Abstract
Speciation is the process by which barriers to gene flow evolve between populations. Although we now know that speciation is largely driven by natural selection, knowledge of the agents of selection and the genetic and genomic mechanisms that facilitate divergence is required for a satisfactory theory of speciation. In this essay, we highlight three advances/problems in our understanding of speciation that have arisen from studies of the genes and genomic regions that underlie the evolution of reproductive isolation. First, we describe how the identification of “speciation” genes makes it possible to identify the agents of selection causing the evolution of reproductive isolation, while also noting that the link between the genetics of phenotypic divergence and intrinsic postzygotic reproductive barriers remains tenuous. Second, we discuss the important role of recombination suppressors in facilitating speciation with gene flow, but point out that the means and timing by which reproductive barriers become associated with recombination cold spots remains uncertain. Third, we establish the importance of ancient genetic variation in speciation, although we argue that the focus of speciation studies on evolutionarily young groups may bias conclusions in favor of ancient variation relative to new mutations.
Keywords: speciation, genetics, natural selection, recombination, standing genetic variation
The modern synthesis paved the way to a unified theory of evolutionary process that could be applied to understanding geographical variation, patterns in the fossil record, and adaptation to environment. But a synthetic theory of speciation is still under development. One reason is that widespread acceptance of the biological species concept—defined by evolved barriers to gene flow between populations (1, 2)—came relatively late. It was already clear that reproductive isolation evolved along with genetic divergence between populations (2). It was also predicted that reduced hybrid fitness, a hallmark of speciation, would be caused mainly by interactions between two or more “complementary genes” (1). However, the mechanisms driving genetic divergence, whether selection or drift, and the types of interactions between genes could only be speculated upon. It was not clear whether genetic differences causing reproductive isolation were mainly in the same genes responsible for phenotypic differences between species or whether, according to Dobzhansky, they represented “a separate category of genetic changes” (3). Thus, with the exception of speciation involving polyploidy, it was not possible at the time of the synthesis to point to even two species in nature and say how they evolved. Recently, appreciation has grown of the outsized importance of selection in the evolution of reproductive isolation (4–6). Yet, identifying agents of selection and their link to the key genetic changes underlying reproductive isolation remains challenging.
In this essay we describe recent progress in addressing three problems in the study of speciation by natural selection. In keeping with the spirit of this PNAS issue, our goal is to celebrate solutions made possible by newfound abilities to identify and sequence “speciation” genes, genomic regions, and their markers. The first problem concerns the causal link between mechanisms of selection, phenotypic divergence between populations, and the loci underlying reproductive isolation. Are the genes that underlie reproductive isolation the same as those underlying adaptive phenotypic differences between populations, or do they belong to a separate category, as Dobzhansky (3) opined? The link between phenotypic divergence and genotype is especially unclear for postzygotic isolation. Hybrid sterility and inviability are predicted to result from negative interactions between uniquely evolved alleles at more than one gene brought together in hybrids. This model of divergence in complementary genes has been well tested and supported (4), although it is unlikely to be universal. A major question is whether incompatibilities evolved via adaptive divergence in phenotypic traits or more intrinsic processes separate from divergent selection on traits.
The second problem concerns the prominent role played by recombination suppressors in reducing effective gene flow between diverging populations. An increasing number of cases are known in which components of reproductive isolation and ecologically relevant phenotypic differences map to chromosomal inversions and other regions of low recombination. What are the evolutionary forces that cause inversions to spread and diverge between populations, and how do they contribute to the evolution of reproductive isolation? Selection may directly favor an inversion because of its immediate phenotypic effects, or inversions may be favored indirectly because they help to overcome antagonistic effects of recombination between coselected genes.
The third problem concerns the source of genetic variation underlying reproductive isolation, and especially the role of standing genetic variation during speciation. Standing genetic variation includes variants maintained by mutation and selection in ancestral populations prior to divergence, as well as variants flowing in from already differentiated populations and closely related species via interbreeding; here we focus on the latter. Cases of adaptation from standing genetic variation are known but contributions to speciation are less clear.
Phenotypic Divergence and the Evolution of Reproductive Isolation
What do we know about the link between the evolution of divergent phenotypes and the build-up of genetic changes that cause reproductive isolation? The answer to this question has two parts. On the one hand, reproductive isolation is frequently a by-product of adaptive divergence between populations in phenotypic traits. This process is far more prevalent than Dobzhansky (3) imagined. Selection on traits favors divergence, and reproductive isolation often evolves as an incidental consequence. On the other hand, genetic changes unrelated to trait divergence may also be a major cause of speciation (7). These mechanisms include genetic conflict and divergent resolution of duplicated genes, which we interpret as belonging to Dobzhansky’s (3) separate category of genetic divergence. Under these mechanisms, selection drives genetic change but doesn’t directly favor divergence. Divergence and speciation happen anyway via evolutionary contingency or the “mutation-order” process (5).
Phenotypic Divergence and the Genetics of Prezygotic Isolation.
Dobzhansky (3) realized that certain adaptive phenotypic differences would contribute to reproductive isolation as an immediate by-product. Perhaps the most straightforward examples are prepollination traits in plants, such as divergence in flowering time. In this case the genes underlying trait differences also cause premating isolation. For example, latitudinal differences in flowering time in Arabidopsis thaliana represent adaptations to variation in day length and length of winter (8). The differences are controlled in part by FRIGIDA and FLOWERING LOCUS C, with divergent populations exhibiting strong temporal premating isolation when raised in a common garden (9). Temporal isolation also evolves in insects adapting to different host plants having contrasting phenologies (10).
Divergence of traits preferred by alternative pollinators has a similar effect on the evolution of prezygotic isolation. Schemske and Bradshaw (11) showed that Mimulus cardinalis and Mimulus lewisii flowers differ in anthocyanin pigment concentrations, which are favored by their alternative pollinators, hummingbirds and bees. Anthocyanin concentration differences are largely controlled by one or more mutations in an R3 MYB transcription factor (12), which thus also contributes to assortative mating. Streisfeld et al. (13) similarly found that a cis-regulatory mutation in R2R3-MYB is responsible for variation in anthocyanin pigmentation between divergent ecotypes of Mimulus aurantiacus having distinct preferred pollinators. Possible circumstances favoring a transition to a new pollen vector include habitat differences in available pollinator species and competition between plant species for pollinators. Other examples of genes underlying differences in pollinator preferences are summarized in refs. 14 and 15. Immigrant inviability (16) is expected to evolve in the same way. Any locus whose alternate alleles are favored in different, spatially distinct local environments will indirectly increase assortative mating if interbreeding is local. The contributions of individual loci to reproductive isolation in this case may be difficult to measure if local adaptation is polygenic.
Of what utility is knowledge of the identity of underlying genes when divergent traits directly cause premating isolation? First, evolutionary response to specific agents of selection on the traits can often be detected most readily by changes in allele frequency at underlying loci, particularly from an experimental test (17–20). Second, the same approach can detect pleiotropic effects of key genes on fitness at life stages even before the causal phenotypic traits are expressed (20). Genes also help to disentangle selection directly on a trait causing prezygotic isolation, such as body size or flowering time, from the confounding effects of general health or “condition” on the same traits (21). Finally, comparative studies of genes underlying prezygotic isolation establishes how often the same genes are reused in independent speciation events, revealing biases and constraints on the evolution of reproductive isolation.
