Abstract
Currently, both pathogenic and commensal viruses are continuously being discovered and acknowledged as ubiquitous components of microbial communities. The advancements of systems microbiological approaches have changed the face of virome research. Here, we focus on viral metagenomic approach to study virus community and their interactions with other microbial members as well as their hosts. This review also summarizes challenges, limitations, and benefits of the current virome approaches. Potentially, the studies of virome can be further applied in various biological and clinical fields.
Keywords: Virome, systems microbiology, virus discovery, virus–host interaction, viral metagenomic
Impact Statement
To address the virus population, virome is a viral metagenomic approach that can be utilized for viral surveillance in both research and clinical practice, with some potential advantageous applications over conventional strategies, especially concerning the information of the whole viral entity in the community. In this review, we describe the viral metagenomic approach to investigate the virus community and interactions with other microbial members as well as their hosts. We also summarize challenges, limitations, and benefits of the current virome approaches along with the potential applications of the viral metagenomic. Therefore, this review provides fundamental knowledge of using a virome approach for both research and clinical practice.
Introduction
Recently, the pandemic of COVID-19 has become a major world problem, emphasizing the importance of virus research. Heretofore, many viruses have been discovered either by laboratory research or by etiological identification in patients; however, huge numbers of viruses remain undiscovered.1,2 Systems microbiology is a multidisciplinary area of microbiology, emphasizing both whole microbiome entities and microbial impact on their hosts or other microbes.3,4
Microbiomes refer to microbial communities in specific areas, and the microbes consist of archaea, bacteria, small eukaryotes, fungi, and viruses. 5 Previously, microbiome research has addressed the bacterial profiling and their relationships to the hosts, while virome studies are small in number. These virome studies have focused on both viral composition in the community and influence of the viruses (i.e., prokaryotic viruses, eukaryotic viruses, and endogenous viruses) on their hosts.6,7 From the clinical perspective, many diseases have been identified as viral infections, and many are unknown; however, most virus infections have no effective treatment or specific drug for treatments.8–10 In future, virome research will play an important role as a resource of knowledge for clinical applications, innovation of novel therapy, and prediction of virus outbreak patterns and of evolutionary processes of viruses in nature.11,12
In this review, we would like to discuss the revolutionization of virome research, including a summary of recent databases and bioinformatic analysis tools, viral interactions to their hosts and other microbes, and potential applications by the integration of high-throughput metagenomic technologies and systems microbiology approaches.
The viral community revealed
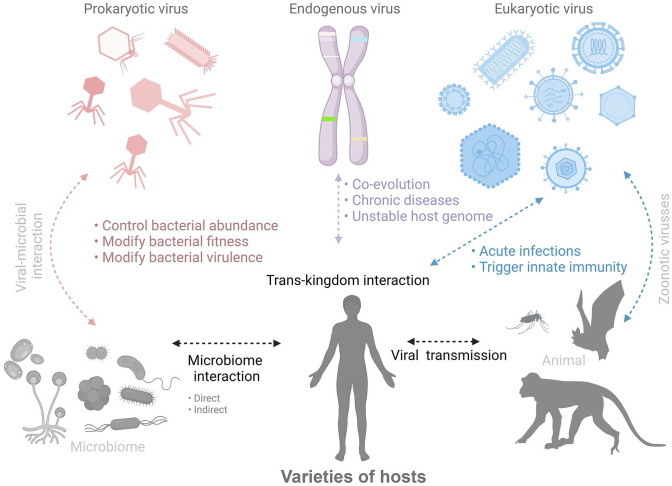
Viruses are the most biologically diverse and biomassive entity on Earth. Currently, 5560 virus species across 150 families have been defined by the International Committee on Taxonomy of Viruses (ICTV). The virus entities have diverse structures in terms of sizes and shapes, ranging from roughly spherical to linear, to amorphic. Furthermore, viral genetic materials are, both chemically and topologically, extremely diverse, consisting of single- or double-stranded DNA or RNA, covalently linked to proteins or chemically modified termini, and existing as a single molecule linear, separate segment linear, or circular forms.8,9,13 Hence, this area of studies has focused on all types of viruses including bacteriophages (prokaryotic-infection viruses), endogenous viruses (viral elements, virus-derived genetic elements, prophages, and endogenous retroviruses in host genomes), and eukaryotic viruses (eukaryotic-infection viruses), as shown in Figure 1.
Figure 1.
Overview of trans-kingdom interaction among viromes and hosts community. Figure created using BioRender (https://biorender.com/). (A color version of this figure is available in the online journal.)
The vast majorities of viruses present on earth are lysogenic (with a latent period in host cells known as prophase) and lytic (infecting and killing the host shortly) phages which influence bacterial communities. With viral metagenomic approaches, bacteriophages were found to represent a large proportion of the microbial community. 11 Indeed, examinations of the human gut virome have revealed that crAssphage and its relatives are the predominant viruses in the gut viral community. 14
The endogenous viruses, such as adenoviruses, herpesviruses, polyomaviruses, and circoviruses, provide invaluable information of ancient viral infections by integrations of the virus genome to their hosts. Generally, these viruses have been revealed to be associated with mammalian evolution (i.e., biological functions, emerging of placentation, immune adaptation and modulation, oncogenesis, and disease progression).15,16 Moreover, approximately 8% of the human genome contains endogenous virus genes, related to many diseases including cancers, amyotrophic lateral sclerosis, multiple sclerosis, and rheumatoid arthritis.17–19
Many infectious diseases in both humans and animals are caused by eukaryotic viruses (e.g., influenza viruses, rotavirus, arboviruses, hepatitis C virus, and HIV). Indeed, these viruses (> 60%) have been revealed to possess the capacity of transmission across host species (zoonotic transmission), leading to major problems around the world (i.e., pandemic, economic loss, and human health issues).20,21 Therefore, the virus metagenomic approaches have been emphasized as novel strategies of virus surveillance, providing information of the entire viral community and having a potential for further applications to be discussed below.22,23
Viral metagenomics
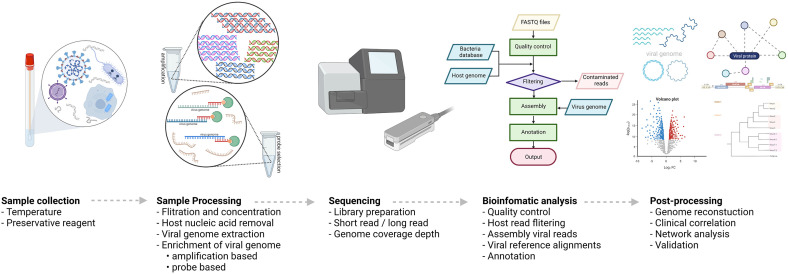
The development of high-throughput next-generation sequencing (NGS) provides both insights and possibility in the accurate and timely detection and sequencing of nearly the full genome of several viruses in both clinical and research areas.11,12,24,25 Fundamentally, there are five main processes for virome research workflow, including (i) sample collection, (ii) sample processing, (iii) sequencing, (iv) bioinformatic analysis, and (v) post-processing, as shown in Figure 2.26–29
Figure 2.
The workflow of virome research based on metagenomic technique. Figure created using BioRender (https://biorender.com/). (A color version of this figure is available in the online journal.)
(i) Sample collection. One of the most important steps is sample collection, since the accuracy and reliability of the analysis outcome can be significantly influenced by this process. Indeed, the main objectives are to preserve specimens (i.e., viral nucleic acids and community integrity) and minimize possible confounding factors for further downstream processes. To these aims, the recommended and considered condition as gold standard for long-term storage is −80°C. Alternatively, preservative reagents may be required for the samples from remote areas (i.e., rural or forest) until access to a −80°C freezer is provided.30,31 In addition to storage temperature, freeze–thaw cycles also impact on the analysis outcome, such as virus and other microbe reads, by reducing the viral nucleic acid integrity in certain viruses.32–34
(ii) Sample processing. Typically, the aim of this process is to minimize host and environmental burdens (i.e., host genomes and cellular debris) and to maximize viral genome as much as possible for the requirement of the sequencing process to be fulfilled.25,35 Of note, the filtration strategies can be carried out by passing the sample through porous filters; however, the size of targeted viruses is of concern. This is because, most often and commercially, the pore size of the filters is 0.45, 0.22, or 0.1 μm, so that large viruses (i.e., giant viruses and vaccinia viruses) may be retained in the filters, reducing the recovery yield, leading to underrepresentation of the viruses.25,36 In this case, pretreatment of the filters with appropriate reagents (i.e., buffers, 10% fetal calf serum, or veal infusion broth) may alleviate the retention of the viruses. 37 A nuclease treatment may also be required for high level of host and bacterial genome contaminations before the viral nucleic acid extraction process.38,39 For instance, samples for study of a whole bacteriophage community are usually contaminated with bacterial host genomes which may affect the downstream analysis due to the phages sharing genes with homology to bacterial genes.40–42 Once the viral particles have been enriched, the appropriate viral genome extraction is required (i.e., formamide, thermal shock, or Phenol:Chloroform:Isoamyl alcohol extraction procedures, depending upon sample types). To enrich the viral genome, retro-transcription and/or amplification strategies, such as SISPA, RP-SISPA, LASL, and MDA, may be carried out before the library preparation.43–46 Alternatively, the recent strategy of probe-capture-based technique for specific viruses (i.e., VirCapSeq-VERT for vertebrate viruses and Twist Respiratory Virus Research Panel for respiratory viruses) may helpfully enrich the targeted viral genomes.47,48
(iii) Sequencing. For virome metagenomic analysis, the sequencing platforms should be carefully selected and conducted, depending on sample types, viral genome yield for minimum concentration requirement of library preparation, the objective of the study, and the influence of the platforms on the number of viral reads and coverages.49,50 In recent years, long-read sequencing platforms, such as Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT), have gradually increased interest in virome research on account of the advantages in the long-read datasets (i.e., detection of methylated sequences within viral genomes and high power of viral discovery for novel viruses).51–53
(iv) Bioinformatic analysis. In microbiome research, bioinformatic analysis pipelines for virome analysis essentially involve the non-viral genome removals based on in silico approaches to reduce computational resources and time-consuming analysis processes. 54 Indeed, there are many bioinformatic tools with different objectives for specific virome research. Moreover, they have shared common procedures, including de novo assembly before being aligned with reference genomes, as well as representative protein annotation from the virus sequences. 55 This review summarizes both commonly used bioinformatic pipelines and tools for virus identification by viral genome alignment and/or assembly, including the detection of all viruses, bacteriophages, and endogenous viruses with integration sites (Table 1), and viral reference databases with their main applications in virome research (Table 2).
