Abstract
Protein-carbohydrate interactions play significant roles in a wide variety of biological systems. Glycan microarrays are commonly utilized to interrogate the selectivity, sensitivity, and breadth of these complex protein-carbohydrate interactions. During the past two decades, numerous distinct glycan microarray platforms have been developed, each assembled from a variety of slide-surface chemistries, glycan-attachment chemistries, glycan presentations, linkers, and glycan densities. Comparative analyses of glycan microarray data have shown that while many protein-carbohydrate interactions behave predictably across microarrays, there are instances when various array formats produce different results. For optimal construction and use of this technology, it is important to understand sources of variances across array platforms. In this study, we performed a systematic comparison of microarray data from 8 lectins across a range of concentrations on the CFG and neoglycoprotein array platforms. While there was good general agreement on the binding specificity of the lectins on the two arrays, there were some cases of large discrepancies. Differences in glycan density and linker composition contributed significantly to variability. The results provide insights for interpreting microarray data and designing future glycan microarrays.
Table of Contents Graphic
Glycan density and linker composition significantly influence binding on glycan microarrays.
Introduction
Glycan-binding proteins are involved in numerous biological processes and are used extensively in basic and clinical research. For example, carbohydrate-binding antibodies and certain lectins are abundant in serum and are an important component of our immune defense system. Carbohydrate-binding monoclonal antibodies and purified plant lectins are used extensively to evaluate carbohydrate expression for both research and diagnostic purposes. In addition, several are in clinical trials for treating diseases. Antibody 3F8 is in clinical trials for the treatment of neuroblastoma and Ch14.18, marketed as Unituxin by United Therapeutics, has been FDA approved for children with high-risk neuroblastoma.1, 2
Information about binding affinity and specificity are critical for understanding the biological roles of glycan-binding proteins, using antibodies and lectins as reagents, developing agonists/antagonists, and pursuing clinical applications. Glycan microarrays provide a high-throughput format to rapidly evaluate binding of glycan-binding proteins to numerous potential glycan ligands in parallel while using only minimal amounts of samples.3–9 Many groups have developed distinct glycan array platforms that use different slide surface chemistry, linkers, immobilization strategies, and/or glycan presentation formats. Several recent studies have compared binding profiles on different glycan array platforms and glycan presentation formats.10–13 While these studies found general agreement across platforms, there were notable differences. To improve this technology and better understand how to interpret glycan array results, it is important to elucidate the factors that contribute to variability between array platforms. In this study, we investigate factors that give rise to differences in binding profiles on glycan microarrays.
Materials and Methods
Lectins.
The following biotinylated lectins were used in this study: Concanavalin A (ConA, Cat# BA-1104–5), Peanut agglutinin (PNA, Cat# BA-2301–1), Ricinus communis agglutinin I (RCA-I, Cat# BA-2001–5), soybean agglutinin (SBA, Cat# 280828–1), and wheat germ agglutinin (WGA, Cat# BA-2101–5) were obtained from EY Labs (San Mateo, CA). Maackia amurensis lectin I (MAL-I, Cat # B1315) and Sambucus nigra Lectin (SNA, Cat# B-1305) were obtained from Vector Laboratories (Burlingame, CA). Helix pomatia agglutinin (HPA, Cat# L6512) was obtained from Sigma-Aldrich (Burlington, MA).
CFG array data.
Data for 5 of the lectins (ConA, HPA, Mal-I, SNA, and WGA) were reported previously.10 Data for the other 3 lectins (PNA, RCA, SBA) were collected at the same time using the same protocols and materials. The data were obtained from the Consortium for Functional Glycomics website (CFG; http://www.functionalglycomics.org/). Detailed experimental procedures for the synthesis of NHS-reactive glycans, fabrication of the microarray, assaying the lectins, and analyzing the data have been reported previously.14
The CFG array is composed of glycans attached to a linker with a terminal amine printed on NHS-activated glass slides containing a hydrogel surface (Nexterion Hydrogel, Schott). The assay details for these data were reported previously.10 Briefly, slides were incubated with freshly diluted biotinylated-lectin in binding buffer (20mM Tris-HCl, pH 7.4 150mM NaCl, 2mM CaCl2, 2mM MgCl2 + 0.05% Tween-20 + 1% BSA) for 1 hour at RT. The slide was washed and then incubated with AlexaFluor®−488 labeled streptavidin for 1 hour in the dark. The microarray slides were extensively washed, dried by centrifugation, imaged using a ProScan Array Scanner (Perkin Elmer), and then processed using Imagene software.
Neoglycoprotein microarray data.
Data for 5 of the lectins (ConA, HPA, Mal-I, SNA, and WGA) were reported previously.10 Data for the other 3 lectins (PNA, RCA, SBA) were collected at the same time using the same batch of slides, assay conditions, and materials. Detailed experimental procedures for the fabrication of the microarray, and the assay and data analysis have been reported previously.15, 16
Our neoglycoprotein microarray is composed of glycans and glycopeptides chemically conjugated to either bovine serum albumin (BSA, A3059, Sigma-Aldrich, St. Louis, MO) or human serum albumin (HSA, A8763, Sigma-Aldrich, St. Louis, MO) to produce neoglycoproteins. The average number of glycans conjugated per molecule of albumin was determined by MALDI-TOF MS. Full details of the preparation and characterization of the neoglycoproteins has been previously published.17 Neoglycoproteins and other glycoproteins are printed onto epoxide-coated glass slides (SuperEpoxy2, ArrayIt, Sunnyvale, CA) to generate microarrays. The assay details for these data were reported previously.10 Briefly, slides were blocked with BSA overnight at 4°C. Next, the were incubated with various concentrations of each biotinylated lectin in binding buffer (20 mM Tris-HCl, 150 mM NaCl, 10 mM CaCl2, 10 mM MgCl2 pH = 7.4 containing 0.05% Tween 20 and 0.1% BSA) for 1 hour at room temperature. After 3 washes, slides were incubated with Cy3-labeled streptavidin for 1 hour in the dark with gentle shaking. The microarray slides were extensively washed, dried by centrifugation, imaged using a GenePix 4000A scanner (Molecular Devices Corporation, Sunnyvale, CA) and analyzed with GenePix Pro 7.0 software (Molecular Devices Corporation, Sunnyvale, CA).
Results
Overview of data used in the study
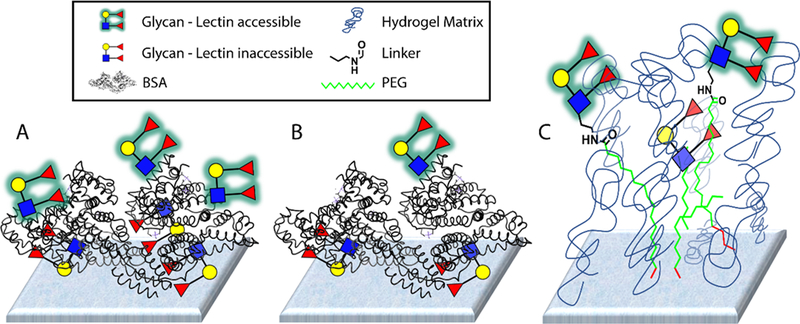
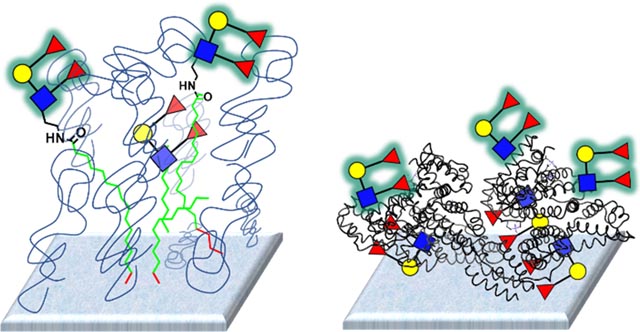
For this study, we used data from 8 different lectins profiled on two different glycan microarray platforms. Our microarray18–20 was composed primarily of neoglycoproteins (NGPs),21 along with about 15 natural glycoproteins, and contained a total of 332 array components. To produce the neoglycoproteins, glycans and glycopeptides were attached via various linkers to bovine serum albumin or human serum albumin.17 In many cases, we conjugated glycans or glycopeptides at a high ratio and a low ratio to produce both high and low-density conjugates. The number after the name represents the average number of determinants (i.e. glycans or glycopeptides) attached per molecule of albumin (e.g. Man-α 05 is a BSA conjugate with an average of 5 molecules of mannose per molecule of BSA). Neoglycoproteins and natural glycoproteins were then printed on epoxide-coated glass microscope slides. The Consortium for Functional Glycomics (CFG) array version 5.0 was composed of 611 glycans that contain an amine terminal linker attached to the reducing end.14 The glycans were printed on an NHS-ester activated glass slide surface, which allows for covalent attachment via amide bond formation. The slides contain a hydrogel coating on the surface. A schematic of the two slide surfaces is shown in Figure 1. Both arrays contained a diverse collection of N-linked glycans, O-linked glycans, glycolipid glycans, and other glycans.
