Abstract
A promoter active in the late phase of the lytic cycle of lactococcal bacteriophage TP901-1 has been identified. The promoter is tightly regulated and requires the product of the phage TP901-1 orf29 for activity. A deletion analysis of the late promoter region showed that a fragment as small as 99 bp contains both the promoter and the region necessary for activation by ORF29. The transcriptional start site of the promoter was identified by primer extension to position 13073 on the TP901-1 genome, thus located 87 bp downstream of orf29 in a 580-bp intergenic region between orf29 and orf30. Furthermore, the region located −85 to −61 bp upstream of the start site was shown to be necessary for promoter activity. During infection, the transcript arising from the late promoter is fully induced at 40 min postinfection, and our results suggest that a certain level of ORF29 must be reached in order to activate transcription of the promoter. Several lactococcal bacteriophages encode ORF29 homologous proteins, indicating that late transcription may be controlled by a similar mechanism in these phages. With the identification of this novel regulator, our results suggest that within the P335 group of lactococcal phages at least two regulatory systems controlling transcription in the late stage of infection exist.
Lactococcus lactis subsp. lactis and L. lactis subsp. cremoris are widely used in the dairy industry for production of fermented milk products. The fermentation processes are highly sensitive to bacteriophage attack, and this problem has a significant economical and practical impact on the utilization of the bacteria. Many naturally occurring phage resistance mechanisms have been identified and characterized. These systems have been used to improve resistance to bacteriophages of commercially important strains with the desired fermentation qualities. Furthermore, in recent years knowledge of lactococcal bacteriophages has emerged, including full genome sequences and assignment of biological functions of genes carried by phages (for a review, see reference 13). Studies of the molecular mechanisms controlling reproduction of bacteriophages during the lytic cycle in the host L. lactis may be used for combating the phage problem by construction of designed phage resistance systems targeting specific components important for proliferation of the infecting phage.
The lactococcal bacteriophage TP901-1 is a small isometric headed phage with a noncontractile tail belonging to the P335 phage species, which contains both virulent and temperate bacteriophages (3, 7). Other members of the P335 phage species, which have been analyzed at the molecular level, are the virulent phage Φ31 and the temperate bacteriophages Tuc2009, ΦLC3, and r1t (for a review, see reference 13).
After infection of the host L. lactis subsp. cremoris 3107, TP901-1 can enter either a lytic cycle or a lysogenic state. A temporal transcriptional analysis of TP901-1 during the lytic cycle revealed sequential clusters of early, middle, and late transcribed regions on the TP901-1 genome (21). The TP901-1 promoters (PL and PR), which are active early in the lytic cycle, are divergently located and the relative activities of the two promoters determine the choice of life cycle (lytic or lysogenic) (21, 22). The PL promoter transcribes the early lytic genes while PR transcribes genes involved in the establishment and maintenance of lysogeny (21). The host RNA polymerase recognizes the early promoters, and initiation of transcription is regulated by the TP901-1 repressor, CI, encoded by orf4 in consort with the modulator of repression, designated MOR, encoded by orf5 (22).
To ensure tight control of gene expression in the later stages of infection, bacteriophages have evolved a variety of mechanisms involving synthesis of a phage-encoded control factor during the early stages of infection. The Escherichia coli phage T7 encodes a single subunit RNA polymerase, which is essential for transcription initiation of late phage genes (29). Many phages such as the Bacillus subtilis phage Φ29 and the E. coli phage P2 encode transcriptional activators that are required for the host RNA polymerase to recognize the late promoters (2, 8, 9). In the case of E. coli phage lambda, late genes are regulated by the phage-encoded antitermination protein Q, which acts at a specific DNA site and modifies the host RNA polymerase to a termination-resistant form, allowing transcription to proceed beyond the termination site and resulting in expression of the late genes (for a review, see reference 14). In E. coli bacteriophage T4, a complex mechanism couples late transcription with DNA replication, since the sliding clamp of the DNA polymerase also acts as a transcriptional activator. Transcription of the T4 late genes is activated through interaction of the DNA-linked activator with two T4-encoded RNA polymerase-binding proteins, a coactivator and a late sigma factor (for a review, see reference 16).
In the virulent bacteriophage Φ31 belonging to the lactococcal P335 phage species, a middle promoter region has been identified. Transcription from this middle promoter is induced by the presence of a Φ31-encoded activator located upstream of the middle promoter on the Φ31 genome (24, 32). The promoter and activator regulating bacteriophage gene expression are conserved between Φ31 and two temperate bacteriophages (r1-t and ΦLC3) that belong to the same phage species as Φ31 and TP901-1 (31). In bacteriophage sk1 that belongs to the lactococcal phage species 936, a middle promoter controlling transcription of four middle genes was identified (5). This promoter was proposed to be induced by a phage-encoded activator. Introduction of mutations in the −10 sequence resulted in abolishment of promoter activity, and the region spanning positions −35 to −55 was shown to be essential for the initiation of transcription (5). Furthermore, a late promoter located upstream of the terminase subunits of lactococcal phage bIL41 of 936 phage species requires phage-encoded proteins for activity (25).
In this work we have examined the regulation of temporal gene expression during the lytic cycle of TP901-1. A two-plasmid system was used to identify a TP901-1 DNA fragment carrying a regulated promoter and a TP901-1-encoded activator. The promoter is active in the late phase of the lytic cycle, and the promoter requires the product of orf29 for activity. Identification of this novel system for regulation of temporal gene expression in lactococcal phages furthermore suggests that within the P335 group of lactococcal phages at least two different regulatory systems control transcription during the late stage of infection.
