Abstract
The response of a complex methanogenic sediment community to 2-chlorophenol (2-CP) was evaluated by monitoring the concentrations of this model contaminant and important metabolic intermediates and products and by using rRNA-targeted probes to track several microbial populations. Key relationships between the evolving population structure, formation of metabolic intermediates, and contaminant mineralization were identified. The nature of these relationships was intrinsically linked to the metabolism of benzoate, an intermediate that transiently accumulated during the mineralization of 2-CP. Before the onset of benzoate fermentation, reductive dehalogenation of 2-CP competed with methanogenesis for endogenous reducing equivalents. This suppressed H2 levels, methane production, and archaeal small-subunit (SSU)-rRNA concentrations in the sediment community. The concentrations of bacterial SSU rRNA, including SSU rRNA derived from “Desulfovibrionaceae” populations, tracked with 2-CP levels, presumably reflecting changes in the activity of dehalogenating organisms. After the onset of benzoate fermentation, the abundance of Syntrophus-like SSU rRNA increased, presumably because these syntrophic organisms fermented benzoate to methanogenic substrates. Consequently, although the parent substrate 2-CP served as an electron acceptor, cleavage of its aromatic nucleus also influenced the sediment community by releasing the electron donors H2 and acetate. Increased methane production and archaeal SSU-rRNA levels, which tracked with the Syntrophus-like SSU-rRNA concentrations, revealed that methanogenic populations in particular benefited from the input of reducing equivalents derived from 2-CP.
Widespread contamination of the environment with anthropogenic chlorinated aromatic compounds has occurred as a result of past industrial and agricultural practices. Anaerobic conditions prevail in many of the natural environments in which these compounds may reside, e.g., flooded soils, sediments, and aquifers, as well as in some engineered biological treatment systems. Therefore, anaerobic biotransformation is a potentially crucial removal mechanism for chlorinated aromatic contaminants. This process is typically initiated by reductive dehalogenation (34). Biotransformation of simple chlorinated aromatics, such as chlorinated phenols and benzoates, frequently leads to the formation of benzoate or a substitution-containing analog as an intermediate (4, 24, 40, 41, 46, 52, 53). Benzoate is converted to methanogenic substrates—acetate, H2, and CO2—by syntrophic organisms under fermentative conditions. As a result, the activities of syntrophic and methanogenic populations are just as critical as the dehalogenating organisms in sustaining the anaerobic biodegradation of simple chlorinated aromatic compounds.
The mineralization of chlorinated aromatic compounds by anaerobic microbial assemblages conforms to one of the defining principles of microbial communities, i.e., that the end product(s) of metabolism by one population is often consumed by another. In microbial communities that mineralize chlorinated aromatic compounds, the potential exists for recycling reducing equivalents derived from the aromatic nucleus to fuel the reductive dehalogenation reaction, thereby creating a cyclic mode of electron flow. For example, the recycling of benzoate-derived electrons in aryl reductive dehalogenation has been demonstrated in a three-member coculture that is able to grow on 3-chlorobenzoate (3-CB) as its sole growth substrate (14, 15). However, these studies have not been verified at the ecophysiological level or with other chlorinated aromatic substrates.
Thus, in order to advance our understanding of the fate of chlorinated aromatic compounds in natural and engineered anaerobic environments, we must be able to track the flow of contaminant-derived carbon and electrons in complex communities. Further, in order to determine how contaminant transformation and evolving community structure are related, this information on system-level processes must be integrated with that on population-level changes. Fortunately, populations in a microbial community can be quantified without the biases of culture-based methods by hybridizing DNA probes with rRNA previously extracted from complex samples (37, 45).
In this study, we used an integrated experimental approach to quantify and interpret system- and population-level phenomena in a methanogenic sediment community that mineralized 2-chlorophenol (2-CP). To do so, we monitored the concentrations of the chlorinated substrate, important metabolic intermediates and products, and characteristic small-subunit (SSU)-rRNA sequences of key microbial populations in a 2-CP-amended sediment slurry batch reactor and an unamended control reactor. These integrated evaluations revealed that the mineralization of a chlorinated aromatic compound can influence the structure and function of an anaerobic microbial community via two mechanisms: (i) by competing with other electron-accepting processes for available electron donors and (ii) by serving as a source of reducing equivalents.
MATERIALS AND METHODS
Establishment of cultures.