Figure 1. Representations of the two glycan microarray surfaces.
A) Depiction of the neoglycoprotein microarray with a high density of glycans. B) Depiction of the neoglycoprotein microarray with a low density of glycans. C) Depiction of the CFG microarray with glycans coupled to an NHS-hydrogel coated slide. For both formats, some glycans might not be accessible for lectin binding.
The 8 lectins included in this study were ConA, RCA, HPA, Mal-I, PNA, SBA, SNA, and WGA. The 8 lectins have varying specificities, which provides comparisons across a diverse range of glycans. Data for 5 of the lectins were published as part of an international glycan microarray comparison.10 Data for the other 3 lectins (PNA, SBA, RCA) were added for this study. Both array groups received lectin aliquots from the same parent lectin stocks, along with aliquots of BSA for use in blocking buffers. Each group incubated the lectins at room temperature in the same buffer with added calcium and magnesium. Incubation times were identical. All other parameters, such as washes, scanning, and processing were at the discretion of the array group. Finally, each of the lectins was profiled at multiple concentrations, which provides more dynamic range and accounts for saturation of binding. Collectively, the study includes a large dataset for analysis.
Analysis of consistency for each array platform
The first step for understanding differences between microarray binding profiles was identifying key differences. We wanted to focus on differences that were (1) large, (2) observed with consistency, and (3) substantially beyond what one would expect based on the inherent variability of the assay. To facilitate analysis of the data, we implemented a floor value of 100 RFU. For both arrays, there is a significant amount of noise for signals below 100, and we did not want to focus on large differences arising from very low signals (e.g. 1 vs 100 RFU). In addition, we log transformed the data using base 2 to simplify comparisons and avoid focusing on differences that have a large RFU value but a small ratio (e.g. 50,000 vs 30,000 RFU gives a difference = 20,000 but the ratio is less than 2-fold; alternatively, 5000 vs 500 RFU has a difference of only 4500 but a ratio of 10-fold).
We started by evaluating the variability of each microarray platform. Ideally, we would compare replicate measurements for each lectin at the same concentration; however, this data was not available. Therefore, we compared datasets for lectins at the nearest possible lectin concentrations. The data for all lectins was grouped into one large analysis, and then Pearson correlation constants were determined. For the CFG array, this included comparisons of 100 μg/mL vs 50 μg/mL, 50 μg/mL vs 10 μg/mL, 10 μg/mL vs 1 μg/mL, 1 μg/mL vs 0.5 μg/mL, and 0.5 μg/mL vs 0.1 μg/mL. Across all 8 lectins, the Pearson correlation constants ranged from 0.80 to 0.98. For our NGP array, we included comparisons of 50 μg/mL vs 10 μg/mL, 10 μg/mL vs 1 μg/mL, 1 μg/mL vs 0.3 μg/mL, and 0.3 μg/mL vs 0.1 μg/mL. Across all 8 lectins, the Pearson correlation constants ranged from 0.85 to 0.96. Overall, both arrays demonstrated a very high degree of consistency.
Next, we identified instances where there was a large difference in signal from one lectin concentration to another for a given array component. Differences in the range of 2 to 10-fold were expected since the comparisons involved 2 to 10-fold differences in lectin concentration. Nevertheless, 20-fold differences only occurred about 1.2% of the time for the CFG array and 1.5% of the time for the NGP array. Differences of greater than 32-fold occurred about 0.5% of the time for both arrays. Differences of greater than 50-fold occurred 0.10% of the time for our array and 0.17% of the time for the CFG array. Differences in lectin concentration likely contribute to these totals. For example, the vast majority of cases where a difference of greater than 50-fold was observed were for comparisons of data points at 10 μg/mL vs 1 μg/mL. Since a 10-fold difference in lectin concentration could easily give rise to a 10-fold difference in signal, a rate of 0.1–0.2% for 50-fold differences was considered very small. Also, one would expect even higher levels of consistency for replicate experiments carried out using the same lectin concentrations. Overall, these results demonstrate a very high degree of consistency for the two array platforms.
Identification of substantial differences between arrays
It is well appreciated that differences in glycan structure can give rise to large differences in lectin binding. Our goal for this study was to evaluate factors other than glycan structure. Therefore, we decided to limit the array-to array comparison to the subset of components that have identical glycans on both array platform. Differences in linker were allowed. This group included 177 comparisons for each lectin, totaling 1416 comparisons across all 8 lectins. The comparisons were carried out for identical or nearly identical lectin concentrations. These data included 6 comparisons: NGP at 50 μg/mL vs CFG at 100 μg/mL, NGP at 50 μg/mL vs CFG at 50 μg/mL, NGP at 10 μg/mL vs CFG at 10 μg/mL, NGP at 1 μg/mL vs CFG at 1 μg/mL, NGP at 0.3 μg/mL vs CFG at 0.5 μg/mL, and NGP at 0.1 μg/mL vs CFG at 0.1 μg/mL. Collectively, a total of 6018 array-to-array comparisons were considered (note: not all lectins were assayed at all concentrations; for example, only 3 lectins were assayed at 50 μg/mL on the CFG array; therefore, the total is not simply 6 × 1416).
Based on our analysis of within-array consistency above, we decided to focus on instances where signals for the same glycan and the same lectin varied by more than 50-fold from one array to the other. Differences of this size would occur very infrequently as a result of inherent variabilities in the assays. In addition, a high cutoff would allow us to avoid focusing on small variations resulting from differences in assay conditions or scanner settings/sensitivity. Finally, 50-fold or greater differences seemed substantial from a general molecular recognition perspective. In addition to the cutoff of 50-fold, we also wanted to focus on cases where the differences were observed consistently. Therefore, we only selected instances where there was a 50-fold or greater difference at one lectin concentration and a 10-fold or greater difference for the same glycan and lectin at one of the other lectin concentrations.
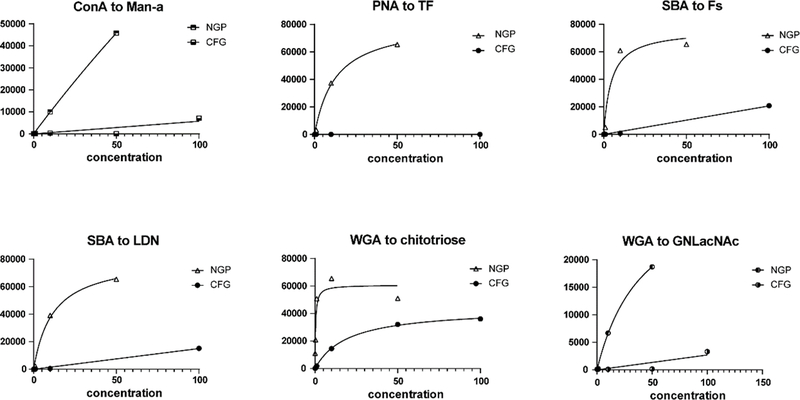
Using these criteria, we identified 101 instances (out of 6018 possible comparisons; ~1.7%) where there was a substantial difference between the arrays (see Figure 2 and Supporting Excel File, sheet “CFG v NGP top 101 differences”). The percentage of instances is much higher than what would be expected based on inherent variability of the assays. In addition, all 101 involved cases where the signal on our neoglycoprotein array was larger than the corresponding signal on the CFG array. Thus, we concluded that these instances are likely real differences between the array platforms and not a result of random variability.
Figure 2. Examples of discrepancies between the CFG and neoglycoprotein (NGP) array.
