Abstract
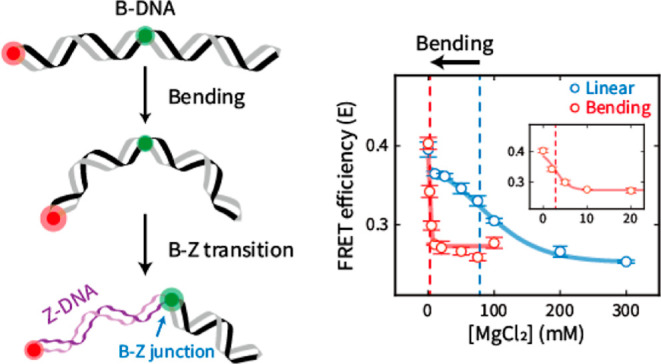
Z-DNA, a noncanonical helical structure of double-stranded DNA (dsDNA), plays pivotal roles in various biological processes, including transcription regulation. Mechanical stresses on dsDNA, such as twisting and stretching, help to form Z-DNA. However, the effect of DNA bending, one of the most common dsDNA deformations, on Z-DNA formation is utterly unknown. Here, we show that DNA bending induces the formation of Z-DNA, that is, more Z-DNA is formed as the bending force becomes stronger. We regulated the bending force on dsDNA by using D-shaped DNA nanostructures. The B–Z transition was observed by single-molecule fluorescence resonance energy transfer. We found that as the bending force became stronger, Z-DNA was formed at lower Mg2+ concentrations. When dsDNA contained cytosine methylations, the B–Z transition occurred at 78 mM Mg2+ (midpoint) in the absence of the bending force. However, the B–Z transition occurred at a 28-fold lower Mg2+ concentration (2.8 mM) in the presence of the bending force. Monte Carlo simulation suggested that the B–Z transition stabilizes the bent form via the formation of the B–Z junction with base extrusion, which effectively releases the bending stress on DNA. Our results clearly show that the bending force facilitates the B–Z transition under physiological salt conditions.
Introduction
Double-stranded DNA (dsDNA) has various conformations, including the canonical right-handed double helical structure (B-DNA) and several noncanonical structures.1 Z-DNA is a noncanonical left-handed double helical structure with Watson–Crick base pairs.2,3 The biological roles of Z-DNA have been studied for decades since its discovery.4−12 It has been well established that Z-DNA plays crucial roles in various biological processes, such as transcription regulation,4,9 genetic instability correlated with DNA damage and repair,8,11 and nucleosome positioning.6,7 Z-DNA is even involved in the regulatory mechanism of extinction memory in mice12 and other diseases.10
The B–Z transition occurs under special circumstances due to the adjacent negatively charged phosphate groups of Z-DNA.2,13−16 The formation of Z-DNA occurs mostly in regions of repeated alternation of purine and pyrimidine bases, especially (CG)n, which is the most preferred sequence.2 High salt concentrations enable the B–Z transition by relieving electrostatic repulsions between phosphate backbone groups.2 Cytosine methylation, one of the most important base modifications,17 contributes to the formation of Z-DNA.13,16,18 As cytosine methylation is involved in various biological processes, such as chromatin compaction19 and transcription regulation,20,21 cytosine methylation may be one of the factors that allow Z-DNA to play a role in cells. Z-DNA binding proteins recognize and bind Z-DNA with high affinity14,15,22 and even induce the B–Z transition.23,24
In addition to salt concentration and chemical modifications, mechanical stresses on DNA have also been key factors in the B–Z transition.25−31 For instance, negative supercoiling, which is induced during transcription,32 stabilizes Z-DNA,26 and torsional stress regulates the level of Z-DNA in cells.27 As a result, the effect of torsion on Z-DNA formation has been extensively studied using bulk supercoiling25−27 and single-molecule tweezer measurements.28,29,31 In addition to torsional stress, tension (stretching force) on DNA also promotes the formation of Z-DNA in negatively supercoiled DNA.28,31 These results imply that the response of DNA to mechanical stresses is an important factor determining the relative stabilities of B- and Z-DNA. However, the effect of DNA bending, one of the most common DNA mechanical deformations observed in various biological phenomena,33−37 on the formation of Z-DNA is entirely unknown. Various DNA–protein interactions induce DNA bending and even generate kinks.33,34 The effect of bending force on Z-DNA formation has not been studied, largely due to the lack of experimental methods to modulate the bending of short dsDNA (<100 bp).38
Herein, we report that DNA bending force induces the formation of Z-DNA. We designed dsDNAs that contained CG repeats as a Z-DNA-forming region and a random sequence as a B-DNA region (Figure 1a,b). To quantitatively investigate the B–Z transition induced by the DNA bending force, we employed D-shaped DNA nanostructures to modulate the dsDNA bending force.39−42 The B–Z transition and the curvature of dsDNA induced by the bending force were detected by single-molecule fluorescence resonance energy transfer (smFRET) (Figures 1c and S1).43−45 We observed that the B–Z transition occurs at lower salt concentrations as the bending force on dsDNA increases. We confirmed that Z-DNA formation under physiological salt conditions (<10 mM Mg2+) is induced by applying a bending force in the presence of cytosine methylation. The melted base pairs at the B–Z junction may release the bending stress and stabilize the Z-DNA form, which is supported by a simulation.
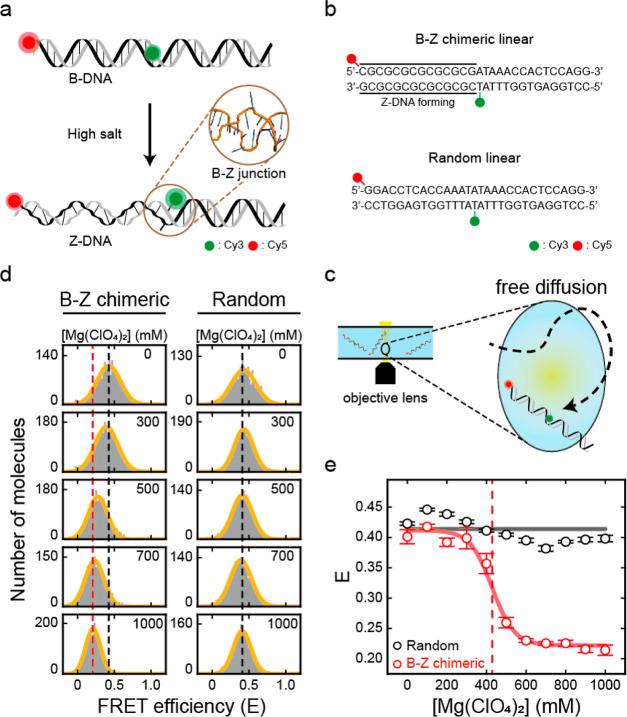
Figure 1.
