Abstract
The quick diagnosis of the novel coronavirus (COVID-19) disease is vital to prevent its propagation and improve therapeutic outcomes. Computed tomography (CT) is believed to be an effective tool for diagnosing COVID-19, however, the CT scan contains hundreds of slices that are complex to be analyzed and could cause delays in diagnosis. Artificial intelligence (AI) especially deep learning (DL), could facilitate and speed up COVID-19 diagnosis from such scans. Several studies employed DL approaches based on 2D CT images from a single view, nevertheless, 3D multiview CT slices demonstrated an excellent ability to enhance the efficiency of COVID-19 diagnosis. The majority of DL-based studies utilized the spatial information of the original CT images to train their models, though, using spectral–temporal information could improve the detection of COVID-19. This article proposes a DL-based pipeline called CoviWavNet for the automatic diagnosis of COVID-19. CoviWavNet uses a 3D multiview dataset called OMNIAHCOV. Initially, it analyzes the CT slices using multilevel discrete wavelet decomposition (DWT) and then uses the heatmaps of the approximation levels to train three ResNet CNN models. These ResNets use the spectral–temporal information of such images to perform classification. Subsequently, it investigates whether the combination of spatial information with spectral–temporal information could improve the diagnostic accuracy of COVID-19. For this purpose, it extracts deep spectral–temporal features from such ResNets using transfer learning and integrates them with deep spatial features extracted from the same ResNets trained with the original CT slices. Then, it utilizes a feature selection step to reduce the dimension of such integrated features and use them as inputs to three support vector machine (SVM) classifiers. To further validate the performance of CoviWavNet, a publicly available benchmark dataset called SARS-COV-2-CT-Scan is employed. The results of CoviWavNet have demonstrated that using the spectral–temporal information of the DWT heatmap images to train the ResNets is superior to utilizing the spatial information of the original CT images. Furthermore, integrating deep spectral–temporal features with deep spatial features has enhanced the classification accuracy of the three SVM classifiers reaching a final accuracy of 99.33% and 99.7% for the OMNIAHCOV and SARS-COV-2-CT-Scan datasets respectively. These accuracies verify the outstanding performance of CoviWavNet compared to other related studies. Thus, CoviWavNet can help radiologists in the rapid and accurate diagnosis of COVID-19 diagnosis.
Keywords: COVID-19, Deep learning, ResNet, Convolutional neural networks, Discrete wavelet transform (DWT), Computed tomography (CT)
1. Introduction
Lately, the world has experienced a pandemic due to the novel coronavirus known as COVID-19. This new disease or infection is induced by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1], [2]. The rates of infection are increasing rapidly across the globe. It has been reported that greater than 108 million individuals have been diagnosed with COVID-19, the mortality rate is more than 2 million 4 hundred cases, and the rate of recovery exceeded 80 million cases [3]. The worldwide rapid spread of the novel coronavirus has placed a huge segment of the globe’s population into quarantine and has devastated several industrial divisions leading to a global economic crisis. Healthcare was one of these sectors that was extremely affected by the novel coronavirus. Due to the large number of cases affected every day, hospitals were not able to cope with this huge number. They experienced an insufficient amount of beds, staff members, medical kits, and supplies. Furthermore, front-line physicians and nurses were at risk of becoming infected, which correspondingly decreased their ability to achieve their work appropriately [4]. Accordingly, it is very important to rapidly diagnose COVID-19 accurately to reduce the number of deaths and prevent the collapse of healthcare structures.
The common symptoms of COVID-19 are dry cough, headache, fever, dyspnea, and myalgia; however, in several situations, no signs are obvious or asymptotic [5], [6]. Asymptotic individuals could infect others, increasing the threat to public health. Currently, the real-time reverse transcription–polymerase chain reaction (RT-PCR) test is considered to be the gold method for identifying COVID-19 [7]. Nevertheless, the prolonged time to reach the result of the test, lack of the number of test kits, and strict lab requirements constrain the quick and efficient examination of suspicious individuals [8], [9]. Moreover, in some cases, the RT-PCR test could encounter false-negative rates which means that the patient is experiencing COVID-19 but the result of the test is negative [10], [11], [12]. Consequently, more effective approaches that have a greater ability to produce a fast and accurate result are required. Among these methods is medical imaging especially computed tomography (CT) modality. CT plays an important role in identifying lung-related anomalies. Various studies indicated its significant ability to achieve a more accurate diagnosis compared to RT-PCR tests [13]. Numerous research articles have proven the ability of the CT modality to identify, screen, and diagnose the novel coronavirus [14], [15]. The visual structure of COVID-19 on CT scans enables them to be used for the diagnosis of COVID-19 [16]. However, due to the similarity between the visual appearance among COVID-19 and other types of pneumonia, the identification process of COVID-19 becomes complex [17]. Additionally, radiologists consume a lot of time in performing diagnosis. Thus, the health industry sector is seeking novel tools to visualize and manage the propagation of such a disease.
Artificial intelligence (AI) is a great example of such a tool. Because of the recent advancements and success of AI technology, they were extensively utilized in health and medical domains to analyze medical data. AI approaches were used in the prognosis and diagnosis of many abnormalities such as gastrointestinal diseases [18], heart abnormalities [19], [20], breast cancer [21], [22], eye diseases [23], brain disorders [24], [25], [26]. It has also been extensively used with ambient and assistive living technologies [27], [28]. This success has encouraged researchers to employ AI techniques, especially deep learning (DL) [29] which is the newest branch of machine learning (ML) to automatically identify COVID-19 in suspected cases [30]. They have shown excellent performance in the diagnosis of the novel coronavirus, being an evidence-based medical method [31], [32] that supports clinicians in achieving an accurate diagnosis and preventing drawbacks of manual diagnosis. The most common DL technique used with medical images is the convolution neural network (CNN) [33]. Thus, several authors utilized various architectures of CNNs for COVID-19 diagnosis using CT images.
Some authors used a single view of CT images with individual CNN networks. The authors in [34] constructed a customized CNN model named CTnet-10 for differentiating COVID-19 and non-COVID-19 cases, reaching an accuracy of 82.1%. In another study, Ardakani et al. [35] introduced a system for identifying COVID-19 and non-COVID-19 using ten individual CNNs of different architectures including SqueezNet, ResNet-50, ResNet-101, ResNet-18, VGG-16, VGG-19, GoogleNet, Xception, and MobileNet. Both ResNet-101 and Xception achieved the highest performance. Xception and ResNet-101 models obtained an accuracy of 99.51% and 99.02%, a sensitivity of 100% and 98.04%, AUC of 99.4% and 87.3%, and specificity of 99.02% and 100% respectively. Zhao et al. [36] proposed a transfer learning (TL) scheme and applied it to a new version of ResNet to discriminate between cases of COVID-19 and non-COVID-19, reaching an accuracy of 99.2%. Another study proposed a system called COVID CT-Net which is a self-developed simple CNN to diagnose CT images in COVID-19 and non-COVID-19. The performance metrics attained using the COVID CT-Net are 95.78%, 95.56%, and 96% for accuracy, specificity, and sensitivity respectively. The main limitation of the previous studies is using CNNs independently to perform classification, however, some research articles verified that merging features of numerous CNNs can improve classification results [21], [37], [38], [39]. Also, they all used CT images of a single view to training their models, nevertheless, training models with multiview CT images demonstrated an excellent ability to enhance the efficiency of COVID-19 diagnosis and alleviate the substantial capacity of radiologists for the primary screening of this type of pneumonia [40].
Furthermore, other authors [41] used multi-view CT images to train ResNet-50 CNN to classify images to COVID-19 and normal and reached a maximum area under the curve (AUC) of 96%. Similarly, Wu et al. [40], employed ResNet-50 trained with multiview CT data to discriminate among COVID-19 and other types of pneumonia and reached an accuracy of 76%, a sensitivity of 81.1%, a specificity of 61.5%. These previous two studies employed only one individual CNN to classify images and one of them was based on small datasets. On the other hand, some authors integrated handcrafted features with deep features, for example, Shankar and Perumal [42] proposed a fusion model called FM-HCF-DLF based on the combination of handcrafted features that include local binary patterns and deep learning features of Inception V3 CNN. The highest performance metrics achieved are 94.08% accuracy, 93.61% sensitivity of 93.61%, 94.56% specificity, and 94.85% precision. Likewise, the authors [43] combined several handcrafted features compromising statistical features, grayscale covariance matrix (GLCM), and the discrete wavelet transform (DWT) with deep features of ResNet-18, ShuffleNet, GoogleNet, and AlexNet. They used several SVM classifiers and reached an accuracy of 99%. The main limitation of the previous two studies is the huge dimension of features used to train the classification models which increases the complexity and classification time. Although the authors in [43] utilized PCA to reduce the dimension of the features extracted from each CNN, the dimension of the principal components when combined with all handcrafted features is still large.
Some other authors employed ensemble DL models, for example; Zhou et al. [44] constructed a framework based on ensemble DL models including AlexNet, GoogleNet, and ResNet to classify CT images as normal, cancerous, and COVID-19. They employed majority voting to fuse the prediction of the three CNNs reaching an accuracy of 98.25%. Similarly, the authors [45] used majority voting to combine the predictions of fifteen CNN models after searching for the best combination of CNNs outputs. The maximum accuracy of 85% is attained. However, the authors in Ref. [46] proposed a system that extracts features from four CNNs including ResNet-18, GoogleNet, AlexNet, and ShuffleNet. Subsequently, principal component analysis (PCA) was used to reduce the dimension of the deep features. Next, the principal components selected from each network are fused in a concatenated manner with other principal components of other networks. These fused principal components are fed to an SVM classifier, obtaining an accuracy of 94.7%, a specificity of 93.7% and sensitivity of 95.6%. Alternatively, Biswas et al. [47] used the stacking ensemble method to fuse predictions of VGG-16, ResNet-50, and Xception CNNs to differentiate between COVID-19 and non-COVID-19 reaching an accuracy of 98.79%. Whereas, in [48], the authors proposed two new CNN structures called CovidResNet and CovidDenseNet to distinguish COVID-19 and non-COVID, or COVID-19, non-COVID, and normal. The predictions of these CNNs are merged using the averaging technique. The results obtained reached 93.87% accuracy, 92.49% sensitivity, 99.13% precision, and 97.73% specificity for binary classification. The earlier methods relied only on spatial information to achieve a diagnosis; however, spatial–temporal information could improve the detection of COVID-19 [49].
