Significance
Recent demonstrations of a genetic basis for variation among hosts in the microbiome leave unresolved the question of how commonly host genetic effects influence individual microbes, and whether these effects impact host fitness. We used replicated field studies in the north and south of Sweden to map host genetic effects in microbial community networks using genome-wide association mapping. By focusing on consistent effects across sites, we found effects of genetic variation on important microbial hubs that contributed to plant fitness in a manner robust to the environment. Our results suggest that ongoing efforts to harness host genotype effects on the microbiome for agricultural purposes can be successful and highlight the value of explicitly considering abiotic variation in those efforts.
Keywords: Arabidopsis thaliana, genome-wide association study, microbiome, fitness, microbial hubs
Abstract
Although complex interactions between hosts and microbial associates are increasingly well documented, we still know little about how and why hosts shape microbial communities in nature. In addition, host genetic effects on microbial communities vary widely depending on the environment, obscuring conclusions about which microbes are impacted and which plant functions are important. We characterized the leaf microbiota of 200 Arabidopsis thaliana genotypes in eight field experiments and detected consistent host effects on specific, broadly distributed microbial species (operational taxonomic unit [OTUs]). Host genetic effects disproportionately influenced central ecological hubs, with heritability of particular OTUs declining with their distance from the nearest hub within the microbial network. These host effects could reflect either OTUs preferentially associating with specific genotypes or differential microbial success within them. Host genetics associated with microbial hubs explained over 10% of the variation in lifetime seed production among host genotypes across sites and years. We successfully cultured one of these microbial hubs and demonstrated its growth-promoting effects on plants in sterile conditions. Finally, genome-wide association mapping identified many putatively causal genes with small effects on the relative abundance of microbial hubs across sites and years, and these genes were enriched for those involved in the synthesis of specialized metabolites, auxins, and the immune system. Using untargeted metabolomics, we corroborate the consistent association between variation in specialized metabolites and microbial hubs across field sites. Together, our results reveal that host genetic variation impacts the microbial communities in consistent ways across environments and that these effects contribute to fitness variation among host genotypes.
Hosts harbor complex microbial communities that are thought to impact health and development (1). Human microbiota has been implicated in a variety of diseases, including obesity and cancer (2). Efforts are thus underway to determine the host factors shaping these communities (3, 4), and to use next-generation probiotics to inhibit colonization by pathogens (5). Similarly, in agriculture, there is great hope that selection on plant traits shaping the composition of the microbiota will help mitigate disease and increase crop yield in a sustainable fashion. Indeed, the Food and Agriculture Organization of the United Nations has made the use of biological control and growth-promoting microbial associations a clear priority for improving food production (6).
Plant-associated microbes can be beneficial in many ways, including improving access to nutrients, activating or priming the immune system, and competing with pathogens. For example, seeds inoculated with a combination of naturally occurring microbes were found to be protected from a sudden-wilt disease that emerged after continuous cropping (7). Thus, it would be advantageous to breed crops that promote the growth of beneficial microbes under a variety of field conditions, a prospect that is made more likely by the demonstration of host genotypic effects on their microbiota (8–11). However, microbial communities are complex entities that are influenced by the combined impact of host factors, the abiotic environment, and microbe–microbe interactions (12). Indeed, several studies have found a strong influence of the environment on estimates of host genotype effects (8, 13, 14). Although most, if not all, studies exploring the influence that host genotype exerts on microbial communities suggest that such plant control could be beneficial to plant performance, almost nothing is known about the relationship between host genotype effects on microbial communities and on plant performance or fitness. Consequently, the extent to which host plants can control microbial communities to their advantage, especially in a consistent manner across multiple environments, remains unclear.
Here, we combine large-scale field experiments in natural environments, extensive microbial community analysis, and genome-wide association mapping to 1) determine how host genotype affects different microbial community members, and thus shapes the overall microbiome; 2) estimate host genotype effects on microbial communities across eight environments and investigate the contribution of those effects to the performance of plant genotypes; and 3) use genome-wide association mapping to identify key pathways that shape the leaf microbial communities across multiple environmental conditions.
Snapshot of Microbial Community Variation
We performed a set of field experiments that included natural inbred lines of Arabidopsis thaliana (hereafter “accessions”) originally collected throughout Sweden, mainly in two climatically contrasted regions of the country (Dataset S1); A. thaliana in the north of Sweden experiences long, snowy winters, and, as a consequence, plants are typically found on south-facing slopes of rocky cliffs. Arabidopsis populations in the south of Sweden, on the other hand, tend to be associated with agricultural or disturbed fields that experience highly variable snow cover over the winter months. We used replicate experiments in four representative Arabidopsis sites, two each in the north (sites NM and NA) and south (sites SU and SR) of Sweden. Experiments were repeated across 2 years, for a total of eight experiments.
Each experiment was organized in a complete randomized block design including 24 replicates of 200 sequenced accessions (15), established as seedlings in a mixture of 10% native and 90% potting soil and timed to coincide with local germination flushes in late summer. Many of the microbiome members from our experiments were also found within the leaves of A. thaliana plants that we collected in the field in southern Sweden in 2017, suggesting that this percentage of native soil was sufficient to seed a representative microbiome (Dataset S2). Immediately upon snowmelt in early spring, we sampled and freeze dried five or six whole rosettes per accession. DNA was extracted from the freeze-dried rosettes, and both the ITS1 portion of the Internal Transcribed Spacer (ITS) and the V5 to V7 regions of the 16S RNA gene were sequenced to characterize the fungal and bacterial communities, respectively (9, 12, 16). The sequences obtained were clustered into operational taxonomic units (OTUs) using Swarm to generate community matrices (17) (see Count Table Filtering). The frequency distributions of OTUs were highly skewed, with the top 10 most common OTUs contributing, on average, 59% of the reads in each experiment (ranging from 45 to 78%). Throughout this study, we chose to focus on the microbes represented by at least 0.01% of the sequencing reads per experiment. While rare microbes may impact host performance and have important ecological roles (18), we would not have had the power to estimate heritability or map host control of these species. Taxonomic assignments indicate that the fungal communities were dominated by Leotimycetes and Dothideomycetes, while the bacterial communities included high proportions of Alphaproteobacteria and Actinobacteria (SI Appendix, Fig. S1).
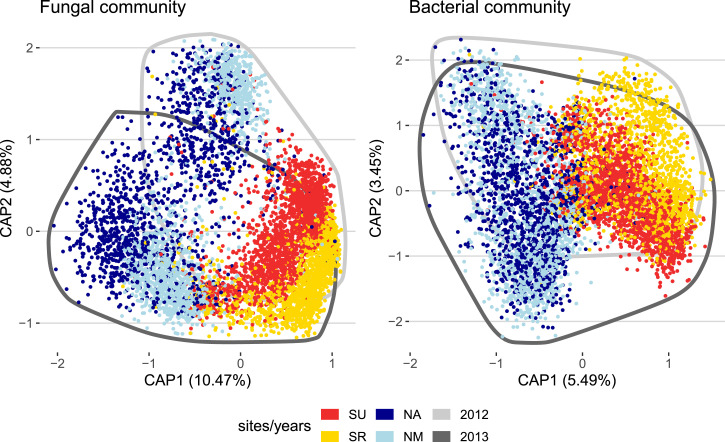
In a principal coordinate (PC) analysis, differences between northern and southern sites explained 10% and 5% of the overall diversity in the fungal and bacterial communities, respectively, while differences between the two consecutive years explained 5% and 3%. This level of differentiation among experiments likely underestimates that present in the native soil, as it has been shown that hosts filter the microbial community to reduce site-to-site differences (19, 20) (Fig. 1). In addition, there may have been a homogenizing effect of using a combination of local and potting soil. Irrespective of how well our treatments mimicked natural microbial communities, our analysis of eight common garden experiments permits assessment of the consistency across time and space of plant genetic effects on their associated microbial communities.
Fig. 1.
Plants grown in different environments have different microbial communities. The plots represent the projection of each sample on the plane defined by the first two constrained components of the fungal and bacterial communities, describing variation among sites and years. The percentages in parentheses are the proportion of the total inertia (square root of the Bray–Curtis dissimilarity) explained by each component. The colors of the points indicate the site from which samples were collected. Experiments from the south are represented in red (SU) and yellow (SR), and experiments from the north are represented in blue (NR) and dark blue (NA). All points from 2012 and 2013 are encircled by a darker and lighter gray line, respectively.
Host Genetic Effects on the Microbiota
Our experiments provided a unique opportunity to investigate associations between host genetic variation and their resident microbiomes, within the context of environmental variation across time and space. We focused on PC from simple unconstrained PC analysis (PCoA) within each experiment in order to summarize the variation among communities including hundreds or thousands of species with a few dimensions, and then calculated the proportion of variance explained by the host genotype (hereafter heritability or H2). Within each experiment, we found significant heritability of PC of the microbial communities (SI Appendix, Table S1), suggesting that genetic variation in the host significantly impacts at least a fraction of the microbiota, in line with results of previous studies (8–10, 12, 21, 22).
