CFTR mutant channels in combination with lumacaftor ± ivacaftor.
a, Exemplar family of basal (top, black), forskolin-activated (middle, red), and VX770-potentiated (bottom, green) currents for 4326delTC mutant channels after 24 hr VX809 treatment (3 µM) and co-expression with nano (left) or enDUB-U21 (right). b, Population I-V curves for basal (black squares), forskolin-activated (red circles), and VX770-potentiated (green triangles) currents from 4326delTC mutants co-expressing nano alone (left; n = 15) or enDUB-U21 (right; n = 14). c, Same format as a, but with N1303K mutants. d, Same format as b, but with N1303K mutants co-expressing nano alone (left; n = 9) or enDUB-U21 (right; n = 11). Data for VX770-potentiated currents adapted from Fig. 4.
Extended Data Fig. 7. Development of NBD1 binders from a yeast surface display nanobody library.
a, On-yeast binding affinity measurements of 9 nanobody clones using serial dilutions of purified FLAG-NBD1. b, Flow cytometric surface labeling assay and cumulative distribution histograms of WT CFTR surface density alone (dotted line) or when co-expressed with nanobody clones.
Extended Data Fig. 8. enDUB-U21CF.E3h deubiquitinates WT CFTR and N1303K.
a, Anti-ubiquitin labeling of WT CFTR and N1303K pulldowns. Lane 1 – untransfected cells; lane 2 – mCherry-CFTR + nanobody; lane 3 – mCherry-CFTR + enDUB-U21CF.E3h; lane 4 – mCherry-N1303K + nanobody; lane 5 – mCherry-N1303K + enDUB-U21CF.E3h. Proteins were pulled down with anti-mCherry and probed with anti-ubiquitin antibody (n = 2 independent experiments). b, Anti-CFTR labeling of WT CFTR and N1303K pulldowns. The top blot was stripped and probed with anti-CFTR antibody.
Extended Data Fig. 9. enDUB-U21 restores functional currents of N1303K and F508del CFTR mutant channels in combination with Trikafta.
a, Population I-V curves for forskolin-activated currents from FRT cells stably expressing WT CFTR (black circles, n = 57) and N1303K + CFP (red squares, n = 8), and forskolin-activated, VX770-potentiated currents from N1303K + CFP treated with VX661 + VX445 (green triangles, n = 9); N1303K + enDUB-U21CF.E3h treated with VX661 + VX445 (blue triangles, n = 9); n cells examined over ≥3 independent experiments (mean ± s.e.m). Data for N1303K + CFP adapted from Fig. 5. b, Population I-V curves for forskolin-activated currents from FRT cells stably expressing WT CFTR (black circles, n = 57) and F508del + CFP (red squares, n = 12), and forskolin-activated, VX770-potentiated currents from F508del + CFP treated with VX661 + VX445 (green triangles, n = 10); F508del + enDUB-U21CF.T2a treated with VX661 + VX445 (blue triangles, n = 9); n cells examined over ≥3 independent experiments (mean ± s.e.m). Data for F508del + CFP adapted from Fig. 6. *p < 0.03, **p < 0.009,***p < 0.0001, two-way ANOVA with Tukey’s multiple comparison test.
Extended Data Fig. 10. Mucociliary differentiation of human bronchial epithelial cells (hBECs) cultured at air-liquid interface (ALI).
a, H&E staining of immature (left; 5 day old) and mature hBEC ALI cultures (middle; 6 weeks), featuring a pseudostratified epithelium with mucin-containing goblet cells (*) (right). Apical (ap) and basal (bs) compartments labeled. b, Immunofluorescence staining of mature ALI cultures, featuring ciliated cells (green; acetylated-tubulin), mucin-containing goblet cells (pink; MUC5AC), and a basal cell layer (red; CK5), and merged image with DAPI staining (blue; nuclei). c, Immunostaining of mature WT hBEC cultures, with anti-CFTR signal (heat map) and anti-ezrin (green) labeling of the apical membrane. White box inset highlights the apical membrane (far right).
Acknowledgements
We thank Ming Chen for technical support, Dr. Manu Ben Johny (Columbia University) for help with the flow-FRET assay and helpful discussions, and Dr. Liza Pon (Columbia University) for guidance in initiating yeast cultures. This work was supported by grants RO1-HL121253 and 1RO1-HL122421 from the NIH (to H.M.C). S.A.K. was supported by a Medical Scientist Training Program grant (T32 GM007367) and NHLBI National Research Service Award (1F30-HL140878). This work was also supported by the TRx Accelerator of the CUMC Irving Institute, as supported by NCATS, NIH (UL1TR001873). Flow cytometry experiments were performed in the CCTI Flow Cytometry Core, supported in part by the NIH (S10RR027050). Confocal images were collected in the HICCC Confocal and Specialized Microscopy Shared Resource, supported by NIH (P30 CA013696).
