Abstract
Broiler chickens from eight commercial farms in Southern Finland were analyzed for the structure of their gastrointestinal microbial community by a nonselective DNA-based method, percent G+C-based profiling. The bacteriological impact of the feed source and in-farm whole-wheat amendment of the diet was assessed by percent G+C profiling. Also, a phylogenetic 16S rRNA gene (rDNA)-based study was carried out to aid in interpretation of the percent G+C profiles. This survey showed that most of the 16S rDNA sequences found could not be assigned to any previously known bacterial genus or they represented an unknown species of one of the taxonomically heterogeneous genera, such as Ruminococcus or Clostridium. The data from bacterial community profiling were analyzed by t-test, multiple linear regression, and principal-component statistical approaches. The percent G+C profiling method with appropriate statistical analyses detected microbial community differences smaller than 10% within each 5% increment of the percent G+C profiles. Diet turned out to be the strongest determinant of the cecal bacterial community structure. Both the source of feed and local feed amendment changed the bacteriological profile significantly, whereas profiles of individual farms with identical feed regimens hardly differed from each other. This suggests that the management of typical Finnish farms is relatively uniform or that hygiene on the farm, in fact, has little impact on the structure of the cecal bacterial community. Therefore, feed compounders should have a significant role in the modulation of gut microflora and consequently in prevention of gastrointestinal disorders in farm animals.
Previous studies showed that there is a relationship between the gastrointestinal (GI) tract microflora and health of animals (6, 36). To date, most attempts to control GI tract microflora have relied on the use of broad-spectrum antibiotics. The recent and widening introduction of bans on the prophylactic use of many antibiotics due to concerns over dissemination of antibiotic resistance traits will likely end most or all such practices. Hence, there is an increasing interest in developing alternative methods of controlling the GI tract microflora. It has been known for a long time that diet can significantly influence the composition and metabolic activity of the GI tract microflora (20, 32, 36). Feed ingredients conducive to the growth of beneficial GI tract bacteria, as well as direct introduction of bacterial populations that favor good health and nutrition in animals (i.e., probiotics), can be used to manage the GI tract microbial community (6, 11, 13, 15, 19, 21). So far, the development of alternate management strategies for the GI tract microflora has been hampered by the lack of practical analytical tools for monitoring of the composition of the total community. Several studies of other animal species and a variety of habitats suggest that only a fraction of the total microbial community is effectively captured by culture-based techniques (2, 8, 10, 14, 18, 28).
Molecular biology tools have been developed to detect the presence of major animal- and food-borne, pathogens, but routine methods for the analysis of the total microbial community structure are rare. We have previously developed methods by which the cecal enterome (the chromosomal DNA of the total bacterial community of the GI tract) can be effectively isolated directly from samples without the biases introduced by cultivation (3, 16). Appropriate analyses of enterome samples should provide better information about the true composition of microbial communities in the GI tract and facilitate the development of new products and strategies to improve animal health and productivity.
Phylogenetic analyses based on 16S rRNA (rDNA) gene sequences that employ universal or more specific PCR primers have previously been used to analyze microbial communities in a variety of environments, including soils, hot springs, epiphytic consortia, subsurface soil, and termite guts (2, 29, 31, 37). However, phylogenetic studies based on 16S rDNA require extensive isolation and analysis of individual sequences to generate a composite view of the total community. This approach can thus be too tedious, expensive, and limited where complex communities or large numbers of samples are involved, as is the case in feeding trials to determine the effects of different dietary formulations on the total intestinal microflora.
In this paper, we present a fast, enterome-based method for the analysis of the cecal microbial community structure of chickens and examine the effect of feed from two different commercial producers on these microbial communities and the variation among eight different farms in southern Finland. This was accomplished through fractionation of the enterome by a method based on the percent G+C content of the component populations. We expand on prior reports describing this technique (3) by demonstrating its accuracy and power in a large-scale comparative analysis as a result of its combination with multiple linear regression (MLR) and principal-component analyses (PCA).
MATERIALS AND METHODS
Experimental design.
Broiler chickens at each of eight commercial farms distributed in southern Finland were raised in floor pens using different commercial wheat-based diets, some with locally added whole wheat. Two farms fed chickens with commercial feed from feed mill A in fall 1997 (hereafter referred to as AControl1 and AControl2). Two other farms fed chickens with feed from the same manufacturer 3 years later, in spring 2000 (hereafter referred to as ALate1 and ALate2). Feed from feed mill B was used at two farms in fall 1997 (hereafter referred to as BControl1 and BControl2), while the remaining two farms used feed from manufacturer B amended with whole wheat 4 months later, in spring 1998 (hereafter referred to as BWheat1 and BWheat2). For each sampling, two to six replicate birds that had been fed their respective diets for 4 to 5 weeks since hatching were killed by cervical dislocation and their ceca were immediately removed and dissected. Cecal contents from individual birds were transferred into 50-ml sterile tubes, kept on ice, and processed for bacterial recovery, cell lysis, and enterome recovery starting within 2 h of collection as described in detail previously (3) and outlined briefly below. Where indicated below, cecal contents from the individual birds on a single feed were pooled to provide a composite sample prior to bacterial fraction recovery, cell lysis, and DNA isolation.
Recovery of bacteria, cell lysis, and enterome isolation.
The bacterial fraction was recovered from cecal contents through multiple rounds of dilution, high-speed centrifugation, and washing as described previously (3). This approach has been shown to effectively recover >95% of the bacteria in the viscous cecal matrix. Bacterial fractions were subsequently lysed in accordance with our previously described protocol, which combines physical (bead beating), chemical (sodium dodecyl sulfate), and enzymatic (lysozyme and proteinase K) steps. This protocol has been shown to lyse more than 99% of the bacteria present (3).
Bacterial community profile analysis.
For profiles of cecal community structure based on percent G+C content, 100 μg of each enterome sample was subjected to cesium chloride-bisbenzimidazole gradient analysis as described previously (3). This approach fractionates the DNA of the component populations of the community based on their characteristic percent G+C content through differential density, which is imposed by the AT-dependent DNA-binding dye bisbenzimidazole (17). Determination of the percent G+C content represented by each gradient fraction was accomplished by regression analysis (r2, >0.99) of data obtained from gradients containing standard DNA samples with known percent G+C compositions (Clostridium perfringens, Escherichia coli, and Micrococcus lysodeikticus). The absorbance value of the percent G+C profile curve was first calculated for each percentage from 20 to 80%. The integral of the percent G+C profiles was then normalized to 100%, and the relative abundance was calculated for each 5% increment of percent G+C for statistical analyses.
Phylogenetic survey based on partial 16S rDNA sequences.
To obtain a sense of the dominant microbial genera represented by the percent G+C profiles, a phylogenetic survey of the total microbial community in the ceca of chickens raised on feed A was performed. Partial 16S/18S sequences of the rDNAs representing the organisms in pooled cecal samples from farms AControl1 and AControl2 were amplified by PCR with universal primers 536f [5′-CAGC(AC)GCCGCGGTAAT(AT)C-3′] and 907r (5′-CCCCGTCAATTCCTTTGAGTTT-3′). These primers are based on highly conserved regions of the 16S and 18S rDNAs of all organisms (hence, they are universal) and are thus suitable for this DNA sequence-based phylogenetic survey (note that our primer 536f is the exact complement of universal primer 519r described in reference 24). PCR mixtures contained 1× PCR buffer (Boehringer Mannheim, Indianapolis, Ind.), 200 μM each deoxynucleoside triphosphate, 20 pmol of each primer, ∼1 ng of community DNA in 10 μl, and 2.5 U of Taq polymerase (Boehringer Mannheim) in a total volume of 50 μl. PCRs were performed by initial denaturation for 5 min at 95°C, followed by 30 cycles of: denaturation for 1 min at 95°C, primer annealing for 1 min at 50°C, and primer extension at 72°C for 3 min. This was followed by a final extension reaction at 72°C for 10 min.