According to the limitations mentioned earlier, this paper proposes an automated pipeline called CoviWavNet based on multiview CT images (3D CT volumes). It considers not only the spatial information included in the original CT images to perform the classification. But uses multilevel DWT to extract the spectral–temporal information by obtaining the heatmaps of the multilevel DWT of the CT slices and using them to train three CNNs including ResNet-18, ResNet-50, and ResNet-101. This spectral–temporal representation is useful and more helpful [50], [51]. It extracts spectral–temporal features from these three CNNs and fuses them with spatial features extracted from the same CNNs trained with the original multiview CT images. Afterward, the proposed pipeline utilizes a feature selection approach to reduce the huge dimension resulting from fusion. The proposed pipeline investigates whether combining the spatial information of CT images with the spectral–temporal information extracted from the multilevel DWT heatmap images could improve the diagnostic accuracy of COVID-19. The article uses a private 3D CT volume dataset called OMNIAHCOV that was acquired in Egypt, where all available online are acquired from China, Europe and the United States. To our knowledge, it is the first study to explore the use of AI models to diagnose COVID-19 in Egyptian patients. For further validation, the proposed pipeline uses another public benchmark dataset that has been extensively used in the literature, known as SARS-COV-2 CT [52].
The novelty and contribution can be summarized as follows:
-
•
Using multiview CT volumes containing CT images at different views and regions of the lung which is not the common case in the literature that uses CT images of a single view.
-
•
Utilizing the heatmaps of the multilevel DWT to train the deep learning models instead of the original CT images.
-
•
Extracting spectral–temporal features from the CNN trained with the heatmaps of the multilevel DWT and integrating it with spatial features extracted from the CNN learned with the original multiview CT images.
-
•
Reducing the huge dimension of features obtained after fusion through a feature selection procedure.
-
•
Validating the proposed model on a large 3D CT volumes dataset acquired in Egypt, where all available online are from China, Europe, and the United States.
The paper is organized as follows. Section 2 describes the methods, the datasets, and the proposed pipeline. Section 3 explains the setup of the experiments, including the adjusted parameters of the CNNs and the evaluation metrics used. Section 4 presents the results. Section 5 discusses the results. Finally, Section 6 concludes the paper.
2. Methods and materials
2.1. Convolutional neural networks
The convolutional neural network (CNN) is a branch of DL methods that are widely used for resolving classification problems of medical images in the field of health informatics [53], [54]. For this reason, in this paper, three CNN architectures are employed. A CNN’s structure depends on perceptron models. These networks automatically obtain features from the image, unlike the conventional artificial neural network, and therefore recently become a hot research topic, especially in medical image processing [55]. The main advantage of CNNs is directly using images to perform the diagnosis process avoiding the unnecessary steps made in traditional machine learning approaches (such as preprocessing, segmentation, and feature extraction) [56]. They could also reduce the complication of the classification models, by utilizing the whole local and global information of a medical image, and for this reason, they have the capacity for vigorous rotation, translation, and scaling. The main three layers of a CNN are the convolutional, pooling, and fully connected (FC) layers. In the former layers, portions of the image are convolved with a small-size filter. Next, a feature map is formed containing the spatial info regarding the pixel values of the original input image. These feature maps are of high dimension; therefore, the key purpose of the pooling layers is to lower this massive dimension through a downsampling procedure. Finally, the FC layers collect input from the previous layers and produce class scores [57]. In this study, three CNNs depending on the residual neural network (ResNet) are used including the ResNet-18 which includes 18 deep layers, ResNet-50 which is 50 layers deep, and finally ResNet-101 which is 101 layers deep.
ResNet is a CNN architecture based on the residual block proposed by He et al. [58]. The residual block proposes shortcut links within the convolution layers, which help the network to hop some convolution layers at a time. This means that the residual block suggests two choices, it can achieve a set of functions on the input, or it may omit this phase entirely. For this reason, ResNet architecture is believed to be more efficient than other CNNs such as AlexNet and GoogleNet. The composition of such crosscut links guarantees the update of the parameters of the CNN’s and prevents the issue of gradient vanishing produced by the backpropagation process. In addition, these shortcuts facilitate CNN optimization.
2.2. Transfer learning
Transfer learning (TL) is a well-known machine learning technique that utilizes current knowledge to handle problems in distinct, although related fields [59]. It employs a fundamental hypothesis in conventional machine learning to transfer current knowledge in order to resolve similar classification learning problems in another or the same field but having few numbers of labeled training data. TL can be used for parameter transfer, instance transfer, feature transfer, and association relationship transfer. In this paper, the first three methods are employed, where parameter transfer is employed to use the same parameters of pre-trained CNNs, instance transfer to employ the same structure of pre-trained models, and feature transfer which uses the pre-trained CNN as a feature extractor.
2.3. Discrete wavelet transform
Discrete wavelet transform (DWT) is a well-known approach to analyzing medical images. DWT offers spectral–temporal presentation by decomposing a medical scan using a set of orthonormal basis functions. It consists of a collection of transforms each with several wavelet basis functions. At the first level of multilevel DWT, the image is analyzed through passing through low- and high-pass filters, resulting in two groups of coefficients. These groups are the approximate coefficients CA1 and three detail coefficients CD1. The detail coefficients involve the vertical, horizontal, and diagonal coefficients, respectively. The multilevel DWT analysis is attained by convolving the approximation values generated in the prior decomposition level into numerous high and low-pass filters [60], [61].
2.4. Experimental datasets
To access and validate the introduced pipeline, two CT datasets are used. The first dataset is a private data called OMNIAHCOV containing 3D CT volumes, while the second one is a publicly available benchmark dataset called SARS-CoV-2 CT-Scan [52].
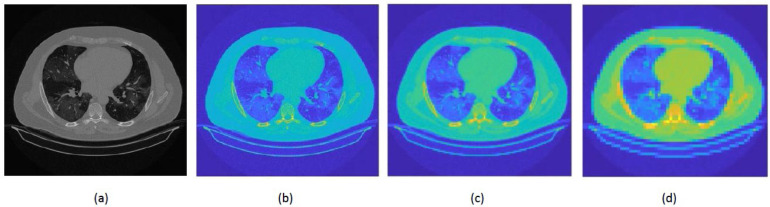
• OMNIAHCOV: The collected dataset contains 5152 normal images and 6012 images of COVID-19. Chest imaging was performed using severe multidetector computed tomography (MDCT) machines. They included the following: (1) SOMATOM Sensation 64, Siemens Medical Systems, Germany, (2) Canon Medical Systems; Toshiba Aquilion 64, USA, and (3) Canon Medical Systems; Toshiba Aquilion CXL/CX 128, USA. The parameters for CT scans were as follows: The slice thickness was 1–1.25 mm, the tube rotation was 0.6–0.9 s, the detector Collimation was 1 mm, the kVp was 120–130, and the mA was 200. All examinations performed did not require intravenous contrast administration. Fig. 1 shows samples of CT scans existing in the dataset. Details regarding the dataset can be found [62].
Fig. 1.
Samples of images available in the OMNIAHCOV dataset.
• SARS-COV-2 CT: is a freely available 2D CT image dataset [52]. The dataset has a total of 2482 CT images, where 1252 of these images are diagnosed as positive CT scans for COVID-19 disease, and the other 1230 are CT scans that were diagnosed as non-COVID-19 cases. The dimension of these CT images changes from 119 × 104 to 416 × 512. Fig. 2 displays samples of CT images existing in the dataset.
Fig. 2.
Samples of images available in SARS-COV-2-CT Scan dataset.
2.5. Proposed CoviWavNet pipeline
The proposed pipeline consists of five steps including image preprocessing, multilevel DWT heatmaps generation, pre-trained ResNets construction and training, feature extraction, integration and selection, and classification steps. In the image pre-processing step, CT slices of the 3D volumes of the OMNIAHCOV dataset are converted from Dicom format to jpg-colored images. The images of both datasets are resized and enhanced. Next, multilevel DWT is applied to these images to display the spatial–temporal information of the images, and then the heatmaps of the DWT levels are generated. Later, three pre-trained CNNs are constructed and trained once using the original CT images and then using the heatmaps of the multilevel DWT images. After that, deep spatial features are extracted from each ResNet trained with the original CT images. Likewise, spectral–temporal deep features are obtained from each ResNet trained with the heatmaps of the multilevel DWT images. Subsequently, deep spatial features and spectral–temporal features extracted from each CNN are combined, and then a feature selection procedure is applied to reduce their huge dimension. Finally, three support vector machine (SVM) classifiers are used to perform diagnosis using the reduced fused set of features. The block diagram of the proposed pipeline is shown in Fig. 3.
Fig. 3.
Proposed CoviWavNet pipeline block diagram.
2.5.1. Image pre-processing
The OMNIAHCOV dataset contains CT slices in Dicom format. These slices are converted to 3D jpg images. The images of both datasets are augmented. This step is done to increase the size of each of the datasets. The augmentation procedure is usually performed to avoid the overfitting that could occur in the CNN training phase [63]. Augmentation also may improve the classification performance of deep learning models [64]. Various augmentation techniques are applied to images of both datasets including scaling (0.85,1.2), shearing (0,50), rotation (−20,20), flipping in the and axes, and random translation in the and directions (−35,35). Next, these images are resized to 227 × 227 × 3 to compromise the input layer size of the ResNets.
2.5.2. Generation of multilevel DWT heatmaps
In this step, 3-levels of DWT are applied to the images. The 3rd level of decomposition is commonly used in several medical applications and is proven to improve classification performance [65], [66]. This study utilized a Haar mother wavelet as the basis function. Haar is chosen because it is an effective approach as indicated in several studies related to the diagnosis of COVID-19 [50], [51], [67]. The approximation coefficients are selected to convert them to heatmaps as the authors of the research article [68] showed that the approximation coefficients obtained from medical data have outstanding performance compared to the detail coefficients. Samples of the heatmaps of the three approximation levels are shown in Fig. 4, Fig. 5 for the OMNIAHCOV and SARS-COV-2-CT Scan datasets respectively.