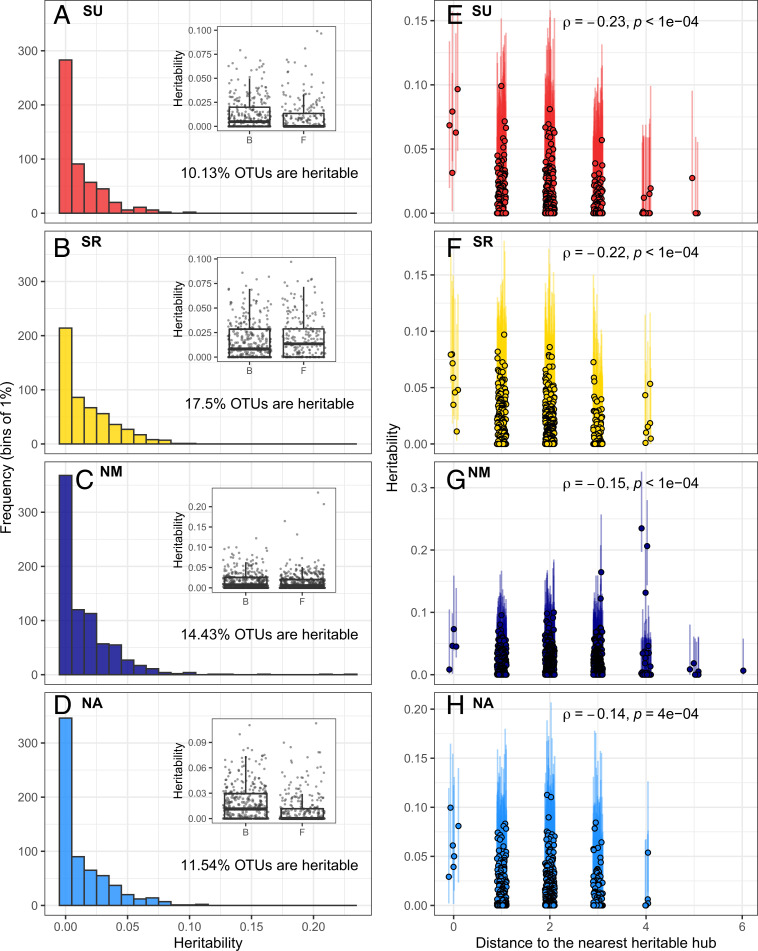
Significant heritability of the resident microbiome could arise from host genotypes exerting weak control over many community members, or by targeting a few microbes that then influence the relative abundance of others through microbe–microbe interactions. In order to investigate these hypotheses, we modeled the log-ratio transformed counts of individual OTUs with random-effect linear models and revealed significant genotypic effects (with the 95% CI of heritability not overlapping zero) for between 10.13% and 21.93% of all OTUs, depending on the site and year (Fig. 2 A–D and SI Appendix, Fig. S2 A–D). The latter explanation thus seems more likely, given that the influence of the host appears focused on relatively few OTUs, although it remains to be investigated whether heritable microbial hubs influence other members of the microbiome (see below). We found no evidence that either fungal or bacterial communities are systematically more impacted by host effects than the other (Fig. 2 A–D and SI Appendix, Fig. S2 A–D), nor that mean relative abundance was strongly correlated with OTU heritability (SI Appendix, Fig. S3).
Fig. 2.
The effect of host genetic variation on the microbial community targets relatively few OTUs and percolates across the network. This figure corresponds to observations in the set of four experiments performed in 2013. The same figure is available for the 2012 experiments in SI Appendix, Fig. S3. (A–D) Each frame presents the distribution of heritability estimates for individual OTUs in one site. In each frame, Inset graph is a box and whiskers plot contrasting the heritability (y axis) of bacterial (B) and fungal (F) OTUs. (E–H) The heritable hubs are represented at a distance of zero (hub). The other points are OTUs connected to heritable hubs, directly (distance = 1) or indirectly (distance > 1). The x axis represents the number of edges in the network separating an OTU and its nearest heritable hub. The correlation coefficients presented are Spearman rank correlations between heritability and distances to the heritable hub(s) (including zero).
Host Genetics Correlate Most Strongly with Ecologically Central Microbes
Having found that host effects are concentrated on a small proportion of OTUs, we investigated the possibility that these heritable OTUs trigger a broader community-level change in the microbiota. First, we computed networks of microbe cooccurrence for each experiment. We explored the ecological importance of heritable OTUs by computing networks of microbe cooccurrence for each experiment using the SPIEC-EASI (SParse InversE Covariance Estimation for Ecological Association Inference) pipeline (23). Although our networks included both fungal and bacterial OTUs, most significant cooccurrences involved OTUs within each domain, with an average of only 7.76% (min = 6.64%, max = 9.91%) of edges connecting fungal and bacterial OTUs. We quantified the ecological importance of OTUs using two common characteristics of nodes in a network (“degree” and “betweenness centrality”) (12), defining ecologically important “hubs” in each network as OTUs in the 95% tail of both of these statistics (SI Appendix, Fig. S4). We identified, on average, 16.5 microbial hubs per experiment (ranging from 11 to 24), representing 78 unique OTUs across all eight experiments (43 bacterial OTUs and 35 fungal OTUs). These hubs were connected to an average of 19.62% (min = 14.50%, max = 25.23%) of the edges in the networks, indicating that they are likely important in structuring the microbial community. In addition, hubs were involved in proportionally more interactions between fungi and bacteria than the rest of the community (SI Appendix, Table S3).
Next, we asked whether heritable OTUs are more likely to be ecologically important hubs, because this could open the door to community-level impacts of host genetic variation. Across all eight experiments, we detected 23 OTUs that were both heritable and hubs at least once (SI Appendix, Table S2 and Dataset S2). This represents a significant enrichment of hub OTUs among heritable OTUs (Wilcoxon rank sum test: n = 8, W = 57, P value = 0.007), suggesting that host effects on the microbiota preferentially influence the relative abundance of ecologically important microbes. In fact, hub OTUs were often among the OTUs with the highest heritability within each experiment; these hub OTUs stand out in that we find no general relationships between heritability and either betweenness or degree (SI Appendix, Fig. S5). To further explore how heritability is distributed among members of microbial communities, we mapped broad-sense heritability onto the ecological network. In six out of eight experiments, we observed a significant negative relationship between heritability and the distance (number of network edges) to the nearest heritable hub (combined P value = 3.96e−25, using Fisher’s method for combining P values) (24) (Fig. 2 E–H and SI Appendix, Fig. S2 E–H). This pattern reveals that host genetic variation impacts the structure of microbial communities, although whether this occurs due to shared host effects on many microbes or host effects on hubs that then percolate in the microbial community through microbe–microbe interactions is unclear.
To discern the contribution of microbe–microbe interactions in the propagation of host genetic effects across the microbial networks, we took advantage of our replicates of each host genotype to permute counts for each OTU. We reasoned that, if microbe–microbe interactions were largely responsible for the patterns of cooccurrence that we observed, then microbial cooccurrences would be diminished by our permutations of replicates within host genotypes. The same diminution would be evident if patterns of microbial cooccurrence were due to microenvironmental variation within experiments, independent of host genotype, although strong microenvironmental effects would have interfered with our ability to detect heritable OTUs. On the other hand, if OTUs tended to cooccur due to shared host genotype effects, then our permutations would have little impact. In the networks computed from the permuted datasets, on average, 91.39% (ranging from 87.67 to 95.2% across our eight experiments) of all OTUs that previously cooccurred with at least one other OTU (with degree > 0) had fewer associations with other microbes. Overall, networks computed from the permuted data had, on average, 75% fewer edges (ranging from 62 to 87%). This indicates that most microbe–microbe associations were not due to shared host genotype effects. Thus, although host genetic variation drove the cooccurrences for a fraction of OTUs, we interpret our empirical networks as consistent with a shared role of host genetics and microbe–microbe interactions, with host genotypes most strongly impacting microbial hubs that then influence other members of the microbial communities.
Not only did the heritable hubs seem to have an impact that percolated through the microbial community, they were widely distributed among accessions, sites, and years. We were able to identify 127 fungal and bacterial OTUs that were found in at least 50% of samples in all experiments. Interestingly, OTUs that were heritable hubs at least once were overrepresented in this core microbiota (χ2 = 51.98, degree of freedom [df] = 1, P value = 5.58e-13). This was not an artifact of their being widespread; significant heritability estimates were detected across the entire range of prevalence. Indeed, prevalence of OTUs explained less than 2.6% of variation in OTU heritability across all experiments (F statistic = 110.66, df = 4176, P value < 2.2e-16; SI Appendix, Fig. S6). Thus, ecologically important OTUs with greatest associations to host genotypes were unusual in being widespread among plants in multiple experiments. Host effects on the fungal OTU #8 (hereafter F8) are especially important; this OTU showed significant heritability (H2 > 0) in five out of the seven experiments in which it was a hub (SI Appendix, Table S2), suggesting that natural variation in A. thaliana influences its microbiota with some consistency across environments. The widespread prevalence of these heritable hubs suggests that variation at particular host genes associates with particular hubs across time and space, potentially providing a means to impact the microbiota in a robust fashion.
