Abstract
The German government initiated the Network University Medicine (NUM) in early 2020 to improve national research activities on the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) pandemic. To this end, 36 German Academic Medical Centers started to collaborate on 13 projects, with the largest being the National Pandemic Cohort Network (NAPKON). The NAPKON’s goal is creating the most comprehensive Coronavirus Disease 2019 (COVID-19) cohort in Germany. Within NAPKON, adult and pediatric patients are observed in three complementary cohort platforms (Cross-Sectoral, High-Resolution and Population-Based) from the initial infection until up to three years of follow-up. Study procedures comprise comprehensive clinical and imaging diagnostics, quality-of-life assessment, patient-reported outcomes and biosampling. The three cohort platforms build on four infrastructure core units (Interaction, Biosampling, Epidemiology, and Integration) and collaborations with NUM projects. Key components of the data capture, regulatory, and data privacy are based on the German Centre for Cardiovascular Research. By April 01, 2022, 34 university and 40 non-university hospitals have enrolled 5298 patients with local data quality reviews performed on 4727 (89%). 47% were female, the median age was 52 (IQR 36–62-) and 50 pediatric cases were included. 44% of patients were hospitalized, 15% admitted to an intensive care unit, and 12% of patients deceased while enrolled. 8845 visits with biosampling in 4349 patients were conducted by April 03, 2022. In this overview article, we summarize NAPKON’s design, relevant milestones including first study population characteristics, and outline the potential of NAPKON for German and international research activities.
Trial registration https://clinicaltrials.gov/ct2/show/NCT04768998. https://clinicaltrials.gov/ct2/show/NCT04747366. https://clinicaltrials.gov/ct2/show/NCT04679584
Supplementary Information
The online version contains supplementary material available at 10.1007/s10654-022-00896-z.
Keywords: COVID-19, SARS-CoV-2, Prospective national cohort, Longitudinal study, Epidemiology, Cross-sectoral, Population-based
Introduction
The pathogen Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) started to spread at the end of 2019 [1, 2], initiating the Coronavirus Disease 2019 (COVID-19) pandemic [3]. As foreshadowed by the 2019 Global Health Security (GHS) report, no country was “fully prepared for epidemics or pandemics, and every country has important gaps to address” [4]. More than two years into the pandemic, by April 01, 2022, the World Health Organization (WHO) reported 485 million positive cases, and over 6.1 million deaths worldwide [5]. For Germany, the Robert Koch Institute (RKI) reported over 21.4 million positive cases and close to 130,000 deaths (as of April 01, 2022) [6].
In its pandemic preparedness program checklist, the WHO dedicates an entire section to “Research and Development”, listing essential and desirable activities for countries. Such activities include the development of study protocols, documentation of the evolution of epidemiological/clinical features, and outbreak investigations [7]. The RKI states similarly that “studies should be planned and prepared in advance of the pandemic so that they can be conducted rapidly at any time” [8]. However, Germany’s latest national pandemic plan from 2017 does not address specific research activities and contains no plans for studies of respective patient collectives [9]. Given the first signs of a pandemic in early 2020, the German Federal Ministry of Education and Research (BMBF) focused on the national establishment and streamlining of COVID-19 related scientific activities. As a result, the BMBF founded the Network University Medicine (NUM) in late March 2020 [10] to coordinate national research activities on SARS-CoV-2/COVID-19 and ensure pandemic preparedness of German academic medical centers in the future. The NUM has thus initiated joint SARS-CoV-2/COVID-19 research activities via 13 complementary projects by leveraging and connecting elements of existing academic research infrastructure in Germany, including all 36 university hospitals and additional collaborating (non-university) health care institutions [11]. The BMBF plans to continue the funding for NUM until 2024 [12].
The NUM’s largest project, the National Pandemic Cohort Network (NAPKON), is aiming to establish a standardized, high-quality data and biosample collection on patients, citizens, and controls with comparator respiratory infections. Next to international activities such as International Severe Acute Respiratory and emerging Infection Consortium (ISARIC) [13], many nations set up COVID-19 cohorts throughout 2020 across the globe [14–24].The NAPKON was initiated in July 2020 as Germany’s most comprehensive COVID-19 cohort. It was delineated from and aligned with three complementary German cohorts: the two already existing Lean European Open Survey on SARS-CoV‑2 infected patients (LEOSS) [25] and the Berlin prospective COVID-19 patient cohort (Pa-COVID-19) [26], as well as the proposal for the Post-COVID-Syndrome Study (COVIDOM) [27].
Here we report in detail on the NAPKON’s objectives, structures, and design. We present relevant milestones achieved and outline the potential of the NAPKON for German and international research activities.
Methods
Study design
The primary aim of the NAPKON is to create a harmonized, expandable, and interoperable network to support both the fight against the current COVID-19 pandemic as well as future pandemics of any origin.
The NAPKON consists of three parallel and complementary prospective cohort platforms that collect data and biosamples of SARS-CoV-2 infected patients, citizens, as well as controls with comparator respiratory infections during the acute phase and follow-up. Objectives for the usage of data and biosamples are to:
Investigate frequency, severity and distinct phenotypes of COVID-19 and Post-COVID-19 Syndrome (PCS) in the German population and identify long-term clinical trajectories of PCS. POP
Identify genomic, epigenomic, transcriptomic, proteomic, and metabolomic signatures predicting course and outcome of acute and post-acute COVID-19.
Decipher further central pathophysiologic mechanisms of specific COVID-19 related pathologies in order to inform development of therapies.
Establish commonalities and differences between COVID-19 and other forms of respiratory tract infections, pneumonia and acute respiratory distress syndrome (ARDS) in detail.
Understand reasons for development of acute or post-acute COVID-19 in SARS-CoV- 2 vaccinated patients.
Each of the three cohort platforms focuses on one or multiple of the above scientific areas of interest. The following paragraphs will introduce the three cohort platforms and supporting infrastructure elements. Cross-sectoral platform
The Cross-Sectoral Platform (Sektorenübergreifende Plattform, SUEP) cohort recruits SARS-CoV-2 infected in- and outpatients of all ages across all departments and performs a comprehensive collection of primary health record data, basic clinical phenotyping (e.g., echocardiography, spirometry with full-body plethysmography) with biosample collection, and patient interviews/patient-reported outcome measures (PROM) (see Tables 1, 2) across all levels of health care providing facilities. This ensures cross-sectoral patient acquisition in the NAPKON. All German university and non-university hospitals and primary care practices can become study sites. In addition, mobile hotspot study teams are planned to cover long-term care and rehabilitation facilities. The cohort is registered at www.clinicaltrials.gov under NCT04768998.
Table 1.
Overview of data collected within NAPKON by cohort platform
| Category | Features | Collected in | ||
|---|---|---|---|---|
| SUEP | HAP | POP | ||
| Socio-demographic data | Age, sex, residence, marital status | x | x | x |
| Educational level and employment status (e.g. general education degree, vocational degree) | x | x | x | |
| Clinical data: pre-infection anamnestic data | Pre-infection lifestyle (e.g. sports activity, dietary pattern) | x | x | |
| Pre-infection smoking and alcohol consumption | x | x | x | |
| Pre-infection health status and functionality (e.g. Barthel Index, care level, Clinical Frailty Scale) | x | x | x | |
| Pre-infection medication | x | x | ||
| Vaccination status | x | x | x | |
| Comorbidities | x | x | x | |
| Directives for medical decisions (e.g. power of attorney, patient decree) | x | x | ||
| Clinical data: parameters in the observational period | Infrastructural treatment context (e.g. health care facility, involved disciplines) | x | x | x |
| Smoking and alcohol consumption | x | x | ||
| Health status and functionality (e.g. Barthel index, care level, Clinical Frailty Scale) | x | x | x | |
| Symptoms, events | x | x | x | |
| Clinically indicated diagnostics (vital signs, pulmonary diagnostics, laboratory parameters, microbiology & virology, radiological findings, functional diagnostics) | x | x | x | |
| Intensive care scores (e. g. SOFA, SAPS) | x | x | ||
| Therapeutic measures (medication, interventions, surgery, complementary medicine) | x | x | x | |
| Pediatric-specific variable extensions (e.g. perinatal medical history, congenital defects, effects on development) | x | |||
| Imaging data | Clinically indicated diagnostic imaging data | x | x | |
| Study-related MRI scans | x | x | ||
| Study related CT-Thorax scans | x | |||
| Study-related echocardiographies | x | x | x | |
| Patient-reported outcome measures (PROM) | Cognitive function (e.g. PROMIS Kognition) | x | x | x |
| Dypsnea (e.g. Modified Medical Research Council Dyspnea Scale, PROMIS Dyspnoe) | x | x | x | |
| Fatigue (e.g. Chalder Fatigue Scale, FACIT-F) | x | x | x | |
| Functional physical status (e.g. Activities of Daily Living) | x | x | x | |
| Mental health (e.g. GAD-7, Brief Resilience Scale) | x | x | x | |
| Pain (e.g. DN2, HIT-6) | x | x | x | |
| Quality of life (e.g. EQ-5D-5L) | x | x | x | |
| Metadata | Study-related metadata (e.g. data quality assessment, protocol deviation) | x | x | x |
| Digital Imaging and Communications in Medicine (DICOM) header information | x | x | x | |
| Biosample accompanying metadata (e.g. regarding transport, processing and storage) | x | x | x | |
Table 2.
Overview of additional study assessments
| Study assessmenta | SUEP | HAP | POP |
|---|---|---|---|
| Abdominal ultrasonography | x | ||
| Additional medical history and recording by study physician | x | x | x |
| Basic endocrinological diagnostics | x | x | |
| Computer tomography chest | x | ||
| Electrocardiography | x | x | x |
| Electroencephalography | x | ||
| Fraction Exspiratory Nitric Oxide | x | ||
| Fundus examination | x | ||
| Home visit | x | ||
| Impulse oscillometry | x | ||
| Long-term ECG | x | ||
| Long-term glucose measurement | x | ||
| Long-term RR | x | ||
| Magnetic resonance imaging brain | x | x | |
| Magnetic resonance imaging heart | x | ||
| Microbiome sampling | x | x | x |
| Myocarditis panel | x | x | |
| Basic neurological examination | x | x | |
| 6-Min walking test | x | ||
| Smell test | x | x | |
| Spiroergometry | x | ||
| Standard laboratory outpatients | x | ||
| Standardized spirometry with bodyplethysmography and diffusion capacity | x | x | x |
| Taste test | x | x | |
| Transthoracic echocardiography | x | x | x |
| Vital sign monitoring | x | x | x |
aModified for patients age < 18.
SUEP, cross-sectoral platform; HAP, high-resolution platform; POP, population-based platform
High-resolution platform
The High-Resolution Platform (Hochauflösende Plattform, HAP) cohort focuses on adult SARS-CoV-2 positive inpatients, especially those with a severe course of COVID-19, i.e., in need of intensive care unit treatment. Within the HAP, data and biosample collection are extended by a multidisciplinary study program of additional clinical examinations, supplementary cytokine profiling, and standardized imaging. The longitudinal biosample collection is of a much higher frequency as compared to the other two cohorts (see Tables 1, 2). The HAP cohort is conducted at ten German university hospitals with an adequate infrastructure for deep phenotyping. The cohort is registered at www.clinicaltrials.gov under NCT04747366.