In other cases, premating reproductive isolation evolves by physical linkage between its underlying genes and those genes causing adaptive phenotypic differences. Linkage between genes for traits and mating preferences behave similarly to pleiotropy in facilitating progress toward speciation by divergent selection, even when there is gene flow. This is because although speciation with gene flow involves the adaptive build-up of linkage disequilibrium between genes under divergent selection and genes controlling mating preferences, progress toward speciation is rapidly eroded by gene flow and recombination unless recombination is slowed by linkage or eliminated by pleiotropy (22). Identifying the underlying genes is the surest way to distinguish between these two possibilities, although this can be challenging if the genes occur in a region of low recombination, such as an inversion (see following section). For example, assortative mating between the Neotropical butterflies Heliconius melpomene and Heliconius cydno maps to a genomic region next to optix, a major locus underlying their color pattern differences (23). In stickleback, divergent mate preferences map to the same set of genomic regions as body size, shape, and niche use. The same genes might underlie assortative mating if, as in stickleback, phenotypic differences between sympatric species are themselves used as mating cues (24). Adaptive beak size differences between Darwin’s finch species are also cues in assortative mating, and the HMGA2 locus is a known contributor to this variation and therefore to assortative mating (25).
Phenotypic Divergence and the Genetics of Postzygotic Isolation.
Postzygotic isolation can evolve indirectly as a by-product of phenotypic adaptation to contrasting environments, but causal links between selection on phenotypes and the resulting genetic changes are still poorly known. For example, a variety of genes responsible for strong hybrid sterility and inviability have been identified in Drosophila (26). Each case involves “complementary genes,” two or more diverged loci whose parental alleles interact antagonistically when combined in hybrids, as predicted by the Bateson–Dobzhansky–Muller model. However, although genetic signatures of selection are often detected at causal genes, none of the cases have yet been tied to known adaptive phenotypic differences between parental populations. This might simply reflect the difficulty of determining the phenotypic effects of alternative alleles in parental species in the wild, but it is also possible that selection on ordinary phenotypic traits played no part in their evolution.
One of the best-known cases in plants is lethality of offspring between Mimulus guttatus adapted to copper mine tailings and off-mine plants. Genomic studies show that the cause of lethality is not the copper tolerance locus itself, Tol1, but rather a mutation tightly linked to it that hitchhiked to high frequency in the mine population as copper tolerance spread (27). Hence, in this case the phenotypic adaptation was coincidentally linked to the genetic changes causing reproductive isolation. In another case, shoot gravitropism, which differs repeatedly between growth forms of Senecio lautus inhabiting contrasting habitats, is associated with partial hybrid sterility linked to variation at one of the underlying genes, ABA3 (28).
Other examples plausibly link the evolution of hybrid inviability to divergent evolution of disease resistance. The plant species Capsella rubella and Capsella orientalis are divergent at the NPR1 immune-response gene. The C. orientalis allele is incompatible with alleles at the RPP5 pathogen-response gene in C. rubella, leading to necrosis in hybrids, an autoimmune response (29, 30). A multitude of cases of hybrid necrosis are known in A. thaliana, caused by interactions between divergent NLR plant-immune genes in interpopulation hybrids (31–33). Molecular evolutionary analyses indicate that alleles underlying hybrid necrosis are typically under balancing selection, yet alleles sporadically diverge rapidly between populations (31). In at least one case, hybrid necrosis results from antagonistic interactions between separate mutations in the same necrosis gene, rather than different genes, leading to heterozygote disadvantage (32), an apparent departure from the usual “complementary genes” model.
Yeast provide another departure from the Bateson–Dobzhansky–Muller complementary genes model, in which all nucleotide differences between strains and species contribute to hybrid spore inviability (34). This is because lack of sequence homology between chromosomes in hybrids impairs crossing over during meiosis and leads to missegregation. Here, all causes of genetic divergence, including those from phenotypic adaptation, contribute to postzygotic isolation.
Measurement of “phenotypic incompatibilities” represents another way to test causal links between phenotypic adaptation and genes causing postzygotic isolation. Hybrids between ecologically divergent parents are partly intermediate in many traits, which is expected to reduce hybrid viability and reproductive success in the absence of a hybrid niche. Such disruptive selection should cause fitness underdominance at causal loci and negative epistasis for fitness between them, although these have not been measured. In addition, and perhaps more importantly, hybrids typically exhibit mismatched phenotype combinations in which values of some traits more closely resemble one of the parent species, but values of other traits are more similar to the other parent, because of dominance and segregation. Arnegard et al. (35) showed that in addition to reduced feeding performance in phenotypic intermediates, many F2 hybrids between benthic and limnetic threespine stickleback species possessed mismatched jaw traits (Fig. 1) that reduce feeding performance. The benthic species has adaptations to generate high suction when feeding on prey attached to vegetation or buried in sediment. The limnetic species is adapted for rapid and distant jaw protrusion when feeding on evasive zooplankton. Hybrids with mismatched combinations of parental traits were predicted to be poor at both functions, chose a different diet, and had lowest growth. Morphological mismatch in F1 hybrid stickleback is similar to that in F2s (36), but backcrosses have not been measured. In a second example, Thompson et al. (37) measured selection on 30 phenotypic traits of field-transplanted F2 hybrids between the sunflowers Helianthus annuus and Helianthus debilis. A multiple regression found that being intermediate in individual traits and being mismatched for pairs of traits independently caused reduced seed set in the hybrids.
Fig. 1.
(Upper) Carbon (C) and nitrogen (N) stable isotope composition of individual F2 hybrids (circles) between limnetic and benthic stickleback species in an experimental pond. “L” individuals in the upper left have a more limnetic-like diet and isotope signature, whereas “B” individuals in the lower right are more benthic-like. “A” individuals in the lower left chose novel food types. Contours estimate mean body size (standard length in millimeters, reflecting feeding performance) of individual fish having different carbon and nitrogen isotope signatures. Parental species possess adaptations to feed on contrasting limnetic and benthic prey types. “A” individuals have mismatched combinations of these parental traits, being limnetic-like in some traits and benthic-like in other traits (Lower). Modified from Arnegard et al. (35).
Since many traits can contribute to overall mismatch of phenotypes, this cause of postzygotic isolation is expected to have a polygenic basis. Parental differences determining phenotype and diet of F2 hybrid stickleback mapped to over half the chromosomes (35). Phenotypic mismatch should lead to antagonistic interactions between pairs of underlying genes, although none have been identified as yet. Polygenic epistasis can nevertheless be detected indirectly. For example, mismatch in F2 populations should generate selection favoring more heterozygous individuals across genomic regions underlying trait differences (38). This prediction has been tested and confirmed in pond experimental F2 hybrid populations of threespine stickleback using a genome-wide measure of ancestry heterozygosity (39). Because the mechanism is extrinsic, this type of postzygotic isolation should be environment-dependent. As predicted, no selection favoring more heterozygous individuals was detected in laboratory stickleback populations (39), which also rules out simple heterosis as an explanation for this pattern.