Variation in Performance of Host Genotypes Explained by Their Influence on Microbial Hubs
The extent to which natural variation among host genotypes in their associated microbes translates into fitness differences has yet to be determined. Our experiments included additional replicates of all genotypes that were left to flower and mature in the field. We harvested mature stems in early summer and used high-throughput image analysis to measure the size of reproductive stems, an estimate of lifetime investment in reproduction in this annual species. This measure encompasses variation in both the number of siliques and their size (which can increase as a function of seed number and seed size) but correlated well with seed production in an independent experiment (SI Appendix, Fig. S7) (24). We thus call our estimate "seed-set" in what follows. We observed that plant seed-set estimates were positively correlated across experiments (SI Appendix, Fig. S8), suggesting fitness variation among accessions was relatively consistent across sites. We therefore asked whether host effects on microbial hubs contributed to some genotypes producing more seeds across all environments investigated. Specifically, we used random intercept models to estimate genotype effects on both heritable microbial hubs and seed-set in a series of analyses that jointly considered all eight experiments and investigated the relationship between these two effects (see Heritable Hubs and Seed-Set across Environments).
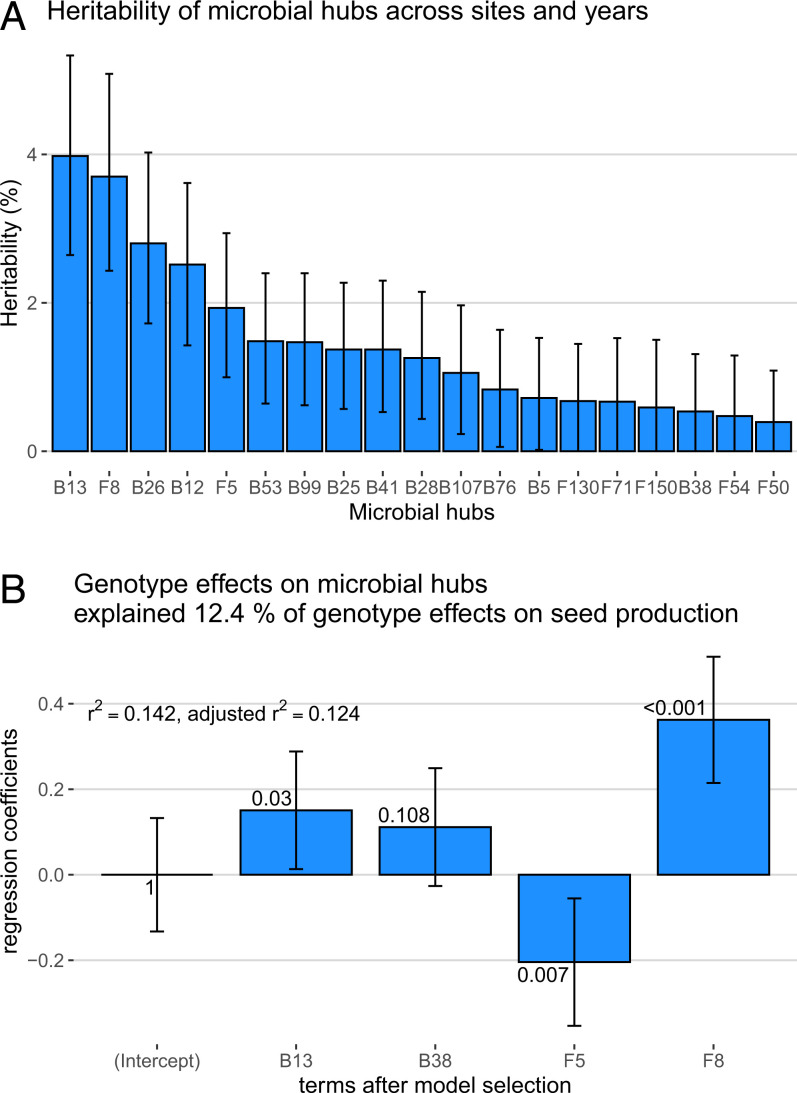
We found that the host genotype explained, on average across experiments, 6.88% (with a 95% CI [5.52, 8.34]) of seed-set. Host genotype effects on the relative abundances of 19 of our 23 heritable microbial hubs, quantified as random intercept deviation, were similarly modest, explaining up to 4% of the variation (Fig. 3A; four heritable hubs were not detected in more than two experiments and were removed for this analysis). We used multiple regression to estimate genetic correlations between host genotype effects on seed-set and on microbial hubs. We detected positive correlations between accession effects on seed-set and accession effects on three heritable hubs, F8, B38, and B13, as well as a negative correlation between accession effects on seed-set and accession effects on F5 (Fig. 3B). The variation explained by host genotype on the relative abundances of microbial hubs explained 12.4% of the host genotype effects on seed-set.
Fig. 3.
Relationship between host genotype seed-set and influence on microbial hubs across sites and years. (A) Proportion of heritable hub relative counts explained by host effects across all sites and years. (B) Coefficients for the linear regression explaining lifetime seed production variation among accessions with accession effects on microbial hubs across experiments (after model selection). The numbers near each bar are the P values associated with each term.
These results reveal that a sizable percentage of genetic variance in seed-set is shared with genetic variation associated with the relative abundance of a few broadly distributed microbial hubs, consistent with a causal relationship between genotype and seed-set mediated by heritable microbial hubs. Of course, the proportion of shared genetic variation between seed-set and heritable microbial hubs is unlikely to be equally important across time and space. In fact, in analyses performed on an experiment-by-experiment basis, we found that relationships between host effects on hubs and on seed-set were stronger in southern Sweden, where we detected significant relationships in both sites and both years (SI Appendix, Table S4).
Overall, our results highlight the importance for plants of controlling their leaf microbial community and suggest that breeding plants for their effects on specific members of microbial communities has the potential to significantly increase plant productivity.
Effect of Hubs on Growth in Controlled Condition
In an effort to confirm that the genetic correlations observed between heritable hubs and plant seed-set were due to an interaction between host and microbial species, we returned to the field to collect wild A. thaliana leaves, cultured ∼3,900 bacterial isolates from within these leaves (25), and sequenced both the 16S RNA gene and gyrase-B. These sequences included 100% matches for 10 of the 43 bacterial hubs, among which 4 were heritable hubs (SI Appendix, Table S2). Among successfully cultured heritable hubs was B38 which appeared to contribute positively to the seed-set of accessions in our field experiments (Fig. 3). This isolate derived from Vårhallarna, in southern Sweden (SI Appendix, Table S5). We subsequently performed shotgun whole genome sequencing of B38 which we identified as Brevundimonas sp. The assembled and annotated genome did not identify putative pathogenic or virulence genetic factors present in the genome.
If there is an interaction between B38 and the host, the growth-promoting effect of B38 could be either direct or indirect, mediated through other members of the community. To test the direct effect of B38 on host growth, we grew Arabidopsis plants of an accession (#6136) from the south of Sweden chosen to have intermediate relative abundance of B38 in the field. Plants were grown under sterile conditions in 1/2 MS media under long-day conditions in the growth chamber, with and without B38 inoculation. Approximately 2 wk after germination, over 600 plants were randomly selected for either drip inoculation with the control or B38 inoculum, and measured for surface area growth over the following 2 wk. Accounting for variation in plant growth among trials and plates within trials, we found that plants treated with B38 grew 5.375 (SE = 1.973) mm2 larger than control plants (F = 7.3981, df = 1, P value = 6.7e−3) between days 7 and 14, corresponding to a 10.22% growth increase.
The microbial hubs could, in principle, influence host fitness directly, for example, by contributing to growth, or indirectly through their influence on other beneficial members of the microbial community (26). Here we show that B38 directly improves host growth over early life stages in isolation from the rest of the microbial community. This result is consistent with our field observations, where we found a positive correlation between genetic variation associated with B38 and with seed-set, suggesting that, in this instance, the correlation is causative. The possibility of additional indirect interactions in the field cannot, of course, be excluded.
Mapping Host Genetic Associations with the Relative Abundances of Microbial Hubs across Experiments
Our observation that host control of the relative abundance of four microbial hubs explains ∼12% of variation in seed-set among Arabidopsis genotypes grown in eight field trials suggests the potential to reveal host genes that can enhance plant performance in the presence of microbes, particularly across environments. Toward this end, we performed genome-wide association mapping for host genotype effects on microbial hubs (n = 19) and seed-set across all experiments. Despite significant differences among accessions, genome-wide association (GWA) analysis yielded few peaks with P values below accepted significance thresholds after correction for multiple testing. Specifically, we found only two significant associations, both for microbial hub B41. The first is located on chromosome 1 at position 29909876 in AT1G79510 annotated as a pseudogene. The second is on chromosome 4 on positions 15704377, 15704472, and 15704478. These consecutive single-nucleotide polymorphisms (SNPs) are located between YUC-1 (AT4G32540), involved in auxin biosynthesis, and LEUNIG (AT4G32551), involved in the development of the leaf blade and floral organs.