The anaerobic sediment inoculum was collected from a previously described site in Lake Michigan (28) using a box corer, transferred to canning jars, and stored at 4°C for approximately 3 months until used. Strict anaerobic and aseptic techniques based on the methods described by Miller and Wolin (33) were used to establish, handle, and sample batch reactor cultures, which were maintained in glass vessels (2-liter or 160-ml) with serum bottle closures. The initial ratio of headspace volume to slurry-phase volume in the reactors was 3:5 (vol/vol). The slurry phase comprised sediment and anaerobic medium (1:9, vol/vol). Sediment was retained in the reactors throughout the experiment. Fitzgerald (19) characterized the total organic carbon content of sediment collected at the sampling site and found that it ranged from approximately 2.3 to 3.6% (wt/wt). In addition to a mineral solution, reducing agents, bicarbonate buffer, resazurin, and trace metals, the anaerobic medium contained a minor amount of yeast extract (45 mg/liter), as previously described (20). Two viable 2-liter and duplicate viable 160-ml sediment slurry reactors were established, as described by Becker et al. (7). A 2-liter sterile control reactor was prepared in the same manner as the viable reactors, except that the sediment inoculum was autoclaved for 1 h on each of three consecutive days before being combined with sterile medium. 2-CP was added to a final concentration of approximately 200 μM to all of the reactors except one 2-liter viable reactor, which served as a no-substrate control. Results obtained with the amended 2-liter and duplicate 160-ml viable reactors were similar. In the interest of brevity, only the results obtained with the 2-liter reactor are reported here. Incubation at 30°C was static except during sampling events, when the reactors were continuously shaken on a platform shaker (100 rpm).
Analytical methods.
All analyses of the reactors were performed at approximately 1-week intervals. The aqueous concentrations of 2-CP and its aromatic metabolites were determined through duplicate analyses using reversed-phase high-performance liquid chromatography with diode array detection, as previously described (7). Measurements of CH4 and H2 in the headspace were performed by gas-chromatographic analysis of either 0.5-ml (CH4) or 0.2- to 1.0-ml (H2) duplicate headspace samples obtained with syringes equipped with push-button valves and sterile needles. CH4 was quantified with a gas chromatograph (model 5890II; Hewlett Packard) equipped with an integrator (model 3396A; Hewlett Packard), a flame ionization detector, and a stainless steel packed column (3.2 mm by 2.44 m; 1% SP-1000 on 60/80 Carbopack-B; Supelco, Inc., Bellefonte, Pa.) with helium carrier gas (40 ml/min) at 60°C. H2 levels were monitored with a mercury reduction gas analyzer (model RGA3; Trace Analytical, Stanford, Calif.) equipped with an integrator (model 3395; Hewlett-Packard). Soluble chemical oxygen demand (COD) measurements were made singly using an Environmental Protection Agency-approved Hach method (method 8000; 1 to 150 mg of COD per liter) (22). Sediment slurry samples analyzed for soluble COD concentration were immediately filtered through glass fiber disks (Whatman grade 934/AH), digested, and analyzed with a spectrophotometer (Spectronic 21; Milton Roy).
RNA extraction.
RNA was extracted by the mechanical disruption–phenol-chloroform extraction process of Stahl et al. (45) as modified by MacGregor et al. (28), except as noted. For each RNA extraction, a 4-ml (sediment) slurry sample was taken and aliquoted (0.5 ml) into screw-cap tubes (2.2 ml; Sarstedt, Inc.), which contained zirconium beads (approximately 0.5 g), buffer-equilibrated phenol (pH 5.1), sodium dodecyl sulfate, and low-pH buffer. The tubes were vortexed, immediately frozen on dry ice, and held at −80°C until the remaining steps in the extraction procedure could be carried out. The nucleic acid pellets were resuspended in 200 μl of RNase-free water.
Hybridization and quantification of extracted RNA.
RNA slot blotting, probe-labeling, prehybridization, and washing were performed as previously described (26, 38, 45). Samples were blotted on MagnaCharge nylon membranes (Micron Separation Inc., Westboro, Mass.) in triplicate, prehybridized at 40°C, and washed at the experimentally determined temperature of dissociation. The rRNA standard for hybridization to the Syntrophus genus probe (Table 1) (S-G-Syn-0424-a-A-18) was transcribed from a clone containing most of the SSU-rRNA gene. The Syntrophus-like SSU-rRNA genes were amplified from the sediment slurry nucleic acid extracts with the primers S-G-Syn-0057-a-S-18 (GTC GTA CGA GAA AAT CCG) and S-G-Syn-1444-a-A-18 (ATA GGG GTT AGC TCA ACG). The PCR amplification was performed using the procedure of Muyzer et al. (36) with slight modifications. Briefly, extracted DNA was diluted (1:10, vol/vol). For each sample amplified, 2 to 5 μl of diluted DNA (100 to 300 ng of template DNA) was added to Taq polymerase buffer (5 μl; 500 mM KCl, 15 mM MgCl2, 100 mM Tris-HCl; Pharmacia, Piscataway, N.J.), 20 pmol each of forward and reverse primers (2 μl), a 0.2 mM solution of deoxyribonucleoside triphosphates (5 μl), 1.5 U of Taq (0.3 μl) (Pharmacia), 500 ng of bovine serum albumin (5 μl; Idaho Technologies, Idaho Falls, Idaho) per μl, and sufficient nuclease-free water to make up a final volume of 50 μl. The amplifications were performed in a thermocycler (model 200; MJ Research, Inc., Watertown, Mass.) with the following program: 3 min at 94°C followed by 30 cycles of 30 s at 94°C, 30 s of annealing at a beginning temperature of 65°C, and 1 min of elongation at 72°C. The annealing temperature was lowered by 1° with each cycle until an annealing temperature of 55°C was reached. The resulting PCR fragments were cloned using a commercially prepared vector (TA cloning system; Invitrogen Corp., San Diego, Calif.). The temperature of dissociation (51°C) for probe S-G-Syn-0424-a-A-18 (CCG ACA GAG CTT TAC GAT) was determined experimentally by a previously described elution method (38). The hybridized membranes were exposed to storage phosphor screens (Molecular Dynamics; Sunnyvale, Calif.), which were scanned with a PhosphorImager (Molecular Dynamics). The resulting digitized images were analyzed with ImageQuant software (Molecular Dynamics). The reference rRNA standards (Table 1) and total rRNA extracted from the sediment slurry samples were hybridized with a universal probe in order to relate the universal and specific probe quantifications and to quantify total rRNA abundance in the sediment slurries, respectively.