Fig. 4.
Samples of COVID-19 heatmaps of the approximation coefficients of the three DWT levels of for the OMNIAHCOV dataset; (a) original, (b) 1st DWT level, (c) 2nd DWT level, and (c) 3rd DWT level.
Fig. 5.
Samples of the heatmaps of the approximation coefficients of the three levels of DWT for the SARS-COV-2-CT Scan dataset; (a) original, (b) 1st DWT level, (c) 2nd DWT level, and (c) 3rd DWT level.
2.5.3. Pretrained ResNets construction and training
TL is utilized in this step to adjust the pre-trained ResNets formerly learned with the ImageNet dataset. Subsequently, the number of output layers of the three ResNets is altered to 2, comparable to the number of classes available in each of the two datasets utilized in this study as an alternative to the 1000 classes of ImageNet. Later, some CNN parameters are changed (demonstrated briefly in the experimental setup section), and then ResNets are trained once using the original CT slices and then using the heatmaps images of the approximation coefficients of the three DWT levels. The filters in the ResNets trained with the heatmaps images of the DWT levels are learned from the spectral–temporal domain, instead of the spatial domain like the ResNet learned from the original CT slices. The low-frequency band of the input image encompasses nearly all of the details and the high-frequency band includes noise info. This nonuniformity of spectral power allows the elimination of high frequencies to achieve a minimum loss to the information from the input image [50]. Thus, spectral–temporal representation is better than spatial demonstration. For this reason, heatmaps of DWT levels are used to train CNNs.
2.5.4. Deep feature extraction, integration, and selection
TL is employed again in this step to obtain spatial deep features from the last average pooling layer of the ResNets trained with the original images. Similarly, TL is used to obtain spectral–temporal features from the last average pooling layer of the ResNets being trained with the heatmap images of the three levels of DWT. The dimension of those features after being extracted is 2048 for ResNet-101 and ResNet-50. Whereas for ResNet-18, the number of features is 512. Afterward, the spatial and spectral–temporal deep features extracted from each ResNet are integrated. The result of this integration is three feature sets of sizes 1024, 4096, and 4096 for ResNet-18, ResNet-50, and ResNet-101, respectively. The dimensions of these feature sets are large; therefore, a feature selection (FS) procedure is required to reduce them.
FS is capable of ignoring redundant irrelevant features that exist in the feature space. Furthermore, it speeds up the classification process and lowers the complexity of the training models [69]. Additionally, FS inhibits overfitting that may possibly occur during the phase of training [70], [71]. Minimum redundancy maximum relevance (mRMR) FS process [72] is utilized in this step. The mRMR procedure chooses a subset of features that are highly correlated with the class (relevant) and minimally correlated to each other (redundant). The criterion used to select features is the minimum redundancy and maximum relevance. Redundancy and relevance are measured using mutual information.
2.5.5. Classification
The classification step is in charge of classifying CT images to COVID-19 and non-COVID-19 or COVID-19 and normal cases utilizing three SVM classifiers. SVM is a robust machine learning technique that employs statistical learning theory to achieve the classification procedure. It is utilized to create the finest hyperplane that has the greatest margin of distinction among the two classes of CT data sets [73]. In this paper, linear, cubic, and quadratic kernel functions are selected because they have reached maximum results. Note that for the OMNIAHCOV dataset, data is split into 75% and 25% for training and testing respectively to ensure that the images belonging to the same patient used in the training phase are not used in the testing phase. Whereas, for the SARS-COV-2-CT Scan dataset, 5- fold cross-validation is used to validate the results.
3. Performance evaluation
The performance of CoviWavNet is accomplished by utilizing several measures. The accuracy (Acc), the area under the receiver operating characteristic (ROC) curve (AUC), specificity (SP), sensitivity (SE), F1-score, precision (Prec), negative predicted value (NPV), and Mathew correlation coefficient (MCC) metrics are employed for accessing the proposed model. These measures are computed as shown below (1)–(7).
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
Where; TN is the entire count of non-COVID-19 scans incorrectly diagnosed. TP is the whole count of COVID-19 scans properly identified. FN is the total count of COVID-19 scans improperly categorized as non-COVID-19. FP is the whole count of non-COVID-19 scans incorrectly identified as COVID-19.
4. Experimental setup
The implementation of the proposed CoviWavNet is done using Matlab 2020a on a 64-bit operating system. Intel(R) Core (TM) i7-10750H (10th generation) processor is used with a processing frequency of 2.6 GHz and 1.512 TB hard disk, Hexa-core processor RAM 16 GB of type DDR4. Experiments are performed on NVIDIA GeForce GTX 1660 with a graphics card of 6 GB size.
The hyperparameters of the CNNs are the mini-batch size, i.e. the amount of data involved in the weight changes of each sub-epoch. As demonstrated in [74], in fact, by using larger batch sizes, CNN models’ quality has declined and their ability to generalize has been assessed by its ability to generalize. Large batch sizes tend to converge to sharp minimizers for both training and test tasks. The sharp minima lead to weak generalizations. In contrast, small batch sizes regularly converge with smooth minimizers and generally achieve the best generalization performance. [75]. Consequently, it is adjusted to 10. Learning rates determine the size of steps in each epoch while moving into a least-loss function. Normally, higher learning speeds make the model learn faster, resulting in inefficient final weights. On the contrary, smaller learning rates can enable models to learn additional optimal weights, or global optimal weights but lead to longer training times. Furthermore, excessive learning rates lead to massive weight changes and model performance (For example, models’ loss on training data sets) varies during the training period. Performance fluctuations are the result of weight differences. On the contrary, too low learning rates may never converge or be trapped in an unbalanced resolution. Therefore, in experiments, the learning rate is selected as 0.001, not too small or too high.
The largest number of epochs is selected as 20 because expanding this number did not enhance performance, just extended the time of computation. Validation frequencies are set to 843 and 173 for the OMNIAHCOV and SARS-COV-2-CT-Scan datasets to compute the validation error only one time at the end of each epoch. The three ResNets networks were trained by stochastic gradient descent with a momentum procedure to improve convergence rates and avoid local minimums during convergence. [76], [77], [78]. To avoid overfitting, a certain CNN parameter has been selected, including batch normalization [79], [80].
5. Results
5.1. ResNets classification results
In this section, a comparison among the classification results of the three ResNets trained with the original CT images and the ResNets trained with the heatmaps images of the three approximation DWT levels is conducted and displayed. Table 1 shows the classification accuracy of the three ResNets trained with the original CT images and the heatmaps images of the three approximation (CA1, CA2, CA3) DWT levels for the OMNIAHCOV and SARS-COV-2-CT Scan datasets. For the OMNIAHCOV dataset, the classification accuracy attained using the ResNets trained with the original CT images are 90.7%, 93.81%, and 94.07% for ResNet-18, ResNet-50, and ResNet-101 respectively. Whereas, for the ResNets trained with the heatmap images of the DWT levels, the classification accuracies of the heatmap images of (CA1) are 91.18%, 94.33%, 94.51% for ResNet-18, ResNet-50, and ResNet-101 respectively. The classification accuracies of the heatmap images of (CA2) are 93.45%, 95.64%, 95.35% for ResNet-18, ResNet-50, and ResNet-101 respectively. The classification accuracies of the heatmap images of (CA3) are 94.77%, 96.45%, and 97.66% for ResNet-18, ResNet-50, and ResNet-101 respectively. These accuracies prove that the heatmap images of the DWT levels improve the classification accuracies of the DL models compared to the original CT images.
Table 1.
Accuracy (%) of ResNet CNNs trained with the original CT images and the heatmaps images of the three approximation coefficients of DWT (CA1, CA2, CA3 ) for OMNIAHCOV and SARS-COV-2-CT Scan datasets.
| Model | Original | CA1 | CA2 | CA3 |
|---|---|---|---|---|
| OMNIAHCOV Dataset | ||||
| ResNet-18 | 90.70 | 91.18 | 93.45 | 94.77 |
| ResNet-50 | 93.81 | 94.33 | 95.64 | 96.45 |
| ResNet-101 | 94.07 | 94.51 | 95.35 | 97.66 |
| SARS-COV-2-CT Scan Dataset | ||||
| ResNet-18 | 70.34 | 74.90 | 77.32 | 84.83 |
| ResNet-50 | 76.51 | 78.39 | 82.82 | 86.04 |
| ResNet-101 | 73.42 | 77.99 | 84.56 | 86.85 |
Similarly, for the SARS-COV-2-CT Scan dataset, the ResNets trained with the original CT images attained accuracies of 70.34%, 76.51%, and 73.42% for ResNet-18, ResNet-50, and ResNet-101 respectively. While for those ResNets learned with the heatmap images of the DWT levels, the classification accuracies achieved for (CA1) are 74.9%, 78.39%, 77.99% for ResNet-18, ResNet-50, and ResNet-101 respectively. The classification accuracies of (CA2) are 77.32%, 82.82%, and 84.56% for ResNet-18, ResNet-50, and ResNet-101 respectively. The classification accuracies of (CA2) are 84.83%, 86.04%, 86.85% for ResNet-18, ResNet-50, and ResNet-101 respectively. Likewise, these results confirm that using the heatmap images of the three levels of DWT is better than using the original CT images, as they enhance the classification accuracies of the three ResNets compared to the original CT images. It can be seen from Table 1 that the heatmaps of the third level of the DWT have the highest performance for the three ResNets.
