Abstract
A collection of 313 motile aeromonads isolated at Danish rainbow trout farms was analyzed to identify some of the genes involved in high levels of antimicrobial resistance found in a previous field trial (A. S. Schmidt, M. S. Bruun, I. Dalsgaard, K. Pedersen, and J. L. Larsen, Appl. Environ. Microbiol. 66:4908–4915, 2000), the predominant resistance phenotype (37%) being a combined oxytetracycline (OTC) and sulphadiazine/trimethoprim resistance. Combined sulphonamide/trimethoprim resistance (135 isolates) appeared closely related to the presence of a class 1 integron (141 strains). Among the isolates containing integrons, four different combinations of integrated resistance gene cassettes occurred, in all cases including a dihydrofolate reductase gene and a downstream aminoglycoside resistance insert (87 isolates) and occasionally an additional chloramphenicol resistance gene cassette (31 isolates). In addition, 23 isolates had “empty” integrons without inserted gene cassettes. As far as OTC resistance was concerned, only 66 (30%) out of 216 resistant aeromonads could be assigned to resistance determinant class A (19 isolates), D (n = 6), or E (n = 39); three isolates contained two tetracycline resistance determinants (AD, AE, and DE). Forty OTC-resistant isolates containing large plasmids were selected as donors in a conjugation assay, 27 of which also contained a class 1 integron. Out of 17 successful R-plasmid transfers to Escherichia coli recipients, the respective integrons were cotransferred along with the tetracycline resistance determinants in 15 matings. Transconjugants were predominantly tetA positive (10 of 17) and contained class 1 integrons with two or more inserted antibiotic resistance genes. While there appeared to be a positive correlation between conjugative R-plasmids and tetA among the OTC-resistant aeromonads, tetE and the unclassified OTC resistance genes as well as class 1 integrons were equally distributed among isolates with and without plasmids. These findings indicate the implication of other mechanisms of gene transfer besides plasmid transfer in the dissemination of antibiotic resistance among environmental motile aeromonads.
The motile aeromonads represent a group of ubiquitous aquatic microorganisms which are not generally considered to be primary human pathogens (15). Some species, however, have been isolated from local or generalized human infections (7, 15, 20). By contrast, many members of the group are recognized as primary pathogens to a wide range of cold-blooded animals, in particular to fish (4, 6, 40). In temperate regions with mainly salmonid production, motile Aeromonas species are not commonly associated with disease outbreaks in aquaculture and are thus not directly targeted by treatment with antimicrobial agents.
However, freshwater fish farming does seem to have an impact on environmental Aeromonas spp., as indicated by a previous investigation of antimicrobial resistance at four Danish rainbow trout farms (35). Water, sediment, and fish samples were examined, and because of their ubiquitous distribution in the freshwater environment, the motile aeromonads were selected as bacterial indicators. In addition, members of the genus Aeromonas readily develop single or multiple antimicrobial resistance phenotypes (10, 12, 18, 23–25), and R-plasmids are commonly found (1, 3, 5, 13, 19, 28). Thus, they were well suited for monitoring the incidence of antibiotic resistance, as well as for investigating the conjugative spread of resistance genes in these settings.
Significantly higher proportions of antibiotic-resistant motile aeromonads were detected in the effluent of fish farms than in their inlets (35). In particular, many of the isolates were resistant to high levels of oxytetracycline (OTC), a combination of sulphadiazine/trimethoprim (S/T), or both. Potentiated sulphonamides and oxolinic acid (OXA) are the only antimicrobials licensed for therapeutic use in Danish aquaculture, while the usage of OTC is restricted, requiring dispensation in every case. The small amounts of OTC used in the fish farms thus did not appear to correlate with the observed high OTC resistance frequencies among environmental Aeromonas isolates (35). By contrast, the administration of S/T in fish farms during bacterial disease outbreaks might have promoted the emergence of S/T resistance in the vicinity of the farms.
Both types of resistance have been reported to be encoded by transferable plasmids within the genus Aeromonas (1, 3, 7, 14). Several classes of tetracycline resistance determinants have been described on R-plasmids within this group of bacteria (1, 2, 5, 8, 9, 30). Tetracycline resistance genes are frequently part of transposons, which are able to change their location within the cell, and thus achieve increased mobility, e.g., by inserting into conjugative plasmids (41, 43). Consequently, we decided to investigate the occurrence and distribution of the tetracycline resistance determinant classes A to E among the motile Aeromonas isolates, comparing OTC-resistant isolates with different plasmid profiles and origins.
Furthermore, the aeromonads were screened for the presence of class 1 integrons in order to elucidate the genetic background of the high S/T resistance level. The 3′ conserved segment of the integron includes the sul1 and qacEΔ1 genes, encoding sulfonamide- and quaternary ammonium compound resistance, respectively. The 5′ conserved segment contains the genes encoding the integration of various numbers of gene cassettes, comprising the variable part of the structure. The gene cassettes usually are antimicrobial resistance genes, and dihydrofolate reductase (dhfr) genes conferring trimethoprim resistance are common (22, 29).
Only few studies have to date addressed the prevalence of class 1 integrons among environmental bacteria (27, 31), and to our knowledge, this is the first report of class 1 integrons within the widely distributed motile Aeromonas species.
Even though class 1 integrons are transposition defective, they are often plasmid borne as they are mobilizable in association with a functional transposon or by transposition proteins supplied in trans (1, 8, 9, 32, 41, 43). Thus, their presence might indicate whether horizontal gene transfer has occurred among the aeromonads found in and around the sampled fish farms, and associated antibiotic resistance genes could explain some of the observed resistance patterns. Subsequent conjugation assays were used to assess the transferability of both tetracycline resistance determinants and class 1 integrons on conjugative plasmids.
MATERIALS AND METHODS
Bacterial isolates and MIC testing.
Between October 1997 and February 1999, we sampled four fish farms situated along a Danish stream on eleven occasions with monthly intervals (35). Farm 1 was located farthest upstream and thus received no effluents from other fish farms. However, the stream had previously received effluents from a sewage treatment plant and some agricultural areas. Farm 2 was situated close to the outlet of the adjacent upstream fish farm, and farm 4 was one of the last farms downstream. Water and sediment samples were collected from the inlet, outlet, and a pond of each farm as earlier described (35). Samples from gills and skin mucus of two to four fish per farm were also included.