The resulting mixture of ∼400-bp PCR products was subsequently cloned into the EcoRV site of the pT7Blue-3 vector with the Perfectly Blunt Cloning Kit (Novagen, Madison, Wis.). Plasmid DNA was purified from the resulting clones with the Qiagen Mini-Prep plasmid purification kit (Qiagen, Valencia, Calif.) in accordance with the manufacturer's specifications. DNA sequence analysis was then performed by using the 16/18S rDNA-specific primers 536f and 907r described above on an ABI 373A automated DNA sequencer (ABI, Foster City, Calif.).
The resulting DNA sequence information was analyzed by using public software (Sequence Match) and sequences available at the Ribosomal Database Project II at http://www.cme.msu.edu/RDP/html/index.html to determine the best-match identification of the organisms present in the broiler cecum (25). The results are presented in the form of S_ab (similarity score a versus b) scores, which are an indication of how well the sequence in question matches a related sequence in the database. Thus, a S_ab score of 1.000 indicates a perfect match to a known sequence while lower scores indicate increasingly unrelated sequences. Detection of potential chimeric products was facilitated by using the Chimera Check feature of the RDP site, and likely chimeras were not considered further.
Statistical analyses. Comparison of percent G+C increments.
The percent G+C profile was divided into 12 5% increments. The proportion of microbes within each increment was obtained by integrating the corresponding area for each sample analyzed.
t test.
The t test was used for pairwise comparison of each percent G+C increment to reveal statistically significant differences (P ≤ 0.05) between the different farms and feed regimens (see Tables 1, 2, and 3).
TABLE 1.
t-test results for differences in percent G+C increments between farms
| Percent G+C increment and feed |
P valuea
|
|||||||
|---|---|---|---|---|---|---|---|---|
| AControl1 | AControl2 | ALate1 | ALate2 | BControl1 | BControl2 | BWheat1 | BWheat2 | |
| 20–24 | ||||||||
| AControl1 | × | × | 0.04 | 0.02 | 0.02 | |||
| AControl2 | × | × | ||||||
| ALate1 | × | × | 0.02 | 0.01 | 0.01 | |||
| ALate2 | × | × | 0.05 | 0.04 | 0.03 | |||
| BControl1 | 0.04 | 0.05 | 0.02 | × | × | |||
| BControl2 | × | × | ||||||
| BWheat1 | 0.02 | 0.04 | 0.01 | × | × | |||
| BWheat2 | 0.02 | 0.03 | 0.01 | × | × | |||
| 25–29 | ||||||||
| AControl1 | × | × | ||||||
| AControl2 | × | × | ||||||
| ALate1 | × | × | 0.04 | |||||
| ALate2 | × | × | 0.03 | 0.05 | ||||
| BControl1 | 0.03 | 0.04 | × | × | ||||
| BControl2 | × | × | ||||||
| BWheat1 | 0.05 | × | × | |||||
| BWheat2 | × | × | ||||||
| 30–34 | ||||||||
| AControl1 | × | × | ||||||
| AControl2 | × | × | ||||||
| ALate1 | × | × | ||||||
| ALate2 | × | × | ||||||
| BControl1 | × | × | ||||||
| BControl2 | × | × | ||||||
| BWheat1 | × | × | ||||||
| BWheat2 | × | × | ||||||
| 35–39 | ||||||||
| AControl1 | × | × | 0.04 | 0.009 | 0.05 | |||
| AControl2 | × | × | 0.02 | 0.0004 | 0.01 | 0.006 | ||
| ALate1 | 0.04 | 0.02 | × | × | 0.003 | |||
| ALate2 | × | × | 0.005 | |||||
| BControl1 | 0.009 | 0.0004 | 0.003 | 0.005 | × | × | 0.04 | |
| BControl2 | 0.01 | × | × | |||||
| BWheat1 | 0.04 | × | × | |||||
| BWheat2 | 0.05 | 0.006 | × | × | ||||
| 40–44 | ||||||||
| AControl1 | × | × | 0.003 | 0.0006 | 0.005 | 0.03 | ||
| AControl2 | × | × | 0.003 | <0.0001 | 0.006 | |||
| ALate1 | 0.003 | 0.003 | × | × | 0.0002 | 0.002 | 0.03 | |
| ALate2 | × | × | 0.006 | 0.01 | ||||
| BControl1 | 0.0006 | <0.0001 | 0.0002 | 0.006 | × | × | 0.03 | 0.05 |
| BControl2 | 0.005 | 0.006 | 0.002 | 0.01 | × | × | ||
| BWheat1 | 0.03 | 0.03 | × | × | ||||
| BWheat2 | 0.03 | 0.05 | × | × | ||||
| 45–49 | ||||||||
| AControl1 | × | × | 0.05 | 0.009 | ||||
| AControl2 | × | × | 0.0002 | |||||
| ALate1 | × | × | 0.006 | |||||
| ALate2 | × | × | 0.03 | 0.01 | ||||
| BControl1 | 0.05 | 0.03 | × | × | ||||
| BControl2 | 0.009 | 0.0002 | 0.006 | 0.01 | × | × | 0.005 | |
| BWheat1 | × | × | ||||||
| BWheat2 | 0.005 | × | × | |||||
| 50–54 | ||||||||
| AControl1 | × | × | 0.005 | 0.0009 | ||||
| AControl2 | × | × | 0.02 | 0.006 | ||||
| ALate1 | × | × | 0.005 | 0.007 | ||||
| ALate2 | × | × | 0.002 | 0.002 | ||||
| BControl1 | × | × | 0.04 | 0.04 | ||||
| BControl2 | × | × | 0.03 | |||||
| BWheat1 | 0.005 | 0.02 | 0.005 | 0.002 | 0.04 | × | × | |
| BWheat2 | 0.0009 | 0.006 | 0.007 | 0.002 | 0.04 | 0.03 | × | × |
| 55–59 | ||||||||
| AControl1 | × | × | 0.01 | 0.03 | 0.02 | 0.009 | ||
| AControl2 | × | × | ||||||
| ALate1 | × | × | ||||||
| ALate2 | × | × | 0.03 | 0.04 | 0.02 | 0.01 | ||
| BControl1 | 0.01 | 0.03 | × | × | ||||
| BControl2 | 0.03 | 0.04 | × | × | ||||
| BWheat1 | 0.02 | 0.02 | × | × | ||||
| BWheat2 | 0.009 | 0.01 | × | × | ||||
| 60–64 | ||||||||
| AControl1 | × | × | 0.05 | 0.05 | ||||
| AControl2 | × | × | 0.007 | |||||
| ALate1 | × | 0.02 | 0.0007 | 0.04 | ||||
| ALate2 | 0.02 | × | 0.02 | |||||
| BControl1 | 0.05 | 0.02 | × | × | 0.01 | |||
| BControl2 | × | × | 0.03 | |||||
| BWheat1 | 0.05 | 0.007 | 0.0007 | 0.01 | 0.03 | × | × | |
| BWheat2 | 0.04 | × | × | |||||
| 65–69 | ||||||||
| AControl1 | × | × | 0.001 | 0.03 | ||||
| AControl2 | × | × | 0.005 | 0.004 | 0.02 | |||
| ALate1 | 0.005 | × | × | <0.0001 | 0.0006 | |||
| ALate2 | × | × | 0.0003 | 0.02 | ||||
| BControl1 | 0.001 | 0.004 | <0.0001 | 0.0003 | × | × | 0.03 | 0.006 |
| BControl2 | 0.03 | 0.02 | 0.0006 | 0.02 | × | × | ||
| BWheat1 | 0.03 | × | × | |||||
| BWheat2 | 0.006 | × | × | |||||
| 70–74 | ||||||||
| AControl1 | × | × | 0.04 | 0.01 | ||||
| AControl2 | × | × | 0.004 | 0.004 | ||||
| ALate1 | 0.004 | × | × | 0.002 | <0.0001 | |||
| ALate2 | × | × | ||||||
| BControl1 | 0.04 | 0.002 | × | × | 0.04 | |||
| BControl2 | 0.01 | 0.004 | <0.0001 | × | × | 0.02 | ||
| BWheat1 | 0.04 | 0.02 | × | × | ||||
| BWheat2 | × | × | ||||||
| 75–79 | ||||||||
| AControl1 | × | × | 0.01 | 0.03 | ||||
| AControl2 | × | × | ||||||
| ALate1 | × | × | 0.005 | 0.02 | ||||
| ALate2 | 0.01 | × | × | 0.0001 | ||||
| BControl1 | × | × | ||||||
| BControl2 | 0.005 | 0.0001 | × | × | 0.004 | |||
| BWheat1 | × | × | ||||||
| BWheat2 | 0.03 | 0.02 | 0.004 | × | × | |||
P values of >0.05 are not shown.