5.2. Classification results of deep feature extraction
Since the results of Table 1 indicated that the heatmap images of the third level of the DWT achieved the maximum accuracy for the three ResNets, the deep spectral–temporal features are extracted from only the third DWT level of the three ResNets. The classification results of the three SVM classifiers learned with these deep spectral–temporal features are discussed in Table 2. The results in Table 2 show that the spectral–temporal features extracted for the three ResNets trained with the heatmap images of the third level of the DWT (CA3) have enhanced the classification performance of CoviWavNet compared to the end-to-end DL classification results displayed in Table 1. This is because, for the OMNIAHCOV dataset, the L-SVM, Q-SVM, and C-SVM classifiers trained with the spectral–temporal features of ResNet-18 attained an accuracy of 95.32%, 95.73%, and 96.18% respectively. These accuracies are greater than the 94.77% accuracy obtained by ResNet-18 trained with CA3 images as illustrated in Table 1. Similarly, an accuracy of 98.24%, 98.24%, and 98.57% is achieved using the L-SVM, Q-SVM, and C-SVM classifiers trained with the spectral–temporal features of ResNet-50. These accuracies are better than the 96.45% reached utilizing ResNet-50 learned with CA3 images as shown in Table 1. Likewise, the same three classifiers trained with the spectral–temporal features of ResNet-101 obtained an accuracy of 98.74%, 98.93%, and 98.76% which is higher than the 97.66% attained with the ResNet-101 learned with CA3 images as shown in Table 1. Furthermore, for the SARS-COV-2-CT Scan dataset, the L-SVM, Q-SVM, and C-SVM classifiers learned with the spectral–temporal features of ResNet-18 reached an accuracy of 95.60%, 96.48%, and 96.72%, respectively. Furthermore, for ResNet-50 and ResNet-101, the L-SVM, Q-SVM, and C-SVM classifiers constructed using the spectral–temporal features reached an accuracy of (97.98%, 97.82%), (98.86%,98.72%), and (98.96%,98.90%) respectively. These accuracies are greater than 70.34%, 76.51% and 73.42% achieved using ResNet-18, ResNet-50 and ResNet-101 CNNs as shown in Table 1. The confusion matrices for the three SVM classifiers trained with spectral–temporal features of ResNet-101 constructed with the CA3 heatmap images for the OMNIAHCOV and SARS-COV-2-CT Scan datasets are shown in Fig. 6, Fig. 7 respectively.
Table 2.
Accuracy (%) of the three SVM classifiers trained with the heatmap images of the third approximation coefficients of DWT extracted from the three ResNets for the OMNIAHCOV and SARS-COV-2-CT Scan datasets.
| Classifier | L-SVM | Q-SVM | C-SVM |
|---|---|---|---|
| Model | |||
| OMNIAHCOV Dataset | |||
| ResNet-18 | 95.32 (0.05) | 95.73 (0.04) | 96.18 (0) |
| ResNet-50 | 98.24 (0.01) | 98.24 (0.05) | 98.57 (0.01) |
| ResNet-101 | 98.74 (0) | 98.93 (0) | 98.76 (0.02) |
| SARS-COV-2-CT-Scan Dataset | |||
| ResNet-18 | 95.60 (0.14) | 96.48 (0.21) | 96.72 (0.17) |
| ResNet-50 | 97.98 (0.07) | 98.86 (0.08) | 98.96 (0.12) |
| ResNet-101 | 97.82 (0.09) | 98.72 (0.07) | 98.90 (0.06) |
Fig. 6.
Confusion matrix for the three SVM classifiers trained with spectral–temporal features of ResNet-101 constructed with the third DWT approximation heatmap images of the OMNIAHCOV dataset, (a) C-SVM, (b) L-SVM, and (c) Q-SVM.
Fig. 7.
Confusion matrix for the three SVM classifiers trained with spectral–temporal features of ResNet-101 constructed with the third DWTapproximation heatmap images for the SARS-COV-2-CT dataset.
5.3. Classification results of deep feature integration and selection
Deep spatial features extracted from each ResNet trained with the original CT slices are integrated with deep spatial–spectral–temporal features of each ResNet learned with the heatmap images of the third level of the DWT (CA3). These deep-based features are reduced using the mRMR FS algorithm. This section illustrates the classification results after the feature integration and selection. Table 3 shows the classification accuracies of the three SVM classifiers trained with the reduced number of integrated spatial–spectral–temporal features compared to the deep spatial features for each ResNet for the OMNIAHCOV dataset. Fig. 8 shows the dimension of the deep spatial features extracted from each ResNet and used to train the three SVM classifier compared to the dimension of the integrated and reduced spatial–spectral–temporal features.
Table 3.
Classification accuracies (%) of the three SVM classifiers trained with the reduced number of integrated spatial–spectral–temporal features compared to the deep spatial features for each ResNet for the OMNIAHCOV dataset.
| Deep Features | L-SVM | Q-SVM | C-SVM |
|---|---|---|---|
| ResNet-18 | |||
| Spatial | 92.7 (0.05) | 92.63 (0.02) | 92.30 (0.07) |
| Spatial-Spectral-Temporal | 98.19 (0.10) | 98.21 (0.02) | 97.98 (0.15) |
| ResNet-50 | |||
| Spatial | 94.17 (0.02) | 94.13 (0.02) | 94.13 (0) |
| Spatial-Spectral-Temporal | 99.23 (0.11) | 99.32 (0.06) | 99.38 (0) |
| ResNet-101 | |||
| Spatial | 95.36 (0) | 95.41 (0.06) | 95.35 (0.04) |
| Spatial-Spectral-Temporal | 99.05 (0) | 98.97 (0.02) | 98.97 (0.02) |
Fig. 8.
Number of deep spatial features extracted from each ResNet and used to train the three SVM classifier compared to the dimension of the integrated and reduced spatial–spectral–temporal features for the OMNIVOV dataset.
As shown in Table 3, for the OMNIAHCOV dataset, using spatial–spectral–temporal features to train the SVM classifiers is better than spatial features alone for the three ResNets. This is clear as the accuracy accomplished using the selected spatial–spectral–temporal features extracted from the ResNet-18 is 98.19%, 98.21%, and 97.98% for the L-SVM, Q-SVM, and C-SVM, respectively. These accuracies are greater than that obtained by the same classifiers trained with the spatial features of ResNet-18 (92.70%,92.63%, and 92.30%). Likewise, the accuracy obtained using the reduced number of the spatial–spectral–temporal features extracted from ResNet-50 is 99.32%, 99.32%, 99.38% for L-SVM, Q-SVM, and C-SVM respectively. Again, these accuracies are higher than the 94.17%, 94.13%, and 94.13% obtained using the same classifiers trained with the spatial features of ResNet-50. Furthermore, the reduced number of integrated spatial–spectral–temporal of ResNet-101 achieved an accuracy of 99.05%, 98.97%, and 98.97% using the L-SVM, Q-SVM, and C-SVM respectively. Such accuracies are better than the 95.36%, 95.41%, and 95.35% achieved using the same classifiers trained with the spatial features of ResNet-101.
It is obvious from Fig. 8 that for the OMNIAHCOV dataset, the mRMR FS approach has successfully reduced the number of integrated spatial–spectral–temporal features used to train the three SVMs. For ResNet-18, the number of selected spatial–spectral features are 20, 21, and 20 for L-SVM, Q-SVM, and C-SVM, respectively. Whereas for ResNet-50, the number of selected integrated spatial–spectral features are 200, 300, and 300 for the same classifiers. On the other hand, for ResNet-101, the number of selected integrated spatial–spectral features are 400, 400, and 400 for L-SVM, Q-SVM, and C-SVM respectively. These reduced feature sets achieved higher accuracy using the same classifiers trained with the spatial features extracted from ResNet-18, ResNet-50, and ResNet-101 having dimensions of 512, 2048, and 2048 features respectively.
Table 4 proves as well that using spatial–spectral–temporal features to learn the SVM classifiers is superior to spatial features alone for the three ResNets. The table shows that for the SARS-COV-2-CT Scan dataset, the SVM classifiers constructed with the reduced number of integrated spatial–spectral–temporal features extracted from ResNet-18 achieved an accuracy of 96.60%,97.56%, and 97.60% for the L-SVM, Q-SVM, and C-SVM classifiers, respectively. Such accuracies are more than that attained using the same classifier trained with spatial features only (95.00%, 96.42%, and 96.72%). Similarly, an accuracy of 99.42%, 99.48%, and 99.62% is obtained with the SVM classifiers learned with the reduced number of integrated spatial–spectral–temporal features mined from ResNet-50. Furthermore, the reduced number of spatial–spectral–temporal features extracted from ResNet-101 enhanced the accuracy of L-SVM (98.46%), Q-SVM (99.02%) and C-SVM (99.34%) compared to 97.4%, 98.2% and 98.3% attained with the same classifier trained with the spatial features of ResNet-101.
Table 4.
Classification accuracies (%) of the three SVM classifiers trained with the reduced number of integrated spatial–spectral–temporal features compared to the deep spatial features for each ResNet for the SARS-COV-2-CT Scan dataset.
| Deep Features | L-SVM | Q-SVM | C-SVM |
|---|---|---|---|
| ResNet-18 | |||
| Spatial | 95.00 (0.13) | 96.42 (0.15) | 96.72 (0.16) |
| Spatial-Spectral-Temporal | 96.60 (0.09) | 97.56 (0.21) | 97.62 (0.09) |
| ResNet-50 | |||
| Spatial | 98.34 (0.10) | 98.78 (0.07) | 98.94 (0.10) |
| Spatial-Spectral-Temporal | 99.42 (0.15) | 99.48 (0.07) | 99.62 (0.04) |
| ResNet-101 | |||
| Spatial | 97.46 (0.01) | 98.30 (0.14) | 98.50 (0.11) |
| Spatial-Spectral-Temporal | 98.46 (0.10) | 99.02 (0.04) | 99.34 (0.08) |
On the other hand, the mRMR FS approach used in CoviWavNet has successfully reduced the dimension of fused spatial–spectral–temporal features of the three ResNets for the SARS-COV-2-CT Scan dataset as shown in Fig. 9. This is clear as for ResNet-18, the size of features after mRMR has decreased to 400, 400, and 400 for L-SVM, Q-SVM, and C-SVM respectively. Also, for ResNet-50, the dimension of features after mRMR has diminished to 1500,1500, and 900 for L-SVM, Q-SVM, and C-SVM respectively. Likewise, for the ResNet-101, the number of features after mRMR has reduced to 1200, 900, and 1300 for L-SVM, Q-SVM, and C-SVM respectively. These reduced sets of features are lower than the spatial features of ResNet-18 (512), ResNet-50 (2048), and ResNet-101 (2048) respectively.
