Significance
Like most human pathogens, the malaria parasite Plasmodium falciparum experiences strong selection pressure from public health interventions such as drug treatment. While most commonly studied in the context of drug targets and related pathways, parasite adaptation to control measures likely extends to phenotypes beyond drug resistance. Here, we use modeling to explore how control measures can reduce levels of within-host competition between P. falciparum genotypes and favor higher rates of sexual investment. We validate these predictions with longitudinally sampled genomic data from French Guiana during a period of malaria decline and find that the most strongly selected genes are enriched for transcription factors involved in commitment to and development of the parasite’s sexual gametocyte form.
Keywords: malaria, sexual commitment, adaptation, modeling, genomics
Abstract
Successful infectious disease interventions can result in large reductions in parasite prevalence. Such demographic change has fitness implications for individual parasites and may shift the parasite’s optimal life history strategy. Here, we explore whether declining infection rates can alter Plasmodium falciparum’s investment in sexual versus asexual growth. Using a multiscale mathematical model, we demonstrate how the proportion of polyclonal infections, which decreases as parasite prevalence declines, affects the optimal sexual development strategy: Within-host competition in multiclone infections favors a greater investment in asexual growth whereas single-clone infections benefit from higher conversion to sexual forms. At the same time, drug treatment also imposes selection pressure on sexual development by shortening infection length and reducing within-host competition. We assess these models using 148 P. falciparum parasite genomes sampled in French Guiana over an 18-y period of intensive intervention (1998 to 2015). During this time frame, multiple public health measures, including the introduction of new drugs and expanded rapid diagnostic testing, were implemented, reducing P. falciparum malaria cases by an order of magnitude. Consistent with this prevalence decline, we see an increase in the relatedness among parasites, but no single clonal background grew to dominate the population. Analyzing individual allele frequency trajectories, we identify genes that likely experienced selective sweeps. Supporting our model predictions, genes showing the strongest signatures of selection include transcription factors involved in the development of P. falciparum’s sexual gametocyte form. These results highlight how public health interventions impose wide-ranging selection pressures that affect basic parasite life history traits.
Due to its clinical relevance, the development of drug resistance is often the focus of studies of malaria parasite evolution, but over the past decade it has become clear that there are multiple ways Plasmodium falciparum (P. falciparum) continues to adapt to humans, mosquitoes, and the environment (1–4). This adaptability means that parasites like Plasmodium are moving targets, whose evolution must be incorporated into control and elimination campaigns. One area of interest is the adaptive potential of life history traits, which could impact growth and virulence within the human host as well as the timing and extent of transmission to mosquitoes (5).
Plasmodium parasites have a complex life cycle that requires two successive hosts. Only the parasite’s sexual gametocyte form can productively infect mosquitoes (the definitive host where reproduction occurs); however, this form cannot further replicate within the human (intermediate) host. A trade-off therefore exists between investing in asexual blood-stage growth, which sustains the parasite’s short-term survival, and producing sexual forms, which are required for reproduction and further human transmission. While this trade-off is expected to vary across hosts and environments (6), it has been difficult to study since accurately assessing the parasite’s sexual investment strategy is challenging. Conversion rate, defined as the proportion of asexual parasites that convert into gametocytes at each round of blood-stage replication, is difficult to quantify (7). Nevertheless, multiple approaches have documented phenotypic variation in gametocyte production among genotypic backgrounds, suggesting that no single sexual investment optimum exists. In artificially induced human infections, estimated conversion rates were highly variable between infections, both in historical studies of malaria-induced neurosyphilis patients (8) and in recent challenge studies (9). In vitro work has demonstrated that phenotypic differences in sexual conversion exist among laboratory-cultured genotypes (10). Recent genomic evidence also points to natural variation among parasite populations in key sexual development genes (1, 4). In particular, transcriptional and genomic profilings of natural infections in Sudan and Kenya have shown that sexual development varies in association with local transmission rates (3), but these results have not been confirmed in other global regions.
Modeling has complemented this empirical research by factoring in the impact of further ecological and intrahost conditions. Greischar et al. (11) showed that higher infectiousness at early versus late infection time points provides different evolutionary advantages to parasites as a function of infection length. Thus the parasite’s optimal gametocyte investment strategy might be impacted by ecological and epidemiological factors that alter infection length (11). On a within-host level, increased investment in asexual parasite proliferation can evade transmission blocking immunity targeting gametocytes and can also provide a competitive advantage against other genotypes in coinfections (12, 13). The latter is of particular interest in a context of declining malaria prevalence, where the fraction of infections containing multiple genotypes is expected to decline, reducing levels of within-host competition (14).
Here, we model within-host and population-level dynamics to explore how intervention-induced population reduction impacts the parasite’s optimal sexual investment strategy. We find that successful intervention strategies create a favorable environment for higher sexual conversion by reducing within-host competition, resulting from a decrease of the fraction of polyclonal infections, and shorter average infection length. We further substantiate these models with empirical analysis of genomic data from French Guiana. French Guiana experienced a dramatic decrease in reported malaria cases from over 3,000/y in 1998 to under 300/y in 2015. We sequenced and analyzed 148 genomes from across this 18-y span and, as our model predicts, found evidence of strong selection on genes regulating sexual commitment and sexual form development. More broadly, the observed genomic patterns demonstrate that selection remained effective, even in a small, declining parasite population. Together, our modeling and empirical results demonstrate how declining parasite prevalence alone can lead to changes in an important transmission-related phenotype. Further understanding this interplay will be key for devising and monitoring successful intervention strategies.
Results
Multiscale Modeling Demonstrates That Reduced within-Host Competition Favors Higher Sexual Investment.
We assessed the competitive advantage of different sexual investment strategies using a mathematical model that included within-host infection and the dynamics of infectiousness, and tracked transmission between human hosts at a population level (transmission model illustrated in SI Appendix, Fig. S1). The deterministic within-host model describes gametocyte density and asexual parasite growth, the latter constrained by both innate and adaptive immune responses that control peak parasitemia and infection length. Different genotypes were defined by the parasite’s daily conversion rate from asexual parasites to gametocytes, zu, set between 0.01 and 0.3. In this model, two distinct genotypes are included and polyclonal infections simultaneously contain both distinct genotypes. We assume a common innate immune response to both genotypes and a partially cross-reactive adaptive immune response between genotypes.