The processing and culturing of the samples are described in detail in a previous paper (35). Random colonies were selected and screened for presumptive Aeromonas isolates with a panel of tests comprising Gram reaction, motility, morphology, catalase and oxidase production, oxidative/fermentative utilization of glucose, and susceptibility to O/129 (16). In order to establish a phenospecies identification of the isolates, the following biochemical tests were performed: beta-hemolysis, arginine dihydrolase, lysine and ornithine decarboxylase, esculin hydrolysis, gas production from glucose, Voges-Proskauer, and acid from sucrose/lactose/salicin/arabinose/cellobiose (modified according to reference 16).
All isolates and transconjugants were tested in a standardized agar dilution assay (26, 35) in order to determine the MICs of several antimicrobial agents presently used in Danish aquaculture against them, including OTC, S/T, and OXA. The following breakpoints were established (35): isolates were considered to be OTC resistant when MICs for them were >8 μg ml−1 (sensitive isolates, 0.125 to 1.0 μg ml−1), S/T resistant when MICs for them were >512 and 102 μg ml−1 (sensitive isolates, 0.5 and 0.1 to 8 and 1.6 μg ml−1), and OXA resistant when MICs for them were >2 μg ml−1 (sensitive isolates, 0.125 to 1.0 μg ml−1).
Detection of tetracycline resistance determinants.
We decided to test all OTC-resistant isolates for tetracycline resistance determinants A to E because they include the classes that are most commonly described in aeromonads (1, 2, 5, 8, 9) and that are frequently associated with R-plasmids (1, 8, 30). A multiplex PCR assay was used according to the method described by Guardabassi et al. (12). Escherichia coli strains containing the respective tetracycline resistance genes were included (class A, NCTC/50078; class B, HB101/pRT11; class C, DO7/pBR322; class D, C600/pSL106; and class E, HB101/pSL1504). Primers are listed in Table 1. Table 2 shows the distribution of identified tetracycline resistance determinants relative to the origin of the isolates.
TABLE 1.
PCR primer sequences, targets, and annealing sites
| Primer | Nucleotide sequence (5′–3′) | Target | Annealing sites | Accession no. (GenBank) |
|---|---|---|---|---|
| TetA Fa | GTA ATT CTG AGC ACT GTC GC | tetA | 4925–4942, complement to 8340–8323 | X00006 |
| TetA R | CTG CCT GGA CAA CAT TGC TT | X00006 | ||
| TetB F | CTC AGT ATT CCA AGC CTT TG | tetB | 2378–2397, complement to 2813–2794 | J01830 |
| TetB R | CTA AGC ACT TGT CTC CTG TT | J01830 | ||
| TetC F | TCT AAC AAT GCG CTC ATC GT | tetC | 92–111, complement to 680–661 | J01749 |
| TetC R | GGT TGA AGG CTC TCA AGG GC | J01749 | ||
| TetD F | ATT ACA CTG CTG GAC GCG AT | tetD | 1551–1570, complement to 2674–2654 | X65876 |
| TetD R | CTG ATC AGC AGA CAG ATT GC | X65876 | ||
| TetE F | GTG ATG ATG GCA CTG GTC AT | tetE | 32–52, complement to 1231–1212 | L06940 |
| TetE R | CTC TGC TGT ACA TCG CTC TT | L06940 | ||
| int 1F | GGC ATC CAA GCA GCA AGC | intI | 4925–4942, complement to 8340–8323 | U12338 |
| int 1R | TAG TCC AGT TCA GAC GAA | qacEΔ | U12338 | |
| qacF | ATC GCA ATA GTT GGC GAA GT | qacEΔ | 8417–8436, complement to 9214–9195 | U12338 |
| sul R | GCA AGG CGG AAA CCC GCG CC | sul1 | U12338 | |
| dhfr F | CTG ATA TTC CAT GGA GTG CCA | dhfr1 | 140–160, complement to 553–573 | AF203818 |
| dhfr R | CGT TGC TGC CAC TTG TTA ACC | AF203818 | ||
| ant F | GCC TGA AGC CAC ACA GTG ATA | ant(3′′)1a | 772–791, complement to 1411–1391 | AF203818 |
| ant R | CTA CCT TGG TGA TCT CGC CTT | AF203818 |
F, forward; R, reverse.
TABLE 2.
Incidence and distribution of OTC and S/T resistance (36) and the corresponding fractions of identified tetracycline (Tet) resistance determinants and class 1 integrons (Int) among motile aeromonads isolated from four Danish rainbow trout farmsd,
| Location (n) | Area within location (No. of isolates) | % of isolates resistant to:
|
% of isolates containing Int | ||
|---|---|---|---|---|---|
| OTC | Tet | S/T | |||
| Sampling site (250) | Inlet (53) | 49a | 17 | 19a | 25a |
| Pond (92) | 71 | 33 | 49 | 47 | |
| Outlet (105) | 77 | 14 | 47 | 45 | |
| Avg | 66 | 22 | 38 | 39 | |
| Fish farm (313) | Farm 1 (69) | 65 | 17 | 61b | 67b |
| Farm 2 (98) | 75 | 30 | 42 | 43 | |
| Farm 3 (53) | 68 | 4c | 25 | 27 | |
| Farm 4 (93) | 67 | 29 | 42 | 37 | |
| Avg | 69 | 20 | 43 | 44 | |
Statistically significantly lower fraction than at other locations on farms (P < 0.05).
Statistically significantly higher proportion than at other farms (P < 0.05).
Statistically significantly lower fraction than at other farms (P < 0.05).
The comparison of different sampling sites (inlets, ponds, and outlets at the four farms) was based on Aeromonas isolates from water and sediment samples, while the comparison of fish farms also included fish isolates.
Analysis of antimicrobial resistance genes associated with integrons.
All isolates and transconjugants were screened for the presence of class 1 integrons with specific primers targeting the conserved 5′ and 3′ segments of the structure as previously described (34). Thus, the size of a PCR product depends on the number and size of the inserted gene cassettes (Fig. 1 and 2). Salmonella enterica serovar Typhimurium DT104 (9616368) was the positive control strain. PCR products were purified (S-400 HR MicroSpin Columns; Amersham Pharmacia Biotech, Uppsala, Sweden) and sequenced. The nucleotide sequence was determined in both senses of the DNA, using cycle sequencer 373A (Applied Biosystems, Perkin-Elmer, Foster City, Calif.) as reported earlier (34). With five different sizes ranging between about 150 bp (no insert) and 2,900 bp (Fig. 1), two to three representatives of each amplicon were sequenced. Subsequently, a suitable DNA restriction enzyme (MboII) was employed to determine whether equally sized integron amplicons contained the same gene cassettes. Restriction cutting and purification of DNA were performed as described (33), and the resulting fragments were separated by agarose gel electrophoresis.