(v) Post-processing. Once viral sequencing data sets have been completely analyzed in the bioinformatic pipeline, this process usually involves data interpretation and validation.55,56 Typically, the assembled viral genomes will be investigated for their genome coverage. In case of poor viral genome constructions, the specific set of primers will be applied for the virus genomes that will be resequenced in order to obtain their complete genomes before other downstream processes. 57 As for novel viruses, validation strategies (i.e., RT-PCR and Sanger sequencing) are usually performed to ensure the findings. In addition, viral communities in the samples such as clinical specimens may often be required in the clinical interpretations. This is through visualization and statistical processes (i.e., heat map, phylogenetic construction, virome profiling, and network analysis) to obtain an insight into the viral community and the specimens.11,16,38
Table 1.
Selected bioinformatic tools and pipeline for virome analysis.
| Tools | Description | Strength | Year | References |
|---|---|---|---|---|
| Viruses | ||||
| METAVIRALSPADES | Identification of viral genomes by using a set of virus-specific hidden Markov models for metagenomic assembly analyzing with variations and coverage depth | • Neural network analysis • Novel virus discovery in diverse metagenomic datasets |
2020 | Antipov et al. 58 |
| virMine | Identification of viral genomes from raw reads representative of viral or mixed (viral and bacterial) communities using an iterative approach for read quality control, assembly, and annotation | • Alternative mode between specific study system and/or feature(s) of interest • Novel species detection |
2019 | Garretto et al. 59 |
| Kraken 2 | Classification and assigning taxonomy of metagenomic sequences with BLAST program in the fastest mode | • Low memory usage • High speed • High sensitivity |
2019 | Wood et al. 60 |
| FastViromeExplorer | Detection and abundance quantification of viruses and phages in large datasets by performing rapid searches with pseudo-alignment tool for RNA-seq data | • RNA-seq data analysis • Rapid mapping of short metagenome reads • Suitable for limited computing power research |
2018 | Tithi et al. 61 |
| VirMAP | Combination of nucleotide and protein metagenomic datasets for taxonomic classification of viral genome reconstructions | • Combinatorial analysis of nucleotide and protein sequences • Virus surveillance capabilities |
2018 | Ajami et al. 62 |
| EZ-Map | Metagenomic analysis of human virome with python-based tools for filtering, alignment, and analysis from cell-free DNA data sets | • Fully automated computational pipeline for both workstations and computing clusters • Suitable for cell-free DNA datasets |
2017 | Czeczko et al. 63 |
| VirusDetect | Using small RNA sequences strategy with homology of reference-alignment and de novo assembly | • Small RNA sequence analysis • Potential novel virus identification • Highly sensitive and efficient identification |
2017 | Zheng et al. 64 |
| VirFinder | Identification of viruses by using machine learning with k-mer based approach for mixed metagenomes containing both viral and host sequences | • A web-based tool • An alignment-free tool using machine learning • High potential to detect novel virus |
2017 | Ren et al. 65 |
| VirusSeeker | BLAST-based NGS data analysis pipeline for both novel virus discovery and virome composition analyses | • False-positive removal • Detection of both RNA and DNA viruses in different families. |
2017 | Zhao et al. 66 |
| Bacteriophage | ||||
| VIBRANT | The hybrid tool using machine-learning and protein-similarity approach for recovery and annotation of viruses and microbes with the curation of predictions, estimation of genome quality, and infection mechanism | • Low false positive • Discovery of phage–microbe interactions |
2020 | Kieft et al. 67 |
| PPR-Meta | Identification of both phage and plasmid fragments from metagenomic using Bi-path convolutional neural network | • Available for a local PC • Identification of phages and plasmids • Novel phage identification |
2019 | Fang et al. 68 |
| MARVEL | Using a random forest machine-learning approach for prediction of double-stranded DNA bacteriophage sequences in metagenomic bins | • High sensitivity • Novel phage identification |
2018 | Amgarten et al. 69 |
| PHASTER | Phage search tool for identifying and annotating prophage sequences within bacterial genomes and plasmids | • Web-based tool • Identification and annotation of prophage sequences |
2016 | Arndt et al. 70 |
| Endogenous virus | ||||
| DeepVISP | Viral integration site prediction using convolutional 6 neural network (CNN) models in the human genome | • Online tool server • Accurate prediction of oncogenic virus integration sites • Identification of biological or regulatory roles with unknown integration site |
2021 | Ren et al. 71 |
| detectedIS | Identification of exogenous DNA integration sites in a plasmid containing transgenes or virus sequences based on a Nextflow workflow combined with a singularity | • Able to use DNA or RNA paired-end sequencing datasets • Accurate and lower computational demand with less execution times. |
2021 | Grassi et al. 72 |
| SurVirus | Viral integration caller with alignment correction of reads for the discovery of integrated sites | • Detection of novel virus integration site with less noise • Quick scan large data sets |
2021 | Rajaby et al. 73 |
| VIcaller | Identification of viral integration events using high-throughput sequencing (HTS) from human dataset through virome-wide screening of clonal integrations under Linux platform. | • Identification of breakpoint of viral integrations in human genome caused cancers • Compatible with whole genome and RNA-seq datasets |
2019 | Chen et al. 74 |
| Seeksv | Detection of somatic structural variants and viral integration using different types of sequencing data | • High efficiency and precision • Identification of breakpoint located in sequence homology regions |
2017 | Liang et al. 75 |
Table 2.
Database of virome.
| Database | Main applications | Source link | References |
|---|---|---|---|
| EBI | European Nucleotide Archive • Viral reference sequences • Viral taxonomy |
https://www.ebi.ac.uk/genomes/virus.html | – |
| HVPC | The human virome protein cluster • Human viral protein database • Diversity • Functional annotation |
https://osf.io/gs4zf/ | Elbehery et al. 76 |
| IMG/VR | Integrated microbial genome/virus • Cultured and uncultured DNA/RNA viral genome sequences • Integrated ecological and evolutionary database |
https://img.jgi.doe.gov/cgi-bin/vr/main.cgi | Paez-Espino et al. 77 |
| MVP | Microbe versus phage • A microbe–phage interaction database |
http://mvp.medgenius.info/home | Gao et al. 78 |
| NCBI virus | National Center for Biotechnology Information Virus • Viral reference sequences • Viral taxonomy |
https://www.ncbi.nlm.nih.gov/labs/virus/vssi/ | Hatcher et al. 79 |
| PhagesDB | Actinobacteriophage database • Genomics of phages • Bacterial hosts • Viral taxonomy |
https://phagesdb.org/ | Kaján et al. 15 |
| pVOGs | Prokaryotic Virus Orthologous Groups • Complete set of orthologous gene families • Multiple complete genomes of bacterial or archaeal viruses |
http://dmk-brain.ecn.uiowa.edu/pVOGs | Grazziotin et al. 80 |
| ViPR | Virus Pathogen Resource • Viral reference sequences • Gene and protein annotations • Immune epitopes • 3D structures • Host factor data |
https://www.viprbrc.org/brc/home.spg?decorator=vipr | Pickett et al. 81 |
| ViralZone | Virus knowledge resource • A comprehensive resource of viral genomic and proteomic sequences • Reference strains • Virion pictures |
https://viralzone.expasy.org/ | Hulo et al. 82 |
| Virus–Host DB | Virus–Host Database • Relationship of viruses and hosts • Taxonomy viruses and their hosts • Complete genomes of reference sequences and host sequences |
https://www.genome.jp/virushostdb/ | Mihara et al. 7 |
| ViruSurf | ViruSurf: an integrated database • Integrated and curated metadata and viral sequences from heterogeneous sources • Analytical dimension characterizes the sequence |
http://gmql.eu/virusurf/ | Canakoglu et al. 83 |
Trans-kingdom interaction
The advancements in sequencing technology have provided the possibility of elucidating the microbial community. To understand how biological events are associated with the clinical outcome/manifestation of the diseases, the studies of viral interactions between other microbes (i.e., bacteria and fungi) as well as viral–host interactions by using combinatorial approaches (i.e., conventional cell biology and NGS technology) can provide the insight of both direct and indirect relationships among trans-kingdom interactions.