B–Z transition in linear dsDNA observed by single-molecule ALEX measurement. (a) Illustration of Z-DNA formation in linear dsDNA under high salt conditions, which is detected by a decrease in FRET efficiency. During the B–Z transition, the B–Z junction, which is a noncanonical structure between B- and Z-DNA with extruded bases (brown circle, PDB code 2ACJ), is generated. (b) DNA sequences used for the B–Z transition experiments. The B–Z chimeric linear sample consists of CG repeats, a Z-DNA forming region, and a random sequence. The random linear sample consists only of a random sequence that does not form Z-DNA. DNAs are labeled with Cy3 and Cy5 as a FRET pair. (c) Brief schematic illustration of the ALEX measurement, which measures the FRET efficiency of freely diffusing single molecules. (d) FRET histograms of the B–Z (chimeric) linear and random linear samples at various concentrations of Mg(ClO4)2 obtained by the ALEX method. Histograms were fitted to a single Gaussian distribution. The black dotted lines denote the average FRET efficiency (E) of each sample in the absence of Mg(ClO4)2. The red dotted line denotes the E of the B–Z linear sample at 1000 mM Mg(ClO4)2. (e) FRET efficiency E of each sample depending on the concentration of Mg(ClO4)2. For the B–Z chimeric linear sample, E decreases due to the formation of Z-DNA, while E of the random linear sample does not change. The red dotted line denotes the midpoint of the B–Z linear sample (430 mM). Error bars were obtained from three independent measurements.
Results and Discussion
Quantitative Observation of Z-DNA Formation by ALEX Measurement
First, we investigated the formation of Z-DNA by observing the differences in FRET efficiencies (Figure 1a). Z-DNA has a shorter diameter and longer pitch than B-DNA.3 The formation of Z-DNA has been successfully observed in previous studies using smFRET.14,28,31,46 Z-DNA is usually formed in purine-pyrimidine repeats, typically in CG repeats, under high salt conditions.2 We prepared linear DNA duplexes: one contained seven CG repeats as a Z-DNA forming sequence and a random sequence (B–Z chimeric linear), and the other consisted only of a random sequence (random linear) (Figure 1b). The sequence of the random portions was designed by using a sequence that has been confirmed not to form Z-DNA under the high salt conditions that we used in this work.47,48 Cy3 and Cy5 were labeled on the flanks of the Z-DNA forming sequence in the B–Z chimeric linear sample (1st and 15th base pairs) and were placed at the same positions in the random linear sample. The formation of Z-DNA was detected by the decrease in FRET efficiency, which was caused by the increase in dsDNA length and formation of the B–Z junction (Figure 1a).14,46 We measured the FRET efficiency of each dsDNA molecule by using alternating-laser excitation FRET (ALEX) (Figures 1c and S1).44,45,49 In the ALEX measurement, the FRET efficiency of freely diffusing single molecules in a buffer solution was measured without surface immobilization (Figure 1c). In addition to the FRET efficiency, ALEX measures the stoichiometric factor, which is used for sorting only FRET pair species (Figure S1b,c).49 The FRET efficiency is inversely proportional to the sixth power of the distance between the donor and the acceptor dyes, which enables FRET efficiency to be used to observe distance changes.50 Thus, the FRET efficiency reports the conformational changes of dsDNA, including the formation of Z-DNA.
Subsequently, we used ALEX to measure the FRET efficiency of two linear samples by varying the salt concentration to compare the conformational changes upon the formation of Z-DNA (Figures 1d,e and S2). To form Z-DNA, we used magnesium perchlorate (Mg(ClO4)2), which stabilizes Z-DNA well.46 Without Mg(ClO4)2, both samples revealed a similar average FRET efficiency (E) of approximately 0.40, corresponding to B-DNA (Figure 1d). As the concentration of Mg(ClO4)2 increased, the E of the B–Z chimeric linear sample decreased from 0.40 to 0.21 due to the formation of Z-DNA (Figure 1d, left panel). However, the E of the random linear sample remained consistent regardless of Mg(ClO4)2, which indicates that Z-DNA was not formed (Figure 1d, right panel). The decrease in E of the B–Z chimeric linear sample was most apparent between 400 and 500 mM Mg(ClO4)2, which indicates the B–Z transition (Figure 1e). The Z-DNA forming sequence transformed fully to Z-DNA at approximately 600 mM Mg(ClO4)2, which is in line with the previous results.46 The midpoint, where B- and Z-DNA were equally probable, was 430 mM (Figure 1e, red dotted line). To confirm the formation of Z-DNA, we also measured the circular dichroism (CD) spectra of the B–Z chimeric linear sample without dye labeling at various Mg(ClO4)2 concentrations (Figure S3). An increase at 255 nm was clearly observed as the salt concentration increased, which is consistent with the reported spectra of B- and Z-DNA, respectively (Figure S3).2 Therefore, ALEX measurement enables us to analyze the formation of Z-DNA quantitatively without surface immobilization.
DNA Bending Force Induces the Formation of Z-DNA
To investigate the effects of DNA bending force on the B–Z transition, we prepared D-shaped DNAs that consisted of a dsDNA portion and a ssDNA string (Figure 2a–c).41,42 In a D-shaped DNA, the bending force is applied by the entropic force from the ssDNA string (Figure 2a).39 D-shaped DNAs enable adjustment of the compressive bending force on the dsDNA portion by varying the length of the ssDNA string (Figure 2a). As the length of the ssDNA string decreases, the bending force on the dsDNA portion increases.41 To construct a D-shaped DNA, we annealed a ring ssDNA with its partially complementary ssDNA (Figures 2c and S4). The length of the dsDNA portion was fixed to 29 bp, and the ssDNA string length varied from 11 to 51 nt (11, 19, 23, 27, and 51 nt). The 51 nt ssDNA string applies nearly no bending force to the dsDNA portion, while the shorter ssDNA strings, such as 23, 19, and 11 nt, apply a bending force to the dsDNA portion, which was confirmed by ALEX (smFRET) measurements (Figure S5). It has been shown that a ssDNA string, of which the contour length is longer than that of the dsDNA portion, can apply a bending force on dsDNA as an entropic string.39,41
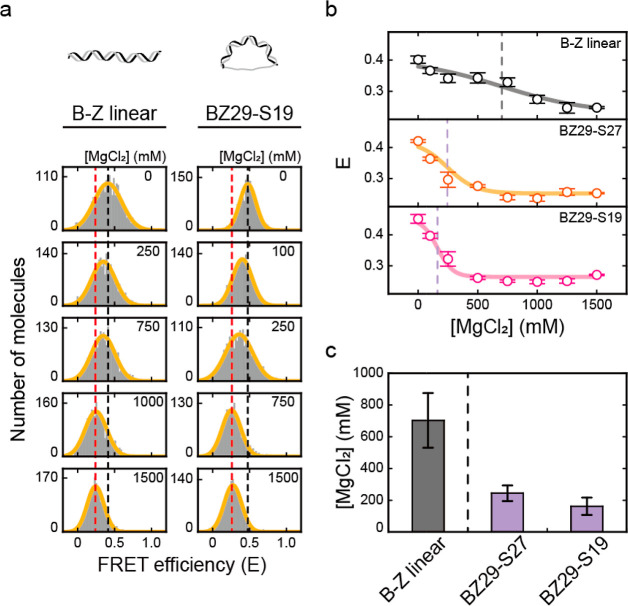
Figure 2.