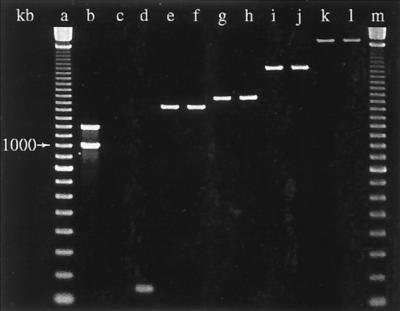
FIG. 1.
Different sizes of PCR products obtained with a primer pair targeting the conserved segments of class 1 integrons in motile aeromonads. Class 1 integrons were—if present—invariably cotransferred to E. coli on OTC resistance plasmids. Lanes a and m, 100-bp DNA marker; lane b, positive control S. enterica serovar Typhimurium DT104 with 1,000- and 1,200-bp amplicons; lane c, negative control; lane d, “empty” integron with no gene inserts between conserved ends; lanes e and f, isolate 1-75 and the corresponding E. coli transconjugant with 1,400-bp amplicons; lanes g and h, isolate 4-229 and transconjugant with 1,550-bp amplicons; lanes i and j, isolate 2-280 and transconjugant with 2,100-bp PCR products; lanes k and l, isolate 4-221 and transconjugant with 2,900-bp products. kb, kilobases.
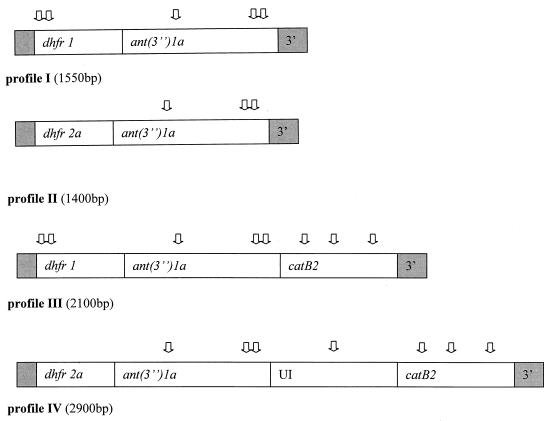
FIG. 2.
Schematic view of MboII restriction sites ( ) used for comparison of gene cassette content of different sizes of amplicons obtained with primers targeting the conserved segments (shaded areas) of class 1 integrons. Out of 141 integron-positive isolates, 74 belonged to profile I, 13 to profile II, 22 to profile III, and 9 to profile IV. Twenty-three isolates had “empty” integrons with no genes inserted into the variable region. UI, unidentified gene insert.
The 1,550-bp amplicons were most prevalent and invariably appeared to contain a dhfr1 insert and a downstream ant(3")1a insert. As these resistance gene cassettes were found alone or in combination in every amplicon type (Fig. 2), a specific PCR assay targeting the dhfr1 and ant(3′′)1a genes was used to detect their presence and order within all PCR products (36) (Table 1).
A separate primer set was used to investigate the 3′ conserved segment of the class 1 integrons (Table 1) containing the qacEΔ and sul1 genes (34) in order to detect defective copies with an incomplete sul1 gene, which were frequent findings in a study of aquatic bacteria by Rosser and Young (31).
Plasmid profiling.
All aeromonads and E. coli transconjugants were screened for their plasmid content by applying the alkaline lysis method described by Kado and Liu (17) followed by agarose gel electrophoresis. We used the 4.0 version of GelCompar (Applied Maths, Sint-Martens-Latem, Belgium) to analyze the resulting plasmid profiles.
Conjugational gene transfer.
All OTC-resistant isolates with large plasmids were included as putative donors in a filter mating assay in order to detect the transfer of R-plasmids to a rifampin-resistant E. coli strain, CSH26Rf (37). Overnight cultures of donor and recipient were adjusted to an optical density at 600 nm of 0.5 with fresh veal infusion broth. Equal volumes (50 μl) of all cultures were mixed on a sterile 0.2-μm-pore-size nitrocellulose filter (Sartorius AG, Göttingen, Germany), which was placed on a veal infusion agar plate (Difco) and incubated at 20°C overnight. Cells were washed off the filter by vortexing in 10-ml sterile 0.9% NaCl solution, and appropriate 10-fold dilutions were prepared. From each dilution, 100-μl aliquots were spread on selective agar plates containing 20 μg of OTC ml−1, 100 μg of rifampin (Bie & Berntsen, Rødovre, Denmark)/ml−1, or both. Donor and recipient were also placed on the double selective plates for mutant detection. All assays were run in duplicate. The identity of transconjugants was confirmed biochemically (oxidase production, amino acids, and Voges-Proskauer reaction), and they were screened for the presence of plasmids as described above.
Statistical methods.
The proportions of antibiotic resistance and the respective detected genes among the Aeromonas isolates were computed for each fish farm and all sampling sites. A logistic regression model was employed (proc genmod in SAS version 6.12; SAS Institute Inc., Cary, N.C.) to detect differences between resistance rates from inlet and outlet as well as differences between fish farms. In a few cases where the logistic regression model did not describe the data well, χ2 tests (with continuity correction) were performed instead.
RESULTS
Resistance phenotypes.
Three hundred thirteen motile, mesophilic aeromonads were investigated. The most prevalent phenospecies identified were Aeromonas hydrophila (35.3%), A. bestiarum (19%), and A. veronii biovar sobria (15.3%). Fifteen percent of the isolates could not be reliably assigned to one phenospecies. Antibiotic resistance patterns did not vary significantly between the identified phenospecies (data not shown).
A total of 216 Aeromonas isolates (69%) were resistant to OTC, while 135 (43%) displayed S/T resistance (35). Sixty-three isolates (20%) were OXA resistant. Multiresistance was common, as 151 isolates (48%) carried at least two additional antibiotic resistance traits besides amoxicillin resistance, which appears to be intrinsic in most Aeromonas species.
