Significance
In early mammalian embryos, enhanced mitochondrial oxidative metabolism is a hallmark essential for postimplantation survival and development. However, resultant mitochondrial oxidative stress would have damaged mitochondrial DNA (mtDNA). Until now, how embryonic mitochondria protect their own genome remains unclear. Herein, we reported that the mitochondrial genome undergoes extensive de novo mtDNA methylation that can protect mtDNA against enhanced oxidative damage during the peri-implantation stage. DNA de novo methyltransferase DNMT3A and DNMT3B enter mitochondria via their own mitochondrial targeting sequences, and then synergistically catalyze de novo mtDNA methylation. Of note, increased mtDNA methylation plays an important role in preventing oxidative stress-induced mtDNA damage. These findings advance our understanding of mitochondrial epigenetics and its role in maintaining mitochondrial homeostasis during early development.
Keywords: mitochondrial DNA, mitochondrial oxidative damage, de novo DNA methylation, DNMT3A/3B, peri-implantation
Abstract
Mitochondrial remodeling during the peri-implantation stage is the hallmark event essential for normal embryogenesis. Among the changes, enhanced oxidative phosphorylation is critical for supporting high energy demands of postimplantation embryos, but increases mitochondrial oxidative stress, which in turn threatens mitochondrial DNA (mtDNA) stability. However, how mitochondria protect their own histone-lacking mtDNA, during this stage remains unclear. Concurrently, the mitochondrial genome gain DNA methylation by this stage. Its spatiotemporal coincidence with enhanced mitochondrial stress led us to ask if mtDNA methylation has a role in maintaining mitochondrial genome stability. Herein, we report that mitochondrial genome undergoes de novo mtDNA methylation that can protect mtDNA against enhanced oxidative damage during the peri-implantation window. Mitochondrial genome gains extensive mtDNA methylation during transition from blastocysts to postimplantation embryos, thus establishing relatively hypermethylated mtDNA from hypomethylated state in blastocysts. Mechanistic study revealed that DNA methyltransferase 3A (DNMT3A) and DNMT3B enter mitochondria during this process and bind to mtDNA, via their unique mitochondrial targeting sequences. Importantly, loss- and gain-of-function analyses indicated that DNMT3A and DNMT3B are responsible for catalyzing de novo mtDNA methylation, in a synergistic manner. Finally, we proved, in vivo and in vitro, that increased mtDNA methylation functions to protect mitochondrial genome against mtDNA damage induced by increased mitochondrial oxidative stress. Together, we reveal mtDNA methylation dynamics and its underlying mechanism during the critical developmental window. We also provide the functional link between mitochondrial epigenetic remodeling and metabolic changes, which reveals a role for nuclear-mitochondrial crosstalk in establishing mitoepigenetics and maintaining mitochondrial homeostasis.
Mitochondrial remodeling is essential for early embryonic survival and development (1). During the transition from blastocysts to postimplantation embryos, a hallmark event is the notable increase in mitochondrial oxidative phosphorylation (OXPHOS), to support the increased energy demands of postimplantation embryos (1, 2). The developmental transition during peri-implantation is very important for successful pregnancy. In humans, ∼75% of failed pregnancies are due to developmental defects within this stage (3). Even in mice, implantation is a critical step in establishment of pregnancy (4). During this critical window, however, enhanced OXPHOS generates excessive reactive oxygen species (ROS), which in turn would have damaged the mitochondrial genome, thus threatening embryonic survival (1, 5, 6). Compared with histone-rich nuclear DNA, naked mitochondrial DNA (mtDNA) lacks histone and is more sensitive to oxidative damage (7, 8). Despite the induction of antioxidant defense systems during peri-implantation (6, 9), how mitochondria protect their own genome during this stage remains unclear. Coinciding with the metabolic remodeling, a remarkable epigenetic change also occurs in mitochondria during this stage: mitochondrial genome may acquire DNA methylation from the hypomethylated blastocysts, to establish relative hypermethylation of mtDNA in postimplantation embryos. Considering the functional role of DNA methylation in maintaining nuclear genome integrity (10, 11), spatiotemporal coincidence of mitochondrial epigenetic and metabolic changes, lead us to ask if increased mtDNA methylation functions to protect the mitochondrial genome against ROS-induced damage.
During the peri-implantation stage, previously published (12–14), and our own high-throughput DNA methylation data (Fig. 1D), implied that mitochondrial genome may undergo a wave of de novo mtDNA methylation from a hypomethylated state in blastocysts, and thus establishing relatively hypermethylated state in postimplantation embryos. Despite the importance of mitochondrial homeostasis during the developmental transition, however, this possibility has never been functionally determined. The complete mitochondrial genome contains 37 genes but does not encode enzymes with the de novo methyltransferase activity (15). Previous studies based on in vitro cellular experiments, demonstrated that nuclear-encoded methyltransferases can enter mitochondria, and thus maintaining or establishing mtDNA methylation (14, 16). By contrast, the essential role of methyltransferases in catalyzing mtDNA methylation, has never been confirmed via in vivo functional analyses. In particular, whether DNA de novo methyltransferases, i.e., DNMT3A and DNMT3B can translocate into mitochondria and catalyze de novo mtDNA methylation during peri-implantation, as well as the underlying mechanism, remains largely illusive.