MATERIALS AND METHODS
DNA technology.
Recombinant plasmid DNA from E. coli was isolated by the alkaline lysis technique. Plasmids were isolated from L. lactis subsp. cremoris by the alkaline lysis technique after incubation with lysozyme (20 mg per ml) for 20 min at 37°C. Preparative portions were further purified with columns as recommended by the supplier (QIAGEN, Hilden, Germany). Pharmacia Biotech supplied restriction endonuclease enzymes, T4 DNA ligase, and buffer systems. All enzymes were used as recommended by the supplier. For PCRs, Pfu DNA polymerase and buffer supplied from Stratagene were used. DNA sequences were determined as previously described (28), with procedures modified according to the manufacturer's directions for the Thermo Sequenase Radiolabeled terminator cycle sequencing kit (Amersham Life Science).
Construction of plasmids.
Plasmids used in this study are listed in Table 1. Plasmid pBf2-1 contains 4.7 kb of the TP901-1 EV2 fragment (7). Plasmid pLB82 was constructed by inserting a purified 1.3-kb AccI fragment of pBf2-1 in the ClaI site of pGEM7-Zf(+). Selection of pLB82 was performed with E. coli DH5α, whereas all the following transformations were performed with L. lactis subsp. cremoris MG1363. To construct pMBP14, a 1.3-kb HindIII-SmaI fragment of pLB82 was inserted in HindIII-SmaI-digested pAK80 containing the lacLM genes (19). By using TP901-1 DNA as a template and primers 28I and 28II, a 0.5-kb PCR product containing orf29 was obtained. Subsequently, the PCR fragment was digested with BamHI and XbaI and inserted into BamHI-XbaI-digested pNZ8010, giving rise to plasmid pMBP22 (11). Plasmids pMAP15, pMAP14, pMAP9, pMAP7, pLB106, pLB115, and pLB116 contain 57, 99, 184, 208, 446, 638, and 880 bp, respectively, and were all constructed by inserting HindIII-BamHI-digested PCR fragments into HindIII-BamHI-digested pAK80 (19). The PCR products were obtained by using pLB82 as a template and primers PL6 and PL1 (pLB106), PL6 and PL9 (pLB115), PL6 and PL10 (pLB116), PL9 and PM6rev (pMAP7), PM4for and PM6rev (pMAP9), PL9 and PM9rev (pMAP14), and PL9 and PM10rev (pMAP15). The nucleotide sequence of all inserts was verified by DNA sequencing.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 | Laboratory strain |
| L. lactis subsp. cremoris | ||
| MG1363 | 15 | |
| NZ3900 | MG1363 pepN::nisRK | 10 |
| 3107 | Indicator strain for TP901-1 | 3 |
| 901 | Lysogenic for TP901-1 | 3 |
| LB560 | NZ3900/pMBP14 + pMBP22 | This study |
| LB562 | NZ3900/pMBP14 + pNZ8010 | This study |
| LB568 | NZ3900/pAK80 + pMBP22 | This study |
| LB570 | NZ3900/pAK80 + pNZ8010 | This study |
| LB709 | NZ3900/pLB106 + pMBP22 | This study |
| LB798 | NZ3900/pLB115 + pMBP22 | This study |
| LB804 | NZ3900/pLB116 + pMBP22 | This study |
| MP86 | NZ3900/pMAP7 + pMBP22 | This study |
| MP88 | NZ3900/pMAP9 + pMBP22 | This study |
| MP103 | NZ3900/pMAP14 + pMBP22 | This study |
| MP105 | NZ3900/pMAP15 + pMBP22 | This study |
| Plasmids | ||
| pGEM7-Zf(+) | E. coli cloning vector | Promega, Madison, Wis. |
| pAK80 | lacLM erm | 19 |
| pNZ8010 | PnisAgusA cat | 11 |
| pEV7 | TP901-1 library clone | 20 |
| pEV8 | TP901-1 library clone | 20 |
| pBf2-1 | TP901-1 library clone | 7 |
| pLB82 | pGEM77-Zf(+)::1.3-kb AccI of pBf2-1 | This study |
| pLB106 | pAK80::446-bp HindIII-BamHI PCR (PL1-PL6) | This study |
| pLB115 | pAK80::638-bp HindIII-BamHI PCR (PL9-PL6) | This study |
| pLB116 | pAK80::880-bp HindIII-BamHI PCR (PL10-PL6) | This study |
| pMBP14 | pAK80::1.3-kb HindIII-SmaI of pLB82 | This study |
| pMBP22 | pNZ8010::0.5-kb BamHI-XbaI PCR(P27I-P27II) | This study |
| pMAP7 | pAK80::208-kb HindIII-BamHI PCR (PL9-PM6rev) | This study |
| pMAP9 | pAK80::184-bp HindIII-BamHI PCR (PM4for-PM6rev) | This study |
| pMAP14 | pAK80::99-bp HindIII-BamHI PCR (PL9-PM9rev) | This study |
| pMAP15 | pAK80::57-bp HindIII-BamHI PCR (PL9-PM10rev) | This study |
Primers used in this study.
For amplification of various regions the following primers were used: PL1 (5′-GGGGGAAGCTTGGCGTGAGTTCGAATCT-3′), PL6 (5′-GGGGGGGAT CCGGCTCATGCCAGAAAT-3′), PL9 (5′-GGGGGAAGCTTGCATGGGTCAAATTGGG-3′), PL10 (5′-GGGGGAAGCTTGATGAGGAATACATCAAACT-3′), P28I (5′-GGGGGGGATCCTAACACAGACGGAGAATTTG-3′), P28II (5′-GGGGGTCTAGATTCGTGCCTTTTTCGTGT CG-3′), PM6rev (5′-GGGGGGGATCCGATTCGAACTCACGCCTCTGC-3′), PM4for (5′-GGGGGAAGCTTCGACACGAAAAAGGCACG-3′), PM9rev (5′-GGGGGGGATCCGCCTTTTACTTCATAATACAAG-3′), and PM10rev (5′ GGGGGGGATCCGCAACACTCCAATTTCGTGCC-3′).