Virus–microbe interaction
To this point, numerous studies have investigated the trans-kingdom relationship between microbes living inside human and animal bodies, especially the viral–bacterial relationship. Fundamentally and essentially, viruses have to bind to bacteria or bacterial products for initiating interactions. The viral–bacterial interactions can be simply classified into two major categories: direct and indirect interaction.11,84,85
For direct interaction, viruses and bacteria directly bind to each other for mutual benefit. Previously, several studies have revealed that the bacteria localized within host organisms promote the viral infections and vice versa.86,87 Indeed, the respiratory bacterial pathogens, such as Streptococcus pneumoniae, Moraxella catarrhalis, and Staphylococcus aureus, have an enhanced adherence capacity to the host epithelial cells when these microbial surfaces are bound with influenza virus, promoting the secondary bacterial infection. Moreover, viral co-infection of different enteric viral strains can be enhanced by the bacterial mediation, and the viral fitness can also be increased by microbes through genetic recombination events of these viruses, reducing the deleterious mutations.88–90
The other benefit of the interaction among viruses and bacteria is virion stability. In this case, bacteria provide the microenvironment of their cell walls (i.e., lipopolysaccharide, peptidoglycan, lipoteichoic acid, and N-acetylglucosamine-containing polysaccharides) to the viruses to enhance the stability of the virion during transmission or environmental exposure.90–93 For example, virion thermal stability of poliovirus, coxsackievirus, human norovirus, and reovirus is promoted in the presence of certain bacteria, while Aichi virus and mengovirus have been discovered to be more vulnerable. Moreover, the interaction among bacteria and viruses has been shown by the increased capacity of bleach.90,93 These studies have revealed, and provided clues concerning, the enhancement of the chance and duration of their active progeny to survive during environmental exposure before binding to their target host cells. From this interaction, it has been intriguing to establish further investigation of the wider aspects of the virome–bacteriome association, to gain more knowledge and understanding of the viral–bacterial relationship.
Furthermore, the viruses and bacteria can be indirectly correlated through bacterial production. In the case of human noroviruses and commensal enteric bacteria, the histo-blood group antigen (HBGA) expressing bacteria may promote the viral infection to the human B cells by some unknown mechanisms. 94 The biotransformations of bacteria are prominent in the gut microbiome. In fact, bile acids can be transformed by gut microbes to secondary bile acids and other products. 95 In the presence of certain bile acids, the receptor binding protruding domain of noroviruses is stabilized and the binding capacity to the host cells is enhanced through corrected conformational rotation of the domain by the electrostabilizing effect between the viral domain and the bile acids.96,97
Although the conventional biological approaches have offered an insight into the viral–bacterial interactions by focusing on a part of the community, the residual microbiota (i.e., bacteriophages, fungi, and other bacteria and viruses) should also be investigated to complete our knowledge of trans-kingdom relationships. Metagenomic approaches to the viral studies provide a different point of view from those of conventional approaches in the focus on the whole community.38,98–100 Nonetheless, in most of the virome studies focusing on the viral–bacterial interaction, the viruses and other microbial relationships (i.e., viral–fungal interaction) have not yet been well studied. Thus, future investigations of virome studies should be established in different aspects of the community, in order to reveal the uncovered parts of the microbiota relationships, and ultimately lead to novel therapeutic strategies.
Virus–host interaction
Through the virome metagenomic approach, several studies have revealed that there are numerous viruses residing in/on the human body, such as skin, oral cavity, lung, gastrointestinal tract, and blood, with both commensal and pathogenic associations with their hosts.11,85
With respect to virus–host interactions, previous studies have shown that host behavior and geographic locations have influences on viruses. The dietary factor is an obvious example, having an effect on viral community structure. The viral infections of infant guts and mortality from viral gastroenteritis are lowered in infants who are received breastfeeding.101–103 In fact, maternal breast milk is composed of many components (i.e., maternal antibodies, oligosaccharides, and lactoferrin) which influence the viral community, especially Adenoviridae, Picornaviridae, Parvoviridae, and Caliciviridae in the early life.11,101,102,104,105 Furthermore, a recent study has demonstrated that food types (i.e., staple foods, side dishes, fruits, and beverages) have been correlated to the virome structure in the human body. 106
In addition to the host’s behavior, host geography is one of the main factors affecting virome structure.106–109 A comparative study of diarrhea-related viruses has shown that Picornaviridae and Adenoviridae are significantly different between 2 different locations in Australia. 109 Moreover, a study of geography affecting virome structure among Chinese cohorts has revealed that viral communities among the cohorts are significantly distinct in the manner of virome diversity, evenness, and richness.106,108 These may indicate that the hosts’ behavior and geography are quite prominent effects on virome structure and could be categorized as a form of interaction between viruses and their hosts.
For some instances, eukaryotic viruses may provide benefits to their hosts. In gnotobiotic mice, murine norovirus (MNV) has been shown to have a beneficial function of compensating for commensal bacterial depletion. Of note, the virus also restored the intestinal morphology and lymphocyte function without any obviously adverse effects. 110 Likewise, a form of viral–host interaction has been discovered between astrovirus and the immunodeficient mouse host, in which the hosts were protected from MNV and rotavirus infection via inducing type III interferon inside the guts. 111
Bacteriophages may indirectly interact with eukaryotic hosts through modification of bacterial composition and genetics or stimulation of host immune responses. 112 One study of Crohn’s disease, for example, has shown the potential of phage therapy in reducing adherent invasive Escherichia coli. 113 In addition, phages may provide the reservoir of mobilizing genetic elements to their bacterial hosts for antimicrobial resistance.43,114,115 This may promote the severity and mortality in some cases through indirect interaction between eukaryotic hosts and bacteriophages. For eukaryotic host immunity, bacteriophages could stimulate the host immune response, without bacterial mediation, through Toll-like receptor (TLR) signaling, in which type I interferon is induced by the phages as well as the production of IL-6, IL-10, IL-12, and IFNγ.116,117
Challenges and limitations
Viruses are the most diverse on Earth with the total number of virus-like particles estimated to be 10 31 but only 1% have been discovered. 118 The advent of high-throughput sequencing technology, particularly metagenomic technologies, allows researchers to access the complexities of microbial communities including bacteriome, mycobiome, and virome. Although more advances of the technologies continually come forward, the virome studies still have their own challenges and limitations related to their sample and downstream analysis processes.27,56,119 The major obstacles include comprehensively defining viral dark matter in the virome data set, and virome–host interaction studies, particularly virome studies in animal model experiments, are still limited for many reasons as described below.
The viral dark matter
Viral dark matter is defined as the nucleotide sequence which originates from viruses that cannot align with any reference nucleotide or amino acid sequence. 120 In previous virome research that uses a purifying viral particles methodology, almost 40% to 90% of sequence were unalienable.121–123 This challenge has been limited for a number of reasons that will be described in the two primary procedures (sample processing and bioinformatics) approaches.
Sample processing
In recent years, many attempts have been efficaciously applied to decrease the amount of viral dark matter. Even though there is no standard protocol for identifying all viruses, specific methods are assigned by specific attributes of a subgroup. Thus, these specific methods are significant when defining all compositions of the virome diversity.
For example, most clinical samples usually suffer from a low abundance of viruses along with a high background of their hosts or other microbes.24,124 Using a filter of 0.22 μm may remove host cellular or bacterial burdens from the viral entities; however, this strategy can also deplete large viruses and reduce the amount of recovered viral DNA by half, establishing biases toward the most abundant of the viral community members. 125 Likewise, some phages have atypical buoyancy; thus, the CsCl gradient ultracentrifugation may promote the bias to specific phage types.33,41 As with other techniques, this method still has some drawbacks including reducing the Virus-like particles (VLPs) and decreasing the sensitivity of viral detection. 126 Furthermore, most commercial genomics extraction kits focus only on either DNA or RNA. This may produce biases toward some type of viruses due to the viral community in virome research containing both DNA and RNA viruses. 127
Although the virome provides culture-independent sequencing for most viruses without group-specific primers to differentiate the viral species like the 16s or ITS region in bacteria and fungi, respectively, the confounding noise from the background still impacts on sequencing datasets. 62 In addition, the metagenomic approach also recommends using the viral mock community aid in assessing biases into a virome pipeline. However, the viral mock communities comprise a limited number of reference materials to cover all types of different genetic materials.
Bioinformatics approaches
The next limitation in virus discovery is that none of the computational methods can always clearly identify viral sequences. Besides, some computational approaches limit the identification of novel viruses, such as nucleotide alignment.