TABLE 2.
t-test results for differences in percent G+C increments between feed regimens
| Percent G+C increment and feed |
P valuea
|
|||
|---|---|---|---|---|
| AControl | ALate | BControl | BWheat | |
| 20–24 | ||||
| AControl | × | × | 0.02 | 0.002 |
| ALate | × | × | 0.004 | 0.0006 |
| BControl | 0.02 | 0.004 | × | × |
| BWheat | 0.002 | 0.0006 | × | × |
| 25–29 | ||||
| AControl | × | 0.02 | ||
| ALate | 0.02 | × | 0.04 | 0.008 |
| BControl | 0.04 | × | × | |
| BWheat | 0.008 | × | × | |
| 30–34 | ||||
| AControl | × | × | ||
| ALate | × | × | ||
| BControl | × | × | ||
| BWheat | × | × | ||
| 35–39 | ||||
| AControl | × | 0.005 | 0.0001 | 0.03 |
| ALate | 0.005 | × | 0.0002 | |
| BControl | 0.0001 | 0.0002 | × | 0.02 |
| BWheat | 0.03 | 0.02 | × | |
| 40–44 | ||||
| AControl | × | 0.005 | <0.0001 | |
| ALate | 0.005 | × | <0.0001 | |
| BControl | <0.0001 | <0.0001 | × | 0.01 |
| BWheat | 0.01 | × | ||
| 45–49 | ||||
| AControl | × | × | 0.002 | |
| ALate | × | × | 0.0008 | |
| BControl | 0.002 | 0.0008 | × | 0.02 |
| BWheat | 0.02 | × | ||
| 50–54 | ||||
| AControl | × | × | <0.0001 | |
| ALate | × | × | <0.0001 | |
| BControl | × | 0.001 | ||
| BWheat | <0.0001 | <0.0001 | 0.001 | × |
| 55–59 | ||||
| AControl | × | × | 0.004 | 0.001 |
| ALate | × | × | 0.04 | 0.005 |
| BControl | 0.004 | 0.04 | × | × |
| BWheat | 0.001 | 0.005 | × | × |
| 60–64 | ||||
| AControl | × | × | 0.01 | 0.04 |
| ALate | × | × | 0.03 | |
| BControl | 0.01 | 0.03 | × | 0.003 |
| BWheat | 0.04 | 0.003 | × | |
| 65–69 | ||||
| AControl | × | × | 0.0005 | |
| ALate | × | × | <0.0001 | |
| BControl | 0.0005 | <0.0001 | × | 0.004 |
| BWheat | 0.004 | × | ||
| 70–74 | ||||
| AControl | × | × | 0.001 | |
| ALate | × | × | 0.0004 | |
| BControl | 0.001 | 0.0004 | × | 0.002 |
| BWheat | 0.002 | × | ||
| 75–79 | ||||
| AControl | × | × | 0.04 | |
| ALate | × | × | 0.009 | |
| BControl | 0.009 | × | 0.02 | |
| BWheat | 0.04 | 0.02 | × | |
P values of >0.05 are not shown.
TABLE 3.
t-test results for differences in percent G+C increments between commercial feeds A and B
| Percent G+C | P valuea for A vs B |
|---|---|
| 20–24 | <0.0001 |
| 25–29 | 0.02 |
| 30–34 | |
| 35–39 | 0.002 |
| 40–44 | |
| 45–49 | 0.001 |
| 50–54 | 0.0007 |
| 55–59 | <0.0001 |
| 60–64 | |
| 65–69 | 0.01 |
| 70–74 | 0.03 |
| 75–79 |
P values of >0.05 are not shown.
MLR analysis.
Differences in percent G+C increments (y variables) were modeled by MLR (for an explanation of MLR technique, see, e.g., reference 7) using SYSTAT (SYSTAT for Windows, Version 5 Edition; SYSTAT, Inc., Evanston, Ill.). MLR models reveal the most significant sources of variation in the relative abundance of bacteria within a given percent G+C increment. The MLR model used in this work was y = c0 + c1 · x1 + c2 · x2 + c3 · x3 + c4 · x4 + c5 · x5 + c6 · x6 + c7 · x7, where x1 to x7 are unitless independent dummy variables and c0 to c7 are the model parameters with the same unit as y (in this case, the unit is percent). x1 is either x11 or x12, where x11 is a dummy variable that receives the value 1 when feed A is given and the value 0 otherwise and x12 is a dummy variable that receives the value 1 when feed B is given and the value 0 otherwise (hence, x11 + x12 = 1); x2 is either x21 or x22, where x21 is a dummy variable that receives the value 1 when feed AControl is given and the value 0 otherwise, and x22 is a dummy variable that receives the value 1 when feed ALate is given and the value 0 otherwise (hence, x21 + x22 = x11); x3 is either x31 or x32, where x31 is a dummy variable that receives the value 1 when feed BControl is given and the value 0 otherwise and x32 is a dummy variable that receives the value 1 when feed BWheat is given and the value 0 otherwise (hence, x31 + x32 = x12); x4 is either x41 or x42, where x41 is a dummy variable that receives the value 1 when the sample is from farm AControl1 and the value 0 otherwise and x42 is a dummy variable that receives the value 1 when the sample is from farm AControl2 and the value 0 otherwise (hence, x41 + x42 = x21); x5 is either x51 or x52, where x51 is a dummy variable that receives the value 1 when the sample is from farm ALate1 and the value 0 otherwise and x52 is a dummy variable that receives the value 1 when sample is from farm ALate2 and the value 0 otherwise (hence, x51 + x52 = x22); x6 is either x61 or x62, where x61 is a dummy variable that receives the value 1 when the sample is from farm BControl1 and the value 0 otherwise and x62 is a dummy variable that receives the value 1 when the sample is from farm BControl2 and the value 0 otherwise (hence, x61 + x62 = x31); and x7 is x71 or x72, where x71 is a dummy variable that receives the value 1 when the sample is from farm BWheat1 and the value 0 otherwise and x72 is a dummy variable that receives the value 1 when the sample is from farm BWheat2 and the value 0 otherwise (hence, x71 + x72 = x32). As there are two possible choices for each of the seven independent variables, we have tested 27 different regressions for each y variable and the most powerful in explaining the variation for each y is reported (see Table 4). Variable x1 tests if the feed type is a significant source of variation, variable x2 tests if temporal difference is a significant source of variation, variable x3 tests if local modification is a significant source of variation, and variables x4 to x7 test if the individual farm is a significant source of variation.