Primer P14 (5′-CGCCTCTGCATTAAAAG-3′) and PM8rev (5′-GGGGGGGATC CGCAGCCACCAATATGAAG-3′) were used in primer extension experiments.
Bacteria and phages.
Bacterial strains used in this study are listed in Table 1. The temperate bacteriophage TP901-1 originates from L. lactis subsp. cremoris 901-1 (3), where it was induced by the use of UV light as previously described (7).
Media and transformation.
E. coli DH5α was grown with agitation at 37°C in Luria-Bertani broth (27) (Difco Laboratories, Detroit, Mich.), and ampicillin was used at a final concentration of 100 μg/ml. E. coli DH5α was made competent with CaCl2 and was transformed as described by Sambrook et al. (27).
Lactococcus strains were propagated without shaking at 30°C in M17 broth (Oxoid Limited, Basingstoke, Hampshire, United Kingdom) containing 0.5% glucose [wt/vol]) (30). Erythromycin and chloramphenicol were both used at final concentrations of 5 μg/ml. L. lactis subsp. cremoris MG1363 and NZ3900 were transformed by electroporation according to the method described by Holo and Nes (18), with 0.03 to 0.5 μg of DNA per electroporation.
The effect of ORF29 on the activity of the late promoter.
Overnight cultures were each diluted in two tubes containing fresh medium (GM17 medium containing 5 μg of erythromycin per ml and 5 μg of chloramphenicol per ml) to an optical density at 600 nm (OD600) of 0.01. After growth for two to three generations, nisin was added to one set of the cultures to a final concentration of 1 ng per ml. After approximately six generations of growth (overnight incubation), OD600, as well as the activities of β-galactosidase and β-glucuronidase, was determined. Unless otherwise stated the experiments were done at least twice. The data represented are from one experiment.
Activity of the late promoter during induction of ORF29 synthesis.
Exponentially growing cells of LB560 and LB562 were diluted in fresh medium (GM17 containing 5 μg of erythromycin per ml and 5 μg of chloramphenicol per ml) to an OD600 of 0.05. At an OD600 of 0.2, nisin was added to the culture to a final concentration of 1 ng per ml. Samples were withdrawn before and after addition of nisin, and the specific activities of β-galactosidase and β-glucuronidase were determined. Unless otherwise stated the experiments were done at least twice. The data represented are from one experiment.
Enzyme assays.
For determination of β-galactosidase activity, cells were permeabilized with sodium dodecyl sulfate (0.1%) and chloroform. Cell debris was removed by high-speed centrifugation. The assays were performed according to Miller (23). The assays for determination of β-glucuronidase activity were performed just as described for determination of β-galactosidase activity, except that PNPG (p-nitrophenyl-β-d-glucuronic acid) was substituted for ONPG (2-nitrophenyl-β-d-galactopyranoside).
Extraction of RNA and primer extensions.
L. lactis subsp. cremoris 3107 was grown to an OD600 of 0.5 and infected with TP901-1 at a multiplicity of infection of 5, and RNA was extracted at time zero and at 10, 20, 30, 40, 50, 60, and 70 min after infection with the RNA Fast Prep Blue kit for bacteria (BIO 101). Furthermore, RNA was extracted from strain LB798 grown for 4 h in the absence or presence of nisin (1 ng per ml). Primers P14 and PMrev8 were phosphorylated using polynucleotide kinase (Roche) and [γ-32P]ATP (Amersham) as described by Sambrook et al. (27). A total of 5 μg of total RNA isolated during infection of L. lactis subsp. cremoris 3107 was mixed with 1 pmol of phosphorylated primer in a volume of 10 μl and incubated at 70°C for 10 min. The mixture was allowed to cool to 37°C over a 1-h period. A total of 0.1 μl of reverse transcriptase M-MuLV (Roche), 0.6 μl of 5× Incubation buffer (Roche), and dATP, dGTP, dCTP, and dTTP to a final concentration of 200 μM were subsequently added, and the mixture was incubated at 40°C for 1 h.
RESULTS
Two-plasmid system for detection of regulated promoters and identification of regulators.
By definition, the middle and late genes of bacteriophages are not expressed immediately after infection. Therefore, it is expected that phage-encoded factors are required for the expression of the TP901-1 late genes. If the middle and late promoters are regulated by positive-control mechanisms they are not expected to follow the consensus for ς70 promoters and are therefore usually not found in a standard sequence analysis. In addition to this, the promoters cannot be found by random cloning of DNA fragments in front of reporter genes, because they are not active in the absence of the phage-encoded activating factor. A two-plasmid system was therefore used to identify this type of promoter-regulator pair. Initially, putative promoter regions were cloned in the promoter probe vector pAK80, which contains the promoter-less reporter genes lacLM encoding a β-galactosidase enzyme (19). TP901-1 genes encoding potential TP901-1 regulators were then placed under control of the nisA promoter in the expression vector pNZ8010 (11). Transcription from the nisA promoter can be induced by the presence of nisin in a L. lactis subsp. cremoris MG1363 derivative (NZ3900) that carries the required nisRK genes (10). The compatible pAK80 and pNZ8010 derivatives were both introduced into NZ3900, and strains containing different combinations of plasmids were examined for β-galactosidase activity in the presence and absence of the inducer nisin.