Population-based platform
The Population-Based Platform (Populationsbasierte Plattform, POP) cohort focuses on describing health consequences of SARS-CoV-2 infection in the general adult population. It is conducted at established epidemiological centers at three university hospitals. Recruitment bias is minimized by contacting a stratified sample of SARS-CoV-2 positive individuals in three locally and structurally distinct German regions. Individuals are identified and contacted via local health authorities that are mandated to register all SARS-CoV-2 infections in their administrative districts. After consenting eligible individuals undergo a telephone interview and are subsequently invited for a baseline visit in the study center and yearly follow-ups. Visits include comprehensive clinical and functional health assessment in distinct organ systems, further interviews/PROM assessment, and biosample collection (see Tables 1, 2). The POP is registered at the Deutsches Register Klinischer Studien (DRKS) under DRKS00023742.
Infrastructure elements of the NAPKON
The NAPKON cohorts rest on a harmonized shared infrastructure provided by four NAPKON core units:
The Interaction Core Unit (ICU) coordinates overall governance, support of the use & access processes, development of datasets, engagement of the scientific community via working parties and a scientific council, age-specific consideration of study aspects via a dedicated Pediatric Core Unit, convening of the general assembly, and other tasks related to internal project management or communication.
The Biosample Core Unit (BCU) selects suitable biosamples together with clinical experts, defines standards of procedure for sampling, processing, storage, quality assurance, as well as regular auditing of biobanks.
The Epidemiology Core Unit (ECU) is responsible for methodological consultation of the project and third parties applying for data/biosamples. It performs an external quality assurance and reporting of collected project data.
The Integration Core Unit (IGCU) designs and manages the integration of external and existing cohort data into the NAPKON.
Since knowledge on COVID-19 was evolving quickly at the time the NAPKON was initiated, it was of highest priority to not only collect a comprehensive set of data and biosamples, but also to be able to change protocols and strategies on the move. This included the option to recontact patients and obtain consent for new and/or additional study interventions, e.g., inclusion in substudies and follow-up of children and adolescents after they attain full age. Central data storage and linkage of data from different sources requires a complex setup with trusted third-party and extensive communication and approval by data protection officers. Such a system was provided by the German Centre for Cardiovascular Research (Deutsches Zentrum für Herz-Kreislauf-Forschung, DZHK), including the following components [28]:
Clinical data management electronic case report form (eCRF) for documentation of all clinical data, including additional tests performed as part of the study.
Imaging data management central storage for clinical routine images and additional images collected by study protocol.
Biosample management central laboratory information and management system (cLIMS).
Ethics coordination coordination of ethical aspects regarding data-infrastructure and governance and professional communication with ethics committees and institutional review boards.
Trusted third party centralized quality assurance and management of (electronic) informed consents, including supporting the invitation of patients to follow-up studies.
Data transfer office provision of datasets and identification of the respective biosamples based on applications approved by the Use & Access committee.
Study population and recruitment
Besides informed consent, the shared inclusion criterion (for primary cases—controls see below) of all three cohorts is a SARS-CoV-2-positive polymerase chain reaction (PCR) of a swab or body fluid.Alternatively, a negative molecular test with a very high clinical suspicion for a SARS-CoV-2 infection is regarded as a positive case (see Table 3 for details). Besides age < 18 for the POP and the HAP no exclusion criteria exist. The SUEP and HAP recruit patients within one week of meeting the inclusion criteria, the POP within 6–12 months after positive testing.
Table 3.
Case and control definitions for the NAPKON.
| Case definition for SARS-CoV-2 infection (= inclusion criteria) | Control definition |
|---|---|
|
Either: Positive polymerase chain reaction (PCR) for SARS-CoV-2 in either Oro/nasopharyngeal swab, BAL, sputum, tracheal secretions, stool, or blooda Or (all of the following): Negative polymerase chain reaction (PCR) for SARS-CoV-2 of a swab or body fluid Definitive infection of the respiratory system Characteristic radiographic imagery A negative test for influenza Exclusion of other potential causes (like chronic diseases of the respiratory system) |
Case definition for SARS-CoV-2 case not applicable Applicable control inclusion criteria for one of the three control strata (pool) Outpatient (e.g., respiratory viral infection) Inpatient (e.g., community-acquired pneumonia) Intensive care unit (e.g., acute respiratory distress syndrome) Capacity for control recruitment with sufficient positive cases in the respective pool over the past eight weeks |
Additional case definitions exist for patients age < 18. No exclusion criteria exist, except for age < 18 for the POP and the HAP
aAntibody testing or rapid tests are no viable alternatives
In total, the NAPKON aims to prospectively recruit and follow 7000 individuals (patients and controls) in the years 2020–2024, starting in November 2020. This sample size is delineated from allocating available funding evenly across high quality data and biosample acquisition in all disease strata (see Suppl Table S1 in the supplements). Study sites include all interested German university hospitals (SUEP & HAP, about 40% of the total study population), non-university hospitals, primary care practices, long-term care/rehabilitation facilities (SUEP, about 15% of the total study population), and patients/citizens contacted via local health authorities in three catchment areas (POP, about 45% of the total study population). Strata for various disease severities exist (see Suppl Table S1) and the recruitment for each cohort is balanced for disease severity. Additional stratification criteria for age, sex, infection date, vaccination status apply depending on the cohort. Given their capacities, the participating study sites are asked to consecutively enroll all eligible patients into the NAPKON. In addition to German, electronic and paper consent forms exist in eight different languages (e.g., English, Arabic, Turkish and French). Dedicated delegation procedures for patients who are incapable of giving consent (e.g., pediatric or unconscious patients) are available at most sites, further reducing selection bias.
A key asset of the NAPKON is the inclusion of 20% controls based on the pool recruitment method [29, 30]. The initial approach to compare COVID-19 with influenza proved not feasible due to very low infection rates by influenza virus in the 2020/21 season. To match comparator conditions effectively with different levels of severity of COVID-19, we therefore defined three different strata of controls (see Table 3).
Visit schedules and follow-up
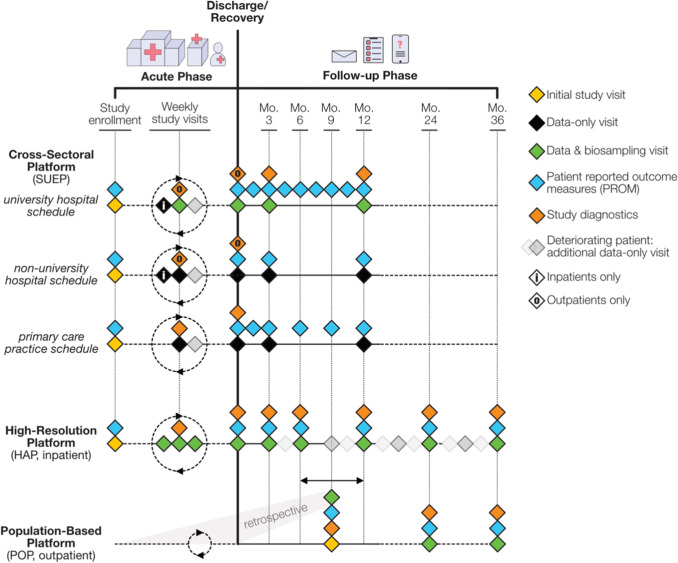
The three NAPKON cohorts follow a harmonized visit schedule. Different intervals and visit types at the study center apply depending on the clinical setting (inpatient/outpatient), study site (university hospital, non-university hospital, primary care practice), patient age (adult vs. pediatric) and course of disease (acute vs. follow-up). Figure 1 juxtaposes the various study schedules of the three NAPKON cohorts. Recruitment of new patients for the SUEP and the HAP are planned until the end of 2023, for the POP until the end of 2022. Follow ups are planned until 2024 for all cohorts. Patients lost to follow-up are not replaced.
Fig. 1.
Visit schedules of the three NAPKON cohort platforms. During the acute phase, data collection and various study diagnostics are scheduled weekly. In case of complications, routine laboratory data and vitals parameters are additionally documented once a week. University hospitals collect biosamples weekly during study visits. Follow-up visits (scheduled in reference to initial diagnosis of SARS-CoV-2 infection) of patients include in-clinic study diagnostics (with biosampling at university hospitals) and questionnaires for PROMs. The POP documents the acute course of its patients retrospectively and performs its comprehensive in-clinic follow-up visits (including biosampling) roughly in yearly intervals [27].
Clinical assessment
During study and follow-up visits, patients undergo age adapted comprehensive clinical assessments according to the respective cohort protocol. All cohort platforms perform extensive laboratory testing, echocardiography, spirometry with full body plethysmography and diffusing capacity, vital sign measurement, and clinical examination. The HAP and the POP perform additional tests of fitness and organ function as well as imaging studies (see Table 2), such as brain magnetic resonance imaging (MRI) and cardio MRI examinations during convalescence, including, amongst others, quantitative multi parametric mapping [31, 32] and stress perfusion and late gadolinium enhancement [33].
Biosample collection
Across the NAPKON cohorts, a common set of quality-assured biosamples is prospectively collected at specific study visits (see Fig. 1 for timing) and processed according to standard operating procedures (SOPs) for subsequent storage in local biobanks. Biosample metadata that fully captures all processing steps are stored in the cLIMS infrastructure to allow central tracking of all biosamples. Patients and/or their legal representatives can decline sampling at any time. Biosample collection primarily involves university sites but can also be conducted by non-university hospitals collaborating with local biobanks. The BCU coordinators train any local biobank staff via tele-education tools for all processes related to collection, processing, storage, and shipment of samples as described in the respective SOPs (recordings and documents available in German at www.bbmri.de/covid-19/nationales-pandemie-kohorten-netz/). Also, the BCU verifies compliance during regular audits at all sites. The NAPKON’s protocols permit study sites to collect further optional biosamples for the site’s own research interests. Systematic molecular phenotyping includes epigenetic, transcriptomic, proteomic, immunogenicity and metabolomic analyses of the patient samples as well as sequencing of all respiratory samples.
Data collection
In all the NAPKON cohorts, study personnel manually transfer primary data and metadata stored in the patient’s health care record at the respective study site or acquired during the study to eCRFs. All data are captured prospectively, apart from anamnestic pre-infection data and the POP’s retrospective assessment of the acute phase. Local study staff collects PROMs as paper-/web-based questionnaires or conducts phone interviews. Quality measures of the clinical data include:
Automatic predefined plausibility and completeness checks in the eCRFs
Local (Review A) and central (Review B) quality assessment
Random source data verification of 10%-20% of the cases.
To further improve quality, the ECU provides a methodological-epidemiological consultation platform on issues related to the planning, conduct, and analysis of the cohorts. Data quality reports on selected indicators, primary coding of core data, definition of plausibility ranges, SOPs, and statistical analysis planning including sample size reviews for use and access procedures are provided as part of this service.