TABLE 1.
Probes and target groups
| Probe | Target group | Reference rRNA | Reference(s) |
|---|---|---|---|
| S-∗-Univ-1392-a-A-15 | All organisms | Escherichia coli | 54 |
| S-D-Bact-0338-a-A-18 | Bacteria | E. coli | 2 |
| S-D-Arch-0915-a-A-20 | Archaea | Methanosarcina acetivorans | 3 |
| S-F-Dsv-0687-a-A-16 | “Desulfovibrionaceae” | Desulfovibrio salexigens | 12 |
| S-G-Syn-0424-a-A-18 | Syntrophus genus | rRNA transcribed from 3-CB-degrading anaerobic sediment community | 5, this study |
The oligonucleotide probes, their target groups, and the reference rRNA used for each probe are listed in Table 1. Crenarchaeotal rRNA constituted only a small fraction of archaeal rRNA in the Lake Michigan sediment (28). Therefore, probe S-D-Arch-0915-a-A-20 is assumed to be specific for methanogenic populations in the sediment community. Probe S-F-Dsv-0687-a-A-16 was used in the characterization of the 2-CP-degrading sediment community primarily because Desulfovibrio-like populations have previously been associated with the anaerobic biotransformation of halogenated aromatic compounds (1, 9, 42, 47). Probe S-G-Syn-0424-a-A-18 was developed for use in the present study based on preliminary characterizations of 2-CP- and 3-CB-degrading communities developed from sediment obtained from the same Lake Michigan site. These studies, which involved PCR amplification and comparative sequencing of bacterial SSU rRNA genes extracted from the 2-CP- and 3-CB-degrading communities, revealed the presence of 16S rRNA gene sequences that shared a high degree of similarity with characterized members of the genus Syntrophus (5, 6; G. Berardesco, J. G. Becker, B. E. Rittmann, and D. A. Stahl, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. Q-40, 1999).
RESULTS
Transformation of 2-CP and detection of aromatic metabolites.
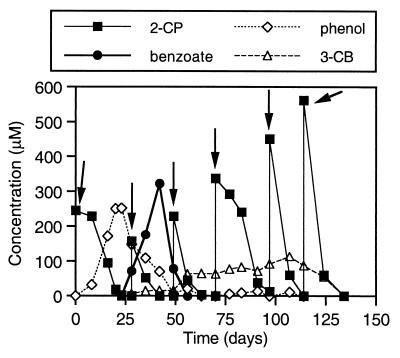
The biotransformation of 2-CP and production of aromatic metabolites by the sediment community have been reported elsewhere (7). Briefly, in the amended 2-liter slurry reactor, the initial dose of 2-CP (∼200 μM) was removed via reductive dehalogenation within 30 days, which resulted in the concomitant accumulation of a stoichiometric amount of phenol (Fig. 1). Following the onset of phenol removal, a second dose of 2-CP was added, and benzoate accumulated transiently, presumably the result of para-carboxylation and subsequent dehydroxylation of phenol (7, 43, 44, 51). Benzoate is subject to fermentation under methanogenic conditions. Cleavage of its aromatic nucleus ultimately releases the electron donors H2 and acetate (25, 35, 48):
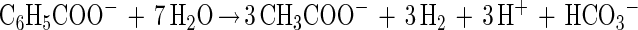 |
1 |
The onset of benzoate degradation apparently occurred between days 42 and 49 in the amended reactor.
FIG. 1.
2-CP transformation and formation of aromatic metabolites (phenol, benzoate, and 3-CB) in a 2-liter batch sediment slurry reactor. Arrows indicate additions of 2-CP.
During the next approximately 85 days, four additions of 2-CP were made at increasing concentrations whenever it was no longer detectable in the slurry, periodically replenishing the supply of 2-CP-derived electron donors to the sediment community. Significant production of 3-CB coincided with the biotransformation of the third addition of 2-CP. It has been suggested that the populations that carry out conversion of phenol to benzoate in anaerobic communities may also be responsible for the biotransformation of 2-CP to 3-CB via para-carboxylation and subsequent dehydroxylation reactions (7, 8).
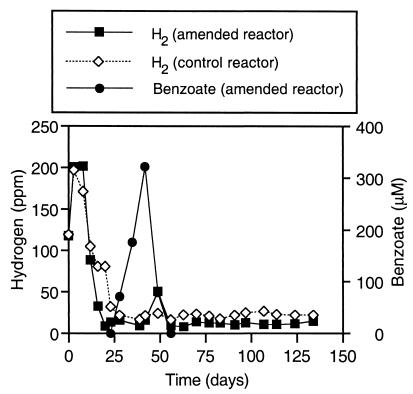
H2 concentrations.