Genetic Conflict and Postzygotic Isolation.
A separate class of mechanisms underlying hybrid inviability and sterility result from gene–gene coevolution without necessarily resulting in phenotypic divergence. One of the best known is genetic conflict between loci over unequal transmission of gene copies to gametes. Mutations causing bias and countermutations restoring fair transmission spread in turn. This occurs independently in different populations, which diverge via the fixation of different mutations despite experiencing overall similar selection pressures. The best-known example in animals occurs between two subspecies of Drosophila pseudoobscura. F1 hybrids are almost completely sterile, and the few fertile individuals produce almost entirely X-chromosome-bearing viable sperm. X-chromosome segregation distortion and F1 hybrid sterility map to the same locus, Ovd (40), linking the evolution of reproductive isolation to genetic conflict.
Conflict between nuclear and cytoplasmic genes over offspring sex allocation is pervasive in plants and a frequent cause of postzygotic isolation in hybrids. Mitochondrial genes are transmitted only via seeds, favoring mitochondrial mutations that divert energy toward seed production and away from pollen in the host plant, producing cytoplasmic male sterility (CMS). Nuclear restorer genes are then favored that restore fair transmission. For example, Case et al. (41) identified a rearrangement upstream of the mitochondrial gene nad6 that caused CMS in a population of M. guttatus. Pollen function is restored in the population by mutations in a pair of tightly linked nuclear pentatricopeptide repeat (PPR) genes. Both sets of mutations are absent in the closely related species Mimulus nasutus. F2 hybrids between the species that carry the M. guttatus CMS gene and the M. nasutus alleles at the PPR loci are male-sterile. Remarkably, mitochondrial CMS-nuclear restorer coevolution appears to be widespread in plants and in the majority of known systems results from structural rearrangements in the mitochondria and restorer mutations in PPR genes (30). This appears to be the most repeatable genetic mechanism of postzygotic isolation known.
Divergent Loss of Duplicated Genes.
Duplicated genes can coevolve in their expression levels within populations if total expression is under natural selection, and one copy might ultimately be silenced. If different copies are silenced by chance in different populations and the genes are not tightly linked within populations, then hybridization between the populations will generate a proportion of recombinant hybrids that inherit two defective copies, causing sterility or inviability. This process is like genetic conflict in that the mechanism involves gene–gene coevolution and can leave little trace in measured phenotypic differences between populations. Individual and whole-genome duplication followed by silencing is common in plants and therefore likely contributes repeatedly to postzygotic isolation between populations and species (30, 42).
Once again, Mimulus provides an example. M. guttatus possesses duplicate copies of the pTAC14 gene, which is involved in chloroplast development in Arabidopsis, on chromosomes 13 and 14. The copy on chromosome 13 is silenced. In the closely related species, M. nasutus, pTAC14 is found only on chromosome 13 either because it is ancestral and unduplicated in this species or because a deletion has removed the copy on chromosome 14. The result is that F2 hybrids that are homozygous for the silenced allele on chromosome 13 and for the M. guttatus null allele on chromosome 14 die from lack of chlorophyll (43). A similar mechanism involving transposition of the gene JYAlpha (likely initially involving a gene duplication) underlies hybrid male sterility in Drosophila simulans and Drosophila melanogaster (44).
Silencing and pseudogenation appear to be the fate of most gene copies (45). Although which copy of a duplicated gene first experiences a silencing mutation is likely random, the mechanism favoring the spread of the silenced copy, whether selection or drift, is uncertain. In Salmonella genomes, pseudogenes are usually deleted and at a rate higher than expected under neutrality, possibly because of costs associated with producing nonfunctional or toxic proteins (46). In Drosophila, relatively young duplicate genes that retain their ancestral function show evidence of negative selection (47). Duplicate gene copies are often pseudogenized after whole-genome duplication, which may happen asymmetrically and be driven by selection (48).
Recessive incompatibilities, such as those generated by the divergent resolution of duplicate genes, will affect only 1 in 16 of F2s (42). Thus, a single such incompatibility does not represent a strong reproductive barrier by itself. However, many recessive postyzygotic incompatibilities can cumulatively create a strong reproductive barrier. Consistent with this, the viability of F2 hybrids is often lower than that of F1s (e.g., ref. 49). Also, many hybrid incompatibilities are not completely recessive (50), and may therefore reduce F1 and backcross fitness as well.
The accumulation of postzygotic incompatibilities is predicted to be slow initially, but to increase faster than linearly over time if individual genes have the potential to develop incompatibilities with two or more other loci (51). This snowball effect has been documented empirically in both plants (52) and animals (53), and may explain why intrinsic genetic incompatibilities become increasingly important at later stages of speciation (54). However, postzygotic incompatibilities appear to play an outsized role in preserving incipient species once they are formed (55, 56).
Role of Recombination Suppressors in Speciation
Recombination is central to speciation (22, 57). Whereas divergent natural selection drives populations apart, gene flow and recombination hold them together. Evolutionary biologists have long been aware of this antagonism. Even prior to the rediscovery of Mendel’s principles of inheritance, Darwin’s contemporaries worried that changes favored by selection would be swamped by mating with nonadapted individuals (58). They also put forward a straightforward solution: geographic isolation (59). If populations were separated by a geographic barrier that kept them from mixing, then differences were expected to accumulate and eventually lead to speciation. This solution became increasingly widely accepted in the 20th century, leading some prominent evolutionists to argue that speciation in sexual species was improbable in the absence of geographic isolation (60, 61).
The main problem with gene flow is that although divergent selection favors the build-up of linkage disequilibrium between alleles favored in local environments and those causing reproductive isolation, gene flow and recombination break the associations apart (22). Nonetheless, theoretical and empirical support for speciation in the face of gene flow began accumulating in the late 20th century (62, 63), with accelerating evidence over the past two decades (64–66). Convincing case studies are now widespread across the animal, fungal, and plant kingdoms, ranging from Rhagoletis fruitflies (67) to Microbotryum fungi (68) to Mimulus monkeyflowers (69). How can it work?
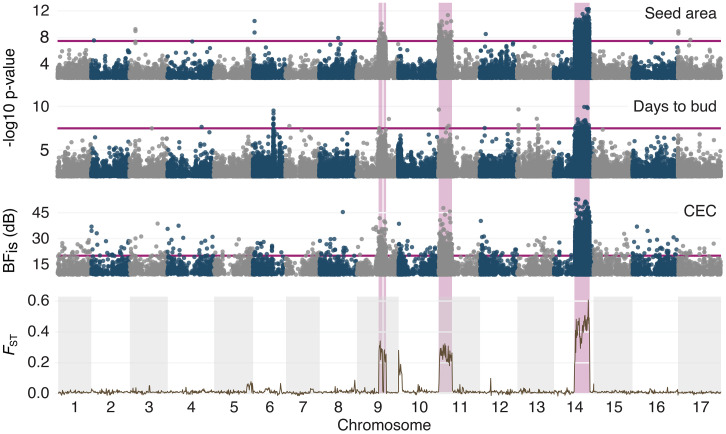
The answer is that in most such cases, traits that contribute to local adaptation and reproduction isolation are found to be associated with chromosomal inversions or other recombination suppressors, thereby resolving the antagonism between selection and recombination (57, 70). This is illustrated in Fig. 2 by parapatric dune and nondune ecotypes of the prairie sunflower (Helianthus petiolaris) (71). Sand dunes have lower soil fertility than the adjacent nondune, and the two habitats differ in their flora and pollinator assemblage. Seed size is strongly locally adapted to habitat. Despite gene flow between them, the adjacent populations are reproductively isolated by multiple mechanisms, including immigrant inviability, extrinsic selection against hybrids, shifts in pollinator assemblage and flowering time, and postpollination assortative mating (72). These barriers combined lead to strong total reproductive isolation (RI > 0.98). Fig. 2 shows that the traits responsible for them, as well as the majority of highly differentiated SNPs, map to chromosomal inversions.