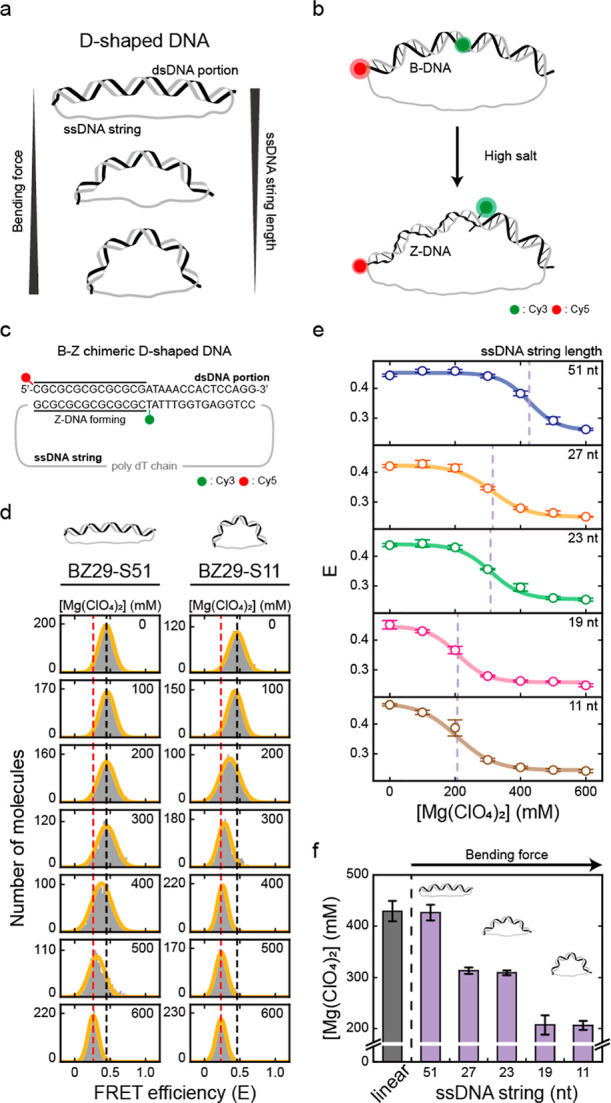
DNA bending force induces Z-DNA formation. The B–Z transition is induced by ssDNA string lengths (BZ29-S51, BZ29-S27, BZ29-S23, BZ29-S19, and BZ29-S11). (a) Illustration of D-shaped DNA nanostructures. In a D-shaped DNA, the bending force on the dsDNA portion increases as the ssDNA string length decreases. (b) Illustration of Z-DNA formation in a D-shaped DNA, which is detected by a decrease in FRET efficiency. (c) DNA sequence used for the B–Z transition experiments. The dsDNA portion has the same sequence as the B–Z chimeric linear sample, and the ssDNA string consists only of thymine. The bending force on the dsDNA portion was modulated by using different lengths of ssDNA string. (d) FRET histograms of BZ29-S51 and BZ29-S11 at various concentrations of Mg(ClO4)2 obtained by the ALEX method. Histograms were fitted to a single Gaussian distribution. The black dotted lines denote the E of each sample in the absence of Mg(ClO4)2. The red dotted lines denote the E of each sample at 600 mM Mg(ClO4)2. (e) FRET efficiency E of each D-shaped DNA sample (BZ29-S51, BZ29-S27, BZ29-S23, BZ29-S19, and BZ29-S11) plotted against the concentration of Mg(ClO4)2. The FRET histograms of each sample are presented in Figure S7. The lavender dotted lines denote the midpoint of each sample. Error bars were obtained from three independent measurements. (f) B–Z transition midpoints of the B–Z linear and D-shaped DNAs. The midpoint of the B–Z linear sample was obtained from Figure 1e (red dotted line). As the bending force increases, the midpoint of the B–Z transition decreases. Error bars were obtained from three independent measurements.
We denote a D-shaped DNA by BZ29-S11, which means a 29 bp B–Z chimeric dsDNA portion and an 11 nt ssDNA string. We designed the dsDNA portion of the D-shaped DNA with the same sequence as the B–Z chimeric linear sample in Figure 1, which contains a Z-DNA forming sequence (Figure 2c). Cy3 and Cy5 were labeled at either end of the Z-DNA forming sequence (1st and 15th base pairs), as in the linear sample (Figure 2c). As a result, the formation of Z-DNA in a D-shaped DNA also leads to a decrease in FRET efficiency (Figure 2b). By measuring multiple distances in a D-shaped DNA, we verified that the structure of D-shaped DNAs was correctly formed as we designed (Figure S6).
We subsequently measured the E values of BZ29-S11 and BZ29-S51 with various concentrations of Mg(ClO4)2 (Figures 2d and S7). As the concentration of Mg(ClO4)2 increased, the E values of both D-shaped DNA samples decreased as much as in the previous linear sample shown in Figure 1d, indicating the formation of Z-DNA. The decrease in E was also saturated at concentrations lower than 600 mM Mg(ClO4)2. The B–Z transition in BZ29-S51 occurred at approximately 420 mM Mg(ClO4)2, which was similar to the midpoint of the B–Z chimeric linear DNA (430 mM). Interestingly, the B–Z transition in BZ29-S11 occurred at approximately 200 mM Mg(ClO4)2, which was significantly lower than that of BZ29-S51 and the linear DNA. This result indicates that strong bending of DNA facilitates the B-to-Z DNA transition. To investigate this result further, we prepared three more D-shaped DNAs (BZ29-S19, BZ29-S23, and BZ29-S27) which have different bending forces on dsDNA. Figure 2e presents the E values for five D-shaped DNAs at various Mg(ClO4)2 concentrations. The midpoints of the B–Z transitions gradually decreased from 430 to 210 mM as the length of the ssDNA string decreased (Figure 2e,f). As a control, we tested the effect of Mg2+ concentrations on the bending of D-shaped DNA (Figure S8), which showed a minimal effect on the bending of the B-form D-shaped DNA (CG29-S23) in our measurement conditions. We also estimated roughly the bending force applied to each D-shaped DNA nanostructure using a worm-like chain model (Figure S9).41 These results indicate that as the bending force on DNA increased, the B–Z transition was facilitated and thus occurred at lower Mg(ClO4)2 concentrations.
