Significance
Epstein-Barr virus (EBV) is a human oncogenic herpesvirus strongly associated with multiple sclerosis. Like all herpesviruses, EBV switches between a productive lytic stage and latent stage. In its latent stage, EBV resides in B cells during its host’s lifetime. The EBV nuclear antigen 2 (EBNA2) exposes a five–amino acid α1-helix on its surface that recruits early B cell factor 1 (EBF1) to activate critical metabolic processes of resting B cells upon infection. Our study demonstrates how EBNA2 exploits EBF1, a key factor of B cell lineage specification, to initiate proliferation and highlights the EBNA2 α1-helix as a potential Achilles heel of the virus at the critical stage when latent infection of the host is established.
Keywords: Epstein-Barr virus, B cell activation, MYC expression, transcription factor, RNA sequencing
Abstract
Epstein-Barr virus (EBV) is a human tumor virus which preferentially infects resting human B cells. Upon infection in vitro, EBV activates and immortalizes these cells. The viral latent protein EBV nuclear antigen 2 (EBNA2) is essential for B cell activation and immortalization; it targets and binds the cellular and ubiquitously expressed DNA-binding protein CBF1, thereby transactivating a plethora of viral and cellular genes. In addition, EBNA2 uses its N-terminal dimerization (END) domain to bind early B cell factor 1 (EBF1), a pioneer transcription factor specifying the B cell lineage. We found that EBNA2 exploits EBF1 to support key metabolic processes and to foster cell cycle progression of infected B cells in their first cell cycles upon activation. The α1-helix within the END domain was found to promote EBF1 binding. EBV mutants lacking the α1-helix in EBNA2 can infect and activate B cells efficiently, but activated cells fail to complete the early S phase of their initial cell cycle. Expression of MYC, target genes of MYC and E2F, as well as multiple metabolic processes linked to cell cycle progression are impaired in EBVΔα1-infected B cells. Our findings indicate that EBF1 controls B cell activation via EBNA2 and, thus, has a critical role in regulating the cell cycle of EBV-infected B cells. This is a function of EBF1 going beyond its well-known contribution to B cell lineage specification.
Epstein-Barr virus (EBV) is a γ-herpesvirus which is associated with diverse malignancies such as nasopharyngeal and gastric carcinoma, posttransplant lymphoproliferative disease, Burkitt lymphoma, and diffuse large B cell, Hodgkin, natural killer, and T cell lymphomas (1). In addition, EBV primary infection in adolescence frequently causes infectious mononucleosis. There is mounting epidemiological evidence that latent EBV infection precedes and is a prerequisite for the development of multiple sclerosis (2, 3). Like other herpesviruses, EBV switches between lytic infection to produce infectious virus and latent infection to maintain a lifelong persistent reservoir in the infected host. EBV can infect epithelial and lymphoid cells, but the viral reservoir of EBV in vivo is nonproliferating memory B cells. To reach this long-lived B cell compartment, the virus initially infects resting human B cells, activates these, and drives their proliferation. This process is considered the essential step to initiate persistent infection in vivo. It can be studied in short-term and long-term cell culture infection models and is referred to as B cell immortalization (4). It requires the timely expression and collaborative action of a group of nuclear viral proteins termed EBV nuclear antigens (EBNAs; specifically, EBNA1, 2, 3A, 3B, 3C, and EBNA-LP), as well as a group of latent membrane proteins (LMPs) termed LMP1 and LMP2.
EBNA2 initiates the immortalization process by directly and immediately activating cellular target genes, including the cellular proto-oncogene MYC (5), as well as by activating certain viral promoters. Several studies have demonstrated that mutant EBVs devoid of EBNA2 cannot immortalize primary human B cells (6–10). Most recently, a panel of viral mutants, deficient for the expression of single latent genes, has been studied during the early phase of the immortalization process. Apart from EBV devoid of EBNA2, all other EBV mutants efficiently supported cell cycle activation (11). This finding highlights the essential role of EBNA2 to initiate the complex process of B cell immortalization.
EBNA2 is a transcription factor that uses cellular, sequence-specific DNA-binding proteins as DNA anchors, since it does not bind DNA directly. EBNA2’s long-known anchor protein is CBF1, a sequence-specific DNA-binding factor which is highly conserved and ubiquitously expressed. EBNA2 mutants incapable of CBF1 binding fail to immortalize B cells in vitro (12). Chromatin immunoprecipitation (ChIP) studies map EBNA2 binding to B cell–specific super enhancers, which are present in primary resting B cells prior to infection (13, 14). Due to the ubiquitous expression of CBF1 in all human cells, it does not confer B cell specificity to EBNA2. Early B cell factor 1 (EBF1) is another DNA-binding protein associated with EBNA2 (15). Interestingly, EBF1 and CBF1 frequently co-occupy high-affinity EBNA2-binding sites, and EBNA2 can enhance EBF1 and CBF1 signal intensities, as demonstrated by ChIP sequencing experiments. This indicates that EBNA2 not only uses transcription factors as DNA anchors but also influences transcription-factor densities at enhancer and superenhancer sites in B cells (16, 17). The EBNA2 N-terminal dimerization (END) domain forms a dimeric globular structure (18). EBF1 promotes the assembly of EBNA2 chromatin complexes in B cells, and the END domain is sufficient to mediate the interaction between EBNA2 and EBF1 (15). The binding site for CBF1 maps to a central region encompassing residues 318 through 327 of EBNA2 and does not involve the END domain (19). CBF1 is not required for EBNA2–EBF1 complex formation (15). In addition, the transcription factor PU.1 is an EBNA2-associated protein (20) that binds to regulatory chromatin sites controlled by EBNA2.
The pioneer factor EBF1 can open chromatin and control B cell–specific gene expression, and it triggers MYC expression and proliferation during mouse early B cell development (21, 22). In addition, EBF1 is required for the generation and survival of distinct mature mouse B cell populations and their proliferation in response to activating signals (23, 24). To date, the function of EBF1 in primary human B cells has not been studied, to our knowledge. In human cell lines, EBF1 expression strongly correlates with transcription of hallmark B cell genes, confirming that EBF1 defines B cell identity in the human system as well. Whether human EBF1 controls B cell proliferation could not be studied in the respective cell-culture model systems (25, 26).
We have previously presented the three-dimensional structure of the END domain. The 58–amino acid residue END domain forms a compact homodimer, which is stabilized by a hydrophobic interface between the two monomers. While interface mutants displayed impaired dimerization, mutagenesis of selected amino acid residues exposed on the hydrophilic surface, H15 and the α1-helix, impaired EBNA2 transactivation without affecting dimerization or binding of CBF1 (18). The goal of this study was to characterize the specific functional impact of EBF1 on the activity of EBNA2 by reverse genetics. Here, we show that an EBNA2 α1-helix deletion mutant (EBNA2Δα1) did not form complexes with EBF1 but bound perfectly well to chromatin regions known to tether EBNA2 via CBF1. EBV mutants carrying the α1-helix deletion in EBNA2 (EBVΔα1) infected and activated primary B cells, but cell cycle progression was severely compromised, and most cells arrested in the early S phase. Long-term cultures of EBVΔα1-infected B cells could only be expanded by continuous CD40 stimulation, indicating that EBF1 is absolutely required to maintain B cells in their EBV-driven immortalized state. RNA-sequencing experiments revealed that EBVΔα1 poorly activated MYC and gene sets known to be targets of MYC and E2F, and failed to promote metabolic processes linked to cell cycle progression; thus explaining the failure of EBVΔα1 to immortalize primary B cells.
