Abstract
Botulism is diagnosed by detecting botulinum neurotoxin and Clostridium botulinum cells in the patient and in suspected food samples. In this study, a multiplex PCR assay for the detection of Clostridium botulinum types A, B, E, and F in food and fecal material was developed. The method employs four new primer pairs with equal melting temperatures, each being specific to botulinum neurotoxin gene type A, B, E, or F, and enables a simultaneous detection of the four serotypes. A total of 43 C. botulinum strains and 18 strains of other bacterial species were tested. DNA amplification fragments of 782 bp for C. botulinum type A alone, 205 bp for type B alone, 389 bp for type E alone, and 543 bp for type F alone were obtained. Other bacterial species, including C. sporogenes and the nontoxigenic nonproteolytic C. botulinum-like organisms, did not yield a PCR product. Sensitivity of the PCR for types A, E, and F was 102 cells and for type B was 10 cells per reaction mixture. With a two-step enrichment, the detection limit in food and fecal samples varied from 10−2 spore/g for types A, B, and F to 10−1 spore/g of sample material for type E. Of 72 natural food samples investigated, two were shown to contain C. botulinum type A, two contained type B, and one contained type E. The assay is sensitive and specific and provides a marked improvement in the PCR diagnostics of C. botulinum.
Clostridium botulinum is a spore-forming bacterium that produces lethal neurotoxin, the causative agent of a paralytic disease known as botulism (28). Based on the toxin type produced, C. botulinum strains are divided in groups I to IV, with groups I and II being the main human pathogens. Group I consists of proteolytic types A, B, and F, and group II consists of nonproteolytic types B, E, and F (5, 30). The two groups are completely different in their phenotypical characteristics, such as temperature requirements, biochemical profile, and production of metabolites (13, 14). The main taxonomic denominator is thus the production of the botulinum neurotoxin (13, 14).
Diagnosis of botulism is obtained by detecting the neurotoxin and C. botulinum cells in a patient or suspected food sample (24, 25). The standard method for detecting the toxin is the mouse bioassay (24), which is time-consuming and expensive and raises ethical concern due to the use of experimental animals. Conventional isolation and identification of C. botulinum is difficult unless the toxicity of the isolates is confirmed by the mouse assay. Commercial biochemical tests have been shown to fail in identifying both group I and II organisms as C. botulinum (20). The isolation of C. botulinum in environmental and food samples is frequently complicated by the presence of proteolytic and nonproteolytic nontoxigenic strains that both phenotypically and genetically resemble C. botulinum and exhibit a high relatedness with their toxigenic counterparts (4, 14, 19, 23).
PCR provides high sensitivity and specificity in detection of a number of pathogenic microorganisms. For C. botulinum, several PCR-based detection methods have been reported during the last decade (1, 3, 6, 10, 11, 12, 17, 29, 31, 32). Following the current taxonomy of C. botulinum, these methods are based on the detection of the botulinum neurotoxin gene (BoNT). Compared to conventional methods, these protocols provide rapid and sensitive detection of C. botulinum. Most of these protocols employ toxin type-specific primers as a single pair in the PCR (12, 17, 29, 31, 32), and not more than one serotype may be detected at a time. Consequently, in an investigation of unknown samples, each C. botulinum type needs to be detected separately, which extends the detection time and increases the reagent costs. Some of the older primer pairs are also poorly designed with regard to their optimal annealing temperatures, resulting in the formation of unspecific amplification products. A different approach is to use a general primer pair common for more than one type of C. botulinum and to differentiate between the toxin types by a type-specific DNA probe (1, 3, 6, 10, 11). In this method, a limited number of essential oligonucleotides may be used, but the probing step required for the complete identification of the serotype extends the detection time.
The multiplex PCR method would provide a more sophisticated approach, enabling a simultaneous and specific detection of more than one serotype of C. botulinum. In general, this method employs more than one pair of specific primers added to the same PCR. Useful applications of multiplex PCR in the detection of other pathogenic bacteria have been previously reported (15, 21, 22), none of these in connection with C. botulinum. The BoNT-specific primers described in earlier papers (12, 17, 29, 31, 32) are highly variable in their melting temperatures and may thus not be added to multiplex reaction mixtures.
In this study, four new pairs of BoNT-specific primers with equal melting temperatures were conducted. Furthermore, a multiplex PCR assay for the simultaneous detection of C. botulinum types A, B, E, and F in foods and fecal material was designed. The assay includes a two-step enrichment, being very sensitive and specific and providing a marked improvement in the PCR diagnostics of C. botulinum.
MATERIALS AND METHODS
Bacterial strains and culturing.
A total of 11 C. botulinum type A, 9 type B, 16 type E, and 7 type F strains and 18 strains of other bacterial species were included in the study (Table 1). Clostridium sporogenes and nonproteolytic nontoxigenic C. botulinum-like strains (further referred to as C. botulinum-like strains) were used as negative controls. These strains have formerly been confirmed to be nontoxigenic by the mouse bioassay (20).
TABLE 1.