Given the harmonized but individual data collection of the three NAPKON cohorts, each maintains its dedicated eCRF. All NAPKON eCRFs contain the German Corona Consensus Dataset (GECCO-83) [34], ensuring syntactic and semantic interoperability for a core dataset via international terminologies (e.g., International Statistical Classification of Diseases and Related Health Problems, 10th revision, German modification (ICD-10-GM) [35], Logical Observation Identifiers Names and Codes (LOINC) [36], the Anatomical Therapeutic Chemical Classification System (ATC) [37], Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) [38]) and defined Health Level 7 (HL7) standard Fast Healthcare Interoperability Resources (FHIR) profiles [39]. The cohorts selected additional data elements by incorporating international data sets (e.g., ISARIC [40]), already established German COVID-19 cohorts (e.g., LEOSS [41], Pa-COVID-19 [42]), and suggestions of scientists (see section “Governance”). By choice of design, the NAPKON’s clinical/imaging data set definitions and biosample panel allow adjustments and extensions via add-on modules. Tables 1 and 2 provide an overview of the data currently collected across the three NAPKON cohorts.
The later presented baseline characteristics of the current NAPKON cohort use descriptive summary statistics for all patients included across the three cohorts that passed Review A. All analyses have been performed with the statistics software R, version 4.0.2.
Study organization
Governance and use and access
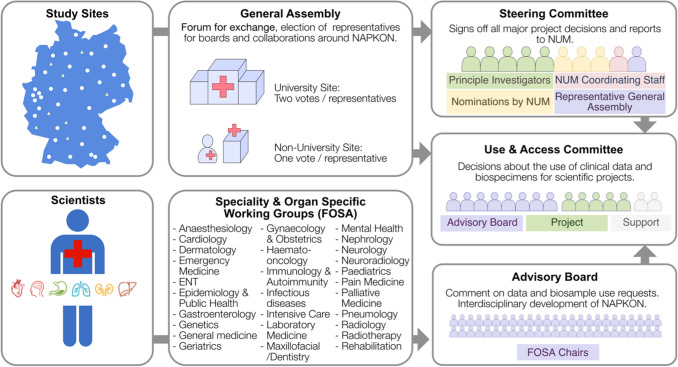
The ICU assumes the overall coordination of the NAPKON. Figure 2 illustrates the stakeholders’ positions in the following governance structures: General Assembly (GA), Advisory Board (AB), Use & Access Committee (UAC), Steering Committee (SC), and Specialty- and Organ-Specific Working Groups (Fach- und Organspezifische Arbeitsgruppen, FOSAs). The SC devises and approves regulating documents for the SC, FOSA, and UAC, as well as the usage regulations and publication regulations. Rules of procedure require gender parity for the FOSAs, AB, and the SC and actively encourage it in the GA.
Fig. 2.
Flow-diagram of the NAPKON governance. Study sites and scientists are prominently included in most governance processes
Groups greater than ten individuals from different university hospitals can establish a FOSA that is open to the general scientific community. The core responsibilities of the FOSAs are to comment on and revise the NAPKON’s data sets and develop subject-specific research questions for analyses. FOSA chairs form the AB, and interdisciplinary advise the NAPKON’s committees and study platforms.
The ICU hosts and supports regular meetings for all governance organs. Scientists can submit applications that outline the reasoning and methodology for their research questions and are also expected to describe how they plan to address limitations of the NAPKON data, e.g., when combining cases across different health care settings or recruitment funnels. Both, the Epidemiology Core Unit and the Advisory Board, participate in the UAC process to extend the UAC’s review. Data privacy & ethics
Study protocols and consent forms detail all patient-related activities for children, adolescents and adults in the NAPKON. The patient documents are harmonized in a way that enables the data from three cohorts to be used in an intersectional manner. All participants are informed about the shared data management and overarching governance and agree to the use of their data for research regarding the description, detection, treatment and prevention of SARS-CoV-2 infection and COVID-19 disease research via the consent form. Further information is provided on the patient information website, for example regarding responsibilities for data processing (https://napkon.de/pat/datenschutz). Patients and/or their legal representatives can withdraw their data or biosample use at any time without giving reasons (https://napkon.de/pat/datenschutz).
The DZHK Trusted Third Party provides centrally managed pseudonyms throughout the NAPKON which prevents identification of individuals by unauthorized persons while allowing linkage of different types of data and biosamples. Scientists working with the NAPKON data or biosamples must adhere to relevant UAC privacy regulations. A restrictive procedure for transmission of any data or biosamples to researchers who are not bound by European data privacy laws is in place. Members of the Use & Access process and individuals who manage incoming research proposals are subject to confidentiality agreements.
Project management infrastructure
The NAPKON self-hosts collaboration and project management solutions under the umbrella term “NAPKON Suite” on its website https://napkon.de. It includes collaborative cloud space, mailing lists, a contact directory, email inboxes, project management software, and administrative tools. Via a single-sign-on and group memberships, all participating scientists, local study teams, and governance members have access to respective services.
The NAPKON Suite provides supporting materials (e.g., SOPs, video recordings, FAQs, flow charts) to all participating study sites. The NAPKON’s homepage bundles all relevant information for different stakeholders, including patient information and short summaries of the UAC-approved scientific projects. Project milestones and deliverables are tracked in an OpenProject [43] implementation as part of the NAPKON Suite.
Study budget
The total budget of the NAPKON from August 06, 2020 until December 31, 2024 is close to 55 Mio €, not including the funding for the DZHK. More than half of the total budget is used for case fees, about 15% site setup costs, with the remainder being allocated towards cohort governance and infrastructure core unit setup/management.
Results
The results presented in the following reflect the status of the cohorts on April 01, 2022, roughly one and a half years after the first patient was recruited.
Governance and use and access
The ICU set up all the NAPKON’s governance structures and reconciled legal documents with the NUM coordination (e.g., data privacy agreement, usage regulations, publication regulations, and others). The NAPKON’s SC convenes biweekly since September 08, 2020. The first digital GA meeting was held on December 17, 2020, the second on November 08, 2021. AB and UAC had their first meeting on February 04, 2021, and March 05, 2021, respectively. ICU held an additional study site & investigator meeting on February 28, 2021. Organized by the ICU, five events titled “FOSA Lectures” have taken place in which FOSAs shared and discussed their current findings on the COVID-19 pandemic.
By April 01, 2021, the NAPKON counts more than 1,500 involved members in the NAPKON Suite. About 70 of these members are part of the general coordination, who generally convene biweekly via video conferencing. Local study groups generally consist of about three to 20 people each. Twenty-eight different FOSAs were formed with over 600 national scientists (Fig. 2 lists all FOSA specialties). Consequently, the AB consists of 56 members. The NAPKON FOSAs were decision-maker in the content definition for three extension modules to the GECCO-83 developed within NAPKON to harmonize focused data collection in cardiology, pediatrics, and vaccination with respect to the pandemic development. Since January 01, 2022 the responsibilities of the FOSA and the AB have been extended across all NUM projects.
The NAPKON’s Use & Access process was tested in March 2021 and opened on April 26, 2021. It relies on the NUM usage regulations, the NAPKON/NUM publication regulations, and the NAPKON usage regulations for biosamples. Interested scientists can access those documents and submit research proposals via https://proskive.napkon.de. The UAC (see Fig. 2) discusses and votes on proposals while respecting efficient processes tailored to the current pandemic situation. Usage regulations grant government agencies and other public health institutions privileged access to the data collected in the NAPKON. The AB members can comment on incoming requests for data and biosamples. The first incoming applications were circulated in the AB on May 03, 2021, and in the UAC on May 12, 2021. By April 01, 2022, 92research proposals have been submitted to the UAC. Of those, the UAC approved 80, declined three with the remaining requests still pending. 24 proposals have received data already, three biosamples. Currently the average time from submission until approval is 12 days.
The ECU's methodological-epidemiological consultancy service has processed more than 20 scientific inquiries to date. ECU also reviews applications to the UAC regarding design and sample size. To describe the content of the cohort data, (center-specific) core analyses have been provided regularly since April 2021.
The integration core unit has developed concepts to incorporate data from cohorts collected prior to the NAPKON. Use cases are being implemented to demonstrate the feasibility of such data integration in the HAP and the SUEP.
Cohorts and study population
All three NAPKON cohorts recruited their first patients in November 2020, 4 months after the initial submission of the NAPKON’s proposal to the NUM. For the SUEP, 28 university hospitals, 17 non-university hospitals, and 23 primary care practices recruit patients, for the HAP and the POP 10 and 3 university hospitals, respectively. 34 of 36 university hospitals in Germany participate in the NAPKON. The extensive protocols feature more than 90 SOPs for clinical tests and diagnostics, multiple imaging follow-ups, and standardized biosample collection. Twenty expressions of interest integrating previous studies’ data or biosamples into the NAPKON infrastructure were solicited by the IGCU, 13 of which met the criteria defined for successful integration. Two initial integration use cases for the HAP and the SUEP, respectively, were launched in April 2021 after a legal opinion helped clarify pending regulatory issues.
The FOSAs have thoroughly reviewed and updated the eCRFs and made many hundred individual suggestions, including about 150 new and 25 removed variables, the definition of plausibility limits, the creation of clinical definitions, the addition of anamnesis checklists, and the revision of the study concept for outpatient practices. The AB led and defined the PROM specification in the NAPKON and was heavily involved in the follow-up visit design. Dedicated committees have worked on add-on modules for cardiology, neurology, dermatology, and pediatrics. The pediatric module, includes specific case definitions for pediatric patients and controls, age-specific plausibility checks, age-adopted medical and social variables. It was activated on May 12, 2021. Other add-on modules (e.g., age-adopted PROMs) are expected to follow soon. The AB members further discussed ideas on maintaining the broadest possible interdisciplinary collaboration between the NUM and the NAPKON in the future. The ideas are part of the “NAPKON Follow-Up Application 2022–2024” that was submitted on June 30, 2021, to the NUM. The development of a generic recruitment infrastructure was finally included in the NUM Clinical Epidemiology and Study Platform (German: NUM Klinische Epidemiologie und Studien Plattform, NUKLEUS) project description, which was approved on December 14, 2021.
By April 01, 2022, the NAPKON has recruited a total of 5298 patients across all cohorts including 17 controls in the SUEP. Study sites have recruited almost 600 patients per month at peak times. Table 4 presents the characteristics of the study population in the style of the ECU’s core analyses for 4727/5298 (89%) SARS-CoV-2 positive patients with a Review A status (local data quality review performed). 2202/4727 (47%) are female, 2524/4727 (53%) are male, 0 are non-binary. The median age was 56 (IQR 42–68), 57 (IQR 47–65), and 46 (IQR 31–68) for the SUEP, the HAP, and the POP respectively. 50 pediatric cases had a positive Review A status at the point of the analysis. 1834/4161 (44%; SUEP: 88%, HAP: 100%, POP: 7%) patients were hospitalized, 611/3997 (15%; SUEP: 33%, HAP: 38%, POP: 2%) admitted to an intensive care unit, and 214/1800 (4%; SUEP: 12%, HAP: 12%, POP: 0%) patients deceased while enrolled. 1741/3581 (49%; SUEP: 49%; HAP: 19%; POP: 55%) had a full vaccination status. 696/1258 (55%), 113/182 (62%), and 256/694 (37%) patients have been followed-up at 3, 6 and 12 months across the cohorts. The POP did baseline visits with 2,346 patients at 6-12 months post primary infection. Pre-existing comorbidity distributions for the SUEP, the HAP and the POP included 50%, 50%, 29% chronic cardiovascular disease; 19%, 21%, and 19% chronic lung disease; 12%, 17%, and 0.5% chronic kidney disease; 13%, 15%, and 25% chronic neurological or psychiatric disease. 130/5,298 (2%) patients withdrew at least parts of their data by March 2022. No information about the completeness of recruitment per center was available at the point of analysis. Missing data due to a pending Review A status or other reasons is indicated in Table 4.