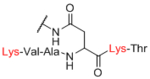
Data are shown for 6 of the 101 instances of large discrepancies between the CFG and NGP array. Data are graphed over all measured concentrations of lectin, with lectin concentration on the x-axis and RFU on the y-axis. Abbreviations are as follows: Man-a (CFG = #3, mannose alpha linked to Sp8; NGP = #275, low density mannose alpha linked to BSA, ~5/BSA), TF [CFG = #141, Galβ1–3GalNAcα-Sp14 (threonine); NGP = #109, Ac-Ser-(Galβ1–3GalNAcα)Ser-Ser-Gly-Hex-BSA, ~16/BSA], Fs (Forssman disaccharide; CFG = #92, GalNAcα1–3GalNAcβ-Sp8; NGP = #108, GalNAcα1–3GalNAcβ-BSA), LDN (LacDiNAc; #100, GalNAcβ1–4GlcNAcβ-Sp8; NGP = #234, GalNAcβ1–4GlcNAcβ-BSA, ~16/BSA), Chitotriose (CFG = #192, GlcNAcβ1–4GlcNAcβ1–4GlcNAcβ-Sp8; NGP = #129, GlcNAcβ1–4GlcNAcβ1–4GlcNAcβ-BSA, ~20/BSA), GNLacNAc (CFG = #183, GlcNAcβ1–3Galβ1–4GlcNAcβ-Sp0; NGP = #255, GlcNAcβ1–3Galβ1–4GlcNAcβ-BSA).
The 101 instances involved some redundancy. When comparing the CFG data to the neoglycoprotein data, there were many cases where there were 2–3 densities of a glycan on the neoglycoprotein array. In these cases, the CFG glycan was compared to each density on the neoglycoprotein separately (e.g. Man-a-Sp8 on the CFG array was compared to Man-α at 5/BSA and 20/BSA on the neoglycoprotein array). In addition, there were cases where a particular glycan was attached to the surface via two different linkers. These situations were also counted as separate comparisons (e.g. GlcNAcβ-Sp0 and GlcNAcβ-Sp8 on the CFG array were both compared to GlcNAcβ-BSA on the neoglycoprotein array). If we only consider the glycan portion, the number of large discrepancies decreased from 101 to 62.
Factors contributing to differences between arrays
There are a variety of factors that could give rise to differences between arrays. Differences in linkers, glycan presentation, and slide surface are likely to have the biggest effects. From an initial qualitative assessment of the 101 cases, both the linker and glycan density appeared to be important factors. Both will be discussed in more detail below in separate sections. In addition, other factors could also influence the binding profiles.
Variability in the experimental conditions is one consideration. As mentioned above, many of the experimental parameters (e.g. batch of lectin, blocking agent, buffer, incubation temperature, incubation times) were held constant between the arrays. Some parameters were different. The washes were similar but not identical. We washed 3 times after the primary incubation and 7 times after the secondary incubation. The CFG washed 8 times after the primary incubation and 12 times after the secondary incubation; 4 of the final washes are with Milli-Q H20 instead of buffer. In our experience, these differences in washes are not expected to have substantial effects. A second difference is the amount of BSA used in the incubation buffers. We used 0.1% while the CFG used 1%. This difference could potentially contribute to the differences in signals, but it was unclear how large the effect would be. Lastly, incubations on the neoglycoprotein microarray contained 10 mM CaCl2 and 10 mM MgCl2, whereas the CFG incubations included 2 mM CaCl2 and 2 mM MgCl2.
In addition to the experimental conditions described above, two other factors could lead to systematic differences: image acquisition/analysis and secondary reagent. For the secondary reagent, we used Cy3-labeled streptavidin and the CFG used AlexaFluor488-labled streptavidin. Different fluorophores can potentially produce different signal intensities, but the relative signal strengths for various glycans would be the same. Additionally, differences in scanner settings/sensitivity would produce systematic variations in the two datasets. If the binding profiles were perfectly correlated but one scanner was more sensitive that the other, one would expect a perfect trendline but with a slope skewed from 1.0. We plotted our data vs the CFG data for each of the identical or nearly identical lectin concentrations mentioned above and then determined the slopes of the trendlines. The average slope of the trendlines was 0.50 (ranging from 0.39 – 0.63). Since our data was plotted on the x-axis, a slope of 0.5 indicates that our signals were on average 2-fold higher than the CFG signals. Thus, we concluded that differences in the scanner sensitivity/settings and/or fluorophore on the secondary reagent produce a systematic variability between the arrays by a factor of about 2. Since 2-fold is relatively small, we did not re-scale or normalize the data to compensate for the difference.
The nature of the lectin could also affect binding profiles measured on arrays. When a lectin binds a glycan on an array surface, there can be a variety of additional interactions, such as interactions with the linker, adjacent molecules of lectin, and the slide surface. Various features of the lectin can influence these additional interactions, such as variations in binding pocket depth (shallow versus deep binding pockets), overall net charge, localized surface charge and/or hydrophobicity around the binding pocket, spacing of binding pockets, and steric accessibility of binding pockets. In our comparison, the 101 instances were not distributed evenly among the lectins. Forty of the 101 cases were signals derived from WGA, and 26 were signals derived from SBA. This over-representation was not simply due to having more abundant strong binding partners for WGA and SBA on the arrays. HPA had a similar number of strong binders, but only 9 of the 101 cases were from HPA data. The variation from one lectin to another highlights the importance of including many lectins/proteins in a cross-platform microarray comparison.
Effects of glycan density
Glycan density can have a large effect on recognition. Monovalent interactions between a glycan and a single lectin binding site are typically weak. Lectins and antibodies achieve tight binding through formation of multivalent complexes. The spacing of glycans on a surface can have a big effect on the ability to form a multivalent interaction. We have observed very large density effects for other lectins and monoclonal antibodies previously on our array.22–25 In this study, 76 of the 101 instances of large differences between the arrays involved comparisons between the CFG array and a high-density glycan on the neoglycoprotein array, indicating that density is a key factor.
To isolate the effects of density, we focused our analysis on just data from our neoglycoprotein microarray. By comparing data from the same array experiments, all other parameters (e.g. incubation times, temperatures, buffers, slide surface, and linker) were identical. Our array has 73 components that are present at both high and low densities. Neoglycoproteins with <8 determinants per molecule of albumin (median = 5) were considered low density. High-density was defined as an average of >10 determinants per molecule of albumin (median = 16). Based on a simplified model of the array surface, 5/BSA would give an estimated spacing of roughly 80 Å from attachment point to attachment point, while 16/BSA would correspond to roughly 40 Å spacing (see Supporting Information). These estimates do not include the linker. We compared binding properties for these 73 components across all 8 lectins giving 584 data points at each of five lectin concentrations.
Our initial assessment involved determining Pearson correlation constants (R) for low-density versus high-density signals at each lectin concentration. The R values ranged from 0.84–0.90 indicating a high degree of correlation between low and high-density glycans. The correlations were slightly better if we compared low-density conjugates at a higher lectin concentration to high-density conjugates at a lower lectin concentration (R values ranged from 0.89–0.92). For example, with both datasets at 1 μg/mL, the R value for low versus high-density conjugates was 0.84. The correlation improved to 0.90 when comparing the low-density conjugates at 1 μg/mL to the high-density conjugates at 0.3 μg/mL.
While the overall correlation was high, there were a number of instances where there were large differences between a high and low-density pair. A summary of these differences is shown in Table 1. Ten-fold differences were observed 61 times at a lectin concentration of 50 μg/mL and 48 times at a lectin concentration of 10 μg/mL. For the vast majority of these cases (>96%), the signal for the high-density conjugate was larger than the low-density. There were 13–15 instances where there was a >50-fold difference in signal. All of the >50-fold differences favored the high-density conjugate and typically involved a strong positive signal at high-density and little or no signal at low-density (e.g. 180 RFU for low-density vs 21,491 RFU at high-density). Thus, differences in glycan density can frequently give rise to 10-fold differences in signal and occasionally result in greater than 50-fold differences. We considered the possibility that larger high-density to low-density ratios for a given glycan pair (20/BSA vs 4/BSA has a ratio of 5, whereas 10/BSA vs 5/BSA has a ratio of 2) could produce larger differences in signals; however, instances of 10-fold differences were not correlated with the high-vs-low ratio. Instances of large density effects were not distributed evenly among the 8 lectins. They were most frequently observed with SBA (31% of large differences were SBA binding).
Table 1.