Fig. 2.
Traits contributing to divergent adaptation and reproductive isolation map to inversions differentiating dune and nondune ecotypes of the prairie sunflower, H. petiolaris. The top three panels display the results from genome-wide association (GWA) analyses of seed size (surface area) and flowering-time (days to bud), as well as genotype–environment association (GEA) analyses of cation exchange capacity (CEC), which is a reflection of soil fertility. Horizontal purple lines represent 5% Bonferroni-corrected significance in the GWA analyses and Bayes factor (BFis) = 20 dB in the GEA analysis. The bottom panel shows genetic differentiation, as measured by FST, in 2-Mbp nonoverlapping sliding windows between dune- and nondune-adapted populations. Inversions underlie significant plateaus that are highlighted in pink and are responsible for the sharp boundaries in the FST plot. Modified from Todesco et al. (71).
Role in Speciation.
Theory predicts that speciation is facilitated by suppressed recombination between loci under divergent selection and those underlying other forms of reproductive isolation (73–75). Empirical studies suggest that this kind of genetic architecture is common. For example, in Ficedula flycatchers, species recognition, species-specific male plumage traits, and genes causing low hybrid fitness map to the Z chromosome, which apparently does not recombine in hybrids (76). An inversion in Anopheles mosquitoes has been shown to contribute to viability selection that varies between habitats, as well as to assortative mating (77). In plants, inversions are frequently reported to affect both locally adapted morphological traits and flowering time (reviewed in ref. 78), which is also locally adapted in many systems.
While recombination suppression between locally adapted alleles and isolation loci represents a straightforward route to speciation (75), bear in mind that the locally adapted traits may themselves act as reproductive barriers. This is illustrated by the dune/nondune sunflowers, in which adaptive divergence in seed size leads to both immigrant inviability and extrinsic selection against hybrids (72). The selective advantage of multiple, tightly linked alleles will be greater than that for individual alleles, permitting adaptive divergence and speciation under higher migration rates than might otherwise be feasible (79, 80).
Inversions appear to contribute more frequently to extrinsic than intrinsic reproductive barriers (78, 81), but observations of reduced fertility in inversion heterozygotes (i.e., underdominance) are not uncommon, especially in interspecific crosses (82). Such reports formed the basis of early models of chromosomal speciation, which suggested that inversions and other structural variants were established via genetic drift (83). However, such models are implausible because of the extreme population bottlenecks required for the establishment of strongly underdominant mutations (84). Alternative explanations include the spread of underdominant inversions via meiotic drive (85) or the accumulation of hybrid incompatibilities after inversion establishment (74, 86). This highlights a general unsolved question: What fraction of the differences mapping to inversions arise before versus after inversion establishment?
Types of Recombination Suppressors.
Of mechanisms known to suppress recombination, inversions have received the most attention because they are relatively easy to detect, especially with new comparative and population genomic methods, and their effects on recombination are well understood (78, 81). Cross-over events within the inversion are suppressed near inversion breakpoints due to interference; when they do occur, gametes often fail to develop or are inviable (87, 88). Inviable gamete production can be costly, with some large inversions causing declines in fertility of up to 50% in heterozygotes (82). However, such strongly underdominant inversions appear to be uncommon, perhaps because they must be established via special genetic mechanisms such as meiotic drive (although see ref, 88).
Local recombination rates can be suppressed by other structural variants, such as deletions, translocations, and mobile element insertions (89). Large multigenic deletions and translocations display strong underdominance in heterozygotes and, as a consequence, they are less likely than inversions to be polymorphic within species (although see ref. 90). Transposable element (TE) abundance is associated with recombination cold spots in many species, likely in part due to insertion bias (91). In addition, there is growing evidence that the silencing of TEs through chromatin modifications and DNA methylation contributes to recombination suppression (92) and may account for a considerable fraction of intra- and interspecific heterogeneity in recombination rates (91). Thus, TE expansions may contribute indirectly to speciation via the spread of recombination suppression.
Recombination may also be suppressed by modifier alleles. Although few such alleles have been characterized, models typically assume that they affect rates of cross-over events between loci that are targeted by selection (93). If a modifier allele is linked to the genes it affects, it can hitchhike to high frequency (94). However, the major recombination modifier gene that has been characterized thus far, PRDM9, affects many recombination hot spots across the genome rather than on the local scale envisioned by models (95). On the other hand, the sequence motifs recognized by PRDM9 could be viewed as modifier alleles and might act as predicted by conventional models (57). In plants, which lack PRDM9, cross-over events mainly occur in open chromatin surrounding gene promotors (96). DNA methylation silences recombination hot spots (97), so the genetic variants controlling such methylation may represent a kind of modifier allele. In maize, nearly half of differentially methylated regions are found to be associated with SNPs close to or within these regions (98), implying that modifiers of DNA methylation can affect recombination rates of nearby genes.
While the prevalence of structural variants versus local modifier alleles is poorly understood, the former spread more easily because they are completely linked to the genes they affect and suppress recombination in heterozygotes only (57). This facilitates divergence by permitting recombination within, but not between diverging populations, while also limiting the accumulation of genetic load. In contrast, allelic modifiers may be only loosely linked to affected loci and will suppress recombination within populations, which may be costly due to deleterious mutation accumulation. Nonetheless, they may be a common but largely overlooked contributor to speciation.
Establishment of Recombination Suppressors.
Models of divergence with gene flow typically assume that recombination suppression evolves by indirect selection favoring individuals possessing all locally adapted alleles (80, 99). For new inversions favored by this mechanism, the rate of spread will depend on the migration rate between populations, as well as on the number and linkage relationships of locally adapted genes captured by the inversion (100). Inversions that capture a larger number of coselected alleles that are loosely linked in the ancestral population will have the greatest selective advantage. The range of conditions for inversion establishment can be further expanded if an allopatric phase is included that aids the initial assembly of cassettes of locally adapted alleles (99).
Because local adaptation in the presence of gene flow appears to be pervasive (66), this model offers a potential explanation for the apparent ubiquity of inversions and (possibly) other recombination suppressors. Furthermore, many inversions have been shown to carry multiple adaptive traits or alleles as required by the model. However, in only a handful of cases has it been demonstrated that inversions have captured preexisting allelic combinations as opposed to recruitment of such alleles after inversion establishment (101).