Table 4.
Description of the study population with review A status (with a total patient population of 5298) by cohort until April 01, 2022
| Variable | N1 | Statistic | HAP, N = 544 | POP, N = 2346 | SÜP, N = 1837 |
|---|---|---|---|---|---|
| Age (numeric) | 4727 | Median (IQR) | 57 (47, 65) | 46 (31, 57) | 56 (42, 68) |
| Age (categorical) | 4727 | ||||
| < 18 | n (%) | 0 (0%) | 0 (0%) | 50 (2.7%) | |
| 18–29 | n (%) | 33 (6.1%) | 501 (21%) | 138 (7.5%) | |
| 30–39 | n (%) | 51 (9.4%) | 446 (19%) | 205 (11%) | |
| 40–49 | n (%) | 83 (15%) | 360 (15%) | 280 (15%) | |
| 50–59 | n (%) | 156 (29%) | 608 (26%) | 368 (20%) | |
| 60–69 | n (%) | 128 (24%) | 260 (11%) | 379 (21%) | |
| 70–79 | n (%) | 69 (13%) | 140 (6.0%) | 255 (14%) | |
| 80+ | n (%) | 24 (4.4%) | 31 (1.3%) | 162 (8.8%) | |
| Gender | 4726 | ||||
| Female | n (%) | 174 (32%) | 1305 (56%) | 723 (39%) | |
| Male | n (%) | 370 (68%) | 1040 (44%) | 1114 (61%) | |
| Non-binary | n (%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| Missing or review A pending | n | 0 | 1 | 0 | |
| Smoking (past or current smoker) | 3867 | ||||
| Yes | n (%) | 28 (6.8%) | 1068 (49%) | 128 (9.9%) | |
| No | n (%) | 381 (93%) | 1100 (51%) | 1162 (90%) | |
| Missing or review A pending | n | 135 | 178 | 547 | |
| Alcohol | 2433 | ||||
| Never | n (%) | 0 (NA%) | 183 (13%) | 516 (50%) | |
| Up to 4 times monthly | n (%) | 0 (NA%) | 676 (48%) | 377 (36%) | |
| Multiple times weekly | n (%) | 0 (NA%) | 536 (38%) | 145 (14%) | |
| Missing or review A pending | n | 544 | 951 | 799 | |
| Obesity at inclusion (BMI ≥ 30 kg/m2) | 4169 | ||||
| No | n (%) | 311 (65%) | 1758 (76%) | 900 (66%) | |
| Yes | n (%) | 167 (35%) | 566 (24%) | 467 (34%) | |
| Missing or review A pending | n | 66 | 22 | 470 | |
| SARS-CoV-2 vaccined | 3581 | ||||
| Yes | n (%) | 93 (19%) | 1189 (55%) | 459 (49%) | |
| No | n (%) | 393 (81%) | 965 (45%) | 482 (51%) | |
| Missing or review A pending | n | 58 | 192 | 896 | |
| In-patient ever | 4161 | ||||
| Yes | n (%) | 544 (100%) | 170 (7.3%) | 1120 (88%) | |
| No | n (%) | 0 (0%) | 2172 (93%) | 155 (12%) | |
| Missing or review A pending | n | 0 | 4 | 562 | |
| Intensive stay ever | 3997 | ||||
| Yes | n (%) | 208 (38%) | 36 (1.5%) | 367 (33%) | |
| No | n (%) | 336 (62%) | 2301 (98%) | 749 (67%) | |
| Missing or review A pending | n | 0 | 9 | 721 | |
| Covid-associated oxygenation | 4619 | ||||
| Invasive/non-invasive ventilation | n (%) | 141 (26%) | 17 (0.7%) | 299 (17%) | |
| O2-therapy only | n (%) | 260 (48%) | 93 (4.0%) | 746 (43%) | |
| No assistance | n (%) | 142 (26%) | 2222 (95%) | 699 (40%) | |
| Missing or review A pending | n | 1 | 14 | 93 | |
| Extracorporeal membrane oxygenation (ECMO) | 4047 | ||||
| Yes | n (%) | 60 (13%) | 1 (<0.1%) | 47 (3.8%) | |
| No | n (%) | 417 (87%) | 2334 (100%) | 1188 (96%) | |
| Missing or review A pending | n | 67 | 11 | 602 | |
| Chronic cardiovascular disease | 3842 | ||||
| Yes | n (%) | 269 (50%) | 601 (29%) | 622 (50%) | |
| No | n (%) | 265 (50%) | 1456 (71%) | 629 (50%) | |
| Missing or review A pending | n | 10 | 289 | 586 | |
| Chronic lung disease | 4013 | ||||
| Yes | n (%) | 109 (21%) | 425 (19%) | 235 (19%) | |
| No | n (%) | 417 (79%) | 1822 (81%) | 1005 (81%) | |
| Missing or review A pending | n | 18 | 99 | 597 | |
| Chronic kidney disease | 4088 | ||||
| Yes | n (%) | 88 (17%) | 8 (0.3%) | 142 (12%) | |
| No | n (%) | 436 (83%) | 2323 (100%) | 1091 (88%) | |
| Missing or review A pending | n | 20 | 15 | 604 | |
| Chronic liver disease | 3722 | ||||
| Yes | n (%) | 44 (8.4%) | 181 (9.2%) | 84 (6.8%) | |
| No | n (%) | 480 (92%) | 1788 (91%) | 1145 (93%) | |
| Missing or Review A pending | n | 20 | 377 | 608 | |
| Rheumatological/immunological disease | 4053 | ||||
| Yes | n (%) | 32 (6.1%) | 219 (9.5%) | 60 (4.9%) | |
| No | n (%) | 492 (94%) | 2075 (90%) | 1175 (95%) | |
| Missing or review A pending | n | 20 | 52 | 602 | |
| Diabetes mellitus | 4001 | ||||
| Yes | n (%) | 110 (21%) | 101 (4.5%) | 266 (21%) | |
| No | n (%) | 418 (79%) | 2129 (95%) | 977 (79%) | |
| Missing or review A pending | n | 16 | 116 | 594 | |
| Solid tumor disease | 4092 | ||||
| Yes | n (%) | 57 (11%) | 39 (1.7%) | 150 (12%) | |
| No | n (%) | 478 (89%) | 2294 (98%) | 1074 (88%) | |
| Missing or review A pending | n | 9 | 13 | 613 | |
| Haematological-oncological disease | 4074 | ||||
| Yes | n (%) | 29 (5.5%) | 7 (0.3%) | 63 (5.2%) | |
| No | n (%) | 498 (94%) | 2323 (100%) | 1154 (95%) | |
| Missing or review A pending | n | 17 | 16 | 620 | |
| HIV infection | 3948 | ||||
| Yes | n (%) | 3 (0.6%) | 2 (<0.1%) | 17 (1.5%) | |
| No | n (%) | 473 (99%) | 2336 (100%) | 1117 (99%) | |
| Missing or review A pending | n | 68 | 8 | 703 | |
| Chronic neurological or psychiatric disease | 3997 | ||||
| Yes | n (%) | 81 (15%) | 570 (25%) | 159 (13%) | |
| No | n (%) | 451 (85%) | 1690 (75%) | 1046 (87%) | |
| Missing or review A pending | n | 12 | 86 | 632 | |
| History of organ transplantation | 4106 | ||||
| Yes | n (%) | 56 (10%) | 8 (0.3%) | 60 (4.9%) | |
| No | n (%) | 479 (90%) | 2327 (100%) | 1176 (95%) | |
| Missing or review A pending | n | 9 | 11 | 601 | |
| General symptoms | 3711 | ||||
| Yes | n (%) | 246 (59%) | 1964 (95%) | 979 (80%) | |
| No | n (%) | 172 (41%) | 110 (5.3%) | 240 (20%) | |
| Missing or Review A pending | n | 126 | 272 | 618 | |
| Respiratory symptoms | 3726 | ||||
| Yes | n (%) | 253 (61%) | 1974 (95%) | 979 (80%) | |
| No | n (%) | 165 (39%) | 110 (5.3%) | 245 (20%) | |
| Missing or review A pending | n | 126 | 262 | 613 | |
| Gastrointestinal symptoms | 2626 | ||||
| Yes | n (%) | 93 (22%) | 900 (89%) | 448 (37%) | |
| No | n (%) | 325 (78%) | 110 (11%) | 750 (63%) | |
| Missing or review A pending | n | 126 | 1336 | 639 | |
| Neurological symptoms | 2829 | ||||
| Yes | n (%) | 94 (22%) | 1094 (91%) | 424 (35%) | |
| No | n (%) | 324 (78%) | 110 (9.1%) | 783 (65%) | |
| Missing or review A pending | n | 126 | 1142 | 630 | |
| Other symptoms | 2750 | ||||
| Yes | n (%) | 93 (22%) | 1019 (90%) | 387 (32%) | |
| No | n (%) | 325 (78%) | 110 (9.7%) | 816 (68%) | |
| Missing or review A pending | n | 126 | 1217 | 634 | |
| Asymptomatic | 4012 | ||||
| Yes | n (%) | 4 (0.8%) | 110 (4.9%) | 62 (5.0%) | |
| No | n (%) | 520 (99%) | 2134 (95%) | 1182 (95%) | |
| Missing or review A pending | n | 20 | 102 | 593 | |
| Early outcome | 1800 | ||||
| Discharged home/ambulatory care | n (%) | 413 (79%) | 0 (NA%) | 834 (65%) | |
| Unknown or no change yet | n (%) | 0 (0%) | 0 (NA%) | 185 (14%) | |
| Transferred to or from another facility | n (%) | 47 (9.0%) | 0 (NA%) | 107 (8.4%) | |
| Deceased | n (%) | 62 (12%) | 0 (NA%) | 152 (12%) | |
| Missing or review A pending | n | 22 | 2346 | 559 | |
| 3M follow-up conducted | 1258 | ||||
| Yes | n (%) | 162 (69%) | 0 (NA%) | 534 (52%) | |
| No | n (%) | 74 (31%) | 0 (NA%) | 488 (48%) | |
| Missing or review A pending | n | 308 | 2346 | 815 | |
| 6M follow-up conducted | 182 | ||||
| Yes | n (%) | 113 (62%) | 0 (NA%) | 0 (NA%) | |
| No | n (%) | 69 (38%) | 0 (NA%) | 0 (NA%) | |
| Missing or review A pending | n | 362 | 2346 | 1837 | |
| 12M follow-up conducted | 694 | ||||
| Yes | n (%) | 37 (35%) | 0 (NA%) | 219 (37%) | |
| No | n (%) | 68 (65%) | 0 (NA%) | 370 (63%) | |
| Missing or review A pending | n | 439 | 2346 | 1248 |
SUEP, cross-sectoral platform; HAP, high-resolution platform; POP, population-based platform
Baseline characteristics for the SUEP and the HAP correspond to the baseline visit during acute infection, for the POP to the first baseline visit 6-12 months after infection
Biosamples
As of April 03, 2022, 4349/5298 (82%) patients had at least one study visit with biosampling, 1442 in the SUEP, 439 in the HAP, and 2,468 in the POP. On average, a patient had 2 study visits with biosampling. This totals in 8845 biosample panels stored in 34 local biobanks (see Table 5 for details). Follow-up samples for month 3, 6, 12, and 24 exist from 740, 1942, 209, and 29 patients, respectively. The BCU performed 34 audits with 79 deviations and 212 recommendations by April 01, 2022.