TABLE 4.
Most significant sources of variation, percentages of change, P values and goodness-of-fit values in the MLR modeling of percent G+C increments
| % G+C increment and determinants | Coefficient | % Change | P value |
|---|---|---|---|
| 20–24 | |||
| Constant | 1.98 | <0.0001 | |
| Feed B | −1.37 | −70 | <0.0001 |
| r2 | 0.48 | <0.0001 | |
| 25–29 | |||
| Constant | 3.06 | <0.0001 | |
| Feed ALate | 2.09 | +68 | 0.0004 |
| r2 | 0.36 | 0.0004 | |
| 30–34 | |||
| Constant | 4.76 | <0.0001 | |
| Feed ALate | 0.94 | +20 | 0.04 |
| r2 | 0.14 | 0.04 | |
| 35–39 | |||
| Constant | 7.63 | <0.0001 | |
| Feed BControl | −1.64 | −21 | 0.0004 |
| Feed AControl | 1.27 | +17 | 0.001 |
| r2 | 0.58 | <0.0001 | |
| 40–44 | |||
| Constant | 12.35 | <0.0001 | |
| Feed BControl | −2.54 | −21 | <0.0001 |
| Farm BWheat1 | 2.28 | +18 | 0.002 |
| Feed AControl | 1.39 | +11 | 0.007 |
| r2 | 0.69 | <0.0001 | |
| 45–49 | |||
| Constant | 12.79 | <0.0001 | |
| Feed BControl | −2.89 | −23 | 0.0002 |
| Farm BWheat1 | −1.58 | −12 | 0.05 |
| r2 | 0.58 | <0.0001 | |
| 50–54 | |||
| Constant | 16.42 | <0.0001 | |
| Feed BWheat | 5.08 | +31 | <0.0001 |
| r2 | 0.69 | <0.0001 | |
| 55–59 | |||
| Constant | 12.63 | <0.0001 | |
| Feed B | 1.91 | +15 | <0.0001 |
| r2 | 0.42 | <0.0001 | |
| 60–64 | |||
| Constant | 10.12 | <0.0001 | |
| Feed BControl | 2.85 | +28 | 0.0004 |
| Farm BWheat1 | −2.37 | −23 | 0.01 |
| Farm ALate1 | 1.68 | +17 | 0.02 |
| r2 | 0.57 | <0.0001 | |
| 65–69 | |||
| Constant | 6.51 | <0.0001 | |
| Feed BControl | 5.37 | +82 | <0.0001 |
| r2 | 0.62 | <0.0001 | |
| 70–74 | |||
| Constant | 2.13 | <0.0001 | |
| Feed BControl | 1.75 | +82 | <0.0001 |
| r2 | 0.51 | <0.0001 | |
| 75–79 | |||
| Constant | 0.44 | <0.0001 | |
| Farm BControl2 | 0.35 | +78 | 0.007 |
| Farm BWheat2 | −0.27 | −61 | 0.01 |
| Farm ALate2 | −0.16 | −37 | 0.04 |
| r2 | 0.47 | 0.0008 |
PCA.
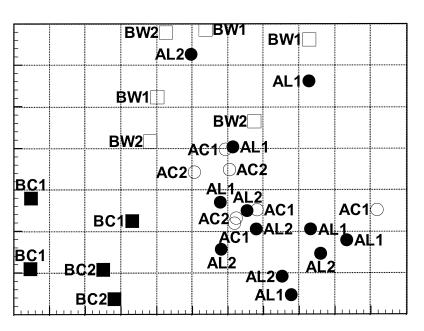
Data were also analyzed by using a multivariate approach wherein linear correlations between all variables were calculated simultaneously (1). Calculations were carried out using SIMCA-P (SIMCA for Windows Version 2.1; Umetri AB). This technique calculates a set of uncorrelated variables called principal components. The number of principal components depends on the set of data, but each principal component is a linear combination of the original variables. A graphical representation with unitless axes can then be constructed. Every point represents all of the data from one individual sample, in this case, one bird. The points that cluster together in the graphical representation have similar overall data patterns (see Fig. 4).
FIG. 4.
PCA of cecal percent G+C profiles of broiler chickens from eight different commercial farms. Positioning of individual broiler chickens is based on the analysis of individual percent G+C profiles using all 12 5% G+C increments collectively and is indicated by the squares (feed B) and circles (feed A). The label next to each symbol indicates the origin of the sample as follows: AC1 and AC2, farms using feed A in 1997; AL1 and AL2, farms using feed A in 2000; BC1 and BC2, farms using feed B in 1997; BW1 and BW2, farms using feed B amended with whole wheat in 1998.
RESULTS
Accuracy of percent G+C profiling.
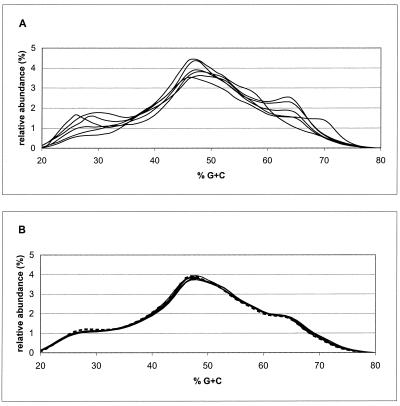
To assess bird-to-bird differences within a single farm (ALate1), cecal enteromes from six individual animals were analyzed by percent G+C profiling. The bacterial community profiles obtained were similar (Fig. 1A) but did display some differences resulting from variation between individual animals, as would be expected in most or all biological systems for any measured physiological parameter.
FIG. 1.
Percent G+C profiles of cecal bacterial communities in broiler chickens from a commercial Finnish farm. (A) Percent G+C profiles of six individual broiler chickens from a farm using commercial feed from feed mill A (farm ALate1). (B) Six replicate percent G+C profiles of a single digesta sample obtained by pooling the six individual digesta samples of panel A (solid lines) and the arithmetic average of the individual profiles shown in panel A (dotted line).
To ensure that the differences observed were true differences between animals and not due to experimental error or lack of reliability of the method, six replicate preparations and analyses (i.e., bacterial fraction recoveries, enterome purification protocols, and percent G+C profiles) were performed on a single composite sample comprising the pooled cecal contents of the same six birds that are shown individually in Fig. 1A. The percent G+C profiles of these replicate analyses showed excellent agreement (Fig. 1B), the calculated average standard deviation for the total normalized profiles being 5.0%. Thus, where differences are observed between profiles in other analyses (e.g., in Fig. 1A), they represent true differences in microbial community composition.
Figure 1B also provides an interesting comparison of the average profile calculated from the profiles of individual animals from Fig. 1A (dotted line) to the replicate profiles obtained from the corresponding pooled sample (solid lines). The close similarity between these profiles is particularly striking considering that the pooled sample represents a “mechanical” averaging of the variation between animals achieved by mixing of the total cecal contents before DNA recovery and analysis, while the mean plot from the individual samples represents an arithmetic averaging of the data from individual animals. Collectively, the data indicate that little variability was introduced into the community profiles as a result of the experimental manipulations. Further, the data show that pooling of samples from replicate individual animals represents a means by which to provide an average representative sample for that treatment. However, to be able to apply appropriate statistical analyses, some replication is needed. As a result, we can distinguish true differences in community composition resulting from different farms or feed regimens from fluctuations resulting from animal-to-animal differences within treatments.