Bacterial strains tested by the multiplex PCR assay
| Species | Type | Strain(s) | Origin | Sourcea | Multiplex PCR resultb |
|---|---|---|---|---|---|
| C. botulinum (group I) | A | ATCC 25763 | Type strain | ATCC | A |
| ATCC 19397 | NKc | ATCC | A | ||
| ATCC 3502 | NK | ATCC | A | ||
| 62A, 69A | Cow liver infarct and stool; USA | Riemann/Lindrothd | A | ||
| SL-2A, SL-3A, SL-4A, SL-6A | NK | Lindroth | A | ||
| RS-3A, RS-4A | Pacific red snapper; USA | Lindroth | A | ||
| B | ATCC 7949 | Canned shallots; USA | ATCC | B | |
| ATCC 17841 | NK | ATCC | B | ||
| SL-1B | NK | Lindroth | B | ||
| 133–4803 | NK | McClung/Lindroth | B | ||
| 126B | NK | IP | B | ||
| F | ATCC 35415 | Liver paste; Denmark | ATCC | F | |
| ATCC 25764 | Crab; UK | ATCC | F | ||
| F4VIF | Blue crab viscera; UK | Lindroth | F | ||
| Crab F | Pasteurized crabmeat; USA | Lindroth | F | ||
| C. botulinum (group II) | B | ATCC 25765 | Marine sediment; USA | ATCC | B |
| ATCC 17844 | NK | ATCC | B | ||
| 2B | Marine sediment; USA | Eklund/Lindroth | B | ||
| 706B | Salted salmon; USA | Hatheway/Lindroth | B | ||
| E | ATCC 9564 | Smoked salmon; Canada | ATCC | E | |
| Beluga E | Fermented white whale flippers; USA | Dolman/Lindroth | E | ||
| 92E | Marine environment; USA | Eklund/Lindroth | E | ||
| 211 E | Pickled herring; Canada | Dolman/Lindroth | E | ||
| 250E | Canned salmon; USA | Crowther/Lindroth | E | ||
| 4062E | Fermented whale blubber; USA | Hatheway/Lindroth | E | ||
| KA-2E | Scola Creek strain; USA | Ricmann/Lindroth | E | ||
| C-51E | Seal meat; Denmark | SSI | E | ||
| C-60E | Dried mutton; Denmark | SSI | E | ||
| K-6E, K-44E, K-45E | Rainbow trout; Finland | DFH | E | ||
| K-115E | Hot-smoked whitefish; Finland and Canada | DFH | E | ||
| S-3E | Mud; Finland | DFH | E | ||
| S-4E, S-6E | Fishpond sediment; Finland | DFH | E | ||
| F | ATCC 23387 | Marine sediment; USA | ATCC | F | |
| 610B8-6F | Salmon; USA | Craig/Lindroth | F | ||
| FT10F | Marine sediment; UK | Hobbs/Lindroth | F | ||
| C. sporogenes | ATCC 19404 | Gas gangrene | ATCC | — | |
| C. botulinum-like | E-like | H11 | Marine sediment; Finland | DFH | — |
| P559 | Marine sediment; Finland | DFH | — | ||
| C. histolyticum | 105 | Finland | EELA | — | |
| C. perfringens | ATCC 3624 | NK | ATCC | — | |
| ATCC 3626 | Intestinal contents of lamb | ATCC | — | ||
| CCUG 2036 | Sheep | CCUG | — | ||
| CCUG 2037 | Sheep | CCUG | — | ||
| CCUG 2038 | NK | CCUG | — | ||
| NCTC 8239 | Boiled salt beef | NCTC | — | ||
| C. septicum | ATCC 12464 | NK | ATCC | — | |
| Listeria innocua | L434 | Processing plant; Finland | DFH | — | |
| L435 | Cheese product; Finland | DFH | — | ||
| Listeria monocytogenes | ATCC 7644 | Human | ATCC | — | |
| NCTC 10527 | Spinal fluid; Germany | NCTC | — | ||
| NCTC 5105 | Human | NCTC | — | ||
| Y. enterocolitica | YTSK 22K2, P102 | Pig tonsil; Finland | DFH | — |
ATCC, American Type Culture Collection; IP, Institute Pasteur, Paris, France; SSI, Statens Serum Institut, Copenhagen, Denmark; DFH, Department of Food and Environmental Hygiene, University of Helsinki; CCUG, Culture Collection, University of Gothenburg, Gothenburg, Sweden; NCTC, National Collection of Type Cultures, London, United Kingdom; EELA, National Veterinary and Food Research Institute, Helsinki, Finland.
A, B, E, or F, BoNT/A, BoNT/B, BoNT/E, or BoNT/F gene-specific amplification product obtained in multiplex PCR assay, respectively. —, no amplification products were obtained by the assay.
NK, not known.
Collected from various sources by Seppo Lindroth, University of California, Davis, Calif. The first name in each pair is the original source.