Initially, the H2 concentrations in both sediment slurry reactors increased from approximately 120 to 200 ppm (Fig. 2). By days 20 and 28, the H2 levels in the 2-CP-amended and control reactors, respectively, had dropped by an order of magnitude. A single burst in the H2 levels in the 2-CP-amended reactor on day 49 coincided with the rapid fermentation of benzoate (Fig. 2) and was not observed in the control reactor. From day 20 on, excluding day 49, the H2 concentrations leveled off at an average value of 12 ppm (standard deviation, 2.5 ppm) in the 2-CP-amended reactor. The amended reactor H2 levels were consistently lower than the H2 concentrations measured in the control reactor, which averaged 22 ppm (standard deviation, 2.8 ppm) from day 28 until the conclusion of the experiment.
FIG. 2.
Headspace hydrogen concentrations in the amended and control reactors and benzoate concentrations in the amended reactor.
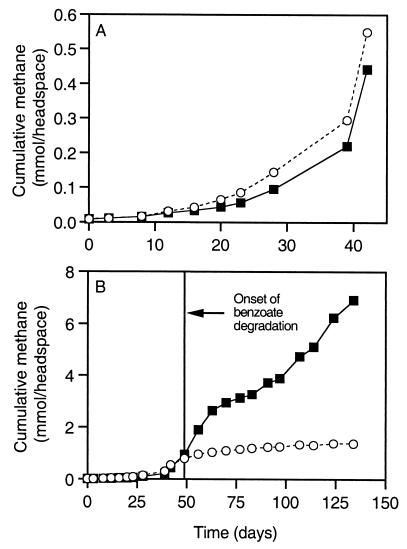
CH4 production.
Before day 49, methane levels in the control consistently exceeded those in the 2-CP-amended reactor by statistically significant amounts after day 8, based on Student's t test (α = 0.05; df = 2) (Fig. 3A). Methane production in the control and 2-CP-amended reactors increased exponentially during this period. Coincident with the onset of benzoate degradation on day 49, methanogenesis continued in the 2-CP-amended reactor but leveled off in the control (Fig. 3B).
FIG. 3.
Cumulative methane production in the 2-CP-amended (squares) and control (circles) reactors. (A) Days 0 to 42; (B) days 0 to 134. Note that different vertical-axis scales are used in the two panels.
“Acetate” concentrations.
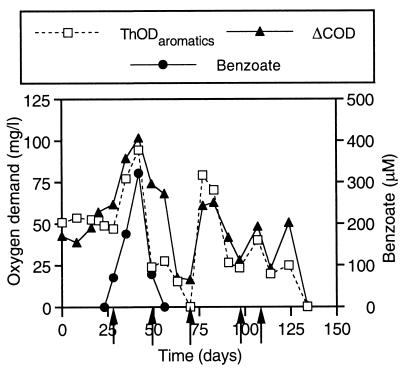
The amount of acetate in the amended reactor was estimated indirectly from oxygen demand measurements as follows. Soluble COD concentrations were monitored in the control and amended reactors. In the control reactor, COD provides an estimate of the abundance of endogenous electron donors in the sediment slurry. In the amended reactor, COD provides a measure of the endogenous substrates, 2-CP and its aromatic transformation products (phenol, benzoate, and 3-CB), and soluble, oxidizable products of benzoate fermentation, i.e., acetate. Therefore, the differences in soluble COD concentrations in the amended reactor and control (CODamended − CODcontrol = ΔCOD) is the total COD exerted by 2-CP, its aromatic transformation products, and acetate. The concentrations of the aromatic compounds were determined using high-performance liquid chromatography, expressed in terms of theoretical oxygen demand (ThOD), and summed ([ThOD2-CP] + [ThODphenol] + [ThODbenzoate] + [ThOD3-CB] = ThODaromatics) in order to determine the fraction of ΔCOD attributable to 2-CP and its aromatic transformation products. Therefore, the difference between ΔCOD and ThODaromatics provides an estimate of the concentration of acetate produced in the amended reactor. However, it is possible that other soluble organic compounds in the amended reactor represent a portion of this difference.
ΔCOD and ThODaromatics are plotted as a function of time in Fig. 4. On the days (28, 49, 70, 97, and 114) when 2-CP was administered to the amended reactor, COD concentrations were analyzed before the 2-CP additions were made. Therefore, those 2-CP spikes were omitted from the ThOD data shown in Fig. 4.
FIG. 4.
Total theoretical oxygen demand of 2-CP and its aromatic metabolites (ThODaromatics) in the 2-CP-amended reactor compared to the difference in the soluble COD concentrations (ΔCOD) in the 2-CP-amended and control reactors. 2-CP was administered on days 28, 49, 70, 97, and 107 (arrows). Because COD concentrations were analyzed before the 2-CP additions were made, 2-CP additions were not included in the ThOD data on those days. On days when the COD value exceeds the corresponding ThOD value, the difference is presumed to be due to acetate, which is produced from benzoate fermentation. Values for benzoate in the 2-CP-amended reactor are shown for reference.