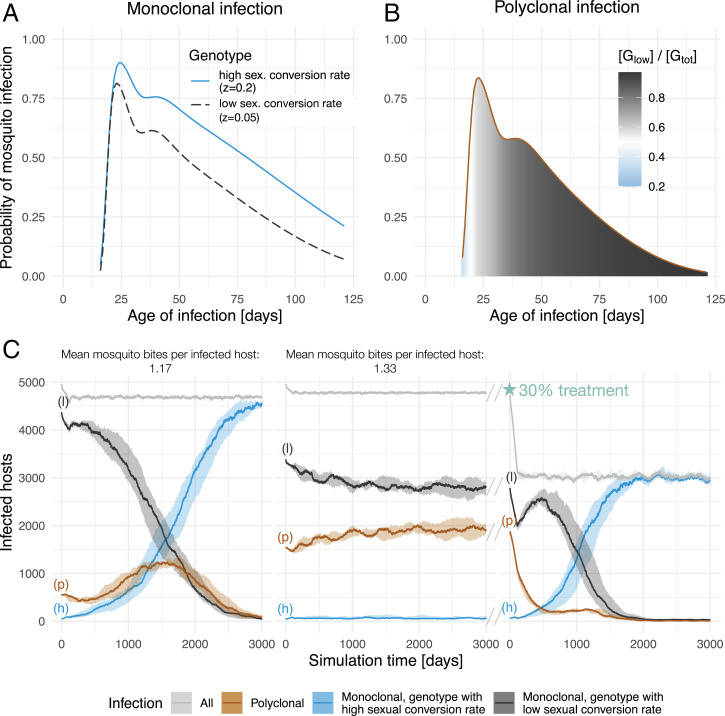
In monoclonal infections—the expected scenario when parasite populations are small—parasites show higher total infectiousness as conversion rate increases (Fig. 1A and SI Appendix, Fig. S2). Within the range of sexual conversion rates investigated, there is always an advantage for higher sexual conversion in monoclonal infections, as asexual growth remains high enough to maintain a blood-stage infection, even with the highest investment into gametocytes. Conversely in polyclonal infections—which are dominant when parasite populations are large—total infectiousness over the course of the infection was higher for the genotype with the lower conversion rate (Fig. 1B), for any conversion rate pairs where zlow < zhigh and (SI Appendix, Fig. S3). In the model, this within-host advantage is driven by the cross-reactive immune response. Considering a polyclonal infection with two genotypes, Slow and Shigh with lower and higher sexual conversion rates, respectively, high levels of Slow asexual parasites elicit an immune response that also affects the parasite density of Shigh parasites. As a higher proportion of Shigh asexual parasites convert into gametocytes, the asexual parasite density is lower for Shigh relative to Slow at the point of immune activation. As a result, over the entire course of the infection, fewer Shigh asexual parasites convert into gametocytes compared to Slow. As this is a direct result of the assumed cross-reactivity between the immune responses against different genotypes, the advantage of Slow in polyclonal infections is lost when assuming little to no cross-reactivity (SI Appendix, Fig. S6). In this model, cross-reactivity levels need to be no less than kCR = 0.7 to observe an advantage of Slow in polyclonal infections (SI Appendix, Fig. S5). Immune competition leads to a trade-off between within-host growth through asexual replication and between-host transmission through gametocyte conversion. The optimal sexual conversion rate is therefore dependent on the presence of distinct genotypes in an infection, with Shigh having highest transmission potential in monoclonal infections and Slow having highest transmission potential in polyclonal infections.
Fig. 1.
Optimal sexual investment strategy in the population defined by within-host competition. (A and B) Infectiousness, as the probability of a feeding mosquito becoming infected, for monoclonal (A) and polyclonal (B) blood-stage infections. In monoclonal blood-stage infections (A), infectiousness is consistently greater for Shigh (dashed line, gray) parasites. In polyclonal blood-stage infections (B), the addition of within-host immune competition changes the optimal sexual investment strategy. Compared to Slow gametocytes (gray), Shigh gametocytes are in the majority in the early days but have lower overall infectiousness over the full course of the infection. The area under the curve is colored by the relative concentration of Slow gametocytes () over the total gametocytes concentration (). Higher Shigh gametocyte concentrations are indicated in blue, versus in gray for higher Slow gametocyte concentrations. (C) Number of infected hosts in the population (y axis), through time (x axis), assuming an average number of infectious mosquito bites per infected host of 1.17 (Left), 1.33 (Center), or 1.33 with 30% treatment (Right). Colors and labels indicate hosts infected with the lower sexually committing parasites (l) in black, higher sexually committing parasites (h) in blue, polyclonal infections (p) in brown, and the overall number of infected hosts in gray. Lines indicate the median and shaded area the minimum and maximum across five simulations.
We next investigated the relative advantage of these sexual investment strategies at the population level by implementing the within-host infection dynamics within an agent-based population model. We fixed the number of infected hosts at n = 5, 000 for all simulations. Although mosquito dynamics were not explicitly modeled, the mosquito distribution between hosts was assumed to follow a negative binomial distribution to account for transmission heterogeneity. Assuming that transmission intensity impacts the number of superinfections found in a population, we varied the average number of infected mosquitoes at each time step, which is defined at the simulation onset and is constant over time. The resulting average number of infectious mosquito bites per newly infected host, referred to as mosquitoes per host for simplicity, was considered a proxy to investigate different transmission intensities (SI Appendix, Fig. S7). Polyclonal infections (infections containing both distinct genotypes) occur in two ways in the model: through superinfection or cotransmission. Superinfection, defined as an infection in a single host arising from two or more mosquitoes, includes both polyclonal infection and superinfection with multiple instances of the same genotype. Cotransmission is defined as infection of a single host from a single mosquito infected with both genotypes. Over the course of a polyclonal infection, the probability of cotransmission depends on the relative concentration of each genotype in the mosquitoes’ blood meal. Cotransmission is very likely within the first 20 d of infection, followed by a decrease in cotransmission probability for the remaining infection days (SI Appendix, Fig. S9). By varying the number of infected mosquitoes across simulations, we modulated the mean number of infected mosquitoes per host and therefore varied the opportunity for polyclonal infections to result from superinfection at baseline. The proportion of polyclonal infections resulting from superinfection in the first year of simulation ranged between 0.29 and 0.40, for average number of infectious bites per host between 1.13 and 1.33 (SI Appendix, Fig. S7). This proportion varies throughout the simulation, as it is dynamically linked to the number of monoclonal infections of both genotypes present, as well as the number of polyclonal infections (SI Appendix, Fig. S7).
At simulation onset Slow was defined as the majority genotype, with Slow comprising 99% of the monoclonal infections and the exact initial genotype frequencies dependent on the initial level of polyclonal infections. Increasing the mean number of infected and infectious mosquitoes per host in the population allows for more polyclonal infections, provided that both genotypes are present (SI Appendix, Fig. S7). Under this model, Slow was maintained as the majority genotype throughout the simulated time (3,000 d) when mosquitoes per host were high (Fig. 1C and SI Appendix, Fig. S7). The Shigh genotype frequency became the majority genotype when mosquitoes per host dropped to 1.26 or lower (SI Appendix, Fig. S7), and by day 3,000, Shigh fully replaced Slow in the parasite population when mosquitoes per host were at 1.17 or lower (Fig. 1C and SI Appendix, Fig. S7). Overall, we found that mosquitoes per host had a large impact on genotype frequencies, with two possible outcomes depending on the number of infectious mosquito bites per newly infected host: When mosquitoes per host were high, maintaining a high level of polyclonal infection, Slow remained the dominant genotype and Shigh was present almost exclusively within polyclonal infections; when mosquitoes per host were low, the Shigh genotype frequency increased until reaching fixation.