Recombination suppressors may spread for other reasons, including direct selection (e.g., due to beneficial breakpoint effects), genetic drift of neutral or weakly deleterious variants, meiotic drive, or as a response to selfish element invasions. There is some evidence for each of these mechanisms. For example, the spread of a 10-Mb nonrecombining region in silverleaf sunflower is due to the gain through introgression of a major flowering-time gene (71). Meiotic drive has been shown to fix pericentric inversions in Drosophila (85). In some instances, multiple mechanisms appear to be involved, such as in Solenopsis fire ants, where benefits of both recombination suppression and inversion breakpoint effects have been documented (102).
A further complication is that many inversions are ancient and their current gene content and mutation profile are likely to be different from when they arose. Inversions that differentiate ecotypes or species are likely to continue to acquire mutations that contribute to local adaptation and reproductive isolation (70). In addition, such inversions will often be homozygous within populations, protecting them from the negative impacts of reduced recombination (103). In contrast, inversions that exist as balanced polymorphisms within populations are likely to accumulate deleterious mutations because of recombination suppression in heterozygotes, leading to associative overdominance (104, 105).
The distribution of inversion lengths within and between species can offer clues to the mechanisms responsible for their spread (106): 1) neutral and underdominant inversions are expected to be small; 2) inversions that are directly beneficial are likely to fix if small, but larger beneficial inversions will carry more deleterious recessive mutations and may be maintained as balanced polymorphisms by associative overdominance; and 3) indirect selection for locally adapted alleles favors intermediate-to-large inversions that encompass multiple adaptive alleles and remain polymorphic because of antagonistic pleiotropy, in which alternative haplotypes are favored in different habitats. Comparative genomic studies that simultaneously estimate inversion ages and lengths offer a potential means for disentangling evolutionary processes acting at the initial stages of establishment from those operating on ancient polymorphisms.
Sources of Genetic Variation Underlying Reproductive Isolation
In what’s known as the “junkyard tornado fallacy,” Hoyle reportedly estimated the probability of complex life emerging via evolution to be comparable to “the chance that a tornado sweeping through a junkyard might assemble a Boeing 747 from the materials therein” (107). Yet, evidence is emerging that old materials of evolution have frequently been reassembled by natural selection to form well-adapted phenotypes and even new species. New mutations are the ultimate source of genetic variation, and many genetic variants underlying the components of reproductive isolation show evidence of unique origins and signatures of selective sweeps consistent with having originated as a single new mutation within the population in which it eventually fixed (4). At the same time, many variants underlying adaptation and possibly reproductive isolation are older than the species in which they presently occur and persist in some form as standing genetic variation. For example, the NPR1 alleles that differ between C. rubella and C. orientalis and interact with RPP5 alleles to cause hybrid necrosis are polymorphic in the progenitor species, Capsella grandiflora (29). Standing variants are often shared between independently derived populations in similar environments that share a common ancestor, with copies often still found at low frequency in extant ancestral populations. Many of these variants occur as lengthy haploblocks and often they are found in genomic regions of low recombination (71, 108, 109). For example, independently evolved dune ecotypes of H. petiolaris in Colorado and Texas possess the same four inversions that underlie reproductive isolation and phenotypic differences from adjacent nondune populations (Fig. 2) and are older than the ecotypes (71).
Freshwater stickleback populations in the northeastern Pacific possess numerous adaptations for freshwater and have diverged rapidly and in parallel from the marine ancestral form since the freshwater bodies formed at the end of the last ice age (110). The low-armor allele at Eda is the best-known case of a single, old variant that was brought to lakes and streams when they were first colonized from the sea 10 to 15 K years ago. The allele resides on a large haploblock of suppressed recombination that has often fixed rapidly and behaves as a single variant (109, 111). The freshwater stickleback genome is dotted with similar haploblocks that have an average age of about 2 Mya (109), which is orders-of-magnitude older than the freshwater populations in which they are fixed, and have an average selective advantage of s = 0.3. The mechanisms of selection are not yet known, but both biotic and abiotic factors contribute. Miller et al. (108) showed that many of these same haploblocks, representing about 2% of the genes in the genome, repeatedly differentiate stickleback populations in small, isolated, and geographically interspersed lakes that differ only in the presence or absence of one other fish species (prickly sculpin). The phenotypic differences between marine and freshwater, and between sculpin and no-sculpin lakes, are associated with some degree of premating isolation (112) but the contribution of individual haplotypes to this process is not yet known.
Similar phenomena are known from other systems. The genomes of the Lake Victoria cichlids are populated by numerous ancient haplotypes that predate the origin of the lake and are present in different combinations in different species (113–115). Individual haploblocks are repeatedly associated with particular ecological types, such as piscivores (114). Similar findings are emerging from the cichlid radiations in the other East African Great Lakes (116, 117). It is likely that old standing genetic variation plays a large part in all cases of very rapid recent adaptive phenotypic diversification, since adaptive radiation entirely by new adaptive mutations is expected to take much longer. Allele-sharing by gene flow is rampant among relatively young species within radiating groups, which also often make use of standing genetic variation that first appeared and was used at earlier stages in the radiation (25, 67, 118–121).
More is known about how ancient variation underlies phenotypic differences than underlies reproductive isolation, which remains a major question. Assortative mating in a young species of seedeater finch is based on plumage traits that derive from older species in the radiation (119). Divergent genomic regions resistant to gene flow between lineages of European sea bass, Dicentrarchus labrax, and hence implicated in reproductive isolation between them, are derived from older extant species in the group (122). Other possible examples are listed in Marques et al. (123).
Reuse of old variation raises the question of how such variation has persisted over time. In most examples above, the differentiated alleles are already fixed or polymorphic in adapted populations and move to new populations by hybridization, spreading when advantageous. In other cases, the variants are maintained in an ancestral population in a different environment despite selection against them. Old variants are likely to be maintained by migration-selection balance rather than mutation-selection balance (124). In threespine stickleback, freshwater-advantageous alleles are found in marine populations in the sea today (109, 125). Adaptation from standing variation maintained by migration-selection balance might help to explain why so many occur in genomic regions of low recombination. Such haploblocks would tend not to be broken up rapidly in the sea by recombination, and this allows multiple linked advantageous variants to persist and then spread rapidly to fixation, when they are once again brought to freshwater by colonizing individuals (108).
Conclusions
A synthetic theory of speciation would include an understanding of the evolutionary agents that drive genetic changes, the genomic features that facilitate change, the types of genes that are most likely to be affected, and how the combination of contingency, selection, and genome structure yield reproductive isolation as a consequence.