Footnotes
Competing interests
S.A.K. and H.M.C have filed a patent application through Columbia University based on this work.
References
- 1.Kullmann DM. Neurological Channelopathies. Annual Review of Neuroscience 33, 151–172, doi: 10.1146/annurev-neuro-060909-153122 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Bohnen MS. et al. Molecular Pathophysiology of Congenital Long QT Syndrome. Physiological Reviews 97, 89–134, doi: 10.1152/physrev.00008.2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cutting GR. Cystic fibrosis genetics: from molecular understanding to clinical application. Nature Reviews Genetics 16, 45–56, doi: 10.1038/nrg3849 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashcroft FM. & Rorsman P. KATP channels and islet hormone secretion: new insights and controversies. Nature Reviews Endocrinology 9, 660, doi: 10.1038/nrendo.2013.166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imbrici P. et al. Therapeutic Approaches to Genetic Ion Channelopathies and Perspectives in Drug Discovery. Frontiers in Pharmacology 7, 121, doi: 10.3389/fphar.2016.00121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wulff H, Christophersen P, Colussi P, Chandy GK. & Yarov-Yarovoy V. Antibodies and venom peptides: new modalities for ion channels. Nature Reviews Drug Discovery, 1, doi: 10.1038/s41573-019-0013-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tester DJ, Will ML, Haglund CM. & Ackerman MJ. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart rhythm 2, 507–517, doi: 10.1016/j.hrthm.2005.01.020 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Wilson AJ, Quinn KV, Graves FM, Bitner-Glindzicz M. & Tinker A. Abnormal KCNQ1 trafficking influences disease pathogenesis in hereditary long QT syndromes (LQT1). Cardiovascular Research 67, 476–486, doi: 10.1016/j.cardiores.2005.04.036 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Haardt M, Benharouga M, Lechardeur D, Kartner N. & Lukacs GL. C-terminal truncations destabilize the cystic fibrosis transmembrane conductance regulator without impairing its biogenesis. A novel class of mutation. The Journal of biological chemistry 274, 21873–21877, doi: 10.1074/jbc.274.31.21873 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Cheng SH. et al. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell 63, 827–834 (1990). [DOI] [PubMed] [Google Scholar]
- 11.Curran J. & Mohler PJ. Alternative Paradigms for Ion Channelopathies: Disorders of Ion Channel Membrane Trafficking and Posttranslational Modification. Annual Review of Physiology 77, 1–20, doi: 10.1146/annurev-physiol-021014-071838 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Huang H. et al. Mechanisms of KCNQ1 channel dysfunction in long QT syndrome involving voltage sensor domain mutations. Sci Adv 4, eaar2631, doi: 10.1126/sciadv.aar2631 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foot N, Henshall T. & Kumar S. Ubiquitination and the Regulation of Membrane Proteins. Physiol Rev 97, 253–281, doi: 10.1152/physrev.00012.2016 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Nalepa G, Rolfe M. & Harper WJ. Drug discovery in the ubiquitin–proteasome system. Nature Reviews Drug Discovery 5, 596–613, doi: 10.1038/nrd2056 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Huang X. & Dixit VM. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Research 26, 484–498, doi: 10.1038/cr.2016.31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jespersen T. et al. The KCNQ1 potassium channel is down-regulated by ubiquitylating enzymes of the Nedd4/Nedd4-like family. Cardiovasc Res 74, 64–74, doi: 10.1016/j.cardiores.2007.01.008 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Mevissen T. et al. OTU Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis. Cell 154, 169–184, doi: 10.1016/j.cell.2013.05.046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothbauer U. et al. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Molecular & cellular proteomics : MCP 7, 282–289, doi: 10.1074/mcp.M700342-MCP200 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Abbott GW. et al. KCNQ1, KCNE2, and Na+-coupled solute transporters form reciprocally regulating complexes that affect neuronal excitability. Sci Signal 7, ra22, doi: 10.1126/scisignal.2005025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aromolaran AS, Subramanyam P, Chang DD, Kobertz WR. & Colecraft HM. LQT1 mutations in KCNQ1 C-terminus assembly domain suppress IKs using different mechanisms. Cardiovasc Res 104, 501–511, doi: 10.1093/cvr/cvu231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peroz D, Dahimène S, Baró I, Loussouarn G. & Mérot J. LQT1-associated Mutations Increase KCNQ1 Proteasomal