Increased Treatment Coverage Favors Higher Sexual Investment.
We next focused on the effect of reduced infection length on transmission, a selection pressure specifically imposed by increased diagnosis, increased treatment coverage, and/or increased treatment efficacy. We assumed infections to be treated when a pyrogenic threshold parasitemia of parasites per microliter was reached, which falls within the range of pyrogenic thresholds observed for the first fever episode (15, 16). In the model, we varied the effect of treatment by varying probability of treatment, resulting in different treatment coverage in the population. When treatment coverage increases, both the number of human infections and the number of infected mosquitoes decrease, and consequently, the fraction of superinfections and hence the fraction of polyclonal infections decrease (Fig. 1C and SI Appendix, Fig. S8). We found that treatment, resulting in shorter blood-stage infections and a decrease in the average number of infectious mosquitoes per newly infected host, led to a decrease in polyclonal infections and favored a shift toward greater sexual investment (Fig. 1C and SI Appendix, Fig. S8). For a given average number of infectious mosquitoes per newly infected host at simulation onset, higher treatment coverage resulted in a faster increase in the frequency of Shigh (Fig. 2A), with the fastest increase found in settings with a low number of mosquitoes per host combined with high treatment levels (Fig. 2B). At an individual level, treatment clears infections shortly after infection onset, thus removing the immune-competitive advantage of Slow over Shigh in treated polyclonal infections (Fig. 1B and SI Appendix, Figs. S3–S5). Timing of treatment impacts the results, however, as the later in the infection treatment occurs, the more time is available for Slow to outcompete Shigh within the host (SI Appendix, Figs. S4 and S5). If infections are treated 16 d after blood-stage infection onset—as a result of a higher pyrogenic threshold or delayed treatment seeking—instead of 10 d as assumed in the transmission model, the advantage of Shigh over Slow is lost in the treated polyclonal infections (SI Appendix, Fig. S4). At a population level, treatment leads to a lower fraction of polyclonal infections (Fig. 2C and SI Appendix, Fig. S8). In summary, increasing treatment coverage selects for higher sexual investment via two mechanisms that reduce within-host competition: fewer polyclonal infections and reduced infection length (Fig. 2D), with the latter expected to be sensitive to timing of treatment.
Fig. 2.
Genotype frequency increase defined by number of infectious mosquitoes per host and treatment coverage in the population. (A) Relative frequency of the higher sexually committing genotype (y axis), through time (x axis), with a frequency of Shigh that equals 0.01 at onset of simulation, for different treatment coverage (colors). (B) Rate of Shigh genotype frequency increase (y axis) as a function of the average number of infectious mosquito bites per newly infected host at onset of simulation (i.e., without treatment) (x axis). Rates of frequency increase were estimated by fitting a logistic growth function and are normalized such that 1 represents the maximum rate across all simulations, and 0 represents the lower limit where no increase is observed during the time of simulation (3,000 d). The three curves represent rates of frequency increase without treatment (gray), with 15% treatment (yellow), and with 40% treatment (blue), with summary statistics showing median, minimum, and maximum across five simulations. Missing estimates indicate simulations for which no increase in genotype frequency was observed, and thus no rate was estimated. (C) Average number of infectious mosquito bites per newly infected host after treatment implementation (mean over the last 300 d of simulation) (y axis) as a function of the fraction of treated infections (x axis). Blue shades indicate the estimated rate of Shigh genotype frequency increase, and simulations where Shigh frequency did not increase are indicated in dark gray. Median across five simulations is shown. (D) Conceptual illustration of the potential mechanisms driving genotype selection, where increased treatment could select for higher sexual investment genotypes by both shortening the infection and decreasing the number of infectious mosquitoes per host and thus reducing polyclonal infections.
French Guiana Parasites Show Increasing Relatedness over a Period of Declining Malaria Prevalence.
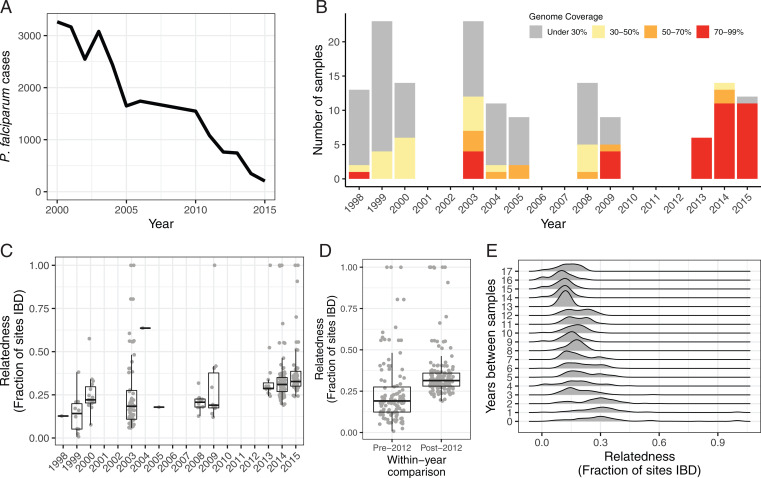
To empirically assess these model predictions, we assembled a longitudinal genomic dataset from French Guiana that spanned from 1998 to 2015. Over these years, French Guiana saw P. falciparum malaria cases decline by an order of magnitude, driven by a large antimalaria campaign that included the introduction of rapid diagnostic tests (RDTs), distribution of insecticide-treated bed nets, and a switch to Artemether-Lumefantrine as the official frontline antimalarial treatment in 2008 (Fig. 3A) (17). Our dataset includes 148 whole-genome sequenced monoclonal samples derived from a combination of direct patient samples and newly culture-adapted clones (<80 d). These mixed sources yielded highly variable amounts of DNA and therefore highly variable genome coverage following Illumina sequencing (Fig. 3B). Even after restricting the analysis to variant calls made with 5× coverage, we noted that a strong reference bias existed in low-coverage samples (SI Appendix, Fig. S10). We analyzed samples individually only if they had at least 30% of the genome at 5× coverage (n = 69).
Fig. 3.
Whole-genome sequencing of longitudinally sampled parasites from French Guiana. (A) P. falciparum malaria cases in French Guiana dropped by an order of magnitude between 1998 and 2015 (19, 20). (B) Whole-genome sequences were obtained for 148 monoclonal P. falciparum infections sampled across these years. (C) Overall, the parasite population shows high relatedness, as measured by pairwise IBD. Median relatedness between samples from the same year increased across the sample period, correlating with the documented decrease in parasite population size. (D) After 2012, parasite pairs collected within the same year showed significantly higher median IBD (0.31) compared to parasite pairs collected within the same year before 2012 (0.19; two-sided Mann–Whitney U test, P < 0.0001) Only high-coverage samples (>30% of the genome at 5 × coverage) were included in the IBD analysis. (E) IBD relationships are also affected by the time between samples, with lower IBD observed between samples with higher temporal separation.