Understanding the link between selection on phenotypes and the genetics of reproductive isolation have made more progress in cases of prezygotic isolation than postzygotic isolation. Many phenotypic traits are themselves both under divergent selection and responsible for reproductive isolation, in which case a shared genetic basis is expected. Selection on phenotypes can also generate immigrant inviability and extrinsic selection against hybrids, whose basis is likely often to be polygenic. Extrinsic selection against hybrids via intermediate and mismatched phenotypes also causes postygotic isolation, and although the genes are not yet known, genetic signatures of the process are detectable. On the other hand, as Dobzhansky (3) proposed, many genetic changes that underlie postzygotic isolation are not the result of divergent selection on phenotypes and result instead from more internal genetic mechanisms involving gene–gene coevolution. Divergent resolution of duplicated genes and conflict over transmission are likely two of the most important coevolution mechanisms. We need more tests to distinguish between drift and selection for the loss of redundant copies of duplicated genes. If truly due to drift, this would be the one case where drift might contribute directly to speciation. However, a drift-driven process might be too slow to play a substantive role in speciation, except perhaps following a whole-genome duplication event, in which many genes are likely to be silenced. However, as far as we are aware, there is no evidence that polyploid lineages accumulate postzygotic incompatibilities faster than diploids.
Recombination suppression appears to play a strong role in the evolution of reproductive isolation when there is gene flow. A big question is whether the reproductive barriers and other differences currently associated with recombination cold spots arose before or after the establishment of recombination suppression. Comparing the effects of young and old inversions could address this question. Another big gap is in our knowledge of the factors causing recombination suppression. We focused on one class of recombination suppressor (inversions), but there are other mechanisms that suppress recombination that may be equally important and that might become established for very different reasons. For example, it could well be that the majority of recombination suppressors in the genome arise as a response to TE invasions (i.e., genetic conflict) and their effects on recombination are an incidental byproduct, with little immediate impact on adaptation and speciation.
Finally, our understanding of the role of standing genetic variation in speciation is incomplete. Most evidence comes from phenotypically diverse and relatively young radiations, which might be biased in their use of standing variation compared to new mutation. Also, most evidence is on the phenotypic effects of standing variation and the effects on reproductive isolation are little known. Besides speed, the consequences of evolution from standing variation are also interesting. Standing variation has a high probability of yielding repeated evolution in different parts of a species range and leading to parallel evolution of phenotypes and even components of reproductive isolation. Parallel evolution is itself a signature of natural selection when it occurs, predictably in association with environmental features, facilitating identification of agents of selection driving the origin of species.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data Availability
There are no data underlying this work.
References
- 1.Dobzhansky T. G., Genetics and the Origin of Species (Columbia University Press, New York, 1937). [Google Scholar]
- 2.Mayr E., Systematics and the Origin of Species (Columbia University Press, New York, 1942). [Google Scholar]
- 3.Dobzhansky T., Speciation as a stage in evolutionary divergence. Am. Nat. 74, 312 (1940). [Google Scholar]
- 4.Coyne J. A., Orr H. A., Speciation (Sinauer Associates, Sunderland, MA, 2004). [Google Scholar]
- 5.Schluter D., Evidence for ecological speciation and its alternative. Science 323, 737–741 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Sobel J. M., Chen G. F., Watt L. R., Schemske D. W., The biology of speciation. Evolution 64, 295–315 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Presgraves D. C., Meiklejohn C. D., Sterility, genetic conflict and complex speciation: Lessons from the Drosophila simulans clade species. Front. Genet. 12, 669045 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caicedo A. L., Stinchcombe J. R., Olsen K. M., Schmitt J., Purugganan M. D., Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. U.S.A. 101, 15670–15675 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lovell J. T., et al. , Pleiotropy of FRIGIDA enhances the potential for multivariate adaptation. Proc. Biol. Sci. 280, 20131043 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feder J. L., Roethele J. B., Wlazlo B., Berlocher S. H., Selective maintenance of allozyme differences among sympatric host races of the apple maggot fly. Proc. Natl. Acad. Sci. U.S.A. 94, 11417–11421 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schemske D. W., Bradshaw H. D. Jr., Pollinator preference and the evolution of floral traits in monkeyflowers (Mimulus). Proc. Natl. Acad. Sci. U.S.A. 96, 11910–11915 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan Y.-W., Sagawa J. M., Young R. C., Christensen B. J., Bradshaw H. D. Jr., Genetic dissection of a major anthocyanin QTL contributing to pollinator-mediated reproductive isolation between sister species of Mimulus. Genetics 194, 255–263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Streisfeld M. A., Young W. N., Sobel J. M., Divergent selection drives genetic differentiation in an R2R3-MYB transcription factor that contributes to incipient speciation in Mimulus aurantiacus. PLoS Genet. 9, e1003385 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rieseberg L. H., Blackman B. K., Speciation genes in plants. Ann. Bot. (Lond.) 106, 439–455 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blackman B. K., “Speciation genes” in Encyclopedia of Evolutionary Biology, Kliman R. M., Ed. (Academic Press, Oxford, 2016), pp. 166–175. [Google Scholar]
- 16.Nosil P., Vines T. H., Funk D. J., Perspective: Reproductive isolation caused by natural selection against immigrants from divergent habitats. Evolution 59, 705–719 (2005). [PubMed] [Google Scholar]
- 17.Schluter D., et al. , Fitness maps to a large-effect locus in introduced stickleback populations. Proc. Natl. Acad. Sci. U.S.A. 118, e1914889118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett R. D. H., et al. , Linking a mutation to survival in wild mice. Science 363, 499–504 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Rennison D. J., Rudman S. M., Schluter D., Genetics of adaptation: Experimental test of a biotic mechanism driving divergence in traits and genes. Evol. Lett. 3, 513–520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korves T. M., et al. , Fitness effects associated with the major flowering time gene FRIGIDA in Arabidopsis thaliana in the field. Am. Nat. 169, E141–E157 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Austen E. J., Rowe L., Stinchcombe J. R., Forrest J. R. K., Explaining the apparent paradox of persistent selection for early flowering. New Phytol. 215, 929–934 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J., Skepticism towards Santa Rosalia, or why are there so few kinds of animals. Evolution 35, 124–138 (1981). [DOI] [PubMed] [Google Scholar]