However, to address the viral sequences, de novo assembly has been considered a major approach in this area. 42 As the complexity of the virome sequencing data increases (i.e., many repeat regions and genomic diversity), the assembly seems to be challenged in many studies, particularly in the validity of the viral annotation. 42 In some cases, during viral genome assembly, mismatches from synonymous single-nucleotide polymorphisms can arise due to limitations of the tools and databases used. One strategy to resolve this issue is utilizing the viral protein amino acid sequences, as in Plass. 128 This use of viral protein amino acid sequences and/or machine-learning analysis has shown potential to predict novel viruses from the sequences and to improve function prediction.42,128,129
The database is a critical module for identifying viral sequences. Although the number of viruses in the database has been rapidly increased, the sequences are largely biased toward mammalian, plant, and bacterial viruses. There is still a small number of fungal protist and archaeal viruses in RefSeq GenBank. 79 In this regard, a number of selected bioinformatics tools and databases are summarized in Tables 1 and 2. Furthermore, the limits of identified viral diversity by Baltimore classification and host relations also possibly encourage the high incidence of viral dark matter.
Unrevealed virome–host interaction studies
Although some virus–host interaction has been elucidated by combinatorial approaches to understand the biological events, two of the biggest remaining challenges are uncultured viruses and animal models.
Uncultured viruses
Most of the viruses discovered through metagenomic approaches are unculturable viruses. This raises the topic of discussion of these viruses in identification of their host ranges, especially bacteriophages. As aforementioned, bacteriophages may indirectly interact with eukaryotic hosts through modification of bacterial entities. Therefore, the determination of their bacterial host range may give an indication of their trans-kingdom relationship. Currently, the culture-independent approaches have been utilized for host range prediction such as viral tagging and in silico predictions.130–133 For viral tagging, this technique utilizes fluorescence-activated cell sorting to separate the labeled phages, binding to their hosts, for further downstream analyses.132–134 Alternatively, in silico predictions may apply the correlation among phages and their bacterial abundances as well as genetic signatures of phages (i.e., genetic homology between phages and bacteria, prophage integration, or CRISPR) to determine the host ranges.35,131
For eukaryotic viruses and bacteriophages, the lack of culture systems is a major limitation for virome research to accomplish Koch’s postulations, the gold standard for microbial disease causality.135–137 Indeed, previous studies have established primary airway epithelial cells to characterize the HCoV-HKU1 and human bocavirus.138,139 To understand the nature of viruses in the gastrointestinal (GI) tract, the development of an organoid may be useful for investigation of the viruses and host interactions and may open the opportunity to establish a novel strategy of mock microbial communities.36,140
Animal models
It is well known that viruses are highly diverse and infect either eukaryotic or prokaryotic cells as obligate parasites. Thus, a specific host to establish specific types of animal models is necessary for virome research in this experiment. In order to manage the practical and ethical constraints, functional studies of the virome in humans should be limited, making animal models an invaluable experimental tool to understand its impact on physiology. 141
Mice have been used as the robust models of virome-associated disorders. Thus, mice are often used as a proxy for human diseases to decipher their pathophysiology or test new therapeutics. Virome diversity correlates with several pathological conditions, such as inflammatory bowel disease, arthritis, and child growth impairment.12,142,143 Macaques are also closely related to humans and are, therefore, a pertinent model to study parameters that influence the human virome. Cynomolgus macaques and Rhesus macaques have recently been used to analyze the effect of aging and chronic diarrhea on the virome, respectively.144,145 Recently, rabbits have been used, leading to the identification of a novel polyomavirus. 146 Finally, swine models have also been used for virome in fecal virome transplants, which have been shown to act against necrotizing enterocolitis in pigs. 147
However, the laboratory animals grow under specific pathogen-free (SPF) conditions, which means they have lower compositions of other viruses and other microbes compared to the natural animal. 148 Overall, the impact of the virome on most animal studies is still unclear; in particular, the phage physiological roles remain unclear in the host (human or animal) and bacterial communities. A further challenge lies in dissociating the effect of viruses from those of bacteria and between individual viruses, with the aim of identifying the mechanism of the virus in human health and disease. 141
Moreover, in terms of data interpretation, one of the major challenges in virome research is determining the relationship between viruses and other factors in the community (i.e., age, genetic, health status, and/or diet of their hosts).149,150 The multiple factor analysis (MFA) technique has been used in this area to examine the multivariate correlation between the viruses and their hosts.151–153
Potential applications of virome research
Although there are many limitations in virome research, several applications of this field are continuously and increasingly being realized in both biological aspects and clinical areas. For biological applications, the virus surveillances using viral metagenomics have shown the possibility to expand virus databases, in which PCR- or panel-based techniques may be limited to the viruses.38,154 Indeed, the virus databases might be utilized as reference genomes of both known and novel viruses as well as viral reference communities for standardized viral metagenomic approaches.
For clinical applications, viral metagenomics allows our understanding of host and microbiota relationships to be enhanced, which leads to possible alternative therapeutic strategies for several diseases. For example, oncolytic therapy based on viruses introduced into cancer cells (e.g., HSV, vaccinia virus, and adenovirus) has recently revealed potential modulations of the tumor microenvironment and cancer-related immunity in anticancer therapy.155,156 Moreover, the efficacy of fecal microbiota transplantation has been demonstrated as a potential treatment for inflammatory bowel disease (IBD), Clostridioides difficile infection, severe colitis associated with graft-versus-host disease following hematopoietic stem cell transplantation, and obesity.55,157–160 In addition, phage-based therapy might offer potential treatments for IBD caused by C. difficile and Escherichia coli, as well as for colorectal cancer, in which the genes associated with cancer are down-regulated after being treated with E. coli bacteriophages. However, there are some issues of concern for the phage treatments, including the appropriate life cycle of phages for effective therapeutic effects, standardization of the protocol and legal frameworks for clinical application, and horizontal gene transfers across kingdoms.64,113,161,162
Diagnostically and prognostically, the virome approach may be applied to identify and reveal other potentially pathogenic viruses in clinical metagenomic fields, such as torque teno virus 7 for Kawasaki disease. 163 Furthermore, respiratory failure, periodontitis, and other illnesses have also been shown to have associations between viruses and symptoms using human virome approaches.28,164 In this regard, the clinical applications of viral metagenomics in parallel with conventional practices may result in novel patient management and therapeutic strategies. 16
Finally, virome research can also be used to address potential zoonotic viral transmissions. Indeed, the recent pandemic caused by SARS-CoV-2 may possibly come from zoonotic transmission, as several studies have demonstrated that viral transmission across species from bats (serving as original reservoirs) to pangolin which may have served as an intermediate host. 165 In addition, a study of fecal virome between wild and captive cynomolgus macaques has revealed three novel macaque viruses and illustrated the potential zoonotic transmissions of rabies and herpes B viruses to humans.38,166,167 These insights could benefit our understanding and management of viral outbreaks in the future, especially in combination with viral metagenomic and artificial intelligence prediction.168,169
Conclusions
In conclusion, the advent of metagenomics has changed the face of virome research through providing an insight into whole viral communities. Utilizing virome approaches, the interactions of both viruses and their hosts, as well as other microbial members in the community, can be accessed. Moreover, combinatorial analyses of virome and other systemic microbiology approaches (i.e., metabolomic, transcriptomic, culturomic, and proteomic) may allow us to establish models and interaction networks, used for specific purposes, such as pandemic preventions, regulation of virus populations, and research applications, based upon the roles of viruses in communities. However, there are many challenges and limitations in current techniques. Therefore, further inventions and investigations are essential for addressing the viral community and their relationships.
Footnotes
Authors’ Contributions: SC and SP contributed to the conception of this review article. SC and PS contributed writing and discussion of this review article. The final version of this article was approved by all authors.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The authors would like to thank the Thailand Government’s Development and Promotion of Science and Technology Talents Project (DPST) for scholarship. This research project is supported by Thailand Science Research and Innovation Fund (TSRI) [Grant Number CU_FRB640001_01_30_4].