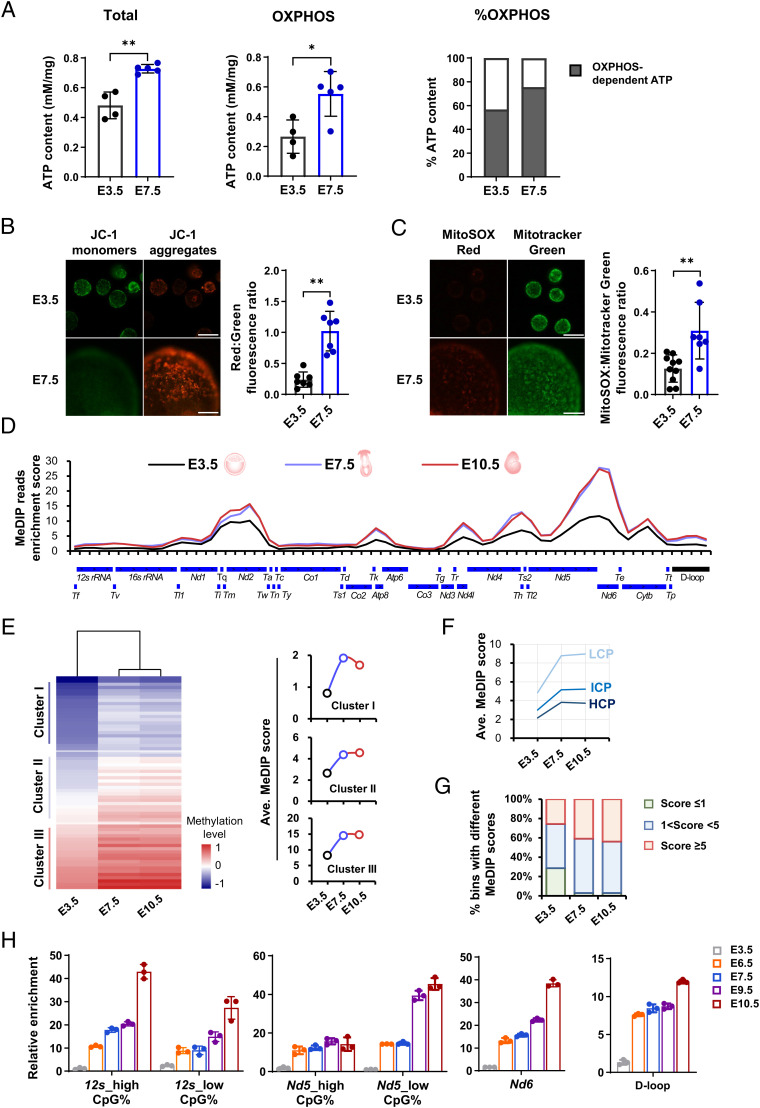
Fig. 1.
Spatiotemporal coincidence of de novo mtDNA methylation and mitochondrial oxidative stress during the peri-implantation window. (A) Comparisons of total ATP levels (Left), as well as OXPHOS-dependent ATP production (Middle) and relative contributions (Right) between E3.5 and E7.5 embryos. (B) Mitochondrial membrane potential in E3.5 and E7.5 embryo. Right: mitochondrial membrane potential by quantifying ratios of red (JC-1 aggregates): green (JC-1 monomer) fluorescence. (Scale bar: 100 μm.) (C) Fluorescent images of intramitochondrial ROS in E3.5 and E7.5 embryos detected by MitoSOX, a mitochondria-specific probe. Right: quantification of intramitochondrial ROS levels. (Scale bar: 100 μm.) (D) Methylation profiles of linearized mtDNA detected using MeDIP-seq across the mouse mitochondrial genome in E3.5, E7.5, and E10.5 embryos. The y axis shows the normalized enrichment of MeDIP-reads in each bin of 500 bp. The annotations below the linearized mtDNA map show the positions of mtDNA elements. (E) Heatmap analysis of all bins of the mitochondrial genome based on their MeDIP enrichment scores at three developmental stages. Right: mtDNA methylation dynamics of three subcategories of bins with different average enrichment scores at E3.5. (F) mtDNA methylation dynamics of three subcategories of bins with relatively high (≥ 2.6%, HCP), intermediate (> 1.2% and < 2.6%, ICP) or low (≤ 1.2%, LCP) CpG density. (G) Fraction of enriched bins with different enrichment scores at three developmental stages. (H) Relative methylation levels detected using MeDIP-qPCR in selected regions during consecutive developmental stages of peri-implantation, including the heavy- and light-strand protein coding gene (Nd5, Nd6), the noncoding gene (12s), and the control region (D-loop) at consecutive developmental stages. Data are presented as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01.
More importantly, enhanced OXPHOS during the transition from blastocysts to postimplantation embryos, results in the accumulation of intramitochondrial ROS (1). Given that the naked mtDNA lacks histone yet exists in an environment with high concentrations of ROS, mtDNA is very sensitive to ROS-induced damage (7, 8, 17). Spatiotemporal coincidence of enhanced mitochondrial oxidative stress and increased mtDNA methylation, is reminiscent of the nontranscriptional function of DNA methylation, i.e., maintaining genome stability (10, 11). It has been indicated that global hypomethylation is associated with increased genome instability in mammalian cells (18–20). Thus, we postulated that de novo mtDNA methylation during the developmental transition, might have a role in protecting the mitochondrial genome against oxidative damage in postimplantation embryos.
Herein, using Dnmt3a and Dnmt3b knockout mouse models, we confirmed the occurrence of de novo mtDNA methylation across the mitochondrial genome during the transition from blastocysts to postimplantation embryos, and identified the underlying mechanism: DNMT3A and DNMT3B can enter mitochondria, via their unique mitochondrial targeting sequences (MTS), and catalyze this process. In vivo and in vitro loss- and gain-of-functional analyses indicated that de novo mtDNA methylation contributes essentially to protect the mitochondrial genome against oxidative damage. Our results reveal the mtDNA methylation dynamics and its physiological role in early embryos, thus presenting insight into the crosstalk between nuclear and mitochondrial genomes that establishes mitoepigenetics, thus maintaining mitochondrial homeostasis.
Results and Discussion
Spatiotemporal Coincidence of De Novo mtDNA Methylation and Mitochondrial Oxidative Stress during the Peri-Implantation Window.
Reanalysis results of our previously published transcriptomic data (6, 21), showed the induction of genes controlling OXPHOS during the developmental transition from preimplantation blastocysts (embryonic day [E] 3.5, E3.5) to postimplantation epiblasts (E7.5) (SI Appendix, Fig. S1 A and B). In line with this, OXPHOS-dependent adenosine triphosphate (ATP) production, and its contribution to total ATP content, significantly increased during this stage (Fig. 1A). Accordingly, a significantly higher mitochondrial membrane potential, which maintains OXPHOS activity, can be detected in E7.5 epiblasts (Fig. 1B). Along with this, intramitochondrial ROS accumulation detected by MitoSOX, a mitochondria-specific probe (Fig. 1C), were significantly elevated during the developmental transition. These observations were in line with the up-regulation of genes related to mtDNA repair (SI Appendix, Fig. S1C), suggesting that mitochondria suffer from considerable oxidative stress during this stage.