Identification of a TP901-1 promoter and a regulator of the promoter.
From the transcriptional analysis of the TP901-1 genome during lytic growth, we expected that middle or late promoters would be located in TP901-1 genomic region present in the library clones pEV7, pEV8, and pBf2-1 (7, 20). This region was therefore investigated for promoters and regulators with the two-plasmid system described above. In this way, we detected a promoter on a 1.3-kb fragment of the library clone pBf2-1(pMBP14) that was active in the presence of orf29(pMBP22). The effect of orf29 expression on promoter activity from the 1.3-kb TP901-1 fragment in pMBP14 was examined by measuring the specific activity of β-galactosidase in the presence and absence of nisin (Table 2). In the absence of ORF29 (strain LB562), no significant promoter activity was seen either in the presence or in the absence of nisin. In contrast, promoter activity was induced 34-fold when expression of orf29 was induced by the presence of nisin (strain LB560). As seen by comparing the specific activities of β-galactosidase of strains LB560 and LB562 grown in the presence of nisin, the activity of the promoter can be induced more than 700-fold by the presence of ORF29. This shows that an inducible promoter is located on the 1.3-kb TP901-1 fragment and that orf29 encodes a positive TP901-1 factor required for activation of this promoter.
TABLE 2.
Identification of a TP901-1 promoter and a regulator of the promotera
| Strain | Plasmids | Promoter fusion to lacLM | Relevant genotype of PnisA plasmid | Sp act of β-galactosidase (U/ml × OD600)
|
|
|---|---|---|---|---|---|
| Without nisin | With nisin | ||||
| LB560 | pMBP14 and pMBP22 | Plate | orf29-gusA | 2.0 | 67 |
| LB562 | pMBP14 and pNZ8010 | Plate | gusA | 0.05 | 0.09 |
| LB568 | pAK80 and pMBP22 | None | orf29-gusA | 0.02 | 0.06 |
| LB570 | PAK80 and pNZ8010 | None | gusA | 0.04 | 0.06 |
Overnight cultures of each strain were diluted in two tubes containing fresh medium (GM17 medium containing 5 μg of erythromycin per ml and 5 μg of chloramphenicol per ml) to an OD600 of 0.01. After growth for two to three generations, nisin was added to one set of the cultures to a final concentration of 1 ng/ml. After approximately six generations of growth (overnight incubation), OD600 (as well as the activities of β-galactosidase and β-glucuronidase) was determined. One representative of two experiments is shown.
Deletion analysis of the inducible promoter of TP901-1.
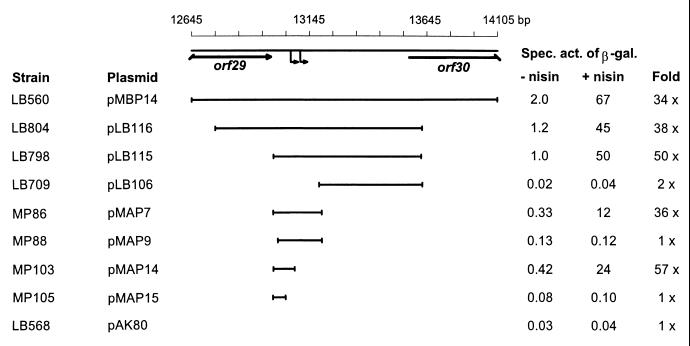
The 1.3-kb TP901-1 fragment present in pMBP14 contains the 3′ end of orf29, the 5′ end of orf30, and a 580-bp intergenic region located between the two genes (4). To localize the position of the promoter within this fragment, a deletion analysis of the fragment was carried out. Strains each containing one of the deletion plasmids, in addition to pMBP22 containing the PnisA-orf29-gusA cassette, were grown in the presence and absence of nisin, and specific β-galactosidase activities were determined (Fig. 1). The 880- and 638-bp fragments present in pLB116 and pLB115 gave rise to promoter activities in the presence of ORF29; however, deletion of an additional 182 bp from the left end of the 638-bp fragment resulted in the loss of promoter activity (pLB106). This suggests that the promoter or at least a part of the promoter is located within the deleted region; as expected, the 208-bp fragment present in pMAP7 showed promoter activity (Fig. 1). From the right end of pMAP7, 110 bp could be deleted and the fragment still retained promoter activity (pMAP14), whereas deletion of additional 42 bp resulted in the loss of promoter activity (pMAP15). This suggests that sequences present in pMAP14 and absent in pMAP15 are important for promoter activity and/or activation by ORF29. Furthermore, deletion of 24 bp from the left end of pMAP7 also resulted in loss of promoter activity (pMAP9), again suggesting that these sequences may be important for promoter activity and/or activation by ORF29. Thus, the smallest fragment showing promoter activity contains 99 bp of the inducible promoter region (pMAP14).
FIG. 1.
Deletion analysis of the TP901-1 promoter region. Numbers refer to the complete TP901-1 genomic sequence present in GenBank under accession number AF304433 (4). The TP901-1 fragments cloned (black lines) indicate a transcriptional fusion to the lacLM genes in pAK80. The specific activities of β-galactosidase of NZ3900 (nisRK) derivatives carrying pMBP22 (orf29) and the indicated plasmids were measured after overnight growth in M17 medium with 0.5% glucose and erythromycin and chloramphenicol (each, 5 μg/ml) in the absence of nisin (−nisin) as well in the presence of nisin (1 ng/ml) (+nisin). Fold, the specific activity of β-galactosidase obtained in the presence of nisin divided by the specific activity of β-galactosidase obtained in the absence of nisin; large black lines, orf genes; small black arrows, mRNA ends identified in primer extension analysis (see Fig. 2).