Pairwise relatedness estimates between samples (as measured by identity-by-descent [IBD]) show a pattern of increased relatedness across the time period (Fig. 3C). Later years show a greater proportion of clonal pairs (IBD 0.99), which could be attributed to the greater sampling depth. More strikingly, however, no parasite pair sampled from the same year after 2012 had an estimated IBD relationship of less than 0.19. Conversely, this same value was the median IBD between all parasite pairs sampled from the same year before 2012 (Fig. 3D). This pattern is consistent with forward simulations, which demonstrate a measurable increase in mean fractional IBD within years of a P. falciparum population bottleneck (18). While not the only driver, this loss of haplotypic diversity is also reflected in the lower relatedness observed between individuals sampled in different years (Fig. 3E). Data artifacts such as undetected polyclonal infections likely had a small effect at most as restricting the analysis to genomes with higher coverage did not alter the observed temporal trend (SI Appendix, Fig. S11).
A Small Subset of P. falciparum Variants Experienced Large Frequency Changes as Malaria Prevalence Declined.
Across our studied time period, this P. falciparum population displayed measurable changes in drug resistance and susceptibility (21). Because of this observed phenotypic evolution, we hypothesized that, in addition to neutral demographic processes, selection has also impacted the genome. We first determined that the population’s increased relatedness did not result from a hard selective sweep on a single genomic background or clonal lineage. Twenty-one parasites had a clonal relationship with at least one other sample, but no clonal cluster contained more than five members or extended beyond 2 y. This contrasts with observations in the Greater Mekong Subregion where highly related clonal lineages rapidly rose to high frequencies after acquiring artemisinin and partner drug resistance (22). We therefore focused on identifying potential selective sweeps that were limited to subregions of the genome. In particular, we leveraged our temporal sampling scheme to investigate potential selection on standing variation present on multiple genetic backgrounds. While such soft sweeps are challenging to detect using genomic data from a single time point, our longitudinal sampling allows a direct assessment of allele frequency changes across the period of interest (23). To calculate allele frequencies in each year, we took a pooled sample approach since the elimination of low-coverage individual samples preferentially impacted early time points (pre-2009).
As expected, most variants showed no appreciable frequency change from 1998 to 2015 (Fig. 4A). Of the 39,007 variants that fit a binomial model, 31,334 had changes that were either under 10% or statistically indistinguishable from 0. Conversely, 974 variants (2.5%) present in or near 447 genes showed a frequency change of 60% or greater. Overall, single-nucleotide polymorphisms (SNPs) in coding regions showed smaller allele frequency changes than SNPs in intergenic or intronic regions (Kolmogorov–Smirnov test, P < 0.00001) and similarly, nonsynonymous SNPs showed smaller frequency changes than synonymous SNPs (Kolmogorov–Smirnov test, P = 0.00068). These observations indicate that purifying selection was effective despite the decreased population size. Allele frequency shifts were not all independent. After 2011, several regions of the genome showed large spikes in mean pairwise IBD (SI Appendix, Fig. S12). This pattern is consistent with the action of strong positive selection, although we cannot discount that these patterns may have been affected by the change in population size. Establishing proper null expectations based on a drift-only model would require population information we currently lack regarding subpopulation connectivity and local outbreak dynamics.
Fig. 4.
Allele frequency changes from 1998 to 2015. (A) Of the 39,007 variants that fit a binomial model (gray), 9,215 were determined to have a nonzero slope (black). Inset breaks down the variants with a nonzero slope by mutational type. The red diamond marks the frequency change observed for a known selected allele (PfCRT C350R) (21). (B) Allele frequency changes were highest among six AP2 genes with well-documented roles in sexual commitment or gametocyte development (ap2-g, ap2-g2, ap2-g3, ap2-g5, PF3D7_1222400, and PF3D7_0613800). Each gene is represented by the nonsynonymous variant or coding indel within it that experienced the greatest allele frequency change across the sampled time period. Only genes containing a detected nonsynonymous SNP or coding indel were included in the analysis. Variants that did not fit a binomial model were coded with an allele frequency change of 0. AP2 gene names are marked in SI Appendix, Fig. S15. *P < 0.05, **P < 0.01, ***P < 0.001, one-sided Mann–Whitney U test. (C) One nonsynonymous variant in ap2-g (blue) and a 36-bp deletion (relative to the 3D7 reference) in ap2-g2 (red) experienced large frequency shifts.
Transcription Factors Involved in Sexual Commitment and Gametocyte Development Contain Variants with Large Frequency Changes.
Our outlier approach successfully captured a previously documented instance of positive selection on a variant implicated in drug resistance (PfCRT C350R) (21). In our dataset, this variant experienced an allele frequency shift of 63%, which places it in the top 11% of all changing variants. While this shows that we are detecting true selection signals, a proportion of outlier loci will have risen in frequency due to drift rather than selection. We therefore focused on gene set enrichment rather than the identity of single variants. We hypothesized that a change in conversion phenotype could be driven by genetic changes in transcription factors as studies in multiple systems have highlighted transcriptional changes as a major mode of local adaptation. The P. falciparum genome contains only 27 known transcription factors, all members of the Apicomplexan-specific ApiAP2 gene family. Across species and individuals, there are marked differences in both the transcription profiles of these AP2 genes and the timing of the life history events they govern (24–27). A recent study of Southeast Asian Plasmodium vivax parasites found evidence that selection on these AP2 transcription factors accompanied a demographic expansion (28), and within P. falciparum specifically, transcriptional changes marked the initial observation of sexual conversion rates differing based on parasite prevalence (3). Overall, among coding variants that had a frequency change 60% (580 coding variants total) transcription factors were enriched, with eight transcription factors (30%) containing a nonsynonymous SNP or coding indel that showed a frequency change 60% (Fisher’s exact test, P = 0.04).