RESULTS
EBNA2-EBF1 Complex Formation Requires the END Domain with Its α1-Helix.
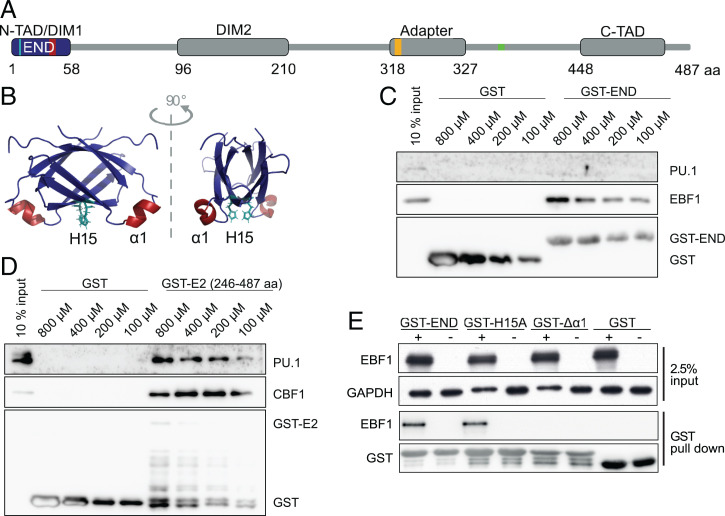
To test if the END domain interacts with PU.1 or CBF1, we performed GST pull-down assays using either the END domain or a C-terminal fragment of EBNA2 as a bait (Fig. 1 A and B). Unlike EBF1, the transcription factor PU.1 did not bind to the END domain but bound a C-terminal fragment of EBNA2 (246 through 487) (Fig. 1C) as reported before (20). The interaction of CBF1 has been mapped to EBNA2 residues 318 through 327 (Fig. 1A) (19) and could be confirmed by GST pull down with the C-terminal fragment of EBNA2 (246 through 487) (Fig. 1D). To identify critical regions within the END domain, we investigated mutants of the END domain in GST pull-down assays with EBF1 containing whole-protein lysates. The single amino acid mutant H15A and a deletion of the α1-helix (E2Δα1) were analyzed (Fig. 1E). While the H15A missense mutant retained most of its EBF1 binding, the END domain mutant lacking the α1-helix (E2Δα1) failed to bind EBF1 (Fig. 1E).
Fig. 1.
The α1-helix on the surface of the END binds EBF1. (A) Schematic representation of functional EBNA2 modules: the END domain (blue) with positions of the α1-helix (amino acid residues 35 through 39; red) and histidine 15 (H15; turquoise), N- and C-terminal transactivation domains (N-TAD and C-TAD, respectively), two dimerization domains (DIM1 and DIM2), and an adaptor region that confers DNA binding via the CBF1 protein. The CBF1 binding region is indicated in orange. The PU.1 binding region is indicated in green. (B) NMR structure of the END domain homodimer, H15 (turquoise), and α1-helix (red). (C and D) GST pull-down experiments and (C) GST-END or (D) C-terminal half (amino acid residues 246 through 487) of EBNA2 (GST-E2). Whole-cell lysates of DG75 cells were incubated with GST–EBNA2 fusion proteins purified from Escherichia coli: the C-terminal half (amino acid residues 246 through 487) of EBNA2 (GST-E2), END domain (GST-END), or GST. (C) Western blot using PU.1-, CBF1-, or GST-specific antibodies. (D) Western blot using PU.1- or EBF1-specific antibodies. (E) GST pull-down experiments with the END domain and EBF1. Whole-cell lysates of DG75 cells transfected with an EBF1 expression plasmid (+) or an empty vector control (−) were incubated with GST-END fusion proteins purified from E. coli: wild-type END (GST-END), END-H15A missense mutant (GST-H15A), END α1-helix deletion mutant proteins (GST-Δα1), or GST. GAPDH served as a loading control for input lysates.
EBVΔα1 Activates Primary B Cells but Does Not Cause Long-Term Proliferation of Infected Cells.
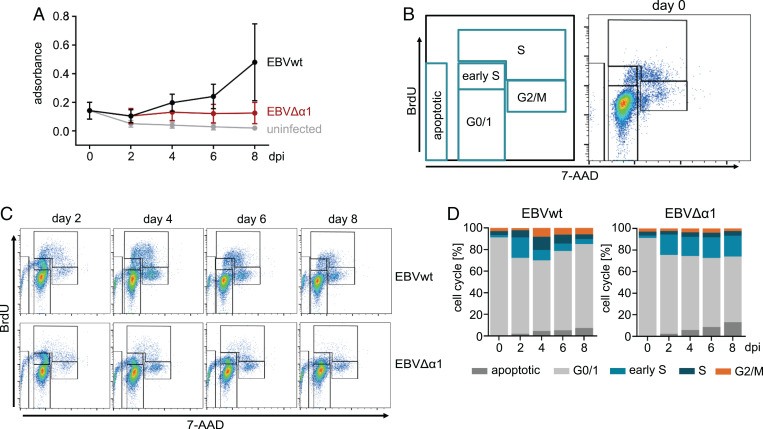
The immortalization of B cells is a complex multistep process. The key function of EBNA2 in the infected B cell is the activation of a cascade of primary and secondary target genes that ultimately drive the proliferation of infected cells. While infection of B cells does not require EBNA2, all subsequent steps including activation and cell cycle progression require the continuous presence of the EBNA2 protein (10). To study the impact of EBNA2–EBF1 complex formation on B cell immortalization, we established a recombinant EBV derivative with a deletion of the α1-helix in the END domain, termed EBVΔα1, and used it to infect primary human B cells. To examine B cell activation and the expansion of the cultures, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays were performed on day 0 prior to infection and on days 2, 4, 6, and 8 postinfection (Fig. 2A). On day 2 postinfection, MTT conversion of wild-type EBV (EBVwt)- and EBVΔα1-infected cell cultures was similar and modestly increased, indicating that these B cells were activated, compared with noninfected control cultures. Starting at day 4 postinfection, MTT conversion of EBVwt-infected cells gradually increased, reflecting the increase of total viable cells. In contrast, MTT conversion of EBVΔα1-infected B cells remained constant at low levels, indicating that these cells remained viable for at least 8 d, but the cultures did not expand. To further characterize the proliferation defect of EBVΔα1-infected B cells, we analyzed the cell cycle phase distribution by bromodeoxyuridine (BrdU) incorporation and 7-amino-actinomycin D staining of total DNA followed by flow cytometry (Fig. 2B–D). Our gating strategy discriminated among G0/G1 (BrdU negative/DNA = 2N), early S (BrdU medium/DNA = 2N), advanced S phase (BrdU high/DNA ≥ 2N), and G2/M phase (BrdU low/DNA ≥ 2N) (Fig. 2B). BrdU incorporation was detected as early as 2 d postinfection. EBVwt-infected B cells progressed through the S phase and G2/M within 4 d and continued to cycle as expected (27). In contrast, EBVΔα1-infected cells incorporated BrdU at low levels and maintained this state for at least 8 d, but cells were mostly arrested in G0/G1 and early S and barely advanced further (Fig. 2C and D). Apoptotic cells accumulated in both EBVwt- and EBVΔα1-infected cultures but were more pronounced in EBVΔα1-infected cultures.
Fig. 2.
