Significance
Pain management represents an unmet healthcare need in many countries. Nav1.8 represents a potential target for developing nonaddictive analgesics. Here we present the cryogenic electron microscopy (cryo-EM) structures of human Nav1.8 alone and bound to a selective pore blocker, A-803467. Unlike reported structures of eukaryotic Nav channels wherein the first voltage-sensing domain (VSDI) is well-resolved in one stable conformation, different conformations of VSDI are observed in the cryo-EM maps of Nav1.8. An extracellular interface between VSDI and the pore domain was identified to be a determinant for Nav1.8’s dependence on higher voltage for activation. A-803467 clenches S6IV within the central cavity. Unexpectedly, the channel selectivity for A-803467 is determined by nonligand coordinating residues through an allosteric mechanism.
Keywords: Nav1.8, cryo-EM structure, voltage sensing, electromechanical coupling, subtype-specific Nav blocker
Abstract
The dorsal root ganglia–localized voltage-gated sodium (Nav) channel Nav1.8 represents a promising target for developing next-generation analgesics. A prominent characteristic of Nav1.8 is the requirement of more depolarized membrane potential for activation. Here we present the cryogenic electron microscopy structures of human Nav1.8 alone and bound to a selective pore blocker, A-803467, at overall resolutions of 2.7 to 3.2 Å. The first voltage-sensing domain (VSDI) displays three different conformations. Structure-guided mutagenesis identified the extracellular interface between VSDI and the pore domain (PD) to be a determinant for the high-voltage dependence of activation. A-803467 was clearly resolved in the central cavity of the PD, clenching S6IV. Our structure-guided functional characterizations show that two nonligand binding residues, Thr397 on S6I and Gly1406 on S6III, allosterically modulate the channel’s sensitivity to A-803467. Comparison of available structures of human Nav channels suggests the extracellular loop region to be a potential site for developing subtype-specific pore-blocking biologics.
Voltage-gated sodium (Nav) channels govern membrane excitability in neurons and muscles (1, 2). Despite high-degree sequence and architectural similarity, different subtypes of Nav channels have specific tissue distributions and distinct voltage dependence and kinetics for activation, inactivation, and recovery (3). Among the nine mammalian Nav channels (SI Appendix, Fig. S1), Nav1.8, a tetrodotoxin (TTX)-resistant subtype encoded by SCN10A, is primarily expressed in the sensory neurons, exemplified by the dorsal root ganglia (DRG) neurons (4–6). Compared with other Nav subtypes, Nav1.8 has several unique biophysical properties, such as activation at more depolarized voltage and slower inactivation with persistent current, which enable the hyperexcitability of the DRG neurons (4, 5, 7–11).
Nav1.8 functions in pain sensation (12–16). Proexcitatory mutations of Nav1.8 were identified in patients with painful small fiber neuropathy (17–19). On the other hand, a natural variant, A1073V, that shifts the voltage dependence of activation to more depolarized direction appeared to ameliorate pain symptoms (20). Specific inhibition of the peripheral Nav1.8 thus represents a potential strategy for developing nonaddictive pain killers (21, 22).
Several Nav1.8-selective blockers, such as VX-150 and PF-06305591, had been tested in clinical trials. However, most of the drug candidates failed to meet the endpoint(s) of phase II trials for various reasons, such as unsatisfactory efficacy or selectivity (21, 23–26). Structures of Nav1.8 bound to lead compounds will shed light on drug optimization for improving potency and selectivity. We focused on A-803467, a Nav1.8-selective blocker, for structural analysis. A-803467 was shown to inhibit Nav1.8 in both the resting state and inactivated state. Despite a wide range of concentration that inhibits response by 50% (IC50) from several nanomolar to 1 μM measured by different groups, A-803467 consistently shows a higher affinity for the inactivated channel (27–31).
In this study, we report the structures of full-length human Nav1.8 alone and bound to A-803467. The first voltage-sensing domain (VSDI) was resolved in multiple conformations. Based on the structural and electrophysiological characterizations, we attempt to address two questions: What underlies the high-voltage activation of Nav1.8, and what determines the subtype specificity of A-803467?
Results
Structural Determination of Human Nav1.8.
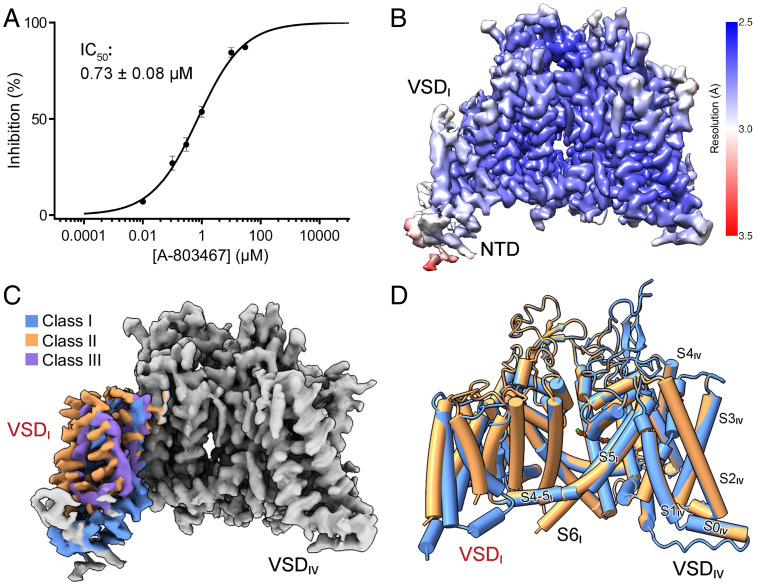
Details of protein expression and purification are presented in Materials and Methods. In our whole-cell recording in ND-7/23 cells, purchased A-803467 blocked Nav1.8 with an IC50 of 0.73 ± 0.08 μM, falling within the reported range (Fig. 1A and SI Appendix, Fig. S2A and Table S1). Based on this, we incubated the blocker with purified Nav1.8 at a final concentration of 10 µM for 1 h before cryo-sample preparation for blocker-bound structure. Following our standard protocol for cryogenic electron microscopy (cryo-EM) data acquisition and processing (32), the three-dimensional (3D) EM reconstructions could be obtained at average resolutions of 2.7 Å and 3.1 Å for Nav1.8 with and without blocker (SI Appendix, Fig. S3).
Fig. 1.
Cryo-EM analysis of human Nav1.8 bound to selective blocker A-803467. (A) Inhibition of Nav1.8 by A-803467. Peak currents of Nav1.8 with A-803467 applied at different concentrations were recorded. Data represent mean ± SEM n = 5, 11, 13, 15, 6, and 6 for A-803467 concentrations at 10 nM, 100 nM, 300 nM, 1 μM, 10 μM, and 30 μM, respectively. Experimental details are presented in SI Appendix, Fig. S2A and Table S1 and Materials and Methods. (B) Heat map for resolution distribution of class I 3D EM reconstitution for human Nav1.8 in the presence of A-803467. Local resolutions were estimated with RELION 3.0 and represented in Chimera (53, 56). (C) Cryo-EM maps of three classes of VSDI. For visual clarity, the identical region of the map is colored gray and the three conformers of VSDI are colored as indicated. (D) VSDI of Nav1.8 was resolved in different conformations. As the resolution of the third class is insufficient for reliable side-chain assignment, we will focus on classes I and II for structural analysis. For visual clarity, sugar moieties, lipids, and a detergent molecule that penetrates the intracellular gate are not shown. All structure figures were prepared with Chimera X or PyMol (60).