A decrease in the midpoint of the B–Z transition by the bending force was also observed by CD measurement (Figure S10). We used BZ29-S23 without dye labeling. As with our single-molecule FRET measurement shown in Figure 2, the decrease in the midpoints induced by the DNA bending force (BZ29-S23 compared with the linear DNA) was clearly observed in the CD measurement. As a result, both the single-molecule ALEX measurement and the CD measurement showed that the DNA bending force facilitates the B–Z transition, and thus, the B–Z transition occurs at lower salt concentrations.
B–Z Transition of Methylated DNAs Occurs at Physiological Salt Concentrations Due to the Bending Force
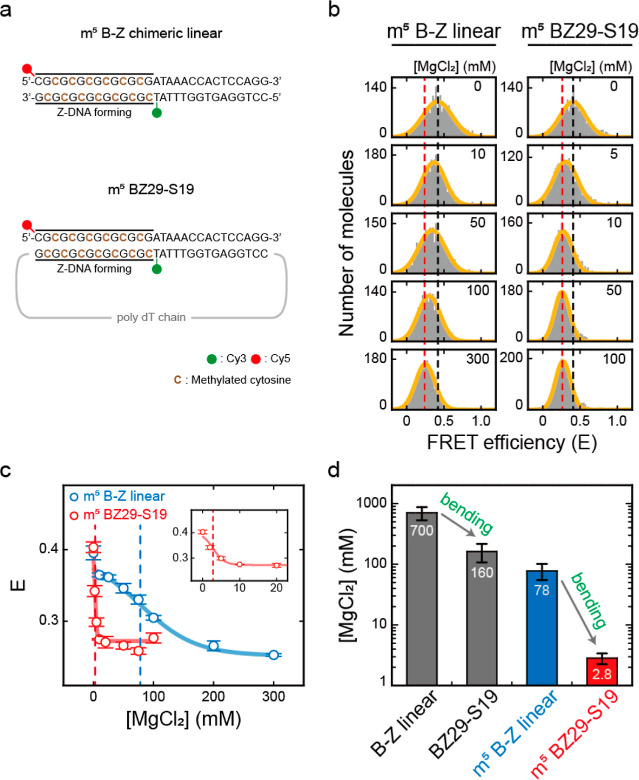
Next, we used magnesium chloride (MgCl2) to form Z-DNA in order to study the formation of Z-DNA under physiological salt conditions. We used the linear and two D-shaped DNA samples (BZ29-S27 and BZ29-S19) which have a weak DNA bending. Using ALEX, we obtained the E values for the linear and D-shaped DNA samples as the concentration of MgCl2 increased (Figures 3a and S11). The results showed that the E values of all DNA samples decreased from approximately 0.40 to 0.25 due to the formation of Z-DNA (Figure 3b). The midpoint of the B–Z linear DNA occurred at 700 mM, while those of BZ29-S27 and BZ29-S19 were observed at 250 and 160 mM, respectively (Figure 3c). Again, the B–Z transition occurred at lower salt concentrations as the bending force became stronger.
Figure 3.
B–Z transition induced by the DNA bending force with different salt MgCl2. (a) FRET histograms of B–Z linear and BZ29-S19 samples at various concentrations of MgCl2 obtained by the ALEX method. Histograms were fitted to a single Gaussian distribution. The black dotted lines denote the E of each sample in the absence of MgCl2. The red dotted lines denote the E of each sample at 1500 mM MgCl2. (b) E of B–Z linear and D-shaped DNA samples (BZ29-S27 and BZ29-S19) plotted against the concentration of MgCl2. The dotted lines denote the midpoint of each sample. Error bars were obtained from three independent measurements. (c) B–Z transition midpoint of each sample (B–Z linear, BZ29-S27, and BZ29-S19). The midpoints decrease as the bending force increases, which shows the same trend as in Figure 2f. Error bars were obtained from three independent measurements.
Here, we note that although the bending force facilitates the B–Z transition to occur at lower salt concentrations, 160 mM is still much higher than the physiological concentration of Mg2+, which is in the 1–20 mM range with 100 mM NaCl.51 Cytosine methylation is an important and frequently occurring DNA modification in cells.17,20,21 Z-DNA formation occurs at lower salt concentrations in the presence of cytosine methylation.13,16,18 Depending on the methylation level and the length of Z-DNA, Z-DNA formation can occur even at 0.5–2 mM Mg2+ by cytosine methylation.28,52 Hence, we prepared B–Z chimeric linear and BZ29-S19 with methylated cytosines in CG repeats, that is, m5 B–Z chimeric linear and m5 BZ29-S19, respectively (Figure 4a). We increased the MgCl2 concentration to form Z-DNA (Figures 4b–d and S11b). Indeed, the midpoint of the B–Z linear DNA decreased sharply from 700 to 78 mM MgCl2 by methylation (Figure 4c, blue dotted line). However, 78 mM MgCl2 is still higher than the physiological concentration. Then, when we applied the bending force of the methylated DNA (m5 BZ29-S19), the midpoint was reduced dramatically to 2.8 mM, that is, a 28-fold lower Mg2+ concentration than that of the methylated linear DNA (Figure 4c, red dotted line). These results indicate that the ability of the bending force to induce Z-DNA formation is enhanced in the methylated DNA samples (Figure 4d). The B–Z transition midpoint (2.8 mM Mg2+) of methylated DNA under the bending force is in the range of physiological salt concentrations. Thus, these results strongly suggested that Z-DNA could be formed by the bending force under physiological conditions in cells when the bending force is applied to methylated DNA. It is to be noted that if the methylation level in CG repeats is high enough, Z-DNA can be formed under physiological conditions without external forces.52
Figure 4.