Determinations of transcription start site.
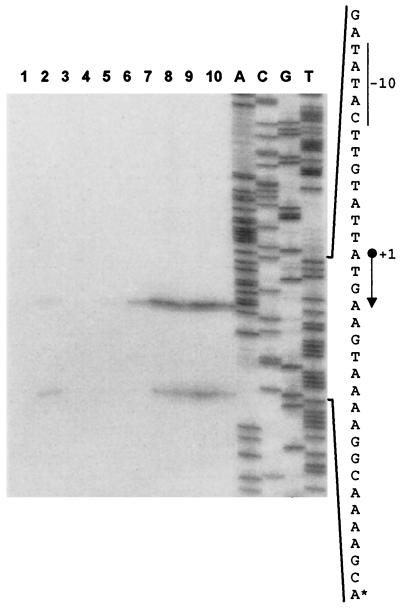
RNA was extracted from strain LB798 containing the promoter fusion plasmid pLB115 as well as the nisin-inducible orf29 expression plasmid (pMBP22), grown in the presence and absence of nisin. Subsequently, primer extension analysis was performed to localize the transcriptional start site of the inducible promoter. In the presence of ORF29, primer extension analysis revealed two putative transcription start sites. The most intensive band could correspond to a transcription start site at nucleotide (nt) 13073, whereas the less-intensive band might correspond to a transcription start site at nt 13093 (Fig. 2, lane 2). The same putative start sites were obtained with a different primer for the analysis (data not shown). In contrast, no primer extension products were observed when ORF29 was not expressed (Fig. 2, lane 1), verifying that promoter activity is tightly regulated and requires ORF29 for activity.
FIG. 2.
Primer extension mapping of the TP901-1 late promoter. Extension reactions were performed with RNA isolated from strain LB798 induced at OD600 of 0.2 with nisin (0 or 1 ng/ml) for 4 h (lanes 1 and 2, respectively). Furthermore, extension reactions were carried out with RNA isolated from L. lactis subsp. cremoris 3107 at 0, 10, 20, 30, 40, 50, 60, and 70 min after infection with TP901-1 (lanes 3 to 10). Lanes marked A, C, G, and T contain the sequence reactions carried out with the primer P14, also used for primer extension analysis. The sense strand sequence flanking the transcription site is shown. The transcription start site marked with +1 corresponds to nt 13073 in the TP901-1 genome. The mRNA end marked with an asterisk (*) corresponds to nt 13093 in the TP901-1 genome. Numbers refer to the complete TP901-1 genomic sequence present in GenBank under accession number AF304433. The arrow indicates the direction of transcription (4).
To determine the time point in the lytic cycle where the identified promoter was active, RNA was isolated from L. lactis subsp. cremoris 3107 infected with bacteriophage TP901-1. Samples were collected every 10 min after infection, and primer extensions were performed. After phage infection, RNA transcripts were observed at 30 min and the band intensity was increased at 40 min; afterward, band intensity remained the same throughout the experiment. Thus, the promoter becomes fully active in the late phase of the lytic cycle and controls transcription of the late expressed region of the TP901-1 genome (4, 21); the promoter was therefore designated the late promoter of TP901-1. In this experiment, two bands were also observed to be identical to the putative transcription sites that were previously identified at nt 13073 and 13093 (Fig. 2, lanes 6 to 10). A −10 region of nearly consensus sequence (TATACT) was present upstream of the nt 13073 start site which also contains the upstream TG motif (Fig. 3A), whereas the predicted −10 region of the 13093 start site is TAAAAG. However, the potential −35 regions (GTGTTG and TGATAT) for both putative promoters deviate significantly from the consensus sequence (Fig. 3A and data not shown).
FIG. 3.
(A) DNA homology of the TP901-1 late promoter region. ClustalV alignment of the 99-bp late promoter fragment of TP901-1 with sequences present in bacteriophage tuc2009 and the abiN operon is shown. Nucleotides preserved between all three sequences are marked with asterisks. The identified transcriptional start site at nt 13073 of the TP901-1 promoter and he −10 and −35 regions are underlined. The gray box indicates the position −85 to −61 bp upstream of the promoter that is absent in pMAP9 and required for ORF29 activation. (B) Homology of the TP901-1 regulator of late transcription. ClustalV alignment of ORF29 encoded by TP901-1 with homologous proteins encoded by bacteriophage tuc2009 (ORF28) and the abiN operon (ORF6) is shown. Amino acids preserved in all three sequences are marked with asterisks, whereas two preserved amino acids and one conservative change in amino acids are marked with dots.
Homology of the TP901-1 promoter region and the regulator.
The smallest fragment showing promoter activity (99 bp in pMAP14) was searched for DNA homology to DNA sequences in GenBank using BLAST, version 2.0.4 (1). Exact or nearly exact matches were found to putative promoter regions of the lactococcal phages bIL309 (100% identity in 99 nt) and bIL286 (98% identity in 99 nt) (data not shown). These temperate phages belong to the P335 group of lactococcal phages and they can be induced by mitomycin C from L. lactis IL-1403 (6). Furthermore, homology was found to regions in the abiN operon (67% identity in 99 nt) and the lactococcal temperate bacteriophage tuc2009 genome (61% identity in 99 nt) (Fig. 3A). A 34-bp sequence covering from +9 to −26 of the late promoter region was fully conserved in the latter sequences (Fig. 3A). No significant homology to phage tuc2009 and the abiN operon was found downstream of the 99-bp region present in pMAP14.