Coinciding with notable changes in mitochondrial metabolism during the peri-implantation window, remarkable epigenetic remodeling also occurs in mitochondria. To systematically characterize its dynamics and features, we profiled mtDNA methylation patterns of mouse E3.5 blastocysts, E7.5 epiblasts, and E10.5 embryos using methylated DNA immunoprecipitation-sequencing (MeDIP-seq). These time points represent the developmental windows of de novo DNA methylation of the nuclear genome (22–24). Notably, distinct from that in the nuclear genome, 5hmC enrichment in the mitochondrial genome is up to 100-fold higher relative to 5mC enrichment (16). Therefore, bisulfite-based methods are thought to be technically unfavorable to detect dynamic changes of 5mC in mtDNA, because 5mC is indistinguishable from 5hmC by bisulfite modification (25). In addition, the accuracy of bisulfite sequencing to detect mtDNA methylation levels remains controversial because of incomplete C-to-T conversion, which might lead to overestimation (26, 27). Based on these considerations, we selected MeDIP-seq using a specific antibody against 5mC, to analyze mtDNA methylation dynamics and features during the critical developmental window.
Based on the technically reliable MeDIP-seq data (SI Appendix, Fig. S2A), we first profiled mtDNA methylation dynamics using reliable and reproducible reads mapping to the mitochondrial genome (SI Appendix, Fig. S2B and Dataset S1). We found that during the developmental transition from blastocysts to postimplantation embryos, the mtDNA methylation levels increased throughout the mitochondrial genome, albeit to varying degrees (Fig. 1D). In line with this, the heatmap showed more detailed information of mtDNA methylation dynamics: nearly all regions, regardless of their basal levels in blastocysts, displayed a notable increase in mtDNA methylation levels (Fig. 1E), and the increase covered not only the coding and noncoding regions, but also the control region (D-loop) of the mitochondrial genome (SI Appendix, Fig. S2C). It is also noteworthy that mtDNA methylation levels appeared to be inversely correlated with CpG density at each developmental stage (SI Appendix, Fig. S2 D and E), thus de novo mtDNA methylation tended to occur within regions with relatively low CpG density (Fig. 1F). The CpG-density-dependent mtDNA methylation pattern is consistent with results reported for the nuclear genome, particularly in postimplantation embryos and somatic cells (22, 28), and accords with the previous finding that mtDNA methylation is more likely to occur within non-CpG nucleotides (29–31). We next examined the fraction of regions that exhibited relatively high, intermediate, or low methylation scores in each developmental stage, and observed a clear increase in the fraction of regions that exhibited high methylation scores from blastocysts to postimplantation embryos, as well as a dramatic decrease in the fraction of regions that exhibited low methylation scores (Fig. 1G).
Finally, we validated the increase in mtDNA methylation from blastocysts to postimplantation embryos using MeDIP-qPCR, using a highly sensitive and specific anti-5mC antibody. During consecutive developmental stages of peri-implantation, all detected regions displayed an increase in the mtDNA methylation level to varying degrees from the hypomethylated state in blastocysts (Fig. 1H). This increase occurred irrespective of the type of mtDNA regions we detected: i.e., the protein coding regions (heavy-strand Nd5, light-strand Nd6), the noncoding region (12s), or the control region (D-loop). We also found that the increase in mtDNA methylation appeared to be asynchronous among different regions, as shown previously for the nuclear genome (22, 24). Collectively, these results demonstrated that the mitochondrial genome undergoes de novo methylation during the developmental transition from blastocysts to postimplantation embryos, which spatiotemporally coincides with enhanced mitochondrial OXPHOS and oxidative stress. Moreover, by reanalyzing the previously published high-throughput DNA methylation data of human early embryos (32) and mouse early embryonic and extraembryonic lineages (33), we found the occurrence of de novo mtDNA methylation, although limited, seems to be common among species and different lineages during the peri-implantation window (SI Appendix, Fig. S3 A–D).
DNMT3A and DNMT3B Translocate into Mitochondria and Catalyze De Novo mtDNA Methylation during the Developmental Transition.
We next attempted to explore the mechanism underlying de novo mtDNA methylation. Given that the mitochondrial genome does not encode enzymes with the de novo methyltransferase activity, we asked if nuclear-encoded DNMT3A and DNMT3B, the main enzymes catalyzing nuclear de novo methylation, are responsible for de novo mtDNA methylation. To this end, we first detected the mitochondrial localization of DNMT3A and DNMT3B via high-quality 3D-microscopic reconstruction using superresolution immunofluorescence analysis. The results indicated that both DNMT3A and DNMT3B were frequently localized to the mitochondrial matrix or membrane by implantation and postimplantation stages, but rarely by preimplantation stage (Fig. 2A and B). The mitochondrial localization of DNMT3A and DNMT3B was also supported by immunoelectron microscopy, in which they appeared as clustered or single colloidal gold particles (Fig. 2C). To further confirm the mitochondrial localization of DNMT3A and DNMT3B, we purified mitochondria. After excluding possible contaminants from the nuclear and cytosolic fractions, in which DNMT3A and DNMT3B preferentially reside, we detected the presence of these two de novo methyltransferases, albeit at relatively low levels, in the purified mitochondria (Fig. 2D). These results demonstrated that DNMT3A and DNMT3B can enter mitochondria during the process of de novo mtDNA methylation.
Fig. 2.