ΔCOD exceeded ThODaromatics for a significant amount of time immediately after the onset of benzoate degradation, i.e., from day 49 to day 63 or 70. Thereafter, benzoate did not accumulate again, and ΔCOD values were very similar to or slightly lower than ThODaromatics except on day 124, when ΔCOD was again significantly greater than ThODaromatics. Between day 114 and day 124, approximately 670 μmol of benzoate was presumably released to the sediment community from the transformation of 2-CP and 3-CB and concomitantly converted to fermentation products.
Quantitative membrane hybridizations.
Hybridizations conducted with domain-, group-, and genus-specific probes and rRNA extracted from the 2-CP-amended and control reactors were performed in order to relate fluctuations in the activities of the targeted populations to 2-CP mineralization. On the days when 2-CP additions were made to the amended reactor, the reactors were sampled for rRNA extractions before the 2-CP amendments were made.
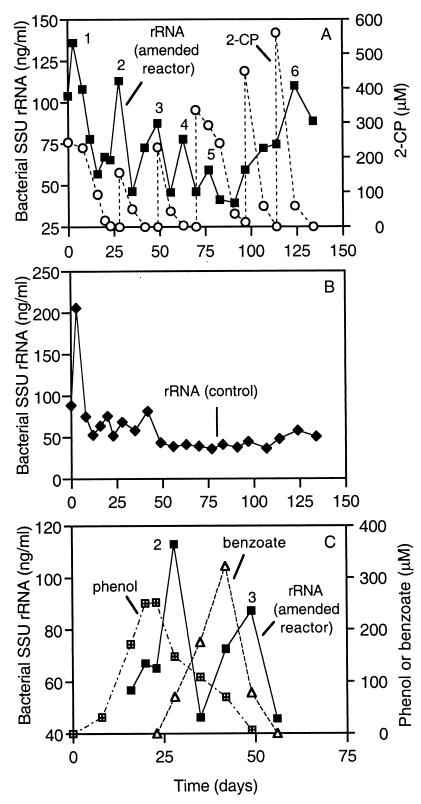
Bacterial SSU-rRNA abundance in the amended reactor is shown in Fig. 5A, along with transformation of 2-CP. In general, the bacterial SSU-rRNA concentrations were higher and fluctuated more in the amended reactor than in the control (Fig. 5B). The relationships between the bacterial SSU rRNA and 2-CP time series and among time series presented below cannot be reliably evaluated by using statistical analyses because they were measured at unequal time intervals. However, the data qualitatively suggest that at least some of the variability in bacterial SSU-rRNA concentrations in the amended reactor is influenced by the mineralization of 2-CP. Peaks in the bacterial SSU-rRNA levels generally coincided with, or lagged slightly behind, removal of 2-CP additions and/or intermediates that accumulated transiently during the mineralization of 2-CP. For example, peaks 2 and 3 in the amended-reactor bacterial SSU rRNA occurred concomitantly with decreases in the concentrations of phenol and benzoate, respectively (Fig. 5C).
FIG. 5.
Bacterial rRNA and transformation of 2-CP in the amended reactor (A), bacterial rRNA in the control reactor (B), and bacterial rRNA, phenol, and benzoate in the amended reactor (C). Numbers designate peaks in rRNA data that are referred to in the text. Note that the horizontal-axis scale in panel C is different from that in panels A and B.
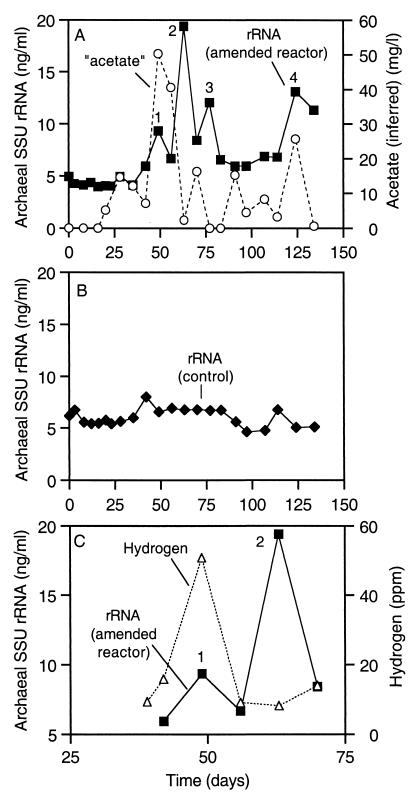
Archaeal SSU-rRNA levels are shown along with the inferred acetate concentrations in the amended reactor in Fig. 6A. Archaeal SSU-rRNA abundance in the control reactor (Fig. 6B) exceeded the levels in the 2-CP-amended reactor for the first 42 days of the experiment. Through day 30, the archaeal SSU-rRNA concentrations in both reactors were very constant. In the amended reactor, peak 1 in the archaeal SSU rRNA occurred on day 49 and coincided with a H2 burst in the amended reactor (Fig. 6C). The greatest increase in the archaeal SSU-rRNA concentrations in the 2-CP-amended reactor (peak 2) occurred between days 56 and 63, immediately following the removal of 300 μM accumulated benzoate. As shown in Fig. 6A, peak 2 in archaeal SSU rRNA occurred concomitantly with a decrease in the inferred acetate concentration in the amended reactor. Peaks 3 and 4 in the amended reactor archaeal SSU rRNA also coincided with, or slightly trailed, spikes in the inferred acetate concentrations.