Summary of large differences between high and low-density glycans
| 10-fold differences | 20-fold differences | 50-fold differences | |
|---|---|---|---|
| 50 μg/mL | 61 | 37 | 15 |
| 10 μg/mL | 48 | 27 | 13 |
| 1 μg/mL | 28 | 15 | 1 |
| 0.3 μg/mL | 8 | 3 | 0 |
| 0.1 μg/mL | 7 | 4 | 0 |
We next compared the CFG data to our low and high-density conjugates separately by determining Pearson correlation constants for each of the identical or nearly identical lectin concentrations (Table 2). For this analysis, we only included array components where the glycan was identical on both arrays. The CFG data correlated much better to our low-density conjugates. For example, at a concentration of 10 μg/mL the Pearson R value was 0.77 for the CFG/low-density comparison but only 0.55 for the CFG/high-density comparison. The analysis suggests that the surface density of glycans on the CFG array is similar to our low-density spots.
Table 2.
Comparison of R values for CFG data vs high or low-density neoglycoproteins
| NGP concentration | CFG concentration | R value for high-density | R value for low-density |
|---|---|---|---|
| 50 μg/mL | 100 μg/mL | 0.58 | 0.80 |
| 50 μg/mL | 50 μg/mL | 0.44 | 0.64 |
| 10 μg/mL | 10 μg/mL | 0.55 | 0.77 |
| 1 μg/mL | 1 μg/mL | 0.75 | 0.87 |
| 0.3 μg/mL | 0.5 μg/mL | 0.81 | 0.88 |
| 0.1 μg/mL | 0.1 μg/mL | 0.65 | 0.85 |
Effects of the linker
Another feature that could potentially have a major impact on differences between array binding profiles is the linker connecting the glycan to the array surface. In fact, 71 of the 101 instances of large differences discussed above involved comparisons with an identical glycan but a different linker. To isolate the effects of linkers, we focused our analysis on data from the CFG microarray. Since the data are all from the same microarrays and experiments, all other parameters (e.g. incubation times, temperatures, buffers, slide surface) were identical. To initiate this analysis, we identified glycans bearing two or more linkers on the CFG array. From this group of 125 array components, there were 57 pairwise comparisons from glycans bearing two linkers. In addition, there were two cases where a glycan was present with 3 different linkers giving rise to 6 pairwise comparisons, and one case where a glycan was present with 4 linkers yielding 6 pairwise comparisons. This combined group of 69 pairwise comparisons was evaluated across 8 lectins at 3–6 concentrations ranging from 0.1 μg/ml to 100 μg/ml, producing a large dataset containing 2484 comparisons. As with previous datasets, we implemented a floor value of 100 RFU and log transformed the data using base 2.
We began by evaluating all pairwise comparisons across the entire dataset. The 2484 comparisons were highly correlated, with a Pearson correlation constant R value of 0.90. Since signals can vary more when there is higher background (e.g. higher lectin concentrations) or low signal strength (e.g. lower lectin concentrations), R values were determined for each lectin concentration separately (Table 3). The correlation constants ranged from 0.88 to 0.97, indicating the lectin concentration does not have a significant effect. In addition to evaluating overall correlations between linkers, we also identified specific cases where there were large differences in signals (Table 4). Ten-fold differences were observed 42 times for this dataset (a rate of 1.69%) and 50-fold differences were observed 10 times (rate =0.4%). The majority (79%) of >10-fold differences occurred in the top 3 lectin concentration groups (100, 50, and 10 μg/ml), presumably because there are more signals and the signals are larger. Taken together, the results show that binding to glycans on different linkers are generally well correlated, but differences in the linker can give rise to 10-fold differences in signal and occasionally produce greater than 50-fold differences.
Table 3.
Pearson correlation constants (R) for glycans with different linkers
| All Lectins | ConA | WGA | SNA | MAL-I | HPA | PNA | RCA | SBA | |
|---|---|---|---|---|---|---|---|---|---|
| All Conc. | 0.90 | 0.95 | 0.77 | 0.93 | 0.92 | 0.92 | 0.66 | 0.85 | 0.63 |
| 100 μg/ml | 0.88 | 0.95 | 0.72 | 0.96 | 0.90 | 0.88 | 0.70 | NA | 0.67 |
| 50 μg/ml | 0.93 | 0.95 | 0.82 | 0.93 | NA | NA | NA | NA | NA |
| 10 μg/ml | 0.88 | 0.94 | 0.86 | 0.77 | 0.93 | 0.91 | 0.27 | 0.88 | 0.48 |
| 1 μg/ml | 0.91 | 0.99 | 0.52 | 0.99 | 0.97 | 0.99 | ND | 0.80 | ND |
| 0.5 μg/ml | 0.97 | 0.93 | ND | 1.00 | NA | NA | NA | NA | NA |
| 0.1 μg/ml | 0.90 | 0.83 | ND | 0.99 | NA | 0.98 | ND | 0.76 | ND |
NA= No data available for analysis. ND= not determined, too few values were above the floor for the given lectin/conc.
Table 4.
Pearson correlations constants for pairwise linker comparisons
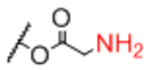
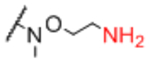
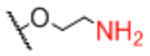
| Linker A | Linker A Structure | Linker B | Linker B Structure | R |
|---|---|---|---|---|
| Sp12 |
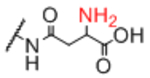
|
Sp13 |
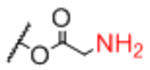
|
0.99 |
| Sp0 |
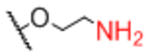
|
Sp9 |
|
0.98 |
| Sp18 |
|
Sp8 |
|
0.98 |
| Sp13 |
|
Sp24 |
|
0.97 |
| Sp13 |
|
Sp21 |
|
0.97 |
| Sp0 |
|
Sp23 |
|
0.96 |
| Sp23 |
|
Sp8 |
|
0.96 |
| Sp10 |
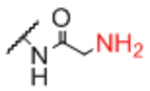
|
Sp12 |
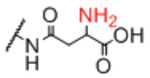
|
0.95 |
| Sp12 |
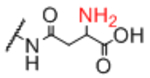
|
Sp21 |
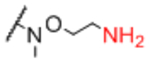
|
0.95 |
| Sp0 |
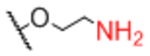
|
Sp8 |
|
0.93 |
| Sp12 |
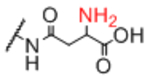
|
Sp24 |
|
0.91 |
| Sp21 |
|
Sp24 |
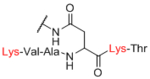
|
0.90 |
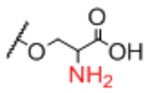
| Sp15 |
|
Sp8 |
|
0.83 |
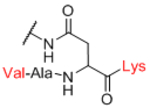
| Sp12 |
|
Sp25 |
|
0.83 |
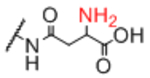
| Sp14 |
|
Sp8 |
|
0.60 |
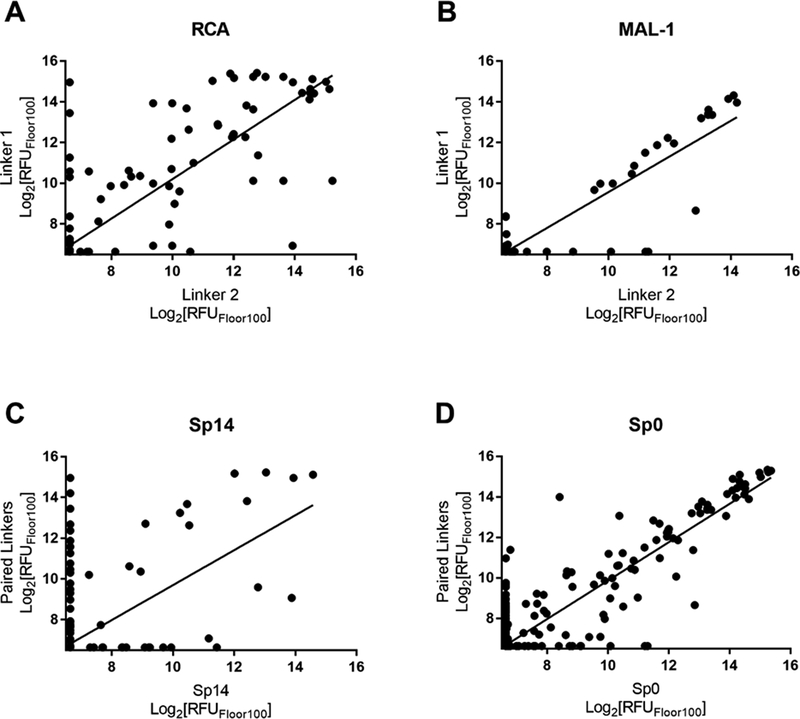
We next evaluated how differences in the linker affected each lectin. We examined the 69 pairwise comparisons against each individual lectin and found a range of R values of 0.63–0.97 (Table 1). PNA and SBA gave the lowest R values of 0.66 and 0.63, respectively. R values obtained for each lectin with concentration dependence (Table 3) showed moderate to good agreement across the dataset, indicating that lectin concentration is not a major source of large differences in this dataset. In addition to overall correlations, we also tabulated specific cases of large differences. For most lectins, the rate of a 10-fold difference was below 2%, but the rate for RCA was 6.3%. The higher rate for RCA was more pronounced at lower concentrations. RCA accounted for 8 of 9 total occurrences in the lowest three concentrations. Although the overall data for RCA were highly correlated (R =0.85), there was considerable scatter about the trendline (see Figure 3A versus 3B). Taken together, RCA appears to be more sensitive to effects of the linker than the other lectins in our study.