Table 5.
Collected number of respective biosamples until April 03, 2022.
| Total | SUEP | HAP | POP | ||
|---|---|---|---|---|---|
| Patients with biosamples | 4349 | 1442 | 439 | 2469 | |
| Visits with biosampling | 8845 | 3915 | 2445 | 2485 | |
| Follow-up visits total | 2920 | 754 | 346 | 1822 | |
| 3 months | 740 | 583 | 155 | 2 | |
| 6 months | 1942 | 3 | 120 | 1819 | |
| 12 months | 209 | 166 | 43 | 0 | |
| 24 months | 29 | 0 | 28 | 1 | |
| Average visit with biosampling per patient | 2 | 3 | 6 | 1 | |
| Intended use | |||||
| EDTA blood | Plasma: proteome, metabolome, biomarker analysis; DNA: genome, epigenome | 10,988 | 3974 | 2470 | 4544 |
| Serum | Clinical and biomarker analysis | 8636 | 3760 | 2435 | 2443 |
| Respiratory samplea | Determination of virus subtype, microbiome | 7077 | 3644 | 950 | 2483 |
| Oro/nasopharyngeal swaba | 2916 | 2358 | 468 | 90 | |
| Salivaa | 4091 | 1217 | 482 | 2392 | |
| ENTAa,b | 65 | 64 | 0 | 1 | |
| BALa,b | 5 | 5 | 0 | 0 | |
| PAXgene RNA | Transcriptome | 8362 | 3627 | 2407 | 2328 |
| Citrate blood | Analysis of coagulation factors, biomarkers | 10,727 | 4187 | 2470 | 4070 |
| PBMC (all variants) | Analysis of cellular immune response | 11,484 | 4701 | 4361 | 2422 |
| CPT | 5918 | 3337 | 936 | 1645 | |
| EDTA | 1139 | 632 | 507 | 0 | |
| Heparine | 4427 | 732 | 2918 | 777 | |
| Urinea | Metabolome, kidney measures | 6358 | 3055 | 959 | 2344 |
BAL, Bronchoalveolar lavage; ENTA, endotracheal aspiration; PBMC, peripheral blood mononuclear cells; CPT, cell preparation tube; SUEP, cross-sectoral platform; HAP, high-resolution platform; POP, population-based platform
aOnly one sample taken per week
bOnly for intensive care patients and clinical indication
Discussion
During the COVID-19 pandemic, many countries pooled relevant health care data on thousands to millions of COVID-19 cases (i.e., United States [15, 44, 45], Scotland [17], United Kingdom [16, 46], Canada [24], China [23], Iran [20], Qatar [21], Middle East [22], Mexico [19] or South Korea [18]). Although dedicated national COVID-19 biobank activities started as early as February 2020 [47] and many large national multi-center observational studies collecting data and biosamples for selected conditions exist [48, 49], the number of national prospective studies following an interdisciplinary approach similar to the NAPKON is still small. This is not particularly surprising, as the pandemic impacted the biobanking activities globally [50], and the multi-site roll-out of interdisciplinary study protocols is exceptionally resource-intensive. Currently, the three comparable studies to the NAPKON approach are Canada’s CANCOV [51], Brazil’s SARS-Brazil [52] and France’s FrenchCOVID [14], targeting longitudinal, multicenter data and biosamples of 2000, 2000, and 5000 patients, respectively.
Launched in February 2020, FrenchCOVID assesses clinical features and pathogen evolution of SARS-CoV-2 infected inpatients daily for 15 days, then weekly up to 100 days, and invites patients for follow-up visits at 3 and 6 months. In addition to clinical data, the study collects biosamples (including blood, urine, stool, respiratory samples, samples from infected sites, and cerebrospinal fluid) of patients of any age. Study sites perform no additional clinical examinations or diagnostics. Recruitment happens at hospitals only (81 sites in total), including many university sites [14]. SARS-Brazil already enrolled more than 1500 of the initially planned 2000 hospitalized adult SARS-CoV-2 infected patients. Biosamples include blood, serum, plasma, and nasal swabs, with a 60 day observational period after inclusion [52]. CANCOV follows a stratified recruitment approach similar to the NAPKON since April 2020, including out- and inpatients of varying disease severity. Thirty-two sites are involved. Its visit schedule collects data and biosamples at baseline, day 7, day 30, 3, 6 and 12 months, including quality of life measures and additional physical examinations [51].
In this context, we highlight several strengths of the NAPKON. The NAPKON overcomes limitations of previous German cohorts (e.g., anonymous recruitment without follow-ups in LEOSS, single-center collection in Pa-COVID-19) and its multi-layered cohort recruitment strategy covers the full SARS-CoV-2 spectrum across all ages, disease severities, and health care sectors. Differentiators include the extensive biosample collection, inclusion of pediatric patients, and collaboration with local health authorities for representative sampling. The visit schedule includes adaptive acute (e.g., continued weekly visits during hospitalization) and detailed follow-up (e.g., continued PROMs and follow-ups up to 3 years) elements in addition to comprehensive study diagnostics. The NAPKON is well equipped to validate previous findings [53, 54], focus on neglected nuances, add to the understanding of new variants of concern [55], and the future effects anticipated from PCS [56].
The most relevant limitations and challenges of the NAPKON include a relatively small number of non-university study sites and unsatisfactory linkage to electronic health care records. Also, generalizability to other less resourced health care systems and non-European-ancestry populations will probably be limited. The deployment of documentation staff allows for far-reaching data collection across IT systems, but sole reliance on manual data transfer is error-prone and cost-intensive. The NAPKON’s extensive infrastructure had to be established in an ongoing pandemic context; thus, it had a delayed start compared to international cohorts, missing out on notable opportunities in the first wave in 2020 (e.g., early contributions to the understanding of diagnostics, pathophysiology, virus subtypes and the treatment). Via the activities of the IGCU this may partly be compensated for. We were not able to compile data on completeness of recruitment/response rates for this article and hope to provide these in upcoming individual in-depth cohort descriptions.
NAPKON has already been remarkably successful although the pace of its development and the circumstances have been challenging. Data and biosamples are heavily requested and the NAPKON established collaborations with consortia such as Connecting European Cohorts to Increase Common and Effective Response to SARS-CoV-2 Pandemic (ORCHESTRA) [57]. With the NAPKON, we established a sustainable and open clinical research network across Germany that through continuous and interdisciplinary development is determined to become a core infrastructure for prospective, interventional clinical research in a consolidated NUM. Complemented by current preparations towards a NAPKON clinical trial platform inspired by the vastly successful RECOVERY study in the United Kingdom [58], this will also allow and expedite conduct of phase II/III clinical trials within the network.. While these infrastructures create opportunity for virtually all major fields of medical research that require such large-scale effort, they create preparedness for handling WHO’s list of priority diseases [59] or novel pathogens of natural or artificial origin.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The National Pandemic Cohort Network (NAPKON) was supported by the German Federal Ministry of Education and Research (BMBF) via the Network University Medicine (NUM) (FKZ: 01KX2021). We express our gratitude to all study teams that supported the NAPKON study: Agaplesion Bethesda Krankenhaus Bergedorf gemeinnützige GmbH (Marc Gregor Bota), Elisabeth-Krankenhaus Essen (Ingo Voigt), Frankfurt Universitätsklinikum (Maria J.G.T. Vehreschild, Jörg J. Vehreschild), Hausarztpraxis Dashti (Hiwa Dashti), Hausarztpraxis Egestorf, Bispingen (Barbara Laumerich), Helios Klinik Bad Saarow (Oliver Pociuli), Helios Klinikum Duisburg (Nikolaus Büchner), Helios Klinikum Erfurt (Sabine Adler), Helios Klinikum Krefeld (Mathias Lehmann), Helios Klinikum Siegburg (Selcuk Tasci), Kinder- und Jugendpraxis Jorczyk, Dresden (Maximilian Jorczyk), KJF Klinik Josefinum gGmbH (Thomas Keller), Klinik Hallerwiese-Cnopfsche Kinderklinik (Michael Schroth), Klinikum Dortmund gGmbH, Medizinische Klinik Nord (Martin Hower), Klinikum Leverkusen (Lukas Eberwein), Klinikum Worms (Tim Zimmermann), Krankenhaus Bethanien - Institut für Pneumologie an der Universität zu Köln (Simon-Dominik Herkenrath), Malteser Krankenhaus St. Franziskus-Hospital (SFH) Flensburg (Milena Milovanovic), MVZ am Isartor, München (Ramona Pauli), MVZ im Altstadt-Caree Fulda GmbH Medizinisches Versorgungszentrum (Jörg Simon), OWL: Evangelisches Klinikum Bethel (Eckard Hamelmann), OWL: Klinikum Bielefeld (Christoph Stellbrink), OWL: Klinikum Lippe (Johannes-Josef Tebbe), Petrus Krankenhaus Wuppertal (Sven Stieglitz), Praxis am Ebertplatz (Christoph Wyen), Praxis Bosch und Renner, Köln (Jan Bosch), Praxis Dilltal, Burbach (Mirko Steinmüller), Praxis Dr. Allerlei, Frankfurt am Main (Christoph Allerlei), Praxis Dr. Böbel, Reutlingen (Markus Böbel), Praxis Dr. Elke Heinitz, Grube (Elke Natascha Heinitz), Praxis Dr. med Ariane Roecken (Ariane Roecken), Praxis Dr. med. Andrea Münkle-Krimly (Andrea Münckle-Krimly), Praxis Dr. med. Christiane Guderian (Christiane Guderian), Praxis im Tennental (Ingmar Silberbaur), SHG-Kliniken Völklingen (Harald Schäfer), Tropenklinik Paul-Lechler-Krankenhaus Tübingen (Claudia Raichle), TUM München (Christoph Spinner), UKGM Standort Marburg (Bernd Schmeck), Uniklinik Carl Gustav Carus Dresden (Heidi Altmann, Nicole Toepfner), Uniklinik der Ruhr-Universität Bochum (Wolfgang Schmidt), Uniklinik Düsseldorf (Björn Jensen), Uniklinik Erlangen (Andreas Kremer), Uniklinik Göttingen (Sabine Blaschke), Uniklinik Halle (Jochen Dutzmann), Uniklinik Hamburg-Eppendorf (Marylyn Addo), Uniklinik Homburg (Robert Bals), Uniklinik Leipzig (Sven Bercker), Uniklinik Münster (Phil-Robin Tepasse), Uniklinik Regensburg (Frank