Effects of DNA bending on the B–Z transitions of methylated (indicated by m5) DNAs. (a) DNA sequences used for the B–Z transition experiments. Methylated cytosines are indicated in brown color. (b) FRET histograms of m5 B–Z linear and m5 BZ29-S19 samples at various concentrations of MgCl2 obtained by the ALEX method. Histograms were fitted to a single Gaussian distribution. The black dotted lines denote the E of each sample in the absence of MgCl2. The red dotted lines denote the E of each sample at the highest concentration of MgCl2 (300 mM for m5 B–Z linear and 100 mM for m5 BZ29-S19). (c) E of m5 B–Z linear and m5 BZ29-S19 samples plotted against the concentration of MgCl2. The blue dotted line denotes the midpoint of the m5 B–Z linear sample (78 mM), and the red dotted line denotes the midpoint of the m5 BZ29-S19 sample (2.8 mM). The inset graph shows the E values of the m5 BZ29-S19 sample at low salt concentrations. Error bars were obtained from three independent measurements. (d) B–Z transition midpoints of each sample (B–Z linear, BZ29-S19, m5 B–Z linear, and m5 BZ29-S19). The midpoints of the B–Z linear and BZ29-S19 samples were obtained from Figure 3c. As the bending force increases, the midpoint decreases. Error bars were obtained from three independent measurements.
Formation of the B–Z Junction May Contribute to the B–Z Transition Induced by the Bending Force
It has been reported that Z-DNA is stiffer than B-DNA, as measured by light scattering: the persistence length of Z-DNA is approximately twice that of B-DNA.53 This means that Z-DNA resists bending stress more than B-DNA. Thus, the formation of Z-DNA itself is not favorable for releasing the tension applied by the bending force, which would result in lower curvature on the dsDNA portion compared with B-DNA. Then, a question arises as to the mechanism by which the DNA bending force induces the B–Z transition. The X-ray crystal structure showed that the B–Z junction generated in the sequence we used has a break in the A-T base pair and extrusion of these bases,47 which was also confirmed by 2-aminopurine (2-AP) fluorescence measurements.48 We confirmed the base extrusion at the B–Z junction using 2-AP fluorescence measurements (Figure S12). Because the bases were extruded from the helix as illustrated in Figure 1a, they were susceptible to cleavage by nucleases and modification by enzymes.30,54,55 The extrusion structure is quite similar to those of the melted base pairs.56 It has been well established that the region of the melted base pairs enables dsDNA to bend more easily than normal dsDNA.41,42 Even the presence of mismatched base pairs in the middle of dsDNA makes dsDNA bend easily upon application of the bending force.41,57 Thus, the formation of the B–Z junction with the extruded bases would stabilize the Z-DNA state compared with B-DNA when a bending force is applied to the dsDNA.
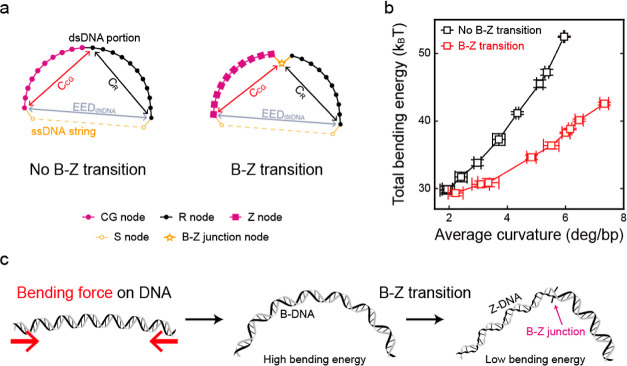
To study the effect of the B–Z junction on the bending force, we performed a simulation on D-shaped DNAs using a semiflexible loop,57,58 a coarse-grained model considering only the bending energy of the dsDNA portion (Figure 5a). Five types of nodes were used: a CG node for a CG base pair, an R node for a base pair of random sequences, a Z node for a base pair of Z-DNA, an S node for a nucleotide of the ssDNA string, and a B–Z junction node for the B–Z junction. The bending energy (B) of each node was calculated as B = Pθ2/2a, where P, θ, and a correspond to the persistence length, the angle between two tangential vectors of the adjacent nodes, and the interval, respectively. From the simulation of D-shaped DNAs with various bending forces, we obtained the end-to-end distance of the dsDNA portion (EEDdsDNA), the chord length of the random portion (CR) and the CG repeat portion (CCG) (Figure S13), and the total bending energy of the dsDNA portion (Figure 5b). Figure 5b presents the total bending energies of the dsDNA portion depending on the average curvature of dsDNA. As the average curvature of dsDNA increased, the total bending energy of dsDNA without the formation of the B–Z junction increased more rapidly than that of the B–Z transition with a B–Z junction. This result indicated that as the bending force increased (high curvature), the difference between the total bending energies of dsDNA portions with and without the B–Z junction also increased significantly. The increased bending energy difference in relation to the bending force will make Z-DNA with the B–Z junction more stable than B-DNA. Thus, the formation of the B–Z junction would stabilize Z-DNA compared with B-DNA under a bending force. Without the formation of the Z-DNA part, the B–Z junction may be unstable compared with the normal B-form DNA (no extrusion of the base-pair). This seems to be the reason Z-DNA retains its form after the formation of the B–Z junction, which effectively releases the bending stress, in the D-shaped DNAs. DNA bending without external torsional stress could have a possibility to induce local torsional stress. The base-stacking could be disrupted by the DNA bending, which also could contribute to the generation of the B–Z junction and Z-DNA.
Figure 5.
Simulation of the total bending energy of the dsDNA portion and a model of the B–Z transition by the bending force. (a) Schematic illustrations of the simulated semiflexible loops, which consist of the dsDNA portion and the ssDNA string. The end-to-end distance of the dsDNA portion (EEDdsDNA) and the chord lengths of the CG repeat portion (CCG) and random portion (CR) were obtained using Monte Carlo simulation (see the Supporting Information for detailed simulation methods). (b) Total bending energy of the dsDNA portion with (red) and without (black) the B–Z junction plotted against the average curvature of the dsDNA portion, that is, the applied bending force. The energy gap between the “no B–Z transition state” and the “B–Z transition state” gradually increases as the average curvature of the dsDNA portion increases. Error bars were obtained from five independent simulations. (c) Illustration of the proposed B–Z transition mechanism by the bending force. When the bending force is applied to the free DNA (red arrows), the entire B-DNA shows smooth bending with a high bending energy. With the B–Z transition, a B–Z junction with extruded base pairs is generated. Then, sharp bending occurs at the B–Z junction, which releases the bending stress and results in a low bending energy state.
Conclusions
In this study, we observed the effects of DNA bending on Z-DNA formation. As the bending force increased, Z-DNA was formed at lower salt concentrations, even at physiological salt concentrations with methylation. Although Z-DNA is stiffer than B-DNA,53 it appeared that the B–Z transition contributes to releasing of the bending stress by forming the B–Z junction with extruded bases47 and stabilizes the bent structure of dsDNA (Figure 5c). Among mechanical stresses, torsional stress and stretching force have been considered to be the factors contributing to the formation of Z-DNA.26−31 Our work demonstrates that the DNA bending force also contributes to the formation of Z-DNA.