We represented each gene by its nonsynonymous SNP or coding indel that showed the largest frequency change and found that larger frequency shifts occurred within transcription factors putatively involved in sexual commitment and gametocyte development (29). Not all transcription factors are well characterized, so we compared the results from three large P. falciparum gametocyte expression studies (30–32). Only three genes were identified as gametocyte associated across all studies (ap2-g, PF3D7_1222400, and PF3D7_0613800), and all three of these genes contained variants with large frequency shifts (SI Appendix, Fig. S15, and Fig. 4B). Of particular note is ap2-g, the master regulator of sexual commitment (33), which contains four nonsynonymous mutations that experienced frequency shifts >0.7 (Fig. 4C and Dataset S2). Additional large frequency shifts are seen in PF3D7_1222400. While not as extensively studied, PF3D7_1222400 is expressed immediately following ap2-g in sexually committed parasites. It is known to evolve loss-of-function mutations in culture, suggesting that it is nonessential in asexual parasites and may play an early role in gametocytogenesis (34). ap2-g and PF3D7_1222400 are within 9 kb of each other on chromosome 12, and both are centrally positioned within a peak of high IBD among post-2011 parasites (SI Appendix, Fig. S12). This could suggest either genetic draft or selection on a haplotype carrying multiple beneficial mutations. We included three additional genes (ap2-g2, ap2-g5, and ap2-g3) in this high-confidence set based on functional studies. Both AP2-G2 and AP2-G5 are required for gametocyte maturation and act as transcriptional repressors in asexual parasites (35, 36), potentially impacting ap2-g expression (36, 37). ap2-g2 is within a high-IBD peak found on chromosome 14 (SI Appendix, Fig. S12) and contains a 36-bp indel that increased in frequency by over 90%, almost fixing in the population by 2015 (Fig. 4C). ap2-g5 contains three nonsynonymous variants that changed frequency by more than 50%. The final gene, ap2-g3, has been shown to be essential for gametocytogenesis (38), but did not contain any variants with unusually large frequency shifts. Together, however, this set of six genes contains larger allele frequency shifts than other AP2s with some evidence of gametocyte involvement (29) (one-sided Mann–Whitney U test, P = 0.027), non–gametocyte-associated AP2s (one-sided Mann–Whitney U test, P = 0.0031), and non-AP2 genes (one-sided Mann–Whitney U test, P = 0.000019; Fig. 4B). As gene length correlates with largest allele frequency shift (Spearman’s rho 0.47, P < 0.00001), we additionally compared each of these AP2s to other genes of comparable length and found that ap2-g, ap2-g2, ap2-g5, PF3D7_1222400, and PF3D7_0613800 were in the 91st, 97th, 82nd, 99th, and 85th percentiles, respectively (SI Appendix, Fig. S13).
An alternative hypothesis is that these commitment-related transcription factors experienced relaxed constraint rather than directional selection as population size decreased. In total, we identified 25 nonsynonymous or indel variants within ap2-g, ap2-g2, ap2-g5, PF3D7_1222400, and PF3D7_0613800 (Dataset S2). Of these, 22 (88%) are present in a large set of global variants sampled from Africa and Asia, with 19 (76%) having a frequency of at least 2% in one other country. The global distribution of these variants suggests that they were segregating in French Guiana prior to the start of our study and that they are unlikely to be strongly deleterious as purifying selection would be more efficacious in these other larger populations.
A second alternative hypothesis is that these patterns arose as culture artifacts. In this study, however, almost all attempted cultures were successful, making any potential culture bias small. Further, while previous studies of laboratory-cultured P. falciparum have found selection for loss-of-function AP2 mutations (34), such mutation events are unlikely to converge on the identical variant with high frequency. Still, we confirmed that our observed pattern reflected population-level allele frequencies by performing PCR and Sanger sequencing of the identified AP2-G2 indel (Fig. 4C) on an overlapping but nonidentical set of 59 French Guiana samples that were extracted directly from patient blood. As with our whole-genome data, the frequency of the AP2-G2 indel increased from 1999 to 2016 in this sample set, reaching apparent fixation by the later sampling periods (SI Appendix, Fig. S14).
Discussion
Through mathematical modeling and empirical analysis of whole-genome sequencing data, we explored links between public health interventions and parasite life history that are independent of canonical intervention-induced evolution like drug resistance. Our model suggests that increased treatment coverage can select for parasites with higher rates of sexual conversion by reducing the fraction of polyclonal infections and shortening infection length of treated infections. Thus, as human interventions cause parasite prevalence to decline, higher sexual investment might be beneficial to the parasite. From a public health perspective, a higher sexual conversion rate could lead to higher transmission potential from human to mosquitoes, resulting in a lower than anticipated impact of interventions. Genomic data from French Guiana support this concept by showing large allele frequency shifts in sexual development regulators across a time period of malaria reduction.
The modeling work presented here conceptualizes potential mechanisms at play that are currently difficult to measure directly in humans. Substantial knowledge gaps in the host–parasite mechanisms behind human infections render within-host models difficult to assess and parameterize (39, 40), and we chose the model of McKenzie and Bossert as it takes a simple approach, limiting the number of parameters and assumptions. However, the results presented here depend heavily on the assumed effect of immune responses, including the effect and level of cross-reactivity of the immune response between genotypes. This framework could be extended to include other forms of within-host competition, such as including multiclone infections with more than two genotypes and multiple infections occurring at different infection time points rather than simultaneously.
Both the within-host mechanisms and the epidemiological dynamics are more complex than modeled. For example, host–pathogen interactions involve more complex immune dynamics, fluctuations around resource limitations and red-blood-cell availability, high variability in infection length, and parasite response to host environmental factors (39, 41). In regions with highly seasonal transmission, varying transmission and exposure-driven immunity will also play a role (42). Under strong seasonality, selection for higher conversion rates might be enhanced during high transmission peaks when most infections are relatively young, and the competitive advantage of the strain with lower conversion might be diminished in older (chronic) infections due to few transmission events in the dry season. In addition, while we focus on immune-driven within-host competition and infection length, studies have highlighted additional connections between intervention strategies and transmission such as the relative proportion of symptomatic versus asymptomatic infections (43). Forces unrelated to demography or host environment—like differential susceptibility to drugs including artemisinin across the parasite life cycle—may also impact sexual development (44, 45). Finally, both mathematical models and empirical work have shown that sexual commitment rates can be plastic and rapidly respond to stress factors such as drug administration, resource limitation, immune response, or competition against other genotypes (7, 46–49). The interplay between plastic and hard-wired sexual development rates will be interesting to explore moving forward.
Plasmodium’s reproductive fitness is not determined solely by conversion rate just as conversion rate affects more than reproductive fitness. Sex ratio and gametocyte density also contribute to reproductive success and are themselves linked (50). In polyclonal infections, these traits have additional evolutionary implications as they affect the likelihood of outcrossing versus clonal propagation (46, 51, 52). Especially in areas with small P. falciparum populations like South America, this balance between clonal persistence and outcrossing can have demographic and evolutionary consequences. Just as we saw for conversion rate, the fitness optima for these other reproductive traits may shift as population size declines and the probability of being in a polyclonal infection decreases. Indeed, we find suggestions of this in the data. In addition to its general role in gametocyte maturation, ap2-g2 expression affects the ratio of male to female gametocytes (37). Three additional AP2 genes are also known to show sex-biased expression in P. falciparum (ap2-o, PF3D7_1107800, and PF3D7_1305200) (53), and two of these (ap2-o and PF3D7_1107800) contained alleles with frequency shifts of 0.76 and 0.69, respectively (SI Appendix, Fig. S15 and Dataset S2). Continued theoretical and empirical explorations are warranted on these topics.