Comparison of farm- and feed-related characteristics of the bacterial profiles by the standard t test.
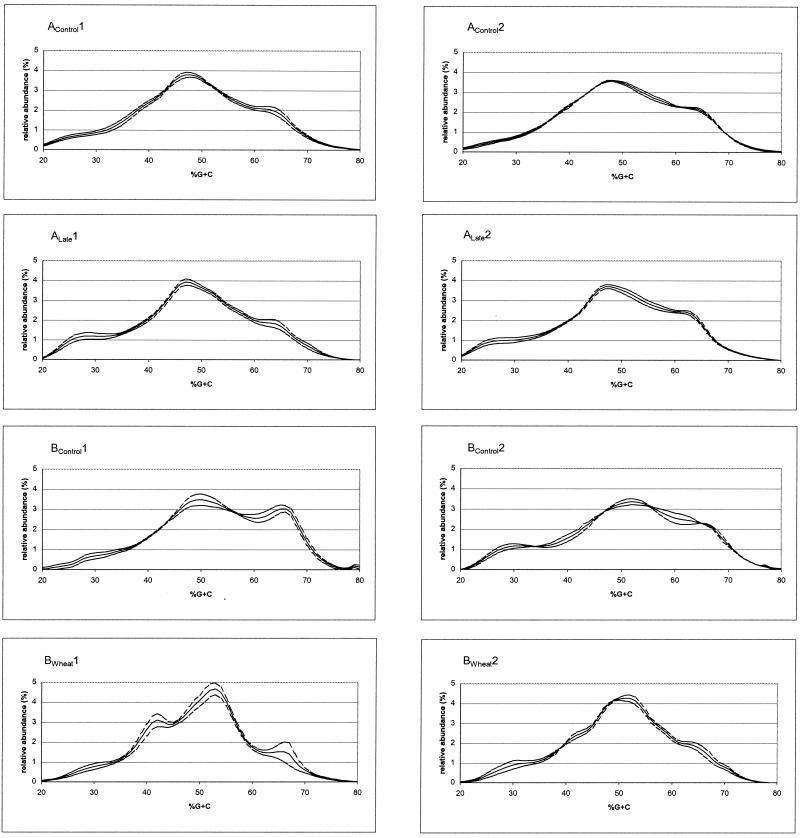
The cecal microflora of individual broiler chickens from each farm were analyzed by the percent G+C profiling approach. The data for individual animals were compared to derive standard error values expressing the variability within each farm or treatment (Fig. 2). Animal-to-animal variability within farms using feed A was lower in the high percent G+C range than that within farms using feed B (a standard F test for equality of variances was run; data not shown). Animal-to-animal variability was especially pronounced in the 40 to 44% G+C range on the farms that locally added whole wheat in feed B (F test; data not shown).
FIG. 2.
Percent G+C profiles of cecal bacterial communities in broiler chickens from eight commercial farms in southern Finland. The profile in each panel shows the arithmetic average of two to six broiler chickens from each farm (solid line) and the standard error (dotted line). The heading of each graph indicates the feed manufacturer, sampling time, and feeding regimen as described in detail in Materials and Methods.
All eight farms were compared to each other pairwise in 5% increments between 20 and 80% G+C. P values for every increment of percent G+C were calculated by using the standard t test. These results are presented in Table 1, where all P values of ≤0.05 for each percent G+C increment are presented to indicate whether individual farms differ significantly from each other for that range of percent G+C content. It is worth noting that P values of ≤0.05 between two farms indicate that the statistical animal-to-animal variability within each farm was significantly smaller than the farm-to-farm variability and, therefore, that the birds from the corresponding farms differ significantly in the relative abundance of microflora. The data in Table 1 show that, when compared pairwise, the chicken cecal GI tract communities on many of the farms differed from each other. However, a closer look revealed that community profiles from any two farms with identical feed regimen and times (AControl1 versus AControl2; ALate1 versus ALate2; BControl1 versus BControl2; or BWheat1 versus BWheat2) did not differ from each other significantly (× symbols in Table 1). Indeed, there was only one instance in which two farms using the same feed at the same time differed significantly from each other, i.e., the 60 to 64% G+C increment of the microbial communities at farms ALate1 and ALate2 (Table 1).
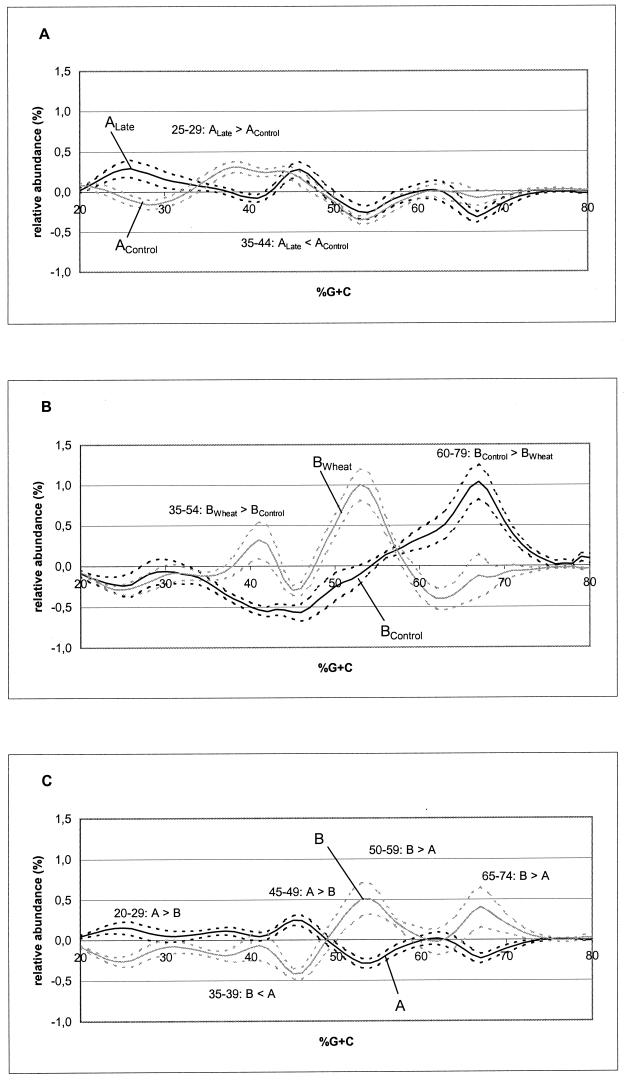
Since we had replicate farms for each feed or time period, we could statistically analyze the relative importance of the commercial feed, temporal distance, and local amendment of the feed on intestinal bacterial community structure in the GI tract. To better illustrate the individual community characteristics that result from feed regimen, the average of bacterial community profiles of all of the individual animals under all of the conditions tested was subtracted from the arithmetic mean of the bacterial community profiles for each of the feed regimens. Figure 3 highlights some characteristics of the feed regimens in pairwise comparisons of treatments through time (AControl versus ALate), local feed amendment (BControl versus BWheat), and feed manufacturer (AControl and ALate versus BControl and BWheat).
FIG. 3.