EBVΔα1-infected B cells arrest in the early S phase. (A) MTT assay of primary B cells infected with EBVwt or EBVΔα1; the assay was performed on days 0, 2, 4, 6, 8 postinfection. The mean of three biological replicates is plotted. Error bars indicate the SD. Day 0 refers to noninfected B cells. (B) Gating strategy for the cell cycle analysis with BrdU and 7-amino-actinomycin D (7-AAD) and one representative FACS plot for noninfected cells (day 0). All cells were gated on lymphocytes and single cells. (C) Results of the BrdU assays of one representative experiment. The assay was performed on days 2, 4, 6, and 8 postinfection with B cells infected with EBVwt or EBVΔα1. (D) Summary of the cell cycle analysis showing the mean results of three biological replicates.
CD40 Activation Rescues Long-Term Proliferation of EBVΔα1-Infected B Cells.
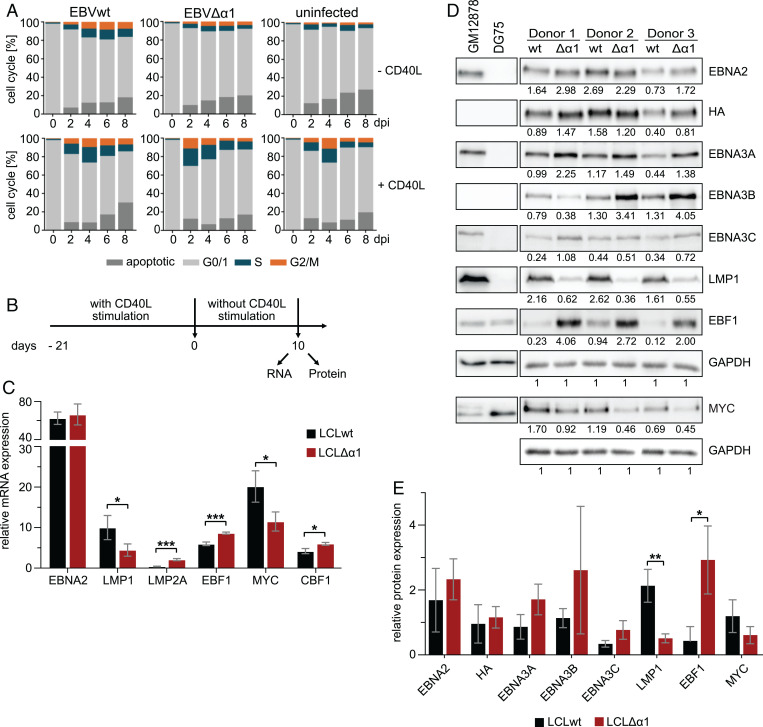
Primary human B cells proliferate in response to CD40 activation in the presence of interleukin-4 (IL-4) (28). CD40 is a costimulatory receptor on the surface of B cells which is triggered by CD40 ligands (CD40L) on helper T cells to activate canonical and noncanonical NFκB signaling, STAT5 phosphorylation, and p38, AKT, and JNK activation (29). Together, these signals resemble, in many aspects, the constitutive signaling of the viral LMP1 protein in EBV-infected B cells and are critical for the survival of lymphoblastoid cell lines (LCLs) evolving from EBV-infected primary B cells in vitro (30). To study the phenotypes of long-term EBVΔα1-infected B cell cultures (LCLΔα1) and to establish them for further biochemical analyses, EBVwt- and EBVΔα1-infected B cells were cultivated in the presence or absence of CD40L-positive murine feeder cells in the absence of IL-4 (31). In contrast to Banchereau et al. (28), our model did not require IL-4 stimulation. Noninfected B cells were used as controls and cell cycle progression was analyzed by flow cytometry of propidium iodide–stained nuclei (Fig. 3A).
Fig. 3.
Long-term cultures of LCLΔα1 established by cocultivation of primary B cells infected with EBVΔα1 on CD40L feeder cells. (A) Cell cycle analyses of primary B cells infected with EBVwt or EBVΔα1. Noninfected or infected primary B cells were either cultured without (−) CD40L feeder cells or with (+) CD40L feeder cells. The cell cycle distribution was analyzed by propidium iodide staining and flow cytometry on days 0, 2, 4, 6, and 8 postinfection. The mean of three biological replicates is plotted. See SI Appendix, Fig. S2 for one representative experiment. (B) Experimental setup for RNA and protein preparation. LCLwt or LCLΔα1 were cultured without CD40L feeder cells for 10 d prior to RNA and protein isolation. (C) Real-time qPCR of viral and cellular genes in LCLwt or LCLΔα1 generated from three donors cultivated for 10 d without CD40L stimulus, as in B. Transcript levels of viral and cellular genes were analyzed and normalized to RNA pol II transcript levels. The mean of three biological replicates is plotted; error bars indicate the SD. See SI Appendix, Fig. S3E for the messenger RNA (mRNA) expression level of RNA pol II. (D) Protein expression of latent viral and cellular proteins in LCLwt or LCLΔα1 generated from three donors after 10 d without CD40L activation. Cell lysates of EBV-infected GM12878 or EBV-negative DG75 cells served as positive or negative controls, respectively. The signals were normalized to the corresponding GAPDH signal and the values are indicated below each Western blot signal. (E) Relative expression of latent viral and cellular proteins. Bar charts were generated with values in D. The mean of three biological replicates is plotted; error bars indicate the SD. *P < 0.05; **P < 0.01; ***P < 0.001. dpi, days postinfection; wt, wild type.
Like noninfected cells, EBVΔα1-infected B cells could not establish long-term cultures without CD40L feeder cells (Fig. 3A). In contrast to feeder-free cultures, all CD40L-stimulated B cells entered the S and G2/M phases rapidly (Fig. 3A). Remarkably, EBVΔα1-infected cells showed a dramatic increase in S phase already on day 2 postinfection and, overall, apoptosis was less pronounced in CD40L-stimulated cells (SI Appendix, Fig. S2). For comparative studies on RNA and protein expression, established pairs of LCLwt and LCLΔα1 derived from the same donor were cultivated in the absence of CD40L feeder cells for 10 d (Fig. 3B). EBVΔα1-infected cells gradually ceased to proliferate but did not die, whereas proliferation of EBVwt-infected cells was independent of CD40L stimulation as expected (SI Appendix, Fig. S3A). EBNA2 RNA abundance was similar in LCLwt and LCLΔα1. LMP1 was significantly decreased, whereas LMP2A was increased in LCLΔα1 (Fig. 3C). Both EBNA2 and HA-tag–specific antibodies detected EBNA2wt and EBNA2Δα1 well (Fig. 3D and E). Protein levels of both EBNA2wt and EBNA2Δα1 varied between individual donors (Fig. 3D). The mean EBNA2Δα1 protein expression across three donors was slightly enriched compared with EBNA2wt (Fig. 3E). Elevated EBNA3 expression levels were observed in two of three LCLΔα1s, indicating that the mutant encodes for a stable protein. EBF1 protein expression was consistently elevated in all LCLΔα1 samples, while MYC and, to an even stronger degree, LMP1 expression was consistently reduced (Fig. 3D and E).
It is known that both EBNA2 (5) and LMP1 (32) induce MYC. We hypothesized that low LMP1 levels could be rate limiting for MYC expression, causing the observed phenotypes in cell proliferation and survival of LCLΔα1 cells. We expressed LMP1 ectopically in EBVΔα1-infected cells but found that LMP1 did not restore their proliferation in the absence of CD40 stimulation (SI Appendix, Fig. S3 C and D). We concluded that elevated LMP1 expression levels are not sufficient to restore the proliferation of EBNA2Δα1-expressing cells. These data demonstrate that permanent CD40 ligation by feeder cells is required for the long-term survival of EBVΔα1-infected B cells. Since we could not substitute CD40 stimulation by LMP1, we conclude that EBNA2–EBF1 complexes have to provide functions beyond LMP1 induction, which might include specific sets of EBNA2 target genes.