On the empirical side, our understanding of the specific factors at play in the French Guiana population is currently incomplete. While we know that transmission declined after 1998, we lack reliable estimates of complexity of infection (COI) decline due to uneven sequencing coverage. The exact phenotypes conferred by these French Guiana AP2 variants also remain unknown as we were unable to successfully generate parasite lines with allelic replacement. Further work is therefore warranted in this and other regions with declining infection rates to explore how specific sexual development phenotypes impact both clinical disease and transmission. This awareness should stimulate discussion on the types of metrics that could be included in surveillance studies to further understand the impact of parasite evolution in regions of low or declining prevalence.
More broadly, the empirical results from this study highlight the insight gained from longitudinal sampling of parasite populations (22, 54, 55). Rather than infer selection from cross-sectional data, a longitudinal analysis permits direct observation of evolution in action. This approach enables tracking of individual alleles rather than large haplotype blocks, and its power is less affected by the strength of selection and initial age of the selected variants. Here, we chose to focus on alleles that showed large frequency changes. This narrowed our scope to alleles that may have experienced strong selection—most commonly at early time points—and placed a ceiling on the potential starting allele frequency of these candidate loci. Thousands of other loci, however, displayed moderate frequency shifts. Distinguishing between selection and drift, determining the temporal onset of selection, and understanding the phenotypic impact of these additional variants will further augment our understanding of parasite evolution. While our study here was retrospective, this approach could be taken in real time and will prove powerful as genomic surveillance efforts expand and longitudinal datasets become more common.
Materials and Methods
Within-Host Model.
The within-host model was adapted from McKenzie and Bossert (12) to include the effect of treatment on the asexual parasites. A full description of the model can be found in SI Appendix. Briefly, the model is a set of delay differential equations that describe the asexual parasite density, the gametocyte density, and the innate and adaptive immune responses. Assumptions include a constant parasite replicate rate [ln(16)/2], a common innate immune response across genotypes, and a genotype-specific but cross-reactive adaptive immune response, assuming that the response specific to one genotype is effective against the other genotype by a factor of 0.75. Genotypes differ by their conversion rate from asexual parasites into gametocytes, zu, which was set at 0.05 and 0.2 for low- and high-transmission–investing genotypes (Slow and Shigh), respectively. These conversion rates fall within the range explored in McKenzie and Bossert (12). The model’s parameters were fitted by McKenzie and Bossert to the malaria therapy dataset (12), which is a collated dataset of patient records for which malaria was used as a therapy to treat patients with tertiary neurosyphilis (56). Treatment is implemented as an additional killing rate of the asexual parasites, ϵ, clears all asexual parasites almost instantly, and is activated the day the asexual parasite density reaches parasites per microliter. This is assumed to represent the pyrogenic threshold parasitemia, which falls within the range observed in the malaria therapy dataset (15, 16). Further exploration on the values of the conversion rates, cross-reactivity of immune response, and timing of treatment is found in SI Appendix. All other parameters have been taken as in McKenzie and Bossert (12) and represent the average infection dynamics observed in human infection.
Transmission Model.
The transmission model was adapted from Chang et al. (57). A full description of the model and parameters can be found in SI Appendix and in SI Appendix, Fig. S1. Simulations start with 5,000 infected human hosts. Two parasite genotypes are considered, with Shigh the genotype with high sexual commitment (proportion of asexuals converting to gametocytes z = 0.2) and Slow the genotype with low sexual commitment (z = 0.05). Infections can be monoclonal, i.e., only one of the two genotypes is present, or polyclonal, both genotypes are present in the same host. Superinfection occurs if more than one infectious mosquito infects the same host. A fraction of initial polyclonal infections and a mean number of infected mosquitoes per day are defined for each scenario. At simulation onset, 1% of monoclonal infections are infected by Shigh. Days since infection onset are evenly distributed among infected hosts at simulation start. At each iteration, infections become 1 d older until cleared naturally or by treatment, and the probability of transmitting to a mosquito is assessed. A random number of hosts, whose asexual parasite density reached the treatment threshold, are sampled to receive treatment. The number of treated hosts in each time step is defined by a Poisson distribution, with mean the defined proportion of treated infections. Each setting was simulated five times to include a stochastic range in the output.
Transmission from Human to Mosquito.
The relationship between the gametocyte density defined by the within-host model and the probability of an infection to be transmitted to a biting mosquito was taken from ref. 58 and is described as , where G is the gametocyte density as a function of the age of infection, a. The number of transmissions from human to mosquitoes follows a Poisson distribution, with the mean given by (the probability of the host being infectious times the number of biting mosquitoes) and a normalizing factor , which is the expected number of naturally cleared infections at each time step divided by the total transmission potential over all infectious hosts at that time step. The normalizing factor ensures that in the absence of treatment the number of infections remains constant throughout the simulations. To allow for a varying mean infectious mosquitoes per newly infected host, the normalizing factor is increased by a factor , with w being the expected fraction of superinfections.
Transmission from Mosquito to Human.
The infected mosquitoes are randomly assigned to bite hosts, following a negative binomial distribution of mosquitoes per host. Ten sporozoites per infected mosquito are inoculated to a new host. The ten sporozoites are drawn from a binomial distribution, with the probability of success to draw a sporozoite from one of the genotypes proportional to its relative gametocyte concentration at time of blood meal. Thus, a polyclonal infection results either from a mosquito inoculating sporozoites from both genotypes or from multiple mosquitoes simultaneously inoculating different genotypes to the same host. For simplicity, superinfections from bites at different time points are not included.
P. falciparum Sample Collection.
P. falciparum samples (n = 201) were collected in French Guiana between 1998 and 2015 from symptomatic patients infected by P. falciparum from only those who sought diagnosis and care. The samples were collected as part of the malaria routine surveillance system implemented in French Guiana. Healthcare facilities were in charge of collecting samples from anonymized P. falciparum-positive cases. Identification of individuals cannot be established. Seventy-six isolates were culture adapted in human red blood cells in enriched RPMI medium containing 10% human serum and were propagated at 37 ∘C in 10% O2, 5% CO2, and 85% N2. The enriched medium was composed of RPMI-1640 (ref. 4130, Sigma Aldrich) with 25 mM Hepes, 5 mM L-glutamine, 22 mM glucose, 25 mM NaHCO35%, 20 mM gentamycin, 0.37 mM hypoxanthine, and 1.6 mM orotic acid. We extracted DNA from whole blood collected in an Ethylenediaminetetraacetic Acid (EDTA) tube using the QIAmp DNA mini kit (Qiagen) and following the manufacturer’s protocol.
Genomic Sequencing and Variant Calling.