Characteristic effects of individual feeding regimens on bacterial community profiles. To reveal specific characteristics of each feeding regimen, the average bacterial community profile of all of the broiler chickens analyzed in this study (grand average) was subtracted from the average bacterial community profile of birds on each feeding regimen. Solid lines show the average profile of each feeding regimen, and dotted lines indicate the corresponding standard error of the mean. (A) Comparison of birds fed feed A in 1997 and 2000 (AControl versus ALate) with indication of those percent G+C increments for which feed AControl and ALate differed from each other according to t-test analysis (Table 2). (B) Comparison of birds fed feed B alone and those given feed B amended with whole wheat (BControl versus BWheat) with indication of those percent G+C increments for which feed BControl and BWheat differed from each other according to t-test analysis (Table 2). (C) Comparison of all birds with feed A in their diet to all birds with feed B in their diet (AControl and ALate versus BControl and BWheat) with indication of those percent G+C increments for which feeds A and B differed from each other according to t-test analysis (Table 3).
The t tests revealed that there were statistically significant differences between the AControl and ALate samples at values lower than 45% G+C. Birds raised on feed A in 2000 had a slightly higher relative abundance of bacteria with 25 to 29% G+C and a lower abundance of bacteria with 35 to 44% G+C than did birds fed given feed A 3 years earlier (Table 2; Fig. 3A). The relative abundance of bacteria with percent G+C contents higher than 45% were essentially identical for all birds on feed A.
The amendment of feed B with whole wheat resulted in a higher abundance of bacteria with 35 to 54% G+C compared to that achieved with feed B with no amendments (Fig. 3B; Table 2). On the other hand, the local amendment of feed B with wheat remarkably decreased the proportion of microbial populations having G+C contents higher than 60%. The magnitude of the effects due to amendment of the feed was several times greater than any effect of the temporal difference illustrated in panel A.
Regardless of the calendar year and local amendments, the data on all of the birds were divided into two pools, depending on the manufacturer of the main commercial feed (A or B). Figure 3C illustrates the major differences in the average cecal microfloral profiles. In general, chickens on feed A had a higher relative abundance of GI tract bacteria in the 20 to 29, 35 to 39, and 45 to 49% G+C range and a lower abundance of bacteria in the 55 to 59 and 65 to 74% G+C increment ranges than did birds on feed B (Fig. 3C; Table 3). It can hence be summarized that the cecal bacterial community favored by feed from mill A was different from that supported by feed from mill B, regardless of prevailing differences in the environment and management on the farm.
Revelation of the main sources of microfloral variation by MLR.
It is worth noting that while the t test compares two selected groups of datum points and shows whether they differ significantly from one another, MLR analysis directly reveals the most important sources of variation for each percent G+C increment.
Table 4 shows the constants, statistically significant determinants (P < 0.05), and r2 values produced by the MLR models used for each percent G+C increment. Constants in the table indicate the abundance of bacteria at a defined percent G+C increment in the absence of the identified significant determinants. Significant determinants either increase or decrease the abundance of bacteria represented by the given percent G+C, as indicated by coefficients and percent change for each percent G+C increment (Table 4). In most cases, the P values for the regressions were low, indicating that the independent variables were truly significant sources of variation for the percent G+C increments. Also, the r2 values associated with low P values indicate that even though there were also unknown sources of variation, in most cases a large proportion of the variation was explained by the variables shown in Table 4.
The individual farm was not the most significant source of variation in any percent G+C increment representing more than 1% of the total bacterial community but increased the explanatory power of the regression model for several percent G+C increments as a second or third variable. The source of feed was the most important source of variation for the 20 to 24 and 55 to 59% G+C increments; feed B expressed 70% lower and 15% higher abundances of bacteria with indicated percent G+C, respectively, than did feed A. The batch of feed and/or feeding time of feed A was the most important source of variation for the 25 to 34% G+C increments; indeed, feed A in 2000 showed a significantly higher abundance of bacteria in the 25 to 29 and 30 to 34% G+C increments, respectively, than any feeding regimen in 1997 and 1998 (Table 4; Fig. 2). Feed B with no wheat amendment (BControl) enriched a bacterial community with significantly lower relative abundance of species with 35 to 49% G+C than did the other feeding regimens and increased those with 60 to 74% G+C (Table 4). Feed B with wheat amendment (BWheat) was the most important source of variation for the bacteria with 50 to 54% G+C, showing a 31% greater abundance of bacteria than feed A or B without wheat amendment. Feed A in 1997 was not the most significant source of variation in any increment of percent G+C but was the second and third most significant source of variation for the 35 to 44% G+C increment and increased the explanatory power of the model. The results of the MLR analysis were in agreement with the t-test procedures shown above; both conclude that feed regimens but not individual farms were the most important sources of variation (Table 4).
Capture of total bacterial divergence by PCA.
The statistical approaches applied as described above revealed differences between farms, feeds, and local feed amendments for individual percent G+C increments. PCA was applied to reveal the total bacterial community divergence between individual broiler chickens, with all percent G+C increments analyzed collectively in a single analysis. It is worth noting that the PCA plot is an abstract representation of the data with unitless axes (Fig. 4).
Figure 4 illustrates how each of the individual broiler chickens was positioned in this analysis. The results showed that the structure of the total bacterial community in the ceca of broiler chickens was strongly feed dependent. Birds from farms BControl1 and BControl2 formed a cluster distinct both from farms BWheat1 and BWheat2 and from the four farms on feed A. Wheat amendment of feed B significantly changed the positioning of the cecal bacterial community in PCA. Most of the broiler chickens on feed A, regardless of the year, clustered together distinct from the birds on feed B (Fig. 4). The PCA results supported the conclusions derived from the t-test and MLR methods employed as described above. The PCA approach gives a visually powerful presentation, encapsulating in a single graph the divergence of the total bacterial community in individual broiler chickens. The detailed features of the differences and the major sources of variation within each percent G+C increment should be looked for in the presentations of the t-test (Tables 1 to 3) and MLR (Table 4) results.
Phylogenetic view into the major bacterial genera in the ceca of broiler chickens.
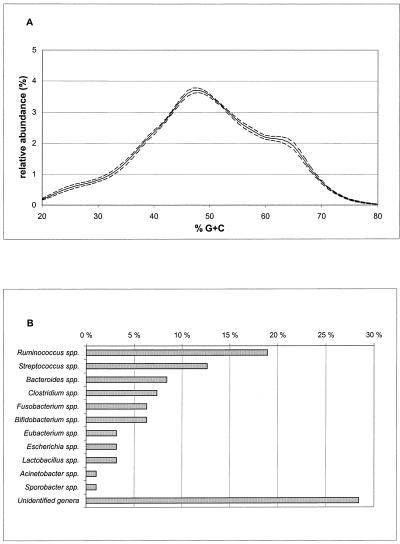
Since total microbial community analysis by percent G+C profiling alone does not reveal the taxonomic identity of the representative bacterial genera, a survey of the cecal community was conducted to provide a phylogenetic context regarding which bacterial genera are represented in the bacterial communities of the chicken GI tract. To accomplish this, we PCR amplified, cloned, and sequenced 100 randomly selected partial 16S rDNA fragments by using universal PCR primers. The pooled cecal enteromes of seven broiler chickens from two farms using feed from the same feed manufacturer (AControl1 and AControl2) were used for this study. Figure 5A shows the average profile and standard error of the mean of the microbial communities present in the ceca of these seven broiler chickens when analyzed by percent G+C profiling. The most abundant peak was centered at 48% G+C content, with additional peaks or shoulders at 25, 38, and 63% G+C.
FIG. 5.
Percent G+C profiling and 16S rDNA analyses of cecal bacterial communities in broiler chickens on two commercial farms in Finland. (A) Average percent G+C profile (solid line) and standard error (dotted lines) of cecal bacterial communities in seven broiler chickens from farms AControl1 and AControl2. (B) Relative abundance of bacterial genera or groups in the samples when analyzed by partial 16S rDNA sequencing. The bacterial genus and group names used are based on best matches to the Ribosomal Database Project II sequence database. A genus name is indicated when the best match (S_ab score) was higher than 0.7 (25).