A potentially more powerful strategy to detect minor quantitative trait loci (QTL) involves computing local association scores along the genome. The assumption underlying this method is that neighboring markers in linkage disequilibrium with causal mutations will also carry association signals; thus, aggregating P values increases power (27). This method identified 344 nonoverlapping loci (hereafter QTLs), with sizes ranging from 93 bp to 150,926 bp, including a total of 25,529 SNPs. Out of the 344 QTLs, only 27 included SNPs associated with multiple traits (Dataset S3).
To investigate functions underlying these associations, we tested pathway and Gene Ontology (GO) term enrichment (biological processes only) (28, 29). Each annotated gene was assigned the highest absolute SNP effect within 5 kb, and we used a combination of methods based on effect sizes accounting for multiple testing, overlapping gene lists, and the potential aggregation of functions and associations along the genome (30–33); we identified 29 enriched GO terms related to biological processes across 16 traits (Datasets S4 and S5), including genes involved in the response to virus (GO:0009615) and nematodes (GO:0009624), hypersensitive response (GO:0009626), and response to chitin (GO:0010200), all of which are related to interactions with other organisms. Three enriched GO terms directly concern auxins and their transport (GO:0009926, GO:0010540, and GO:0009734); auxins have previously been documented to contribute to shaping plant interactions with beneficial bacteria (34, 35). Specialized metabolites also appear to be involved in shaping the relative abundance of microbial hubs. Indeed, hub B107 is associated with genes in the geranylgeranyl diphosphate metabolism (GO:0033385), the universal precursor of diterpenes, which include carotenoids, gibberellins, and hormones such as abscisic acid. In addition, loci associated with B76 are enriched in genes related to specialized metabolite biosynthesis (GO:0044550) and genes involved in the synthesis of sinapoyl glucose and sinapoyl malate (PWY-3301), a side branch in the synthesis of phenylpropanoids. Genes involved in the synthesis of glucosinolates from phenylalanine (like glucotropaeolin in ref. 36, PWY-2821) and hexahomomethionine [specifically, 8-(methylsulfinyl)octyl-glucosinolate (36), PWYQT-4475] are also enriched in loci associated with B5 and F71, respectively.
The functions highlighted by our analysis are in line with other studies suggesting the involvement of specialized metabolites, auxins, and the immune system in influencing the leaf microbial communities (37, 38). Our analysis also highlights less obvious functions, like fatty acid and brassinosteroids biosynthesis (Dataset S5). This is especially true for beneficial members of the community. For example, loci associated with the relative abundance of the beneficial microbial hub B38 are enriched for transition metal ion transport (GO:0000041), response to carbohydrates (GO:0009743), and fatty acid biosynthesis (PWY-4381).
Plant Specialized Metabolites Correlated with Microbial Hub Abundance
Our biological processes and pathway enrichment analysis suggest that specialized metabolites are involved in shaping microbial hubs. To support this result, we quantified 20 compounds using untargeted metabolomics in a subset of the field samples in which we characterized the rosette microbiome. These compounds were chosen to be abundant, allowing annotation, while limiting the number of tests required to explore their association with microbial hubs. We found that the relative abundance of 14 out of 19 hubs was significantly correlated with at least one of 11 specialized metabolites (after correction for multiple testing), 6 of which displayed significant heritability across field sites ranging from 1 to 38% (SI Appendix, Fig. S9 A and B).
The molecule 8-(methylsulfinyl)octyl-glucosinolate (36) (260_GSL_8MSO in SI Appendix, Fig. S9 and Table S6) displayed the strongest relationship with multiple microbial hubs in the field (SI Appendix, Fig. S9A and Table S6), as well as significant heritability under field conditions (SI Appendix, Fig. S9B). The variation among accessions of this abundant glucosinolate was less evident in the greenhouse and in sterile conditions (SI Appendix, Fig. S9B), however, leaving open the possibility that the correlation is induced by one or more of the microbial hubs. In contrast, other molecules significantly related to the abundance of microbial hubs in the field across experiments (354_C_Cy-GRGF_785 and 358_F_R-K-R_577; SI Appendix, Table S6) are heritable in all conditions, and variation among accessions in the field is positively correlated with the variation among accessions in the greenhouse. This suggests that these flavonoids are constitutively and consistently produced by accessions and influence microbial hubs in a manner that is robust to heterogeneity among field experiments.
Conclusion
In this study, we show that, not only does host genetic variation influence the microbiome, it does so consistently. Host genotype effects are centered on ecologically important hub species, and appear to percolate through the microbial community, at least in part as a result of microbe–microbe interactions. Our replicate field experiments were instrumental in allowing us to reveal consistent host effects on the leaf microbiome via common and widespread hub species.
Furthermore, we found that the influence of host genetics on a handful of prevalent microbial hubs has a far-reaching impact on the community, and is associated with a substantial fraction of the variation in our fitness estimates among accessions. Although these relationships are correlational, a causal relationship is plausible (39), and, indeed, we were able to culture one of the identified hubs and confirm a direct positive effect on host fitness experimentally.
Understanding how host performance or fitness components are influenced by their ability to shape microbial communities could provide a basis for breeding crops favoring microbes that are beneficial to both growth and resistance to pathogens. We successfully mapped variation in host microbe interactions using genome-wide association, and our results suggest that natural and artificial selection can act on plant traits such as leaf specialized metabolites, auxins, and the immune system to improve plant performance through effects on microbial communities (40, 41). In addition, we found that at least some plant metabolites are expressed in a consistent manner that is robust to variation among our experiments and correlates with the relative abundance of microbial hubs. Our results therefore suggest that ongoing efforts to harness host genotype effects on the microbiome for agricultural purposes can be successful, and highlight the value of explicitly considering abiotic variation in those efforts.
Materials and Methods
Field Experiments.
This study uses a set of 200 diverse accessions (inbred lines; SI Appendix, Table S1) that were previously resequenced (15). The seeds were produced simultaneously in the greenhouse of the University of Chicago under long-day conditions, except for a 12-wk vernalization period at 4 °C, required to induce flowering. The seeds for the common garden experiments were cold stratified in water at 4 °C for 3 d before being planted in trays of 66 open-bottom wells, each measuring 4 cm in diameter. For each experiment, trays were filled with a mix of 90% standard greenhouse soil and 10% local soil. The local soil was collected at the site where each experiment was established, within 2 d of seeds being planted in each year. The standard greenhouse soil was bought in a single order for the four experiments each year. The sites chosen for the experiments were as follows:
SU: Ullstorp (agricultural field, lat: 56.067, long: 13.945)
SR: Ratchkegården (agricultural field, lat: 55.906, long: 14.260)
NM: Ramsta (agricultural field, lat: 62.85, long: 18.193)
NA: Ådal (south-facing slope, lat: 62.862, long 18.331)
The sites were chosen to be Arabidopsis habitats and located near known natural populations. Each experiment included three complete randomized blocks, including eight replicates per accession. Experiments were sown in pairs (two in the north and two in the south) over 6 d, corresponding to the sowing of one block a day, alternating between the two experiments (between 7 and 12 August in the north, and between 31 August and 5 September in the south, in both years). The trays were placed in a common garden the morning after sowing under row tunnels to avoid disturbance by precipitation and to favor germination (on the campus of Mid Sweden University in the north and Lund University in the south). Trays were watered as needed, and missing seedlings were transplanted between cells within blocks and then thinned to one per cell after 9 d. Seventeen days after sowing, trays were laid in the field in their final location over tilled soil. For each experiment, the blocks were laid across the most obvious environmental gradient (exposition, shading, slope, soil humidity, …). The pierced bottom of the cells allowed the roots to grow through and reach the soil, as was verified upon harvest. The same protocol was followed in 2011 and 2012.
Sample Collection and DNA Extractions.
The rosettes used to characterize the microbial community were harvested in the spring of 2012 and 2013 only a few days after the plants were exposed, following snowmelt. We harvested two randomly selected replicates per accession in each experimental block. Upon harvest, rosettes were placed in sealed paper envelopes, placed on dry ice, and then kept at −80 °C until lyophilized (S1 Appendix, Supplementary Methods). DNA extractions were performed on powdered lyophilized rosette tissue. The protocol used included two enzymatic digestions to maximize yield from gram-negative bacteria (42) but otherwise followed (43). Further details about sample processing and DNA extractions are given in SI Appendix, DNA Extraction.
PCR and Sequencing.
To describe the microbial communities, we amplified and sequenced fragments of the taxonomically informative genes 16S and ITS for bacteria and fungi, respectively. For bacteria, we amplified the hypervariable regions V5, V6, and V7 of the 16S gene using the primers 799F (5′-AACMGGATTAGATACCCKG-3′) and 1193R (5′-ACGTCATCCCCACCTTCC-3′) (9, 44). For fungi, we amplified the ITS-1 region using the primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) (16) and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′) (45). The sequencing was performed using 11 MiSeq 500 cycle V2 kits following ref. 46. Primer design (47), PCR conditions (48), and sequencing methods (49, 50) are presented in more detail in SI Appendix, PCR and Sequencing.
Sequence Processing and Clustering.