DNMT3A and DNMT3B translocate into mitochondria and catalyze de novo mtDNA methylation during the developmental transition. (A and B) High-quality 3D-microscopic reconstruction of the interactions between TOM20-labeled mitochondria (ring-shaped cross section) and DNMT3A (A) or DNMT3B (B) in E3.5, E7.5, and E10.5 embryos using superresolution immunofluorescence analysis. (Scale bar: 250 nm.) Right: Fluorescence intensity profiles cross the white line in the merged picture. (C) Mitochondrial localization of DNMT3A and DNMT3B (revealed by clustered or single colloidal gold particles) detected using immunoelectron microscopy with gold-coupled antibodies in E7.5 embryos, (Scale bar: 100 nm.) (D) Western blotting analysis of DNMT3A and DNMT3B in purified mitochondria from E9.5 and E10.5 embryos (using earlier stage embryos was technically unfeasible because of the high number of embryos required). Nuclear (N), cytosolic (C), and mitochondrial (M) protein fractions were detected with antibodies against nuclear (H3K27me3)-, mitochondrial (voltage-dependent anion carrier, VDAC)-, and cytosolic (β-tubulin)-specific markers, respectively, to exclude the possible contamination of purified mitochondria with the nuclear or cytosolic fraction. (E and F) Chromatin immunoprecipitation analysis of the DNMT3A-mtDNA (E) and DNMT3B-mtDNA (F) physical interactions in selected regions during the peri-implantation stage. (G) Relative methylation levels detected using MeDIP-qPCR in selected regions in wild-type, Dnmt3a−/−, and Dnmt3b−/− E10.5 embryos. Data show the means ± SD of three independent experiments. **P < 0.01.
Physical binding of DNMT3A or DNMT3B to the hypomethylated DNA region is a prerequisite for catalyzing de novo methylation. Therefore, we next detected whether DNMT3A and DNMT3B could interact directly with mtDNA after entering the mitochondrial matrix. Using high-sensitivity immunoprecipitation, we detected the binding of DNMT3A and DNMT3B peaked by postimplantation stage (Fig. 2E and F), which accords with the relatively hypermethylated mtDNA observed in postimplantation embryos (Fig. 1H). From this stage onwards, most of the binding tended to decline (Fig. 2E and F). Collectively, we proved, using multiple methodologies, that DNMT3A and DNMT3B can translocate into mitochondria and bind to mtDNA during the process of de novo mtDNA methylation. Although DNMT3A and DNMT3B were previously reported to be absent in mitochondria from mouse embryonic fibroblasts and HCT116 human colon carcinoma cells (16), follow-up studies provided evidence that DNMT3A and DNMT3B were localized in the mitochondria or detectable in the mitochondrial protein fraction in human and mouse central nervous tissues, skeletal muscle, as well as in HeLa and 3T3-L1 cells (29, 34, 35). Our results, together with these findings, suggest that DNMT3A and DNMT3B translocate into mitochondria in a cell type- and developmental stage-dependent manner.
To finally determine the role of DNMT3A or DNMT3B in catalyzing de novo mtDNA methylation, we measured mtDNA methylation levels in Dnmt3a−/− and Dnmt3b−/− E10.5 embryos, in which relatively stable mtDNA methylation patterns should been established. Deficiency of either Dnmt3a or Dnmt3b had no effect on mitochondrial mass (SI Appendix, Fig. S4 A and B); however, it did result in severe mtDNA hypomethylation, irrespective of the type of region detected (Fig. 2G), suggesting that DNMT3A and DNMT3B might catalyze de novo mtDNA methylation in a synergistic manner. These results, together with mitochondrial location of DNMT3A and DNMT3B, suggest that de novo mtDNA methylation depends on both DNMT3A and DNMT3B. This may be explained by the mitochondrial genome-wide distribution of DNMT3A or DNMT3B-preferred sites (SI Appendix, Fig. S4C). In agreement with this, previous studies have demonstrated that not only Dnmt3a knockout alone, but also Dnmt3a/Dnmt3b double knockout and Dnmt1/Dnmt3a/Dnmt3b triple knockout mouse embryonic stem cells showed a significant reduction in regional or global mtDNA methylation levels (14, 29). Similarly, a recent study clarified that knockdown of each DNMT, especially DNMT3B, in human breast epithelial cells markedly reduced mtDNA methylation levels (31), suggesting DNMT3A and DNMT3B might display functional synergism in catalyzing de novo mtDNA methylation. This notion is somewhat different to that occurring in the nuclear genome. It has been well-established that DNMT3B is the main enzyme responsible for nuclear de novo methylation around the implantation period, and DNMT3A cooperates with DNMT3B to methylate its targets (23, 24). More recently, however, a study revealed that a unique proportion of nuclear genomic regions showed both DNMT3A and DNMT3B-dependent DNA methylation (36). In line with this, earlier studies, focusing on specific sites, also showed the synergistic function of DNMT3A and DNMT3B in methylating the promoters of Oct4, Nanog, and hematopoietic stem cell-specific genes (37, 38), similar to our observations in the mitochondrial genome.
DNMT3A and DNMT3B Contain a Functional MTS that Promotes Efficient Mitochondrial Translocation.
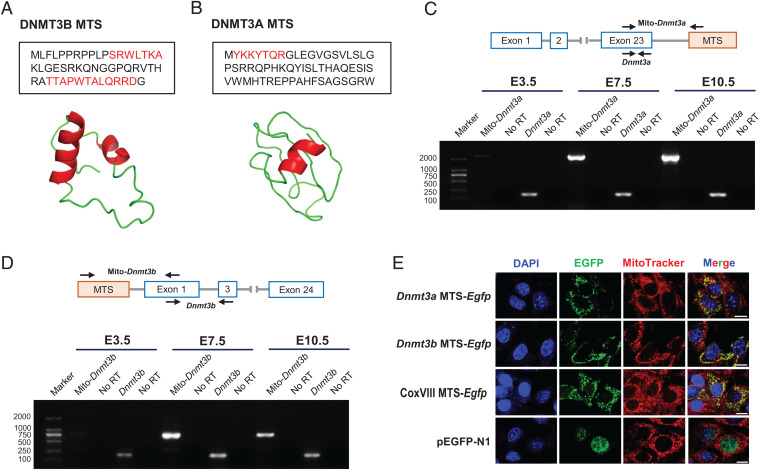
We next asked how DNMT3A and DNMT3B enter mitochondria. Nuclear-encoded mitochondrial proteins usually possess an MTS in their N terminus. This MTS forms a cleavable amphiphilic helical structure that facilitates the import of precursor proteins into the mitochondrial matrix (39). Although the mitochondrial location of DNMT3A and DNMT3B has been reported (29, 34, 35), the mechanism responsible for the location has never been identified (14). By examining the 5′-flanking regions of the published coding sequence of mouse Dnmt3a and Dnmt3b, we found an in-frame potential MTS coding region equivalent to 50 codons, starting with the ATG, which might encode a peptide that forms an amphiphilic helix, in the 5′-flanking region of Dnmt3b (Fig. 3A and SI Appendix, Fig. S4A), but not Dnmt3a. Of note, compared with exclusive mitochondrial proteins, mitochondrial proteins that can be located in different subcellular compartments, especially those preferentially residing in the nucleus with a conditional distribution in the mitochondria, always harbor an unconventional MTS. These unconventional MTSs are usually difficult to predict using bioinformatic analysis because they are likely to have lower prediction scores and might be located in the C- terminus (40, 41). Thus, we next examined the 3′-flanking regions, and detected three low-scoring potential MTSs in the 3′-flanking region of Dnmt3a. Among these, an MTS equivalent to 60 codons was predicted to have significant helical amphiphilicity (Fig. 3B and SI Appendix, Fig. S5A). In addition, the detection of transcripts containing the predicted MTS preferentially in postimplantation embryos (Fig. 3 C and D and SI Appendix, Fig. S5B), as well as the results that these transcripts were initially activated during the transition from blastocysts to epiblasts (SI Appendix, Fig. S5C), not only gives an explanation for the stage-dependent mitochondrial translocation of DNMT3A and DNMT3B isoforms, but also may explain why the mitochondrial genome gain DNA methylation during this developmental transition.