ORCID iD: Sunchai Payungporn  https://orcid.org/0000-0003-2668-110X
https://orcid.org/0000-0003-2668-110X
References
- 1. Greninger AL. A decade of RNA virus metagenomics is (not) enough. Virus Res 2018;244:218–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Santiago-Rodriguez TM, Hollister EB. Human virome and disease: high-throughput sequencing for virus discovery, identification of phage-bacteria dysbiosis and development of therapeutic approaches with emphasis on the human gut. Viruses 2019;11:656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vieites JM, Guazzaroni M-E, Beloqui A, Golyshin PN, Ferrer M. Metagenomics approaches in systems microbiology. FEMS Microbiol Rev 2008;33:236–55 [DOI] [PubMed] [Google Scholar]
- 4. Fondi M, Liò P. Multi-omics and metabolic modelling pipelines: challenges and tools for systems microbiology. Microbiol Res 2015;171:52–64 [DOI] [PubMed] [Google Scholar]
- 5. Malard F, Dore J, Gaugler B, Mohty M. Introduction to host microbiome symbiosis in health and disease. Mucosal Immunol 2021;14:547–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guo G, Ye L, Shi X, Yan K, Huang J, Lin K, Xing D, Ye S, Wu Y, Li B, Chen C, Xue X, Zhang H. Dysbiosis in peripheral blood mononuclear cell virome associated with systemic lupus erythematosus. Front Cell Infect Microbiol 2020;10:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mihara T, Nishimura Y, Shimizu Y, Nishiyama H, Yoshikawa G, Uehara H, Hingamp P, Goto S, Ogata H. Linking virus genomes with host taxonomy. Viruses 2016;8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Virgin Herbert W. The virome in mammalian physiology and disease. Cell 2014;157:142–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wylie KM, Weinstock GM, Storch GA. Emerging view of the human virome. Transl Res 2012;160:283–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li N, Ma WT, Pang M, Fan QL, Hua JL. The commensal microbiota and viral infection: a comprehensive review. Front Immunol 2019;10:1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang G, Bushman FD. The human virome: assembly, composition and host interactions. Nat Rev Microbiol 2021;19:514–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khan Mirzaei M, Xue J, Costa R, Ru J, Schulz S, Taranu ZE, Deng L. Challenges of studying the human virome—relevant emerging technologies. Trends Microbiol 2021;29:171–81 [DOI] [PubMed] [Google Scholar]
- 13. Cantalupo PG, Pipas JM. Detecting viral sequences in NGS data. Curr Opin Virol 2019;39:41–8 [DOI] [PubMed] [Google Scholar]
- 14. Koonin EV, Yutin N. The crAss-like phage group: how metagenomics reshaped the human virome. Trends Microbiol 2020;28:349–59 [DOI] [PubMed] [Google Scholar]
- 15. Kaján GL, Doszpoly A, Tarján ZL, Vidovszky MZ, Papp T. Virus–host coevolution with a focus on animal and human DNA viruses. J Mol Evol 2020;88:41–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet 2019;20:341–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chiu ES, VandeWoude S. Endogenous retroviruses drive resistance and promotion of exogenous retroviral homologs. Ann Rev Animal Biosci 2021;9:225–48 [DOI] [PubMed] [Google Scholar]
- 18. Garcia-Montojo M, Doucet-O’Hare T, Henderson L, Nath A. Human endogenous retrovirus-K (HML-2): a comprehensive review. Crit Rev Microbiol 2018;44:715–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gröger V, Cynis H. Human endogenous retroviruses and their putative role in the development of autoimmune disorders such as multiple sclerosis. Front Microbiol 2018;9:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gumi B, Schelling E, Berg S, Firdessa R, Erenso G, Mekonnen W, Hailu E, Melese E, Hussein J, Aseffa A, Zinsstag J. Zoonotic transmission of tuberculosis between pastoralists and their livestock in South-East Ethiopia. Ecohealth 2012;9:139–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huber C, Finelli L, Stevens W. The economic and social burden of the 2014 Ebola outbreak in West Africa. J Infect Dis 2018;218:S698–704 [DOI] [PubMed] [Google Scholar]
- 22. Jonas O, Seifman R. Do we need a global virome project. Lancet Glob Health 2019;7:e1314–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carlson CJ, Farrell MJ, Grange Z, Han BA, Mollentze N, Phelan AL, Rasmussen AL, Albery GF, Bett B, Brett-Major DM, Cohen LE, Dallas T, Eskew EA, Fagre AC, Forbes KM, Gibb R, Halabi S, Hammer CC, Katz R, Kindrachuk J, Muylaert RL, Nutter FB, Ogola J, Olival KJ, Rourke M, Ryan SJ, Ross N, Seifert SN, Sironen T, Standley CJ, Taylor K, Venter M, Webala PW. The future of zoonotic risk prediction. Philos T Royal Soc B: Biol Sci 2021;376:20200358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haynes M, Rohwer F. The human virome. In: Nelson KE. (ed.) Metagenomics of the human body. Berlin: Springer, 2011, pp.63–77 [Google Scholar]
- 25. Kleiner M, Hooper LV, Duerkop BA. Evaluation of methods to purify virus-like particles for metagenomic sequencing of intestinal viromes. BMC Genom 2015;16:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barzon L, Lavezzo E, Militello V, Toppo S, Palù G. Applications of next-generation sequencing technologies to diagnostic virology. Int J Mol Sci 2011;12:7861–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol 2012;10:607–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Noell K, Kolls JK. Further defining the human virome using NGS: identification of Redondoviridae. Cell Host Microbe 2019;25:634–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Van Rijn-Klink A, De Vries JJ, Claas EC. Next-generation sequencing in clinical virology. In: Moran-Gilad J, Yagel Y. (eds) Application and integration of omics-powered diagnostics in clinical and public health microbiology. Berlin; Heidelberg: Springer, 2021, pp.89–110. [Google Scholar]
- 30. Kohl C, Wegener M, Nitsche A, Kurth A. Use of RNALater® preservation for virome sequencing in outbreak settings. Front Microbiol 2017;8:1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nel Van Zyl K, Whitelaw AC, Newton-Foot M. The effect of storage conditions on microbial communities in stool. PLoS ONE 2020;15: e0227486–1022786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hirst JC, Hutchinson EC. Single-particle measurements of filamentous influenza virions reveal damage induced by freezing. J Gen Virol 2019;100:1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shkoporov AN, Ryan FJ, Draper LA, Forde A, Stockdale SR, Daly KM, McDonnell SA, Nolan JA, Sutton TD, Dalmasso M. Reproducible protocols for metagenomic analysis of human faecal phageomes. Microbiome 2018;6:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Granados A, Petrich A, McGeer A, Gubbay JB. Measuring influenza RNA quantity after prolonged storage or multiple freeze/thaw cycles. J Virol Methods 2017;247:45–50 [DOI] [PubMed] [Google Scholar]
- 35. Sathiamoorthy S, Malott RJ, Gisonni-Lex L, Ng SHS. Selection and evaluation of an efficient method for the recovery of viral nucleic acids from complex biologicals. NPJ Vaccines 2018;3:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ly M, Jones MB, Abeles SR, Santiago-Rodriguez TM, Gao J, Chan IC, Ghose C, Pride DT. Transmission of viruses via our microbiomes. Microbiome 2016;4:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ver BA, Melnick JL, Wallis C. Efficient filtration and sizing of viruses with membrane filters. J Virol 1968;2:21–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sawaswong V, Fahsbender E, Altan E, Kemthong T, Deng X, Malaivijitnond S, Payungporn S, Delwart E. High diversity and novel enteric viruses in fecal viromes of healthy wild and captive Thai cynomolgus macaques (Macaca fascicularis). Viruses 2019;11:971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kumar R, Nagpal S, Kaushik S, Mendiratta S. COVID-19 diagnostic approaches: different roads to the same destination. Virusdisease 2020;31:97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Edwards RA, McNair K, Faust K, Raes J, Dutilh BE. Computational approaches to predict bacteriophage–host relationships. FEMS Microbiol Rev 2016;40:258–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Callanan J, Stockdale SR, Shkoporov A, Draper LA, Ross RP, Hill C. RNA phage biology in a metagenomic era. Viruses 2018;10:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sutton TDS, Hill C. Gut bacteriophage: current understanding and challenges. Front Endocrinol 2019;10:784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fancello L, Desnues C, Raoult D, Rolain JM. Bacteriophages and diffusion of genes encoding antimicrobial resistance in cystic fibrosis sputum microbiota. J Antimicrob Chemother 2011;66:2448–54 [DOI] [PubMed] [Google Scholar]
- 44. Miranda JA, Culley AI, Schvarcz CR, Steward GF. RNA viruses as major contributors to Antarctic virioplankton. Environ Microbiol 2016;18:3714–27 [DOI] [PubMed] [Google Scholar]