The α1-Helix Assists in EBF1-Dependent EBNA2 Chromatin Binding.
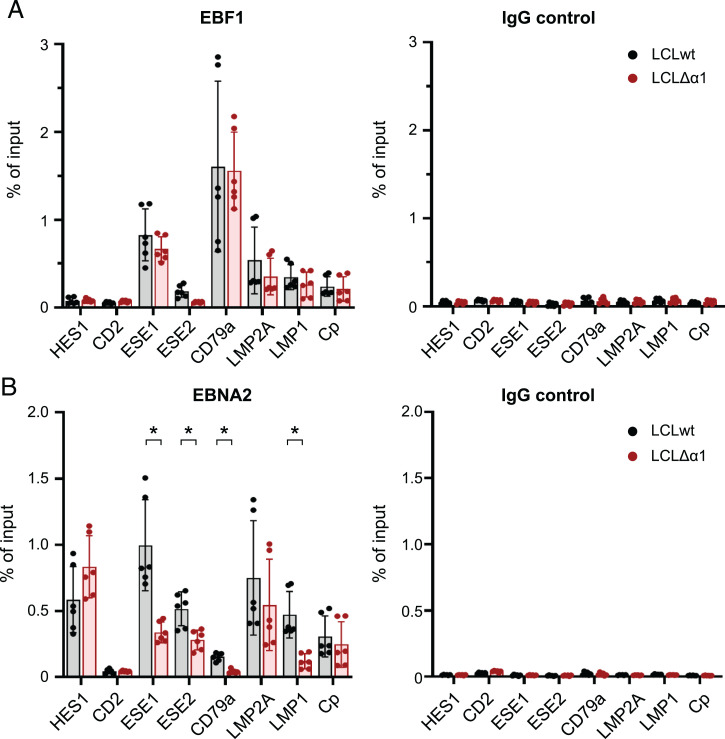
We have previously shown that EBNA2 can bind to chromatin in CBF1 knock-out cell lines (15). This finding indicates that EBF1 and CBF1 may act independently to recruit EBNA2 to chromatin, although the majority of EBNA2 binding sites encompass DNA sequence motifs that are bound by both EBF1 and CBF1. To study how the deletion of EBNA2’s α1-helix impairs the access of EBNA2 to chromatin, we performed ChIP followed by qPCR on selected genomic regions using either EBF1- or EBNA2-specific antibodies. LCLs infected with EBVwt or EBVΔα1 were cultivated on CD40L feeder cells and were removed from feeder-cell layers 1 d prior to chromatin isolation. During normal hematopoietic development, EBF1 is exclusively expressed in B cells. As a pioneer factor, EBF1 directs the cell fate of lymphocyte precursor into the B cell lineage and prevents the development of T cells (33). The HES1 gene, a well-characterized, canonical Notch target gene in T cells, carries two paired CBF1 binding sites within its promoter. They facilitate dimerization of intracellular Notch (34) as Notch, similar to EBNA2, uses CBF1 as a DNA anchor (35, 36). Since EBNA2 can activate HES1 expression (37, 38), we speculated that HES1 activation by EBNA2 in B cells might also be mediated solely by CBF1 and is independent of EBF1. Lu et al. (17) recently reported that HES1 expression is perturbed by CBF1 knockdown in LCLs. We show that EBF1 did not bind to the HES1 promoter, but EBNA2wt and EBNA2Δα1 did, indicating that both proteins use canonical CBF1 sites within the HES promoter to bind chromatin (Fig. 4A and B). The results of ChIP–qPCR experiments also were in good accordance with our finding that the deletion of the α1-helix in EBNA2 cannot affect CBF1 binding (SI Appendix, Fig. S1B). CD2, a T cell marker gene, served as a negative control. As expected, neither EBF1 nor EBNA2 bound to the CD2 locus.
Fig. 4.
EBNA2’s α1-helix assists in EBF1-dependent EBNA2-chromatin binding. (A and B) ChIP-qPCR for (A) EBF1 or immunoglobulin G (IgG) control and (B) EBNA2 or IgG control in established LCLwt and LCLΔα1 at cellular and viral genomic regions. LCLwt and LCLΔα1 were cultured without CD40L-expressing feeder cells for 1 d prior to chromatin preparation. CD2 was used as a negative control. Mean of three biological replicates is plotted; error bars indicate the SD. *P < 0.05.
EBV superenhancer (ESE) 1 and ESE2 control MYC expression in EBV-infected B cells (16). EBF1 and EBNA2wt were recruited well to ESE1, but EBNA2Δα1 binding was significantly impaired, indicating that EBNA2 uses EBF1 to interact with ESE1 (Fig. 4A and B). EBNA2Δα1 recruitment to ESE2 was less pronounced compared with ESE1 but also was significantly impaired compared with EBNA2wt. CD79a is an EBF1 target gene that is well characterized during B cell development (39). In LCLs, EBF1 bound strongly to the CD79a promoter. EBNA2 binding to the CD79a promoter was weak in comparison with other loci but strictly required the α1-helix within the END domain (Fig. 4A and B).
In addition, we tested the binding of EBNA2 at latent viral, EBNA2-responsive promoters. Activation of the LMP2A and the viral C-promoter by EBNA2 is well characterized (35) and strictly dependent on CBF1 (40), although EBF1 can be detected at these promoters. Both promoters C-promoter and LMP2A bound EBNA2Δα1 and EBNA2wt equally well and, thus, EBNA2 binding was independent of EBF1. These findings, as well as elevated LMP2A expression levels in LCLΔα1 cells (Fig. 3), are consistent with those of previous reports that have described enhanced LMP2A expression in EBF1-depleted but diminished LMP2A expression in CBF1-depleted LCLs (17). Thus, despite elevated EBF1 levels in LCLΔα1 cells (Fig. 3) and equal binding to the LMP2A promotor (Fig. 4A), regulation of LMP2A was independent of EBF1. As reported previously (13, 41), EBF1 was recruited to the LMP1 promoter, but efficient EBNA2 binding was clearly impaired in the absence of the α1-helix (Fig. 4B). In summary, EBNA2Δα1 is a highly specific loss-of-function mutant that selectively abolishes EBF1-dependent functions of EBNA2.
EBVΔα1 Fails to Efficiently Initiate the Expression of Critical Cellular Genes upon B cell Infection.