Considering that Z-DNA is found on nucleosomal sequences,7 which are highly bent DNA regions, and affects the position of nucleosomes,6,7 Z-DNA formation induced by the bending force may play an important role in chromatin remodeling. Supercoiled DNAs also contain highly bent DNA structures at the apex of plectoneme.59 DNA bending is usually induced by various DNA–protein interactions and thus occurs ubiquitously over the whole genome, as one of the most common types of mechanical deformation in cells.33−37 For example, architectural DNA-binding proteins, such as the eukaryotic high mobility group B (HMGB) and the heat unstable (HU) protein of bacteria, induce strong bending of DNA.60 Although HU binds to DNA non-specifically, other nucleoid-associated proteins, such as IHF and Fis, bind to specifically regulatory sequences to induce DNA bending.61 Transcription factors, such as catabolite activator protein, also induce DNA bending, regulating transcription activity.62 DNA bending is also found in the processes of DNA mismatch repair by MutS and MutL.63 In addition, DNA loops, which accompany DNA bending structures, are also generated during many biological processes, such as replication, transcription, and recombination.64−66 For example, enhancers and mediators bound at separate DNA sites are brought to RNA polymerases by the formation of DNA loops in eukaryotes.67 In prokaryotes, repressors of gal, lac, and deo operons regulate transcription levels by forming DNA loops.65,66 Protein complexes for chromosome organization and segregation, such as condensin, also contribute to the formation of loop structures.68,69 Thus, our work suggests that the B–Z transition induced by the DNA bending force may be involved in more biological processes than we expected.
Acknowledgments
This work was supported by the Creative-Pioneering Researchers Program of Seoul National University and NRF-2019R1A2C2090896 and NRF-2020R1A5A1019141 of the National Research Foundation of Korea.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c02466.
Experiments, simulations, materials and methods, including DNA sequences, brief descriptions of the ALEX microscope and D-shaped DNA, FRET histograms of the normal and methylated DNAs depending on the salt concentrations, circular dichroism measurements of linear and D-shaped DNAs, proving the bending of the dsDNA, FRET efficiencies of linear and D-shaped DNAs with a CG-rich region, midpoints depending on the bending force, 2-AP fluorescence measurements, and reduced chord lengths depending on the reduced end-to-end distance (PDF)
Author Contributions
† J.Y. and S.Y. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Choi J.; Majima T. Conformational changes of non-B DNA. Chem. Soc. Rev. 2011, 40, 5893–5909. 10.1039/c1cs15153c. [DOI] [PubMed] [Google Scholar]
- Pohl F. M.; Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: Equilibrium and kinetic studies with poly(dG-dC). J. Mol. Biol. 1972, 67, 375–396. 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Wang A. H. J.; Quigley G. J.; Kolpak F. J.; Crawford J. L.; van Boom J. H.; van der Marel G.; Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 1979, 282, 680–686. 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wittig B.; Wölfl S.; Dorbic T.; Vahrson W.; Rich A. Transcription of human c-myc in permeabilized nuclei is associated with formation of Z-DNA in three discrete regions of the gene. EMBO J. 1992, 11, 4653–4663. 10.1002/j.1460-2075.1992.tb05567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A.; Zhang S. Z-DNA: the long road to biological function. Nat. Rev. Genet. 2003, 4, 566–572. 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- Wong B.; Chen S.; Kwon J.-A.; Rich A. Characterization of Z-DNA as a nucleosome-boundary element in yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2007, 104, 2229. 10.1073/pnas.0611447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland N.; Xu Y.; Sugiyama H.; Zhao K. SWI/SNF-mediated chromatin remodeling induces Z-DNA formation on a nucleosome. Cell Biosci. 2012, 2, 3. 10.1186/2045-3701-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; Vasquez K. M. Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair 2014, 19, 143–151. 10.1016/j.dnarep.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Mundo I. M. A.; Zewail-Foote A.; Kerwin M.; Vasquez S. M.; Vasquez K. M. Alternative DNA structure formation in the mutagenic human c-MYC promoter. Nucleic Acids Res. 2017, 45, 4929–4943. 10.1093/nar/gkx100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A. Z-DNA and Z-RNA in human disease. Commun. Biol. 2019, 2, 7. 10.1038/s42003-018-0237-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney J. A.; Wang G.; Mukherjee A.; Christensen L.; Subramanian S. H. S.; Zhao J.; Vasquez K. M. Distinct DNA repair pathways cause genomic instability at alternative DNA structures. Nat. Commun. 2020, 11, 236. 10.1038/s41467-019-13878-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall P. R.; Zhao Q.; Li X.; Wei W.; Periyakaruppiah A.; Zajaczkowski E. L.; Leighton L. J.; Madugalle S. U.; Basic D.; Wang Z.; Yin J.; Liau W.-S.; Gupte A.; Walkley C. R.; Bredy T. W. Dynamic regulation of Z-DNA in the mouse prefrontal cortex by the RNA-editing enzyme Adar1 is required for fear extinction. Nat. Neurosci. 2020, 23, 718–729. 10.1038/s41593-020-0627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behe M.; Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc. Natl. Acad. Sci. 1981, 78, 1619. 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S.; Kim D.; Kim K. K.; Kim Y.-G.; Hohng S. Intrinsic Z-DNA Is Stabilized by the Conformational Selection Mechanism of Z-DNA-Binding Proteins. J. Am. Chem. Soc. 2011, 133, 668–671. 10.1021/ja107498y. [DOI] [PubMed] [Google Scholar]
- Lee Y.-M.; Kim H.-E.; Park C.-J.; Lee A.-R.; Ahn H.-C.; Cho S. J.; Choi K.-H.; Choi B.-S.; Lee J.-H. NMR Study on the B–Z Junction Formation of DNA Duplexes Induced by Z-DNA Binding Domain of Human ADAR1. J. Am. Chem. Soc. 2012, 134, 5276–5283. 10.1021/ja211581b. [DOI] [PubMed] [Google Scholar]