Hanses), Uniklinik RWTH Aachen (Dirk Müller-Wieland), Uniklinik Schleswig-Holstein, Kiel (Anette Friedrichs), Uniklinik Schleswig-Holstein, Lübeck (Jan Rupp), Uniklinik Tübingen (Siri Göpel), Uniklinik Würzburg (Jens Maschmann), Universitätsklinikum Augsburg (Christine Dhillon), Universitätsklinikum Bonn (Jacob Nattermann), Universitätsklinikum Essen (Ingo Voigt), Universitätsklinikum Magdeburg (Wilfred Obst), Universitätsmedizin der Johannes Gutenberg-Universität Mainz (Martin Franz Sprinzl), Universitätsmedizin Greifswald (Christian Scheer), Universitätsmedizin Mannheim (Andreas Teufel), Universitätsmedizin Oldenburg (Ulf Günther), Charité—Universitätsmedizin Berlin (Martin Witzenrath, Thomas Keil, Thomas Zoller, Sein Schmidt, Michael Hummel, Lilian Krist, Julia Fricke, Maria Rönnefarth, Denise Treue, Ludie Kretzler, Chantip Dang-Heine, Paul Triller, Andreas Jooß, Jenny Schlesinger, Natalja Liseweski, Christina Pley, Carmen Scheibenbogen), Hannover Medizinische Hochschule (Marius Hoeper), Jena (Philipp A. Reuken), Klinikum der Universität München (Michael von Bergwelt), UKSH-Kiel (Rainer Noth), UKSH-Lübeck (Daniel Drömann), Universitätsklinikum Frankfurt (Maria J.G.T. Vehreschild), Universitätsklinikum Freiburg (Siegbert Rieg), Universitätsklinikum Giessen / Marburg (Istvan Vadasz), Universitätsklinikum Köln (Philipp A. Koehler), Universtitästklinikum Heidelberg (Uta Merle), Kiel Universitätsklinikum (Stefan Schreiber), Würzburg Universitätsklinikum (Peter Heuschmann, Stefan Störk). The following individuals have supported governance bodies of the NAPKON: Use & Access Committee Anette Friedrichs, Astrid Petersmann, Claudia Ellert, Georg Schmidt, Janne Vehreschild, Katrin Milger, Marie von Lilienfeld, Martin Witzenrath, Oliver Witzke, Patrick Meybohm, Peter Heuschmann, Sabine Blaschke, Sandra Frank, Stefan Schreiber, Thomas Illig. Advisory Board Alexander Hein, Andrea Wittig, Andreas Simm, Anette Friedrichs, Anke Reinacher-Schick, Anna Frey, Antonella Iannaccone, Astrid Petersmann, Benjamin Maasoumy, Benjamin Waschki, Bimba Hoyer, Brigitt van Oorschot, Carolina van Schaik, Christina Lemhöfer, Christina Polidori, Christine Klein, Daniel Medenwald, Eva Christina Schulte, Eva Grill, Felix Meinel, Folke Brinkmann, Ghazal Arabi, Heike Bickeböller, Holger Lindner, Ildiko Gagyor, Jessica Hassel, Jürgen Deckert, Katrin Milger-Kneidinger, Kerstin Ludwig, Marcus Dörr, Marie von Lilienfeld-Toal, Martin Möckel, Martin Weigl, Matthias Nauck, Miriam Banas, Muenevver Demir, Nicole Lindenberg, Nora Hettich, Norma Jung, Oliver Witzke, Orlando Guntinas-Lichius, Patrick Meybohm, Reinhard Berner, Sabine Blaschke, Samuel Knauss, Sandra Frank, Sebastian Baumeister, Sebastian Dolff, Selma Ugurel, Sophia Stöcklein, Stefanie Joos, Winfred Häuser. The coordination and infrastructure of the NAPKON was conducted by: ICU: Jörg Janne Vehreschild, Maximilian Schons, Sina Hopff, Markus Brechtel, Cristina Schmidt-Ibanez, Johannes Schneider, Carolin Jakob, Franziska Voß. BCU: Inga Bernemann, Sonja Kunze, Maike Tauchert, Thomas Illig, Gabriele Anton. ECU: Cornelia Fiessler, Mirjam Kohls, Olga Miljukov, Steffi Jiru-Hillmann, Jens-Peter Reese, Peter Heuschmann. IGCU: Jens-Peter Reese, Peter Heuschmann, Anna-Lena Hofmann, Julia Schmidt, Kathrin Ungethüm, Anna Horn, Michael Krawczak. SUEP: Jörg Janne Vehreschild, Melanie Stecher, Lisa Pilgram, Clara Brünn, Ramsia Geisler, Margarete Scherer, Kristina Seibel, Marina Hagen. POP: Thomas Bahmer, Wolfgang Lieb, Daniel Pape, Stefan Schreiber, Anne Hermes, Irene Lehmann, Corina Maetzler, Lukas Tittmann. HAP: Lena Johanna Lippert, Sein Schmidt, Fridolin Steinbeis, Martin Witzenrath, Florian Kurth, Charlotte Thibeault. DZHK: Roberto Lorbeer, Bettina Lorenz-Depiereux, Monika Kraus, Christian Schäfer, Jens Schaller, Mario Schattschneider, Dana Stahl, Heike Valentin, Dagmar Krefting, Matthias Nauck. Pediatric core unit: Nicole Toepfner, Reinhard Berner. GECCO & Interoperability Team: Christof von Kalle, Sylvia Thun, Alexander Bartschke, Liudmila Lysyakova, Stefanie Rudolph, Julian Sass. Kardiology team: Eike Nagel, Valentina Püntmann. We acknowledge the dedicated support of clinical research support staff of the Institute of Experimental and Translational Cardiovascular Imaging, including Tammy Wolf, Thourier Azdad, Franziska Weis, Ira Krückemeier, Simon Bohlender, MSc, Deniz Desik, BA, and Layla Laghchioua, MSc. Coordination NUM: Ralf Heyder, Silke Wiedmann
Funding
Open Access funding enabled and organized by Projekt DEAL. The project National Pandemic Cohort Network (NAPKON) is part of the Network University Medicine (NUM) and was funded by the German Federal Ministry of Education and Research (BMBF) (FKZ: 01KX2021). Parts of the infrastructure of the Würzburg study site were supported by the Bavarian Ministry of Research and Art to support Corona research projects. Parts of the NAPKON project suite and study protocols of the Cross-Sectoral cohort platform are based on projects funded by the German Center for Infection Research (DZIF). Part of the infrastructure for the Population-Based Platform received funding of the State of Schleswig Holstein (COVIDOM) and DFG Exzellenzcluster.
Data availability
Data is available under the regulations of NAPKON’s Use & Access process regulations (https://proskive.napkon.de).
Code availability
The code of the analysis or NAPKON Suite portal is available from corresponding author upon reasonable request.
Declarations
Conflict of interest
Dr. Heuschmann reports grants from German Ministry of Research and Education, during the conduct of the study; research grants from German Ministry of Research and Education, European Union, Charité—Universitätsmedizin Berlin, Berlin Chamber of Physicians, German Parkinson Society, University Hospital Würzburg, Robert Koch Institute, German Heart Foundation, Federal Joint Committee (G-BA) within the Innovationfond, German Research Foundation, Bavarian State (ministry for science and the arts), German Cancer Aid, Charité—Universitätsmedizin Berlin (within Mondafis; supported by an unrestricted research grant to the Charité from Bayer), University Göttingen (within FIND-AF randomized; supported by an unrestricted research grant to the University Göttingen from Boehringer-Ingelheim), University Hospital Heidelberg (within RASUNOA-prime; supported by an unrestricted research grant to the University Hospital Heidelberg from Bayer, BMS, Boehringer-Ingelheim, Daiichi Sankyo), outside the submitted work. Dr. Bahmer reports grants from the German Center for Lung Research (DZL) and fees for lecturing, advice and travel expenses from AstraZeneca, GlaxoSmithKline. Novartis, Chiesi, Boehringer Ingelheim, MSD, Roche. Dr. Bellinghausen reports grants or contracts from Gilead Sciences. Dr. Hanses reports having received fees for presentations and lectures for DGINA, Akademie für Infektionsmedizin, and participation on the advisory boards for MSD, GSK and Sobi. Dr. Herold reports grants or contracts from the German Research Foundation (DFG), Ministry of Research and Education (BMBF), consulting fees from Janssen, Sanofi and AstraZeneca, participation on a data safety monitoring board or advisory Board for Atriva. Dr. Keil reports grants from European Union; German Research Foundation; German Federal Ministry of Research and Education; Innovationsfonds by the Federal Joint Committee (G-BA); and the Bavarian State Ministry of Health and Care. Dr. Pape reports having received support for attending ECCMID 2021 from Advanz Pharma Germany. Dr. Püntmann reports honoraria for presentations for Byer AG and Siemens AG, and being involved in the ACC Taskforce on long-term sequela of COVID-19 infection. Dr. Störk reports grants from Federal Ministry of Education and Research for the Comprehensive Heart Failure Center Würzburg. Dr. Nagel reports grants from Byer AG and NeoSoft Ltd., consulting fees from Bayer AG, honoraria for presentations for Byer AG, Pfizer AG and Siemens Healthineers. He has received payments for expert testimony by Bayer AG, participates in the Advisory board CMR-ICD: NCT04558723, has patents issued and received equipment from MEDIS and NeoSoft. Dr. Thibeault reports grants from the Deutsche Forschungsgemeinschaft. Dr. Witzenrath reports grants from Deutsche Forschungsgemeinschaft, Bundesministerium für Bildung und Forschung, Deutsche Gesellschaft für Pneumologie, European Respiratory Society, Marie Curie Foundation, Else Kröner Fresenius Stiftung, Capnetz Stiftung, International Max Planck Research School, Vaxxilon, Actelion, Bayer Health Care, Biotest, Boehringer Ingelheim. He received consulting fees from Noxxon, Pantherna, Vaxxilon, Aptarion, Glaxo Smith Kline, Sinoxa, Biotest, Thieme, and received honoraria for presentations from Astra Zeneca, Berlin Chemie, Chiesi, Novartis, Actelion, Boehringer Ingelheim, Glaxo Smith Kline, Biotest, Bayer Health Care. He has issued several patents. Dr. Vehreschild reports to have received honoraria for presentations from Merck / MSD, Gilead, Pfizer, Astellas Pharma, Basilea, German Centre for Infection Research (DZIF), University Hospital Freiburg/Congress and Communication, Academy for Infectious Medicine, University Manchester, German Society for Infectious Diseases (DGI), Ärztekammer Nordrhein, University Hospital Aachen, Back Bay Strategies, German Society for Internal Medicine (DGIM), Shionogi, Molecular Health, Netzwerk Universitätsmedizin, Janssen, NordForsk. He received research funding from Merck / MSD, Gilead, Pfizer, Astellas Pharma, Basilea, German Centre for Infection Research (DZIF), German Federal Ministry of Education and Research (BMBF), (PJ-T: DLR), University of Bristol, Rigshospitalet Copenhagen. All other authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript, except for the Network University Medicine.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publication
No identifying information is included in this article.