The existence of ORF29 homologous proteins was investigated by comparing the ORF29 amino acid sequence with sequences present in GenBank with BLAST, version 2.0.4 (1). In this search we found that ORF29 showed homology to ORF28 of lactococcal bacteriophage tuc2009 (82% identity in 140 amino acids [aa]) and to ORF6 of the abiN operon of L. lactis subsp. cremoris S114 (81% identity in 133 aa) (Fig. 3B). No function has been assigned to either putative protein, but the presence of abiN located downstream of orf6 in the abiN operon was shown to give rise to an abortive infection phenotype (26). The temperate phages bIL309 and bIL286, which contain sequences highly homologous to the TP901-1 late promoter region, encode proteins almost identical to ORF29 (99 and 97%, respectively). In addition, less-similar proteins were found in the temperate bacteriophages bIL310 (72% identity in 133 aa) and bIL285 (54% identity in 70 aa) (data not shown) that also are mitomycin C inducible from L. lactis IL-1403 (6).
Regulation of the late promoter of TP901-1 by ORF29.
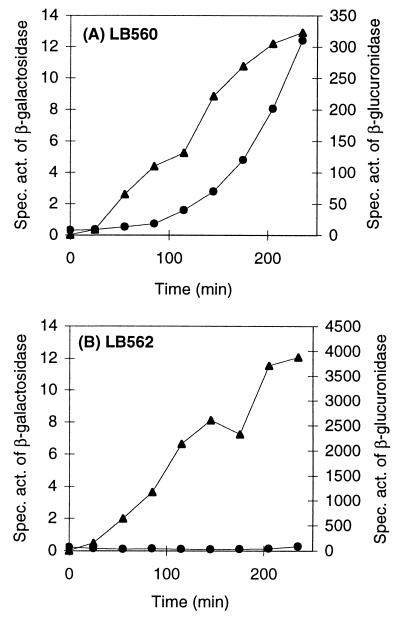
As shown in Table 2, the late promoter of TP901-1 requires the presence of ORF29 to be active. To further analyze the relation between promoter activity and ORF29 concentration, we examined the level of β-galactosidase during nisin induction of ORF29 production in strain LB560 (Fig. 4A). At the same time, the transcription of the orf29 gene could be measured as β-glucuronidase activity, because the gusA gene located downstream of orf29 is cotranscribed. A protein band with a size corresponding to the predicted size of ORF29 was observed only in strain LB560 induced by nisin, thus verifying expression of ORF29 (data not shown). As a control, strain LB562 carrying plasmids pMBP14 and pNZ8010 (the vector of pMBP22) was used (Fig. 4B). We found that immediately after addition of nisin, the specific activity of β-glucuronidase increased in both strains, indicating an immediate induction of the nisin promoter (Fig. 4) and thus transcription of orf29 in strain LB560 (Fig. 4A). The approximately 10-fold-lower activity of β-glucuronidase in strain LB560 than that in LB562 is most likely due to some termination downstream of the cloned orf29 gene. After approximately 50 min of growth in the presence of nisin, the specific activity of β-galactosidase increases significantly above the baseline level in strain LB560 (Fig. 4A) but not in strain LB562 (Fig. 4B). This shows that even though transcription of orf29 is induced immediately after addition of nisin, activation of the late phage promoter is delayed, indicating that a certain level of ORF29 may be necessary for efficient activation of transcription initiation.
FIG. 4.
Activity of the late promoter during induction of ORF29 synthesis. Strains LB560 containing pMBP14(Plate-lacLM) and pMBP22(PnisA-orf29-gusA) (A) and LB562 containing pMBP14(Plate-lacLM) and pNZ8010(PnisA-gusA) (B) were grown exponentially in M17 medium with 0.5% glucose and erythromycin and chloramphenicol (each, 5 μg/ml). At 5 min, nisin was added to a final concentration of 1 ng/ml. The specific activities of β-galactosidase (circles) and β-glucuronidase (triangles) were measured.
DISCUSSION
The late promoter and activator of lactococcal bacteriophage TP901-1.
In this work, we have used a two-plasmid system to identify a phage TP901-1 promoter and the phage-encoded activator of this promoter. In general this system can be used to detect promoters that are active only in the presence of an activator and may thus be a useful tool to identify promoter-activator pairs in L. lactis. The TP901-1 promoter region is located in a 580-bp intergenic space between orf29 and orf30 on the phage genome. Since orf30 encodes the putative small terminase subunit of TP901-1 (4), the promoter potentially controls synthesis of proteins responsible for packaging of TP901-1 genomes. The promoter is active during the late phase of the lytic cycle and requires the product of orf29 for activity. Characterization of the promoter-activator pair of TP901-1 shows that expression of ORF29 can induce transcription from the promoter more than 700-fold. The slightly increased activity of the TP901-1 promoter in the absence of nisin in strain LB560 compared to that in strain LB562 is probably due to a leaky nisA promoter, leading to some production of ORF29 even in the absence of nisin. Thus, the identified TP901-1 promoter is tightly regulated and requires ORF29 for activity.