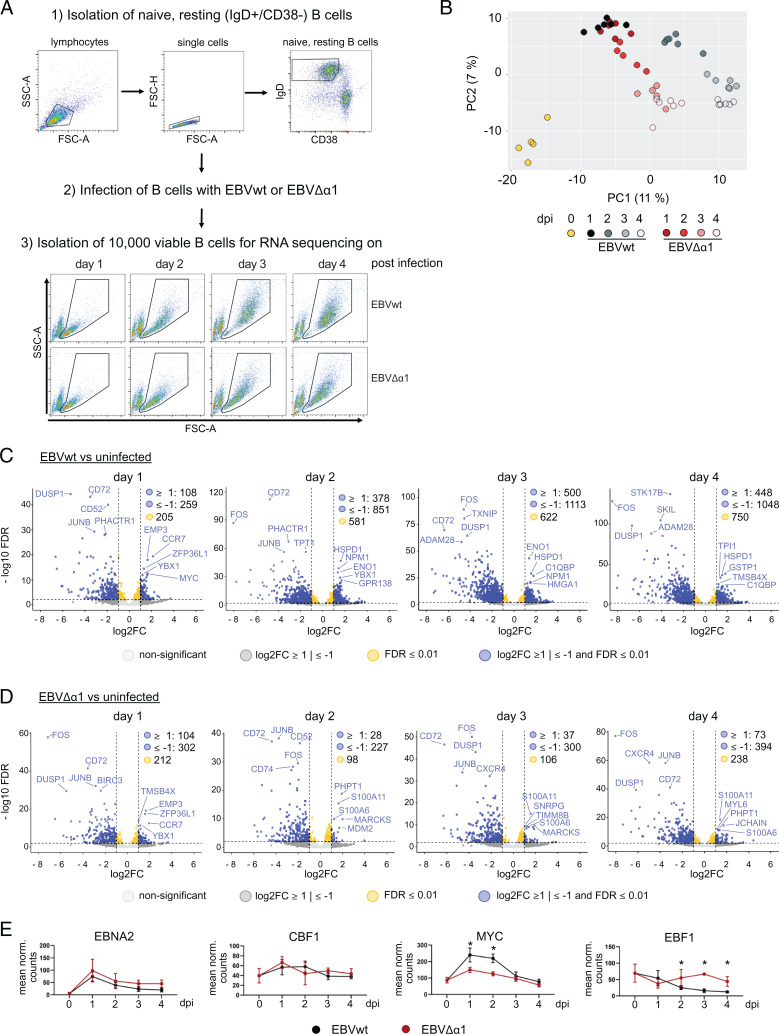
As illustrated in Fig. 2, EBVΔα1-infected B cells started to enter the S phase of the cell cycle on day 2 postinfection, but progression through the S and G2/M phases in the following days was strongly impaired. To characterize the defect of this mutant in detail, we analyzed the gene expression patterns of naïve resting B cells prior to infection (day 0) and on days 1, 2, 3, and 4 postinfection with either EBVwt or EBVΔα1. To this end, adenoid human B cells were isolated and sorted for naïve resting B cells (IgD+/CD38−) by flow cytometry (Fig. 5A) (27). Forward and side scatter of all cells indicated an increase in cell size and granularity on day 2 postinfection, but these morphological changes were more pronounced in EBVwt-infected B cells (Fig. 5A).
Fig. 5.
Dynamic changes of gene expression patterns in B cells infected with EBVwt or EBVΔα1. (A) Workflow for cell preparation and infection. Naïve resting B cells (IgD+/CD38−) were isolated from adenoids and infected with EBVwt or EBVΔα1. A total of 10,000 noninfected, naïve, resting B cells were collected to serve as the day 0 sample. On days 1, 2, 3, and 4 after infection, 10,000 viable cells were isolated by flow cytometry sorting and collected for RNA sequencing. Flow cytometry plots of one representative experiment are shown. (B) Principal component analysis of all protein-coding genes. The percentage of variance explained by the first and second principal components (PCs; PC1 and PC2, respectively) are shown in parentheses. (C and D) Volcano plots of DE protein-coding genes comparing (C) EBVwt- or (D) EBVΔα1-infected B cells on days 1, 2, 3, and 4 postinfection with noninfected samples (day 0). Dotted lines indicate log2 fold change = 1 and FDR = 0.01. Gene names of the top five up- and down-regulated genes according to the FDR are indicated. € Mean normalized expression of EBNA2, CBF1, MYC, and EBF1 in EBVwt- and EBVΔα1-infected B cells on days 0, 1, 2, 3, and 4 postinfection. Error bars indicate SD, and asterisks indicate FDR < 0.001 calculated by DESeq2. dpi, days postinfection.
Total cellular RNA was isolated on individual days and high-quality complementary DNA libraries for RNA-sequencing analyses were generated (SI Appendix, Fig. S4) using the bulk RNA-sequencing method prime-sEq. (42). Principal component analysis of RNA-sequencing results for protein-coding genes confirmed the considerable switch of gene-expression patterns caused by EBVwt and EBVΔα1 infection (Fig. 5B). Moreover, the principal component analysis illustrated a substantial overlap of the samples obtained with EBVwt and EBVΔα1 infection on day 1 and a stronger divergence starting from day 2 and thereafter (Fig. 5B). Overall, the regulation of gene expression in B cells was affected to a greater degree upon EBVwt infection compared with EBVΔα1 infection on day 2 and thereafter (Fig. 5C and D). Surprisingly, we detected a significant loss of transcripts over time in infected, compared with noninfected, cells, which might have been caused by repression of specific genes or loss of transcript stability in activated cells (Fig. 5C and D). Regarding viral genes, only 4,920 reads could be mapped to the viral genome, of which 3,095 were mapped to EBNA2. This, unfortunately, provided us with little information about viral gene expression, and EBNA2 was the only gene with meaningful expression levels. Expression of EBNA2 in EBVwt and EBVΔα1 infections was similar during the 4 d of the experiment (Fig. 5E). Also, no significant differential expression of CBF1 was detected between viral strains over time (Fig. 5E). MYC induction peaked in both infection conditions 1 d postinfection but was significantly less induced in EBVΔα1-infected cells (Fig. 5E). In contrast, EBF1 transcript levels were significantly higher than in EBVwt-infected cells on days 2, 3, and 4 (Fig. 5E). Taken together, the data show that gene expression was altered upon infection of B cells with EBVwt or EBVΔα1. However, the degree of gene regulation was strongly impaired in EBVΔα1-infected B cells, although EBNA2 expression levels were comparable to those of EBVwt-infected B cells.
EBVΔα1 Fails to Initiate Important Processes Required for Cell Cycle Progression in Infected B Cells.
We performed gene clustering with subsequent gene ontology (GO) term analysis on differentially expressed (DE) genes with a false discovery rate (FDR) < 0.1 and a fold change of ≥2 (27) to study the dynamics of gene regulation and their functions. We could identify dynamic gene clusters with associated GO terms in EBVwt-infected (SI Appendix, Fig. S5 B–F) and EBVΔα1-infected (SI Appendix, Fig. S5 G–J) cells. The gene cluster associated with the cell cycle and division had a similar dynamic (SI Appendix, Fig. S5 B and G), but it was larger in the EBVwt-infected (n = 1,033 genes) than in EBVΔα1-infected (n = 732 genes) cells. Another interesting observation was that genes associated with RNA metabolism (“wt dark blue” and “Δα1 light blue” in SI Appendix, Fig. S5 C and H) peaked on day 1 postinfection but tended to decrease more markedly in EBVΔα1-infected cells toward levels seen in noninfected cells (day 0). Importantly, the “wt dark red cluster” in SI Appendix, Fig. S5D, strongly associated with biosynthesis and mitochondrial processes, did not show up in EBVΔα1-infected cells. In fact, most of the genes in this cluster were either not DE in the mutant (74.6%) or they tended to be only transiently activated (i.e., 17.4% of those genes are in the “Δα1 light blue” cluster in SI Appendix, Fig. S5H).