All clostridial strains were cultured in 10 ml of tryptose-peptone-glucose-yeast extract (TPGY) medium (Difco Laboratories, Detroit, Mich.) and incubated under anaerobic conditions (MK3 Anaerobic Work Station; Don Whitley Scientific Ltd., West Yorkshire, United Kingdom) at 37°C (C. botulinum group I, C. sporogenes, Clostridium histolyticum, Clostridium perfringens, and Clostridium septicum) or 30°C (C. botulinum group II and C. botulinum-like strains) for 24 to 48 h, followed by overnight culturing of 20 h at respective temperatures. Listeria spp. and Yersinia enterocolitica strains were cultured on blood agar plates at 30°C for 24 h before template preparation.
Template preparation.
Cells from 1 ml of each clostridial overnight culture were washed with 1 ml of TE buffer (0.01 M Tris-HCl, 0.001 M EDTA) for 1 h at 37°C and suspended in 1 ml of distilled water. One to five typical colonies of Listeria spp. and Y. enterocolitica strains were picked from agar plates, washed with 100 μl of distilled water, and suspended in 100 μl of distilled water. In addition to the individual cell suspensions of each bacterial strain, three mixed suspensions containing proteolytic C. botulinum types A (ATCC 25763), B (126B), and F (ATCC 25764), the nonproteolytic types B (Eklund 2B), E (Dolman Beluga E), and F (Craig 610B8-6B), or all four serotypes were prepared by mixing the individual cell suspensions. All suspensions were heated at 99°C for 10 min to break up the cells and release the bacterial DNA and were centrifuged for 5 min at 10,000 × g. A volume of 1 μl of each supernatant was used as template in the PCR mixture.
Primers.
Based on published DNA sequences of the BoNT gene (2, 8, 9, 27, 33, 34, 35), four new primer pairs with each being specific for either C. botulinum type A, B, E, or F were designed (Table 2). The primers were selected from the nonhomologous regions of the BoNT types A, B, E, and F gene by using the Primer 3 software (S. Rozen and H. J. Skaletsky, Primer 3, Whitehead Institute for Biomedical Research, Cambridge, Mass. [http://www-genome.wi.mit.edu/genome_software/other/primer3.html], 1998).
TABLE 2.
Primers for multiplex PCR detection of C. botulinum types A, B, E, and F
| Typea | Primer | Sequence (5′-3′) | Product size (bp) | Location on gene (coding region) | Temp (°C) | GC content (%) |
|---|---|---|---|---|---|---|
| Af | CBMLA1 | AGC TAC GGA GGC AGC TAT GTT | 782 | 1788–1808 | 63.9 | 52 |
| Ar | CBMLA2 | CGT ATT TGG AAA GCT GAA AAG G | 2569–2548 | 63.4 | 41 | |
| Bf | CBMLB1 | CAG GAG AAG TGG AGC GAA AA | 205 | 434–453 | 64.3 | 50 |
| Br | CBMLB2 | CTT GCG CCT TTG TTT TCT TG | 638–619 | 64.5 | 45 | |
| Ef | CBMLE1 | CCA AGA TTT TCA TCC GCC TA | 389 | 156–175 | 63.7 | 45 |
| Er | CBMLE2 | GCT ATT GAT CCA AAA CGG TGA | 544–525 | 63.6 | 43 | |
| Ff | CBMLF1 | CGG CTT CAT TAG AGA ACG GA | 543 | 185–194 | 64.1 | 50 |
| Fr | CBMLF2 | TAA CTC CCC TAG CCC CGT AT | 727–708 | 63.3 | 55 |
Subscript f, forward primer; subscript r, reverse primer.
PCR.
PCR was performed with 50 μl of reaction mixture containing 1 μl of template, 0.3 μM concentrations of each primer (Sigma-Genosys Ltd., Cambridgeshire, United Kingdom), 220 nM concentrations of each deoxynucleotide triphosphate (dATP, dCTP, dGTP, and dTTP; dNTP Mix; Finnzymes, Espoo, Finland), 32 mM Tris-HCl (pH 8.4), 80 mM KCl, 4.8 mM MgCl, and 2 U of DNA polymerase (DynaZyme; Finnzymes). The reaction mixture was overlaid with mineral oil before adding the template and amplification (PTC-200 Peltier Thermocycler; MJ Research Inc., Watertown, Mass.). Each PCR cycle consisted of denaturation at 95°C for 30 s, annealing at 60°C for 25 s, and extension at 72°C for 1 min 25 s and was repeated 27 times. Final extension at 72°C for 3 min followed. The amplified PCR products were visualized in 2% agarose gels (I.D.NA agarose; BioWhittaker Molecular Applications, Rockland, Maine) stained with ethidium bromide. Standard DNA fragments (DNA molecular weight marker VI; Boehringer Mannheim, Mannheim, Germany) were used as molecular weight markers to indicate the sizes of the amplification products.
Inhibition of PCR by sample material.