Ethics approval
NAPKON’s study protocols have been approved at institutional review boards/ethics committees of all participating study sites.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Maximilian Schons, Email: nuep@mxschons.com.
Lisa Pilgram, Email: lisa.pilgram@charite.de.
Jens-Peter Reese, Email: Reese_J@ukw.de.
Melanie Stecher, Email: melanie.stecher@uk-koeln.de.
Gabriele Anton, Email: gabriele.anton@helmholtz-muenchen.de.
Katharina S. Appel, Email: katharina.appel@kgu.de
Thomas Bahmer, Email: Thomas.Bahmer@uksh.de.
Alexander Bartschke, Email: alexander.bartschke@bih-charite.de.
Carla Bellinghausen, Email: carla.bellinghausen@kgu.de.
Inga Bernemann, Email: Bernemann.Inga@mh-hannover.de.
Markus Brechtel, Email: markus.brechtel@uk-koeln.de.
Folke Brinkmann, Email: folke.brinkmann@rub.de.
Clara Brünn, Email: cl.bruenn@gmail.com.
Christine Dhillon, Email: christine.dhillon@uk-augsburg.de.
Cornelia Fiessler, Email: Fiessler_C@ukw.de.
Ramsia Geisler, Email: ramsia.geisler@kgu.de.
Eckard Hamelmann, Email: Eckard.Hamelmann@uni-bielefeld.de.
Stefan Hansch, Email: stefan.hansch@stud.uni-regensburg.de.
Frank Hanses, Email: frank.hanses@ukr.de.
Sabine Hanß, Email: sabine.hanss@med.uni-goettingen.de.
Susanne Herold, Email: Susanne.Herold@innere.med.uni-giessen.de.
Ralf Heyder, Email: ralf.heyder@charite.de.
Anna-Lena Hofmann, Email: Hofmann_A9@ukw.de.
Sina Marie Hopff, Email: sina.hopff@uk-koeln.de.
Anna Horn, Email: horn_a2@ukw.de.
Carolin Jakob, Email: carolin.jakob2@uk-koeln.de.
Steffi Jiru-Hillmann, Email: E_Hillmann_S@ukw.de.
Thomas Keil, Email: thomas.keil@charite.de.
Yascha Khodamoradi, Email: yascha.khodamoradi@kgu.de.
Mirjam Kohls, Email: Kohls_M@ukw.de.
Monika Kraus, Email: monika.kraus@helmholtz-muenchen.de.
Dagmar Krefting, Email: dagmar.krefting@med.uni-goettingen.de.
Sonja Kunze, Email: sonja.kunze@helmholtz-muenchen.de.
Florian Kurth, Email: florian.kurth@charite.de.
Wolfgang Lieb, Email: wolfgang.lieb@epi.uni-kiel.de.
Lena Johanna Lippert, Email: lena.lippert@charite.de.
Roberto Lorbeer, Email: Roberto.Lorbeer@med.uni-muenchen.de.
Bettina Lorenz-Depiereux, Email: lorenz-depiereux@helmholtz-muenchen.de.
Corina Maetzler, Email: c.maetzler@neurologie.uni-kiel.de.
Olga Miljukov, Email: Miljukov_O@ukw.de.
Matthias Nauck, Email: Matthias.Nauck@med.uni-greifswald.de.
Daniel Pape, Email: Daniel.Pape@uksh.de.
Valentina Püntmann, Email: valentina.puntmann@kgu.de.
Lennart Reinke, Email: LennartMichel.Reinke@uksh.de.
Christoph Römmele, Email: christoph.roemmele@uk-augsburg.de.
Stefanie Rudolph, Email: stefanie.rudolph@bih-charite.de.
Julian Sass, Email: julian.sass@bih-charite.de.
Christian Schäfer, Email: Christian.Schaefer@med.uni-greifswald.de.
Jens Schaller, Email: jens.schaller@charite.de.
Mario Schattschneider, Email: Mario.Schattschneider@med.uni-greifswald.de.
Christian Scheer, Email: Christian.Scheer@med.uni-greifswald.de.
Margarete Scherer, Email: margarete.scherer@kgu.de.
Sein Schmidt, Email: sein.schmidt@bih-charite.de.
Julia Schmidt, Email: Schmidt_J12@ukw.de.
Kristina Seibel, Email: kristina.seibel@uk-koeln.de.
Dana Stahl, Email: dana.stahl@uni-greifswald.de.
Fridolin Steinbeis, Email: fridolin.steinbeis@charite.de.
Stefan Störk, Email: Stoerk_S@ukw.de.
Maike Tauchert, Email: maiketauchert@t-online.de.
Johannes Josef Tebbe, Email: johannes-josef.tebbe@klinikum-lippe.de.
Charlotte Thibeault, Email: charlotte.thibeault@charite.de.
Nicole Toepfner, Email: Nicole.Toepfner@uniklinikum-dresden.de.
Kathrin Ungethüm, Email: Ungethuem_K@ukw.de.
Istvan Vadasz, Email: Istvan.Vadasz@innere.med.uni-giessen.de.
Heike Valentin, Email: heike.valentin@med.uni-greifswald.de.
Silke Wiedmann, Email: silke.wiedmann@charite.de.
Thomas Zoller, Email: thomas.zoller@charite.de.
Eike Nagel, Email: eike.nagel@cardiac-imaging.org.
Michael Krawczak, Email: krawczak@medinfo.uni-kiel.de.
Christof von Kalle, Email: christof.kalle@bih-charite.de.
Thomas Illig, Email: Illig.Thomas@mh-hannover.de.
Stefan Schreiber, Email: s.schreiber@mucosa.de.
Martin Witzenrath, Email: martin.witzenrath@charite.de.
Peter Heuschmann, Email: E_Heuschma_P@ukw.de.
Jörg Janne Vehreschild, Email: janne.vehreschild@uk-koeln.de.
References
- 1.Bchetnia M, Girard C, Duchaine C, Laprise C. The outbreak of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a review of the current global status. J Infect Public Health. 2020;13:1601–10. doi: 10.1016/j.jiph.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet (Elsevier) 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Timeline: WHO’s COVID-19 response [Internet]. 2021 [cited 2021 Jul 18]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/interactive-timeline
- 4.Elisabeth E. Cameron, Jennifer B. Nuzzo, Jessica A. Bell, et al. Global Health Security Index 2019 [Internet]. 2019. Available from: https://www.ghsindex.org/report-model/
- 5.WHO. WHO Coronavirus (COVID-19) Dashboard April 01 2022 [Internet]. 2022 [cited 2022 Apr 20]. Available from: https://web.archive.org/web/20220401050337/https://covid19.who.int/
- 6.Robert Koch-Institut. Coronavirus Disease 2019 (COVID-19) Daily Situation Report by the Robert Koch Institute 01/04/2022-current status for Germany. 2022.
- 7.World Health Organization. A checklist for pandemic influenza risk and impact management: building capacity for pandemic response [Internet]. Geneva; 2018. Available from: https://apps.who.int/iris/bitstream/handle/10665/259884/9789241513623-eng.pdf;jsessionid=4858E315F5380ADDE7A40B84BE42A2E7?sequence=1
- 8.Robert-Koch-Institut. RKI - Influenza-Pandemieplanung - Antworten auf häufig gestellte Fragen zu Pandemieplanung [Internet]. 2016 [cited 2021 Jul 18]. Available from: https://www.rki.de/SharedDocs/FAQ/Pandemieplanung/FAQ_Liste_Pandemieplanung.html;jsessionid=C1871BC9C65ABDFEA278437C0314211D.internet082
- 9.Robert-Koch-Institut. Nationaler Pandemieplan Teil 1 - Strukturen und Maßnahmen [Internet]. Berlin; 2017 Feb p. 74. Available from: https://www.gmkonline.de/documents/pandemieplan_teil-i_1510042222_1585228735.pdf
- 10.BMBF-Internetredaktion. Karliczek: Wir fördern Nationales Netzwerk der Universitätsmedizin im Kampf gegen Covid-19 - BMBF [Internet]. Bundesministerium für Bildung und Forschung - BMBF. 2021 [cited 2021 Jul 18]. Available from: https://www.bmbf.de/de/karliczek-wir-foerdern-nationales-netzwerk-der-universitaetsmedizin-im-kampf-gegen-covid-11230.html
- 11.Netzwerk Universitätsmedizin. Projekte | Netzwerk Universitätsmedizin [Internet]. 2021 [cited 2021 Jul 19]. Available from: https://www.netzwerk-universitaetsmedizin.de/projekte
- 12.Deutscher Bundestag. Bundeshaushalt 2021 beschlossen [Internet]. Deutscher Bundestag. 2020 [cited 2021 Jul 19]. Available from: https://www.bundestag.de/presse/hib/810198-810198
- 13.Group ICC, Baillie JK, Baruch J, Beane A, Blumberg L, Bozza F, et al. ISARIC clinical data report issued: 14 July 2021. medRxiv. Cold Spring Harbor Laboratory Press; 2021.