The predominant multiresistance phenotype was OTC-S/T resistance (89 isolates, 28%), followed by OTC-OXA (31 isolates, 10%) and OTC-OXA-S/T (28 isolates, 9%). Most resistance patterns were equally likely to occur in all farms. One exception was OTC-S/T, farm 3 isolates being less likely to have this phenotype (11%) than isolates from farms 1 (38%), 2 (33%), and 4 (25%). However, this difference was nonsignificant at a 5% level of significance. Twenty-four percent of the aeromonads (75 isolates) were sensitive to all of the four remaining antibiotics. The results from the statistical analysis of the overall resistance data were earlier described in detail (35), and only a few significant effects are included here (Table 2).
Antibiotic resistance genes.
Table 2 sums up the distribution of the identified tetracycline resistance determinants and OTC-resistant Aeromonas isolates. It appeared that, while 69% of the aeromonads were OTC resistant, only 30% were carrying one or two of the five tetracycline resistance determinants included in the screening (Table 2). A total of 19 tetA-positive, 39 tetE-positive, and 6 tetD-positive isolates was found. Three isolates contained two tetracycline resistance determinants: tetA and -E, tetA and -D, and tetE and -D. Tetracycline resistance determinants B and C were not detected.
A statistically significant increase of overall OTC resistance levels occurred among isolates from ponds and outlets compared to those from inlets (35) (Table 2). However, it appeared that the tetA, tetD, and tetE genes were evenly distributed among isolates from different sample sites, except among pond isolates, where more isolates with unclassified determinants were detected. When OTC-resistant motile aeromonads from the four fish farms were compared, farm 3 isolates differed from other isolates, as only 4% of the involved tetracycline resistance determinants were classified (χ2 test; P < 0.005) (Table 2). Conversely, overall OTC resistance was evenly distributed among farms.
Table 2 also shows the distribution of S/T resistance and integrons among the aeromonads collected from water and sediments. A total of 141 isolates, including all 135 S/T-resistant isolates, contained class 1 integrons with different resistance gene inserts (Fig. 2) and an intact sul1 gene (data not shown). Six integron-positive isolates without integrated gene cassettes did not express S/T resistance phenotypically. The detected integrons were evenly distributed within all identified phenospecies (data not shown).
As S/T resistance thus was closely correlated to the presence of class 1 integrons, the observed statistical effects were similar: farm 1 isolates were likelier to contain integrons than isolates from other farms (logistic regression; P = 0.028) (Table 2), and aeromonads from inlets of the four fish farms were less frequently integron positive than those from ponds and outlets (logistic regression, P = 0.013). One exception was farm 2, where the proportion of S/T-resistant, integron-positive Aeromonas isolates was higher at the inlet (44%) than in the pond (31%) or at the outlet (31%).
A schematic overview of the observed integron structures and their content of antimicrobial resistance genes is given in Fig. 2. Restriction enzyme profiles correlated well with the respective sizes of the PCR products (Fig. 2), indicating that the gene content of a certain amplicon corresponded to the sequenced amplicons of the same magnitude.
A considerable number of integron-positive isolates (23 of 141) were “empty” with no gene cassettes inserted between the conserved segments of the integron. As a common feature, all integron inserts included an ant(3′′)1a gene downstream of a trimethoprim resistance gene. Two types of dhfr genes were found, dhfr1 and dhfr2a. The most prevalent amplicon (74 isolates) was the 1,550-bp PCR product, containing dhfr1 and ant(3′′)1a inserts. Moreover, 22 2,100-bp products, 13 1,400-bp products, and 9 2,900-bp products were found among the S/T-resistant isolates. The gene inserts in the 2,100-bp amplicon were identical with those of the 1,550-bp product, with an additional downstream chloramphenicol resistance gene, catB2.
Likewise, in the 2,900-bp amplicon, the two upstream gene cassettes, dhfr2a and ant(3′′)1a, were the same inserts as in the 1,400-bp amplicons. A catB2 cassette was identified as the last downstream gene, while the remaining insert did not yield satisfactory nucleotide sequences for identification, despite repeated purification and sequencing procedures.
As shown in Table 3, there was a positive correlation between the content of large plasmids and the presence of tetA among OTC-resistant motile aeromonads (χ2 test; P < 0.005). Correspondingly, tetA was the predominant tet determinant detected in transconjugants (10 of 17), despite the comparatively low overall incidence (Tables 3 and 4).
TABLE 3.
Correlation of plasmid profiles and tetracycline (Tet) resistance determinants among 216 OTC-resistant and 97 OTC-sensitive Aeromonas isolates and the association of tetracycline resistance determinants with self-transferable plasmids, as established in filter mating assays with E. coli recipients
| Characterization of sample | No. of isolates that
are
|
||||
|---|---|---|---|---|---|
| OTC resistant
|
Sensitive | ||||
| TetA | TetD | TetE | NCb | ||
| Plasmid profile Aa | 13 | 2 | 5 | 20 | 10 |
| Plasmid profiles B, C, and D | 1 | 3 | 18 | 68 | 29 |
| No plasmids | 5 | 1 | 16 | 64 | 58 |
| Total | 19 | 6 | 39 | 152 | 97 |
| Donor | 13 | 2 | 5 | 20 | |
| Transconjugant | 11 | 1 | 0 | 5 | |
Profile A, one or several large plasmids (>30 kb); and profiles B, C, and D, small to medium-sized plasmids (2.3 to 25 kb).
NC, resistance determinant, not classified.
TABLE 4.