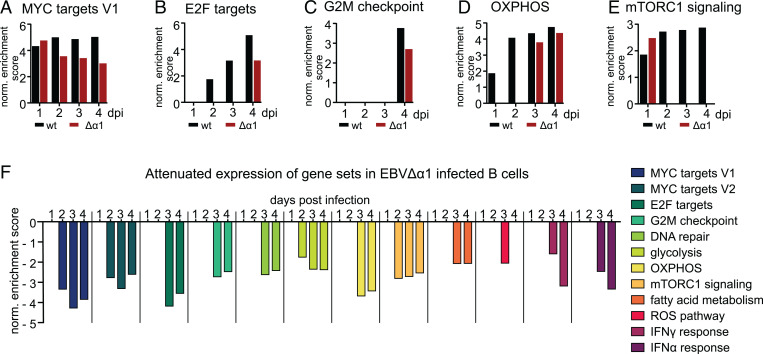
Next, we performed a gene set enrichment analysis (GSEA) with DE, protein-coding genes (FDR < 0.01) to identify cellular processes affected in EBVΔα1-infected B cells at each time point. We included the molecular signature–database hallmark gene sets for the GSEA, which encompassed two clusters for MYC-regulated genes (MYC targets V1 and V2) (43). MYC targets V1 defines a broad set of downstream targets, whereas MYC targets V2 is more stringently defined. Notably, several gene sets were exclusively enriched in EBVwt infections and included the MYC targets V2 as well as cellular and metabolic gene sets such as DNA repair, fatty acid metabolism, glycolysis, or reactive oxygen species pathways (SI Appendix, Fig. S5K). Gene sets significantly enriched in cells infected with either virus on at least 1 d postinfection included MYC targets V1, E2F targets, oxidative phosphorylation (OXPHOS), the G2M checkpoint, and mTORC1 signaling (Fig. 6A–E). Interestingly, the enrichment for MYC V1 was comparable in EBVwt- or EBVΔα1-infected cells on day 1, but the normalized enrichment scores declined in EBVΔα1-infected B cells afterward. The E2F targets, OXPHOS, and G2M gene sets were only enriched in the EBVΔα1 infection at later time points, indicating a delayed induction of these gene sets. mTORC1 is an important sensor for nutrients, redox state, and energy supply. Interestingly, mTORC1 signaling was enriched on day 1 postinfection but lost in the following days in EBVΔα1 infections. This indicates that an important signal activating messenger RNA transcription and protein translation was initiated in cells infected with both viruses but not maintained in cells infected with EBVΔα1. GSEA with DE genes identified by comparing EBVΔα1 and EBVwt infections revealed that further important processes required for a successful establishment of latency were more strongly enriched in EBVwt- than in EBVΔα1-infected cells (Fig. 6F). MYC is an important target gene of EBNA2 (5), and a stronger correlation of MYC targets V1/V2 with EBVwt infection could indicate insufficient levels of MYC in EBVΔα1-infected cells. E2F targets and the G2M checkpoint include genes that are important for the cell cycle, which has been reported to be initiated between days 3 and 4 postinfection (27), but we could detect an earlier cell cycle onset. Metabolic processes such as glycolysis, OXPHOS, fatty acid metabolism, reactive oxygen species pathways, and mTORC1 signaling are pathways up-regulated upon B cell activation (44, 45). Overall, RNA sequencing revealed that changes in gene expression, especially starting on day 2 and onward, were less pronounced in EBVΔα1- than in EBVwt-infected B cells and partially reminiscent of noninfected cells. Additionally, important target genes and metabolic pathways were not induced or delayed in EBVΔα1-infected cells.
Fig. 6.
EBVΔα1 cannot efficiently induce critical cellular and metabolic processes. GSEA was performed with DE protein-coding genes with a P value of ≤0.01, as calculated by DESeq2. Gene sets with an FDR < 0.05 were considered significant. (A–E) Gene sets that are present in both EBVwt- and EBVΔα1-infected B cells on at least 1 d postinfection. Bar graphs show the normalized enrichment score (NES) at each day postinfection. DE genes of the analysis of EBVwt vs. noninfected or EBVΔα1 vs. noninfected were included for these GSEA. (F) GSEA with DE genes defined by the analysis EBVΔα1 vs. EBVwt. The bar graph shows the NES for each gene set at each day postinfection. A negative NES correlates with an enrichment in EBVwt-infected B cells over EBVΔα1-infected B cells. dpi, days postinfection; norm., normalized; ROS, reactive oxygen species.
DISCUSSION
EBVΔα1 Can Activate Primary Human B Cells but Cell Cycle Progression Is Aborted in the Early S Phase.
Upon infection of primary resting B cells, EBV establishes a complex, specific, cellular and viral gene expression program. Eventually, a limited number of viral latent nuclear antigens (e.g., EBNAs) and LMPs cooperate to establish the long-term proliferation of infected B cell cultures. EBV devoid of EBNA2 (11) or EBV mutants with EBNA2 alleles incapable of interacting with CBF1 (12) cannot activate infected B cells, demonstrating the importance of EBNA2 and its association with CBF1 for the immortalization process. EBNA2 is a multifunctional viral oncogene that is associated with a plethora of cellular proteins (11). While the core of the EBNA2 binding site of CBF1 has been carefully mapped to a central double-tryptophan motif (47), information on the exact binding motif of further EBNA2-binding partners is limited and recombinant viral mutants, which lack specific binding sites, have not been reported, to our knowledge.
We have shown previously that EBNA2 requires its END to build stable complexes with EBF1, a candidate factor to confer B cell specificity to EBNA2 (15). Here, we characterize an EBNA2 mutant (EBNA2Δα1) lacking the α1-helix, which consists of five amino acids, within the END domain, and show that EBNA2Δα1 is deficient for EBF1 binding. To study the impact of the association of EBF1 with EBNA2 on B cell immortalization, we generated viral mutants and found that a mutant EBV encoding EBNA2Δα1 (EBVΔα1) failed to immortalize primary human B cells. B cells infected with EBVΔα1 became activated and entered the S phase but failed to complete the cell cycle (Fig. 2). The growth arrest was rescued by CD40 stimulation (Fig. 3A). The phenotype of EBVΔα1-infected B cells differs dramatically from B cells infected with an EBNA2 knockout EBV. B cells infected with EBNA2 knockout EBV do not survive beyond 3 d postinfection and fail to synthesize cellular DNA.
EBNA2Δα1 Selectively Impairs EBF1-Related Functions of EBNA2.
In EBV-infected B cells, EBNA2 directly drives the expression of LMP1 (5, 20, 48–50). In LCLs, roughly 50% of LMP1 promoter activation by EBNA2 depends on CBF1 (12). Multiple studies (13, 17, 41) have demonstrated that EBF1 is critical for EBNA2-dependent LMP1 activation. Using the EBF1-binding–deficient EBVΔα1 mutant virus, we demonstrate that the activation of LMP1 by EBNA2 requires the interaction with EBF1 via the α1-helix of the END domain. Recruitment of EBNA2Δα1 to the LMP1 promotor was reduced (Fig. 4), leading to reduced LMP1 expression levels in cells infected with the EBNA2Δα1-mutant EBV (Fig. 3). In contrast, EBNA2Δα1 bound equally well to the CBF1-dependent viral C-, LMP2A, and cellular HES1 promoters, compared with EBNA2wt. Thus, EBVΔα1 carries a specific loss-of-function mutation in EBNA2 that does not affect functions related to the CBF1 interaction but selectively affects EBNA2’s transactivation capacity linked to EBF1 (Fig. 4).
Expression of MYC and Basic Cellular Processes Are Impaired in EBVΔα1-Infected B Cells.
EBNA2 directly activates MYC expression and thus is involved in multiple events triggering B cell activation, proliferation, and survival (5). The central role of MYC in LCLs is well documented, as ectopic expression of MYC can drive the proliferation of latently infected B cells even when EBNA2 is inactivated (51). EBNA2 regulates MYC by inducing chromatin loops that bridge two enhancers located at –556kb (ESE1) and –428kb (ESE2) upstream of the MYC transcriptional start site to the MYC promoter (13, 14, 52, 53). EBF1 is recruited to both enhancers, although ESE1 is more active than ESE2 (16). Here, we confirm this differential binding pattern of EBF1 (Fig. 4). Importantly, EBNA2Δα1 binding was significantly impaired at ESE1 and ESE2, demonstrating that the α1-helix, consisting of five amino acid residues, substantially contributes to EBNA2 binding to both B cell–specific MYC enhancers. This nicely explains significantly reduced MYC levels 1 d postinfection in primary B cells and in established LCLΔα1.