The two maps can be almost completely overlaid. Unexpectedly, VSDI in both reconstructions is barely discernible. This observation is in contrast to the cryo-EM maps of all other human Nav channels, wherein all four VSDs are well-resolved (33–38). Because a much larger dataset for Nav1.8 treated with A-803467 was obtained at this stage, we focused on this one for further classification of the region masked for VSDI. Three conformations, designated classes I, II, and III, were attained (Fig. 1B and C and SI Appendix, Figs. S3–S6 and Table S2). While VSDI in classes I and II could be resolved to ∼3 Å for reliable model building, the resolution of VSDI in class III was around 4 Å, insufficient for accurate side-chain assignment. In the following, we will focus on classes I and II for structural analysis (Fig. 1D and SI Appendix, Fig. S5A).
VSDI Exhibits Multiple Conformations.
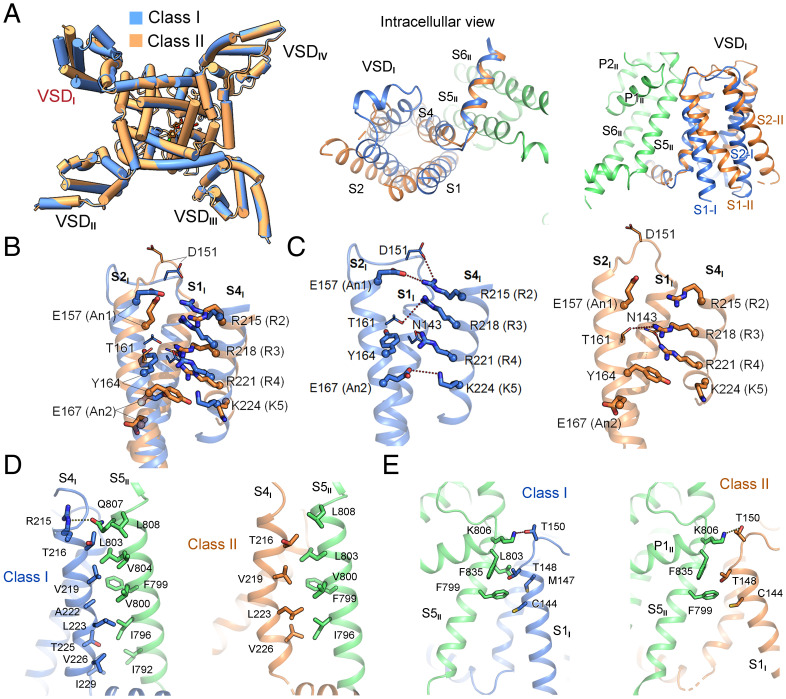
In the structure of Nav1.8, VSDs in repeats II to IV all exhibit the typical “up” (or depolarized) conformation, as observed in other human Nav structures (SI Appendix, Fig. S7A) (33–38). VSDI in the two classes undergoes a rotation of ∼12° around the central pore, pivoting the S4 segment (Fig. 2A, Left). However, individual domain comparison shows that VSDI does not move as a rigid body. In fact, the four segments undergo unconcerted displacement (Fig. 2A, Middle and Right). Despite the pronounced shift, three gating charge residues, R2 to R4, are above the occluding residue Tyr164 on S2I in both VSDI structures, representing the up states. However, the gating charge residues are coordinated differently in these two conformers (Fig. 2B and C).
Fig. 2.
Distinct conformations of VSDI. (A) Domainwise rotation of VSDI between the two classes. (Left) An intracellular view of the superimposed Nav1.8 structures, which deviate only in VSDI. (Middle and Right) Enlarged views of VSDI superimposed in the context of the overall structures. The PD segments in repeat II, which are identical in the two classes, are colored green. (B) Intradomain structural shifts between the two classes of VSDI. When compared individually, the two VSDI structures differ dramatically with a root-mean-square deviation of 2.8 Å over 88 Cα atoms. Gating charge residues, R2–K5, and the conserved residues on S2 and S3 that facilitate charge transfer are shown as thick sticks. Cα atoms of these residues are shown as spheres. (C) VSDI exhibits depolarized (up) conformation in both classes. The R2–R4 gating charge residues are above the occluding Tyr164 in both classes. The potential polar interactions, direct or indirect, are indicated by dashed lines. (D) S4I in class I has more extensive interactions with the neighboring S5II than that in class II. The interface residues between S4I and S5II are shown as sticks. (E) Altered interface between S1I and S5II in the two classes as a result of the ∼12° domainwise rotation of VSDI and swing of S1I.
In the structures of human Nav1.1, 1.2, 1.4, 1.5, and 1.7 (33–37, 39), VSDI exhibits similar, if not identical, conformations. Whereas class I VSDI is similar to them, the loosely packed class II VSDI represents an outlier to the existing gallery of VSD structures. In class I, R2 simultaneously interacts with Asp151 on the S1–2 loop and the conserved An1 residue Glu157 on S2I. R3 forms hydrogen bond (H-bond) with Thr161 on S2I, and R4 is H-bonded to Asn143 on S1I. Below the upward-pointing occluding residue Tyr164, K5 is coordinated by the An2 residue Glu167 (Fig. 2C, Left). In the class II conformer, R2 and K5 are out of reach of any of the charge transfer-facilitating residues or nearby polar residues. R3 and R4 may respectively interact with Thr161 on S2I and Asn143 on S1I. The strongest binding comes from the cation–π interaction between R4 and Tyr164, whose aromatic ring swings downward (Fig. 2C, Right).
In addition to the interior structural variations, a major difference between the two classes occurs to the interface of VSDI and the pore domain (PD) mediated by the S1 and S4 segments in VSDI and the S5II segment in the PD (Fig. 2D and E). The VSDI–PD interface is more extensive in class I, involving seven residues on S4I that associate with eight residues on S5II via both hydrophobic and polar interactions (Fig. 2D, Left). In class II, however, S4I is curved, resulting in only four residues involved in van der Waals contacts with five hydrophobic residues on S5II (Fig. 2D, Right). S1I also engages residues on its extracellular terminus to interact with S5II and P1II. As S1I in class II swings away from the PD on the cytosolic side, the distances between the corresponding residues are in general larger than those in class I (Fig. 2E). Similar to the observation in our accompanying paper on Nav1.7 (40), the extracellular contact between S1I and the PD represents a pivot for the structural shift of VSDI.
The Nonconserved VSDI–PD Interface in Part Underlies High Voltage Dependence for Nav1.8 Activation.