Equal volumes of the overnight cultures of C. botulinum types A (Riemann 62A), B (126B), E (Beluga E), and F (ATCC 25764) were mixed, and 21 Eppendorf tubes were filled with 1 ml of the mixture. Raw minced beef, hot-smoked whitefish, and pig feces were each added to seven tubes at levels of 0.5, 0.25, 0.1, 0.05, 0.025, 0.01, and 0.005 g/ml of the overnight culture, followed by the cell wash and PCR as described. The final concentrations of the sample materials were estimated to be correspondingly 500, 250, 100, 50, 25, 10, and 5 μg of sample material per 50-μl PCR mixture.
Sensitivity of the PCR.
The overnight cultures of C. botulinum types A (Riemann 62A), B (Eklund 2B), E (Dolman Beluga E), and F (Craig 610B8-6F) were quantified by the five-tube most probable number (MPN) method (26). The cultures were serially diluted in peptone water, followed by the cell wash and resuspension in 1 ml of distilled water. Each dilution was heated and used as a template in the PCR.
Detection limit of multiplex PCR assay in inoculated food and feces samples.
In order to test the applicability of the multiplex PCR protocol in investigation of food and fecal specimens, 10-g samples of raw minced beef, hot-smoked whitefish, and pig feces were inoculated with 10-fold dilutions of individual spore suspensions of either C. botulinum type A (ATCC 25763), type B (proteolytic strain 126B, or nonproteolytic strain Eklund 2B), type E (Dolman Beluga E), or type F (proteolytic strain ATCC 25764 or nonproteolytic strain Craig 610B8-6B) to yield final spore counts of 10−2 to 103 spores/g of sample. In addition, one 10-g sample of minced beef was inoculated with a mixture of the above-mentioned strains, containing equal numbers of each of the four types at the level of 103 spores/g of minced beef.
All samples were homogenized (Stomacher 400 Laboratory Blender; Seward Medical Ltd., London, United Kingdom) in sterile pouches with 90 ml (wt/vol [1:9]) of peptone water for 120 s. A total of two 5-g samples of each homogenate were then transferred to tubes containing 45 ml of TPGY broth and incubated at 30 and 37°C under anaerobic conditions for 5 days. The samples were incubated for 1 day (samples inoculated with individual strains), 3 days (all samples), or 5 days (samples inoculated with individual strains), and anaerobic overnight culturing in 10 ml of TPGY followed at respective temperatures. Templates were prepared and PCR was performed as described earlier. The final concentration of each sample material in the PCR was estimated to be approximately 1 μg/reaction mixture. The experiment was repeated once.
Investigation of natural food samples.
A number of 36 vegetable sausages, 11 cans of Finnish wild deer meat, and 25 whitefish heads collected from a local fishery were investigated for the presence of C. botulinum. A total of 10 g of each sausage and deer sample was transferred to bottles containing 100 ml of TPGY medium, and the whitefish heads were each placed in tubes containing 45 ml of TPGY broth. The bottles and tubes were incubated anaerobically at 30°C for 3 days, followed by overnight culturing at 30°C. Cell washing and PCR were performed as described. The final concentration of sample material in the PCR was estimated to be 10 μg.
RESULTS
Multiplex PCR of bacterial cell suspensions.
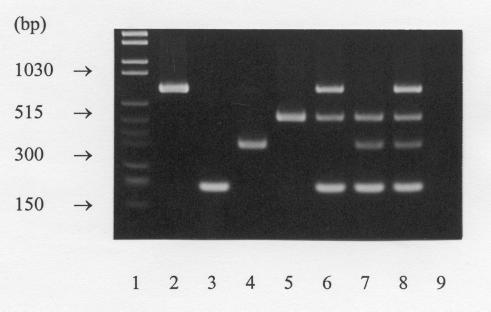
C. botulinum types A, B, E, and F alone yielded the expected amplification products (Table 1): type A, 782 bp; type B, 205 bp; type E, 389 bp; and type F, 543 bp (Fig. 1). The mixed-cell suspensions yielded the corresponding DNA fragments (Fig. 1). None of the C. sporogenes or C. botulinum-like strains or other bacterial species yielded a PCR product by this assay (Table 1). The PCR products were clearly visualized in agarose gels; a 150-to-200-bp difference in the size of each amplification product enabled an easy distinction between the fragments without the use of high-resolution agarose (Fig. 1).
FIG. 1.
Multiplex PCR detection of C. botulinum. Lanes: 1, molecular weight marker; 2, C. botulinum type A; 3, C. botulinum type B; 4, C. botulinum type E; 5, C. botulinum type F; 6, proteolytic C. botulinum types A, B, and F; 7, nonproteolytic C. botulinum types B, E, and F; 8, C. botulinum types A, B, E, and F; and 9, negative control.
Inhibition of PCR by sample material.
Of the three sample materials tested, only minced beef inhibited the PCR at higher concentrations: the levels of 500 and 250 μg/reaction mixture yielded no PCR products, whereas 100 μg/reaction mixture allowed for the detection of type B only, and 50 and 25 μg/reaction mixture yielded types B and E (Table 3). At lower concentrations of minced beef, all four types were detected. Feces at the level of 250 μg/reaction mixture inhibited only the amplification of the type A-specific DNA fragment.