Fig. 9.
Number of deep spatial features extracted from each ResNet and used to train the three SVM classifier compared to the dimension of the integrated and reduced spatial–spectral–temporal features for the SARS-COV-2-CT-Scan dataset.
Performance metrics including sensitivity, specificity, precision, F1-scores, NPV, and MCC are calculated for the L-SVM classifier trained with the reduced set of fused spatial–spectral–temporal features of the three ResNets and shown in Table 5. The table shows that for the OMNIAHCOV dataset, the L-SVM classifier learned with the reduced set of fused spatial–spectral–temporal features attained a sensitivity of 97.96%, 99.27%, 98.92%, specificity of 98.54%, 99.19%, 99.26%, F1-score 98.39%, 99.32%, 99.16% for ResNet-18, ResNet-50, and ResNet-101 respectively. Furthermore, the precision obtained is 98.84%, 99.35%, 99.40%, NPV is 97.44%, 99.1%, 98.66%, and MCC is 96.38%, 98.46%, 98.12% for the L-SVM classifier learned with the reduced set of fused spatial–spectral–temporal features extracted from ResNet-18, ResNet-50, and ResNet-101 respectively.
Table 5.
Performance metrics (%) of the L-SVM classifier trained with the reduced number of integrated spatial–spectral–temporal features for OMINICOV and SARS-COV-2-CT Scan datasets.
| Spatial-Spectral-Temporal CNN Model | SE | SP | F1-score | Prec | NPV | MCC |
|---|---|---|---|---|---|---|
| OMNIAHCOV Dataset | ||||||
| ResNet-18 | 97.96 (0.19) | 98.54 (0.06) | 98.39 (0.07) | 98.84 (0.06) | 97.44 (0.25) | 96.38 (0.16) |
| ResNet-50 | 99.27 (0) |
99.19 (0.28) | 99.32 (0.12) | 99.35 (0.23) | 99.10 (0) | 98.46 (0.26) |
| ResNet-101 | 98.92 (0.03) | 99.26 (0) |
99.16 (0.02) | 99.40 (0) |
98.66 (0.04) | 98.12 (0.03) |
| SARS-COV-2-CT-Scan Dataset | ||||||
| ResNet-18 | 97.72 (0.21) | 95.48 (0.10) | 96.59 (0.05) | 95.46 (0.11) | 97.77 (0.24) | 93.24 (0.12) |
| ResNet-50 | 99.37 (0.14) | 99.58 (0.16) | 99.39 (0.15) | 99.60 (0.16) | 99.17 (0.15) | 98.77 (0.30) |
| ResNet-101 | 98.44 (0.06) | 98.50 (0.22) | 98.48 (0.10) | 98.53 (0.21) | 98.40 (0.06) | 96.94 (0.21) |
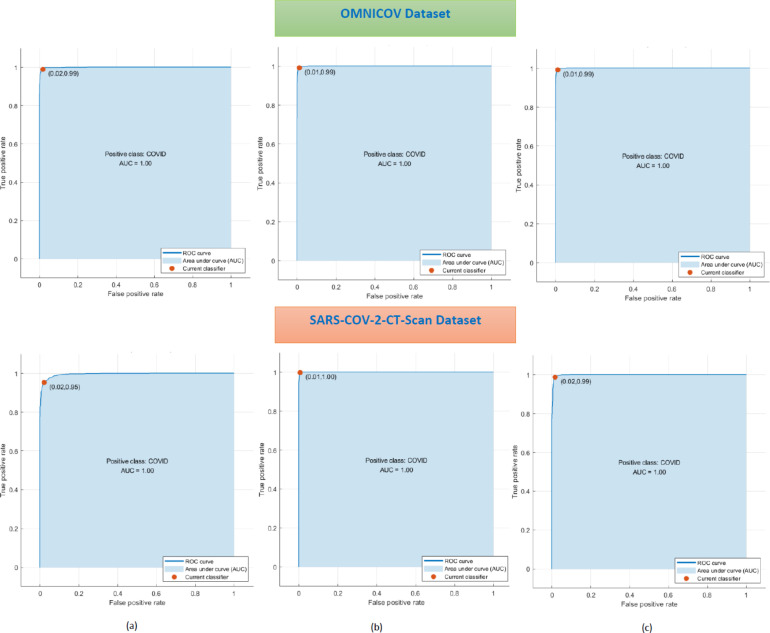
On the other hand, for the SARS-COV-2-CT Scan dataset, the L-SVM classifier learned with the same features reached a sensitivity of 97.72%, 99.37%, 98.44%, specificity of 95.48%, 99.58%, 98.50%, and F1-score of 96.59%, 99.39%, 98.48% for ResNet-18, ResNet-50, and ResNet-101 respectively. While, the precision achieved is 95.46%, 99.60%, 98.53%, NPV is 97.77%, 99.17%, 98.40%, and MCC is 93.24%, 98.77%, 96.94%. The ROC curves for the L-SVM classifier trained with the reduced integrated spatial–spectral–temporal features of the three ResNets for the OMNIAHCOV and SARS-COV-2-CT Scan datasets are shown in Fig. 10. It can be seen from Fig. 10 that the AUCs for L-SVM trained with the reduced set of fused spatial–spectral–temporal features of the three ResNets are 1 for both datasets.
Fig. 10.
ROC curves for the L-SVM classifier trained with the reduced integrated spatial–spectral–temporal features of (a) ResNet-18, (b) ResNet-50, (c) ResNet-101 for the OMNIAHCOV and SARS-COV-2-CT-Scan datasets.
To evaluate and verify the statistical significance of ML classification performance, the one-way variance analysis (ANOVA) test is performed to classifier results after a repeated 5-fold cross-validation procedure. This analysis is applied to the accuracy of the classifiers after the FS process. The ANOVA test results are revealed in Table 6, Table 7 for the OMNIAHCOV and SARS-COV-2-CT-Scan datasets. As observed from Table 6, Table 7, the p-values reached using this test are lesser than , where for all ResNets of both datasets. Thus, statistical differences in classification accuracies of the classifiers can be concluded.
Table 6.
One-way analysis of variance test details for classifiers after FS using the OMNIAHCOV data set.
| Source of Variation | SS | df | MS | F | p Value |
|---|---|---|---|---|---|
| ResNet-18 | |||||
| Columns | 0.166 | 2 | 0.083 | 6.03 | 0.0154 |
| Error | 0.165 | 12 | 0.0137 | ||
| Total | 0.332 | 14 | |||
| ResNet-50 | |||||
| Columns | 0.006 | 2 | 0.03 | 4.15 | 0.0427 |
| Error | 0.0868 | 12 | 0.007 | ||
| Total | 0.147 | 14 | |||
| ResNet-101 | |||||
| Columns | 0.021 | 2 | 0.011 | 37.49 | 0.001 |
| Error | 0.003 | 12 | 0.0003 | ||
| Total | 0.025c | 14 | |||
Table 7.
One-way analysis of variance test details for classifiers after FS using the SARS-COV-2-CT-Scan dataset.
| Source of Variation | SS | df | MS | F | p Value |
|---|---|---|---|---|---|
| ResNet-18 | |||||
| Columns | 3.276 | 2 | 1.638 | 65.52 | 0.001 |
| Error | 0.3 | 12 | 0.025 | ||
| Total | 3.576 | 14 | |||
| ResNet-50 | |||||
| Columns | 0.1053 | 2 | 0.053 | 4.39 | 0.0371 |
| Error | 0.144 | 12 | 0.012 | ||
| Total | 0.2493 | 14 | |||
| ResNet-101 | |||||
| Columns | 1.984 | 2 | 0.992 | 129.39 | 0.001 |
| Error | 0.092 | 12 | 0.0077 | ||
| Total | 2.076 | 14 | |||
5.4. Comparison with other related studies
To verify the competitiveness of CoviWavNet, its performance is first compared with methods based on 3D Chest CT datasets comparable to OMNIAHCOV dataset. The results of this comparison are shown in Table 8. Moreover, the results of CoviWavNet are compared with relevant studies based on SARS-COV-2-CT- Scan dataset; a benchmark CT dataset for COVID-19 diagnosis. The results of this comparison are shown in Table 9. The results demonstrated in Table 8 show the competitive ability of CoviWavNet compared to other methods based on other 3D chest CT datasets. This is because the accuracy, sensitivity, specificity, and AUC are 0.9923, 0.9927, 0.9919, and 1. These performance measures are superior to the accuracy (0.76), sensitivity (0.811), specificity (0.615), and AUC (0.819) achieved in the study conducted in [40] that used ResNet-50. Furthermore, the performance of CoviWavNet is greater than that of the study [41] which achieved accuracy (0.96, 0.93), sensitivity (1,1), specificity (0.9,0.92) and AUC (0.9,0.97) using ResNet-50 and ResNet-101 respectively with voting algorithms. Moreover, the performance CoviWavNet is higher than the accuracy (0.9752), sensitivity (0.9799), and specificity (0.9749). The main reason for CoviWavNet’s outstanding performance is fusing spatial and spectral–temporal information from CT images and the heatmaps of DWT images, whereas other studies depend only on spatial information of the original CT slices only.
Table 8.
Comparison of the results attained with CoviWavNet using OMNIAHCOV dataset and those of the related studies based on relevant 3D datasets for binary classification.
| Article | Dataset | # Images | Method | Acc | SE | SP | AUC |
|---|---|---|---|---|---|---|---|
| [40] | Multicenter | Non-COVID 254 COVID-19 365 |
ResNet-50 | 0.7600 | 0.8110 | 0.615 | 0.8190 |
| [41] | Mosmed-1110 | Normal 65 COVID-19 46 |
ResNet-50+MVa | 0.9600 | 1.000 | 0.96 | 0.9000 |
| [41] | CCAP | Normal 65 COVID-19 46 |
ResNet-101+MPb | 0.9300 | 1.000 | 0.92 | 0.9700 |
| [81] | COVID-CTset | Normal 48,260 COVID-19 15,589 |
ResNet-50 | 0.9752 | 0.9799 | 0.9749 | – |
| Proposed | OMNIAHCOV | Normal 5152 COVID-19 6012 |
ResNet-50 trained with DWT heatmaps and CT images + L-SVM | 0.9923 | 0.9927 | 0.9919 | 1.000 |
MV is the majority vote ensemble method.