- 45. Gong Z, Liang Y, Wang M, Jiang Y, Yang Q, Xia J, Zhou X, You S, Gao C, Wang J, He J, Shao H, McMinn A. Viral diversity and its relationship with environmental factors at the surface and deep sea of Prydz Bay, Antarctica. Front Microbiol 2018;9:2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yau S, Seth-Pasricha M. Viruses of polar aquatic environments. Viruses 2019;11:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Briese T, Kapoor A, Mishra N, Jain K, Kumar A, Jabado OJ, Lipkin WI. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. Mbio 2015;6:e01491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim KW, Deveson IW, Pang CNI, Yeang M, Naing Z, Adikari T, Hammond JM, Stevanovski I, Beukers AG, Verich A. Respiratory viral co-infections among SARS-CoV-2 cases confirmed by virome capture sequencing. Sci Rep 2021;11:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tanaka N, Takahara A, Hagio T, Nishiko R, Kanayama J, Gotoh O, Mori S. Sequencing artifacts derived from a library preparation method using enzymatic fragmentation. PLoS ONE 2020;15:e0227427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pérez-Cataluña A, Cuevas-Ferrando E, Randazzo W, Sánchez G. Bias of library preparation for virome characterization in untreated and treated wastewaters. Sci Tot Environ 2021;767:144589. [DOI] [PubMed] [Google Scholar]
- 51. Marine RL, Magaña LC, Castro CJ, Zhao K, Montmayeur AM, Schmidt A, Diez-Valcarce M, Ng TFF, Vinjé J, Burns CC, Nix WA, Rota PA, Oberste MS. Comparison of Illumina MiSeq and the Ion Torrent PGM and S5 platforms for whole-genome sequencing of picornaviruses and caliciviruses. J Virol Methods 2020;280:113865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cao J, Zhang Y, Dai M, Xu J, Chen L, Zhang F, Zhao N, Wang J. Profiling of human gut virome with oxford nanopore technology. Med Microecol 2020;4:100012 [Google Scholar]
- 53. Sevim V, Lee J, Egan R, Clum A, Hundley H, Lee J, Everroad RC, Detweiler AM, Bebout BM, Pett-Ridge J. Shotgun metagenome data of a defined mock community using Oxford Nanopore, PacBio and Illumina technologies. Sci Data 2019;6:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tangherlini M, Dell’Anno A, Zeigler Allen L, Riccioni G, Corinaldesi C. Assessing viral taxonomic composition in benthic marine ecosystems: reliability and efficiency of different bioinformatic tools for viral metagenomic analyses. Sci Rep 2016;6:28428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang L, Fang X, Liao H, Zhang Z, Zhou X, Han L, Chen Y, Qiu Q, Li SC. A comprehensive investigation of metagenome assembly by linked-read sequencing. Microbiome 2020;8:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Carding SR, Davis N, Hoyles L. The human intestinal virome in health and disease. Aliment Pharmacol Ther 2017;46:800–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Capobianchi MR, Giombini E, Rozera G. Next-generation sequencing technology in clinical virology. Clin Microbiol Infect 2013;19:15–22 [DOI] [PubMed] [Google Scholar]
- 58. Antipov D, Raiko M, Lapidus A, Pevzner PA. MetaviralSPAdes: assembly of viruses from metagenomic data. Bioinformatics 2020;36:4126–9 [DOI] [PubMed] [Google Scholar]
- 59. Garretto A, Hatzopoulos T, Putonti C. virMine: automated detection of viral sequences from complex metagenomic samples. PeerJ 2019;7:e6695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Gen Biol 2019;20:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tithi SS, Aylward FO, Jensen RV, Zhang L. FastViromeExplorer: a pipeline for virus and phage identification and abundance profiling in metagenomics data. PeerJ 2018;6:e4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ajami NJ, Wong MC, Ross MC, Lloyd RE, Petrosino JF. Maximal viral information recovery from sequence data using VirMAP. Nat Commun 2018;9:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Czeczko P, Greenway SC, de Koning AJ. EzMap: a simple pipeline for reproducible analysis of the human virome. Bioinformatics 2017;33:2573–4 [DOI] [PubMed] [Google Scholar]
- 64. Zheng Y, Gao S, Padmanabhan C, Li R, Galvez M, Gutierrez D, Fuentes S, Ling KS, Kreuze J, Fei Z. VirusDetect: an automated pipeline for efficient virus discovery using deep sequencing of small RNAs. Virology 2017;500:130–8 [DOI] [PubMed] [Google Scholar]
- 65. Ren J, Ahlgren NA, Lu YY, Fuhrman JA, Sun F. VirFinder: a novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome 2017;5:1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhao G, Wu G, Lim ES, Droit L, Krishnamurthy S, Barouch DH, Virgin HW, Wang D. VirusSeeker, a computational pipeline for virus discovery and virome composition analysis. Virology 2017;503:21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kieft K, Zhou Z, Anantharaman K. VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome 2020;8:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fang Z, Tan J, Wu S, Li M, Xu C, Xie Z, Zhu H. PPR-Meta: a tool for identifying phages and plasmids from metagenomic fragments using deep learning. Gigascience 2019;8:giz066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Amgarten D, Braga LPP, da Silva AM, Setubal JC. MARVEL, a tool for prediction of bacteriophage sequences in metagenomic bins. Front Genet 2018;9:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 2016;44:W16–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ren J, Song K, Deng C, Ahlgren NA, Fuhrman JA, Li Y, Xie X, Poplin R, Sun F. Identifying viruses from metagenomic data using deep learning. Quant Biol 2020;8:64–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Grassi L, Harris C, Zhu J, Hardman C, Hatton D. DetectIS: a pipeline to rapidly detect exogenous DNA integration sites using DNA or RNA paired-end sequencing data. Bioinformatics 2021;37:4230–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rajaby R, Zhou Y, Meng Y, Zeng X, Li G, Wu P, Sung W-K. SurVirus: a repeat-aware virus integration caller. Nucleic Acids Res 2021;49:e33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chen X, Kost J, Sulovari A, Wong N, Liang WS, Cao J, Li D. A virome-wide clonal integration analysis platform for discovering cancer viral etiology. Genome Res 2019;29:819–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liang Y, Qiu K, Liao B, Zhu W, Huang X, Li L, Chen X, Li K. Seeksv: an accurate tool for somatic structural variation and virus integration detection. Bioinformatics 2016;33:184–91 [DOI] [PubMed] [Google Scholar]
- 76. Elbehery AHA, Feichtmayer J, Singh D, Griebler C, Deng L. The human virome protein cluster database (HVPC): a human viral metagenomic database for diversity and function annotation. Front Microbiol 2018;9:1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Paez-Espino D, Roux S, Chen I-MA, Palaniappan K, Ratner A, Chu K, Huntemann M, Reddy TBK, Pons JC, Llabrés M. IMG/VR v. 2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res 2019;47:D678–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gao NL, Zhang C, Zhang Z, Hu S, Lercher MJ, Zhao X-M, Bork P, Liu Z, Chen W-H. MVP: a microbe–phage interaction database. Nucleic Acids Res 2017;46:D700–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hatcher EL, Zhdanov SA, Bao Y, Blinkova O, Nawrocki EP, Ostapchuck Y, Schäffer AA, Brister JR. Virus variation resource: improved response to emergent viral outbreaks. Nucleic Acids Res 2017;45:D482–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Grazziotin AL, Koonin EV, Kristensen DM. Prokaryotic Virus Orthologous Groups (pVOGs): a resource for comparative genomics and protein family annotation. Nucleic Acids Res 2016;49:D491–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pickett BE, Greer DS, Zhang Y, Stewart L, Zhou L, Sun G, Gu Z, Kumar S, Zaremba S, Larsen CN, Jen W, Klem EB, Scheuermann RH. Virus pathogen database and analysis resource (ViPR): a comprehensive bioinformatics database and analysis resource for the coronavirus research community. Viruses 2012;4:3209–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hulo C, de Castro E, Masson P, Bougueleret L, Bairoch A, Xenarios I, Le Mercier P. ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res 2011;39:D576–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Canakoglu A, Pinoli P, Bernasconi A, Alfonsi T, Melidis Damianos P, Ceri S. ViruSurf: an integrated database to investigate viral sequences. Nucleic Acids Res 2020;49:D817–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Neu U, Mainou BA. Virus interactions with bacteria: partners in the infectious dance. PLoS Pathog 2020;16:e1008234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Koonin EV, Dolja VV, Krupovic M. The healthy human virome: from virus–host symbiosis to disease. Curr Opin Virol 2021;47:86–94 [DOI] [PubMed] [Google Scholar]
- 86. El Ahmer OR, Raza MW, Ogilvie MM, Weir DM, Blackwell CC. Binding of bacteria to HEp-2 cells infected with influenza A virus. FEMS Immunol Med Microbiol 1999;23:331–41 [DOI] [PubMed] [Google Scholar]