TABLE 3.
Inhibition of multiplex PCR by different concentrations of food and fecal material
| Concn of sample materiala (μg/PCR mixture) |
C. botulinum type(s) detected
|
||
|---|---|---|---|
| Minced beef | Fish | Feces | |
| 500 | A, B, E, F | A, B, E, F | |
| 250 | A, B, E, F | B, E, F | |
| 100 | B | A, B, E, F | A, B, E, F |
| 50 | B, E, F | A, B, E, F | A, B, E, F |
| 25 | B, E | A, B, E, F | A, B, E, F |
| 10 | A, B, E, F | A, B, E, F | A, B, E, F |
| 5 | A, B, E, F | A, B, E, F | A, B, E, F |
The actual level of sample material in the PCR was estimated to be 1 μg with a dilution (1:9) step in peptone water and was 10 μg without the dilution.
Sensitivity of multiplex PCR.
The sensitivity of the multiplex PCR for C. botulinum types A, E, and F was approximately 102 cells/reaction mixture, and for type B it was 10 cells/reaction mixture.
Detection limit of assay in inoculated food and feces.
The detectable C. botulinum spore concentration in the individually inoculated food and fecal samples varied from 10−2 to 103 spores/g of sample material, depending on the inoculated C. botulinum strain, sample material, and enrichment time and temperature (Table 4). The optimal enrichment times ranged from 1 to 5 days followed by overnight culturing (Table 4), but with all strains being detectable within 3 days. In the minced beef sample inoculated with a mixture of C. botulinum types A, B, E, and F and enriched for 3 days, the type B, E, and F strains were detected at 30°C and type B and F strains were detected at 37°C. The type A strain was not detected in this sample after the 3-day incubation period at either temperature.
TABLE 4.
Detection limit and optimal enrichment time of multiplex PCR assay for C. botulinum types A, B, E, and F in food and fecal samples
| C. botulinum type | Detection limit in three sample materials ([spores/g of sample material], optimal enrichment time [days])
|
||
|---|---|---|---|
| Minced beef | Hot-smoked whitefish | Feces | |
| A | 10−2, 5 | 10−1, 3 | 10, 3 |
| B | 10−2, 1 | 10−2, 3 | 10−1, 3 |
| E | 10−1, 3 | 10−1, 1 | 103, 5 |
| F | 10−1, 3 | 10−2, 5 | 10, 3 |
Presence of C. botulinum in natural food samples.
One vegetable sausage and one can of deer meat were shown to contain C. botulinum type B. Type E was found in one vegetable sausage and, unexpectedly, type A was found in two fish heads.
DISCUSSION
A multiplex PCR assay for the simultaneous detection and identification of C. botulinum types A, B, E, and F in foods and fecal material was developed. The method is based on a primer cocktail consisting of four new pairs of oligonucleotide primers, each being specific for the botulinum neurotoxin type A, B, E, or F gene. This method provides a marked improvement in the PCR diagnostics of C. botulinum, since the previously described methods require more than one step for the complete detection and identification of several C. botulinum types (1, 3, 6, 10, 11, 12, 17, 29, 31, 32). The total time required by the multiplex PCR assay, including a two-step enrichment, was 2 to 6 days, depending on the sample material. The detection limit was 10−2 to 103 spores/g of sample material. All C. botulinum cultures yielded the expected amplification products that, due to differences of 150 to 200 bp in the product size, were easily differentiated in low-resolution agarose gels.
The oligonucleotide primers were designed for the nonhomologous regions of the botulinum neurotoxin types A, B, E, and F genes, and a limited variation in the selection criteria between the eight primers was allowed. This was not the case in the earlier studies on PCR detection of C. botulinum (12, 29, 31, 32), as the melting temperatures of those primers varied by up to 20°C, resulting in unspecific amplification. The melting temperatures of the present primers were almost equal, enabling the optimal annealing of all the eight primers at 60°C (Table 2). Amplification of unspecific products was thus predominantly avoided in the samples tested.
The sensitivity of the PCR varied from 10 to 102 cells/reaction mixture, corresponding to 104 to 105 vegetative cells/ml of bacterial culture. The PCR seemed to be the most sensitive for type B, which could be due to the smallest size (205 bp) of the four amplification fragments. The above-mentioned concentrations were easily obtained by the two-step enrichment employed in the present study, as was observed with the inoculated samples; 10−2 C. botulinum spores/g of sample material could be detected. Previous reports on PCR protocols designed for a single type of C. botulinum provide a variety of sensitivities, from 2.5 pg of purified DNA (32) to 104 cells/g of sample material (1), depending on the method of DNA purification and enrichment conditions of the target cells. In the latter study (1), they found that with an enrichment step of 18 h, they could detect as few as 1 cell/10 g of food sample when the DNA was recovered by a purification procedure. The detection limit in the present method was determined from washed and heated cell lysates, and it is likely that the sensitivity of this method could be further improved by DNA purification.