MP maximum probability ensemble method.
On the other hand, Table 7 shows the remarkable performance of CoviWavNet. This is because it attained an accuracy of 99.62%, a sensitivity of 99.54%, a specificity of 99.69%, an F1-score of 99.62%, and a precision of 99.70%, which are higher than all other methods shown in Table 7. Soares et al. [52] achieved a lower accuracy of 97.38%, an F1 score of 97.31%, a precision of 99%, and a sensitivity of 95.53% using an explainable deep learning model. Whereas, the authors of [82] introduced a Bi-LSTM (bidirectional long–short-term memory) combined with a mixture density network (DBM) model and obtained an accuracy of 98.37%, a sensitivity of 98.87%, a precision of 98.74%, and an F1 score of 98.14%. Also, the authors in [48] obtained a lower accuracy of 86.88%, sensitivity 87.41%, specificity of 85.92%, F1-score of 89.52%, and precision of 91.76% using a model called CoviDenseNet based on TL and DenseNet. Furthermore, the authors in [52] proposed a customized CNN with a few layers and attained an accuracy, sensitivity, and specificity of 95.78%, 96%, and 95.56%. While the study [83] constructed a customized CNN model and utilized gradient weighted class activation mapping to envisage and map class activations. The model of [83] reached a 95% accuracy, 96% sensitivity, 95% F1-score, and 95% precision, which are lower than those of CoviWavNet. The main limitation of studies [52], [83] is not using pre-trained models, where the authors instead constructed a model from scratch and finetuned all of its parameters, which is a complex task. Besides, the two studies [43], [84] attained an accuracy of 99%, a sensitivity of 99%, and an F1-score of 99%. Although the study [43] tried to lower the huge dimension of the features extracted from each of the 4 CNNs used in their model with PCA, they still have a large number of features produced after fusing the deep features with other hand-crafted features. Also, the study [84] suffered from the high computational load generated by the genetic algorithm to select features extracted from the CNN. Moreover, the authors of [85] combine the prediction of three CNNs using fuzzy ranking to achieve accuracy, sensitivity, specificity, F1 score, precision of 98.93%, 99.08%, 99%, 98.93%, and 98.93% which is lower than the performance of CoviWavNet. The main reason for all of the above studies to have lower performance than CoviWavNet is that they depend only on the spatial or temporal information extracted from the original CT while CoviWavNet utilizes the spatial–spectral–temporal information.
Table 9.
Comparison of the results (%) attained with CoviWavNet and those of the related studies based on the SARS-COV-2-CT-Scan dataset.
| Article | Method | Acc | SE | SP | F1-score | Prec |
|---|---|---|---|---|---|---|
| [83] | Customized CNN | 95.00 | 96.00 | – | 95.00 | 95.00 |
| [48] | CoviDenseNet | 86.88 | 87.41 | 85.92 | 89.53 | 91.76 |
| [52] | x-DNN3 | 97.38 | 95.53 | – | 97.31 | 99.00 |
| [86] | Customized Simple CNN | 95.78 | 96.00 | 95.56 | – | – |
| [82] | Bi-LSTM | 98.37 | 98.87 | – | 98.14 | 98.74 |
| [47] | VGG-16+ResNet-50+Xception+Majority voting | 98.79 | 98.79 | 98.79 | 98.79 | 98.79 |
| [85] | Fuzzy Ranking + VGG-11, ResNet-50-2, and Inception v3 |
98.93 | 99.08 | 99.00 | 98.93 | 98.93 |
| [84] | Customized CNN+Genetic Algorithm+XBoost | 99.00 | 99.00 | – | 99.00 | 99.10 |
| [43] | ResNet18+ShuffleNet+AlexNet+ GoogleNet+DWT+GLCM+ Statistical features+PCA+SVM |
99.00 | 99.00 | 99.00 | 99.00 | 99.00 |
| Proposed | ResNet-50 trained with DWT heatmaps and CT images + C-SVM | 99.62 | 99.54 | 99.69 | 99.62 | 99.70 |
x-DNN is an explainable deep neural network.
6. Discussion
The paper proposed a pipeline for the automatic diagnosis of COVID-19 called CoviWavNet. The proposed pipeline is based on multi-view 3D volumes, not a single view. It used a dataset collected in Egypt based on 3D CT volumes called OMNIAHCOV. In order to validate the performance of CoviWavNet, it also used a benchmark CT dataset. First, CoviWavNet used 3-levels of DWT to analyze CT slices and then converted the approximation coefficients to heatmaps and used them to train three ResNets. These heatmap images of the three DWT levels represent the spectral–temporal information included in the images. CoviWavNet compared the classification accuracy of the three ResNet trained with the heatmap of the three DWT levels with the same ResNets trained with the original CT images. The results of this comparison are shown in Table 1. Table 1 revealed that the three ResNets trained with the heatmaps images of the three DWT levels performed better than ResNets trained with the original CT images. This proves that spectral–temporal information is superior to spatial information during ResNet training.
Table 1 indicated that the ResNets trained with the heatmaps of the third approximation coefficient level have the greatest performance among all other levels. Therefore, CoviWavNet extracted deep spectral–temporal features from each ResNet by TL and used them to train three SVM classifiers. Table 2 showed that spectral–temporal features mined with TL from the three ResNets have superior performance compared to end-to-end DL classification results shown in Table 1. Afterward, the spectral–temporal features mined from each CNN were fused with the spatial features extracted from the same ResNet trained with the original CT images. A reduction step was performed using the mRMR FS method to reduce the huge dimension of features after the fusion step. The results of the extraction, integration, and reduction of features are shown in Table 3, Table 4 for the OMNIAHCOV and SARS-COV-2CT-Scan datasets. These results were compared with those obtained using spatial features extracted from the same ResNet trained with the original CT images and shown also in Table 3, Table 4 for the OMNIAHCOV and SARS-COV-2CT-Scan datasets. Table 3, Table 4 verified that using spatial–spectral–temporal information is better than using only spatial information for diagnosing COVID-19. Furthermore, Fig. 8, Fig. 9 proved the success of the mRMR FS method in reducing the number of fused spatial–spectral–temporal while maintaining superior performance in terms of number of features and accuracy compared to using only deep spatial features extracted from the original CT images.
Some performance metrics are computed for the performance of CoviWavNet to further evaluate its performance and were displayed in Table 5. As discussed in [87], [88], [89], [90], for a medical diagnostic system to be reliable, precision and specificity must exceed 95%, while the sensitivity should increase by 80%. According to the results shown in Table 5 for both datasets, both specificity and precision attained using CoviWavNet are greater than 95% for the OMNIAHCOV and SARS-COV-2CT-Scan datasets. Furthermore, the sensitivity attained with CoviWavNet is more than 80% for the OMNIAHCOV and SARS-COV-2CT-Scan datasets. These results confirm that CoviWavNet is a reliable pipeline that can be used to assist the radiologist in the automatic diagnosis of COVID-19 while avoiding the challenges of manual diagnosis.
Also, the performance of CoviWavNet was compared with methods based on datasets similar to OMNIAHCOV, and the results of this comparison are shown in Table 8. The results demonstrated in Table 8 verified the outstanding performance of CoviWavNet compared to other methods based on comparable datasets. Furthermore, to further validate the performance of CoviWavNet, its performance was compared with relevant studies based on the benchmark dataset called SARS-COV-2-CT-Scan. This comparison was illustrated in Table 9 and the results of this table proved the superior performance of CoviWavNet compared to other studies based on either spatial or temporal information. The results confirmed that utilizing spatial–spectral–temporal information is better than using either spatial or temporal information alone.
Finally, the performance of CoviWavNet is compared with baseline DL models. The result of this comparison is shown in Fig. 11. It can be noted from Fig. 11 that the accuracy of CoviWavNet is greater than that achieved by other baseline DL models including DenseNet-201, MobileNet, Shuffle, Squeeze, Inception, ResNet-18, ResNet-50, ResNet-101, DarkNet-19, and DarkNet-53 for the OMNIAHCOV and SARS-COV-2-CT-Scan datasets. This proves the superiority of CoviWavNet over other baseline DL models.
Fig. 11.
Performance comparison between CoviWavNet and baseline DL networks for the OMNIAHCOV and SARS-COV-CT-SCAN datasets.