Figure 3. Effects of linkers.
Data shown for all linker pairwise comparisons with RCA (A) and MAL-1 (B). RCA linker pairwise comparisons are broadly distributed when compared to linker pairwise comparisons with MAL-1. Data are graphed over all measured concentrations of lectin from each of the 69 pairwise comparisons. Data shown for all pairwise comparisons of the poorly correlated linker Sp14 (C; R=0.60) as compared to well correlated linker Sp0 (D; R=0.93). Data are graphed over all measured concentrations of all lectins.
Next, we evaluated effects of specific linkers. We determined correlations for all pairwise comparison that involved the same linker pair (e.g. all pairwise comparisons of Sp0 to Sp8; Table 4). There are 20 linker-paired comparisons, but only 15 had sufficient data to determine an R value. The R values in this analysis ranged from 0.60–0.99 (Table 4). Examination of R values for both individual linkers and paired-linkers identified Sp14 as a statistically significant source of difference on the CFG array (compare Figure 3C vs 3D). The worst correlations were for Sp14 versus Sp8 with an R value of 0.60. In addition to general correlations, we also examined specific cases of large differences. The rate of 10-fold differences for individual linkers ranged from 0.00% - 5.56% (Table 5). Amino acids Sp14 and Sp15 stand out as linkers contributing to sources of large differences (2.98%−5.56%). Additionally, the linker Sp21 had one of the highest rates of large difference at 5.56%. Interestingly, all the large differences associated with Sp21 were within the RCA dataset and in all cases Sp21 gave a larger signal than the matched peptide or amino acid linkers (Sp24, Sp12, and Sp13). Sp24 and Sp25 also had large differences of 4.8% and 4.2%, respectively. Not surprisingly, the specific comparisons of Sp14 vs. Sp8 were a prominent source of large differences with a rate of 3.2% (Table S2). Comparisons of Sp15 vs. Sp8 also had a high rate (5.6%). Sp15 (serine), and Sp14 (threonine) both have a free carboxylic acid on the linker which could have negative interactions with the lectins. Overall, the results suggest that the peptide portion of glycopeptides substantially influence recognition of the glycan portion on the array.
Table 5.
Large differences for pairwise comparisons across lectins
| Lectin | 10-fold differences (rate) | 50-fold differences (rate) |
|---|---|---|
| ConA | 4 (0.97%) | 1 (0.24%) |
| WGA | 7 (1.69%) | 3 (0.72%) |
| SNA | 5 (1.21%) | none |
| MAL-I | 4 (1.93%) | none |
| HPA | 4 (1.45%) | none |
| PNA | 3 (1.09%) | 2 (0.72%) |
| RCA | 13 (6.28%) | 3 (1.45%) |
| SBA | 2 (0.72%) | 1 (0.36%) |
| All Lectins | 42 (1.69%) | 10 (0.40%) |
Estimation of other effects
From the results above, it was apparent that glycan density and linker structure can have major effects on glycan microarray data. It was not known how much other factors, such as slide surface and wash conditions, affect the data. To estimate the effects of other factors, we focused on comparing data obtained on the two arrays for the subgroup of array components with identical glycans and the fewest differences in linker composition and glycan density. Therefore, we limited this part of the analysis to 42 glycans we had obtained from the CFG. For these array components, the glycan portion and the core portion of the linker (O-CH2-CH2-N) were identical; however, the conjugation chemistry used to attach the linker to the surface or to albumin was different. Therefore, the stereochemistry of the glycosidic linkage and the first few atoms of the linker are identical. Moreover, the composition of the glycan and any impurities would be the same. We also limited this part of the study to our conjugates with low density, to best match the estimated density on the CFG array surface.
Of the 101 instances of large differences between our array and the CFG, only 10 came from the subgroup containing the 42 CFG glycans at low density. Thus, the vast majority of large discrepancies between arrays likely derive from differences in linkers and glycan density. Furthermore, 9 of the 10 involved binding by WGA, suggesting that certain lectins are much more sensitive to factors such as the slide surface, wash conditions, CaCl2/MgCl2 concentrations, and/or amount of BSA in the incubation buffer.
Discussion
Glycan microarrays have emerged as the primary tool used to evaluate binding properties of lectins and carbohydrate-binding antibodies.3–9 In addition, they are frequently being used to study immune responses and identify biomarkers for various diseases.26, 27 The experiments are rapid and provide considerable information about relationships between structure and recognition. There are a variety of microarray platforms that have been developed, such as glycans on hydrogel modified surfaces,14 neoglycolipid microarrays,5 neoglycoprotein microarrays,20 dendrimer microarrays,28–35 glycopolymer microarrays.36–38 microbial/pathogen glycan microarrays,39–42 natural/shotgun microarrays,43–47 liposome microarrays,48 surfactant vesicle arrays,49 nanoarrays,50 and bead arrays.51–54 These platforms use a variety of different chemistries to immobilize glycans, including non-covalent adsorption, amide bond formation, azide plus alkyne cycloaddition click chemistry, photoactivation with carbene/nitrene insertion, Michael additions, and epoxide opening. While different platforms provide good general agreement about binding properties of lectins, there can be key differences. Efforts to understand factors that give rise to these differences are important for interpreting data and optimal use of this technology. In addition, these efforts could lead to better microarray construction and improved assays.
There are several challenges when trying to understand factors that contribute to differences in microarray profiles. First, differences can arise from experimental issues, such as different sources of protein or different buffers. Second, the content on different arrays is often similar but not identical. For example, glycans on different arrays may vary by a single monosaccharide residue at the reducing end, such as Man9 attached to one GlcNAc residue versus Man9 attached to two GlcNAc residues. In addition, some molecules on different arrays will have identical glycans but different linkers. Features of presentation, such as glycan density, can also vary. Thus, it is difficult to determine if differences in binding profiles are due to the array platform, linker, glycan structure, and/or glycan density. Third, there is often insufficient data to observe general trends. For example, a given lectin or antibody may only bind a handful of glycans that are present on two different array platforms. Finally, there is some level of variability inherent in any assay. Thus, our first goal was to design a study that would address these challenges.
We selected two arrays for our study: our neoglycoprotein microarray and the CFG microarray. The CFG array is one of the largest and most widely used arrays in the world.14 Since its inception, there have been thousands of array experiments run on the CFG array, and the data are publicly accessible via their website, providing a tremendous resource for the community. Our neoglycoprotein microarray provides a distinct platform with unique presentation of glycans.18–20 Both arrays contain a diverse collection of glycans with representation from many different glycan families, such as N-linked and O-linked, glycolipid glycans, non-human glycans, and Lewis/blood group antigens.