The primer extension analysis identified two phage-inducible RNA ends corresponding to positions 13073 and 13093. Thus, one possibility is that the late transcription is initiated from a tandem promoter. However, mRNA ends may also arise from processing of the primary transcript; furthermore, the reverse transcriptase may produce premature termination during the elongation process. In accordance with the suggestion that the mRNA starting at position 13073 originates from a promoter start site, the deletion analysis clearly shows that the start site at nt 13073 alone (pMAP14) is sufficient to induce ORF29-activated transcription. This was further verified by loss of promoter activity in the deletion plasmid pMAP15, in which the transcription start site at nt 13073 and its −10 region is deleted. Thus, the promoter having a start site at position 13073 is verified by deletion analysis. The region upstream of position 13093 shows only weak identity to a −10 consensus region. Since we have no further evidence for a second promoter in this region, we suggest that the band at position 13093 is due to processing of the primary mRNA or an artifact of the reverse transcriptase reaction. A tandem mRNA signal was also observed for the middle promoter of the virulent lactococcal phage Φ31. Here, the upstream start site was verified by promoter cloning to be a functional promoter on its own (32), while no promoter activity could be found for the second mRNA signal located downstream of the identified promoter.
Interaction between TP901-1 DNA and ORF29 has not be demonstrated in this work. However, the deletion analysis shows that the region located from −85 to −61 bp upstream of the position 13073 transcriptional start site is necessary for activation by ORF29, since pMAP9 containing the transcriptions start site and a 60-bp upstream region did not show any promoter activity in the presence of ORF29 (Fig. 3A). This region may therefore interact directly with ORF29 during the activation of transcription initiation.
The features of the regulatory system of TP901-1 late transcription resemble the well-studied regulatory system of E. coli phage P2. The P2 late promoters show poor sequence similarity to the ς70 consensus promoter sequence and show almost no basal expression in the absence of an activator (8, 9). Furthermore, transcription of the late promoters of P2 is under positive control of a P2-encoded protein, which binds just upstream of the position predicted for RNA polymerase binding (17). In contrast, the activator of virulent lactococcal bacteriophage Φ31 is suggested to bind to sequences overlapping the −35 region of the promoter; deletion analysis showed that removal of the region from −54 to −45 eliminated promoter activity (32). In bacteriophage sk1, a deletion analysis of the middle promoter region demonstrated that the region from −46 to −55 was necessary for promoter activity, and mutagenesis further showed that bases in the region from −36 to −55 were necessary for activity (5).
Regulation of temporal gene expression during lytic growth of TP901-1.
To produce new phage particles during lytic growth, the bacteriophage-encoded proteins are needed at specific time periods and a strict control of expression of phage genes is therefore very important. For bacteriophage TP901-1, ORF29 is identified as a regulatory protein that activates transcription from a late TP901-1 promoter. During the lytic cycle of TP901-1, a 10-kb mRNA produced in the early phase of the cycle is likely also to contain the orf29 gene (21). Even though ORF29 protein may already be present early in the lytic cycle, a transcript arising from the late promoter is not observed until 30 min postinfection, and the promoter does not become fully active before 40 min postinfection. In agreement with this, we found that when the activator and the late promoter are plasmid borne, transcription initiation of the late promoter is also delayed compared to the induction of the regulator. This may indicate that a certain level of ORF29 must be reached in order to activate transcription initiation of the promoter.
Control of temporal gene expression in lactococcal bacteriophages.
In contrast to the variety of mechanisms found in other bacteriophages, activators are the only examples of delayed gene expression control in lactococcal bacteriophages identified so far. The genetic organization of the promoter and regulator was similar in temperate phage TP901-1 and virulent phage Φ31; however, no homology at the DNA or amino acid level was found. In contrast, homology searches revealed that an ORF29-homologous protein and a sequence homologous to the late promoter region are present in several genomes of temperate lactococcal bacteriophages. This indicates that the same control unit (promoter and regulator) performs regulation of temporal gene expression in these bacteriophages as in TP901-1. Interestingly, regions homologous to the TP901-1 late promoter and activator were found only in temperate lactococcal bacteriophages belonging to the P335 group. No homology was found to phages belonging to other lactococcal phage groups or to phages infecting bacteria other than L. lactis, suggesting a phylogenetically limited distribution of this control unit. In summary, our results suggest that within the P335 group of lactococcal phages at least two regulatory systems controlling transcription in the late stage of infection exist, the TP901-1 like (TP901-1, tuc2009, bIL285, bIL286, and bIL309) and the Φ31 like (Φ31, r1t, and ΦLC3) (31). A phage resistance mechanism based on the Φ31 middle promoter and activator has already been constructed (12). The identification of a second regulatory system among the P335 phages may be exploited in newly designed phage resistance mechanisms effective against a larger group of the P335 phages.
ACKNOWLEDGMENTS
Martin Bastian Pedersen is acknowledged for construction of pMBP14 and pMBP22, and we sincerely appreciate the expert technical assistance of Lise Sørensen and Jeannette de Sparra Lundin. We are grateful to Mogens Kilstrup for discussions and critical reading of the manuscript.