- 23.Merrill R. M., et al. , Genetic dissection of assortative mating behavior. PLoS Biol. 17, e2005902 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bay R. A., et al. , Genetic coupling of female mate choice with polygenic ecological divergence facilitates stickleback speciation. Curr. Biol. 27, 3344–3349.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamichhaney S., et al. , A beak size locus in Darwin’s finches facilitated character displacement during a drought. Science 352, 470–474 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Orr H. A., The genetic basis of reproductive isolation: Insights from Drosophila. Proc. Natl. Acad. Sci. U.S.A. 102 (suppl. 1), 6522–6526 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright K. M., Lloyd D., Lowry D. B., Macnair M. R., Willis J. H., Indirect evolution of hybrid lethality due to linkage with selected locus in Mimulus guttatus. PLoS Biol. 11, e1001497 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilkinson M. J., et al. , Adaptive divergence in shoot gravitropism creates hybrid sterility in an Australian wildflower. Proc. Natl. Acad. Sci. U.S.A. 118, e2004901118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sicard A., et al. , Divergent sorting of a balanced ancestral polymorphism underlies the establishment of gene-flow barriers in Capsella. Nat. Commun. 6, 7960 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fishman L., Sweigart A. L., When two rights make a wrong: The evolutionary genetics of plant hybrid incompatibilities. Annu. Rev. Plant Biol. 69, 707–731 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Bomblies K., et al. , Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5, e236 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chae E., et al. , Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell 159, 1341–1351 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phadnis N., Malik H. S., Speciation via autoimmunity: A dangerous mix. Cell 159, 1247–1249 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Ono J., Greig D., Boynton P. J., Defining and disrupting species boundaries in Saccharomyces. Annu. Rev. Microbiol. 74, 477–495 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Arnegard M. E., et al. , Genetics of ecological divergence during speciation. Nature 511, 307–311 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chhina A. K., Thompson K. A., Schluter D., Adaptive divergence and the evolution of hybrid trait mismatch in threespine stickleback. Evol. Lett. 6, 34–45 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson K. A., Urquhart-Cronish M., Whitney K. D., Rieseberg L. H., Schluter D., Patterns, predictors, and consequences of dominance in hybrids. Am. Nat. 197, E72–E88 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Simon A., Bierne N., Welch J. J., Coadapted genomes and selection on hybrids: Fisher’s geometric model explains a variety of empirical patterns. Evol. Lett. 2, 472–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson K. A., et al. , Genetic evidence for environment-dependent hybrid incompatibilities in threespine stickleback. PLoS Biol. 20, e3001469 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phadnis N., Orr H. A., A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323, 376–379 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Case A. L., Finseth F. R., Barr C. M., Fishman L., Selfish evolution of cytonuclear hybrid incompatibility in Mimulus. Proc. Biol. Sci. 283, 20161493 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lynch M., Force A. G., The origin of interspecific genomic incompatibility via gene duplication. Am. Nat. 156, 590–605 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Zuellig M. P., Sweigart A. L., Gene duplicates cause hybrid lethality between sympatric species of Mimulus. PLoS Genet. 14, e1007130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masly J. P., Jones C. D., Noor M. A., Locke J., Orr H. A., Gene transposition as a cause of hybrid sterility in Drosophila. Science 313, 1448–1450 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Gout J.-F., Lynch M., Maintenance and loss of duplicated genes by dosage subfunctionalization. Mol. Biol. Evol. 32, 2141–2148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuo C.-H., Ochman H., The extinction dynamics of bacterial pseudogenes. PLoS Genet. 6, e1001050 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang X., Assis R., Natural selection drives rapid functional evolution of young Drosophila duplicate genes. Mol. Biol. Evol. 34, 3089–3098 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gillard G. B., et al. , Comparative regulomics supports pervasive selection on gene dosage following whole genome duplication. Genome Biol. 22, 103 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stelkens R. B., Schmid C., Seehausen O., Hybrid breakdown in cichlid fish. PLoS One 10, e0127207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tao Y., Hartl D. L., Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. III. Heterogeneous accumulation of hybrid incompatibilities, degree of dominance, and implications for Haldane’s rule. Evolution 57, 2580–2598 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Orr H. A., The population genetics of speciation: The evolution of hybrid incompatibilities. Genetics 139, 1805–1813 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moyle L. C., Nakazato T., Hybrid incompatibility “snowballs” between Solanum species. Science 329, 1521–1523 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Matute D. R., Butler I. A., Turissini D. A., Coyne J. A., A test of the snowball theory for the rate of evolution of hybrid incompatibilities. Science 329, 1518–1521 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Christie K., Strauss S. Y., Along the speciation continuum: Quantifying intrinsic and extrinsic isolating barriers across five million years of evolutionary divergence in California jewelflowers. Evolution 72, 1063–1079 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Futuyma D. J., On the role of species in anagenesis. Am. Nat. 130, 465–473 (1987). [Google Scholar]
- 56.Irwin D. E., Assortative mating in hybrid zones is remarkably ineffective in promoting speciation. Am. Nat. 195, E150–E167 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Ortiz-Barrientos D., Engelstädter J., Rieseberg L. H., Recombination rate evolution and the origin of species. Trends Ecol. Evol. 31, 226–236 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Romanes G. J., Physiological selection; an additional suggestion on the origin of species. Zool. J. Linn. Soc. 19, 337–411 (1886). [Google Scholar]
- 59.Gulick J. T., On the variation of species as related to their geographical distribution, illustrated by the Achatinellinae. Nature 6, 222–224 (1872). [Google Scholar]
- 60.Mayr E., Animal Species and Evolution (Belknap Press of Harvard University Press, Cambridge, 1963). [Google Scholar]
- 61.Futuyma D. J., Mayer G. C., Non-allopatric speciation in animals. Syst. Biol. 29, 254–271 (1980). [Google Scholar]
- 62.Bush G. L., Sympatric speciation in animals: New wine in old bottles. Trends Ecol. Evol. 9, 285–288 (1994). [DOI] [PubMed] [Google Scholar]
- 63.Dieckmann U., Doebeli M., On the origin of species by sympatric speciation. Nature 400, 354–357 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Gavrilets S., Fitness Landscapes and the Origin of Species (Monographs in population biology 41, Princeton University Press, Princeton, NJ, 2004). [Google Scholar]
- 65.Papadopulos A. S. T., et al. , Speciation with gene flow on Lord Howe Island. Proc. Natl. Acad. Sci. U.S.A. 108, 13188–13193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nosil P., Ecological Speciation (Oxford University Press, 2012). [Google Scholar]
- 67.Feder J. L., et al. , Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc. Natl. Acad. Sci. U.S.A. 100, 10314–10319 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hartmann F. E., et al. , Higher gene flow in sex-related chromosomes than in autosomes during fungal divergence. Mol. Biol. Evol. 37, 668–682 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hall M. C., Lowry D. B., Willis J. H., Is local adaptation in Mimulus guttatus caused by trade-offs at individual loci? Mol. Ecol. 19, 2739–2753 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faria R., Johannesson K., Butlin R. K., Westram A. M., Evolving inversions. Trends Ecol. Evol. 34, 239–248 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Todesco M., et al. , Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature 584, 602–607 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Ostevik K. L., Andrew R. L., Otto S. P., Rieseberg L. H., Multiple reproductive barriers separate recently diverged sunflower ecotypes. Evolution 70, 2322–2335 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Trickett A. J., Butlin R. K., Recombination suppressors and the evolution of new species. Heredity 73, 339–345 (1994). [DOI] [PubMed] [Google Scholar]
- 74.Navarro A., Barton N. H., Accumulating postzygotic isolation genes in parapatry: A new twist on chromosomal speciation. Evolution 57, 447–459 (2003). [DOI] [PubMed] [Google Scholar]
- 75.Dagilis A. J., Kirkpatrick M., Prezygotic isolation, mating preferences, and the evolution of chromosomal inversions. Evolution 70, 1465–1472 (2016). [DOI] [PubMed] [Google Scholar]