- Wang S.; Long Y.; Wang J.; Ge Y.; Guo P.; Liu Y.; Tian T.; Zhou X. Systematic Investigations of Different Cytosine Modifications on CpG Dinucleotide Sequences: The Effects on the B-Z Transition. J. Am. Chem. Soc. 2014, 136, 56–59. 10.1021/ja4107012. [DOI] [PubMed] [Google Scholar]
- Schübeler D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- Zacharias W.; Jaworski A.; Wells R. D. Cytosine methylation enhances Z-DNA formation in vivo. J. Bacteriol. 1990, 172, 3278–3283. 10.1128/jb.172.6.3278-3283.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings C. K.; Anderson J. N. Links between DNA methylation and nucleosome occupancy in the human genome. Epigenet. Chromatin 2017, 10, 18. 10.1186/s13072-017-0125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature 1986, 321, 209–213. 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Maunakea A. K.; Nagarajan R. P.; Bilenky M.; Ballinger T. J.; D’Souza C.; Fouse S. D.; Johnson B. E.; Hong C.; Nielsen C.; Zhao Y.; Turecki G.; Delaney A.; Varhol R.; Thiessen N.; Shchors K.; Heine V. M.; Rowitch D. H.; Xing X.; Fiore C.; Schillebeeckx M.; Jones S. J. M.; Haussler D.; Marra M. A.; Hirst M.; Wang T.; Costello J. F. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010, 466, 253–257. 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz T.; Rould M. A.; Lowenhaupt K.; Herbert A.; Rich A. Crystal Structure of the Zα Domain of the Human Editing Enzyme ADAR1 Bound to Left-Handed Z-DNA. Science 1999, 284, 1841. 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- Kang Y.-M.; Bang J.; Lee E.-H.; Ahn H.-C.; Seo Y.-J.; Kim K. K.; Kim Y.-G.; Choi B.-S.; Lee J.-H. NMR Spectroscopic Elucidation of the B–Z Transition of a DNA Double Helix Induced by the Zα Domain of Human ADAR1. J. Am. Chem. Soc. 2009, 131, 11485–11491. 10.1021/ja902654u. [DOI] [PubMed] [Google Scholar]
- Park C.; Zheng X.; Park C. Y.; Kim J.; Lee S. K.; Won H.; Choi J.; Kim Y.-G.; Choi H.-J. Dual conformational recognition by Z-DNA binding protein is important for the B–Z transition process. Nucleic Acids Res. 2020, 48, 12957–12971. 10.1093/nar/gkaa1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K.; Klysik J.; Stirdivant S. M.; Wells R. D. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature 1982, 299, 312–316. 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- Rahmouni A. R.; Wells D. Stabilization of Z DNA in Vivo by Localized Supercoiling. Science 1989, 246, 358–363. 10.1126/science.2678475. [DOI] [PubMed] [Google Scholar]
- Wittig B.; Dorbic T.; Rich A. The level of Z-DNA in metabolically active, permeabilized mammalian cell nuclei is regulated by torsional strain. J. Cell Biol. 1989, 108, 755–764. 10.1083/jcb.108.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.; Kim S. H.; Hong S.-C. Minute negative superhelicity is sufficient to induce the B-Z transition in the presence of low tension. Proc. Natl. Acad. Sci. 2010, 107, 4985. 10.1073/pnas.0911528107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass F. C.; Fernandes L. E.; Bryant Z. Torque measurements reveal sequence-specific cooperative transitions in supercoiled DNA. Proc. Natl. Acad. Sci. 2012, 109, 6106. 10.1073/pnas.1113532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Cui Y.; An R.; Liang X.; Li Q.; Wang H.; Wang H.; Fan Y.; Dong P.; Li J.; Cheng K.; Wang W.; Wang S.; Wang G.; Xue C.; Komiyama M. Topologically Constrained Formation of Stable Z-DNA from Normal Sequence under Physiological Conditions. J. Am. Chem. Soc. 2019, 141, 7758–7764. 10.1021/jacs.8b13855. [DOI] [PubMed] [Google Scholar]
- Kim S. H.; Jung H. J.; Lee I.-B.; Lee N.-K.; Hong S.-C. Sequence-dependent cost for Z-form shapes the torsion-driven B–Z transition via close interplay of Z-DNA and DNA bubble. Nucleic Acids Res. 2021, 49, 3651–3660. 10.1093/nar/gkab153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F.; Wang J. C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. 1987, 84, 7024. 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalov P. L.; Dragan A. I.; Crane-Robinson C. The cost of DNA bending. Trends Biochem. Sci. 2009, 34, 464–470. 10.1016/j.tibs.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Harteis S.; Schneider S. Making the Bend: DNA Tertiary Structure and Protein-DNA Interactions. Int. J. Mol. Sci. 2014, 15, 12335. 10.3390/ijms150712335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irobalieva R. N.; Fogg J. M.; Catanese D. J.; Sutthibutpong T.; Chen M.; Barker A. K.; Ludtke S. J.; Harris S. A.; Schmid M. F.; Chiu W.; Zechiedrich L. Structural diversity of supercoiled DNA. Nat. Commun. 2015, 6, 8440. 10.1038/ncomms9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; Wang M. D. DNA supercoiling during transcription. Biophys. Rev. 2016, 8, 75–87. 10.1007/s12551-016-0215-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A.; Bobrovnikov D. G.; Ha T. DNA mechanics and its biological impact. J. Mol. Biol. 2021, 433, 166861. 10.1016/j.jmb.2021.166861. [DOI] [PubMed] [Google Scholar]
- Yeou S.; Lee N. K. Single-Molecule Methods for Investigating the Double-Stranded DNA Bendability. Mol. Cells 2022, 45, 33–40. 10.14348/molcells.2021.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff H.; Reinhard B. M.; Siu M.; Agarwal H.; Spakowitz A.; Liphardt J. Biocompatible Force Sensor with Optical Readout and Dimensions of 6 nm3. Nano Lett. 2005, 5, 1509–1514. 10.1021/nl050875h. [DOI] [PubMed] [Google Scholar]
- Qu H.; Wang Y.; Tseng C.-Y.; Zocchi G. Critical Torque for Kink Formation in Double-Stranded DNA. Phys. Rev. X 2011, 1, 021008. 10.1103/physrevx.1.021008. [DOI] [Google Scholar]