Degradation Independently of Derlin-1. Journal of Biological Chemistry 284, 5250–5256, doi: 10.1074/jbc.M806459200 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Mattmann ME. et al. Identification of (R)-N-(4-(4-methoxyphenyl)thiazol-2-yl)-1-tosylpiperidine-2-carboxamide, ML277, as a novel, potent and selective Kv7.1 (KCNQ1) potassium channel activator. Bioorganic & Medicinal Chemistry Letters 22, 5936–5941, doi: 10.1016/j.bmcl.2012.07.060 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veit G. et al. From CFTR biology toward combinatorial pharmacotherapy: expanded classification of cystic fibrosis mutations. Molecular biology of the cell 27, 424–433, doi: 10.1091/mbc.E14-04-0935 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boeck K. & Amaral MD. Progress in therapies for cystic fibrosis. The Lancet Respiratory Medicine 4, 662–674, doi: 10.1016/S2213-2600(16)00023-0 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Wainwright CE. et al. Lumacaftor–Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del CFTR. The New England Journal of Medicine 373, 220–231, doi: 10.1056/NEJMoa1409547 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goor F. et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proceedings of the National Academy of Sciences 108, 18843–18848, doi: 10.1073/pnas.1105787108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goor F. et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proceedings of the National Academy of Sciences 106, 18825–18830, doi: 10.1073/pnas.0904709106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farinha CM. & Matos P. Repairing the basic defect in cystic fibrosis – one approach is not enough. FEBS Journal 283, 246–264, doi: 10.1111/febs.13531 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Faesen AC. et al. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem Biol 18, 1550–1561, doi: 10.1016/j.chembiol.2011.10.017 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Liu F, Zhang Z, Csanády L, Gadsby DC. & Chen J. Molecular Structure of the Human CFTR Ion Channel. Cell 169, 85–910065408, doi: 10.1016/j.cell.2017.02.024 (2017). [DOI] [PubMed] [Google Scholar]
- 31.McMahon C. et al. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nature Structural & Molecular Biology 25, 289–296, doi: 10.1038/s41594-018-0028-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galietta LV, Jayaraman S. & Verkman AS. Cell-based assay for high-throughput quantitative screening of CFTR chloride transport agonists. American journal of physiology. Cell physiology 281, 42, doi: 10.1152/ajpcell.2001.281.5.C1734 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Durmowicz AG, Lim R, Rogers H, Rosebraugh CJ. & Chowdhury BA. The U.S. Food and Drug Administration’s Experience with Ivacaftor in Cystic Fibrosis. Establishing Efficacy Using In Vitro Data in Lieu of a Clinical Trial. Annals of the American Thoracic Society 15, 1–2, doi: 10.1513/AnnalsATS.201708-668PS (2018). [DOI] [PubMed] [Google Scholar]
- 34.Han ST. et al. Residual function of cystic fibrosis mutants predicts response to small molecule CFTR modulators. JCI Insight 3, doi: 10.1172/jci.insight.121159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goor F, Yu H, Burton B. & Hoffman BJ. Effect of ivacaftor on CFTR forms with missense mutations associated with defects in protein processing or function. Journal of Cystic Fibrosis 13, 29–36, doi: 10.1016/j.jcf.2013.06.008 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Middleton PG. et al. Elexacaftor-Tezacaftor-Ivacaftor for Cystic Fibrosis with a Single Phe508del Allele. N Engl J Med 381, 1809–1819, doi: 10.1056/NEJMoa1908639 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukacs GL. & Verkman AS. CFTR: folding, misfolding and correcting the ΔF508 conformational defect. Trends in Molecular Medicine 18, 81–91, doi: 10.1016/j.molmed.2011.10.003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okiyoneda T. et al. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science (New York, N.Y.) 329, 805–810, doi: 10.1126/science.1191542 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sigoillot M. et al. Domain-interface dynamics of CFTR revealed by stabilizing nanobodies. Nature Communications 10, doi:ARTN 2636 10.1038/s41467-019-10714-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kreda SM. et al. Characterization of wild-type and deltaF508 cystic fibrosis transmembrane regulator in human respiratory epithelia. Mol Biol Cell 16, 2154–2167, doi: 10.1091/mbc.e04-11-1010 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Meegen MA, Terheggen-Lagro SW, Koymans KJ, van der Ent CK. & Beekman JM. Apical CFTR expression in human nasal epithelium