The multiplex PCR assay ensured a sensitive and specific tool for the detection of C. botulinum in the inoculated food and fecal samples. The optimal enrichment time varied from 1 to 5 days, depending on the inoculated C. botulinum strain and sample material (Table 4). In general, by extending the enrichment time by up to 5 days, the assay had an increased sensitivity for the types A and F strains, whereas the type B strains in all samples and type E strain in beef and fish were easily detected in 1 to 3 days. The optimal enrichment temperatures seemed to vary by the sample material rather than by the inoculated group I and II strains; optimal enrichment of the beef was obtained at 37°C, while that of the fish and feces was at 30°C. The lowest detection limits were observed in minced beef and hot-smoked fish, where 10−2 to 10−1 spore/g of sample material was detected. These spore counts correspond to the natural contamination level of C. botulinum in foods (7). For feces, the detection limit was higher; 10−1 to 103 spores/g of feces were found, with the highest detection level being that for the type E strain. A number of sample materials, including feces and fatty foods, are generally known to cause failure in the PCR detection of microorganisms due to the inhibition of the DNA polymerase enzyme. However, it seems that this is not the case in this study, as the concentrations of all sample materials in the PCR were shown to be below the inhibitory level (Table 3).
The likelihood of PCR inhibition seemed to correlate with the size of the amplification product (Table 3). Relatively low concentrations of beef (25 to 100 μg) readily inhibited the amplification of types A and F, whereas higher concentrations (250 to 500 μg) were required to inhibit the amplification of types B and E. This may suggest that the DNA polymerase enzyme had a limited activity in the presence of lower concentrations of minced beef, being able to amplify the smallest fragments but not the larger ones.
C. botulinum type E is naturally highly prevalent in aquatic environments and fish (16, 17, 18), but not as frequently found in meats or fecal material. In this study, the type E strain was more rapidly detected in fish samples than in beef and feces, which may indicate that beef and feces are not natural niches for type E. Furthermore, it is even possible that the germination and growth rate of the C. botulinum type E strain were reduced by the presence of these sample materials.
In the minced beef inoculated with the mixture of types A, B, E, and F spores, types B and F were detected at both incubation temperatures and type E was detected at 30°C. According to a common understanding of the optimal growth temperatures of group I and II organisms, the types B and F strains detected at 30°C were assumed to be the nonproteolytic ones, whereas those detected at 37°C were most probably the proteolytic strains. The type A strain did not seem to reach the detectable level in minced beef within 3 days at either temperature, which may be explained by a relatively slow growth rate observed in beef inoculated by the individual type A strain (Table 4). It can thus be expected that type A is better detected in meat by extending the incubation time to 5 days.
As for the natural food samples, C. botulinum type A was detected in fish within 3 days, which is in agreement with the individually inoculated food samples (Table 4). The presence of type A in fish was not expected, and it remains unknown whether the contamination occurred from the surroundings of the processing plant. More expected were the findings with type B being detected in a can of deer meat and types B and E being detected in a vegetable sausage.
As the primary sources of C. botulinum types A, B, E, and F are different, this assay may also be modified for the detection of either a single type of C. botulinum or the proteolytic or the nonproteolytic types alone (Fig. 1). Combining the types A-, B-, and F-specific primers with the incubation temperatures of 35 to 40°C would probably ensure the detection of the proteolytic types more likely than that of the nonproteolytics. Similarly, B-, E-, and F-specific primers in combination with a slightly lower incubation temperature would result in optimal detection of the nonproteolytic types (Fig. 1). This shows a high flexibility and usefulness of the assay in the detection of different types of C. botulinum in various types of sample materials.
In conclusion, the multiplex PCR assay in combination with the two-step enrichment is a sensitive and specific method to simultaneously detect several C. botulinum types present in food or clinical material. The sensitivity of the assay enables the detection of low numbers of spores present in natural samples. The study also demonstrated that a careful consideration of the appropriate enrichment conditions for each type of sample material is required to obtain optimal results. The multiplex PCR assay provides high flexibility in detection of several types of C. botulinum present in various types of environments. The assay thus markedly improves the PCR diagnostics of C. botulinum.
ACKNOWLEDGMENTS
We thank Kirsi Ristkari for excellent laboratory assistance.
This study was supported by the Finnish Research Programme on Environmental Health 1998-2001 and the National Technology Agency.