Characterization of 40 OTC-resistant, motile Aeromonas isolates with large (>30 kb) plasmids from a freshwater fish farming environment, including presumptive biochemical identification and detection of antibiotic resistance genesd
| Isolate no. | Date (mo/yr) | Source | Identification | Resistance phenotype | Tet determinant | Class 1 integrons/gene inserts | Tc status |
|---|---|---|---|---|---|---|---|
| 1-75 | 11/97 | Water/inlet | A. hydrophila | OTC, S/T | A | dhfr2a-ant(3′′)1a | + |
| 1-78 | 11/97 | Water/pond | A. hydrophila | OTC, OXA, S/T | E | NDc | |
| 1-163 | 4/98 | Rainbow trout | A. hydrophila | OTC, S/T | A | dhfr1-ant(3′′)1a-catB2 | |
| 1-231 | 6/98 | Sediment/outlet | A. hydrophila | OTC, OXA, S/T | D | ND | + |
| 1-250 | 7/98 | Rainbow trout | NIa | OTC, S/T | NCb | dhfr1-ant(3′′)1a | + |
| 1-318 | 10/98 | Sediment/pond | A. hydrophila | OTC | E | ND | |
| 1-337 | 10/98 | Water/pond | A. veronii biovar sobria | OTC, S/T | A | dhfr1-ant(3′′)1a-orf-catB2 | + |
| 1-358 | 10/98 | Sediment/outlet | A. veronii biovar sobria | OTC, S/T | A | dhfr1-ant(3′′)1a-orf-catB2 | + |
| 1-372 | 12/98 | Sediment/outlet | A. hydrophila | OTC, OXA, S/T | NC | ND | |
| 2-41 | 10/97 | Rainbow trout | A. bestiarum | OTC, S/T | NC | dhfr1-ant(3′′)1a | + |
| 2-62 | 11/97 | Sediment/outlet | A. hydrophila | OTC, S/T | A | dhfr2a-ant(3′′)1a | + |
| 2-85 | 1/98 | Water/pond | A. hydrophila | OTC, S/T | A | dhfr1-ant(3′′)1a-orf-catB2 | + |
| 2-155 | 4/98 | Water/pond | A. hydrophila | OTC, OXA | NC | ND | + |
| 2-189 | 6/98 | Water/outlet | NI | OTC, S/T | NC | dhfr1-ant(3′′)1a | + |
| 2-197 | 6/98 | Rainbow trout | NI | OTC, S/T | NC | dhfr1-ant(3′′)1a-catB2 | |
| 2-219 | 6/98 | Rainbow trout | A. hydrophila | OTC, S/T | NC | ND | |
| 2-280 | 9/98 | Sediment/pond | A. hydrophila | OTC, S/T | A | dhfr1-ant(3′′)1a-catB2 | + |
| 2-282 | 9/98 | Sediment/outlet | A. hydrophila | OTC | NC | ND | |
| 2-321 | 9/98 | Sediment/outlet | A. hydrophila | OTC, OXA | NC | ND | |
| 2-333 | 10/98 | Sediment/outlet | NI | OTC, OXA | NC | ND | |
| 2-410 | 2/99 | Sediment/pond | A. bestiarum | OTC, S/T | A | dhfr1-ant(3′′)1a | |
| 3-30 | 10/97 | Sediment/outlet | A. hydrophila | OTC, S/T | NC | dhfr1-ant(3′′)1a | |
| 3-130 | 3/98 | Sediment/outlet | A. hydrophila | OTC, OXA, S/T | NC | ND | |
| 3-135 | 4/98 | Water/inlet | A. hydrophila | OTC | NC | ND | |
| 3-217 | 7/98 | Water/inlet | A. hydrophila | OTC | NC | ND | |
| 3-247 | 9/98 | Sediment/outlet | NI | OTC | NC | ND | |
| 3-372 | 2/99 | Water/pond | A. hydrophila | OTC, OXA, S/T | NC | dhfr2a-ant(3′′)1a | + |
| 4-90 | 11/97 | Rainbow trout | A. hydrophila | OTC, S/T | E | dhfr1-ant(3′′)1a | |
| 4-97 | 1/98 | Water/outlet | A. hydrophila | OTC, S/T | E | dhfr1-ant(3′′)1a-catB2 | |
| 4-206 | 6/98 | Sediment/pond | A. hydrophila | OTC, S/T | AE | dhfr1-ant(3′′)1a-catB2 | |
| 4-221 | 6/98 | Rainbow trout | A. veronii biovar sobria | OTC, S/T | A | dhfr1-ant(3′′)1a-orf-catB2 | + |
| 4-229 | 7/98 | Rainbow trout | NI | OTC, OXA, S/T | A | dhfr1-ant(3′′)1a | + |
| 4-241 | 7/98 | Rainbow trout | A. veronii biovar sobria | OTC, S/T | NC | dhfr1-ant(3′′)1a-orf-catB2 | |
| 4-280 | 9/98 | Rainbow trout | A. bestiarum | OTC, OXA, S/T | NC | dhfr1-ant(3′′)1a-orf-catB2 | + |
| 4-302 | 9/98 | Sediment/inlet | A. hydrophila | OTC | NC | No inserts | |
| 4-307 | 9/98 | Rainbow trout | A. veronii biovar veronii | OTC, OXA, S/T | A | dhfr1-ant(3′′)1a-orf-catB2 | + |
| 4-348 | 9/98 | Rainbow trout | A. hydrophila | OTC, S/T | NC | dhfr1-ant(3′′)1a | |
| 4-353 | 9/98 | Sediment/inlet | A. hydrophila | OTC | NC | No inserts | |
| 4-354 | 9/98 | Sediment/inlet | A. bestiarum | OTC | E | No inserts | |
| 4-440 | 2/99 | Sediment/pond | A. hydrophila | OTC, S/T | A | dhfr1-ant(3′′)1a-orf-catB2 | + |
NI, no reliable identification of Aeromonas phenospecies.
NC, tetracycline resistance determinant not classified.
ND, not detected.
Tc + indicates the detection of transconjugants in filter mating assays, where Aeromonas donors transferred both their tetracycline (Tet) resistance determinant and class 1 integron on R-plasmids to E. coli recipients.
In contrast, the occurrence of class 1 integrons was not related to the respective plasmid profiles, and they were just as prevalent among Aeromonas isolates without plasmids. Twenty-seven out of 40 OTC-resistant donors harbored different class 1 integrons (Table 4), and they were invariably cotransferred to the E. coli recipient in those cases where plasmid transfer was detected (15 of 17).
Plasmid profiles and conjugative transfer of resistance genes.
The plasmid content of the 313 aeromonads did not seem to vary between the different phenospecies (data not shown). One hundred forty-four (46%) isolates did not contain any plasmids, while 16% did harbor at least one large (>30 kb) plasmid (profile A) (Table 3). The GelCompar analysis yielded three additional clusters: profile B with one small to medium-sized plasmid (2.3 to 20 kb; 42 isolates), profile C with two plasmids between 6.5 and 15 kb (43 isolates), and profile D with three to nine plasmids between 3 and 25 kb (34 isolates). The different plasmid profiles did not seem to vary according to sample matter (water, sediment, and fish) or origin of the isolate (farms or sample site) (data not shown).