- 87. Rowe HM, Meliopoulos VA, Iverson A, Bomme P, Schultz-Cherry S, Rosch JW. Direct interactions with influenza promote bacterial adherence during respiratory infections. Nat Microbiol 2019;4:1328–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Robinson CM, Jesudhasan PR, Pfeiffer JK. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe 2014;15:36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Erickson AK, Jesudhasan PR, Mayer MJ, Narbad A, Winter SE, Pfeiffer JK. Bacteria facilitate enteric virus co-infection of mammalian cells and promote genetic recombination. Cell Host Microbe 2018;23:77–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Aguilera ER, Nguyen Y, Sasaki J, Pfeiffer JK. Bacterial stabilization of a panel of picornaviruses. Msphere 2021;4:e00183–10019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Li D, Breiman A, le Pendu J, Uyttendaele M. Binding to histo-blood group antigen-expressing bacteria protects human norovirus from acute heat stress. Front Microbiol 2015;6:659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Almand EA, Moore MD, Outlaw J, Jaykus LA. Human norovirus binding to select bacteria representative of the human gut microbiota. PLoS ONE 2017;12:e0173124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Berger AK, Yi H, Kearns DB, Mainou BA. Bacteria and bacterial envelope components enhance mammalian reovirus thermostability. PLoS Pathog 2017;13:e1006768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinjé J, Tibebetts SA, Wallet SM, Karst SM. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014;346:755–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Guzior DV, Quinn RA. Review: microbial transformations of human bile acids. Microbiome 2021;9:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hofmann AF, Hagey LR. Key discoveries in bile acid chemistry and biology and their clinical applications: history of the last eight decades. J Lipid Res 2014;55:1553–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kilic T, Koromyslova A, Hansman GS. Structural basis for human norovirus capsid binding to bile acids. J Virol 2019;93: e01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Monaco Cynthia L, Gootenberg David B, Zhao G, Handley Scott A, Ghebremichael Musie S, Lim Efrem S, Lankowski A, Baldridge Megan T, Wilen Craig B, Flagg M, Norman Jason M, Keller Brian C, Luévano Jesús M, Wang D, Boum Y, Martin Jeffrey N, Hunt Peter W, Bangsberg David R, Siedner Mark J, Kwon Douglas S, Virgin Herbert W. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe 2016;19:311–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Choi S, Sohn KH, Jung JW, Kang MG, Yang MS, Kim S, Choi JH, Cho SH, Kang HR, Yi H. Lung virome: new potential biomarkers for asthma severity and exacerbation. J Allergy Clin Immunol 2021;148:1007–15 [DOI] [PubMed] [Google Scholar]
- 100. Kullberg RFJ, Hugenholtz F, Brands X, Kinsella CM, Peters-Sengers H, Butler JM, Deijs M, Klein M, Faber DR, Scicluna BP, Van der Poll T, Van der Hoek L, Wiersinga WJ, Haak BW. Rectal bacteriome and virome signatures and clinical outcomes in community-acquired pneumonia: an exploratory study. Eclinicalmedicine 2021;39:101074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lamberti LM, Fischer Walker CL, Noiman A, Victora C, Black RE. Breastfeeding and the risk for diarrhea morbidity and mortality. BMC Publ 2011;11:S15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Turin CG, Ochoa TJ. The role of maternal breast milk in preventing infantile diarrhea in the developing world. In: Okhuysen PC. (ed), Current tropical medicine reports. Berlin: Springer, 2014, pp.97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Liang G, Zhao C, Zhang H, Mattei L, Sherrill-Mix S, Bittinger K, Kessler LR, Wu GD, Baldassano RN, DeRusso P, Ford E, Elovitz MA, Kelly MS, Patel MZ, Mazhani T, Gerber JS, Kelly A, Zemel BS, Bushman FD. The stepwise assembly of the neonatal virome is modulated by breastfeeding. Nature 2020;581:470–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Quigley MA, Carson C, Sacker A, Kelly Y. Exclusive breastfeeding duration and infant infection. Eur J Clin Nutr 2016;70:1420–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Pandolfi E, Gesualdo F, Rizzo C, Carloni E, Villani A, Concato C, Linardos G, Russo L, Ferretti B, Campagna I, Tozzi A. Breastfeeding and respiratory infections in the first 6 months of life: a case control study. Front Pediatr 2019;7:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zuo T, Sun Y, Wan Y, Yeoh YK, Zhang F, Cheung CP, Chen N, Luo J, Wang W, Sung JJY, Chan PKS, Wang K, Chan FKL, Miao Y, Ng SC. Human-gut-DNA virome variations across geography, ethnicity, and urbanization. Cell Host Microbe 2020;28:741–514 [DOI] [PubMed] [Google Scholar]
- 107. Rampelli S, Turroni S, Schnorr SL, Soverini M, Quercia S, Barone M, Castagnetti A, Biagi E, Gallinella G, Brigidi P, Candela M. Characterization of the human DNA gut virome across populations with different subsistence strategies and geographical origin. Environ Microbiol 2017;19:4728–35 [DOI] [PubMed] [Google Scholar]
- 108. Liu S, Huang S, Chen F, Zhao L, Yuan Y, Francis SS, Fang L, Li Z, Lin L, Liu R, Zhang Y, Xu H, Li S, Zhou Y, Davies RW, Liu Q, Walters RG, Lin K, Ju J, Korneliussen T, Yang MA, Fu Q, Wang J, Zhou L, Krogh A, Zhang H, Wang W, Chen Z, Cai Z, Yin Y, Yang H, Mao M, Shendure J, Wang J, Albrechtsen A, Jin X, Nielsen R, Xu X. Genomic analyses from non-invasive prenatal testing reveal genetic associations, patterns of viral infections, and Chinese population history. Cell 2018;175:347–59.e14 [DOI] [PubMed] [Google Scholar]
- 109. Holtz LR, Cao S, Zhao G, Bauer IK, Denno DM, Klein EJ, Antonio M, Stine OC, Snelling TL, Kirkwood CD, Wang D. Geographic variation in the eukaryotic virome of human diarrhea. Virology 2014;468–470:556–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature 2014;516:94–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ingle H, Lee S, Ai T, Orvedahl A, Rodgers R, Zhao G, Sullender M, Peterson ST, Locke M, Liu TC, Yokoyama CC, Sharp B, Schultz-Cherry S, Miner JJ, Baldridge MT. Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon-λ. Nat Microbiol 2019;4:1120–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Neil JA, Cadwell K. The intestinal virome and immunity. J Immunol 2018;201:1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Galtier M, Sordi LD, Sivignon A, de Vallée A, Maura D, Neut C, Rahmouni O, Wannerberger K, Darfeuille-Michaud A, Desreumaux P, Barnich N, Debarbieux L. Bacteriophages targeting adherent invasive Escherichia coli strains as a promising new treatment for Crohn’s disease. J Crohn’s Colitis 2017;11:840–7 [DOI] [PubMed] [Google Scholar]
- 114. Poole K. Resistance to β-lactam antibiotics. In: Eichmann K (ed), Cellular and molecular life sciences. Berlin: Springer, 2004, pp.2200–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Taylor VL, Fitzpatrick AD, Islam Z, Maxwell KL. Chapter one: the diverse impacts of phage morons on bacterial fitness and virulence. In: Kielian M, Mettenleiter TC, Roossinck MJ. (eds) Advances in virus research. Cambridge, MA: Academic Press, 2019, pp.1–31 [DOI] [PubMed] [Google Scholar]
- 116. Gogokhia L, Buhrke K, Bell R, Hoffman B, Brown DG, Hanke-Gogokhia C, Ajami NJ, Wong MC, Ghazaryan A, Valentine JF, Porter N, Martens E, O’Connell R, Jacob V, Scherl E, Crawford C, Stephens WZ, Casjens SR, Longman RS, Round JL. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe 2019;25:285–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Sweere JM, Van Belleghem JD, Ishak H, Bach MS, Popescu M, Sunkari V, Kaber G, Manasherob R, Suh GA, Cao X, de Vries CR, Lam DN, Marshall PL, Birukova M, Katznelson E, Lazzareschi DV, Balaji S, Keswani SG, Hawn TR, Secor PR, Bollyky PL. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 2019;363:eaat9691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, Rubin E, Ivanova NN, Kyrpides NC. Uncovering Earth’s virome. Nature 2016;536:425–30 [DOI] [PubMed] [Google Scholar]
- 119. Shkoporov AN, Hill C. Bacteriophages of the human gut: the “known unknown” of the microbiome. Cell Host Microbe 2019;25:195–209 [DOI] [PubMed] [Google Scholar]
- 120. Krishnamurthy SR, Wang D. Origins and challenges of viral dark matter. Virus Res 2017;239:136–42 [DOI] [PubMed] [Google Scholar]
- 121. Fawaz M, Vijayakumar P, Mishra A, Gandhale PN, Dutta R, Kamble NM, Sudhakar SB, Roychoudhary P, Kumar H, Kulkarni DD, Raut AA. Duck gut viral metagenome analysis captures snapshot of viral diversity. Gut Pathog 2016;8:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Handley Scott A, Desai C, Zhao G, Droit L, Monaco Cynthia L, Schroeder Andrew C, Nkolola Joseph P, Norman Megan E, Miller Andrew D, Wang D, Barouch Dan H, Virgin Herbert W. SIV infection-mediated changes in gastrointestinal bacterial microbiome and virome are associated with immunodeficiency and prevented by vaccination. Cell Host Microbe 2016;19:323–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Reyes A, Blanton LV, Cao S, Zhao G, Manary M, Trehan I, Smith MI, Wang D, Virgin HW, Rohwer F, Gordon JI. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc Natl Acad Sci USA 2015;112:11941–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Rascovan N, Duraisamy R, Desnues C. Metagenomics and the human virome in asymptomatic individuals. Annu Rev Microbiol 2016;70:125–41 [DOI] [PubMed] [Google Scholar]
- 125. Hoyles L, McCartney AL, Neve H, Gibson GR, Sanderson JD, Heller KJ, van Sinderen D. Characterization of virus-like particles associated with the human faecal and caecal microbiota. Res Microbiol 2014;165: 803–12 [DOI] [PubMed] [Google Scholar]
- 126. Warwick-Dugdale J, Solonenko N, Moore K, Chittick L, Gregory AC, Allen MJ, Sullivan MB, Temperton B. Long-read viral metagenomics captures abundant and microdiverse viral populations and their niche-defining genomic islands. PeerJ 2019;7:e6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wang D. 5 challenges in understanding the role of the virome in health and disease. PLoS Pathog 2020;16:e1008318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Steinegger M, Mirdita M, Söding J. Protein-level assembly increases protein sequence recovery from metagenomic samples manyfold. Nat Methods 2019;16:603–6 [DOI] [PubMed] [Google Scholar]
- 129. Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, Tammadoni S, Nosrat B, Conrad D, Rohwer F. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE 2009;4:e7370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Deng L, Ignacio-Espinoza JC, Gregory AC, Poulos BT, Weitz JS, Hugenholtz P, Sullivan MB. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature 2014;513:242–5 [DOI] [PubMed] [Google Scholar]