The sequences obtained were compared to those deposited in the Ribosomal Database Project II website (http://www.cme.msu.edu/RDP/html/index.html) (25). Since the S_ab scores of the best matches were, in most cases, lower than 0.9, we were not confident in designating these beyond genus level identification (Fig. 5B). Nearly 30% of the sequences showed S_ab scores lower than 0.7 compared with any sequence deposited in the Ribosomal Database Project II database, which indicates poorer than genus level identification (Fig. 5B; unidentified genera). This group comprised 11 different taxonomic units with S_ab scores of <0.7 with respect to one another. Bacteria with 16S sequence homology to species of the genus Ruminococcus comprised nearly 20% of the total bacterial community in the ceca of the chickens analyzed. The second most abundant genus was Streptococcus, followed by Bacteroides, Clostridium, Fusobacterium, and Bifidobacterium (Fig. 5B). The other bacterial genera detected each comprised less than 5% of the total population. No eucaryotic sequences were found, suggesting that 18S represented a minor proportion (<5%) of the total 16/18S rDNA.
DISCUSSION
The method of bacterial community analysis used here detected bacterial shifts accurately when combined with the relevant mathematical analyses. It is among the few techniques truly capable of depicting the total microbial community in a single analysis. The average standard deviation of the method itself was 5%, and therefore, it is theoretically possible to detect differences of greater than 7% between individual treatments at a 95% confidence level with six replicates. The percent G+C technique used gives the relative abundance of bacterial groups as defined by their respective percent G+C contents. The method therefore provides taxonomically relevant information on the bacterial members of the community. However, in complex bacterial communities with many unknown members, the diagnostic value of the percent G+C profiling method is limited; when used alone, it often does not identify the specific bacterial taxa.
In the present work, we carried out a small phylogenetic study to get an idea of the component bacterial genera in the chicken cecum. Sequencing of samples from one farm is not proof of their presence in other farms. However, it is most likely that the same major taxa were also present in the broiler chickens from other locations but at different relative abundances. In this survey, species related to ruminococci and streptococci were the most abundant bacteria present; percent G+C contents reported for the known species of Streptococcus and Ruminococcus range from 34 to 46% (4, 22, 35). In that particular area, there was a major shoulder in the corresponding percent G+C profile (Fig. 5A). The genus known to have the highest percent G+C content of those identified in this study was Bifidobacterium, ranging from 58 to 65% (34). Also, in this percent G+C range, a shoulder could be seen in the percent G+C profile (Fig. 5A). The profile of Fig. 5A shows that the most abundant bacteria in the chicken cecum represented species with 40 to 55% G+C, the highest peak appearing at 48% G+C. Due to the large proportion of the representatives of unidentified genera and the taxonomic heterogeneity of major genera such as Bacteroides, Eubacterium, and Clostridium, it is not possible to reliably reconstruct the entire percent G+C profile from the 16S rDNA sequencing data. However, due to its nonselective nature, percent G+C profiling provides a good foundation for more intensive molecular and culture-based studies when unknown microbial assemblages are being analyzed for the first time.
Culture-based techniques can be very selective, effectively detecting minority populations, but likely never capture the total microbial community of complex anaerobic habitats such as the avian GI tract. Similarly, there are effective molecular methods for detecting specific bacterial species (5, 23, 33). However, these methods are often dependent on specific probes and primers to detect selected populations and groups and are therefore limited when studying complex and previously undefined microbial ecosystems. Denaturing gradient gel electrophoresis of 16S rDNA fragments has been used to estimate the complexity of microbial communities in a number of ecosystems (9, 12, 26, 27, 30). An advantage of the denaturing gradient gel electrophoresis approach is that it generates a fingerprint of the total microbial community, but the drawback is that, alone, it provides no additional information on the taxonomic identity of the component bacterial groups (e.g., percent G+C).
In this study, we used three different independent statistical analyses for the data (t test, MLR, and PCA) and the outputs of all of them showed that feed was the strongest individual determinant of the total microbial community structure in the ceca of broiler chickens. This is a significant finding, since conventional wisdom holds that the environment in which birds are raised is at least as important as their feed in determining the microbial communities within their intestinal tracts. In fact, in this study, individual farms were not among the most significant determinants of the cecal microflora. In Finnish broiler farms, management practices are relatively uniform and well controlled and perhaps that is why farm-to-farm differences in the present study were found to be minor. Also, there did not seem to be significant seasonal variation since, e.g., the four farms on feed A represented two different seasons but showed almost identical percent G+C profiles (Fig. 4).
These data suggest that the feed manufacturer has the greatest influence on the intestinal microflora. The effect of feed A was studied first in 1997 and then again in 2000. The bacterial profiles of birds fed these feeds displayed some differences in the low percent G+C area (Fig. 3), but in the comprehensive PCA, all of the birds on feed A clustered practically together (Fig. 4). Most likely, the two batches of feed A differed in the source and variety of raw materials and perhaps also in the presence and relative proportions of some of the ingredients used in diet manufacture, which may account for the small changes observed in microbial populations. Indeed, it was remarkable that the microfloral populations did not differ more substantially when similar feeds were compared over time. The process of feed manufacturing, in itself, may be as important as the ingredients; i.e., conditioning process temperature, steam pressure, and dwell time, coupled with die size, may be responsible for the signature of the microbes from feed mill A compared with that of those from feed mill B.
Due to the reduced usage of prophylactic antibiotics in the feed industry, new undiagnosed enteric diseases have arisen in Europe. In agreement with some earlier studies applying culture-based methods and various host animals, we show here that diet affects the composition of the intestinal bacterial community significantly (20, 32, 36). This points to the importance of the feed manufacturer in determining the intestinal community structure and in this way reducing the risk of emerging diseases. According to our study, the farmer can also dramatically affect the composition of the cecal bacterial community. On-farm modification of a feeding regimen by whole-wheat amendment was the most powerful single factor affecting the GI tract microflora. Addition of wheat to the diet strongly increased bacteria with 50 to 55% G+C and equally dramatically suppressed those with 60 to 69% G+C, most likely bifidobacteria (Fig. 3; Table 4). The presence of whole wheat tended to increase the variation between birds (Fig. 4). This fits with the likely fact that when birds are offered a choice between pellets and whole grain, there will be significant differences in preference between individuals. As a result, their nutrient intake will vary more markedly than that of birds fed a homogeneous ration, and hence, bacterial substrate presentation to the intestinal lumen will vary accordingly.
It is not possible to assess the physiological significance of all of the bacterial shifts detected by the methods described above. However, the availability of relatively fast methods for monitoring of total bacterial communities is a prerequisite for informative epidemiological surveys in the future. The type of data shown in this paper could be correlated with other parameters reflecting animal performance and health (e.g., productivity, immune status, and bacterial metabolites) and together would greatly improve our understanding of GI interactions and the importance of the microbial community structure.
ACKNOWLEDGMENTS
This work was financially supported by the Finnish Technology Development Center.
We gratefully acknowledge Seppo Peuranen for sampling arrangements and useful comments. Linda Schimmelpfennig is acknowledged for contributing to cloning and sequence analysis. We also thank Osmo Siikanen, Brita Mäki, Jaana Oksanen, and Hilkka Heikkinen for excellent technical assistance in percent G+C profiling.