Tables 3 and 4 summarize the results of the filter mating assays, where 17 of 40 OTC-resistant donors with plasmid profile A transferred an R-plasmid to E. coli. All transconjugants had received a large plasmid between 110 and 160 kb.
During the course of this study, we successfully transferred OTC resistance plasmids to susceptible Aeromonas field isolates in addition to the E. coli strain (data not shown). In another set of experiments, the role of conjugation under simulated natural conditions was investigated, including environmental donors and recipients (M. S. Bruun, A. S. Schmidt, I. Dalsgaard, and J. L. Larsen, unpublished data).
DISCUSSION
The high OTC resistance levels (69%) detected among the aeromonads in this study were unexpected, considering that this agent has been rarely used for therapeutic purposes in Danish aquaculture since a change in legislation in 1994. DePaola et al. (8) found similarly high proportions of OTC-resistant aeromonads from catfish and their environments (58 to 83%), where the drug was routinely used in medicated feed. Other comparable investigations of motile aeromonads from different freshwater environments report considerably lower tetracycline resistance levels (7, 10, 11, 23, 28, 42). One explanation may be that, once acquired, the resistance genes are maintained within the population, protecting the bacteria from tetracyclines produced by other members of the microflora or residues in agricultural or domestic effluents. Goni-Urriza et al. (10) detected an increase of tetracycline resistance levels of Aeromonas isolates from 0 to 27% in a stream before and after the stream passed a wastewater discharge point. However, this resistance appeared to be entirely chromosomally mediated (10).
More than three different tetracycline resistance determinants occurred among OTC-resistant isolates, and in a few instances more than one determinant was detected in a single strain. DePaola et al. (8) classified over 90% of tetracycline-resistant A. hydrophila isolates from catfish farms as either tetA or tetE positive, and TetA, TetD, and TetE are considered to be the main tetracycline resistance determinants among motile aeromonads (1, 5, 8, 9). However, the majority of OTC-resistant isolates in this study (70%) did not belong to classes A to E (Tables 2 and 3). Thus, the genetic background of OTC resistance among motile aeromonads from this habitat appeared to be rather diverse and to vary locally, possibly as a response to various physical conditions or differences in local genetic exchange processes. Although probably not the only mechanism of horizontal resistance gene transfer, the transfer of R-plasmids is thought to play a major role in the dissemination of OTC resistance in the fish farming environment (5, 30, 39, 40, 41). In our study, there was a positive correlation between OTC-resistant motile aeromonads harboring large plasmids and the presence of tetA. Moreover, the majority of successfully transferred R-plasmids carried the tetA resistance gene. This contrasts with DePaola's findings, in which most donors in successful matings had unidentified tetracycline resistance determinants (8, 9).
Human as well as environmental Aeromonas isolates do often contain conjugative R-plasmids, some of which have been assigned to incompatibility groups C and U (14, 30). Both groups have wide host ranges, yet IncU plasmids have only been detected occasionally in genera other than Aeromonas (14, 44).
Rhodes et al. (30) reported that IncU OTC resistance plasmids in mesophilic aeromonads from hospital effluents and fish farms were closely related to IncU OTC resistance plasmids isolated from the fish pathogen Aeromonas salmonicida and a human E. coli strain. The predominant tetracycline resistance determinant determinant was tetA, and the presence of a complete or truncated form of tetracycline resistance transposon Tn1721 was demonstrated on several of the R-plasmids, thus illuminating yet another mechanism of horizontal dissemination of antimicrobial resistance in these settings (30, 36, 41). The present work demonstrated the common occurrence of class 1 integrons and their resistance gene cassettes on OTC resistance plasmids. Still, there was no evidence of a correlation between the occurrence of integrons and a certain plasmid type or tetracycline resistance determinant. Conversely, there was a close association of class 1 integrons and S/T resistance, as all S/T-resistant isolates contained an integron with a sul1 gene within the 3′ conserved region and a dhfr gene cassette insert. Aeromonads from the inlets compared to those from the ponds and outlets of the fish farms were less likely to be OTC, OXA, and S/T resistant (35) and consequently integron positive. Furthermore, Aeromonas isolates from farm 1 were significantly more likely to be S/T resistant and integron positive than isolates from the farms farther downstream, perhaps due to a more frequent use of the drug at this farm during the trial period (35). It may be speculated that the distribution of these genetic elements is enhanced by the frequent use of potentiated sulphonamides in the fish farming environment, including occasional additional resistance gene cassettes like ant(3′′)1a or catB2. If such integrons are mobilized onto R-plasmids, indirect selection could contribute to the maintenance of tetracycline resistance genes within the population.
Only few researchers so far have addressed the incidence and spread of class 1 integrons and integron-associated resistance genes in environmental microorganisms (27, 31, 41). Even in clinical settings, the epidemiology of integrons and gene cassettes is not resolved (22, 29, 32, 38, 43). Rosser and al. (31) detected the incidence of class 1 integrons to be 3.6% among gram-negative bacteria from an estuarine environment. Unlike in this study, almost half of the integrons lacked a sul1 gene and about the same proportion did not have any genes inserted into the variable region. Rosser et al. proposed that gene cassettes are excised from the structure in the absence of antibiotic selective pressure or that “empty” integrons represent ancestral elements which have not yet acquired gene cassette inserts.
The abundance of class 1 integrons and the inserted dhfr genes among the motile aeromonads correlates with transient selective pressures exerted by the administration of combined sulphonamide/trimethoprim drugs in freshwater fish farms. Aminoglycosides and chloramphenicol, on the other hand, are not used in Danish aquaculture. Possibly, aquatic motile aeromonads have acquired the gene cassettes by interacting with other microorganisms in soil or domestic effluents (31, 41). Aminoglycoside resistance gene cassettes may also be more stably integrated within integrons, explaining their frequent occurrence and persistence in many bacterial species and habitats (33).
In conclusion, our results point towards a significant effect of aquaculture on the motile aeromonads in and around the investigated fish farms, leading to an increase of OTC, S/T, and OXA resistance levels within this group. High levels of multiresistance (48%) indicated that the horizontal spread of resistance genes has contributed to the dissemination of, in particular, OTC-S/T resistance phenotypes (35), and the finding of class 1 integrons in 45% of the isolates further supported this hypothesis. The diversity of the isolates and characterized genes does not support the clonal spread of antibiotic-resistant aeromonads in and around the fish farms.