FIG. 6.
Relationships between archaeal rRNA and methanogenic substrates. (A) Archaeal rRNA and inferred acetate concentrations in the 2-CP-amended reactor; (B) archaeal rRNA in the control; (C) archaeal rRNA and hydrogen in the 2-CP-amended reactor. Determination of inferred acetate concentrations is explained in the legend to Fig. 4. Negative “acetate” values are reported as zero. Numbers designate peaks in rRNA data that are referred to in the text. Note that the horizontal-axis scale in panel C is different from that in panels A and B.
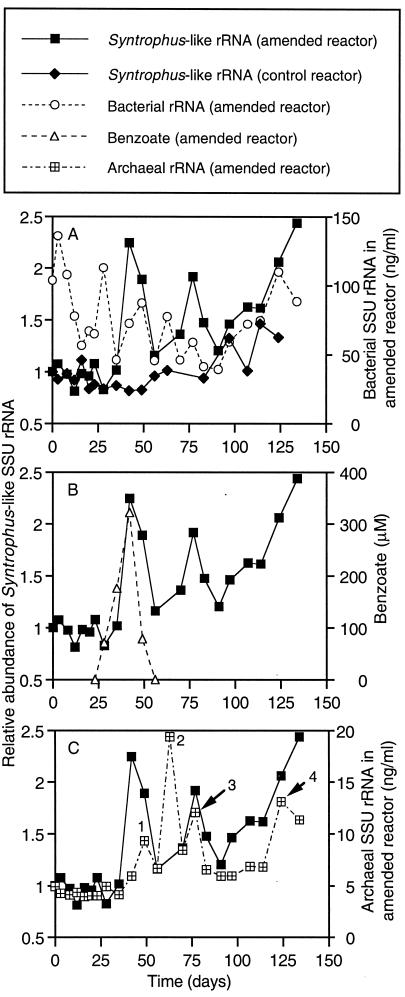
The amount of rRNA in the control and amended reactors that hybridized with the probe targeting the genus Syntrophus did not exceed 25 ng per ml of slurry; however, the concentrations could not be quantified absolutely, in part because the use of in vitro-transcribed reference rRNA instead of native rRNA has been shown to underestimate population abundance in quantitative hybridizations (32). Therefore, for this probe only, the raw hybridization data are normalized to the time zero values for the corresponding reactor.
A clear demarcation in the pattern of Syntrophus-like SSU-rRNA concentrations in the 2-CP-amended reactor is observed at approximately 42 days (Fig. 7A), which is when transiently accumulated benzoate reached its maximum concentration in the amended reactor (Fig. 7B). Prior to day 42, the levels of Syntrophus-like SSU rRNA in the 2-CP-amended reactor were very steady, unlike the concentrations of bacterial SSU rRNA in the same reactor (Fig. 7A). After day 42, greater fluctuations in the Syntrophus-like SSU-rRNA content of the amended reactor were observed, and for the most part, they tracked with the bacterial SSU-rRNA levels. In contrast, Syntrophus-like SSU-rRNA levels in the control reactor were relatively constant even after 42 days.
FIG. 7.
Relationships of Syntrophus-like rRNA in the 2-CP-amended reactor with bacterial rRNA in the 2-CP-amended reactor and Syntrophus-like rRNA in the control reactor (A), benzoate in the 2-CP-amended reactor (B), and archaeal rRNA in the 2-CP-amended reactor (C). Relative values were obtained through normalization to the time zero value for the corresponding reactor. Numbers designate peaks in rRNA data that are referred to in the text.
As shown in Fig. 7B, in the amended reactor, rising levels in the Syntrophus-like SSU-rRNA concentrations between days 23 and 42 paralleled increasing benzoate concentrations. When the initial accumulation of benzoate was subsequently depleted, the Syntrophus-like SSU-rRNA concentrations in the amended reactor also dropped off. Although benzoate was not detected in the amended reactor after day 49, it was presumably supplied to the 2-CP-degrading sediment community in equimolar amounts whenever 2-CP was administered and subsequently biotransformed. The biotransformation of 3-CB after day 107 also presumably produced benzoate as an intermediate.
As shown in Fig. 7C, after the onset of benzoate degradation, archaeal and Syntrophus-like SSU-rRNA concentrations in the amended reactor followed similar trends. Peaks 1 and 2 in the archaeal SSU-rRNA concentrations followed closely behind the first peak in the Syntrophus-like SSU rRNA and the concomitant decline in benzoate concentrations (Fig. 7B). Archaeal SSU-rRNA peaks 3 and 4 also coincided with increasing Syntrophus-like SSU-rRNA levels (Fig. 7C).