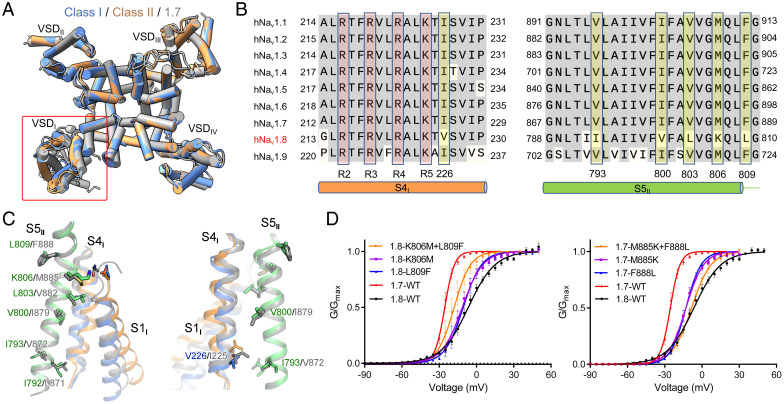
The structures of VSDs II, III, and IV and the PD in Nav1.8 are nearly identical to those in Nav1.7 (Protein Data Bank [PDB] ID code: 7W9K) with a root-mean-square deviation of 0.66 Å over 803 Cα atoms (Fig. 3A). As VSDI in Nav1.8, with its multiple conformations, appears to be the only major variation between the structures of Nav1.8 and other Nav channels, there should be a molecular basis for the unprecedented conformation of VSDI in Nav1.8.
Fig. 3.
The interface between VSDI and S5II in part accounts for the high-voltage activation of Nav1.8. (A) Structures of the transmembrane domains of human Nav1.8 and Nav1.7 mainly deviate in VSDI. Shown here is an extracellular view of the superimposed structures of Nav1.7 (PDB ID code: 7W9K) and the two classes of Nav1.8. ECLs are omitted for clarity. (B) Nav1.8-unique residues on S4I and S5II. Several S4I–S5II interface residues are invariant in all Nav subtypes except Nav1.8. These residues are shaded yellow. Refer to SI Appendix, Fig. S1 for complete sequence alignment of the four repeats of the nine human Nav channels. (C) Less-conserved VSDI–S5II interface between Nav1.8 and Nav1.7. Varied residues between Nav1.8 and Nav1.7 on the VSDI–S5II interface are shown as sticks. Shown here are two opposite side views to highlight the interface residues between S1I and S5II (Left) and between S4I and S5II (Right). The structures are superimposed as in A. (D) The Nav1.8-unique residues on the interface of VSDI and S5II are in part responsible for its dependence on more depolarized voltage of activation. Swapping the corresponding VSDI–S5II interface residues between Nav1.8 and Nav1.7 led to respective left and right shifts of voltage dependence in the activation of the two channels. Analysis of the corresponding mutations of Nav1.5 is presented in SI Appendix, Fig. S9. Refer to SI Appendix, Tables S3 and S4 for measured parameters for these Nav1.8, Nav1.7 and Nav1.5 variants.
Structure-guided sequence analysis led to an interesting discovery: Among the interfaces between the four VSDs and the PD, only that between VSDI and S5II contains Nav1.8-unique residues, while the other interfaces are nearly identical across the nine subtypes (Fig. 3B and SI Appendix, Fig. S1). Reciprocally, among the residues on S4I and S5II, all Nav1.8-unique residues are exclusively mapped to their interface (Fig. 3B and C). These sequence variations may underlie the conformational heterogeneity of VSDI in the structure of Nav1.8. It is known that depolarization-elicited movements of VSDs I to III are associated with channel activation (41–44). As Nav1.8 has the unique property of activation at more depolarized voltage, we speculated that the variation of the VSDI–PD interface may in part account for the higher voltage required for Nav1.8 activation.
To test this hypothesis, we substituted the varied residues in Nav1.8 with the corresponding ones in other channels and examined the voltage-dependent activation of these mutants (Fig. 3D). It is noted that Val-226 on S4I and Ile793 on S5II in Nav1.8, which directly interact with each other in both classes, represent a swap of Ile and Val in other Nav channels (Fig. 3B and C). We did not alter these two residues.
Among the tested mutants, V800I had negligible effect on activation. L803V shifted the activation curve slightly to the depolarization direction, moving the V1/2 from −6.5 mV to −3.3 mV (SI Appendix, Table S3). When Lys806 and Leu809 were individually replaced by Met and Phe, the resulting V1/2 values were left-shifted to −11.4 mV and −10.7 mV, respectively. When the two point mutations were combined, the activation curve was further shifted toward more polarized direction, with V1/2 reaching −17.9 mV (Fig. 3D, Left and SI Appendix, Fig. S8A and Table S3).
We then examined whether reverse mutations would move the activation curve of Nav1.7 to the depolarized direction. Indeed, individual point mutations, M885K and F888L, both shifted the V1/2 of the voltage-dependent activation curve of Nav1.7 from −25.5 mV to −12.9 mV, and a combined double point mutation further moved the activation curve rightward with V1/2 at −8.4 mV, close to that of Nav1.8 (Fig. 3D, Right and SI Appendix, Fig. S8A and Table S3).
Nav1.7 is a TTX-sensitive subtype. To further support the above analysis, we performed parallel analysis of Nav1.5, another TTX-resistant subtype. Single point mutations M858K and F861L shifted the V1/2 of the voltage-dependent activation of Nav1.5 from −40.9 mV to −32.1 mV and −38.0 mV, respectively, and combining the two point mutations resulted in a V1/2 of the further right-shifted activation curve to −24.6 mV (SI Appendix, Figs. S8A and S9B and Table S3).
We also examined the impact of these mutations on inactivation and channel kinetics (tau) (SI Appendix, Figs. S8B and S9). There is no consistent reciprocal change in these properties between the mutants and the corresponding wild-type channels (SI Appendix, Fig. S9 and Table S4). The characterizations of swapping mutations between Nav1.5/Nav1.7 and Nav1.8 thus provide strong support that the extracellular interface of VSDI and S5II contributes to the requirement of more depolarized voltage for Nav1.8 activation.
Nav1.8 Blockade by A-803467.
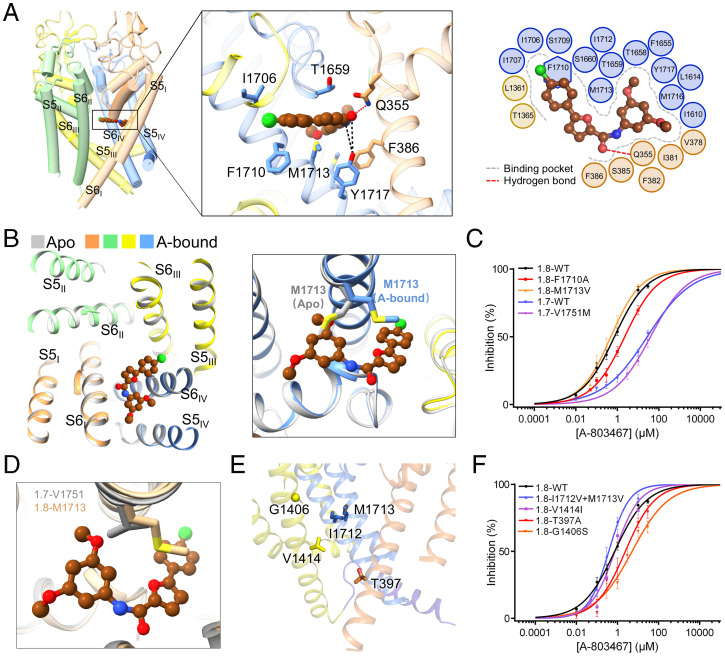
The density for the bound A-803467 was unambiguously resolved in the central cavity of the PD (SI Appendix, Fig. S5B). The backbone of the blocker is parallel to the central permeation axis and right beneath the selectivity filter (SF), directly obstructing the permeation path in the central cavity like quinidine to Nav1.5 (39). The overall molecule is positioned to clench S6IV (Fig. 4A). The dimethoxybenzene group of the blocker inserts into the fenestration enclosed by repeats I and IV (Fig. 4A, Inset). A-803467 is far off the central axis of the PD, affording a plausible interpretation for its relatively modest potency as a pore blocker (Fig. 1A).