REFERENCES
- 1.Aranda E, Rodríguez M M, Asensio M A, Córdoba J J. Detection of Clostridium botulinum types A, B, E and F in foods by PCR and DNA probe. Lett Appl Microbiol. 1997;25:186–190. doi: 10.1046/j.1472-765x.1997.00204.x. [DOI] [PubMed] [Google Scholar]
- 2.Binz T, Kurazono H, Wille M, Frevert J, Wernars K, Niemann H. The complete sequence of botulinum neurotoxin type A and comparison with other clostridial neurotoxins. J Biol Chem. 1990;265:9153–9158. [PubMed] [Google Scholar]
- 3.Braconnier A, Broussolle V, Nguyen-The C, Carlin F. Screening for Clostridium botulinum type A, B, and E in cooked chilled foods containing vegetables and raw material using polymerase chain reaction and molecular probes. J Food Prot. 2001;64:201–207. doi: 10.4315/0362-028x-64.2.201. [DOI] [PubMed] [Google Scholar]
- 4.Broda D M, Boerema J A, Bell R G. A PCR survey of psychrotrophic Clostridium botulinum-like isolates for the presence of BoNT genes. Lett Appl Microbiol. 1998;27:219–223. doi: 10.1046/j.1472-765x.1998.00420.x. [DOI] [PubMed] [Google Scholar]
- 5.Buchanan R E, Gibbons N E. Bergey's manual of determinative bacteriology. 8th ed. Baltimore, Md: The Williams and Wilkins Company; 1974. [Google Scholar]
- 6.Campbell K D, Collins M D, East A K. Gene probes for identification of the botulinal neurotoxin gene and specific identification of neurotoxin types B, E, and F. J Clin Microbiol. 1993;31:2255–2262. doi: 10.1128/jcm.31.9.2255-2262.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dodds K L. Clostridium botulinum in foods. In: Hauschild A H W, Dodds K L, editors. Clostridium botulinum. Ecology and control in foods. New York, N.Y: Marcel Dekker, Inc.; 1993. pp. 53–68. [Google Scholar]
- 8.East A K, Richardson P T, Allaway D, Collins M D, Roberts T A, Thompson D E. Sequence of the gene encoding type F neurotoxin of Clostridium botulinum. FEMS Microbiol Lett. 1992;96:225–230. doi: 10.1016/0378-1097(92)90408-g. [DOI] [PubMed] [Google Scholar]
- 9.Elmore M J, Hutson R A, Collins M D, Bodsworth N J, Whelan S M, Minton N J. Nucleotide sequence of the gene coding for proteolytic (Group I) Clostridium botulinum type F neurotoxin: genealogical comparison with other clostridial neurotoxins. Syst Appl Microbiol. 1995;18:23–31. [Google Scholar]
- 10.Fach P, Hauser D, Guillou J P, Popoff M R. Polymerase chain reaction for the rapid identification of Clostridium botulinum type A strains and detection in food samples. J Appl Bacteriol. 1993;75:234–239. doi: 10.1111/j.1365-2672.1993.tb02771.x. [DOI] [PubMed] [Google Scholar]
- 11.Fach P, Gibert M, Griffais R, Guillou J P, Popoff M R. PCR and gene probe identification of botulinum neurotoxin A-, B-, E-, F-, and G-producing Clostridium spp. and evaluation in food samples. Appl Environ Microbiol. 1995;61:389–392. doi: 10.1128/aem.61.1.389-392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franciosa G, Ferreira J L, Hatheway C L. Detection of type A, B, and E botulism neurotoxin genes in Clostridium botulinum and other Clostridium species by PCR: evidence of unexpressed type B toxin genes in type A toxigenic organisms. J Clin Microbiol. 1994;32:1911–1917. doi: 10.1128/jcm.32.8.1911-1917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatheway C L. Toxigenic clostridia. Clin Microbiol Rev. 1990;3:66–98. doi: 10.1128/cmr.3.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatheway C L. Clostridium botulinum and other clostridia that produce botulinum neurotoxin. In: Hauschild A H D, Dodds K L, editors. Clostridium botulinum—ecology and control in foods. New York, N.Y: Marcel Dekker; 1992. pp. 3–20. [Google Scholar]
- 15.Herman L M F, deRidder H R M, Vlaenmynck F M M. A multiplex PCR method for the identification of Listeria sp. and Listeria monocytogenes in dairy samples. J Food Prot. 1995;58:867–872. doi: 10.4315/0362-028X-58.8.867. [DOI] [PubMed] [Google Scholar]
- 16.Hielm S, Hyytiä E, Andersin A-B, Korkeala H. A high prevalence of Clostridium botulinum type E in Finnish freshwater and Baltic Sea sediment samples. J Appl Microbiol. 1998;84:133–137. doi: 10.1046/j.1365-2672.1997.00331.x. [DOI] [PubMed] [Google Scholar]
- 17.Hielm S, Hyytiä E, Ridell J, Korkeala H. Detection of C. botulinum in fish and environmental samples using polymerase chain reaction. Int J Food Microbiol. 1996;31:357–365. doi: 10.1016/0168-1605(96)00984-1. [DOI] [PubMed] [Google Scholar]