Conjugation assays demonstrated that different types of class 1 integrons were cotransferred to E. coli recipients on OTC resistance plasmids. However, it was evident that other genes and transfer mechanisms besides conjugation are probably involved, which should be further investigated in the future. The significance of antibiotic-resistant environmental bacteria has been much disputed (21, 30, 31, 41). Although ubiquitous in freshwater, motile Aeromonas species are not commonly retrieved from human or animal disease in temperate climate zones (15). However, extensive antimicrobial resistance within this group might provide a pool of resistance genes capable of transfer to other water-borne bacteria or fish pathogens. A better understanding of such processes in natural environments is crucial in order to assess the risk of antibiotic resistance among ubiquitous aquatic bacteria such as the motile aeromonads.
ACKNOWLEDGMENTS
This study was supported by the Danish Ministry of Food, Agriculture and Fisheries.
We thank Farah Bahrani for excellent technical assistance and Frank M. Aarestrup, Lars Bogø Jensen, and Dorthe Sandvang at the Danish Veterinary Laboratory, Copenhagen, Denmark, for various aspects of the initial sequencing work. We are also grateful to Ib Skovgaard and Bo M. Bibby of the Department of Mathematics and Physics, The Royal Veterinary and Agricultural University, for their assistance with the statistical analysis.
REFERENCES
- 1.Adams C A, Austin B, Meaden P G, McIntosh D. Molecular characterization of plasmid-mediated oxytetracycline resistance in Aeromonas salmonicida. Appl Environ Microbiol. 1998;64:4194–4201. doi: 10.1128/aem.64.11.4194-4201.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen S R, Sandaa R-A. Distribution of tetracycline resistance determinants among gram-negative bacteria isolated from polluted and unpolluted marine sediments. Appl Environ Microbiol. 1994;60:908–912. doi: 10.1128/aem.60.3.908-912.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki T. Drug-resistant plasmids from fish pathogens. Microbiol Sci. 1988;5:219–223. [PubMed] [Google Scholar]
- 4.Aoki T, Egusa S, Ogata Y, Watanabe T. Detection of resistance factors in fish pathogen Aeromonas liquefaciens. J Gen Microbiol. 1971;65:343–349. doi: 10.1099/00221287-65-3-343. [DOI] [PubMed] [Google Scholar]
- 5.Aoki T, Takahashi A. Class D tetracycline resistance determinants of R plasmids from the fish pathogens Aeromonas hydrophila, Edwardsiella tarda, and Pasteurella piscicida. Antimicrob Agents Chemother. 1987;31:1278–1280. doi: 10.1128/aac.31.8.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Austin B, Adams C. Fish pathogens. In: Austin B, Altwegg M, Gosling P J, Joseph S, editors. The genus Aeromonas. New York, N.Y: John Wiley & Sons; 1996. pp. 197–229. [Google Scholar]
- 7.Chaudhury A, Nath G, Shukla B N, Sanyal S C. Biochemical characterisation, enteropathogenicity and antimicrobial resistance plasmids of clinical and environmental Aeromonasisolates. J Med Microbiol. 1996;44:434–437. doi: 10.1099/00222615-44-6-434. [DOI] [PubMed] [Google Scholar]
- 8.DePaola A, Flynn P A, McPhearson R M, Levy S B. Phenotypic and genotypic characterization of tetracycline- and oxytetracycline-resistant Aeromonas hydrophila from cultured channel catfish (Ictalurus punctatus) and their environments. Appl Environ Microbiol. 1988;54:1861–1863. doi: 10.1128/aem.54.7.1861-1863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DePaola A, Hill W E, Harrell F M. Oligonucleotide probe determination of tetracycline-resistant bacteria isolated from catfish ponds. Mol Cell Probes. 1993;7:345–348. doi: 10.1006/mcpr.1993.1051. [DOI] [PubMed] [Google Scholar]
- 10.Goñi-Urriza M, Capdepuy M, Arpin C, Raymond N, Caumette P, Quentin C. Impact of an urban effluent on antibiotic resistance of riverine Enterobacteriaceae and Aeromonasspp. Appl Environ Microbiol. 2000;66:125–132. doi: 10.1128/aem.66.1.125-132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goñi-Urriza M, Pineau L, Capdepuy M, Roques C, Caumette P, Quentin C. Antimicrobial resistance of mesophilic Aeromonasspp. isolated from two European rivers. J Antimicrob Chemother. 2000;46:297–301. doi: 10.1093/jac/46.2.297. [DOI] [PubMed] [Google Scholar]
- 12.Guardabassi L, Dijkshoorn L, Collard J M, Olsen J E, Dalsgaard A. Distribution and in-vitro transfer of tetracycline resistance determinants in clinical and aquatic Acinetobacterstrains. J Med Microbiol. 2000;49:929–936. doi: 10.1099/0022-1317-49-10-929. [DOI] [PubMed] [Google Scholar]
- 13.Hassani L, Imziln B, Boussaid A, Gauthier M J. Seasonal incidence of and antibiotic resistance among Aeromonasspecies isolated from domestic wastewater before and after treatment in stabilization ponds. Microb Ecol. 1992;23:227–237. doi: 10.1007/BF00164098. [DOI] [PubMed] [Google Scholar]
- 14.Hedges R W, Smith P, Brazil G. Resistance plasmids of Aeromonads. J Gen Microbiol. 1985;131:2091–2095. [Google Scholar]
- 15.Janda J M, Abbott S L. Human pathogens. In: Austin B, Altwegg M, Gosling P J, Joseph S, editors. The genus Aeromonas. New York, N.Y: John Wiley & Sons; 1996. pp. 151–173. [Google Scholar]
- 16.Joseph S W, Carnahan A. The isolation, identification, and systematics of the motile Aeromonasspecies. Annu Rev Fish Dis. 1994;4:315–343. [Google Scholar]
- 17.Kado C I, Liu F T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelch W J, Lee J S. Antibiotic resistance patterns of gram-negative bacteria isolated from environmental sources. Appl Environ Microbiol. 1978;36:450–456. doi: 10.1128/aem.36.3.450-456.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein B U, Siesenop U, Boehm K H. Investigations on transferable antibiotic resistance through R-plasmids between obligate and facultative fish pathogenic bacteria. Bull Eur Assoc Fish Pathol. 1996;16:138–142. [Google Scholar]