Fig. 4.
Structural basis for specific blockade of Nav1.8 by A-803467. (A) A-803467 clenches S6IV of Nav1.8 beneath the selectivity filter. (Inset) Coordination of A-803467, shown as brown ball and sticks, by pore residues. Direct and water-mediated hydrogen bonds (H-bonds) are indicated by red and black dashed lines, respectively. (Right) Plane diagram of residues constituting the binding pocket within the cutoff distance from the ligand of 6 Å. The binding pocket and H-bond are indicated by gray dashed contour and red dashed lines, respectively. (B) The only change in the PD between the apo and A-803467 bound Nav1.8 structures is the swing of the side chain of Met1713. Shown on the left is an extracellular view of the superimposed PD of the apo (silver) and A-803467 bound (domain colored) structures of Nav1.8. (C) The only Nav1.8-unique blocker-binding residue Met1713 is not responsible for the subtype specificity. Among the Nav1.8 single-point mutants that were designed to validate the observed ligand binding, Ala substitution of Gln355, Phe386, and Thr1659 resulted in dramatically reduced peak currents, which were too small to support reliable analysis of pore blockage. Refer to SI Appendix, Table S1 for detailed parameters. (D) The variation between Met1713 in Nav1.8 and the corresponding Val1751 in Nav1.7 does not alter the local structure. Shown here is an enlarged view of the structures of class I Nav1.8 and Nav1.7 superimposed as in Fig. 3A. (E) Only very few residues lining the fenestrations and the intracellular gate are variant in Nav1.8. The side chains of Nav1.8-unique residues are shown as sticks and Cα of G1406 is shown as a sphere. Repeat II is omitted for clarity. (F) Allosteric modulation of A-803467 potency by Thr397, a S6I residue below the intracellular gate, and Gly1406, a fenestration residue on S6III. Single point mutations T397A and G1406S reduced the sensitivity to the blocker, with the IC50 shifted to 2.23 ± 0.51 µM and 5.12 ± 0.86 µM, respectively. Refer to SI Appendix, Fig. S2 and Tables S1 and S5 and Materials and Methods for experimental details and measured parameters.
To examine whether blocker binding induces any conformational changes of the PD, we compared the structure with apo Nav1.8 (SI Appendix, Figs. S3, S4, and S6 and Table S2). Superimposition of the PD of the apo and blocker-bound Nav1.8 shows identical backbone structures (SI Appendix, Fig. S7B and C). The only difference occurs in Met1713, whose side chain has to swing to avoid clash with the blocker (Fig. 4B and SI Appendix, Fig. S6). The structural observation is consistent with the reported preference of the blocker for an inactivated channel (27), as the structures of Nav1.8 reported here, characterized with up VSDs, fenestration-containing nonconductive PD, and the IFM motif wedged into the accepter site in the PD, most likely represent an inactivated state.
The compound is coordinated through both polar and hydrophobic interactions by residues primarily from S6I, S6IV, and the SF loops (defined as the P loops) in repeats I and IV (Fig. 4A). Gln355 on the PI loop is directly H-bonded to the oxide of A-803467, establishing the only direct H-bond. Tyr1717, a key gating residue on S6IV, may be able to form water-mediated H-bonds with the peptide bond of A-803467. A number of hydrophobic residues encompass the carbon backbones of the blocker, with Phe386 on S6I, Phe1710, and Met1713 on S6IV and Thr1659 on PIV loop in close proximity. A-803467 thus likely obstructs ion permeation by presenting a hydrophobic barrier beneath the SF (Fig. 4A).
To validate the binding pose of A-803467 revealed by the structure, we generated a number of Nav1.8 variants each containing a single residue substitution. Unfortunately, small current recorded for Nav1.8 mutants containing single Ala substitution, Q355A, F386A, or T1659A, could not be used for accurate blockage analysis. Consistent with a previous report (28), replacement of Phe1710 by Ala resulted in a reduced potency, resulting in the IC50 to 2.28 ± 0.31 µM (Fig. 4C and SI Appendix, Fig. S2A and Table S1).
We then set out to identify the residues responsible for the subtype specificity. Among all the blocker-coordinating residues, Met1713 is the only one that is unique to Nav1.8. In all other subtypes, this locus is occupied by Val (Fig. 4D and SI Appendix, Fig. S1). When Met1713 was mutated to Val, the potency was slightly enhanced to an IC50 of 0.47 ± 0.07 µM (Fig. 4C and SI Appendix, Fig. S2A and Table S1), contradicting what had been expected. Supporting this observation, reverse mutation of the corresponding locus Val1751 in Nav1.7 to Met did not increase its sensitivity and even caused a subtle drop from 27.68 ± 3.89 µM to 39.82 ± 11.27 µM (Fig. 4C and SI Appendix, Fig. S2A and Table S1).
Our studies suggest that the Nav1.8 selectivity of A-803467 is not directly related to the blocker binding site. As the blocker prefers the inactivated state over the resting state, we reasoned that the selectivity might be related to the conformational changes associated with the fenestrations or the intracellular gate. We thereby mutated the limited number of nonconserved residues that are mapped to these functional regions (Fig. 4E and SI Appendix, Fig. S1).
Single mutation V1414I did not alter IC50, and double mutation I1712V/M1713V even increased the potency to an IC50 of 0.38 ± 0.04 µM (Fig. 4F and SI Appendix, Fig. S2A and Table S1). Gly1406 on S6III, a residue mapped to the π helical bulge encompassing the III–IV fenestration but ∼11 Å away from A-803467, is unique to Nav1.8. The corresponding locus is occupied by a Ser in all other subtypes (SI Appendix, Fig. S1). G1406S reduced the potency of the blocker on Nav1.8, resulting in an IC50 of 5.12 ± 0.86 µM. Notably, this mutation has even more prominent effect than F1710A (Fig. 4F and SI Appendix, Fig. S2 and Tables S1 and S5). An unexpected observation came from Ala replacement of Thr397, which is one helical turn below the intracellular gate and far away from the blocker binding site. A-803467 was less potent on Nav1.8-T397A with an IC50 of 2.23 ± 0.51 µM, similar to that of Nav1.8-F1710A (Fig. 4F and SI Appendix, Fig. S2 and Tables S1 and S5).
Discussion
Development of subtype-specific Nav blockers is of great value for drug discovery. An advanced understanding of the molecular determinants for the subtype selectivity is required to optimize lead compounds. Our present characterizations show that the Nav1.8 specificity of A-803467 is remotely modulated by nonligand binding residues. Among the two identified residues, Thr397 is immediately beneath the intracellular gate and Gly1406 is on the edge of the III–IV fenestration. As the potency of A-803467 is channel state-dependent, the reduced sensitivities of the two variants may result from altered channel properties, such as voltage dependence for activation or inactivation. Interestingly, the two single point mutations, T397A and G1406S, have opposite effects on the voltage dependence of the activation curve, T397A to the more polarized and G1406S to the depolarized directions. Neither of them alters the voltage dependence of inactivation (SI Appendix, Fig. S2B and Table S5). It is noted that while Gly1406 is unique to Nav1.8, Thr397 is conserved in Nav1.9, which is less sensitive to A-803467 (SI Appendix, Fig. S1), suggesting that this locus is only one of the determining factors for subtype specificity.