We utilized several experimental design strategies to facilitate our investigation. First, we started with data and protocols from a recent international comparison of glycan microarray platforms.10 In that study, each array group received lectin aliquots from the same parent lectin stocks, along with aliquots of BSA for use in blocking buffers. Lectins were incubated for the same length of time at the same temperature in the same buffer. Salt concentrations were the same, and both groups used buffers supplemented with MgCl2 and CaCl2, albeit at somewhat difference concentrations. Therefore, most of the experimental conditions were identical or very similar. Second, we used a relatively large dataset in order to observe general trends. The data included 8 different lectins profiled on 2 different microarrays at 5–6 different concentrations producing over 36,000 data points. The lectins had differing specificities, allowing us to assess binding to different glycan structural families. In addition, we could evaluate effects from proteins with different architectures and amino acid compositions. To improve the overlap in glycan content between the array platforms, we obtained 42 glycans from the CFG. For these array components, the glycan portion and the core atoms of the linker were identical on both arrays.
While there was good agreement in the general binding properties of the lectins on different arrays, some significant differences were also observed. We focused on instances where the differences were large (>50-fold), consistent, and well beyond what one might expect from inherent variability of the assays. This group included 101 instances of sizeable differences from one array to the other. We then analyzed effects of various factors that could potential contribute to the differences.
One key element of recognition by lectins is glycan density.23–25, 55–69 We isolated the effects of glycan density by focusing analyses on data derived from our neoglycoprotein array where the same glycan was present at two different densities. By using this approach, all other factors (e.g. slide surface, linker composition, experiment conditions) were held constant. Ultimately, we found that differences in glycan density can frequently give rise to 10-fold differences in signal and sometimes produce greater than 50-fold differences. Therefore, glycan density can have a major effect on glycan microarray binding profiles, even within the same array platform.
We also compared the CFG data to our high and low-density components separately. The CFG data compared much better to our low-density conjugates. Therefore, we postulate that glycans on the surface of the CFG array are presented at low density, or at a density that is comparable to our low density. Of the 101 instances of big differences between array platforms, 76 involved comparing a CFG signal for a particular glycan to the corresponding high-density component on our array. Thus, variations in glycan density could account for a large proportion of the differences in binding profiles between our array and the CFG array.
At present, the optimal density or spacing of glycans on an array surface is not known. Our estimated spacings for glycans on our surface were ~40 Å for high density and ~80 Å for low density. These distances are on par with glycan spacings one might find in natural systems, such as glycans on a protein or cell surface. In addition, the binding sites of many lectins and antibodies are in the range of 30–100 Å apart. Thus, distances of 40–80 Å would allow formation of multivalent complexes for many natural glycan-binding proteins. That being said, the optimal density depends on the particular protein(s) being studied and likely varies for different studies. Therefore, variations in glycan density can be useful when construction glycan microarrays.
In addition to glycan density, the composition of the linker is also an important factor to consider.12, 13, 70–72 In our study, differences in linker could give rise to 10-fold variations in signal and occasionally 50-fold differences. Linkers with similar structure tended to produce very highly correlated signals. Linkers composed of amino acids had weaker correlations with other linkers and gave rise to higher rates of large discrepancies. The effects could be due to charge. Sp14 and Sp15 contain a free carboxylic acid in close proximity with the reducing end of the glycan. Interactions with this charged moiety could have a substantial effect on recognition. A prior study on binding to the CFG array also noted strong effects from linker Sp14 and suggested it was due to distinct conformational preferences.13 In that study, Sp14 was found to give more false negative results, and molecular modeling indicated that this linker orients glycans down towards the surface making them less accessible for recognition.13 Our results and prior results indicate that presentation of a glycan on a linear, flexible linker is quite different than presentation on a peptide. Thus, incorporation of glycopeptides on glycan microarrays can provide useful and distinct chemical diversity.
The optimal linker likely varies for different array platforms. Effects of linkers may differ depending on the surface composition and chemistry. For example, the CFG hydrogel arrays display glycans at the end of a long PEG chain, which may provide considerable flexibility and distance from the glass surface. Platforms where the linker attachment site is closer to the surface may have more pronounced effects. For example, short linkers that place the glycan in close proximity to a surface may hinder access by lectins and antibodies. Rigid linkers that limit conformational flexibility may also impede certain glycan-binding proteins.
When choosing a linker, the application is a key consideration. For establishing binding specificities or discovering new lectin, one may want a linker that is suitable for many glycan-binding proteins. For example, a long, flexible linker can allow glycans to move and adapt to accommodate different spacings, orientations, and steric environments of binding sites. In other cases, one may want glycan-linker pairs that can selectively capture lectins or glycan-binding antibodies. For example, when profiling lectins or antibodies from human serum, one needs to capture specific subpopulations of antibodies or specific lectins in the presence of a complex mixture of other glycan-binding proteins. Both the glycan and linker can contribute to selectivity. Arrays with variations in linkers could allow one to rapidly screen different glycan-linker pairings and identify ones that provide appropriate selectivity.
Overall, this study provides insights for interpreting glycan microarray data and designing better arrays. Both glycan density and the linker can have a substantial effect on recognition, a fact that is important to consider when analyzing both positive and negative signals. An absence of signal could easily arise if the glycan density is not well matched to the binding sites of the glycan-binding protein or the linker impedes recognition. When interpreting positive signals, glycans presented on a long flexible linker may provide different binding data than natural glycans. In nature, glycan determinants are often attached to a carrier glycan chain. For example, the blood group A determinant can be presented at the non-reducing termini of an N-linked glycan. Like other “linkers”, the carrier chain may hinder or prevent binding. Alternatively, a glycan determinant may be displayed on a cell membrane via connection to a lipid. The lipid linker and close proximity to the membrane may provide a very different local environment than a long flexible linker on an array surface. In addition to interpretation, our results provide insights for designing future arrays. For example, array constructions with variations in glycan density and linker composition can provide a unique opportunity to enhance diversity in a way that complements variations in glycan composition. Lastly, the results indicate that different glycan microarray platforms are useful for the community. The ability of different microarray formats to provide distinct presentation of glycans enhances our understanding of glycan recognition and affords complementary, rather than redundant, information. In the future, it may be advantageous to design and assemble microarrays that combine desirable features of various microarray platforms and presentation formats.
Supplementary Material
Acknowledgements
We thank the Consortium for Functional Glycomics (GM62116; The Scripps Research Institute), X. Huang (Michigan State University), Lai-Xi Wang (University of Maryland), and J. Barchi (National Cancer Institute) for generously contributing glycans for the array. This work was supported by the Intramural Research Program of the National Cancer Institute, NIH.
Footnotes
Conflicts of interest
There are no conflicts to declare.