- 20.Ko W-C, Lee H-C, Chuang Y-C, Liu C-C, Wu J-J. Clinical features and therapeutic implications of 104 episodes of monomicrobial Aeromonasbacteraemia. J Infect. 2000;40:267–273. doi: 10.1053/jinf.2000.0654. [DOI] [PubMed] [Google Scholar]
- 21.Kruse H, Sørum H. Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl Environ Microbiol. 1994;60:4015–4021. doi: 10.1128/aem.60.11.4015-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Freijo P, Fluit A C, Schmitz F-J, Verhoef J, Jones M E. Many class I integrons comprise distinct stable structures occurring in different species of Enterobacteriaceaeisolated from widespread geographic regions in Europe. Antimicrob Agents Chemother. 1999;43:686–689. doi: 10.1128/aac.43.3.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNicol L A, Aziz K M, Huq I, Kaper J B, Lockman H A, Remmers E F, Spira W M, Voll M J, Colwell R R. Isolation of drug-resistant Aeromonas hydrophilafrom aquatic environments. Antimicrob Agents Chemother. 1980;17:477–483. doi: 10.1128/aac.17.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McPhearson R M, DePaola A, Zywno S R, Motes M L, Jr, Guarino A M. Antibiotic resistance in gram-negative bacteria from cultured catfish and aquaculture ponds. Aquaculture. 1991;99:203–211. [Google Scholar]
- 25.Motyl M R, McKinley G, Janda J M. In vitro susceptibilities of Aeromonas hydrophila, Aeromonas sobria, and Aeromonas caviaeto 22 antimicrobial agents. Antimicrob Agents Chemother. 1985;28:151–153. doi: 10.1128/aac.28.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Tentative standard. NCCLS Document M31-T. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 27.Petersen A, Guardabassi L, Dalsgaard A, Olsen J E. Class I integrons containing a dhfr trimethoprim resistance gene cassette in aquatic Acinetobacterspp. FEMS Microbiol Lett. 2000;183:73–76. doi: 10.1111/j.1574-6968.2000.tb08876.x. [DOI] [PubMed] [Google Scholar]
- 28.Pettibone G W, Mear J P, Sampsell B M. Incidence of antibiotic and metal resistance and plasmid carriage in Aeromonas isolated from brown bullhead (Ictalurus nebulosus) Lett Appl Microbiol. 1996;23:234–240. [Google Scholar]
- 29.Recchia G D, Hall R M. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 1997;5:389–394. doi: 10.1016/S0966-842X(97)01123-2. [DOI] [PubMed] [Google Scholar]
- 30.Rhodes G, Huys G, Swings J, McGann P, Hiney M, Smith P, Pickup R W. Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721in dissemination of the tetracycline resistance determinant TetA. Appl Environ Microbiol. 2000;66:3883–3890. doi: 10.1128/aem.66.9.3883-3890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosser S J, Young H-K. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J Antimicrob Chemother. 1999;44:11–18. doi: 10.1093/jac/44.1.11. [DOI] [PubMed] [Google Scholar]
- 32.Sallen B, Rajoharison A, Desvarenne S, Mabilat C. Molecular epidemiology of integron-associated antibiotic resistance genes in clinical isolates of enterobacteriaceae. Microb Drug Resist. 1995;1:195–202. doi: 10.1089/mdr.1995.1.195. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Sandvang D, Aarestrup F M, Jensen L B. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella entericaTyphimurium DT104. FEMS Microbiol Lett. 1997;157:177–181. doi: 10.1111/j.1574-6968.1997.tb12770.x. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt A S, Bruun M S, Dalsgaard I, Pedersen K, Larsen J L. Occurrence of antimicrobial resistance in fish-pathogenic and environmental bacteria associated with four Danish rainbow trout farms. Appl Environ Microbiol. 2000;66:4908–4915. doi: 10.1128/aem.66.11.4908-4915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt A S, Bruun M S, Dalsgaard I, Larsen J L. Characterization of class 1 integrons associated with R-plasmids in clinical Aeromonas salmonicidaisolates from various geographical areas. J Antimicrob Chemother. 2001;47:735–743. doi: 10.1093/jac/47.6.735. [DOI] [PubMed] [Google Scholar]
- 37.Sengeløv G, Sørensen S J. Methods for detection of conjugative plasmid transfer in aquatic environments. Curr Microbiol. 1998;37:273–280. doi: 10.1007/s002849900378. [DOI] [PubMed] [Google Scholar]
- 38.Seward R J, Lambert T, Towner K J. Molecular epidemiology of aminoglycoside resistance in Acinetobacterspp. J Med Microbiol. 1998;47:455–462. doi: 10.1099/00222615-47-5-455. [DOI] [PubMed] [Google Scholar]
- 39.Shotts E B J, Vanderwork V L, Campbell L M. Occurrence of R factors associated with Aeromonas hydrophilaisolates from aquarium fish and waters. J Fish Res Board Can. 1976;33:736–40. [Google Scholar]
- 40.Son R, Rusul G, Sahilah A M, Zainuri A, Raha A R, Salmah I. Antibiotic resistance and plasmid profile of Aeromonas hydrophila isolates from cultured fish, Telapia (Telapia mossambica) Lett Appl Microbiol. 1997;24:479–482. doi: 10.1046/j.1472-765x.1997.00156.x. [DOI] [PubMed] [Google Scholar]
- 41.Sørum H. Mobile drug resistance genes among fish bacteria. APMIS. 1998;84(Suppl.):74–76. doi: 10.1111/j.1600-0463.1998.tb05652.x. [DOI] [PubMed] [Google Scholar]
- 42.Spanggaard B, Jørgensen F, Gram L, Huss H H. Antibiotic resistance in bacteria isolated from three freshwater fish farms and an unpolluted stream in Denmark. Aquaculture. 1993;115:195–207. [Google Scholar]
- 43.Sundstrøm L. The potential of integrons and connected programmed rearrangements for mediating horizontal gene transfer. APMIS. 1998;84(Suppl.):37–42. doi: 10.1111/j.1600-0463.1998.tb05646.x. [DOI] [PubMed] [Google Scholar]
- 44.Tschaepe H, Tietze E, Koch C. Characterization of conjugative R plasmids belonging to the new incompatibility group, IncU. J Gen Microbiol. 1981;127:155–160. doi: 10.1099/00221287-127-1-155. [DOI] [PubMed] [Google Scholar]