- 131. Roux S, Tournayre J, Mahul A, Debroas D, Enault F. Metavir 2: new tools for viral metagenome comparison and assembled virome analysis. BMC Bioinformatics 2014;15:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Džunková M, D’Auria G, Moya A. Direct sequencing of human gut virome fractions obtained by flow cytometry. Front Microbiol 2015;6:955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Labonté JM, Swan BK, Poulos B, Luo H, Koren S, Hallam SJ, Sullivan MB, Woyke T, Wommack KE, Stepanauskas R. Single-cell genomics-based analysis of virus–host interactions in marine surface bacterioplankton. ISME J 2015;9:2386–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Roux S, Adriaenssens EM, Dutilh BE, Koonin EV, Kropinski AM, Krupovic M, Kuhn JH, Lavigne R, Brister JR, Varsani A, Amid C, Aziz RK, Bordenstein SR, Bork P, Breitbart M, Cochrane GR, Daly RA, Desnues C, Duhaime MB, Emerson JB, Enault F, Fuhrman JA, Hingamp P, Hugenholtz P, Hurwitz BL, Ivanova NN, Labonté JM, Lee KB, Malmstrom RR, Martinez-Garcia M, Mizrachi IK, Ogata H, Páez-Espino D, Petit MA, Putonti C, Rattei T, Reyes A, Rodriguez-Valera F, Rosario K, Schriml L, Schulz F, Steward GF, Sullivan MB, Sunagawa S, Suttle CA, Temperton B, Tringe SG, Thurber RV, Webster NS, Whiteson KL, Wilhelm SW, Wommack KE, Woyke T, Wrighton KC, Yilmaz P, Yoshida T, Young MJ, Yutin N, Allen LZ, Kyrpides NC, Eloe-Fadrosh EA. Minimum information about an uncultivated virus genome (MIUViG). Nat Biotechnol 2019;37:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Fredricks DN, Relman DA. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin Microbiol Rev 1996;9:18–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Chiu CY, Greninger AL, Chen EC, Haggerty TD, Parsonnet J, Delwart E, Derisi JL, Ganem D. Cultivation and serological characterization of a human Theiler’s-like cardiovirus associated with diarrheal disease. J Virol 2010;84:4407–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Janowski AB, Bauer IK, Holtz LR, Wang D. Propagation of astrovirus VA1, a neurotropic human astrovirus, in cell culture. J Virol 2017;91: e00740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Dijkman R, Koekkoek SM, Molenkamp R, Schildgen O, van der Hoek L. Human bocavirus can be cultured in differentiated human airway epithelial cells. J Virol 2009;83:7739–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Pyrc K, Sims AC, Dijkman R, Jebbink M, Long C, Deming D, Donaldson E, Vabret A, Baric R, van der Hoek L, Pickles R. Culturing the unculturable: human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. J Virol 2010;84:11255–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng X-L, Qu L. Replication of human noroviruses in stem cell–derived human enteroids. Science 2016;353:1387–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Chaffringeon L, Lamy-Besnier Q, Debarbieux L, De Sordi L. The intestinal virome: lessons from animal models. Curr Opin Virol 2021;51:141–8 [DOI] [PubMed] [Google Scholar]
- 142. Clooney AG, Sutton TDS, Shkoporov AN, Holohan RK, Daly KM, O’Regan O, Ryan FJ, Draper LA, Plevy SE, Ross RP, Hill C. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 2019;26:764–85 [DOI] [PubMed] [Google Scholar]
- 143. Mangalea MR, Paez-Espino D, Kieft K, Chatterjee A, Chriswell ME, Seifert JA, Feser ML, Demoruelle MK, Sakatos A, Anantharaman K, Deane KD, Kuhn KA, Holers VM, Duerkop BA. Individuals at risk for rheumatoid arthritis harbor differential intestinal bacteriophage communities with distinct metabolic potential. Cell Host Microbe 2021;29:726–39.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Zhao G, Droit L, Gilbert MH, Schiro FR, Didier PJ, Si X, Paredes A, Handley SA, Virgin HW, Bohm RP, Wang D. Virome biogeography in the lower gastrointestinal tract of rhesus macaques with chronic diarrhea. Virology 2019;527:77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Tan X, Chai T, Duan J, Wu J, Zhang H, Li Y, Huang Y, Hu X, Zheng P, Song J, Ji P, Jin X, Zhang H, Xie P. Dynamic changes occur in the DNA gut virome of female cynomolgus macaques during aging. Microbiologyopen 2021;10:e1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Xiao Y, Wang H, Feng L, Pan J, Chen Z, Wang H, Yang S, Shen Q, Wang X, Shan T, Zhang W. Fecal, oral, blood and skin virome of laboratory rabbits. Arch Virol 2020;165:2847–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Brunse A, Deng L, Pan X, Hui Y, Castro-MejÃ-a JL, Kot W, Nguyen DN, Secher JB, Nielsen DS, Thymann T. Fecal filtrate transplantation protects against necrotizing enterocolitis. ISME J 2022;16:686–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Hansen AK, Hansen CHF. The microbiome and rodent models of immune mediated diseases. Mamm Genome 2021;32:251–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Pavlopoulos GA, Oulas A, Iacucci E, Sifrim A, Moreau Y, Schneider R, Aerts J, Iliopoulos I. Unraveling genomic variation from next generation sequencing data. Biodata Min 2013;6:1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Thompson J, Johansen R, Dunbar J, Munsky B. Machine learning to predict microbial community functions: an analysis of dissolved organic carbon from litter decomposition. PLoS ONE 2019;14:e0215502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Buttigieg PL, Ramette A. A guide to statistical analysis in microbial ecology: a community-focused, living review of multivariate data analyses. FEMS Microbiol Ecol 2014;90:543–50 [DOI] [PubMed] [Google Scholar]
- 152. Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res 2017;45:W180–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Ho DW, Sze KM, Ng IO. Virus-Clip: a fast and memory-efficient viral integration site detection tool at single-base resolution with annotation capability. Oncotarget 2015;6:20959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Mishra N, Fagbo SF, Alagaili AN, Nitido A, Williams SH, Ng J, Lee B, Durosinlorun A, Garcia JA, Jain K, Kapoor V, Epstein JH, Briese T, Memish ZA, Olival KJ, Lipkin WI. A viral metagenomic survey identifies known and novel mammalian viruses in bats from Saudi Arabia. PLoS ONE 2019;14:e0214227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Wang L, Guo W, Shen H, Guo J, Wen D, Yu Y, Wu W. Plasma microbial cell-free DNA sequencing technology for the diagnosis of sepsis in the ICU. Front Mol Biosci 2021;8:659390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Turkington CJ, Varadan AC, Grenier SF, Grasis JA. The viral Janus: viruses as aetiological agents and treatment options in colorectal cancer. Front Cell Infect Microbiol 2021;10:814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Kao D, Roach B, Silva M, Beck P, Rioux K, Kaplan GG, Chang H-J, Coward S, Goodman KJ, Xu H. Effect of oral capsule–vs colonoscopy-delivered fecal microbiota transplantation on recurrent Clostridium difficile infection: a randomized clinical trial. JAMA 2017;318:1985–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Wu X, Zhang T, Chen X, Ji G, Zhang F. Microbiota transplantation: targeting cancer treatment. BMC Publ Health 2019;452:144–51 [DOI] [PubMed] [Google Scholar]
- 159. Ser HL, Letchumanan V, Goh BH, Wong SH, Lee LH. The use of fecal microbiome transplant in treating human diseases: too early for poop. Front Microbiol 2021;12:519836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Rasmussen TS, Mentzel CMJ, Kot W, Castro-Mejía JL, Zuffa S, Swann JR, Hansen LH, Vogensen FK, Hansen AK, Nielsen DS. Faecal virome transplantation decreases symptoms of type 2 diabetes and obesity in a murine model. Gut 2020;69:2122–30 [DOI] [PubMed] [Google Scholar]
- 161. Schneider CL. Bacteriophage-mediated horizontal gene transfer: transduction. Bacterioph: Biol Technol Ther 2021:151–92 [Google Scholar]
- 162. Maronek M, Link R, Ambro L, Gardlik R. Phages and their role in gastrointestinal disease: focus on inflammatory bowel disease. Cells 2020;9:1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. L’Huillier AG, Brito F, Wagner N, Cordey S, Zdobnov E, Posfay-Barbe KM, Kaiser L. Identification of viral signatures using high-throughput sequencing on blood of patients with Kawasaki disease. Front Pediatr 2019;7:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Tschöpe C, Ammirati E, Bozkurt B, Caforio ALP, Cooper LT, Felix SB, Hare JM, Heidecker B, Heymans S, Hübner N, Kelle S, Klingel K, Maatz H, Parwani AS, Spillmann F, Starling RC, Tsutsui H, Seferovic P, Van Linthout S. Myocarditis and inflammatory cardiomyopathy: current evidence and future directions. Nat Rev Cardiol 2021;18:169–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Zhang T, Wu Q, Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol 2020;30:1346.e2–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Riesland NJ, Wilde H. Expert review of evidence bases for managing monkey bites in travelers. J Travel Med 2015;22:259–62 [DOI] [PubMed] [Google Scholar]
- 167. Eberle R, Jones-Engel L. Understanding primate herpesviruses. J Emerg Dis Virol 2017;3:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Bragazzi NL, Dai H, Damiani G, Behzadifar M, Martini M, Wu J. How big data and artificial intelligence can help better manage the COVID-19 pandemic. Int J Environ Res Publ Health 2020;17:3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Allam Z, Dey G, Jones DS. Artificial intelligence (AI) provided early detection of the coronavirus (COVID-19) in China and will influence future urban health policy internationally. AI 2020;1:156–65 [Google Scholar]