7. Conclusion
The pandemic caused by the novel coronavirus has overwhelmed the world and has led to considerable losses and challenges globally. Additionally, a huge deal of matter has evolved because of the advent of COVID-19 variants. With the rapid spread of COVID-19 worldwide, it is very difficult for radiologists to perform a rapid and accurate diagnosis of the novel coronavirus under the enormous workload. AI technology; in specific DL techniques have the potential to help radiologists in the primary screening of COVID-19. Such techniques could reduce the excess workload of radiologists, improve diagnostic accuracy, and produce a fast diagnosis. Therefore, this article proposed a pipeline called CoviWavNet based on DL techniques for the diagnosis of COVID-19. This study used a large CT dataset acquired in Egypt called OMNIAHCOV. This dataset consists of 3D CT volumes of multiview slices, while most of the methods in the literature are based on single-view 2D images. To further validate the capacity of CoviWavNet, a publicly available benchmark dataset was also utilized called the SARS-COV-2-CT-Scan dataset. First, CoviWavNet analyzed CT slices with three levels of DWT and transformed approximation coefficients into heatmap images. Then, it used these images to train three ResNets. The results proved that using the heatmap images of the approximation coefficients of DWT is better than using the original CT slices. Second, CoviWavNet extracted deep spectral–temporal features from the three ResNets and used them as input to three SVM classifiers. CoviWavNet verified that deep spectral–temporal features are superior to deep spatial features obtained from the ResNets trained with the original CT images. Finally, CoviWavNet fused spatial and spectral–temporal features and reduced them using the mRMR FS algorithm and demonstrated that the reduced set of spatial–spectral–temporal features had enhanced the diagnostic accuracy. The proposed pipeline achieved promising performance, reaching a final accuracy of 99.23% and 99.62% for the OMNIAHCOV and SARS-COV-2-CT-Scan datasets, respectively, which are superior to related studies. The results have demonstrated that mRMR FS method provided a smaller feature set compared to the deep features extracted from each ResNet individually and achieved great performance in terms of the metrics utilized. Therefore, CoviWavNet could be used as an element of a computer-aided diagnostic system and may provide a fast and automated tool to assist radiologists in COVID-19 diagnosis. This study did not consider differentiating COVID-19 from other types of pneumonia which will be addressed in future work. Furthermore, more feature selection algorithms and deep learning techniques will be investigated.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., et al. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Islam M.M., Karray F., Alhajj R., Zeng J. A review on deep learning techniques for the diagnosis of novel coronavirus (covid-19) IEEE Access. 2021;9:30551–30572. doi: 10.1109/ACCESS.2021.3058537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leite H., Lindsay C., Kumar M. COVID-19 outbreak: Implications on healthcare operations. TQM J. 2020 [Google Scholar]
- 5.Siordia JA., Jr. Epidemiology and clinical features of COVID-19: A review of current literature. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishiura H., Kobayashi T., Miyama T., Suzuki A., Jung S., Hayashi K., et al. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19) Int. J. Infect. Dis. 2020;94:154. doi: 10.1016/j.ijid.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rani D., Bajaj H., Singh R. SARS-COV-2 (COVID-19) and role of real time reverse transcription polymerase chain reaction (RT-PCR) in its diagnosis. Res. J. Pharm. Technol. 2021;14:3437–3440. [Google Scholar]
- 8.Wang X., Tan L., Wang X., Liu W., Lu Y., Cheng L., et al. Comparison of nasopharyngeal and oropharyngeal swabs for SARS-CoV-2 detection in 353 patients received tests with both specimens simultaneously. Int. J. Infect. Dis. 2020;94:107–109. doi: 10.1016/j.ijid.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Attallah O. ECG-BiCoNet: An ECG-based pipeline for COVID-19 diagnosis using Bi-layers of deep features integration. Comput. Biol. Med. 2022 doi: 10.1016/j.compbiomed.2022.105210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296:E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J., Wu R., Huang H., Zheng W., Ren X., Wu N., et al. Computed tomographic imaging of 3 patients with coronavirus disease 2019 pneumonia with negative virus real-time reverse-transcription polymerase chain reaction test. Clin. Infect. Dis. 2020;71:2–850. doi: 10.1093/cid/ciaa207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Attallah O. An intelligent ECG-based tool for diagnosing COVID-19 via ensemble deep learning techniques. Biosensors. 2022;12:299. doi: 10.3390/bios12050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rousan L.A., Elobeid E., Karrar M., Khader Y. Chest x-ray findings and temporal lung changes in patients with COVID-19 pneumonia. BMC Pulm. Med. 2020;20:1–9. doi: 10.1186/s12890-020-01286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hani C., Trieu N.H., Saab I., Dangeard S., Bennani S., Chassagnon G., et al. COVID-19 pneumonia: a review of typical CT findings and differential diagnosis. Diagn. Interv. Imaging. 2020;101:263–268. doi: 10.1016/j.diii.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attallah O. A computer-aided diagnostic framework for coronavirus diagnosis using texture-based radiomics images. Digit. Health. 2022;8 doi: 10.1177/20552076221092543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi H., Han X., Zheng C. Evolution of CT manifestations in a patient recovered from 2019 novel coronavirus (2019-nCoV) pneumonia in Wuhan, China. Radiology. 2020;295:20. doi: 10.1148/radiol.2020200269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai H.X., Hsieh B., Xiong Z., Halsey K., Choi J.W., Tran T.M.L., et al. Performance of radiologists in differentiating COVID-19 from non-COVID-19 viral pneumonia at chest CT. Radiology. 2020;296:E46–54. doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Attallah O., Sharkas M. GASTRO-CADx: a three stages framework for diagnosing gastrointestinal diseases. PeerJ Comput. Sci. 2021;7 doi: 10.7717/peerj-cs.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karthikesalingam A., Attallah O., Ma X., Bahia S.S., Thompson L., Vidal-Diez A., et al. An artificial neural network stratifies the risks of reintervention and mortality after endovascular aneurysm repair; a retrospective observational study. PLoS One. 2015;10 doi: 10.1371/journal.pone.0129024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Attallah O., Ma X. Bayesian neural network approach for determining the risk of re-intervention after endovascular aortic aneurysm repair. Proc. Instit. Mech. Eng. Part H: J. Eng. Med. 2014;228:857–866. doi: 10.1177/0954411914549980. [DOI] [PubMed] [Google Scholar]
- 21.Attallah O., Anwar F., Ghanem N.M., Ismail M.A. Histo-CADx: duo cascaded fusion stages for breast cancer diagnosis from histopathological images. PeerJ Comput. Sci. 2021;7 doi: 10.7717/peerj-cs.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anwar F., Attallah O., Ghanem N., Ismail M.A. Automatic breast cancer classification from histopathological images. 2019 International Conference on Advances in the Emerging Computing Technologies; AECT; IEEE; 2020. pp. 1–6. [Google Scholar]
- 23.Attallah O. DIAROP: Automated deep learning-based diagnostic tool for retinopathy of prematurity. Diagnostics. 2021;11:2034. doi: 10.3390/diagnostics11112034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Attallah O. MB-AI-His: Histopathological diagnosis of pediatric medulloblastoma and its subtypes via AI. Diagnostics. 2021;11:359–384. doi: 10.3390/diagnostics11020359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Attallah O. CoMB-deep: Composite deep learning-based pipeline for classifying childhood medulloblastoma and its classes. Front. Neuroinform. 2021;15 doi: 10.3389/fninf.2021.663592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Attallah O., Zaghlool S. AI-based pipeline for classifying pediatric medulloblastoma using histopathological and textural images. Life. 2022;12:232. doi: 10.3390/life12020232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayman A., Attalah O., Shaban H. An efficient human activity recognition framework based on wearable imu wrist sensors. 2019 IEEE International Conference on Imaging Systems and Techniques; IST; IEEE; 2019. pp. 1–5. [Google Scholar]
- 28.Attallah O., Abougharbia J., Tamazin M., Nasser A.A. A BCI system based on motor imagery for assisting people with motor deficiencies in the limbs. Brain Sci. 2020;10:864–888. doi: 10.3390/brainsci10110864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alyasseri Z.A.A., Al-Betar M.A., Doush I.A., Awadallah M.A., Abasi A.K., Makhadmeh S.N., et al. Review on COVID-19 diagnosis models based on machine learning and deep learning approaches. Expert Syst. 2021 doi: 10.1111/exsy.12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaishya R., Javaid M., Khan I.H., Haleem A. Artificial intelligence (AI) applications for COVID-19 pandemic. Diabetes Metab. Syndr. Clin. Res. Rev. 2020 doi: 10.1016/j.dsx.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naudé W. 2020. Artificial intelligence against COVID-19: An early review. [Google Scholar]
- 32.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021 doi: 10.1109/TCBB.2021.3065361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ezzat D., Hassanien A.E., Ella H.A. An optimized deep learning architecture for the diagnosis of COVID-19 disease based on gravitational search optimization. Appl. Soft Comput. 2021;98 doi: 10.1016/j.asoc.2020.106742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah V., Keniya R., Shridharani A., Punjabi M., Shah J., Mehendale N. Diagnosis of COVID-19 using CT scan images and deep learning techniques. Emerg. Radiol. 2021;28:497–505. doi: 10.1007/s10140-020-01886-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao W., Jiang W., Qiu X. Deep learning for COVID-19 detection based on CT images. Sci. Rep. 2021;11:1–12. doi: 10.1038/s41598-021-93832-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amin S.U., Alsulaiman M., Muhammad G., Mekhtiche M.A., Hossain M.S. Deep learning for EEG motor imagery classification based on multi-layer CNNs feature fusion. Future Gener. Comput. Syst. 2019;101:542–554. [Google Scholar]
- 38.Xu Q., Wang Z., Wang F., Gong Y. Multi-feature fusion CNNs for drosophila embryo of interest detection. Physica A. 2019;531 [Google Scholar]
- 39.Zhang Q., Li H., Sun Z., Tan T. Deep feature fusion for Iris and periocular biometrics on mobile devices. IEEE Trans. Inf. Forensics Secur. 2018;13:2897–2912. doi: 10.1109/TIFS.2018.2833033. [DOI] [Google Scholar]