References
- 1.Yu AL; Gilman AL; Ozkaynak MF; London WB; Kreissman SG; Chen HX; Smith M; Anderson B; Villablanca JG; Matthay KK; Shimada H; Grupp SA; Seeger R; Reynolds CP; Buxton A; Reisfeld RA; Gillies SD; Cohn SL; Maris JM; Sondel PM Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N. Engl. J. Med 2010, 363, 1324–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheung NKV; Cheung IY; Kushner BH; Ostrovnaya I; Chamberlain E; Kramer K; Modak S Murine anti-GD2 monoclonal antibody 3F8 combined with granulocyte- macrophage colony-stimulating factor and 13-cis-retinoic acid in high-risk patients with stage 4 neuroblastoma in first remission. J. Clin. Oncol 2012, 30, 3264–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geissner A; Seeberger PH Glycan Arrays: From Basic Biochemical Research to Bioanalytical and Biomedical Applications. Ann. Rev. Anal. Chem 2016, 9, 223–247. [DOI] [PubMed] [Google Scholar]
- 4.Song X; Heimburg-Molinaro J; Smith DF; Cummings RD Glycan microarrays of fluorescently-tagged natural glycans. Glycoconj. J 2015, 32, 465–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palma AS; Feizi T; Childs RA; Chai W; Liu Y The neoglycolipid (NGL)-based oligosaccharide microarray system poised to decipher the meta-glycome. Curr. Opin. Chem. Biol 2014, 18, 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park S; Gildersleeve JC; Blixt O; Shin I Carbohydrate Microarrays. Chem. Soc. Rev 2013, 42, 4310–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D Carbohydrate antigen microarrays. In Methods Mol Biol, 2012; Vol. 808, pp 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rillahan CD; Paulson JC Glycan microarrays for decoding the glycome. Annu. Rev. Biochem 2011, 80, 797–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyun JY; Pai J; Shin I The Glycan Microarray Story from Construction to Applications. Acc. Chem. Res 2017, 50, 1069–1078. [DOI] [PubMed] [Google Scholar]
- 10.Wang L; Cummings R; Smith D; Huflejt M; Campbell C; Gildersleeve JC; Gerlach JQ; Kilcoyne M; Joshi L; Serna S; Reichardt NC; Pera NP; Pieters R; Eng W; Mahal LK Cross-Platform Comparison of Glycan Microarray Formats. Glycobiology 2014, 24, 507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padler-Karavani V; Song X; Yu H; Hurtado-Ziola N; Huang S; Muthana S; Chokhawala HA; Cheng J; Verhagen A; Langereis MA; Kleene R; Schachner M; De Groot RJ; Lasanajak Y; Matsuda H; Schwab R; Chen X; Smith DF; Cummings RD; Varki A Cross-comparison of protein recognition of sialic acid diversity on two novel sialoglycan microarrays. J. Biol. Chem 2012, 287, 22593–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kilcoyne M; Gerlach JQ; Kane M; Joshi L Surface chemistry and linker effects on lectin-carbohydrate recognition for glycan microarrays. Analytical Methods 2012, 4, 2721–8. [Google Scholar]
- 13.Grant OC; Smith HM; Firsova D; Fadda E; Woods RJ Presentation, presentation, presentation! Molecular-level insight into linker effects on glycan array screening data. Glycobiology 2014, 24, 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blixt O; Head S; Mondala T; Scanlan C; Huflejt ME; Alvarez R; Bryan MC; Fazio F; Calarese D; Stevens J; Razi N; Stevens DJ; Skehel JJ; van Die I; Burton DR; Wilson IA; Cummings R; Bovin N; Wong CH; Paulson JC Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci 2004, 101, 17033–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell CT; Zhang Y; Gildersleeve JC Construction and use of glycan microarrays. Current Protocols in Chemical Biology 2010, 2, 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia L; Gildersleeve JC The Glycan Array Platform as a Tool to Identify Carbohydrate Antigens. Methods in Molecular Biology 2015, 1331, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y; Gildersleeve JC General Procedure for the Synthesis of Neoglycoproteins and Immobilization on Epoxide-Modified Glass Slides. In Carbohydrate Microarrays: Methods and Protocols, Chevolot Y, Ed. Humana Press: 2012; Vol. 808, pp 155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manimala JC; Roach TA; Li Z; Gildersleeve JC High-throughput carbohydrate microarray profiling of 27 antibodies demonstrates widespread specificity problems. Glycobiology 2007, 17, 17C–23C. [DOI] [PubMed] [Google Scholar]
- 19.Manimala JC; Roach TA; Li ZT; Gildersleeve JC High-throughput carbohydrate microarray analysis of 24 lectins. Angew. Chem. Int. Ed 2006, 45, 3607–3610. [DOI] [PubMed] [Google Scholar]
- 20.Manimala J; Li Z; Jain A; VedBrat S; Gildersleeve JC Carbohydrate array analysis of anti-Tn antibodies and lectins reveals unexpected specificities: Implications for diagnostic and vaccine development. ChemBioChem 2005, 6, 2229–2241. [DOI] [PubMed] [Google Scholar]
- 21.Stowell CP; Lee YC Neoglycoproteins: the preparation and application of synthetic glycoproteins. Adv. Carbohydr. Chem. Biochem 1980, 37, 225–281. [DOI] [PubMed] [Google Scholar]
- 22.Gildersleeve JC; Wright WS Diverse molecular recognition properties of blood group A binding monoclonal antibodies. Glycobiology 2016, 26, 443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y; Li Q; Rodriguez LG; Gildersleeve JC An array-based method to identify multivalent inhibitors. J. Am. Chem. Soc 2010, 132, 9653–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y; Campbell CT; Li Q; Gildersleeve JC Multidimensional Glycan Arrays for Enhanced Antibody Profiling. Mol. BioSyst 2010, 6, 1583–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oyelaran OO; Li Q; Farnsworth DF; Gildersleeve JC Microarrays with Varying Carbohydrate Density Reveal Distinct Subpopulations of Serum Antibodies. J. Proteome Res 2009, 8, 3529–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muthana S; Gildersleeve JC Glycan Microarrays: Powerful Tools for Biomarker Discovery. Disease Markers 2014, 14, 29–41. [DOI] [PubMed] [Google Scholar]
- 27.Arthur CM; Cummings RD; Stowell SR Using glycan microarrays to understand immunity. Curr. Opin. Chem. Biol 2014, 18, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narla SN; Nie H; Li Y; Sun XL Multi-dimensional glycan microarrays with glyco-macroligands. Glycoconj. J 2015, 32, 483–495. [DOI] [PubMed] [Google Scholar]
- 29.Li X; Gao J; Liu D; Wang Z Studying the interaction of carbohydrate-protein on the dendrimer-modified solid support by microarray-based plasmon resonance light scattering assay. Analyst 2011, 136, 4301–4307. [DOI] [PubMed] [Google Scholar]
- 30.Ciobanu M; Huang KT; Daguer JP; Barluenga S; Chaloin O; Schaeffer E; Mueller CG; Mitchell DA; Winssinger N Selection of a synthetic glycan oligomer from a library of DNA-templated fragments against DC-SIGN and inhibition of HIV gp120 binding to dendritic cells. Chem. Comm 2011, 47, 9321–9323. [DOI] [PubMed] [Google Scholar]
- 31.Parera Pera N; Branderhorst HM; Kooij R; Maierhofer C; Van Der Kaaden M; Liskamp RMJ; Wittmann V; Ruijtenbeek R; Pieters RJ Rapid screening of lectins for multivalency effects with a glycodendrimer microarray. ChemBioChem 2010, 11, 1896–1904. [DOI] [PubMed] [Google Scholar]
- 32.Zhou X; Turchi C; Wang D Carbohydrate cluster microarrays fabricated on three-dimensional dendrimeric platforms for functional glycomics exploration. J. Proteome Res 2009, 8, 5031–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukuda T; Onogi S; Miura Y Dendritic sugar-microarrays by click chemistry. Thin Solid Films 2009, 518, 880–888. [Google Scholar]
- 34.Wang S-K; Liang P-H; Astronomo RD; Hsu T-L; Hsieh S-L; Burton DR; Wong C-H Targeting the carbohydrates on HIV-1: Interaction of oligomannose dendrons with human monoclonal antibody 2G12 and DC-SIGN. Proc. Natl. Acad. Sci 2008, 105, 3690–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Branderhorst HM; Ruijtenbeek R; Liskamp RMJ; Pieters RJ Multivalent carbohydrate recognition on a glycodendrimer-functionalized flow-through chip. ChemBioChem 2008, 9, 1836–1844. [DOI] [PubMed] [Google Scholar]