We created Illumina sequencing libraries using a Nextera XT low-input library kit. We sequenced the libraries on an Illumina HiSeq 2500 instrument using a 200-cycle run mode, generating 100-bp paired-end reads, and indexing 92 libraries per lane. Raw sequence data are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive as BioProject PRJNA336113 (59). Reads were aligned with BWA-mem to the PlasmoDB P. falciparum 3D7 v. 28 reference. Variants within the “core” genome [as defined by Miles et al. (60)] were called with GATK v.3 Haplotype Caller using a set of genotyped pedigreed crosses for variant and base quality score recalibration. We estimated COI using the likelihood-based method COIL (61) and removed samples with a COI > 1. We additionally removed any samples that were called monoclonal by the COIL approach but that had unusually high proportions of heterozygous variant calls. The remaining set of 148 putatively monoclonal samples was used for downstream analysis. Due to the low genomic coverage for early time points and the different sampling methods (direct sequencing from blood vs. culturing), we were not able to reliably estimate COI changes through time.
Analyses were limited to calls with five or more reads, but even after implementing this filter, we observed a strong reference bias in low-coverage samples (SI Appendix, Fig. S10). We therefore analyzed samples individually only if they had at least 30% of the genome covered at 5× (n = 69). For the longitudinal allele frequency analysis, we downsampled the sequences from each individual monoclonal sample to ∼1× mean genome coverage and then merged all downsampled BAM files from the same year. We called variants on pooled BAMs with GATK v.3 Haplotype Caller and used the variant call read ratios to calculate allele frequency estimates for each time point. Global allele frequencies were obtained from the MalariaGEN Pf3k release 5.0 data (https://www.malariagen.net/projects/pf3k).
Genomic Analysis.
We calculated pairwise identity-by-descent using hmmIBD (62) for the 69 monoclonal samples with at least 30% of the genome covered at 5×. Allele frequency trajectories were estimated for all biallelic variants by performing a logistic regression with the estimated reference allele frequency at each available time point. A 95% confidence interval for the slope of this regression was calculated using the confint function in R. Variants with fewer than 8-y–based allele frequency estimates showed an upward bias in the frequency change estimates and so were excluded from further analysis (SI Appendix, Fig. S16).
Supplementary Material
Acknowledgments
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, NIH, Department of Health and Human Services, under Grant U19AI110818 to the Broad Institute. This work was supported by an Investissement d’Avenir grant managed by Agence Nationale de la Recherche Centre d’Etude pour la Biodiversité Amazonienne (CEBA[ref. ANR-10-LABX-25-01]), the Santé publique France as National Reference Centre for Malaria, and the French Ministry for Research. F.C. and C.O.B. are supported by a Maximizing Investigators’ Research Award for Early-Stage Investigators (R35 GM-124715). S.P. received funding support from European Commission Grant REGPOT-CT-2011-285837-430 STRonGer. L.M.C. is supported by NSF Standard Grant 1902214.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122165119/-/DCSupplemental.
Data Availability
The scripts for the within-host and transmission models are available at https://github.com/flaviaCa/Amazomics and short-read sequencing data have been deposited in the NCBI Sequence Read Archive as BioProject PRJNA336113 (59).
References
- 1.Mobegi V. A., et al., Genome-wide analysis of selection on the malaria parasite Plasmodium falciparum in West African populations of differing infection endemicity. Mol. Biol. Evol. 31, 1490–1499 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molina-Cruz A., et al., Plasmodium evasion of mosquito immunity and global malaria transmission: The lock-and-key theory. Proc. Natl. Acad. Sci. U.S.A. 112, 15178–15183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rono M. K., et al., Adaptation of Plasmodium falciparum to its transmission environment. Nat. Ecol. Evol. 2, 377–387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahouidi A., et al.; MalariaGEN, An open dataset of Plasmodium falciparum genome variation in 7,000 worldwide samples. Wellcome Open Res. 6, 42 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reece S. E., Ramiro R. S., Nussey D. H., Plastic parasites: Sophisticated strategies for survival and reproduction? Evol. Appl. 2, 11–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stearns S. C., Life history evolution: Successes, limitations, and prospects. Naturwissenschaften 87, 476–486 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Greischar M. A., Mideo N., Read A. F., Bjørnstad O. N., Quantifying transmission investment in malaria parasites. PLOS Comput. Biol. 12, e1004718 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichner M., et al., Genesis, sequestration and survival of Plasmodium falciparum gametocytes: Parameter estimates from fitting a model to malariatherapy data. Trans. R. Soc. Trop. Med. Hyg. 95, 497–501 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Cao P., et al., Modeling the dynamics of Plasmodium falciparum gametocytes in humans during malaria infection. eLife 8, e49058 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prajapati S. K., et al., The transcriptome of circulating sexually committed Plasmodium falciparum ring stage parasites forecasts malaria transmission potential. Nat. Commun. 11, 6159 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greischar M. A., Beck-Johnson L. M., Mideo N., Partitioning the influence of ecology across scales on parasite evolution. Evolution 73, 2175–2188 (2019). [DOI] [PubMed] [Google Scholar]
- 12.McKenzie F. E., Bossert W. H., An integrated model of Plasmodium falciparum dynamics. J. Theor. Biol. 232, 411–426 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Mideo N., Day T., On the evolution of reproductive restraint in malaria. Proc. Biol. Sci. 275, 1217–1224 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tusting L. S., Bousema T., Smith D. L., Drakeley C., “Measuring changes in Plasmodium falciparum transmission. Precision, accuracy and costs of metrics” in Advances in Parasitology (Academic Press, 2014), vol. 84, pp. 151–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeffery G. M., Young M. D., Burgess R. W., Eyles D. E., Early activity in sporozoite-induced Plasmodium falciparum infections. Ann. Trop. Med. Parasitol. 53, 51–58 (1959). [DOI] [PubMed] [Google Scholar]
- 16.Gatton M. L., Cheng Q., Evaluation of the pyrogenic threshold for Plasmodium falciparum malaria in naive individuals. Am. J. Trop. Med. Hyg. 66, 467–473 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Musset L., et al., Malaria on the Guiana Shield: A review of the situation in French Guiana. Mem. Inst. Oswaldo Cruz 109, 525–533 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendry J. A., Kwiatkowski D., McVean G., Elucidating relationships between P. falciparum prevalence and measures of genetic diversity with a combined genetic-epidemiological model of malaria. PLOS Comput. Biol. 17, e1009287 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO, World Malaria Report 2008 (World Health Organization, Geneva, Switzerland, 2008).
- 20.WHO, World Malaria Report 2016 (World Health Organization, Geneva, Switzerland, 2016).