correlates with lung disease in cystic fibrosis. PLoS One 8, e57617, doi: 10.1371/journal.pone.0057617 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukuda R. & Okiyoneda T. Peripheral Protein Quality Control as a Novel Drug Target for CFTR Stabilizer. Front Pharmacol 9, 1100, doi: 10.3389/fphar.2018.01100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meacham GC, Patterson C, Zhang W, Younger JM. & Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol 3, 100–105, doi: 10.1038/35050509 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Delisle BP. et al. Biology of Cardiac Arrhythmias. Circulation Research 94, 1418–1428, doi: 10.1161/01.RES.0000128561.28701.ea (2004). [DOI] [PubMed] [Google Scholar]
- 45.Schapira M, Calabrese MF, Bullock AN. & Crews CM. Targeted protein degradation: expanding the toolbox. Nat Rev Drug Discov 18, 949–963, doi: 10.1038/s41573-019-0047-y (2019). [DOI] [PubMed] [Google Scholar]
- 46.Kanner SA, Morgenstern T. & Colecraft HM. Sculpting ion channel functional expression with engineered ubiquitin ligases. eLife 6, doi: 10.7554/eLife.29744 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caussinus E, Kanca O. & Affolter M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol 19, 117–121, doi: 10.1038/nsmb.2180 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Mullard A. First targeted protein degrader hits the clinic. Nat Rev Drug Discov, doi: 10.1038/d41573-019-00043-6 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Wu T. et al. Targeted protein degradation as a powerful research tool in basic biology and drug target discovery. Nat Struct Mol Biol 27, 605–614, doi: 10.1038/s41594-020-0438-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi D. & Grossman SR. Ubiquitin becomes ubiquitous in cancer: emerging roles of ubiquitin ligases and deubiquitinases in tumorigenesis and as therapeutic targets. Cancer Biol Ther 10, 737–747, doi: 10.4161/cbt.10.8.13417 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isaacson MK. & Ploegh HL. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 5, 559–570, doi: 10.1016/j.chom.2009.05.012 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 52.Fridy PC. et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nature Methods 11, 1253–1260, doi: 10.1038/nmeth.3170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sekine-Aizawa Y. & Huganir RL. Imaging of receptor trafficking by using alpha-bungarotoxin-binding-site-tagged receptors. Proc Natl Acad Sci U S A 101, 17114–17119, doi: 10.1073/pnas.0407563101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao S. et al. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol Cell 36, 457–468, doi: 10.1016/j.molcel.2009.09.043 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peters KW. et al. CFTR Folding Consortium: methods available for studies of CFTR folding and correction. Methods Mol Biol 742, 335–353, doi: 10.1007/978-1-61779-120-8_20 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galietta LJ, Haggie PM. & Verkman AS. Green fluorescent protein-based halide indicators with improved chloride and iodide affinities. FEBS letters 499, 220–224, doi: 10.1016/s0014-5793(01)02561-3 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Gentzsch M. et al. Pharmacological Rescue of Conditionally Reprogrammed Cystic Fibrosis Bronchial Epithelial Cells. Am J Respir Cell Mol Biol 56, 568–574, doi: 10.1165/rcmb.2016-0276MA (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang C. et al. Long-Term In Vitro Expansion of Epithelial Stem Cells Enabled by Pharmacological Inhibition of PAK1-ROCK-Myosin II and TGF-beta Signaling. Cell Rep 25, 598–610 e595, doi: 10.1016/j.celrep.2018.09.072 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rayner RE, Makena P, Prasad GL. & Cormet-Boyaka E. Optimization of Normal Human Bronchial Epithelial (NHBE) Cell 3D Cultures for in vitro Lung Model Studies. Sci Rep 9, 500, doi: 10.1038/s41598-018-36735-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanner SA, Jain A. & Colecraft HM. Development of a High-Throughput Flow Cytometry Assay to Monitor Defective Trafficking and Rescue of Long QT2 Mutant hERG Channels. Frontiers in Physiology 9, 397, doi: 10.3389/fphys.2018.00397 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee S-R, Sang L. & Yue DT. Uncovering Aberrant Mutant PKA Function with Flow Cytometric FRET. Cell Reports 14, 3019–3029, doi: 10.1016/j.celrep.2016.02.077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Extended Data files). All supporting data are available from the corresponding author upon reasonable request. CFTR structure was reproduced from PDB (PDB: 5UAK). Nanobody structure was reproduced from PDB (PDB: 3K1K).