In the absence of a structure of Nav channel in the resting state, there can be potentially multiple interpretations for the observation. One possible explanation is that these residues may directly or indirectly affect the entrance or exit path of A-803467 to the PD binding site. While the molecular mechanism for the modulation of blocker sensitivity by these two remote loci awaits future investigation, the discovery that the subtype specificity of a pore blocker can be affected by allosteric sites presents both challenges and opportunities for drug development targeting Nav channels. It highlights the importance of resolving structures of Nav channels in multiple working states, which will reveal the mechanism for the allosteric modulation.
Structural comparison of all available human Nav channels identifies the extracellular loops (ECLs) above the PD to be the most versatile region. The recently resolved structure of the human N-type Cav2.2 channel in complex with Food and Drug Administration–approved pain killer ziconotide reveals the subtype-specific coordination of the peptide blocker by the channel’s ECLs (45), demonstrating the feasibility of exploring the ECLs for drug development.
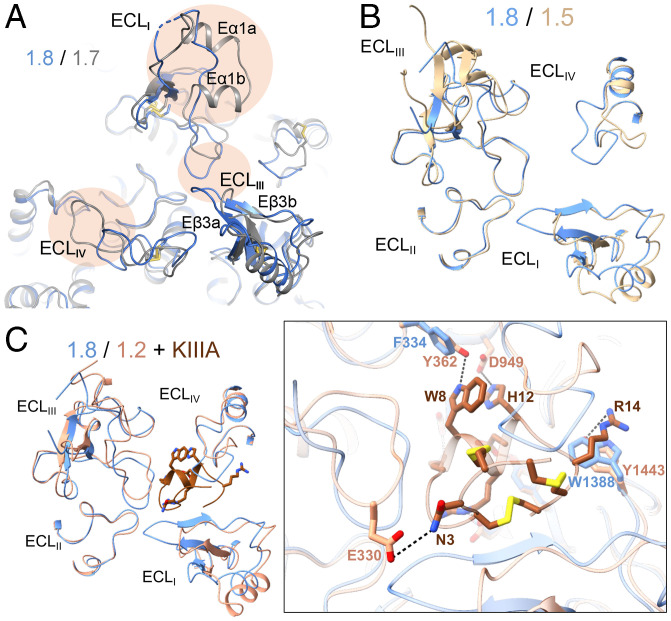
The ECLs of Nav1.8 cannot be completely aligned with those in any of the available Nav structures (Fig. 5A and SI Appendix, Fig. S1). Compared to Nav1.7, the ECLs in Nav1.8 lack the helical hairpin, Eα1a and Eα1b, in repeat I, and the loop between the β hairpin Eβ3a and Eβ3b in repeat III is longer (Fig. 5A). Both being TTX-resistant subtypes, Nav1.8 and Nav1.5 have substantially different loop structures in ECLI and ECLIV (Fig. 5B). This can be instrumental for avoiding cardiac-adverse effects when targeting Nav1.8 for drug discovery.
Fig. 5.
The ECLs above the PD may be exploited for the development of Nav1.8-specific suppressors. (A) The ECLs exhibit marked structural differences between Nav1.8 and Nav1.7. Shown here is a tilted extracellular view of the superimposed overall structures of class I Nav1.8 (blue) and Nav1.7 (gray) (PDB ID code: 7W9K). The structural distinctions of the ECL above the PD are highlighted with oval shades. (B) Nav1.8 and Nav1.5, both TTX-resistant subtypes, also deviate substantially in the ECL region. Shown here is an extracellular view of the ECLs from the superimposed overall structures of human Nav1.8 and Nav1.5 (PDB ID code: 6LQA). (C) The different structures of the ECLs underlie the distinct sensitivities of Nav1.8 and Nav1.2 to the peptide pore blocker µ-conotoxin KIIIA (KIIIA). (Inset) An extracellular view of the ECLs in the superimposed structures of Nav1.8 and KIIIA-bound Nav1.2 (PDB ID code: 6J8E). The elongated ECLI in Nav1.8 may interfere with KIIIA binding.
Supporting the importance of ECLs in developing subtype-specific blockers, μ-conotoxin KIIIA (KIIIA) shows strong, weak, and nearly no blockade on Nav1.2, 1.5, and 1.8, respectively (46). The structures provide a clear molecular interpretation. ECL residues in repeats I to III participate in KIIIA binding. Nav1.8 lacks three key coordinating residues on ECLI and ECLII, while its elongated ECLIII may present a potential steric clash with KIIIA (Fig. 5C). The sequence and structural differences of these loop regions determine Nav’s distinct sensitivities to KIIIA (34), thereby representing a potential site for developing Nav1.8-specific peptide pore blockers and biologics.
Another surprise is the distinct conformations of VSDI, which guided us to explore the nonconserved VSDI–PD interface of Nav1.8. Although this interface is characterized to be a molecular element for the high voltage dependence of Nav1.8 activation (Fig. 3D and SI Appendix, Table S3), our findings do not exclude the presence of other determinants that are yet to be identified. Furthermore, the molecular basis for the slower inactivation of Nav1.8 remains to be elucidated.
Notwithstanding the many outstanding questions to be addressed, the structural and functional analyses reported here provide important insight into the molecular basis for the distinct biophysical properties of Nav1.8 and serve as a framework for future investigations.
Materials and Methods
Transient Coexpression of Human Nav1.8 and β1.
The optimized coding DNA for human Nav1.8 (UniProt: Q9Y5Y9) was cloned into the pCAG vector (47) with twin Strep-tag and one FLAG tag in tandem at the amino terminus. Human β1 (UniProt: Q07699) was subcloned into the pCAG vector without affinity tag. HEK293F cells (Invitrogen) were cultured in SMM 293T-II medium (Sino Biological Inc.) under 5% CO2 in a Multitron-Pro shaker (Infors, 130 rpm) at 37 °C and transfected with plasmids when the cell density reached 2.0 × 106 cells per mL. For transfection, ∼2.0 mg plasmids (1.5 mg for Nav1.8 and 0.5 mg for β1) were preincubated with 4 mg 25-kDa linear polyethylenimines (Polysciences) in 25 mL fresh medium for 20 min before adding into 1 L cell culture. Transfected cells were cultured for 48 h before harvesting.
Protein Purification.
Protein purification was carried out following our optimized protocol (35). Twenty-eight liters of cells cotransfected with Nav1.8 and β1 were harvested by centrifugation at 800 × g and resuspended in the lysis buffer containing 25 mM Tris⋅HCl (pH 7.5) and 150 mM NaCl. The suspension was supplemented with 1% (wt/vol) n-dodecyl-β-d-maltopyranoside (DDM, Anatrace), 0.1% (wt/vol) cholesteryl hemisuccinate Tris salt (Anatrace), and protease inhibitor mixture containing 2 mM phenylmethylsulfonyl fluoride (VWR), 6.5 μg/mL aprotinin (MCE), 3.5 μg/mL pepstatin (Sigma), and 25 μg/mL leupeptin (Sigma). After incubation at 4 °C for 2 h, cell lysate was ultracentrifuged at 20,000 × g for 45 min, and the supernatant was applied to anti-Flag M2 affinity gel (Sigma) by gravity at 4 °C. Sample-loaded resin was rinsed four times using the W buffer that contains 25 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 0.05% glycol-diosgenin (Anatrace), and the protease inhibitor mixture. Proteins were eluted with W buffer plus 200 μg/mL FLAG peptide (Sigma). The eluent was applied to Strep-Tactin Sepharose (IBA) and the purification protocol was similar to the previous steps except the elution buffer, which contains 2.5 mM d-desthiobiotin (IBA) in addition to the W buffer. The eluted proteins were concentrated using a 100-kDa cutoff Centricon (Millipore) and applied to a Superose-6 column (GE Healthcare) that was preequilibrated in the W buffer.