- 18.Hyytiä E, Hielm S, Korkeala H. Prevalence of Clostridium botulinum type E in Finnish fish and fishery products. Epidemiol Infect. 1998;120:245–250. doi: 10.1017/s0950268898008693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee W H, Riemann H. Correlation of toxic and non-toxic strains of Clostridium botulinum by DNA composition and homology. J Gen Microbiol. 1979;60:117–123. doi: 10.1099/00221287-60-1-117. [DOI] [PubMed] [Google Scholar]
- 20.Lindström M, Jankola H, Hielm S, Hyytiä E, Korkeala H. Identification of Clostridium botulinum with API 20 A, Rapid ID 32 A and RapID ANA II. FEMS Immunol Med Microbiol. 1999;24:267–274. doi: 10.1111/j.1574-695X.1999.tb01293.x. [DOI] [PubMed] [Google Scholar]
- 21.Meer R R, Songer J G. Multiplex polymerase chain reaction assay for genotyping Clostridium perfringens. Am J Vet Res. 1997;58:702–705. [PubMed] [Google Scholar]
- 22.Nakajima H, Inoue M, Mori T, Itoh K-I, Arakawa E, Watanabe H. Detection and identification of Yersinia pseudotuberculosis and pathogenic Yersinia enterocolitica by an improved polymerase chain reaction method. J Clin Microbiol. 1992;30:2484–2486. doi: 10.1128/jcm.30.9.2484-2486.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura S, Okado I, Nakashio S, Nishida S. Clostridium sporogenes isolates and their relationship to C. botulinum based on deoxyribonucleic acid reassociation. J Gen Microbiol. 1977;100:395–401. doi: 10.1099/00221287-100-2-395. [DOI] [PubMed] [Google Scholar]
- 24.Nordic Committee on Food Analysis. Botulinum toxin. Detection in foods, blood and other test materials. Method no. 79. 2nd ed. Espoo, Finland: Nordic Committee on Food Analysis; 1991. [Google Scholar]
- 25.Nordic Committee on Food Analysis. Clostridium botulinum. Detection in foods and other test materials. Method no. 80. 2nd ed. Espoo, Finland: Nordic Committee on Food Analysis; 1991. [Google Scholar]
- 26.Oblinger J L, Koburger J A. The most probable number technique. In: Speck M L, editor. Compendium of methods for the microbiological examination of foods. 2nd ed. Washington, D.C.: American Public Health Association, Inc.; 1984. [Google Scholar]
- 27.Poulet S, Hauser D, Quantz M, Niemann H, Popoff M R. Sequences of the botulinal neurotoxin E derived from Clostridium botulinum type E (strain Beluga E) and Clostridium butyricum (strains ATCC 43181 and ATCC 43755) Biochem Biophys Res Commun. 1992;183:107–113. doi: 10.1016/0006-291x(92)91615-w. [DOI] [PubMed] [Google Scholar]
- 28.Prévot A R. Rapport d'introduction du président du sous-comitéClostridium pour l'unification de la nomenclature des types toxigéniques de C. botulinum. Int Bull Bacterial Nomencl. 1953;3:120–123. [Google Scholar]
- 29.Sciacchitano C J, Hirshfield I N. Molecular detection of Clostridium botulinum type E neurotoxin gene in smoked fish by polymerase chain reaction and capillary electrophoresis. J AOAC Int. 1996;79:861–865. [PubMed] [Google Scholar]
- 30.Smith L D S, Sugiyama H. Botulism: the organism, its toxins, the disease. Springfield, Ill: Charles C. Thomas; 1988. [Google Scholar]
- 31.Szabo E A, Pemberton J M, Gibson A M, Thomas R J, Pascoe R R, Desmarchelier P M. Application of PCR to a clinical and environmental investigation of a case of equine botulism. J Clin Microbiol. 1994;32:1986–1991. doi: 10.1128/jcm.32.8.1986-1991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeshi K, Fujinaga Y, Inoue K, Nakajima H, Oguma K, Ueno T, Sunagawa H, Ohyama T. Simple method for detection of Clostridium botulinum type A to F neurotoxin genes by polymerase chain reaction. Microbiol Immunol. 1996;40:5–11. doi: 10.1111/j.1348-0421.1996.tb03310.x. [DOI] [PubMed] [Google Scholar]
- 33.Thompson E D, Brehm J K, Oultram J D, Swinfield T-J, Shone C C, Atkinson T, Melling J, Minton N P. The complete amino acid sequence of the Clostridium botulinum type A neurotoxin, deduced by nucleotide analysis of the encoding gene. Eur J Biochem. 1990;189:73–81. doi: 10.1111/j.1432-1033.1990.tb15461.x. [DOI] [PubMed] [Google Scholar]
- 34.Whelan S M, Elmore M J, Bodsworth N J, Brehm J K, Atkinson T, Minton N P. Molecular cloning of the Clostridium botulinum structural gene encoding the type B neurotoxin and determination of its entire nucleotide sequence. Appl Environ Microbiol. 1992;58:2345–2354. doi: 10.1128/aem.58.8.2345-2354.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whelan S M, Elmore M J, Bodsworth N J, Atkinson T, Minton N P. The complete amino acid sequence of the Clostridium botulinum type-E neurotoxin, derived by nucleotide-sequence analysis of the encoding gene. Eur J Biochem. 1992;203:657–667. doi: 10.1111/j.1432-1033.1992.tb16679.x. [DOI] [PubMed] [Google Scholar]