REFERENCES
- 1.Afifi A A, Clark V. Principal component analysis. In: Afifi A A, Clark V, editors. Computer-aided multivariate analysis. New York, N.Y: Chapman & Hall; 1990. pp. 371–394. [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apajalahti J H A, Särkilahti L K, Mäki B R E, Heikkinen J P, Nurminen P H, Holben W E. Effective recovery of bacterial DNA and percent-guanine-plus-cytosine-based analysis of community structure in the gastrointestinal tract of broiler chickens. Appl Environ Microbiol. 1998;64:4084–4088. doi: 10.1128/aem.64.10.4084-4088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brock T D, Madigan M T. Biology of microorganisms. Englewood Cliffs, N.J: Prentice Hall; 1991. [Google Scholar]
- 5.Burr M D, Josephson K L, Pepper I L. An evaluation of ERIC PCR and AP PCR fingerprinting for discriminating Salmonella serotypes. Lett Appl Microbiol. 1998;27:24–30. doi: 10.1046/j.1472-765x.1998.00378.x. [DOI] [PubMed] [Google Scholar]
- 6.Cresci A, Orpianesi C, Silvi S, Mastrandrea V, Dolara P. The effect of sucrose or starch-based diet on short-chain fatty acids and faecal microflora in rats. J Appl Microbiol. 1999;86:245–250. doi: 10.1046/j.1365-2672.1999.00668.x. [DOI] [PubMed] [Google Scholar]
- 7.Draper N, Smith H. Applied regression analysis, second edition. New York, N.Y: John Wiley & Sons, Inc.; 1981. [Google Scholar]
- 8.Felske A, Akkermans A D L, De Vos W M. In situ detection of an uncultured predominant bacillus in Dutch grassland soils. Appl Environ Microbiol. 1998;64:4588–4590. doi: 10.1128/aem.64.11.4588-4590.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fritsche T R, Horn M, Seyedirashti S, Gautom R K, Schleifer K-H, Wagner M. In situ detection of novel bacterial endosymbionts of Acanthamoeba spp. phylogenetically related to members of the order Rickettsiales. Appl Environ Microbiol. 1999;65:206–212. doi: 10.1128/aem.65.1.206-212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garriga M, Pascual M, Monfort J M, Hugas M. Selection of lactobacilli for chicken probiotic adjuncts. J Appl Microbiol. 1998;84:125–132. doi: 10.1046/j.1365-2672.1997.00329.x. [DOI] [PubMed] [Google Scholar]
- 12.Gelsomino A, Keijzer-Wolters A C, Cacco G, van Elsas J D. Assessment of bacterial community structure in soil by polymerase chain reaction and denaturing gradient gel electrophoresis. J Microbiol Methods. 1999;38:1–15. doi: 10.1016/s0167-7012(99)00054-8. [DOI] [PubMed] [Google Scholar]
- 13.Gusils C, Chaia A P, Gonzalez S, Oliver G. Lactobacilli isolated from chicken intestines: potential use as probiotics. J Food Prot. 1999;62:252–256. doi: 10.4315/0362-028x-62.3.252. [DOI] [PubMed] [Google Scholar]
- 14.Hanson R S, Hanson T E. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hock E, Halle I, Matthes S, Jeroch H. Investigations on the composition of the ileal and caecal microflora of broiler chicks in consideration to dietary enzyme preparation and zinc bacitracin in wheat-based diets. Agribiol Res Z Agrarbiol Agrikult Oekol. 1997;50:85–95. [Google Scholar]
- 16.Holben W E. Isolation and purification of bacterial community DNA from environmental samples. In: Hurst C H, Knudsen G R, McInerny M J, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C.: ASM Press; 1997. pp. 431–436. [Google Scholar]
- 17.Holben W E, Harris D. DNA based monitoring of total bacterial community structure in environmental samples. Mol Ecol. 1995;4:627–631. doi: 10.1111/j.1365-294x.1995.tb00263.x. [DOI] [PubMed] [Google Scholar]
- 18.Holben W E, Noto K, Sumino T, Suwa Y. Molecular analysis of bacterial communities in a three-compartment granular activated sludge system indicates community-level control by incompatible nitrification processes. Appl Environ Microbiol. 1998;64:2528–2532. doi: 10.1128/aem.64.7.2528-2532.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izat A L, Hierholzer R E, Kopek J M, Adams M H, Reiber M A, McGinnis J P. Effects of d-mannose on incidence and levels of salmonellae in ceca and carcass samples of market age broilers. Poult Sci. 1990;69:2244–2247. doi: 10.3382/ps.0692244. [DOI] [PubMed] [Google Scholar]
- 20.Jensen B B. The impact of feed additives on the microbial ecology of the gut in young pigs. J Anim Feed Sci. 1998;7:45–64. [Google Scholar]
- 21.Jin L Z, Ho Y W, Abdullah N, Ali M A, Jalaludin S. Effects of adherent lactobacillus cultures on growth, weight of organs and intestinal microflora and volatile fatty acids in broilers. Anim Feed Sci Technol. 1998;70:197–209. [Google Scholar]
- 22.Krieg N R, Holt J G. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. [Google Scholar]
- 23.Kwang J, Littledike E T, Keen J E. Use of the polymerase chain reaction for salmonella detection. Lett Appl Microbiol. 1996;22:46–51. doi: 10.1111/j.1472-765x.1996.tb01106.x. [DOI] [PubMed] [Google Scholar]
- 24.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maidak B L, Cole J R, Lilburn T G, Parker C T, Jr, Saxman P R, Farris R J, Garrity G M, Olsen G J, Schmidt T M, Tiedje J M. The RDP-II (Ribosomal Database Project) Nucleic Acids Res. 2001;29:173–174. doi: 10.1093/nar/29.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie van Leeuwenhoek. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- 28.Ohkuma M, Kudo T. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:461–468. doi: 10.1128/aem.62.2.461-468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olsen G J, Lane D J, Giovannoni S J, Pace N R, Stahl D A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 30.Overmann J, Coolen M J, Tuschak C. Specific detection of different phylogenetic groups of chemocline bacteria based on PCR and denaturing gradient gel electrophoresis of 16S rRNA gene fragments. Arch Microbiol. 1999;172:83–94. doi: 10.1007/s002030050744. [DOI] [PubMed] [Google Scholar]
- 31.Pace N R, Stahl D A, Lane D J, Olsen G J. The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microb Ecol. 1986;9:1–55. [Google Scholar]
- 32.Pluske J R, Durmic Z, Pethick D W, Mullan B P, Hampson D J. Confirmation of the role of rapidly fermentable carbohydrates in the expression of swine dysentery in pigs after experimental infection. J Nutr. 1998;128:1737–1744. doi: 10.1093/jn/128.10.1737. [DOI] [PubMed] [Google Scholar]
- 33.Rasmussen H N, Olsen J E, Jorgensen K, Rasmussen O F. Detection of Campylobacter jejuni and Camp. coli in chicken faecal samples by PCR. Lett Appl Microbiol. 1996;23:363–366. doi: 10.1111/j.1472-765x.1996.tb00209.x. [DOI] [PubMed] [Google Scholar]
- 34.Sgorbati B, Biavati B, Palenzona D. The genus Bifidobacterium. In: Wood B J B, Holzapfel W H, editors. The genera of lactic acid bacteria. Glasgow, Scotland: Chapman & Hall; 1995. pp. 279–306. [Google Scholar]
- 35.Stewart C S. The rumen bacteria. In: Hobson P N, editor. The rumen microbial ecosystem. London, England: Elsevier Applied Science; 1988. pp. 21–75. [Google Scholar]
- 36.Wagner D D, Thomas O P. Influence of diets containing rye or pectin on the intestinal flora of chicks. Poult Sci. 1987;57:971–975. doi: 10.3382/ps.0570971. [DOI] [PubMed] [Google Scholar]
- 37.Ward D M, Weller R, Bateson M M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]