The demultiplexed fastq files generated by MiSeq reporter for the first read of each run were quality filtered and truncated to remove potential primer sequences and low-quality base calls using the program cutadapt (51). The reads were then further filtered and converted to fasta files using the FASTX-Toolkit (-q 30 -p 90 -Q33). The fasta files for each run were then dereplicated using AWK code provided in the swarm git repository (https://github.com/torognes/swarm) (17). The resulting dereplicated fasta files were filtered for PCR chimeras using the vsearch uchime_denovo command (https://github.com/torognes/vsearch). The dereplicated fasta files for each run were then combined and further dereplicated at the study level. The fasta files were then used as input for OTU clustering using swarm (-t 4 -c 20000). The clustering identified 150,412 and 251,065 OTUs for the fungal and bacterial communities, respectively. The output files were combined into two separate community matrices using a custom python script (available at GitLab, https://forgemia.inra.fr/bbrachi/microbiota_paper) (52). The taxonomy of each OTU was determined using the quiime2 2019.1 v8 feature classifier trained on the UNITE V8 and SILVA 1.32 database for bacteria and fungi, respectively (53, 54).
Count Table Filtering.
The count tables obtained for both the bacterial and fungal communities were filtered in successive steps by removing the following:
-
1)
samples corresponding to empty wells and additional plant genotypes present in the experiments sampled by mistake (leaving 7,476 and 7,240 samples for the fungal and bacterial count tables, respectively)
-
2)
samples with less than 1,000 reads (leaving 6,678 and 6,819 samples for the fungal and bacterial count tables, respectively)
-
3)
OTUs not represented in at least 10 reads in at least five samples (leaving 1,381 and 993 OTUs for the fungal and bacterial count tables, respectively)
-
4)
for the bacterial community, OTUs assigned to plant mitochondria (leaving 993 OTUs in the bacterial count table, no OTUs assigned to plant mitochondria)
-
5)
for a second time, samples with less than 1,000 reads (leaving 6,656 and 6,783 samples for the fungal and bacterial count tables, respectively).
The final count tables used in the study included 993 OTUs and 6,793 samples for the bacterial communities and 1,381 OTUs and 6,656 samples for the fungal community.
The counts for the bacterial community included between 570 and 1,051 samples per experiment. The counts for the fungal community included between 530 and 996 samples per experiment.
Differentiation of the Microbial Communities among Sites and Years.
This analysis was performed for the fungal and bacterial communities independently, including all samples and only OTUs with read counts above 0.01% of total read counts (after the filtering described above) across sites and years. To investigate how the microbial communities differed among sites and years, we performed a constrained ordination on log-transformed read counts using the capscale function in the R-package Vegan (55) and following ref. 56. The log transformation offers the advantage of removing large differences in scale among variables. The capscale function performs canonical analysis of PC, an analysis similar to redundancy analysis (rda), but based on the decomposition of a Bray–Curtis dissimilarity matrix among samples (instead of Euclidean distance in the case of rda). This allows identification of the dimension that maximized the variance explained by components, while discriminating groups of samples, here sites and years, with the formula “Y ≈ site + year + site * year” where Y is the count matrix normalized to 1,000 reads and transformed with log(x + 1) (23, 56).
Core Microbiota.
In order to define a core microbiota, we counted, for each OTU, the number of site/year combinations in which it was prevalent. We defined “prevalent” as being present in at least 50% of the samples in a given site/year. We performed this analysis using count tables for each experiment with the filtering described in the previous paragraph. Therefore, for an OTU to be designated as a member of the core microbiota, it needed to have nonzero counts in more than 50% of the samples within each site/year combination and, due to previously described filtering, needed to be represented by at least 10 reads in five of those samples across all site/year combinations (see Count Table Filtering).
Heritability of the Microbiota.
In this analysis, count tables were split per site and year before filtering for OTUs represented by more than 0.01% of the reads (after the filtering described in Count Table Filtering) for each of the bacterial and fungal communities. The resulting 16 counts tables were normalized to 1,000 reads per sample and used to calculate 16 Bray–Curtis pairwise dissimilarity matrices among samples. Count tables were not rarefied. Relative abundances were multiplied by the minimum depth of 1,000 reads. These matrices were then decomposed into 10 PC. For each component, we estimated broad sense heritability (hereafter H2), that is, the proportion of variance explained by a random intercept effect capturing the identity of the accessions present in the experiment (plate effects had limited impact on H2 estimates but were included in the models) in models following
| [1] |
where was one of the 10 PC, is the effect of the plate, Plate is the design matrix capturing the assignation the ith sample to the jth plate, and is the random intercept term capturing the effect of the kth accession and captures the residual variance. Heritability (H2) was estimated as the percentage of variance explained by the random accession intercept,
| [2] |
Mixed models were fitted using the function lmer in the lme4 R package (57). We computed 95% confidence intervals (CIs) using 1,000 bootstraps, and components were considered to have significant H2 when their CIs did not overlap zero (lower bound of the CI ≥ 0.01).
Heritability of Individual OTUs.
This analysis was also performed per site, year, and community, as in the microbiota H2 estimation analysis. In this analysis, counts were transformed to centered log-ratios (CLR; after adding one to all counts to handle zeros) using a dedicated function in the R package mixOmics (58, 59). Individual transformed OTU counts were modeled with a model following Eq. 3,
| [3] |
where is the vector of transformed counts for one OTU, and is the random intercept term capturing the effect of the kth accession. captures the residual variance. H2 estimates and CIs were computed as the proportion variance explained by the accession term for each OTU (Eq. 2). We computed 95% CIs using 1,000 bootstraps, and OTUs were considered to have significant heritability when their CIs did not overlap zero (lower bound of the confidence interval ≥ 0.01). H2 estimates for our estimate of seed-set (see below) were estimated the same way using a Box–Cox transformation.
Microbe–Microbe Cooccurrence Networks.
Microbe–microbe cooccurrence networks were computed for the fungal and bacterial communities together, using the count tables per site/year and filtering OTUs represented by less than 0.01% of the reads within each community. The count tables were then combined into the same table and analyzed using the SPIEC-EASI (v1.1) pipeline (23). This method computes sparse microbial ecological networks in a fashion robust to compositional bias and uses conditional independence to identify true ecological interactions, meaning that a connection between two OTUs will be significant when one provides information about the other, given the state of all other OTUs in the network. This means that covariance among OTUs induced by microenvironmental and host genetic variation is controlled. SPIEC-EASI was run using the neighborhood selection framework, and model selection was regularized with parameters set to a minimum lambda ratio of 1e−2 and a sequence of 50 lambda values (see documentation for SPIEC-EASI and the huge R package, which provides regularization functions) (60).
Network Statistics.
The inferences of microbe–microbe ecological interactions inferred using SPIEC-EASI were passed to the igraph package (61), which was used for enforcing simplicity of graphs (no edges that connect vertices to themselves or duplicated edges), computing degree and betweenness centrality of vertices, computing distances between vertices, and plotting. With each of the eight networks thus computed, hubs were defined as OTUs with degree and betweenness centrality both in the 5% tail of their respective distributions. We then checked the overlap between heritable OTUs and hubs, and the overrepresentation of heritable OTUs among hubs was tested using a simple χ2 test across all site/year combinations. The relationship between distances to heritable hubs (OTUs that are both hubs and have significant H2) and heritability was investigated using Spearman’s rank correlation coefficient. Distances were calculated as the number of edges between OTUs and the closest heritable hub in the network. OTUs not connected to heritable hubs were assigned a distance equal to one more than the maximum distance observed for OTUs connected to heritable hubs.
In order to investigate whether the microbe–microbe associations detected in the networks were mostly due to host genetic effects shared among microbes, we performed permutations of the count tables for each site and year as follow:
-
1)
Compute read counts per sample.
-
2)
Perform a log-ratio transformation of the count table (count + 1).
-
3)
Compute heritability estimates for each OTU (H2; see Heritability of Individual OTUs).
-
4)
For each OTU, and for each Arabidopsis genotype, resample the log-ratio transformed counts without replacement across samples. This permutation scheme maintains shared host effects on OTUs but breaks up correlations among OTUs that are independent of the host genotype.
-
5)
Compute new heritability estimates on the permuted data for each OTU (H2P), which is equal to H2.
-
6)
Transform the nonpermuted and the permuted log-ratio transform count tables back to proportions using the softmax function (https://rpubs.com/FJRubio/softmax) and then back to counts using the counts per sample computed in step 1 above.
-
7)
Infer interaction networks from both these new count tables using SPIEC-EASI (see Microbe–Microbe Cooccurrence Networks).
Estimation of Seed-Set.
The experiments each included eight replicates per block per accession (24 replicates per experiment). While we harvested two replicates per block (six replicates per experiment) for microbiota analysis, the remaining plants were left to grow, flower, and produce seeds in the field. We harvested the mature stems of all remaining plants at the end of the spring, when all plants had finished flowering and siliques were mature, and stored them flat in individual paper envelopes. We estimated lifetime seed production (seed-set) by the size of the mature stems. After removing remaining traces of roots and rosettes, each mature plant was photographed on a black background, using a digital single-lens reflex camera (Nikon 60D) mounted on a copy-stand and equipped with a 60-mm macro lens (Nikon 60mm). The photographs were segmented [using custom scripts in R based on the EBimage package (62)] to isolate plants from the image background and estimate the total surface of the image they occupied.