The purified protein was pooled for cryo-EM and mass spectrometric (MS) analysis. Although our preliminary trials showed that cotransfection with β1 could improve the protein yield, β1 protein was barely detected in the MS. It is possible that the potentially weak association between Nav1.8 and β1 cannot survive the purification steps, similar to what has been observed for Nav1.5–β1 coexpression (36, 39). To obtain the structure of blocker-bound channel, A-803467 (Meilunbio) was added to the purified protein at a final concentration of 10 μM for incubation at 4 °C for 1 h. The protein samples with or without the blocker were both concentrated to ∼1 mg/mL for cryo-EM sample preparation.
Whole-Cell Electrophysiology.
Nav1.8 cannot be well-recorded in HEK293T cells due to small peak currents. In our study, Nav1.8 variants were all recorded in ND-7/23 cells (National Collection of Authenticated Cell Cultures). However, ND-7/23 cells have a high background of endogenous TTX-sensitive Na+ currents. Therefore, Nav1.7 variants were recorded in HEK293T cells. Both cell lines could be used for Nav1.5 recording. We recorded Nav1.5 variants in HEK293T cells for their cleaner background.
Both ND-7/23 cells and HEK293T cells were cultured in Dulbecco's modified Eagle’s medium (BI) supplemented with 4.5 mg/mL glucose and 10% (vol/vol) fetal bovine serum (BI). For patch-clamp recordings, the cells were plated onto glass coverslips and transiently cotransfected with the expression plasmids for indicated channel variants and enhanced green fluorescent protein in the presence of lipofectamine 2000 (Invitrogen). Cells with green fluorescence were selected for patch-clamp recording 18 to 36 h after transfection. All experiments were performed at room temperature. No authentication was performed for the commercially available cell line. Mycoplasma contamination was not tested.
The whole-cell Na+ currents were recorded in ND-7/23 cells or HEK293T cells using an EPC10-USB amplifier with Patchmaster software v2*90.2 (HEKA Elektronic), filtered at 3 kHz (low-pass Bessel filter) and sampled at 50 kHz. The borosilicate pipettes (Sutter Instrument) used in all experiments had a resistance of 2 to 4 MΩ and the electrodes were filled with the internal solution composed of (in millimolar) 105 CsF, 40 CsCl, 10 NaCl, 10 EGTA, 10 Hepes, pH 7.4 with CsOH. The bath solutions contained (in millimolar) 140 NaCl, 4 KCl, 10 Hepes, 10 d-glucose, 1 MgCl2, 1.5 CaCl2, pH 7.4 with NaOH. Four hundred nanomolar TTX was added to the bath solution during recording of the ND-7/23 cells in order to completely block endogenous TTX-sensitive sodium current. Data were analyzed using Origin (OriginLab) and GraphPad Prism (GraphPad Software).
The voltage dependence of ion current (I-V) was analyzed using a protocol consisting of steps from a holding potential of −120 mV (for 200 ms) to voltages ranging from −90 mV to +80 mV for 50 ms in 5-mV increments. The linear component of leaky currents and capacitive transients were subtracted using the P/4 procedure. In the activation property analysis and conductance density calculation, we used the equation G = I/(V − Vr), where Vr (the reversal potential) represents the voltage at which the current is zero. For the activation curves, conductance (G) was normalized and plotted against the voltage from −90 mV to +20 mV∼+50 mV as appropriate. For voltage dependence of inactivation, cells were clamped at a holding potential of −90 mV and were applied to step prepulses from −120 mV (−140 mV for Nav1.5) to +20 mV for 1,000 ms with an increment of 5 mV. Then, the Na+ currents were recorded at the test pulse of 0 mV for 50 ms. The peak currents under the test pulses were normalized and plotted against the prepulse voltage. Activation and inactivation curves were fitted with a Boltzmann function to obtain V1/2 and slope values. Time course of inactivation data from the peak current at 0 mV was fitted with a single exponential equation: y = A1 exp(−x/τinac) + y0, where A1 was the relative fraction of current inactivation, τinac was the time constant, x was the time, and y0 was the amplitude of the steady-state component.
To investigate the blockage of Nav1.8 variants by A-803467, cells were held at −120 mV and stepped to +10 mV (0 mV for Nav1.7 variants) for 50 ms. A-803467 was dissolved in dimethyl sulfoxide (final concentration less than 0.1%, Sigma) to make a stock solution of 100 mM. Solutions with indicated A-803467 concentrations were freshly prepared and perfused to the recording cell for up to 10 min to get to the maximal block using a multichannel perfusion system (VM8, ALA). Prior to A-803467 perfusion, cells were recorded for 5 min to establish stable peak current. Concentration-response curve was fitted with Y = Bottom + (Top − Bottom)/(1 + 10^((LogIC50 − X) * Hill Slope)), where IC50 is the concentration of A-8034687 that blocks 50% of the current and X is log of A-803467 concentration and Hill Slope is the slope factor. All data points are presented as mean ± SEM and n is the number of experimental cells from which recordings were obtained. Statistical significance was assessed using an unpaired t test with Welch’s correction, one-way ANOVA analysis, and extra sum-of-squares F test.
Cryo-EM Data Acquisition.
Aliquots of 3.5 μL freshly purified Nav1.8 were placed on glow-discharged holey carbon grids (Au 300 mesh, R1.2/1.3, Quantifoil). Grids were blotted for 3.0 s and plunge-frozen in liquid ethane cooled by liquid nitrogen with Vitrobot Mark IV (Thermo Fisher). Electron micrographs were acquired on a Titan Krios electron microscope (Thermo Fisher) operating at 300 kV and equipped with a Gatan K3 detector and GIF Quantum energy filter. A total of 16,934 movie stacks for A-803467–bound and 16,287 movie stacks for apo Nav1.8 were automatically collected using AutoEMation (48) with a slit width of 20 eV on the energy filter and a preset defocus ranging from −1.8 µm to −1.5 µm in superresolution mode at a nominal magnification of 81,000×. Each stack was exposed for 2.56 s with 0.08 s per frame, resulting in 32 frames per stack. The total dose rate was 50 e−/Å2 for each stack. The stacks were motion-corrected with MotionCor2 (49) and binned twofold, resulting in 1.08 Å per pixel. Dose weighting was performed for both data (50). The defocus values were estimated using Gctf (51).
Image Processing.