Fig. 3.
DNMT3A and DNMT3B contain a functional MTS that promotes efficient mitochondrial translocation. (A and B) Ribbon models of the representative α-helical structure (red) and corresponding residues (red text) in the MTS of DNMT3B (A) and DNMT3A (B). The structure was modeled using the I-TASSER server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). (C and D) Agarose gel electrophoresis of RT-PCR products amplified from mature transcripts capable of encoding the MTS of DNMT3A (C) and DNMT3B (D) in E3.5, E7.5, and E10.5 embryos. No RT, no reverse transcriptase control. (E) Mitochondrial translocation of EGFP directed by the predicted MTS of DNMT3A and DNMT3B in transiently transfected NIH/3T3 cells. MitoTracker Red was used to label the mitochondria. The commercialized vector containing an MTS from the COX subunit VIII and pEGFP-N1 were used as the positive control and negative control, respectively. (Scale bar: 10 μm.)
Finally, we functionally determined the efficacy of predicted MTS. When cloned in-frame into pEGFP-N1 with the C-terminal enhanced green fluorescent protein (EGFP), both potential Dnmt3a and Dnmt3b MTS, similar to the well-established CoxVIII MTS, could successfully direct EGFP to the mitochondria in either NIH/3T3 or HEK293T cell, as revealed by the colocalization of MitoTracker Red with EGFP (Fig. 3E and SI Appendix, Fig. S5D). Thus, these results indicated that both DNMT3A and DNMT3B have their own unconventional, but efficient, MTSs, which are responsible for the mitochondrial translocation of DNMT3A and DNMT3B during the process of de novo mtDNA methylation.
De Novo mtDNA Methylation Functions to Protect the Mitochondrial Genome against Oxidative Damage.
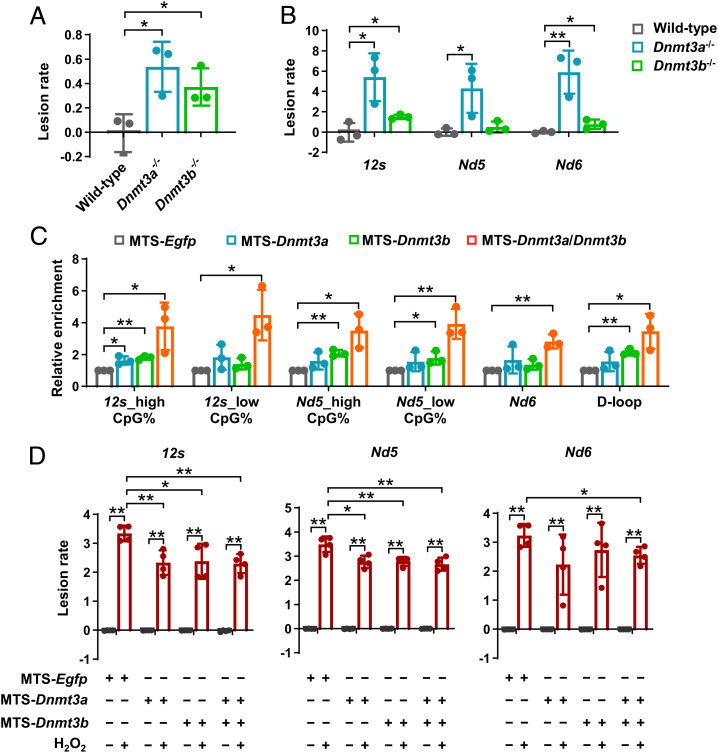
Having demonstrated the essential role of DNMT3A and DNMT3B in catalyzing de novo mtDNA methylation and its underlying mechanism, we next investigated the developmental significance of de novo mtDNA methylation during early embryogenesis. Many previous studies have focused on the functional link between mtDNA methylation and gene expression or copy number; however, the results varied markedly or even contradicted each other among different studies. This is reminiscent of the important role of DNA methylation in maintaining genome stability (10, 11). Our data have shown that postimplantation embryos underwent an increase in mitochondrial aerobic metabolism, as well as intramitochondrial ROS accumulation arising from enhanced OXPHOS (Fig. 1 A–C and SI Appendix, Fig. S1 A and B). Given that mitochondria are both the major source and the sensitive target of ROS, mtDNA by this stage will suffer from significant oxidative stress that might induce mtDNA damage. This notion is also supported by the result of exogenous H2O2 treatment that mimics mitochondrial oxidative stress, which significantly induced mtDNA damage in postimplantation embryos (SI Appendix, Fig. S6 A and B). Thus, spatiotemporal coincidence of de novo mtDNA methylation and mitochondrial oxidative stress, led us to postulated that de novo mtDNA methylation might participate in protecting the mitochondrial genome against oxidative damage in postimplantation embryos. Quantification of mtDNA damage scores supported our hypothesis: compared with their wild-type counterparts, both Dnmt3a−/− and Dnmt3b−/− embryos showed significant increases in mtDNA damage throughout the mitochondrial genome (Fig. 4 A and B). Of note, increased mtDNA damage was not due to the effect of Dnmts deficiency per se. Possible changes that may lead to mtDNA damage were excluded: i.e., OXPHOS was inhibited (SI Appendix, Fig. S6 C–E) in Dnmt3b-deficient postimplantation embryos. In addition, Dnmt3b deficiency did not induce mitochondrial oxidative stress (SI Appendix, Fig. S6F).