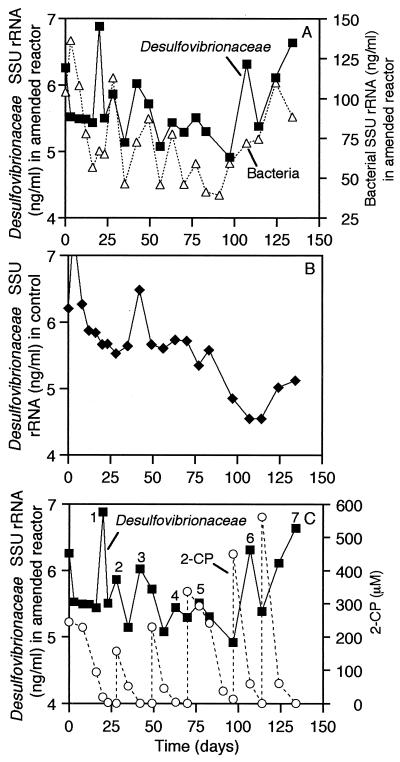
“Desulfovibrionaceae” and bacterial SSU-rRNA levels in the amended reactor are shown in Fig. 8A. Through day 20, the patterns in these measurements were not very similar, but after day 20, they followed the same general trends. In contrast, “Desulfovibrionaceae” SSU-rRNA concentrations in the control reactor remained relatively constant or decreased during most of the 134-day experiment. In particular, the “Desulfovibrionaceae” levels associated with peaks 1, 6, and 7 in the amended reactor were significantly higher than in the control and occurred concomitantly with the biotransformation of 2-CP additions (Fig. 8C).
FIG. 8.
“Desulfovibrionaceae” and bacterial rRNA in the amended reactor (A), “Desulfovibrionaceae” rRNA in the control (B), and “Desulfovibrionaceae” rRNA and transformation of 2-CP in the amended reactor (C). Numbers designate peaks in rRNA data that are referred to in the text.
DISCUSSION
Although the chlorophenol-degrading sediment community evaluated in this study was highly complex, we were able to quantify and interpret system-level processes—reductive dehalogenation, fermentation, and methanogenesis—and relate this information to the abundances of key populations—desulfovibrios, syntrophic benzoate degraders, and methanogens—that were involved in the mineralization of 2-CP. This allowed us to identify several important relationships between the evolving population structure of the sediment community and contaminant transformation.
The basis of one of these relationships was the competition of reductive dehalogenation with other reductive processes for available electron donors. Analysis of the aromatic metabolites of 2-CP biotransformation revealed that significant benzoate removal began between days 42 and 49, which is of particular significance because it demarcated a change in the nature of electron donors available to the 2-CP-transforming community. Before the onset of benzoate transformation, electrons consumed in reductive processes were necessarily derived from substrates that were endogenous to the sediment inoculum or from the minor amount of yeast extract that was initially supplied in the anaerobic medium, because no other exogenous electron donors were provided. After day 49, the electron-accepting processes occurring in the sediment community could also obtain electron donors from metabolites of 2-CP biotransformation.
Several lines of evidence suggest that prior to the onset of benzoate degradation, the addition of the chlorinated substrate led to the consumption of electron donors by dehalogenating populations at the expense of other members of the 2-CP-degrading sediment community. In presenting this evidence, it is assumed that H2 served as the ultimate electron donor for reductive dehalogenation, while endogenous organic compounds served mainly as precursors of the production of H2 via fermentations. This is analogous to models of tetrachloroethene dehalogenation in anaerobic systems supplied with organic substrates (18, 50).
Several terminal electron-accepting processes may compete for H2 in an anaerobic environment like the freshwater sediment used in this study. However, nitrate and sulfate were not detected in the amended and control reactors, as described elsewhere (5). The occurrence of Mn(IV) and Fe(III) reduction in the sediment community cannot be ruled out. However, 2-CP was repeatedly added to the amended reactor in increasingly larger doses, without any decreases in the rate of removal. This suggests that 2-CP served as a terminal electron acceptor in a form of anaerobic respiration that is sometimes referred to as dehalorespiration (23). Furthermore, growth of several isolates via ortho dechlorination of phenolic compounds has been previously demonstrated (10, 21, 29, 39, 47). CO2 was also added to the amended reactor, which was methanogenic. Thus, methanogenesis was probably the most important sink competing with reductive dehalogenation for reducing equivalents in the 2-CP-degrading sediment community.
A number of studies have evaluated competition among different H2-consuming populations (reference 50 and references therein), including hydrogenotrophic dehalogenating populations (18, 50). In general, each terminal electron-accepting process appears to establish a characteristic H2 threshold. The H2 thresholds are functions of the physiological properties of the H2-consuming populations and of the redox potential of the electron acceptor couple (50). As a rule, a higher redox potential for the electron acceptor couple yields a lower H2 threshold.
The standard half-reduction couple at pH 7 (Eo′) for the 2-CP–phenol couple is +0.399 V (13), compared to an Eo′ value of −0.24 V for the reduction of CO2 to CH4 (30). Excluding the spike on day 49, from day 20 on, the H2 concentrations (Fig. 2) leveled off in the 2-CP-amended reactor at an average value of 12 ppm (Fig. 2). A higher average H2 level (22 ppm) was maintained in the methanogenic control reactor. Thus, consistent with other studies, it appears that dehalogenating populations were able to maintain a lower H2 threshold than methanogenic members of the sediment community. Prior to the onset of benzoate degradation between days 42 and 49, the lower H2 threshold presumably enabled the dehalogenating organisms to successfully compete with hydrogenotrophic methanogens for the endogenous reducing equivalents required to reductively dehalogenate 2-CP.