This work was supported by grants from the EC BIOTECH program (BIO4-96-0402) and the Carlsberg Foundation.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barthelemy I, Lazaro J M, Mendez E, Mellado R P, Salas M. Purification in an active form of the phage phi 29 protein p4 that controls the viral late transcription. Nucleic Acids Res. 1987;15:7781–7793. doi: 10.1093/nar/15.19.7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun V, Hertwig S, Neve H, Geis A, Teuber M. Taxonomic differentiation of bacteriophages of Lactococcus lactis by electron microscopy, DNA-DNA hybridization, and protein profiles. J Gen Microbiol. 1989;135:2551–2560. [Google Scholar]
- 4.Brøndsted L, Østergaard S, Pedersen M, Hammer K, Vogensen F K. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: Evolution, structure, and genome organization of lactococcal bacteriophages. Virology. 2001;283:93–109. doi: 10.1006/viro.2001.0871. [DOI] [PubMed] [Google Scholar]
- 5.Chandry P S, Moore S C, Boyce J D, Davidson B E, Hillier A J. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol Microbiol. 1997;26:49–64. doi: 10.1046/j.1365-2958.1997.5491926.x. [DOI] [PubMed] [Google Scholar]
- 6.Chopin A, Bolotin A, Sorokin A, Ehrlich S D, Chopin M-C. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 2001;29:644–651. doi: 10.1093/nar/29.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christiansen B, Johnsen M G, Stenby E, Vogensen F K, Hammer K. Characterization of the lactococcal temperate phage TP901-1 and its site-specific integration. J Bacteriol. 1994;176:1069–1076. doi: 10.1128/jb.176.4.1069-1076.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie G E, Calendar R. Bacteriophage P2 late promoters. Transcription initiation sites for two late mRNAs. J Mol Biol. 1983;167:773–790. doi: 10.1016/s0022-2836(83)80110-7. [DOI] [PubMed] [Google Scholar]
- 9.Christie G E, Calendar R. Bacteriophage P2 late promoters. II. Comparison of the four late promoter sequences. J Mol Biol. 1985;181:373–382. doi: 10.1016/0022-2836(85)90226-8. [DOI] [PubMed] [Google Scholar]
- 10.de Ruyter P G G A, Kuipers O P, Beerthuyzen M M, van Alen-Boerrigter I J, de Vos W M. Functional analysis of promoters in the nisin cluster of Lactococcus lactis. J Bacteriol. 1996;178:3434–3439. doi: 10.1128/jb.178.12.3434-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Ruyter P G G A, Kuipers O P, de Vos W M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djordjevic G M, O'Sullivan D J, Walker S A, Conkling M A, Klaenhammer T R. A triggered-suicide system designed as a defense against bacteriophages. J Bacteriol. 1997;179:6741–6748. doi: 10.1128/jb.179.21.6741-6748.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forde A, Fitzgerald G F. Bacteriophage defense systems in lactic acid bacteria. Antonie Leeuwenhoek. 1999;76:89–113. [PubMed] [Google Scholar]
- 14.Friedman D I, Court D L. Transcription antitermination: the lambda paradigm updated. Mol Microbiol. 1995;18:191–200. doi: 10.1111/j.1365-2958.1995.mmi_18020191.x. [DOI] [PubMed] [Google Scholar]
- 15.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geiduschek E P, Fu T-J, Kassavetis G A, Sanders G M, Tinker-Kulberg R L. Transcriptional activation by topologically linkable protein: forging a connection between replication and gene activity. In: Eckstein F, Lilley D M J, editors. Nucleic acids and molecular biology. Vol. 11. Heidelberg, Germany: Springer-Verlag; 1997. pp. 135–150. [Google Scholar]
- 17.Grambow N J, Birkeland N K, Anders D L, Christie G E. Deletion analysis of a bacteriophage P2 late promoter. Gene. 1990;95:9–15. doi: 10.1016/0378-1119(90)90407-i. [DOI] [PubMed] [Google Scholar]
- 18.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Israelsen H, Madsen S M, Vrang A, Hansen E B, Johansen E. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl Environ Microbiol. 1995;61:2540–2547. doi: 10.1128/aem.61.7.2540-2547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madsen P L. Transcription of the lactococcal temperate phage TP901-1. Ph.D. thesis. Lyngby, Denmark: Technical University of Denmark; 1996. [Google Scholar]
- 21.Madsen P L, Hammer K. Temporal transcription of the lactococcal temperate phage TP901-1 and DNA sequence of the early promoter region. Microbiology. 1998;144:2203–2215. doi: 10.1099/00221287-144-8-2203. [DOI] [PubMed] [Google Scholar]
- 22.Madsen P L, Johansen A H, Hammer K, Brøndsted L. The genetic switch regulating activity of early promoters of the temperate lactococcal bacteriophage TP901-1. J Bacteriol. 1999;181:7430–7438. doi: 10.1128/jb.181.24.7430-7438.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 24.O'Sullivan D J, Walker S A, West S G, Klaenhammer T R. Development of an expression strategy using a lytic phage to trigger explosive plasmid amplification and gene expression. Bio/Technology. 1996;14:82–87. doi: 10.1038/nbt0196-82. [DOI] [PubMed] [Google Scholar]
- 25.Parreira R, Valyasevi R, Lerayer A L, Ehrlich S D, Chopin M-C. Gene organization and transcription of a late-expressed region of a Lactococcus lactis phage. J Bacteriol. 1996;178:6158–6165. doi: 10.1128/jb.178.21.6158-6165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prévots F, Tolou S, Delpech B, Kaghad M, Daloyau M. Nucleotide sequence and analysis of the new chromosomal abortive infection gene abiN of Lactococcus lactis subsp. cremoris S114. FEMS Microbiol Lett. 1998;159:331–336. doi: 10.1111/j.1574-6968.1998.tb12879.x. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Summers W C, Siegel R B. Transcription of late phage RNA by T7 RNA polymerase. Nature. 1970;228:1160–1162. doi: 10.1038/2281160a0. [DOI] [PubMed] [Google Scholar]
- 30.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker S A, Dombroski C S, Klaenhammer T R. Common elements regulating gene expression of temperate and lytic bacteriophages of Lactococcus species. Appl Environ Microbiol. 1998;64:1147–1152. doi: 10.1128/aem.64.3.1147-1152.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker S A, Klaenhammer T R. Molecular characterization of a phage-inducible middle promoter and its transcriptional activator from the lactococcal bacteriophage Φ31. J Bacteriol. 1998;180:921–931. doi: 10.1128/jb.180.4.921-931.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]