We validated this method with mature plants harvested from a previous experiment that was planted in NM in fall 2010, and that included the 200 accessions used in this study. We counted siliques and estimated the average silique size for 1,607 mature stems that were also photographed. The total silique length produced per plant (number * average size) was highly correlated with our size estimates based on image analysis (Spearman’s rho = 0.84) and displayed a clear linear relationship.
Relationship between Host Effects on Microbial Hubs and Seed-Set.
To investigate the relationship between host genotype effects on heritable hubs and seed-set in each experiment, we computed estimates of accession effects (best unbiased linear predictors [BLUPs]) for both log-ratio transformed heritable hubs and Box–Cox transformed seed-set estimates. We then fitted multiple regressions for each site/year combination aiming to explain seed-set variation among accessions with their influence over microbial hubs and following Eq. 4.
| [4] |
where is the seed-set estimate of the ith accession (BLUP), and is the effect of the ith accession on the jth hub. is the regression coefficient for the jth hub, and is the regression coefficient for the jth hub squared. captures residual variance per accession. We then performed forward/backward model selection to obtain the final models presented in SI Appendix, Table S4.
Heritable Hubs and Seed-Set across Environments.
We next investigated host effects on heritable hubs and seed-set across all eight experiments. Similarly to previous analyses, count tables were split per site and year before filtering for OTUs represented by more than 0.01% of the reads (after the filtering described in Count Table Filtering) for each of the bacterial and fungal communities. The resulting 16 count tables were then transformed (CLR) and combined into one before fitting a mixed model following Eq. 5,
| [5] |
where is the vector of transformed counts for one OTU, is the effect of the experiment j, is the design matrix capturing the assignation the ith sample to the jth experiment, n = 8), and is the random intercept term capturing the effect of the kth accession. .
Seed-set data were analyzed the same way, except we performed Box–Cox transformation of the data. The lambda parameter for the Box–Cox transformation was estimated using the same model, but without the random accession term. Heritability was calculated according to Eq. 2.
For both heritable microbial hubs and seed-set, we retrieved random intercept accession effects (BLUPs) and fitted a multiple linear regression following Eq. 6,
| [6] |
where is the effect of the ith accession (n = 200) on seed-set (across all experiments), is the effect of accession i on hub j across all experiments, and is the squared effect of accession i on hub j. and are the corresponding regression coefficient for hub j and captures the residual variance per accession. The final model was obtained after backward/forward model selection based on AIC.
Isolation, Culture, and Identification of Microbial Hubs.
Bacteria sampling from wild A. thaliana plants.
We collected two leaves from 10 plants at five locations in Sweden (SI Appendix, Table S5) which we stored in 20% glycerol at −20 °C. Wild A. thaliana microbial isolates were collected using modified methods that were previously described (25), using six distinct media selected to capture a diverse set of bacterial isolates (63). After isolating and cultivating colonies, we performed DNA extraction and identified over 3,900 isolates using 16S and gyraseB sequencing [SI Appendix, Bacteria Sampling from Wild A. thaliana Plants (64)]. Matches to our experimental OTUs are indicated in Dataset S2. Of the isolates identified, we focused on the heritable hub, B38, which appears to contribute to seed-set in the field.
B38 Whole Genome Assembly.
We used a low-input method for Illumina library preparation (65). Briefly, ∼2 ng of extracted DNA was used in a reduced volume (5 µL) tagmentation reaction with TDE1 (incubate 55ºC for 10 mins, room temperature for 5 mins). The tagmentation reaction was added to a 15-μL PCR, adding the Illumina adapters (Kapa HiFi Hotstart PCR kit KK502, standard Illumina adapters and cycling). The library was cleaned with 0.8× volume SPRI (solid-phase reversible immobilization) beads, quantified on the Bioanalyzer, and run on the MIseq2500 using paired end 300 chemistry. Reads were trimmed for adapters (BBDuk, ktrim = r, k = 23, mink = 11, hdist = 1 tbo) and quality across a sliding window (k = 4, trimq = 20) (66). Reads were assembled using SPAdes (using the settings --isolate -k 21,33,55,77) and annotated with the software Prokka designed for rapid prokaryotic genome annotation (67, 68).
Plant Growth Assays with B38.
Plant growth.
A. thaliana accession 6136 from Southern Sweden was used in the growth assays. In our field experiments, it displayed average relative counts for B38 (rank 102 of 199). The plant assay used slightly modified methods as previously described (69). The seeds were exposed to chlorine gas for sterilization: In a bell jar with dessicant, an open 1.5-mL tube with seeds was placed next to a 50-mL beaker with 40 mL of Chlorox bleach and 1 mL of hydrochloric acid, sealed with parafilm, and incubated for 4 h. Sterilized seeds were subsequently sown on 24-well tissue plates containing 1.5 mL of 1/2 MS media (Murashige & Skoog medium including Nitsch vitamins, bioWORLD) containing 500 mg/L MES (2-Morpholinoethanesulfonic acid hydrate), pH 5.7 to 5.8. Plates were wrapped in parafilm and vernalized in the dark at 4 °C for 4 d. The plates were individually wrapped with micropore tape to prevent environmental contamination and transferred to a growth chamber with 16 h of light at 16 °C. The plants were treated with either B38 or control inoculum between days 13 and 15 postvernalization. The plates were returned to the chamber to grow for another 14 d.
B38 inoculation.
The B38 isolate grew in R2A liquid media in an orbital shaker for approximately 3 days, until the optical density at a wave length of 600 (OD600) reached 0.2. To ensure no environmental contamination, a portion of the inoculum was saved for DNA extraction and subsequent 16S Sanger sequencing verification. The liquid cultures were pelleted by centrifuging at 1,800 relative centrifugal force (RCF) at 18ºC for 7 min, decanted, and resuspended in 0.1 M MgSO4. The plants in each 24-well plate were randomly selected to receive the infection (B38 + 0.1 M MgSO4) or control (0.1 M MgSO4) treatment. Each plant was drip inoculated using pipettes with 180 µL of the selected treatment. The plates were rewrapped in micropore tape and returned to the growth chamber.
Measuring plant growth.
We performed three trials of 11, 28, and 23 plates, totaling 62 twenty-four-well plates. Plants were not treated and were removed from the experiment if they had less than three true leaves, cracked agar, or failed to germinate, resulting in a total of 1,094 plants. The plants were individually photographed immediately before inoculation, then again at 7 and 14 d postinoculation. The images were processed using a custom script employing cv2 in Python (70), which quantified plant surface area in each well by scaling based on the wells’ size, converting images into binary images, and measuring nonwhite pixels within each well (i.e., plant surface area). The output images were manually inspected, and any images which failed to be accurately processed were manually measured using the same pipeline described above, but using Image J.
Due to the high humidity of the plates and the drip inoculation, 422 plants showed signs of waterlog stress. Plants were scored for symptoms of stress induced by waterlogging (blindly with regard to B38 inoculation) as categorized by translucent/white leaves or stunted growth, and were removed from the experiment.
We used a linear mixed model (Eq. 4) accounting for variation in plant growth among trials and plates within trials to estimate the effect of B38 inoculation.
| [7] |
In Eq. 4, is the growth of ith plant in the jth plate/assay combination. is the estimate of the treatment effect compared to the controls (intercept), and is the treatment (inoculation with a B38 or control solution); is the random intercept effect capturing variation among plates in assays (n = 62 plates across three trials). captures the residual variances.
Genome-Wide Association Mapping.
Single polymorphism calling and filtering.
SNPs used in this study were generated in the context of the 1001 Genome Project (71) and published in Long et al. (15). As pipelines evolved, we reran SNP calling to ensure optimal quality (SI Appendix, Single Polymorphism Calling and Filtering).
Phenotype preparation and association analysis.
Association mapping analyses were performed for the 11 heritable microbial hubs for which we estimated host genotype effects across experiment and accession seed-set estimates. Association analyses were performed using a classical one-trait mixed model accounting for genetic relatedness among accessions (kinship) (71).
In order to take advantage of linkage disequilibrium and gain power by grouping association statistics in contiguous markers, we computed local association scores (27). We followed the instructions provided by the authors and defined the parameter Xi as the 0.999 quantile of the distribution of rounded to the closest integer for each trait investigated (19 microbial hubs and seed-set). The approach highlights regions, which we call QTLs.
The null association model (without fixed SNP effect) from Gemma allows us to estimate SNP-based heritability or pseudoheritability (72), which is the proportion of variance explained by the random accession effect, accounting for the genetic similarity among accessions. To investigate whether the regions highlighted by the local score approach included true positives, we computed SNP-based heritability for each trait, each time using three sets of SNPs to compute the kinship matrix: 1) all the SNPs in the genome over 10% frequency, 2) all the SNPs within QTLs identified by the local score approach, and 3) all SNPs not included in the QTLs identified by the local score approach.