For simplicity, the dataset with A-803467 will be referred to as the A-bound dataset in this section. Data processing for the A-bound and apo datasets will be described in parallel in the form of A-bound/apo. A diagram for data processing is presented in SI Appendix, Fig. S3. A total of 8,336,754/6,310,392 particles were automatically picked using Gautomatch. After two-dimensional classification, a total of 3,267,310/2,771,855 good particles were selected; these particles were subject to one round of nonuniform refinement in cryoSPARC (52), resulting in 3D reconstructions at 2.9/3.9 Å. The resulting particles were subject to one additional round of heterogeneous refinement in cryoSPARC, with the number of classes set to 5/9. A total of 21,109,468/410,478 particles were selected and applied to nonuniform refinement, resulting in final reconstructions at overall resolutions of 2.7/3.1 Å. At this stage of data processing, both datasets exhibit considerable conformational heterogeneity as judged by the poorly resolved VSDI (SI Appendix, Fig. S3). The resolved regions of the A-bound and apo reconstruction appear identical. Since the apo dataset had much smaller particle number compared to the A-bound one, we focused on the A-bound dataset to improve the local map quality of the VSDI region.
First, the data star from the auto-refinement procedure was fed into the SGD initial model generation procedure of RELION-3.1 (K = 1) (53), which was initiated from random initial seeds and terminated after only a few iterations (typically between 3 and 10). Second, the reference map of each iteration before termination was substituted with the 2.7-Å overall refined EM density, and the SGD process was then resumed using the optimizer star file generated in RELION. Through this procedure, we attempted to explore the high-dimensional manifold in vicinity to the consensus refinement result using a batch-based stepwise model update routine. Third, the updated EM map after different SGD update epochs was fed into a principle component analysis (PCA) script to look for large variations within the population. The solved principle components were then added onto the consensus refinement result after proper scaling of the eigenvector solved from PCA. The used linear coefficients were sampled in the observed range using a fixed interval. Using one eigenvector solved from PCA, 10 different initial models were generated from the consensus refinement result. Finally, the 10 different initial models were used as seeds for the maximum-likelihood based 3D-classification procedure in RELION-3.1, enabling resolution of VSDI with different conformations. Upon identification of three conformational states, the entire dataset consisting of 21,109,468 particles were subjected to further 3D classification through local search and classification. The guided 3D-classification procedure (54) was used and the three reconstructions with different VSDI conformations were used as the first three classes, whereas empty micelles were used for the other starting models to draw out “bad particles.” The particles belonging to the three individual classes were separated and imported into cryoSPARC, wherein local refinement of these three classes yielded reconstructions at 2.7 Å (class I), 3.2 Å (class II), and 3.0 Å (class III) resolutions out of 237,316, 141,848, and 274,882 particles, respectively. The local resolutions of the VSDI region were sufficient for model building and side-chain assignment for class I and class II and main chain for class III (SI Appendix, Figs. S3–S6 and Table S2).
The apo structure was merely used as a control to examine the conformational changes of the PD upon A-803467 binding. Therefore, no further deep classification was performed for the dataset, which reached an overall resolution of 3.1 Å, with VSDI barely visible (SI Appendix, Figs. S3, S4, and S6 and Table S2).
Model Building and Structure Refinement.
Model building was performed based on the maps of Nav1.8 in the presence of A-803467 or not. The starting model of Nav1.8, which was built in SWISS-MODEL (55) based on the structure of Nav1.4 α subunit (PDB ID code 6AGF), was fitted into the EM maps by Chimera (56). All Nav1.8 residues, lipids, and sugar moieties were manually checked in Coot (57). The chemical properties of amino acids were taken into consideration during model building. There was no density belonging to β1, which is consistent with MS. In A-bound Nav1.8, more than 1,100 residues were assigned with side chains, A-803467, sugar moieties and lipid molecules were modeled due to the densities (SI Appendix, Table S2). The unmodelled segments include the N-terminal residues and the S2–3I linker, a short fragment in ECLI (residues 283 to 288), the intracellular I-II linker (residues 408 to 650), II–III linker (residues 896 to 1135), and the C-terminal sequences after Ala1728. In apo Nav1.8, VSDI was unmodelled besides the segments mentioned above due to the lack of density (SI Appendix, Fig. S3).
Structural refinement was performed using the phenix.real_space_refine application in PHENIX (58) real space with secondary structure and geometry restraints. Overfitting of the overall model was monitored by refining the model in one of the two independent maps from the gold-standard refinement approach and testing the refined model against the other map (59). Statistics of the map reconstruction and model refinement can be found in SI Appendix, Table S2.
Supplementary Material
Acknowledgments
We thank Xiaomin Li, Fan Yang, and Jianlin Lei for technical support during EM image acquisition. We thank the Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for providing the cryo-EM facility support. We thank the Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for computational facility support. This work was funded by Beijing Nova Program (Z191100001119127 to X.P.) from Beijing Municipal Science and Technology Commission. N.Y. has been supported by the Shirley M. Tilghman endowed professorship from Princeton University since 2017.
Footnotes
Reviewers: L.F., Stanford University; and V.Y.-Y., University of California, Davis.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2208211119/-/DCSupplemental.
Data Availability
Structural models and EM maps of human Nav1.8 with or without A-803467 have been deposited in PDB and Electron Microscopy Data Bank, respectively: PDB 7WE4 and EMD-32439 for class I; PDB 7WEL and EMD-32451 for class II; PDB 7WFR and EMD-32475 for class III; and PDB 7WFW and EMD-32476 for apo Nav1.8.