- 36.Godula K; Rabuka D; Nam Ki T.; Bertozzi CR Synthesis and Microcontact Printing of Dual End-Functionalized Mucin-like Glycopolymers for Microarray Applications. Angew. Chem. Int. Ed 2009, 48, 4973–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Godula K; Bertozzi CR Synthesis of glycopolymers for microarray applications via ligation of reducing sugars to a poly(acryloyl hydrazide) scaffold. J. Am. Chem. Soc 2010, 132, 9963–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang ML; Cohen M; Fisher CJ; Schooley RT; Gagneux P; Godula K Determination of receptor specificities for whole influenza viruses using multivalent glycan arrays. Chem. Comm 2015, 51, 5326–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D; Liu S; Trummer BJ; Deng C; Wang A Carbohydrate Microarrays for the Recognition of Cross-Reactive Molecular Markers of Microbes and Host Cells. Nat. Biotechnol 2002, 20, 275–81. [DOI] [PubMed] [Google Scholar]
- 40.Stowell SR; Arthur CM; McBride R; Berger O; Razi N; Heimburg-Molinaro J; Rodrigues LC; Gourdine JP; Noll AJ; Von Gunten S; Smith DF; Knirel YA; Paulson JC; Cummings RD Microbial glycan microarrays define key features of host-microbial interactions. Nat. Chem. Biol 2014, 10, 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geissner A; Reinhardt A; Rademacher C; Johannssen T; Monteiro J; Lepenies B; Thépaut M; Fieschi F; Mrázková J; Wimmerova M; Schuhmacher F; Götze S; Grünstein D; Guo X; Hahm HS; Kandasamy J; Leonori D; Martin CE; Parameswarappa SG; Pasari S; Schlegel MK; Tanaka H; Xiao G; Yang Y; Pereira CL; Anish C; Seeberger PH Microbe-focused glycan array screening platform. Proc. Natl. Acad. Sci 2019, 116, 1958–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knirel YA; Gabius HJ; Blixt O; Rapoport EM; Khasbiullina NR; Shilova NV; Bovin NV Human tandem-repeat-type galectins bind bacterial non-βGal polysaccharides. Glycoconj. J 2014, 31, 7–12. [DOI] [PubMed] [Google Scholar]
- 43.De Boer AR; Hokke CH; Deelder AM; Wuhrer M Serum antibody screening by surface plasmon resonance using a natural glycan microarray. Glycoconj. J 2008, 25, 75. [DOI] [PubMed] [Google Scholar]
- 44.De Boer AR; Hokke CH; Deelder AM; Wuhrer M General microarray technique for immobilization and screening of natural glycans. Anal. Chem 2007, 79, 8107–13. [DOI] [PubMed] [Google Scholar]
- 45.Song X; Heimburg-Molinaro J; Cummings RD; Smith DF Chemistry of natural glycan microarrays. Curr. Opin. Chem. Biol 2014, 18, 70–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lonardi E; Deelder AM; Wuhrer M; Balog CIA Microarray technology using glycans extracted from natural sources for serum antibody fluorescent detection. In Methods in Molecular Biology, 2012; Vol. 808, pp 285–302. [DOI] [PubMed] [Google Scholar]
- 47.Song X; Lasanajak Y; Xia B; Heimburg-Molinaro J; Rhea JM; Ju H; Zhao C; Molinaro RJ; Cummings RD; Smith DF Shotgun glycomics: A microarray strategy for functional glycomics. Nat. Methods 2011, 8, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma Y; Sobkiv I; Gruzdys V; Zhang H; Sun XL Liposomal glyco-microarray for studying glycolipid-protein interactions. Anal. Bioanal. Chem 2012, 404, 51–58. [DOI] [PubMed] [Google Scholar]
- 49.Pond MA; Zangmeister RA Carbohydrate-functionalized surfactant vesicles for controlling the density of glycan arrays. Talanta 2012, 91, 134–139. [DOI] [PubMed] [Google Scholar]
- 50.Bian S; He J; Schesing KB; Braunschweig AB Polymer pen lithography (PPL)-induced site-specific click chemistry for the formation of functional glycan arrays. Small 2012, 8, 2000–2005. [DOI] [PubMed] [Google Scholar]
- 51.Adams EW; Ueberfeld J; Ratner DM; O’Keefe BR; Walt DR; Seeberger PH Encoded fiber-optic microsphere arrays for probing protein-carbohydrate interactions. Angew. Chem. Int. Ed 2003, 42, 5317–5320. [DOI] [PubMed] [Google Scholar]
- 52.Liang R; Yan L; Loebach J; Ge M; Uozumi Y; Sekanina K; Horan N; Gildersleeve J; Thompson C; Smith A; Biswas K; Still WC; Kahne D Parallel synthesis and screening of a solid phase carbohydrate library. Science 1996, 274, 1520–1522. [DOI] [PubMed] [Google Scholar]
- 53.Purohit S; Li T; Guan W; Song X; Song J; Tian Y; Li L; Sharma A; Dun B; Mysona D; Ghamande S; Rungruang B; Cummings RD; Wang PG; She JX Multiplex glycan bead array for high throughput and high content analyses of glycan binding proteins. Nat. Comm 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamamoto K; Ito S; Yasukawa F; Konami Y; Matsumoto N Measurement of the carbohydrate-binding specificity of lectins by a multiplexed bead-based flow cytometric assay. Anal. Biochem 2005, 336, 28–38. [DOI] [PubMed] [Google Scholar]
- 55.Smith EA; Thomas WD; Kiessling LL; Corn RM Surface Plasmon Resonance Imaging Studies of Protein-Carbohydrate Interactions. J. Am. Chem. Soc 2003, 125, 6140–6148. [DOI] [PubMed] [Google Scholar]
- 56.Houseman BT; Mrksich M Carbohydrate arrays for the evaluation of protein binding and enzymatic modification. Chem. Biol 2002, 9, 443–54. [DOI] [PubMed] [Google Scholar]
- 57.Ngundi MM; Taitt CR; McMurry SA; Kahne D; Ligler FS Detection of bacterial toxins with monosaccharide arrays. Biosens. Bioelectron 2006, 21, 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chevolot Y; Bouillon C; Vidal S; Morvan F; Meyer A; Cloarec JP; Jochum A; Praly JP; Vasseur JJ; Souteyrand E DNA-based carbohydrate biochips: A platform for surface glyco-engineering. Angew. Chem. Int. Ed 2007, 46, 2398–2402. [DOI] [PubMed] [Google Scholar]
- 59.Liang PH; Wang SK; Wong CH Quantitative Analysis of Carbohydrate-Protein Interactions Using Glycan Microarrays: Determination of Surface and Solution Dissociation Constants. J. Am. Chem. Soc 2007, 129, 11177–11184. [DOI] [PubMed] [Google Scholar]
- 60.Mercey E; Sadir R; Maillart E; Roget A; Baleux F; Lortat-Jacob H; Livache T Polypyrrole oligosaccharide array and surface plasmon resonance imaging for the measurement of glycosaminoglycan binding interactions. Anal. Chem 2008, 80, 3476. [DOI] [PubMed] [Google Scholar]
- 61.Song X; Xia B; Lasanajak Y; Smith DF; Cummings RD Quantifiable fluorescent glycan microarrays. Glycoconj. J 2008, 25, 15–25. [DOI] [PubMed] [Google Scholar]
- 62.Grant CF; Kanda V; Yu H; Bundle DR; McDermott MT Optimization of immobilized bacterial disaccharides for surface plasmon resonance imaging measurements of antibody binding. Langmuir 2008, 24, 14125–14132. [DOI] [PubMed] [Google Scholar]
- 63.Tian X; Pai J; Shin I Analysis of density-dependent binding of glycans by lectins using carbohydrate microarrays. Chemistry - An Asian Journal 2012, 7, 2052–2060. [DOI] [PubMed] [Google Scholar]
- 64.Godula K; Bertozzi CR Density Variant Glycan Microarray for Evaluating Cross-Linking of Mucin-like Glycoconjugates by Lectins. J. Am. Chem. Soc 2012, 134, 15732–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mortel KH; Weatherman RV; Kiessling LL Recognition Specificity of Neoglycopolymers Prepared by Ring-Opening Metathesis Polymerization. J. Am. Chem. Soc 1996, 118, 2297–8. [Google Scholar]
- 66.Cairo CW; Gestwicki JE; Kanai M; Kiessling LL Control of multivalent interactions by binding epitope density. J. Am. Chem. Soc 2002, 124, 1615–1619. [DOI] [PubMed] [Google Scholar]
- 67.Horan N; Yan L; Isobe H; Whitesides GM; Kahne D Nonstatistical binding of a protein to clustered carbohydrates. Proc. Natl. Acad. Sci 1999, 96, 11782–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mammen M; Choi SK; Whitesides GM Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. Engl 1998, 37, 2754–94. [DOI] [PubMed] [Google Scholar]
- 69.Bashir S; Leviatan Ben Arye S; Reuven EM; Yu H; Costa C; Galinanes M; Bottio T; Chen X; Padler-Karavani V Presentation Mode of Glycans Affect Recognition of Human Serum anti-Neu5Gc IgG Antibodies. Bioconjugate Chem. 2019, 30, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chandrasekaran A; Srinivasan A; Raman R; Viswanathan K; Raguram S; Tumpey TM; Sasisekharan V; Sasisekharan R Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat. Biotech 2008, 26, 107–113. [DOI] [PubMed] [Google Scholar]
- 71.Lewallen DM; Siler D; Iyer SS Factors affecting protein-glycan specificity: Effect of spacers and incubation time. ChemBioChem 2009, 10, 1486–1489. [DOI] [PubMed] [Google Scholar]
- 72.Dam TK; Oscarson S; Roy R; Das SK; Pagé D; Macaluso F; Brewer CF Thermodynamic, kinetic, and electron microscopy studies of concanavalin A and Dioclea grandiflora lectin cross-linked with synthetic divalent carbohydrates. J. Biol. Chem 2005, 280, 8640–8646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.