- 21.Pelleau S., et al., Adaptive evolution of malaria parasites in French Guiana: Reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc. Natl. Acad. Sci. U.S.A. 112, 11672–11677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amato R., et al., Origins of the current outbreak of multidrug-resistant malaria in Southeast Asia: A retrospective genetic study. Lancet Infect. Dis. 18, 337–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feder A. F., Pennings P. S., Petrov D. A., The clarifying role of time series data in the population genetics of HIV. PLoS Genet. 17, e1009050 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hott A., et al., Artemisinin-resistant Plasmodium falciparum parasites exhibit altered patterns of development in infected erythrocytes. Antimicrob. Agents Chemother. 59, 3156–3167 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Modrzynska K., et al., A knockout screen of ApiAP2 genes reveals networks of interacting transcriptional regulators controlling the Plasmodium life cycle. Cell Host Microbe 21, 11–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunnik E. M., et al., Changes in genome organization of parasite-specific gene families during the Plasmodium transmission stages. Nat. Commun. 9, 1910 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akkaya M., et al., A single-nucleotide polymorphism in a Plasmodium berghei ApiAP2 transcription factor alters the development of host immunity. Sci. Adv. 6, eaaw6957 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parobek C. M., et al., Selective sweep suggests transcriptional regulation may underlie Plasmodium vivax resilience to malaria control measures in Cambodia. Proc. Natl. Acad. Sci. U.S.A. 113, E8096–E8105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Josling G. A., Williamson K. C., Llinás M., Regulation of sexual commitment and gametocytogenesis in malaria parasites. Annu. Rev. Microbiol. 72, 501–519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poran A., et al., Single-cell RNA sequencing reveals a signature of sexual commitment in malaria parasites. Nature 551, 95–99 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brancucci N. M. B., et al., Probing Plasmodium falciparum sexual commitment at the single-cell level. Wellcome Open Res. 3, 70 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Biljon R., et al., Hierarchical transcriptional control regulates Plasmodium falciparum sexual differentiation. BMC Genomics 20, 920 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kafsack B. F., et al., A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature 507, 248–252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claessens A., Affara M., Assefa S. A., Kwiatkowski D. P., Conway D. J., Culture adaptation of malaria parasites selects for convergent loss-of-function mutants. Sci. Rep. 7, 41303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh S., et al., The PfAP2-G2 transcription factor is a critical regulator of gametocyte maturation. Mol. Microbiol. 115, 1005–1024 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shang X., et al., A cascade of transcriptional repression determines sexual commitment and development in Plasmodium falciparum. Nucleic Acids Res. 49, 9264–9279 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Y., et al., PfAP2-G2 is associated to production and maturation of gametocytes in Plasmodium falciparum via regulating the expression of PfMDV-1. Front. Microbiol. 11, 631444 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikadai H., et al., Transposon mutagenesis identifies genes essential for Plasmodium falciparum gametocytogenesis. Proc. Natl. Acad. Sci. U. S. A. 110, E1676–E1684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Childs L. M., Buckee C. O., Dissecting the determinants of malaria chronicity: Why within-host models struggle to reproduce infection dynamics. J. R. Soc. Interface 12, 20141379 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camponovo F., et al., Mechanistic within-host models of the asexual Plasmodium falciparum infection: A review and analytical assessment. Malar. J. 20, 309 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meibalan E., Marti M., Biology of malaria transmission. Cold Spring Harb. Perspect. Med. 7, a025452 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrade C. M., et al., Increased circulation time of Plasmodium falciparum underlies persistent asymptomatic infection in the dry season. Nat. Med. 26, 1929–1940 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Barry A., et al., Higher gametocyte production and mosquito infectivity in chronic compared to incident Plasmodium falciparum infections. Nat. Commun. 12, 2443 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Portugaliza H. P., et al., Artemisinin exposure at the ring or trophozoite stage impacts Plasmodium falciparum sexual conversion differently. eLife 9, 1–22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witmer K., et al., Transmission of artemisinin-resistant malaria parasites to mosquitoes under antimalarial drug pressure. Antimicrob. Agents Chemother. 65, e00898-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider P., Reece S. E., The private life of malaria parasites: Strategies for sexual reproduction. Mol. Biochem. Parasitol. 244, 111375 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider P., et al., Adaptive plasticity in the gametocyte conversion rate of malaria parasites. PLoS Pathog. 14, e1007371 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greischar M. A., Mideo N., Read A. F., Bjørnstad O. N., Predicting optimal transmission investment in malaria parasites. Evolution 70, 1542–1558 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carter L. M., et al., Stress and sex in malaria parasites: Why does commitment vary? Evol. Med. Public Health 2013, 135–147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitri C., Thiery I., Bourgouin C., Paul R. E., Density-dependent impact of the human malaria parasite Plasmodium falciparum gametocyte sex ratio on mosquito infection rates. Proc. Biol. Sci. 276, 3721–3726 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Childs L. M., Prosper O. F., Simulating within-vector generation of the malaria parasite diversity. PLoS One 12, e0177941 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong W., Wenger E. A., Hartl D. L., Wirth D. F., Modeling the genetic relatedness of Plasmodium falciparum parasites following meiotic recombination and cotransmission. PLOS Comput. Biol. 14, e1005923 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walzer K. A., Kubicki D. M., Tang X., Chi J. T. A., Single-cell analysis reveals distinct gene expression and heterogeneity in male and female Plasmodium falciparum gametocytes. MSphere 3, e00130-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson T. J., et al., Population parameters underlying an ongoing soft sweep in Southeast Asian malaria parasites. Mol. Biol. Evol. 34, 131–144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cerqueira G. C., et al., Longitudinal genomic surveillance of Plasmodium falciparum malaria parasites reveals complex genomic architecture of emerging artemisinin resistance. Genome Biol. 18, 78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collins W. E., Jeffery G. M., A retrospective examination of sporozoite- and trophozoite-induced infections with Plasmodium falciparum: Development of parasitologic and clinical immunity during primary infection. Am. J. Trop. Med. Hyg. 61 (suppl. 1), 4–19 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Chang H. H., Childs L. M., Buckee C. O., Variation in infection length and superinfection enhance selection efficiency in the human malaria parasite. Sci. Rep. 6, 26370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stepniewska K., et al., Plasmodium falciparum gametocyte dynamics in areas of different malaria endemicity. Malar. J. 7, 249 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Broad Institute, Plasmodium falciparum genome sequencing. NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA336113. Deposited 2 August 2016. [Google Scholar]
- 60.Miles A., et al., Indels, structural variation, and recombination drive genomic diversity in Plasmodium falciparum. Genome Res. 26, 1288–1299 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galinsky K., et al., COIL: A methodology for evaluating malarial complexity of infection using likelihood from single nucleotide polymorphism data. Malar. J. 14, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaffner S. F., Taylor A. R., Wong W., Wirth D. F., Neafsey D. E., hmmIBD: Software to infer pairwise identity by descent between haploid genotypes. Malar. J. 17, 196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scripts for the within-host and transmission models are available at https://github.com/flaviaCa/Amazomics and short-read sequencing data have been deposited in the NCBI Sequence Read Archive as BioProject PRJNA336113 (59).