Fig. 4.
De novo mtDNA methylation functions to protect the mitochondrial genome against oxidative damage. (A and B) Quantitative measurement of mtDNA damage within the long-range region (A) and selected regions (B) in wild-type, Dnmt3a−/−, and Dnmt3b−/− postimplantation embryos. MtDNA damage is expressed relative to the wild-type (Zero-class lesions). (C) Effect of mitochondria-targeted co-overexpression of Dnmt3a and Dnmt3b, as well as overexpression of Dnmt3a or Dnmt3b alone, on the relative methylation levels in selected regions in NIH/3T3 cells. Mitochondria-targeted EGFP was transfected in parallel as the control. (D) Effect of mitochondria-targeted co-overexpression of Dnmt3a and Dnmt3b, as well as overexpression of Dnmt3a or Dnmt3b alone, on protecting mtDNA against H2O2-induced oxidative damage in NIH/3T3 cells. mtDNA damage (lesion rate) is expressed relative to the control (normalized to zero-class lesions). Data are presented as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01.
Next, we attempted to further confirmed the role of elevated mtDNA methylation levels in protecting the mitochondrial genome against oxidative damage using an in vitro model. To this end, we established the model via mitochondria-targeted co-overexpression of Dnmt3a and Dnmt3b, as well as targeted overexpression of Dnmt3a or Dnmt3b alone in NIH/3T3 fibroblasts (SI Appendix, Fig. S7 A–C), which are sensitive to H2O2-induced mitochondrial oxidative stress and mtDNA damage (SI Appendix, Fig. S7 D and E). Our results indicated that hypermethylated mtDNA showed significantly reduced mtDNA damage under H2O2-induced oxidative stress (Fig. 4 C and D). Note that co-overexpression of Dnmt3a and Dnmt3b was more effective in elevating mtDNA methylation levels and protecting against mtDNA damage, compared with overexpressing Dnmt3a or Dnmt3b alone (Fig. 4 C and D), which further supported the concept of functional synergism between DNMT3A or DNMT3B in catalyzing de novo mtDNA methylation.
Additionally, the protective role of de novo mtDNA methylation against oxidative damage, raised the possibility that nuclear de novo DNA methylation may also participate in the protection of nuclear genome. Our results from knockout embryos and in vitro model, indicated that elevated nuclear DNA methylation is also important for maintaining genome stability in the nucleus (SI Appendix, Fig. S8 A and B). In line with this, we noticed that glutathione antioxidant system and DNA repair pathway were notably activated during the peri-implantation window (SI Appendix, Fig. S8 C and D), implying that embryos must face a comprehensive intracellular oxidative stress during the critical developmental transition, and a series of redundant mechanisms may be activated by this stage and participate in the protection of both the mitochondrial and nuclear genomes. Although the mechanism responsible for the contribution of cytosine methylation to genome stability remains controversial, possible explanations include increased DNA condensation and flexibility (42–44). In particular, mtDNA methylation may increase the binding affinity of mitochondrial transcription factor A (TFAM), and thus forming the TFAM-DNA complex, which in turn, alters the packaging and deformability of mtDNA (45, 46).
The presence of 5mC in mtDNA is a hallmark finding that advances the knowledge of mitochondrial biology and supports the novel concept of mitoepigenetics (16). Follow-up studies also confirmed mtDNA methylation among myriads of cell types (12, 16, 29, 47, 48), and evidence from clinical patients and mouse models further suggested that changes in mtDNA methylation are associated with pathogenesis of many diseases (31, 34, 49–52) and aging (53, 54). Despite these findings, the physiological role of mtDNA methylation, especially its developmental significance, remains unknown before the present study. Our results from the in vitro and in vivo models, suggested that de novo mtDNA methylation contributes to protecting the mitochondrial genome against oxidative damage. Our finding is in line with previous observations in the nuclear genome, in which global or regional hypomethylation are associated with an increased frequency of DNA damage of antioncogenes, and are implicated in carcinogenesis (55, 56).
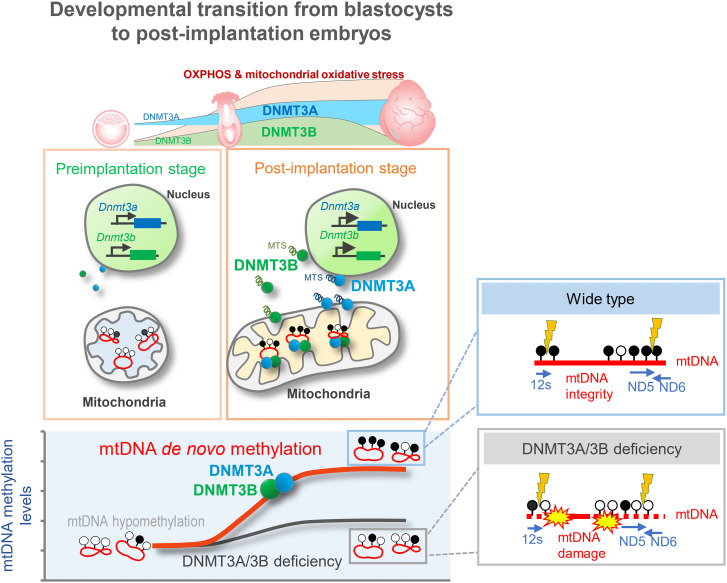
In summary, our data provide direct evidence that the mitochondrial genome undergoes a wave of extensive de novo methylation during the developmental transition from blastocysts to postimplantation embryos. During this developmental window, DNMT3A and DNMT3B translocate into the mitochondrial matrix and bind mtDNA to catalyze de novo methylation of the hypomethylated mitochondrial genome. We also identified that DNMT3A and DNMT3B contain their own functional MTSs that promote efficient mitochondrial translocation. Our results also suggested that established mtDNA methylation in postimplantation embryos functions to protect the mitochondrial genome against oxidative damage (Fig. 5), provide the functional link between mitochondrial epigenetic remodeling and metabolic changes. Given that nearly all previous related studies only focused on mtDNA methylation in differentiated or adult somatic cells, our current findings, focusing on early embryos, answer the question regarding the origins of previously identified 5mC in mtDNA. Furthermore, focusing on the essential role of nuclear encoded epigenetic enzymes on mitoepigenetics and mtDNA stability, the present study advances current knowledge of the crosstalk that couples the nuclear and mitochondrial genomes, and the role of this crosstalk in maintaining mitochondrial homeostasis.