Pathway enrichment analysis.
To investigate biological functions associated with seed-set of accessions or their influence over microbial hubs, we searched for enrichment in annotated pathways (in the BIOCYC database) and GO categories (biological processes only) in A. thaliana. Gene-set enrichment methods are designed for assays that directly assign P values or effects to individual genes (i.e., RNA sequencing experiments). Here, for each trait, each gene was attributed the largest absolute SNP effect within a distance of 5 kb on each side and followed the setRank procedure that accounts for overlapping categories and multiple testing. We set the parameter “setPCutoff” to 0.01 and set the “fdrCutoff” to 0.05 (30). To account for specificities of gene-set enrichment in the context of association mapping, we also tested the enrichment of the gene groups identified by setRank using a weighted Kolmogorov–Smirnov score (31) and a permutation scheme accounting for the nonindependence of marker effects due to linkage disequilibrium along the genome, as well as the potential clustering of genes with similar function (32, 33). Briefly, enrichment was calculated using a weighted Kolmogorov sum using gene effect rank (and not a gene effect significance threshold) (31). Enrichments were then tested against an empirical distribution generated from 1e5 permutations. For each permutation, chromosomes are randomly reordered and reoriented, and the whole genome is shifted (or “rotated”) by a random number, before reassigning SNP effects to genes and calculating enrichment for the groups of genes of interest. We considered only categories with empirical P values below 0.05.
Untargeted Metabolomics.
Plant material and sample preparation.
This analysis uses three sets of samples. The first are samples collected from the experiments in Sweden and correspond to a subset of those used for the microbial community. In particular, we chose samples from the four experiments established in 2012 and focused on a subset of 50 accessions selected to span the genetic variation among hosts in our mapping population. The second set of samples correspond to six replicates of the same 50 genotypes grown in the University of Chicago greenhouse during the summer 2014 under long-day conditions (16-h light period), in standard culture soil. After 28 d, plants were vernalized for 3 wk at 4 °C, and leaf samples were collected after vernalization, immediately flash frozen in liquid nitrogen, freeze-dried, and stored at room temperature. The third set corresponds to three replicates of the same 50 genotypes, grown on sterile agar medium (Murashige & Skoog with Nitsch vitamins) in individual well plates in a growth chamber with a 16-h light period (long-day condition). Seeds were sterilized by a 70% ethanol bath for 10 min, and manipulated under a sterile hood. Samples were collected after 28 d of growth, flash frozen, freeze-dried, and stored at room temperature.
Dried samples from the three sets were coarsely ground, and distributed in 18 ninety-six-well plates with two ceramic grinding beads per well (10 mg per well ± 2 mg). Samples were randomized across all plates to limit confounding of biological effects. In addition, each plate included 16 random samples (1/6) from each experimental unit (greenhouse, sterile, and the four field experiments).
Specialized Metabolite Extraction and Liquid Chromatography–MS Analysis.
The extraction protocol was designed to extract polar compounds such as glucosinolates and flavonoids. Samples in plates were ground using a Geno/Grinder (SPEX SamplePrep 2010) at 1,750 rpm for 2 min. The extraction buffer (70% methanol, 30% water, internal standard: quercetin, 0.0708 mM) was added using a Tecan pipetting robot (100 μL per milligram of dry material). Samples were shaken at room temperature for 2 h and filtered on 96-well filter plates (0.45 μm) on a vacuum manifold. The flow-through was collected in 96-well plates and stored at 4 °C.
Samples were autoinjected through a Zorbax SB-C18 2.1 × 150 mm, 3.5-μm column on an Agilent Q-TOF liquid chromatography–MS with dual electrospray ionisation (ESI, Agilent 6520) with the following parameters: 325 °C gas temperature, 6 L⋅min−1 drying gas, 35-eV fixed collision energy, 35 psig nebulizer, 68-V skimmer voltage, 750-V OCT 1 RF Vpp, 170-V fragmentor, and 3,500-V capillary voltage. Mass accuracy was within 2 ppm to 5 ppm. Samples were eluted with 0.1% formic acid in water (A) and 100% acetonitrile (B) using the following separation gradient: 95% A injection followed by a gradient to 90% A at 1 min, 45% A at 6 min, and 100% B at 6.5 min with 4-min hold and 3-min equilibration. An external standard (sinigrin, 1 mM) was run four times before each plate and one time every 20 samples to monitor and maintain run quality. Compounds were characterized using retention times and fragmentation patterns of chromatograms with automatic agile integration in Agilent Mass Hunter Software (Qualitative Analysis B6 2012), and fragments were compared to online databases, massbank (massbank.jp) and plantCyc (plantcyc.org). The XCMS package for peak detection in R (cran.r-project.org) was used to align chromatograms, adjust retention times, and group the peaks. For every molecule, a “barcode” peak was chosen to have a unique retention time and mass to charge ratio (m/z) combination. The size of these peaks relative to the internal standard, Quercetin, was used to quantify each molecule in every sample.
Statistical analysis.
The peaks’ intensities relative to the internal standard were used to capture molecule concentration variation. Standardized intensities were square root transformed before analysis. Heritability of individual compounds in the three conditions was performed using random intercept models identical to those used to estimate OTU heritability. A fixed “site” effect was added for the field samples. In the greenhouse and sterile conditions, a simple random accession term was used to quantify heritability and estimate accession effects (BLUPs). Those accession effects were used to estimate genetic correlation between specialized metabolites in the field and the greenhouse. We used Pearson’s correlation coefficient and corrected the corresponding P values for false discovery rate (FDR; n = 20).
For the field samples, we modeled the relationships between the relative abundances of 19 microbial hubs and the relative intensity of 20 compounds (SI Appendix, Table S6) using a linear model following Eq. 8,
| [8] |
where are the log-ratio transformed counts of one of the 19 microbial hubs used for mapping, are the four site effects, is the design matrix assigning sample i to site s, is the effect of one of the 20 molecules identified in our untargeted screen, and is the relative intensity of the molecules measured in sample i. are site-specific regression coefficients (interactions between the site and molecule effects). captures the residual variances. We fitted 380 models (19 hubs and 20 molecules) and used F tests to estimate term significance. All P values corresponding to the molecule effect were corrected for FDR (n = 380).
Supplementary Material
Acknowledgments
Thanks go to Mia Holm for her hospitality and wonderful dinners after hard work in the field as well as help during harvesting; to Einar Holm for helping with field work and taking photos of harvested plants; to Torbjörn Säll for assistance with sampling and providing greenhouse space in Lund; and, finally, to the Kleen family, the Öhman family, Nils Jönsson, and the Rathckegården farm for allowing us to install our experiments on their land. Thanks go to Timothée Flutre and Talia Karasov for helpful discussions on previous versions of the manuscript. Thanks go to Man Yu from the C.D. lab, who helped generate stem images used for seed-set estimates and manual seed-set estimates. This work was funded by a grant from the National Health Institute (Grant R01 GM 083068) to J.B., M.N., and C.D.; by a Dropkin Foundation Fellowship to B.B.; and with support from the University of Chicago and New York University (to J.B.). B.B. has received the support of the European Union in the framework of the Marie-Curie FP7 COFUND People Programme, through the award of an AgreenSkills/AgreenSkills+ fellowship (under Grant Agreement 267196). P.D., M.L.M., and P.L.G. are students in the Magistère de Génétique Graduate Program at Université de Paris. Computing resources and storage were provided by the Center for Research Informatics, funded by the Biological Sciences Division at the University of Chicago with additional funding provided by the Institute for Translational Medicine; CTSA Grant UL1 TR000430 from the NIH; the genotoul bioinformatics platform Toulouse Occitanie, France (Bioinfo Genotoul, http://bioinfo.genotoul.fr); and Bordeaux Bioinformatics Center at the University of Bordeaux, France.
Footnotes
Reviewers: P.J.B., Agricultural Research Service, US Department of Agriculture; T.M.-O., Duke University; and V.S., University of California, Davis.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201285119/-/DCSupplemental.
Repeatability of Analysis and Data Availability
All scripts used to perform the analyses presented in this paper, as well as nonessential but complementary figures, are available in the GitLab repository https://forgemia.inra.fr/bbrachi/microbiota_paper (52).
Data tables for OTU counts, seed-set estimates, and plant growth data for the B38 experiment are also available in a Zenodo repository (73).
Metabarcoding Illumina sequences (ITS and 16S amplicons) and the B38 sequence data have been deposited in National Center for Biotechnology Information under BioProject PRJNA707473 (74).
References
- 1.van Opstal E. J., Bordenstein S. R., Rethinking heritability of the microbiome. Science 349, 1172–1173 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Vétizou M., et al. , Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdul-Aziz M. A., Cooper A., Weyrich L. S., Exploring relationships between host genome and microbiome: New insights from genome-wide association studies. Front. Microbiol. 7, 1611 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodrich J. K., et al. , Human genetics shape the gut microbiome. Cell 159, 789–799 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pamer E. G., Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science 352, 535–538 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.United Nations Food and Agriculture Organization, “Sustainable agriculture for biodiversity-biodiversity for sustainable agriculture” (Food and Agriculture Organization of the United Nations report I9577EN/1/05.2018, 2018, https://www.fao.org/3/I6602E/i6602e.pdf).