- 40.Wu X., Hui H., Niu M., Li L., Wang L., He B., et al. Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: A multicentre study. Eur. J. Radiol. 2020;128 doi: 10.1016/j.ejrad.2020.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serte S., Demirel H. Deep learning for diagnosis of COVID-19 using 3D CT scans. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shankar K., Perumal E. A novel hand-crafted with deep learning features based fusion model for COVID-19 diagnosis and classification using chest X-ray images. Complex Intell. Syst. 2021;7:1277–1293. doi: 10.1007/s40747-020-00216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ragab D.A., Attallah O. FUSi-CAD: Coronavirus (COVID-19) diagnosis based on the fusion of CNNs and handcrafted features. PeerJ Comput. Sci. 2020;6 doi: 10.7717/peerj-cs.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou T., Lu H., Yang Z., Qiu S., Huo B., Dong Y. The ensemble deep learning model for novel COVID-19 on CT images. Appl. Soft Comput. 2021;98 doi: 10.1016/j.asoc.2020.106885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shalbaf A., Vafaeezadeh M. Automated detection of COVID-19 using ensemble of transfer learning with deep convolutional neural network based on CT scans. Int. J. Comput. Assist. Radiol. Surg. 2021;16:115–123. doi: 10.1007/s11548-020-02286-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Attallah O., Ragab D.A., Sharkas M. MULTI-DEEP: A novel CAD system for coronavirus (COVID-19) diagnosis from CT images using multiple convolution neural networks. PeerJ. 2020;8 doi: 10.7717/peerj.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biswas S., Chatterjee S., Majee A., Sen S., Schwenker F., Sarkar R. Prediction of covid-19 from chest ct images using an ensemble of deep learning models. Appl. Sci. 2021;11:7004. [Google Scholar]
- 48.Alshazly H., Linse C., Abdalla M., Barth E., Martinetz T. 2021. COVID-nets: Deep CNN architectures for detecting COVID-19 using chest CT scans. MedRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jingxin L., Mengchao Z., Yuchen L., Jinglei C., Yutong Z., Zhong Z., et al. 2021. COVID-19 lesion detection and segmentation–a deep learning method. Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh K.K., Singh A. Diagnosis of COVID-19 from chest X-ray images using wavelets-based depthwise convolution network. Big Data Min. Anal. 2021;4:84–93. [Google Scholar]
- 51.Verma A.K., Vamsi I., Saurabh P., Sudha R., Sabareesh G.R., Rajkumar S. Wavelet and deep learning-based detection of SARS-nCoV from thoracic X-ray images for rapid and efficient testing. Expert Syst. Appl. 2021;185 doi: 10.1016/j.eswa.2021.115650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soares E., Angelov P., Biaso S., Froes M.H., Abe D.K. 2020. SARS-CoV-2 CT-scan dataset: A large dataset of real patients CT scans for SARS-CoV-2 identification. MedRxiv. [Google Scholar]
- 53.Jin S., Wang B., Xu H., Luo C., Wei L., Zhao W., et al. 2020. AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system in four weeks. MedRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ravì D., Wong C., Deligianni F., Berthelot M., Andreu-Perez J., Lo B., et al. Deep learning for health informatics. IEEE J. Biomed. Health Inf. 2016;21:4–21. doi: 10.1109/JBHI.2016.2636665. [DOI] [PubMed] [Google Scholar]
- 55.Yao X., Wang X., Wang S.-H., Zhang Y.-D. A comprehensive survey on convolutional neural network in medical image analysis. Multimedia Tools Appl. 2020:1–45. [Google Scholar]
- 56.Sarvamangala D.R., Kulkarni R.V. Convolutional neural networks in medical image understanding: a survey. Evol. Intell. 2021:1–22. doi: 10.1007/s12065-020-00540-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu W., Wang Z., Liu X., Zeng N., Liu Y., Alsaadi F.E. A survey of deep neural network architectures and their applications. Neurocomputing. 2017;234:11–26. [Google Scholar]
- 58.He K., Zhang X., Ren S., Sun J. IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition. [DOI] [Google Scholar]
- 59.Lu J., Behbood V., Hao P., Zuo H., Xue S., Zhang G. Transfer learning using computational intelligence: A survey. Knowl.-Based Syst. 2015;80:14–23. [Google Scholar]
- 60.MALLAT SG, A Theory for Multiresolution Signal Decomposition: The Wavelet Representation n.d.:20.
- 61.Anitha V., Murugavalli S. Brain tumour classification using two-tier classifier with adaptive segmentation technique. IET Comput. Vis. 2016;10:9–17. doi: 10.1049/iet-cvi.2014.0193. [DOI] [Google Scholar]
- 62.Sabri Y.Y., Nassef A.A., Ibrahim I.M.H., Abd El Mageed MR., Khairy M.A. CT chest for COVID-19, a multicenter study—experience with 220 Egyptian patients. Egypt. J. Radiol. Nucl. Med. 2020;51:1–15. [Google Scholar]
- 63.Shorten C., Khoshgoftaar T.M. A survey on image data augmentation for deep learning. J. Big Data. 2019;6:1–48. doi: 10.1186/s40537-021-00492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mikolajczyk A., Grochowski M. Data augmentation for improving deep learning in image classification problem. 2018 International Interdisciplinary PhD Workshop; IIPhDW; IEEE; 2018. pp. 117–122. [Google Scholar]
- 65.Arumugam M., Sangaiah A.K. Arrhythmia identification and classification using wavelet centered methodology in ECG signals. Concurr. Comput.: Pract. Exper. 2020;32 [Google Scholar]
- 66.Castro B., Kogan D., Geva A.B. 21st IEEE Convention of the Electrical and Electronic Engineers in Israel. IEEE; 2000. ECG feature extraction using optimal mother wavelet; pp. 346–350. (Proceedings (Cat. No. 00EX377)). [Google Scholar]
- 67.Saraswati N.W.S., Wardani N.W., Indradewi I. Detection of Covid chest X-ray using wavelet and support vector machines. Int. J. Eng. Emerg. Technol. 2020;5:116–121. [Google Scholar]
- 68.Enamamu T., Otebolaku A., Marchang J., Dany J. Continuous m-health data authentication using wavelet decomposition for feature extraction. Sensors. 2020;20:5690. doi: 10.3390/s20195690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Attallah O., Karthikesalingam A., Holt P.J., Thompson M.M., Sayers R., Bown M.J., et al. Feature selection through validation and un-censoring of endovascular repair survival data for predicting the risk of re-intervention. BMC Med. Inform. Decis. Mak. 2017;17:115–133. doi: 10.1186/s12911-017-0508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chizi B., Rokach L., Maimon O. Encyclopedia of Data Warehousing and Mining. second ed. IGI Global; 2009. A survey of feature selection techniques; pp. 1888–1895. [Google Scholar]
- 71.Attallah O., Karthikesalingam A., Holt P.J., Thompson M.M., Sayers R., Bown M.J., et al. Using multiple classifiers for predicting the risk of endovascular aortic aneurysm repair re-intervention through hybrid feature selection. Proc. Instit. Mech. Eng. H. 2017;231:1048–1063. doi: 10.1177/0954411917731592. [DOI] [PubMed] [Google Scholar]
- 72.Radovic M., Ghalwash M., Filipovic N., Obradovic Z. Minimum redundancy maximum relevance feature selection approach for temporal gene expression data. BMC Bioinformatics. 2017;18:1–14. doi: 10.1186/s12859-016-1423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wagh K.P., Vasanth K. Innovations in Electronics and Communication Engineering. Springer; 2019. Electroencephalograph (EEG) based emotion recognition system: A review; pp. 37–59. [Google Scholar]
- 74.Keskar N.S., Mudigere D., Nocedal J., Smelyanskiy M., Tang PTP. 2016. On large-batch training for deep learning: Generalization gap and sharp minima. ArXiv Preprint ArXiv:160904836. [Google Scholar]
- 75.M. Li, T. Zhang, Y. Chen, A.J. Smola, Efficient mini-batch training for stochastic optimization, in: Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2014, pp. 661–670.
- 76.Chee J., Li P. Understanding and detecting convergence for stochastic gradient descent with momentum. 2020 IEEE International Conference on Big Data; Big Data; IEEE; 2020. pp. 133–140. [Google Scholar]
- 77.Liu Y., Gao Y., Yin W. An improved analysis of stochastic gradient descent with momentum. Adv. Neural Inf. Process. Syst. 2020;33:18261–18271. [Google Scholar]
- 78.Loizou N., Richtárik P. Momentum and stochastic momentum for stochastic gradient, newton, proximal point and subspace descent methods. Comput. Optim. Appl. 2020;77:653–710. [Google Scholar]
- 79.Garbin C., Zhu X., Marques O. Dropout vs batch normalization: an empirical study of their impact to deep learning. Multimedia Tools Appl. 2020;79:12777–12815. [Google Scholar]
- 80.Ioffe S., Szegedy C. International Conference on Machine Learning. PMLR; 2015. Batch normalization: Accelerating deep network training by reducing internal covariate shift; pp. 448–456. [Google Scholar]
- 81.Rahimzadeh M., Attar A., Sakhaei S.M. A fully automated deep learning-based network for detecting covid-19 from a new and large lung ct scan dataset. Biomed. Signal Process. Control. 2021;68 doi: 10.1016/j.bspc.2021.102588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pathak Y., Shukla P.K., Arya K.V. Deep bidirectional classification model for COVID-19 disease infected patients. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020 doi: 10.1109/TCBB.2020.3009859. [DOI] [PubMed] [Google Scholar]
- 83.Panwar H., Gupta P.K., Siddiqui M.K., Morales-Menendez R., Bhardwaj P., Singh V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-scan images. Chaos Solitons Fractals. 2020 doi: 10.1016/j.chaos.2020.110190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carvalho E.D., Silva R.R., Araújo F.H., de AL Rabelo R., de Carvalho Filho AO. An approach to the classification of COVID-19 based on CT scans using convolutional features and genetic algorithms. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kundu R., Basak H., Singh P.K., Ahmadian A., Ferrara M., Sarkar R. Fuzzy rank-based fusion of CNN models using gompertz function for screening COVID-19 CT-scans. Sci. Rep. 2021;11:1–12. doi: 10.1038/s41598-021-93658-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Swapnarekha H., Behera H.S., Nayak J., Naik B. Covid CT-net: A deep learning framework for COVID-19 prognosis using CT images. J. Interdiscip. Math. 2021;24:327–352. [Google Scholar]
- 87.Attallah O. An effective mental stress state detection and evaluation system using minimum number of frontal brain electrodes. Diagnostics. 2020;10:292–327. doi: 10.3390/diagnostics10050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Colquhoun D. An investigation of the false discovery rate and the misinterpretation of p-values. R. Soc. Open Sci. 2014;1 doi: 10.1098/rsos.140216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ellis P.D. Cambridge University Press;; 2010. The Essential Guide To Effect Sizes: Statistical Power, Meta-Analysis, and the Interpretation of Research Results. [Google Scholar]
- 90.Ragab D.A., Attallah O., Sharkas M., Ren J., Marshall S. A framework for breast cancer classification using multi-DCNNs. Comput. Biol. Med. 2021;131 doi: 10.1016/j.compbiomed.2021.104245. [DOI] [PubMed] [Google Scholar]