Fig. 5.
A model illustrating the dynamics, mechanism, and functional consequence of de novo mtDNA methylation that occurs during the developmental transition from blastocysts to postimplantation embryos.
Materials and Methods
Animal Studies and Ethical Approval.
All female ICR mice used in this study were obtained from SPF Biotechnology Co. Ltd. Heterozygous Dnmt3a+/− and Dnmt3b+/− mice that carry one mutant locus of Dnmt3a and Dnmt3b, respectively (23), were obtained from the NIH mutant mouse research and resource center (MMRRC, NIH). All mice were maintained in a climate-controlled room on a 12-h light/dark cycle and allowed food and water ad libitum. The China Agricultural University Institutional Animal Care and Use Committee approved this study, which was performed in accordance with the committee’s guidelines. All efforts were made to minimize animal suffering.
Preparation and Collection of Mouse Embryos.
Embryos were obtained by natural breeding. ICR female mice (7–8 wk old) were cocaged individually with ICR males (10–12 wk old). The morning of the appearance of a vaginal plug was designated E0.5. Embryos were collected at selected stages. The criteria for sampling embryos were based on the developmental progress and morphology. Embryos showing typical morphological features according to the well-established landmarks were sampled for further analyses, as reported by the well-established guidance (57) and previous studies (21, 24, 58). At E3.5, well-developed late-cavitating blastocysts were obtained by flushing the uterus with M2 medium (Sigma-Aldrich). At E6.5 and E7.5, the conceptuses covered with the decidual mass were gently teased away from the uterus. The decidua in which the conceptus embedded was peeled off, and the parietal yolk sac was opened to expose the visceral yolk sacendoderm layer. The well-developed epiblast was separated from extraembryonic tissues using microdissecting watchmaker’s forceps under a stereomicroscope. To ensure accurate sample collection and avoid cross-contamination between embryonic and extraembryonic tissues, pre-experiments have been performed using qRT-PCR and immunostaining for markers specific to the epiblast (59) before large-scale sampling. All sampled embryos were washed serially using phosphate-buffered saline (PBS; Gibco) containing 0.1% polyvinyl alcohol (PBS-PVA) (Sigma-Aldrich) and stored immediately in liquid nitrogen for further use.
Statistical Analysis.
Statistical tests were performed on SPSS 26.0 Windows version of software. Experiments were independently replicated for a minimum of three times unless otherwise specified, and all data are reported as the mean ± SD. Student’s t test (for two groups) was used to analyze the significant difference between the treatment and control groups. For multiple comparisons, data were analyzed by one-way ANOVA followed by Tukey’s multiple comparison test to compare selected pairs of experimental groups. Statistically significant differences were defined as P < 0.05.
Supplementary Material
Acknowledgments
We thank Zhiyuan Chen (Boston Children’s Hospital) for reading the manuscript and providing helpful comments. We thank Dr. Jinyu Wang (Tsinghua University) for technical assistance in super-resolution immunofluorescence analysis. We also thank E-GENE (Shenzhen) for technical assistance of bisulfite sequence data analyzing. This work was supported by grants from the National Natural Science Foundation of China (grant Number 31930103), Chinese Universities Scientific Fund (Grant Number 2022TC019), National Key R&D Program (Grant Numbers 2017YFD0501901 and 2017YFD0501905), and the Beijing Innovation Consortium of Agriculture Research System.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201168119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Mathieu J., Ruohola-Baker H., Metabolic remodeling during the loss and acquisition of pluripotency. Development 144, 541–551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J., et al. , Metabolism in pluripotent stem cells and early mammalian development. Cell Metab. 27, 332–338 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Norwitz E. R., Schust D. J., Fisher S. J., Implantation and the survival of early pregnancy. N. Engl. J. Med. 345, 1400–1408 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Dorsch M., Wittur I., Garrels W., Success of embryo transfer in mice with freshly collected and cryopreserved two-cell embryos with different genetic backgrounds correlated with the number of transferred embryos: A 5-year retrospective analysis. Lab. Anim. 53, 577–586 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Ramalho-Santos J., et al. , Mitochondrial functionality in reproduction: From gonads and gametes to embryos and embryonic stem cells. Hum. Reprod. Update 15, 553–572 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Ren L., et al. , High-resolution profiles of gene expression and DNA methylation highlight mitochondrial modifications during early embryonic development. J. Reprod. Dev. 63, 247–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yakes F. M., Van Houten B., Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 94, 514–519 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dakubo G. D., Parr R. L., Costello L. C., Franklin R. B., Thayer R. E., Altered metabolism and mitochondrial genome in prostate cancer. J. Clin. Pathol. 59, 10–16 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen J. M., Harris C., Glutathione during embryonic development. Biochim. Biophys. Acta 1850, 1527–1542 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Senner C. E., The role of DNA methylation in mammalian development. Reprod. Biomed. Online 22, 529–535 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Weber M., Schübeler D., Genomic patterns of DNA methylation: Targets and function of an epigenetic mark. Curr. Opin. Cell Biol. 19, 273–280 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Sirard M. A., Distribution and dynamics of mitochondrial DNA methylation in oocytes, embryos and granulosa cells. Sci. Rep. 9, 11937 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi H., et al. , Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 8, e1002440 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dou X., et al. , The strand-biased mitochondrial DNA methylome and its regulation by DNMT3A. Genome Res. 29, 1622–1634 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harvey A., Gibson T., Lonergan T., Brenner C., Dynamic regulation of mitochondrial function in preimplantation embryos and embryonic stem cells. Mitochondrion 11, 829–838 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Shock L. S., Thakkar P. V., Peterson E. J., Moran R. G., Taylor S. M., DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl. Acad. Sci. U.S.A. 108, 3630–3635 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savu O., et al. , Stability of mitochondrial DNA against reactive oxygen species (ROS) generated in diabetes. Diabetes Metab. Res. Rev. 27, 470–479 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Dodge J. E., et al. , Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J. Biol. Chem. 280, 17986–17991 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Karpf A. R., Matsui S., Genetic disruption of cytosine DNA methyltransferase enzymes induces chromosomal instability in human cancer cells. Cancer Res. 65, 8635–8639 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Xu G. L., et al. , Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402, 187–191 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Ren L., et al. , Dynamic comparisons of high-resolution expression profiles highlighting mitochondria-related genes between in vivo and in vitro fertilized early mouse embryos. Hum. Reprod. 