In LCLs, LMP1 enhances MYC expression in the presence of EBNA2 (32) and thereby contributes to the proliferation of LCLs. We tested if ectopic expression of LMP1 in long-term LCLΔα1 cultures (Fig. 3 and SI Appendix, Fig. S3) could substitute for CD40 stimulation (SI Appendix, Fig. S3) but found that ectopic LMP1 expression was not sufficient to promote the proliferation of LCLΔα1 cultures. We conclude that cellular pathways downstream of CD40 signaling, which are not provided by LMP1, are important to maintain continuous proliferation of these cultures. Since we know that high levels of MYC protein will certainly rescue any EBNA2 deficiency in LCLs (51), we could not test if ectopic expression of MYC might rescue the proliferation of LCLΔα1. Such experiments would not provide new mechanistic insights into the specific function of EBNA2–EBF1 complexes. In summary, we conclude that full MYC activation by EBNA2 is achieved through the interaction with EBF1 allowing EBNA2 the access to specific MYC enhancers early after infection of naïve B cells as well as in established LCLs. Since EBNA2 (52, 54) and EBF1 (55, 56) interact with the chromatin remodeler Brg1, we speculate that this interaction might contribute to the regulation of the EBV-specific MYC enhancers and, thereby, might enhance MYC activation.
To characterize the defects of EBVΔα1-infected B cells in detail, a time-course RNA-sequencing experiment was performed surveying noninfected cells and the subsequent early phase until day 4 postinfection. Similar patterns of gene expression changes were grouped into clusters for EBNA2wt- and EBNA2Δα1-expressing cells and a GO term analysis was performed for each cluster (SI Appendix, Fig. S5 B–J). Cell cycle and RNA processing functions as well as multiple biosynthetic pathways were attenuated in EBVΔα1-infected cells. According to the GSEA, several enriched gene sets were exclusively found in EBVwt- but not in EBVΔα1-infected B cells (SI Appendix, Fig. S5K). Induction of MYC and E2F target genes, both critical for G1 phase cell cycle progression, was severely impaired in EBVΔα1-infected B cells already at 1 d postinfection. Also, glycolysis, OXPHOS, and fatty acid metabolism, which are known to be closely linked to distinct phases of the cell cycle (57), were attenuated in these cells. In summary, the phenotype of EBVΔα1-infected B cells revealed a broad spectrum of alterations in gene expression that can be well explained by low levels of MYC in these cells. MYC is a pleiotropic transcription factor that not only directly activates target gene expression by binding to specific DNA sequences of regulatory genomic sites but, in addition, accumulates at active promoter sites and amplifies the transcriptional activity of a global spectrum of target genes (58). In cancer cells, oncogenic MYC levels are implicated in many metabolic pathways to regulate and fulfill the increased demand for energy and compounds for cell growth and proliferation (59). Since hyperactivation of MYC is absent in EBVΔα1-infected B cells (Fig. 5E), the metabolic defects in these cells are most likely caused by the inefficient activation of MYC. We have characterized global temporal changes in gene expression by GSEA and cluster analyses and inferred functional physiological changes based on GO terms. To pinpoint the quantitative contribution of single genes or gene sets to the EBVΔα1 phenotype, it will be important to elaborate technologies that allow the timely expression of RNA interference and messenger RNA libraries during the course of infection of highly purified and, thus, size-limited B cell populations for further studies.
During development, EBF1 acts as a pioneer factor that alters the chromatin signature of lymphocyte precursors, directing them into the B cell linage (60). In murine pro-B cells, EBF1 activates MYC and promotes proliferation of these cells (61). Our study highlights how EBF1, in cooperation with EBNA2, supports the activation of basic metabolic processes in human peripheral mature B cells. In conclusion, the protruding α1-helix on the surface of the EBNA2 N-terminal dimerization domain is a key structural element of EBNA2 that confers B cell proliferation at a critical first step toward establishment of latency by hijacking developmental signaling cues. Life-long persistence of EBV is a risk factor that contributes to the pathogenesis of malignant, nonmalignant, and autoimmune diseases. The results of our study can help develop new strategies to interfere with the etiology of EBNA2-driven disease by targeting a specific time window of the EBV immortalization process postactivation but prior to the establishment of latency.
Material and Methods
Human primary B cells were isolated from adenoids and infected with viral supernatants to generate lymphoblastoid cell lines. EBV mutants were generated by viral mutagenesis, and viral titers were determined by infection of Raji cells with subsequent fluorescence activated cell sorting analysis of reporter gene expression. Protein biochemical analysis of the EBNA2–EBF1 complex was performed by affinity capture assays with recombinant GST-tagged protein, sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and Western blot. Functional analysis of the EBNA2–EBF1 complex was performed by infection of primary B cells with wild-type or mutant EBV, with subsequent cell cycle analysis, ChIP, gene expression analysis via qPCR, and transcriptome analysis via RNA sequencing. Detailed description of all applied methods is provided in the SI Methods.
Supplementary Material
Acknowledgments
We thank Ezgi Akidil, Dagmar Pich, and Adam Yen-Fu Chen (Helmholtz Zentrum München [HMGU]) for valuable experimental support, Laboratory for Functional Genome Analysis (Ludwig Maximilians University) for the excellent sequencing service, and the monoclonal antibody core facility (HMGU) for providing antibodies. We also thank Rudolf Grosschedl (Max Planck Institute of Immunobiology and Epigenetics) and Mikael Sigvardsson (Lund University) for insightful discussions and sharing of expression vectors. L.E.W. and W.E. were supported by the Deutsche Forschungsgemeinschaft through the SFB1243 (Subproject A14) and the Cyliax foundation. X.Z. was supported by the China Scholarship Council (Grant 201603250052).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200512119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information. RNA sequencing data have been deposited in ArrayExpress (E-MTAB-11350) (62).