- 14.Institut National de la Santé Et de la Recherche Médicale, France. Clinical Characterisation Protocol for Severe Emerging Infections, French COVID Cohort (FrenchCOVID) [Internet]. clinicaltrials.gov; 2020 May. Report No.: NCT04262921. Available from: https://clinicaltrials.gov/ct2/show/NCT04262921
- 15.Haendel MA, Chute CG, Bennett TD, Eichmann DA, Guinney J, Kibbe WA, et al. The National COVID Cohort Collaborative (N3C): rationale, design, infrastructure, and deployment. J Am Med Inform Assoc. 2020;28:427–43. doi: 10.1093/jamia/ocaa196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dennis JM, McGovern AP, Vollmer SJ, Mateen BA. Improving survival of critical care patients with coronavirus disease 2019 in England: A national cohort study, March to June 2020*. Crit Care Med. 2021;49:209–14. doi: 10.1097/CCM.0000000000004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simpson CR, Robertson C, Vasileiou E, McMenamin J, Gunson R, Ritchie LD, et al. Early pandemic evaluation and enhanced surveillance of COVID-19 (EAVE II): protocol for an observational study using linked Scottish national data. BMJ Open; 2020;10:e039097. [DOI] [PMC free article] [PubMed]
- 18.Jung C-Y, Park H, Kim DW, Choi YJ, Kim SW, Chang TI. Clinical characteristics of asymptomatic patients with COVID-19: a nationwide cohort study in South Korea. Int J Infect Dis. 2020;99:266–8. doi: 10.1016/j.ijid.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kammar-García A, Vidal-Mayo J, Vera-Zertuche JM, Lazcano-Hernández M, Vera-López O, Segura-Badilla O, et al. Impact of comorbidities in Mexican SARS-COV-2-positive patients: a retrospective analysis in a national cohort. Rev Invest Clin. 2020;72:151–8. doi: 10.24875/RIC.20000207. [DOI] [PubMed] [Google Scholar]
- 20.Jalili M, Payandemehr P, Saghaei A, Sari HN, Safikhani H, Kolivand P. Characteristics and mortality of hospitalized patients with COVID-19 in Iran: a national retrospective cohort study. Ann Intern Med Am Coll Phys. 2020;174:125–7. doi: 10.7326/M20-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omrani AS, Almaslamani MA, Daghfal J, Alattar RA, Elgara M, Shaar SH, et al. The first consecutive 5000 patients with Coronavirus Disease 2019 from Qatar; a nation-wide cohort study. BMC Infect Dis. 2020;20:777. doi: 10.1186/s12879-020-05511-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallah SI, Abdulrahman A, Alawadhi AI, AlQahtani MM. Clinical and epidemiological characteristics of COVID-19 in a multi-national cohort in the Middle East. J Epidemiol Global Health [Internet]. Atlantis Press; 2021 [cited 2021 Apr 5]; Available from: https://www.atlantis-press.com/journals/jegh/125953894 [DOI] [PMC free article] [PubMed]
- 23.Liang W-H, Guan W-J, Li C-C, Li Y-M, Liang H-R, Zhao Y, et al. Clinical characteristics and outcomes of hospitalised patients with COVID-19 treated in Hubei (epicentre) and outside Hubei (non-epicentre): a nationwide analysis of China. Eur Respir J. 2020;55:2000562. doi: 10.1183/13993003.00562-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murthy S, Archambault PM, Atique A, Carrier FM, Cheng MP, Codan C, et al. Characteristics and outcomes of patients with COVID-19 admitted to hospital and intensive care in the first phase of the pandemic in Canada: a national cohort study. CMAJO Canadian Medical Association; 2021;9:E181–8. [DOI] [PMC free article] [PubMed]
- 25.Jakob CEM, Borgmann S, Duygu F, Behrends U, Hower M, Merle U, et al. First results of the “Lean European Open Survey on SARS-CoV-2-infected patients (LEOSS)”. Infection. 2021;49:63–73. doi: 10.1007/s15010-020-01499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurth F, Roennefarth M, Thibeault C, Corman VM, Müller-Redetzky H, Mittermaier M, et al. Studying the pathophysiology of coronavirus disease 2019: a protocol for the Berlin prospective COVID-19 patient cohort (Pa-COVID-19) Infection. 2020;48:619–26. doi: 10.1007/s15010-020-01464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horn A, Krist L, Lieb W, Montellano FA, Kohls M, Haas K, et al. Long-term health sequelae and quality of life at least 6 months after infection with SARS-CoV-2: design and rationale of the COVIDOM-study as part of the NAPKON population-based cohort platform (POP). Infection [Internet]. 2021 [cited 2021 Oct 21]; Available from: 10.1007/s15010-021-01707-5 [DOI] [PMC free article] [PubMed]
- 28.Deutsches Zentrum für Herz- und Kreislaufforschung. Clinical Research Platform: DZHK [Internet]. 2021 [cited 2021 Jul 26]. Available from: https://dzhk.de/en/research/clinical-research/clinical-research-platform/
- 29.Grobbee DE, Hoes AW. Clinical epidemiology: principles, methods, and applications for clinical research. Jones & Bartlett Learning; 2009.
- 30.Haas K, Purrucker JC, Rizos T, Heuschmann PU, Veltkamp R. Rationale and design of the Registry of Acute Stroke Under Novel Oral Anticoagulants-prime (RASUNOA-prime) Eur Stroke J. 2019;4:181–8. doi: 10.1177/2396987318812644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiskopf N, Suckling J, Williams G, Correia M, Inkster B, Tait R, et al. Quantitative multi-parameter mapping of R1, PD*, MT, and R2* at 3T: a multi-center validation. Front Neurosci [Internet]. 2013 [cited 2022 Apr 20]; 7. 10.3389/fnins.2013.00095 [DOI] [PMC free article] [PubMed]
- 32.Frontiers | Quantitative Multi-Parameter Mapping Optimized for the Clinical Routine | Neuroscience [Internet]. [cited 2022 Apr 20]. Available from: 10.3389/fnins.2020.611194/full [DOI] [PMC free article] [PubMed]
- 33.Society for Cardiovascular Magnetic Resonance (SCMR) recommended CMR protocols for scanning patients with active or convalescent phase COVID-19 infection | Journal of Cardiovascular Magnetic Resonance | Full Text [Internet]. [cited 2022 Apr 20]. Available from: 10.1186/s12968-020-00656-6 [DOI] [PMC free article] [PubMed]
- 34.Sass J, Bartschke A, Lehne M, Essenwanger A, Rinaldi E, Rudolph S, et al. The German Corona Consensus Dataset (GECCO): a standardized dataset for COVID-19 research in university medicine and beyond. BMC Med Inform Decis Mak. 2020;20:341. doi: 10.1186/s12911-020-01374-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.ICD-10-GM [Internet]. [cited 2021 Aug 16]. Available from: https://www.dimdi.de/dynamic/en/classifications/icd/icd-10-gm
- 36.McDonald CJ, Huff SM, Suico JG, Hill G, Leavelle D, Aller R, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem. 2003;49:624–33. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 37.WHOCC - Home [Internet]. [cited 2021 Aug 16]. Available from: https://www.whocc.no/
- 38.SNOMED - Home | SNOMED International [Internet]. [cited 2021 Aug 16]. Available from: https://www.snomed.org/
- 39.Index - FHIR v4.0.1 [Internet]. [cited 2021 Aug 16]. Available from: https://hl7.org/FHIR/
- 40.World Health Organization. ISARIC CORE COVID-19 Case Report Form [Internet]. ISARIC. 2021 [cited 2021 Jul 26]. Available from: https://isaric.org/research/covid-19-clinical-research-resources/covid-19-crf/
- 41.LEOSS study group. LEOSS: Lean European Open Survey on SARS-CoV-2 Infected Patients - Portal für Medizinische Datenmodelle (MDM-Portal) [Internet]. 2021 [cited 2021 Jul 26]. Available from: https://medical-data-models.org/42476
- 42.Thibeault C, Mühlemann B, Helbig ET, Mittermaier M, Lingscheid T, Tober-Lau P, et al. Clinical and virological characteristics of hospitalised COVID-19 patients in a German tertiary care centre during the first wave of the SARS-CoV-2 pandemic: a prospective observational study. Infection. 2021. [DOI] [PMC free article] [PubMed]
- 43.OpenProject - Open Source Projektmanagement-Software [Internet]. 2021 [cited 2021 Oct 21]. Available from: https://www.openproject.org/de/
- 44.Rentsch CT, Kidwai-Khan F, Tate JP, Park LS, King JT, Skanderson M, et al. Covid-19 by Race and Ethnicity: A National Cohort Study of 6 Million United States Veterans. medRxiv [Internet]. 2020 [cited 2021 Apr 5]; Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7273292/
- 45.Bennett TD, Moffitt RA, Hajagos JG, Amor B, Anand A, Bissell MM, et al. The National COVID cohort collaborative: clinical characterization and early severity prediction. medRxiv. 2021.
- 46.Wood A, Denholm R, Hollings S, Cooper J, Ip S, Walker V, et al. Linked electronic health records for research on a nationwide cohort of more than 54 million people in England: data resource. BMJ; 2021;373:n826. [DOI] [PMC free article] [PubMed]
- 47.Huang S-F, Huang Y-C, Chang F-Y, Lin J-C, Chiu C-H, Chen C-W, et al. Rapid establishment of a COVID-19 biobank in NHRI by National Biobank Consortium of Taiwan. Biomed J. 2020;43:314–7. doi: 10.1016/j.bj.2020.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.National Cancer Institute (NCI). NCI COVID-19 in Cancer Patients Study (NCCAPS): a longitudinal natural history study [Internet]. clinicaltrials.gov; 2021 Aug. Report No.: NCT04387656. Available from: https://clinicaltrials.gov/ct2/show/NCT04387656
- 49.Sengpiel V. COPE - COVID-19 in pregnancy and early childhood—a study protocol for a prospective multicentre cohort study [Internet]. clinicaltrials.gov; 2021 Jun. Report No.: NCT04433364. Available from: https://clinicaltrials.gov/ct2/show/NCT04433364
- 50.Henderson MK, Kozlakidis Z, Fachiroh J, Wiafe Addai B, Xu X, Ezzat S, et al. The responses of biobanks to COVID-19. Biopreserv Biobank. 2020;18:483–91. doi: 10.1089/bio.2020.29074.mkh. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.CANCOV Study Group. CANCOV—The Canadian COVID-19 Prospective Cohort Study [Internet]. 2021 [cited 2021 Jul 27]. Available from: https://cancov.net/
- 52.Hospital Israelita Albert Einstein. Brazilian COVID-19 Registry for Clinical Presentation of Individuals With COVID-19: a National, Multicentre, Prospective Observational Study (SARS-Brazil) [Internet]. clinicaltrials.gov; 2021 Apr. Report No.: NCT04479488. Available from: https://clinicaltrials.gov/ct2/show/NCT04479488
- 53.Pearson H. How COVID broke the evidence pipeline. Nature. 2021;593:182–5. doi: 10.1038/d41586-021-01246-x. [DOI] [PubMed] [Google Scholar]
- 54.Scales D, Gorman J, Jamieson KH. The Covid-19 infodemic—applying the epidemiologic model to counter misinformation. New Engl J Med; 2021;0:null. [DOI] [PubMed]
- 55.Abdool Karim SS, de Oliveira T. New SARS-CoV-2 variants—clinical, public health, and vaccine implications. New Engl J Med. 2021;384:1866–8. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phillips S, Williams MA. Confronting our next national health disaster—long-haul covid. New Engl J Med; 2021;0:null. [DOI] [PubMed]
- 57.Connecting European Cohorts to Increase Common and Effective Response to SARS-CoV-2 Pandemic: ORCHESTRA | ORCHESTRA Project | H2020 | CORDIS | European Commission [Internet]. [cited 2021 Aug 17]. Available from: https://cordis.europa.eu/project/id/101016167
- 58.ISRCTN - ISRCTN50189673: A randomised trial of treatments to prevent death in patients hospitalised with COVID-19 (coronavirus) [Internet]. [cited 2021 Jul 27]. Available from: https://www.isrctn.com/ISRCTN50189673
- 59.World Health Organization. Prioritizing diseases for research and development in emergency contexts [Internet]. 2021 [cited 2021 Jul 18]. Available from: https://www.who.int/activities/prioritizing-diseases-for-research-and-development-in-emergency-contexts
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available under the regulations of NAPKON’s Use & Access process regulations (https://proskive.napkon.de).
The code of the analysis or NAPKON Suite portal is available from corresponding author upon reasonable request.