30, 2892–2911 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Smith Z. D., et al. , A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484, 339–344 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okano M., Bell D. W., Haber D. A., Li E., DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Borgel J., et al. , Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 42, 1093–1100 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Huang Y., et al. , The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 5, e8888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owa C., Poulin M., Yan L., Shioda T., Technical adequacy of bisulfite sequencing and pyrosequencing for detection of mitochondrial DNA methylation: Sources and avoidance of false-positive detection. PLoS One 13, e0192722 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B., et al. , CpG methylation patterns of human mitochondrial DNA. Sci. Rep. 6, 23421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith Z. D., et al. , DNA methylation dynamics of the human preimplantation embryo. Nature 511, 611–615 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellizzi D., et al. , The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res. 20, 537–547 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bianchessi V., et al. , Methylation profiling by bisulfite sequencing analysis of the mtDNA Non-Coding Region in replicative and senescent Endothelial Cells. Mitochondrion 27, 40–47 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Patil V., et al. , Human mitochondrial DNA is extensively methylated in a non-CpG context. Nucleic Acids Res. 47, 10072–10085 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu P., et al. , Single-cell DNA methylome sequencing of human preimplantation embryos. Nat. Genet. 50, 12–19 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Smith Z. D., et al. , Epigenetic restriction of extraembryonic lineages mirrors the somatic transition to cancer. Nature 549, 543–547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong M., Gertz B., Chestnut B. A., Martin L. J., Mitochondrial DNMT3A and DNA methylation in skeletal muscle and CNS of transgenic mouse models of ALS. Front. Cell. Neurosci. 7, 279 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chestnut B. A., et al. , Epigenetic regulation of motor neuron cell death through DNA methylation. J. Neurosci. 31, 16619–16636 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Z., Yin Q., Inoue A., Zhang C., Zhang Y., Allelic H3K27me3 to allelic DNA methylation switch maintains noncanonical imprinting in extraembryonic cells. Sci. Adv. 5, eaay7246 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Challen G. A., et al. , Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell 15, 350–364 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J. Y., et al. , Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol. Cell. Biol. 27, 8748–8759 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neupert W., Herrmann J. M., Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Li M., et al. , Identification and characterization of mitochondrial targeting sequence of human apurinic/apyrimidinic endonuclease 1. J. Biol. Chem. 285, 14871–14881 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dinur-Mills M., Tal M., Pines O., Dual targeted mitochondrial proteins are characterized by lower MTS parameters and total net charge. PLoS One 3, e2161 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoo J., Kim H., Aksimentiev A., Ha T., Direct evidence for sequence-dependent attraction between double-stranded DNA controlled by methylation. Nat. Commun. 7, 11045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang H., et al. , Sequence-dependent DNA condensation as a driving force of DNA phase separation. Nucleic Acids Res. 46, 9401–9413 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Y. J., et al. , Cytosine Methylation Enhances DNA Condensation Revealed by Equilibrium Measurements Using Magnetic Tweezers. J. Am. Chem. Soc. 142, 9203–9209 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Dostal V., Churchill M. E. A., Cytosine methylation of mitochondrial DNA at CpG sequences impacts transcription factor A DNA binding and transcription. Biochim. Biophys. Acta. Gene Regul. Mech. 1862, 598–607 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larsson N. G., et al. , Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 18, 231–236 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Ghosh S., Sengupta S., Scaria V., Comparative analysis of human mitochondrial methylomes shows distinct patterns of epigenetic regulation in mitochondria. Mitochondrion 18, 58–62 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Devall M., et al. , Regional differences in mitochondrial DNA methylation in human post-mortem brain tissue. Clin. Epigenetics 9, 47 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pirola C. J., et al. , Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut 62, 1356–1363 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Baccarelli A. A., Byun H. M., Platelet mitochondrial DNA methylation: A potential new marker of cardiovascular disease. Clin. Epigenetics 7, 44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blanch M., Mosquera J. L., Ansoleaga B., Ferrer I., Barrachina M., Altered Mitochondrial DNA Methylation Pattern in Alzheimer Disease-Related Pathology and in Parkinson Disease. Am. J. Pathol. 186, 385–397 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Gao J., Wen S., Zhou H., Feng S., De-methylation of displacement loop of mitochondrial DNA is associated with increased mitochondrial copy number and nicotinamide adenine dinucleotide subunit 2 expression in colorectal cancer. Mol. Med. Rep. 12, 7033–7038 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Mawlood S. K., Dennany L., Watson N., Dempster J., Pickard B. S., Quantification of global mitochondrial DNA methylation levels and inverse correlation with age at two CpG sites. Aging (Albany NY) 8, 636–641 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dzitoyeva S., Chen H., Manev H., Effect of aging on 5-hydroxymethylcytosine in brain mitochondria. Neurobiol. Aging 33, 2881–2891 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki K., et al. , Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell 9, 199–207 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Kim Y. I., et al. , Folate deficiency in rats induces DNA strand breaks and hypomethylation within the p53 tumor suppressor gene. Am. J. Clin. Nutr. 65, 46–52 (1997). [DOI] [PubMed] [Google Scholar]
- 57.Behringer R., Gertsenstein M., Nagy K. V., Nagy A., Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2014). [Google Scholar]
- 58.Tan K., et al. , Impaired imprinted X chromosome inactivation is responsible for the skewed sex ratio following in vitro fertilization. Proc. Natl. Acad. Sci. U.S.A. 113, 3197–3202 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li L., et al. , Location of transient ectodermal progenitor potential in mouse development. Development 140, 4533–4543 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.