- 76.Saether S. A., et al. , Sex chromosome-linked species recognition and evolution of reproductive isolation in flycatchers. Science 318, 95–97 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Ayala D., Guerrero R. F., Kirkpatrick M., Reproductive isolation and local adaptation quantified for a chromosome inversion in a malaria mosquito. Evolution 67, 946–958 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Huang K., Rieseberg L. H., Frequency, origins, and evolutionary role of chromosomal inversions in plants. Front. Plant Sci. 11, 296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rieseberg L. H., Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16, 351–358 (2001). [DOI] [PubMed] [Google Scholar]
- 80.Kirkpatrick M., Barton N., Chromosome inversions, local adaptation and speciation. Genetics 173, 419–434 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wellenreuther M., Bernatchez L., Eco-evolutionary genomics of chromosomal inversions. Trends Ecol. Evol. 33, 427–440 (2018). [DOI] [PubMed] [Google Scholar]
- 82.King M., Species Evolution: The Role of Chromosome Change (Cambridge University Press, 1995). [Google Scholar]
- 83.White M. J. D., Modes of Speciation (W. H. Freeman and Co, San Francisco, 1978). [Google Scholar]
- 84.Hedrick P. W., The establishment of chromosomal variants. Evolution 35, 322–332 (1981). [DOI] [PubMed] [Google Scholar]
- 85.Lindholm A. K., et al. , The ecology and evolutionary dynamics of meiotic drive. Trends Ecol. Evol. 31, 315–326 (2016). [DOI] [PubMed] [Google Scholar]
- 86.Noor M. A. F., Grams K. L., Bertucci L. A., Reiland J., Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. U.S.A. 98, 12084–12088 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Coyne J. A., Meyers W., Crittenden A. P., Sniegowski P., The fertility effects of pericentric inversions in Drosophila melanogaster. Genetics 134, 487–496 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee Y. W., Fishman L., Kelly J. K., Willis J. H., A segregating inversion generates fitness variation in yellow monkeyflower (Mimulus guttatus). Genetics 202, 1473–1484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rowan B. A., et al. , An ultra high-density Arabidopsis thaliana crossover map that refines the influences of structural variation and epigenetic features. Genetics 213, 771–787 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Villoutreix R., et al. , Large-scale mutation in the evolution of a gene complex for cryptic coloration. Science 369, 460–466 (2020). [DOI] [PubMed] [Google Scholar]
- 91.Kent T. V., Uzunović J., Wright S. I., Coevolution between transposable elements and recombination. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160458 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Slotkin R. K., Martienssen R., Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285 (2007). [DOI] [PubMed] [Google Scholar]
- 93.Nei M., Modification of linkage intensity by natural selection. Genetics 57, 625–641 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Charlesworth D., Charlesworth B., Selection on recombination in clines. Genetics 91, 581–589 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baudat F., et al. , PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Choi K., et al. , Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 45, 1327–1336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yelina N. E., et al. , DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis. Genes Dev. 29, 2183–2202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Eichten S. R., et al. , Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 25, 2783–2797 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Feder J. L., Gejji R., Powell T. H. Q., Nosil P., Adaptive chromosomal divergence driven by mixed geographic mode of evolution. Evolution 65, 2157–2170 (2011). [DOI] [PubMed] [Google Scholar]
- 100.Charlesworth B., Barton N. H., The spread of an inversion with migration and selection. Genetics 208, 377–382 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Villoutreix R., et al. , Inversion breakpoints and the evolution of supergenes. Mol. Ecol. 30, 2738–2755 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yan Z., et al. , Evolution of a supergene that regulates a trans-species social polymorphism. Nat. Ecol. Evol. 4, 240–249 (2020). [DOI] [PubMed] [Google Scholar]
- 103.Huang K., et al. , High homozygosity of inversions in sunflower species largely averts accumulation of deleterious mutations. bioRxiv [Preprint] (2022). https://www.biorxiv.org/content/10.1101/2022.01.06.475294v1 (Accessed 27 May 2022).
- 104.Mérot C., Llaurens V., Normandeau E., Bernatchez L., Wellenreuther M., Balancing selection via life-history trade-offs maintains an inversion polymorphism in a seaweed fly. Nat. Commun. 11, 670 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jay P., et al. , Mutation load at a mimicry supergene sheds new light on the evolution of inversion polymorphisms. Nat. Genet. 53, 288–293 (2021). [DOI] [PubMed] [Google Scholar]
- 106.Connallon T., Olito C., Natural selection and the distribution of chromosomal inversion lengths. Mol. Ecol., 10.1111/mec.16091 (2021). [DOI] [PubMed] [Google Scholar]
- 107."Hoyle on evolution." Nature 294, 105 (1981).29451265 [Google Scholar]
- 108.Miller S. E., Roesti M., Schluter D., A single interacting species leads to widespread parallel evolution of the stickleback genome. Curr. Biol. 29, 530–537.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Roberts Kingman G. A., et al. , Predicting future from past: The genomic basis of recurrent and rapid stickleback evolution. Sci. Adv. 7, eabg5285 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fang B., Merilä J., Matschiner M., Momigliano P., Estimating uncertainty in divergence times among three-spined stickleback clades using the multispecies coalescent. Mol. Phylogenet. Evol. 142, 106646 (2020). [DOI] [PubMed] [Google Scholar]
- 111.Jones F. C., et al. ; Broad Institute Genome Sequencing Platform & Whole Genome Assembly Team, The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484, 55–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McKinnon J. S., et al. , Evidence for ecology’s role in speciation. Nature 429, 294–298 (2004). [DOI] [PubMed] [Google Scholar]
- 113.Meier J. I., et al. , Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 8, 14363 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McGee M. D., et al. , The ecological and genomic basis of explosive adaptive radiation. Nature 586, 75–79 (2020). [DOI] [PubMed] [Google Scholar]
- 115.Nakamura H., et al. , Genomic signatures for species-specific adaptation in Lake Victoria cichlids derived from large-scale standing genetic variation. Mol. Biol. Evol. 38, 3111–3125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Svardal H., et al. , Ancestral hybridization facilitated species diversification in the Lake Malawi cichlid fish adaptive radiation. Mol. Biol. Evol. 37, 1100–1113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Irisarri I., et al. , Phylogenomics uncovers early hybridization and adaptive loci shaping the radiation of Lake Tanganyika cichlid fishes. Nat. Commun. 9, 3159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Choi J. Y., et al. , Ancestral polymorphisms shape the adaptive radiation of Metrosideros across the Hawaiian Islands. Proc. Natl. Acad. Sci. U.S.A. 118, e2023801118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Turbek S. P., et al. , Rapid speciation via the evolution of pre-mating isolation in the Iberá Seedeater. Science 371, eabc0256 (2021). [DOI] [PubMed] [Google Scholar]
- 120.Moest M., et al. , Selective sweeps on novel and introgressed variation shape mimicry loci in a butterfly adaptive radiation. PLoS Biol. 18, e3000597 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Richards E. J., Martin C. H., Adaptive introgression from distant Caribbean islands contributed to the diversification of a microendemic adaptive radiation of trophic specialist pupfishes. PLoS Genet. 13, e1006919 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Duranton M., et al. , The contribution of ancient admixture to reproductive isolation between European sea bass lineages. Evol. Lett. 4, 226–242 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Marques D. A., Meier J. I., Seehausen O., A combinatorial view on speciation and adaptive radiation. Trends Ecol. Evol. 34, 531–544 (2019). [DOI] [PubMed] [Google Scholar]
- 124.Schluter D., Conte G. L., Genetics and ecological speciation. Proc. Natl. Acad. Sci. U.S.A. 106 (suppl. 1), 9955–9962 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Colosimo P. F., et al. , Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307, 1928–1933 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no data underlying this work.