- 7.Santhanam R., et al. , Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc. Natl. Acad. Sci. U.S.A. 112, E5013–E5020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner M. R., et al. , Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 7, 12151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horton M. W., et al. , Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat. Commun. 5, 5320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peiffer J. A., et al. , Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 6548–6553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lundberg D. S., et al. , Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agler M. T., et al. , Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14, e1002352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rochefort A., et al. , Influence of environment and host plant genotype on the structure and diversity of the Brassica napus seed microbiota. Phytobiomes J. 3, 326–336 (2019). [Google Scholar]
- 14.Veach A. M., et al. , Rhizosphere microbiomes diverge among Populus trichocarpa plant-host genotypes and chemotypes, but it depends on soil origin. Microbiome 7, 76 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long Q., et al. , Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 45, 884–890 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardes M., Bruns T. D., ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118 (1993). [DOI] [PubMed] [Google Scholar]
- 17.Mahé F., Rognes T., Quince C., de Vargas C., Dunthorn M., Swarm: Robust and fast clustering method for amplicon-based studies. PeerJ 2, e593 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jousset A., et al. , Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 11, 853–862 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beilsmith K., Perisin M., Bergelson J., Natural bacterial assemblages in Arabidopsis thaliana tissues become more distinguishable and diverse during host development. MBio.12, 2020.03.04.958165 (2021). [DOI] [PMC free article] [PubMed]
- 20.Bulgarelli D., et al. , Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Bergelson J., Mittelstrass J., Horton M. W., Characterizing both bacteria and fungi improves understanding of the Arabidopsis root microbiome. Sci. Rep. 9, 24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng S., et al. , Genome wide association study reveals plant loci controlling heritability of the rhizosphere microbiome. ISME J.15, 3181–3194 (2021). [DOI] [PMC free article] [PubMed]
- 23.Kurtz Z. D., et al. , Sparse and compositionally robust inference of microbial ecological networks. PLOS Comput. Biol. 11, e1004226 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roux F., Gasquez J., Reboud X., The dominance of the herbicide resistance cost in several Arabidopsis thaliana mutant lines. Genetics 166, 449–460 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai Y., et al. , Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Farrar K., Bryant D., Cope-Selby N., Understanding and engineering beneficial plant-microbe interactions: Plant growth promotion in energy crops. Plant Biotechnol. J. 12, 1193–1206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonhomme M., et al. , A local score approach improves GWAS resolution and detects minor QTL: Application to Medicago truncatula quantitative disease resistance to multiple Aphanomyces euteiches isolates. Heredity 123, 517–531 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller L. A., Zhang P., Rhee S. Y., AraCyc: A biochemical pathway database for Arabidopsis. Plant Physiol. 132, 453–460 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schläpfer P., et al. , Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 173, 2041–2059 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simillion C., Liechti R., Lischer H. E. L., Ioannidis V., Bruggmann R., Avoiding the pitfalls of gene set enrichment analysis with SetRank. BMC Bioinformatics 18, 151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charmpi K., Ycart B., Weighted Kolmogorov Smirnov testing: An alternative for Gene Set Enrichment Analysis. Stat. Appl. Genet. Mol. Biol. 14, 279–293 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Brachi B., et al. , Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 6, e1000940 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atwell S., et al. , Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465, 627–631 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donoso R., et al. , Biochemical and genetic bases of indole-3-acetic acid (auxin phytohormone) degradation by the plant-growth-promoting rhizobacterium paraburkholderia phytofirmans PsJN. Appl. Environ. Microbiol. 83, e01991-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ganin H., et al. , Indole derivatives maintain the status quo between beneficial biofilms and their plant hosts. Mol. Plant Microbe Interact. 32, 1013–1025 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Fahey J. W., Zalcmann A. T., Talalay P., The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56, 5–51 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Huang A. C., et al. , A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364, eaau6389 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Castrillo G., et al. , Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.French E., Kaplan I., Iyer-Pascuzzi A., Nakatsu C. H., Enders L., Emerging strategies for precision microbiome management in diverse agroecosystems. Nat. Plants 7, 256–267 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Finkel O. M., Castrillo G., Herrera Paredes S., Salas González I., Dangl J. L., Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 38, 155–163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foster K. R., Schluter J., Coyte K. Z., Rakoff-Nahoum S., The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgan J. L., Darling A. E., Eisen J. A., Metagenomic sequencing of an in vitro-simulated microbial community. PLoS One 5, e10209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amani J., Kazemi R., Abbasi A. R., Salmanian A. H., A simple and rapid leaf genomic DNA extraction method for polymerase chain reaction analysis. Iran. J. Biotechnol. 9, 69–71 (2011). [Google Scholar]
- 44.Chelius M. K., Triplett E. W., The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 41, 252–263 (2001). [DOI] [PubMed] [Google Scholar]
- 45.White T. J., Bruns S., Lee S., Taylor J., “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR Protocols: A Guide to Methods and Applications, Innis M. A., Gelfand D. H., Sninsky J. J., White T. J., Eds. (Academic, 1990), pp. 315–322. [Google Scholar]
- 46.Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., Schloss P. D., Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walters W. A., et al. , PrimerProspector: De novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics 27, 1159–1161 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samarakoon T., Wang S. Y., Alford M. H., Enhancing PCR amplification of DNA from recalcitrant plant specimens using a trehalose-based additive. Appl. Plant Sci. 1, 1200236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohland N., Reich D., Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22, 939–946 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caporaso J. G., et al. , Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10 (2011). [Google Scholar]
- 52.Brachi B., microbiota_paper, data for "Plant genetic effects on microbial hubs impact fitness across field trials." GitLab. https://forgemia.inra.fr/bbrachi/microbiota_paper. Deposited 10 August 2020.
- 53.Kõljalg U., et al. , Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Quast C., et al. , The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dixon P., VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003). [Google Scholar]
- 56.Anderson M. J., Willis T. J., Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology 84, 511–525 (2003). [Google Scholar]
- 57.Bates D., Maechler M., Bolker B., Walker S., Fitting linear mixed-effects models using lme4. J. Stat. Softw. (2015). [Google Scholar]
- 58.Aitchison J., The Statistical analysis of compositional data. J. R. Stat. Soc. B 44, 365–374 (1982). [Google Scholar]
- 59.Lê Cao K.-A. K.-A., González I., Déjean S., González I., Unravelling “omics” Data with the R Package mixOmics (HAL, 2012). [Google Scholar]
- 60.Zhao T., Liu H., Roeder K., Lafferty J., Wasserman L., The huge package for high-dimensional undirected graph estimation in R. J. Mach. Learn. Res. 13, 1059–1062 (2012). [PMC free article] [PubMed] [Google Scholar]
- 61.Csárdi G., Nepusz T., The igraph software package for complex network research. InterJournal Complex Syst. 1695, 1–9 (2006). [Google Scholar]
- 62.Pau G., Fuchs F., Sklyar O., Boutros M., Huber W., EBImage—An R package for image processing with applications to cellular phenotypes. Bioinformatics 26, 979–981 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCaig A. E., Grayston S. J., Prosser J. I., Glover L. A., Impact of cultivation on characterisation of species composition of soil bacterial communities. FEMS Microbiol. Ecol. 35, 37–48 (2001). [DOI] [PubMed] [Google Scholar]
- 64.Bartoli C., et al. , In situ relationships between microbiota and potential pathobiota in Arabidopsis thaliana. ISME J. 12, 2024–2038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baym M., et al. , Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10, e0128036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bushnell B., BBDuk. Jt Genome Inst. https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-userguide/bbduk-guide/. Accessed 25 August 2020.
- 67.Seemann T., Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Bankevich A.et al., . SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karasov T. L., et al. , Arabidopsis thaliana and Pseudomonas pathogens exhibit stable associations over evolutionary timescales. Cell Host Microbe 24, 168–179.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bradski G., The OpenCV Library. Dr. Dobbs J. Softw. Tools 120, 122–125 (2000). [Google Scholar]
- 71.Zhou X., Stephens M., Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44, 821–824 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang J., et al. , Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 43, 519–525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brachi B., Whitehurst H., Data for “Plant genetic effects on microbial hubs impact host fitness in repeated field trials.” Zenodo. 10.5281/zenodo.6783090. Deposited 30 June 2022. [DOI]
- 74.Brachi B., Whitehurst H., Microbial sequence data for “Plant genetic effects on microbial hubs impact host fitness in repeated field trials.” NCBI BioProject. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA707473. Deposited 30 June 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scripts used to perform the analyses presented in this paper, as well as nonessential but complementary figures, are available in the GitLab repository https://forgemia.inra.fr/bbrachi/microbiota_paper (52).
Data tables for OTU counts, seed-set estimates, and plant growth data for the B38 experiment are also available in a Zenodo repository (73).
Metabarcoding Illumina sequences (ITS and 16S amplicons) and the B38 sequence data have been deposited in National Center for Biotechnology Information under BioProject PRJNA707473 (74).