- Kim C.; Lee O. C.; Kim J. Y.; Sung W.; Lee N. K. Dynamic Release of Bending Stress in Short dsDNA by Formation of a Kink and Forks. Angew. Chem., Int. Ed. Engl. 2015, 54, 8943–8947. 10.1002/anie.201502055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee O. c.; Kim C.; Kim J.-Y.; Lee N. K.; Sung W. Two conformational states in D-shaped DNA: Effects of local denaturation. Sci. Rep. 2016, 6, 28239. 10.1038/srep28239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T.; Enderle T.; Ogletree D. F.; Chemla D. S.; Selvin P. R.; Weiss S. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. 1996, 93, 6264. 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanidis A. N.; Lee N. K.; Laurence T. A.; Doose S.; Margeat E.; Weiss S. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. 2004, 101, 8936. 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellenkamp B.; Schmid S.; Doroshenko O.; Opanasyuk O.; Kühnemuth R.; Rezaei Adariani S.; Ambrose B.; Aznauryan M.; Barth A.; Birkedal V.; Bowen M. E.; Chen H.; Cordes T.; Eilert T.; Fijen C.; Gebhardt C.; Götz M.; Gouridis G.; Gratton E.; Ha T.; Hao P.; Hanke C. A.; Hartmann A.; Hendrix J.; Hildebrandt L. L.; Hirschfeld V.; Hohlbein J.; Hua B.; Hübner C. G.; Kallis E.; Kapanidis A. N.; Kim J.-Y.; Krainer G.; Lamb D. C.; Lee N. K.; Lemke E. A.; Levesque B.; Levitus M.; McCann J. J.; Naredi-Rainer N.; Nettels D.; Ngo T.; Qiu R.; Robb N. C.; Röcker C.; Sanabria H.; Schlierf M.; Schröder T.; Schuler B.; Seidel H.; Streit L.; Thurn J.; Tinnefeld P.; Tyagi S.; Vandenberk N.; Vera A. M.; Weninger K. R.; Wünsch B.; Yanez-Orozco I. S.; Michaelis J.; Seidel C. A. M.; Craggs T. D.; Hugel T. Precision and accuracy of single-molecule FRET measurements—a multi-laboratory benchmark study. Nat. Methods 2018, 15, 669–676. 10.1038/s41592-018-0085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S.; Son H.; Kim Y.-G.; Hohng S. Z-DNA stabilization is dominated by the Hofmeister effect. Phys. Chem. Chem. Phys. 2013, 15, 15829–15832. 10.1039/c3cp52047a. [DOI] [PubMed] [Google Scholar]
- Ha S. C.; Lowenhaupt K.; Rich A.; Kim Y.-G.; Kim K. K. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature 2005, 437, 1183–1186. 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- Subramani V. K.; Ravichandran S.; Bansal V.; Kim K. K. Chemical-induced formation of BZ-junction with base extrusion. Biochem. Biophys. Res. Commun. 2019, 508, 1215–1220. 10.1016/j.bbrc.2018.12.045. [DOI] [PubMed] [Google Scholar]
- Lee N. K.; Kapanidis A. N.; Wang Y.; Michalet X.; Mukhopadhyay J.; Ebright R. H.; Weiss S. Accurate FRET Measurements within Single Diffusing Biomolecules Using Alternating-Laser Excitation. Biophys. J. 2005, 88, 2939–2953. 10.1529/biophysj.104.054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryer L.; Haugland R. P. Energy transfer: a spectroscopic ruler. Proc. Natl. Acad. Sci. U.S.A. 1967, 58, 719–726. 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani A. M. P. Cellular magnesium homeostasis. Arch. Biochem. Biophys. 2011, 512, 1–23. 10.1016/j.abb.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H.; Jung H. J.; Hong S.-C. Z-DNA as a Tool for Nuclease-Free DNA Methyltransferase Assay. Int. J. Mol. Sci. 2021, 22, 11990. 10.3390/ijms222111990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T. J.; Bloomfield V. A. Chain flexibility and hydrodynamics of the B and Z forms of pory(dG-dC).pory(dG-dC). Nucleic Acids Res. 1983, 11, 1919–1930. 10.1093/nar/11.6.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K.; Klysik J.; Wells R. D. Conformational flexibility of junctions between contiguous B- and Z-DNAs in supercoiled plasmids. Proc. Natl. Acad. Sci. 1983, 80, 2447. 10.1073/pnas.80.9.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajjugal Y.; Rathinavelan T. Sequence dependent influence of an A...A mismatch in a DNA duplex: An insight into the recognition by hZαADAR1 protein. J. Struct. Biol. 2021, 213, 107678. 10.1016/j.jsb.2020.107678. [DOI] [PubMed] [Google Scholar]
- Vargas-Lara F.; Starr F. W.; Douglas J. F. Molecular rigidity and enthalpy–entropy compensation in DNA melting. Soft Matter 2017, 13, 8309–8330. 10.1039/c7sm01220a. [DOI] [PubMed] [Google Scholar]
- Jeong J.; Kim H. D. Base-Pair Mismatch Can Destabilize Small DNA Loops through Cooperative Kinking. Phys. Rev. Lett. 2019, 122, 218101. 10.1103/physrevlett.122.218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X.; Vologodskii A. Theoretical Analysis of Disruptions in DNA Minicircles. Biophys. J. 2009, 96, 1341–1349. 10.1016/j.bpj.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlicek J. W.; Oussatcheva E. A.; Sinden R. R.; Potaman V. N.; Sankey O. F.; Lyubchenko Y. L. Supercoiling-Induced DNA Bending. Biochemistry 2004, 43, 10664–10668. 10.1021/bi0362572. [DOI] [PubMed] [Google Scholar]
- Peters J. P.; Maher L. J. DNA curvature and flexibility in vitro and in vivo. Q. Rev. Biophys. 2010, 43, 23–63. 10.1017/s0033583510000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwade J. E.; Leonard A. C. Blocking, Bending, and Binding: Regulation of Initiation of Chromosome Replication During the Escherichia coli Cell Cycle by Transcriptional Modulators That Interact With Origin DNA. Front. Microbiol. 2021, 12, 732270. 10.3389/fmicb.2021.732270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S. C.; Shields G. C.; Steitz T. A. Crystal Structure of a CAP-DNA Complex: The DNA Is Bent by 90(Degrees). Science 1991, 253, 1001. 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Iyer R. R.; Pluciennik A.; Burdett V.; Modrich P. L. DNA Mismatch Repair: Functions and Mechanisms. Chem. Rev. 2006, 106, 302–323. 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- Saiz L.; Vilar J. M. G. DNA looping: the consequences and its control. Curr. Opin. Struct. Biol. 2006, 16, 344–350. 10.1016/j.sbi.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Hofmann A.; Heermann D. W. The role of loops on the order of eukaryotes and prokaryotes. FEBS Lett. 2015, 589, 2958–2965. 10.1016/j.febslet.2015.04.021. [DOI] [PubMed] [Google Scholar]
- Hao N.; Sullivan A. E.; Shearwin K. E.; Dodd I. B. The loopometer: a quantitative in vivo assay for DNA-looping proteins. Nucleic Acids Res. 2021, 49, e39 10.1093/nar/gkaa1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrascheck M.; Escher D.; Mahmoudi T.; Verrijzer C. P.; Schaffner W.; Barberis A. DNA looping induced by a transcriptional enhancer in vivo. Nucleic Acids Res. 2005, 33, 3743–3750. 10.1093/nar/gki689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji M.; Shaltiel A.; Bisht S.; Kim E.; Kalichava A.; Haering H.; Dekker C. Real-time imaging of DNA loop extrusion by condensin. Science 2018, 360, 102–105. 10.1126/science.aar7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M. R.; Hochwagen A.; Ercan S. Condensin action and compaction. Curr. Genet. 2019, 65, 407–415. 10.1007/s00294-018-0899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.