References
- 1.Shannon-Lowe C., Rickinson A. B., Bell A. I., Epstein-Barr virus-associated lymphomas. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Läderach F., Münz C., Epstein Barr virus exploits genetic susceptibility to increase multiple sclerosis risk. Microorganisms 9, 2191 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjornevik K., et al. , Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science 375, 296–301 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Münz C., Latency and lytic replication in Epstein-Barr virus-associated oncogenesis. Nat. Rev. Microbiol. 17, 691–700 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Kaiser C., et al. , The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 73, 4481–4484 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen J. I., Wang F., Mannick J., Kieff E., Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. U.S.A. 86, 9558–9562 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammerschmidt W., Sugden B., Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature 340, 393–397 (1989). [DOI] [PubMed] [Google Scholar]
- 8.Cohen J. I., Wang F., Kieff E., Epstein-Barr virus nuclear protein 2 mutations define essential domains for transformation and transactivation. J. Virol. 65, 2545–2554 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kempkes B., Pich D., Zeidler R., Hammerschmidt W., Immortalization of human primary B lymphocytes in vitro with DNA. Proc. Natl. Acad. Sci. U.S.A. 92, 5875–5879 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kempkes B., et al. , B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 14, 88–96 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pich D., et al. , First days in the life of naive human B lymphocytes infected with Epstein-Barr virus. MBio 10, e01723-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yalamanchili R., et al. , Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology 204, 634–641 (1994). [DOI] [PubMed] [Google Scholar]
- 13.Zhao B., et al. , Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc. Natl. Acad. Sci. U.S.A. 108, 14902–14907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang S., et al. , The Epstein-Barr virus regulome in lymphoblastoid cells. Cell Host Microbe 22, 561–573.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaser L. V., et al. , EBF1 binds to EBNA2 and promotes the assembly of EBNA2 chromatin complexes in B cells. PLoS Pathog. 13, e1006664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou H., et al. , Epstein-Barr virus oncoprotein super-enhancers control B cell growth. Cell Host Microbe 17, 205–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu F., et al. , EBNA2 drives formation of new chromosome binding sites and target genes for B-cell master regulatory transcription factors RBP-jκ and EBF1. PLoS Pathog. 12, e1005339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friberg A., et al. , The EBNA-2 N-terminal transactivation domain folds into a dimeric structure required for target gene activation. PLoS Pathog. 11, e1004910 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling P. D., Hayward S. D., Contribution of conserved amino acids in mediating the interaction between EBNA2 and CBF1/RBPJk. J. Virol. 69, 1944–1950 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johannsen E., et al. , Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by J kappa and PU.1. J. Virol. 69, 253–262 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boller S., Li R., Grosschedl R., Defining B., Cell chromatin: Lessons from EBF1. Trends Genet. 34, 257–269 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Sigvardsson M., Molecular regulation of differentiation in early B-lymphocyte development. Int. J. Mol. Sci. 19, 1928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Györy I., et al. , Transcription factor Ebf1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes Dev. 26, 668–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vilagos B., et al. , Essential role of EBF1 in the generation and function of distinct mature B cell types. J. Exp. Med. 209, 775–792 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bullerwell C. E., et al. , EBF1 drives hallmark B cell gene expression by enabling the interaction of PAX5 with the MLL H3K4 methyltransferase complex. Sci. Rep. 11, 1537 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bohle V., Döring C., Hansmann M. L., Küppers R., Role of early B-cell factor 1 (EBF1) in Hodgkin lymphoma. Leukemia 27, 671–679 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Mrozek-Gorska P., et al. , Epstein-Barr virus reprograms human B lymphocytes immediately in the prelatent phase of infection. Proc. Natl. Acad. Sci. U.S.A. 116, 16046–16055 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banchereau J., De Paoli P., Vallé A., Garcia E., Rousset F., Long-term human B cell lines dependent on interleukin-4 and antibody to CD40. Science (80-.) 251, 70–72 (1990). [DOI] [PubMed] [Google Scholar]
- 29.Elgueta R., et al. , Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol. Rev. 229, 152–172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorley-Lawson D. A., Epstein-Barr virus: Exploiting the immune system. Nat. Rev. Immunol. 1, 75–82 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Wiesner M., et al. , Conditional immortalization of human B cells by CD40 ligation. PLoS One 3, e1464 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dirmeier U., et al. , Latent membrane protein 1 of Epstein-Barr virus coordinately regulates proliferation with control of apoptosis. Oncogene 24, 1711–1717 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Rothenberg E. V., Transcriptional control of early T and B cell developmental choices. Annu. Rev. Immunol. 32, 283–321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnett K. L., et al. , Structural and mechanistic insights into cooperative assembly of dimeric Notch transcription complexes. Nat. Struct. Mol. Biol. 17, 1312–1317 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strobl L. J., et al. , Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-J κ. Immunobiology 198, 299–306 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Zimber-Strobl U., Strobl L. J., EBNA2 and Notch signalling in Epstein-Barr virus mediated immortalization of B lymphocytes. Semin. Cancer Biol. 11, 423–434 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Maier S., et al. , Cellular target genes of Epstein-Barr virus nuclear antigen 2. J. Virol. 80, 9761–9771 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohlhof H., et al. , Notch1, Notch2, and Epstein-Barr virus-encoded nuclear antigen 2 signaling differentially affects proliferation and survival of Epstein-Barr virus-infected B cells. Blood 113, 5506–5515 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Hagman J., Travis A., Grosschedl R., A novel lineage-specific nuclear factor regulates mb-1 gene transcription at the early stages of B cell differentiation. EMBO J. 10, 3409–3417 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimber-Strobl U., et al. , Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 13, 4973–4982 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murata T., et al. , Induction of Epstein-Barr virus oncoprotein LMP1 by transcription factors AP-2 and early B cell factor. J. Virol. 90, 3873–3889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janjic A., et al. , Prime-seq, efficient and powerful bulk RNA-sequencing. bioRxiv [Preprint] (2021). [DOI] [PMC free article] [PubMed]
- 43.Liberzon A., et al. , The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akkaya M., Pierce S. K., From zero to sixty and back to zero again: The metabolic life of B cells. Curr. Opin. Immunol. 57, 1–7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egawa T., Bhattacharya D., Regulation of metabolic supply and demand during B cell activation and subsequent differentiation. Curr. Opin. Immunol. 57, 8–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kempkes B., Ling P. D., EBNA2 and its coactivator EBNA-LP. Curr Top Microbiol Immunol. 391, 35–59 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Ling P. D., Rawlins D. R., Hayward S. D., The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc. Natl. Acad. Sci. U.S.A. 90, 9237–9241 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laux G., Adam B., Strobl L. J., Moreau-Gachelin F., The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-J kappa interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 13, 5624–5632 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grossman S. R., Johannsen E., Tong X., Yalamanchili R., Kieff E., The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J κ recombination signal binding protein. Proc. Natl. Acad. Sci. U.S.A. 91, 7568–7572 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waltzer L., et al. , The human J kappa recombination signal sequence binding protein (RBP-J kappa) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 13, 5633–5638 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Polack A., et al. , c-myc Activation renders proliferation of Epstein-Barr virus (EBV)-transformed cells independent of EBV nuclear antigen 2 and latent membrane protein 1. Proc. Natl. Acad. Sci. U.S.A. 93, 10411–10416 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wood C. D., et al. , MYC activation and BCL2L11 silencing by a tumour virus through the large-scale reconfiguration of enhancer-promoter hubs. eLife 5, e18270 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McClellan M. J., et al. , Modulation of enhancer looping and differential gene targeting by Epstein-Barr virus transcription factors directs cellular reprogramming. PLoS Pathog. 9, e1003636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu D. Y., Kalpana G. V., Goff S. P., Schubach W. H., Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J. Virol. 70, 6020–6028 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao H., et al. , Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc. Natl. Acad. Sci. U.S.A. 106, 11258–11263 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y., et al. , A prion-like domain in transcription factor EBF1 promotes phase separation and enables B cell programming of progenitor chromatin. Immunity 53, 1151–1167.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Icard P., Fournel L., Wu Z., Alifano M., Lincet H., Interconnection between metabolism and cell cycle in cancer. Trends Biochem. Sci. 44, 490–501 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Nie Z., et al. , c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong Y., Tu R., Liu H., Qing G., Regulation of cancer cell metabolism: Oncogenic MYC in the driver’s seat. Signal Transduct. Target. Ther. 5, 124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boller S., et al. , Pioneering activity of the C-terminal domain of EBF1 shapes the chromatin landscape for B cell programming. Immunity 44, 527–541 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Somasundaram R., et al. , EBF1 and PAX5 control pro-B cell expansion via opposing regulation of the Myc gene. Blood 137, 3037–3049 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.S. Beer et al., E-MTAB-11350 - RNA-seq of EBVwt or EBVmutant infected human primary B cells on day 1, 2, 3, and 4 post-infection. ArrayExpress. https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-11350/. Deposited 4 July 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. RNA sequencing data have been deposited in ArrayExpress (E-MTAB-11350) (62).