References
- 1.Hille B., Ion Channels of Excitable Membranes (Sinauer Associates, Sunderland, MA, ed. 3, 2001). [Google Scholar]
- 2.Hodgkin A. L., Huxley A. F., Resting and action potentials in single nerve fibres. J. Physiol. 104, 176–195 (1945). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett D. L., Clark A. J., Huang J., Waxman S. G., Dib-Hajj S. D., The role of voltage-gated sodium channels in pain signaling. Physiol. Rev. 99, 1079–1151 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Elliott A. A., Elliott J. R., Characterization of TTX-sensitive and TTX-resistant sodium currents in small cells from adult rat dorsal root ganglia. J. Physiol. 463, 39–56 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akopian A. N., Sivilotti L., Wood J. N., A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature 379, 257–262 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Sangameswaran L., et al. , Structure and function of a novel voltage-gated, tetrodotoxin-resistant sodium channel specific to sensory neurons. J. Biol. Chem. 271, 5953–5956 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Dib-Hajj S. D., et al. , Two tetrodotoxin-resistant sodium channels in human dorsal root ganglion neurons. FEBS Lett. 462, 117–120 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Vijayaragavan K., O’Leary M. E., Chahine M., Gating properties of Na(v)1.7 and Na(v)1.8 peripheral nerve sodium channels. J. Neurosci. 21, 7909–7918 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.John V. H., et al. , Heterologous expression and functional analysis of rat Nav1.8 (SNS) voltage-gated sodium channels in the dorsal root ganglion neuroblastoma cell line ND7-23. Neuropharmacology 46, 425–438 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Rush A. M., et al. , A single sodium channel mutation produces hyper- or hypoexcitability in different types of neurons. Proc. Natl. Acad. Sci. U.S.A. 103, 8245–8250 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Browne L. E., Clare J. J., Wray D., Functional and pharmacological properties of human and rat NaV1.8 channels. Neuropharmacology 56, 905–914 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Khasar S. G., Gold M. S., Levine J. D., A tetrodotoxin-resistant sodium current mediates inflammatory pain in the rat. Neurosci. Lett. 256, 17–20 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Akopian A. N., et al. , The tetrodotoxin-resistant sodium channel SNS has a specialized function in pain pathways. Nat. Neurosci. 2, 541–548 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Matthews E. A., Wood J. N., Dickenson A. H., Na(v) 1.8-null mice show stimulus-dependent deficits in spinal neuronal activity. Mol. Pain 2, 5 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joshi S. K., et al. , Involvement of the TTX-resistant sodium channel Nav 1.8 in inflammatory and neuropathic, but not post-operative, pain states. Pain 123, 75–82 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Zimmermann K., et al. , Sensory neuron sodium channel Nav1.8 is essential for pain at low temperatures. Nature 447, 855–858 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Faber C. G., et al. , Gain-of-function Nav1.8 mutations in painful neuropathy. Proc. Natl. Acad. Sci. U.S.A. 109, 19444–19449 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han C., et al. , The G1662S NaV1.8 mutation in small fibre neuropathy: Impaired inactivation underlying DRG neuron hyperexcitability. J. Neurol. Neurosurg. Psychiatry 85, 499–505 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Huang J., et al. , Small-fiber neuropathy Nav1.8 mutation shifts activation to hyperpolarized potentials and increases excitability of dorsal root ganglion neurons. J. Neurosci. 33, 14087–14097 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coates M. D., et al. , Impact of the NaV1.8 variant, A1073V, on post-sigmoidectomy pain and electrophysiological function in rat sympathetic neurons. J. Neurophysiol. 122, 2591–2600 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alsaloum M., Higerd G. P., Effraim P. R., Waxman S. G., Status of peripheral sodium channel blockers for non-addictive pain treatment. Nat. Rev. Neurol. 16, 689–705 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Horishita T., et al. , Antidepressants inhibit Nav1.3, Nav1.7, and Nav1.8 neuronal voltage-gated sodium channels more potently than Nav1.2 and Nav1.6 channels expressed in Xenopus oocytes. Naunyn Schmiedebergs Arch. Pharmacol. 390, 1255–1270 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Hijma H. J., Siebenga P. S., de Kam M. L., Groeneveld G. J., A phase 1, randomized, double-blind, placebo-controlled, crossover study to evaluate the pharmacodynamic effects of VX-150, a highly selective NaV1.8 inhibitor, in healthy male adults. Pain Med. 22, 1814–1826 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Payne C. E., et al. , A novel selective and orally bioavailable Nav 1.8 channel blocker, PF-01247324, attenuates nociception and sensory neuron excitability. Br. J. Pharmacol. 172, 2654–2670 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shields S. D., Butt R. P., Dib-Hajj S. D., Waxman S. G., Oral administration of PF-01247324, a subtype-selective Nav1.8 blocker, reverses cerebellar deficits in a mouse model of multiple sclerosis. PLoS One 10, e0119067 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kort M. E., et al. , Discovery and biological evaluation of 5-aryl-2-furfuramides, potent and selective blockers of the Nav1.8 sodium channel with efficacy in models of neuropathic and inflammatory pain. J. Med. Chem. 51, 407–416 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Jarvis M. F., et al. , A-803467, a potent and selective Nav1.8 sodium channel blocker, attenuates neuropathic and inflammatory pain in the rat. Proc. Natl. Acad. Sci. U.S.A. 104, 8520–8525 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Browne L. E., Blaney F. E., Yusaf S. P., Clare J. J., Wray D., Structural determinants of drugs acting on the Nav1.8 channel. J. Biol. Chem. 284, 10523–10536 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinckley C. A., et al. , Characterization of vixotrigine, a broad-spectrum voltage-gated sodium channel blocker. Mol. Pharmacol. 99, 49–59 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Yang T., et al. , Blocking Scn10a channels in heart reduces late sodium current and is antiarrhythmic. Circ. Res. 111, 322–332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X., Priest B. T., Belfer I., Gold M. S., Voltage-gated Na+ currents in human dorsal root ganglion neurons. eLife 6, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen H., et al. , Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science 355, eaal4326 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Pan X., et al. , Structure of the human voltage-gated sodium channel Nav1.4 in complex with β1. Science 362, eaau2486 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Pan X., et al. , Molecular basis for pore blockade of human Na+ channel Nav1.2 by the μ-conotoxin KIIIA. Science 363, 1309–1313 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Shen H., Liu D., Wu K., Lei J., Yan N., Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins. Science 363, 1303–1308 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Li Z., et al. , Structure of human Nav1.5 reveals the fast inactivation-related segments as a mutational hotspot for the long QT syndrome. Proc. Natl. Acad. Sci. U.S.A. 118, e2100069118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan X., et al. , Comparative structural analysis of human Nav1.1 and Nav1.5 reveals mutational hotspots for sodium channelopathies. Proc. Natl. Acad. Sci. U.S.A. 118, e2100066118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang G., et al. , High-resolution structures of human Nav1.7 reveal gating modulation through α-π helical transition of S6IV. Cell Rep. 39, 110735 (2022). [DOI] [PubMed] [Google Scholar]
- 39.Li Z., et al. , Structural basis for pore blockade of the human cardiac sodium channel Nav 1.5 by the antiarrhythmic drug quinidine*. Angew. Chem. Int. Ed. Engl. 60, 11474–11480 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Huang G., et al. , Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Nav1.7. Proc. Natl. Acad. Sci. U.S.A. 119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horn R., Ding S., Gruber H. J., Immobilizing the moving parts of voltage-gated ion channels. J. Gen. Physiol. 116, 461–476 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chanda B., Bezanilla F., Tracking voltage-dependent conformational changes in skeletal muscle sodium channel during activation. J. Gen. Physiol. 120, 629–645 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lacroix J. J., Campos F. V., Frezza L., Bezanilla F., Molecular bases for the asynchronous activation of sodium and potassium channels required for nerve impulse generation. Neuron 79, 651–657 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilchrist J., Bosmans F., Using voltage-sensor toxins and their molecular targets to investigate NaV 1.8 gating. J. Physiol. 596, 1863–1872 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao S., Yao X., Yan N., Structure of human Cav2.2 channel blocked by the painkiller ziconotide. Nature 596, 143–147 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang M. M., et al. , Structure/function characterization of micro-conotoxin KIIIA, an analgesic, nearly irreversible blocker of mammalian neuronal sodium channels. J. Biol. Chem. 282, 30699–30706 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Matsuda T., Cepko C. L., Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. U.S.A. 101, 16–22 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lei J., Frank J., Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J. Struct. Biol. 150, 69–80 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grant T., Grigorieff N., Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife 4, e06980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Scheres S. H., Processing of structurally heterogeneous Cryo-EM Data in RELION. Methods Enzymol. 579, 125–157 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Zhang X., et al. , An atomic structure of the human spliceosome. Cell 169, 918–929.e14 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Waterhouse A., et al. , SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46 (W1), W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amunts A., et al. , Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeLano W. L., The PyMOL Molecular Graphics System (2002).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural models and EM maps of human Nav1.8 with or without A-803467 have been deposited in PDB and Electron Microscopy Data Bank, respectively: PDB 7WE4 and EMD-32439 for class I; PDB 7WEL and EMD-32451 for class II; PDB 7WFR and EMD-32475 for class III; and PDB 7WFW and EMD-32476 for apo Nav1.8.