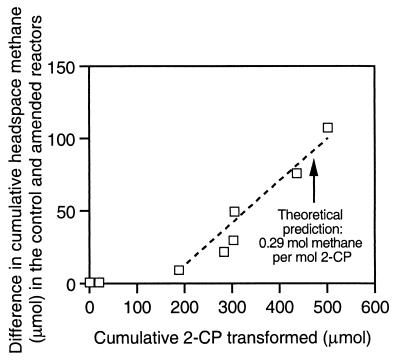
The ability of dehalogenating organisms to maintain a lower H2 threshold than that of methanogens suggests that, under H2-limiting conditions, the diversion of a portion of the limited supply of electron donors away from methanogenesis to dehalorespiration should decrease CH4 production. Further, the decrease in CH4 production should be related to the cumulative amount of chlorinated substrate that has undergone reductive dehalogenation. A theoretical prediction of the impact of 2-CP reductive dehalogenation on CH4 production was obtained using the stoichiometric method developed by McCarty (31), which is based on thermodynamic and bioenergetic principles. In implementing McCarty's approach, it was assumed that methanogenic and dehalogenating organisms utilized not only the same electron donor (H2) but also the same carbon source (CO2) and nitrogen source (NH4+). Further, it was assumed that 2-CP and CO2 served as the electron acceptors for dehalogenating and methanogenic organisms, respectively. A theoretical requirement of 1.22 mol of H2 per mol of 2-CP to meet the electron donor requirements of the dehalogenating organisms for energy and synthesis was calculated. The predicted CH4 yield is 0.24 mol of CH4 per mol of H2 consumed for the energy and synthesis needs of hydrogenotrophic methanogens. Thus, CH4 levels should theoretically be diminished by 0.29 mol per mol of 2-CP transformed, based on the stated assumptions.
This theoretical prediction was compared to experimental evidence of the impact of reductive dehalogenation on methanogenesis prior to the onset of benzoate degradation. The difference between the cumulative CH4 produced in the control and amended reactors on days 0 through 42 is plotted as a function of the cumulative mass of 2-CP transformed in the amended reactor (Fig. 9). Through day 12, H2 levels in the amended and control reactors were not significantly different, based on Student's t test (α = 0.05; df = 2), or were higher in the amended reactor than in the control (Fig. 2). This explains why the first two data points in Fig. 9, which correspond to observations made on days 0 and 8, reveal no significant difference (based on Student's t test; α = 0.05; df = 2) in the cumulative amount of methane produced in the amended and control reactors. After day 12, the 2-CP-degrading community maintained H2 levels that were significantly and consistently lower than in the control, with the exception of the H2 burst on day 49. Thus, by day 16, which corresponds to the third data point in Fig. 9 and the transformation of 190 μmol of 2-CP, a significantly lower cumulative CH4 production was observed in the amended reactor than in the control. Through day 42, the difference in the cumulative CH4 produced in the two reactors grew with increases in the cumulative amount of 2-CP dehalogenated in the amended reactor. As shown in Fig. 9, the theoretical estimate of the impact of reductive dehalogenation on methanogenesis provided a good prediction of the experimental observations, once the availability of reducing equivalents became limiting in the 2-CP-degrading community. Of course, combinations of substrates other than those assumed above could be used by dehalogenating and methanogenic organisms. Although different substrate combinations would result in somewhat different theoretical estimates of the decrease in CH4 production resulting from 2-CP-reductive dehalogenation, the general trends would be the same.
FIG. 9.
Difference in cumulative methane produced in the control and 2-CP-amended reactors as a function of the cumulative amount of 2-CP transformed in the amended reactor. Squares represent data obtained before the onset of benzoate degradation (days 0 to 42). Methane levels in the control were higher than in the 2-CP-amended reactor during this period. The assumptions made in determining the theoretical impact of 2-CP reductive dehalogenation on methanogenesis are described in the text.
Archaeal SSU-rRNA levels in the control reactor also exceeded the levels in the 2-CP-amended reactor for the first 42 days of the experiment (Fig. 6). This trend is consistent with higher CH4 production in the control than in the amended reactor during this period (Fig. 3A).
It should be noted that 2-CP has been shown to inhibit aceticlastic methanogenesis (11); therefore, it is also possible that toxicity effects contributed to the initial reduction of methanogenesis in the 2-CP-amended reactor. In addition, the theoretical calculations do not take into account the potential impact of subsequent steps in the mineralization of 2-CP, e.g., reductive dehydroxylation of p-hydroxybenzoate, on methane production.
Qualitative evaluation of the data suggests that the biodegradation of 2-CP had a direct influence on the amount of rRNA targeted by probe S-F-Dsv-0687-a-A-16 in the amended reactor (Fig. 8C). Although it has recently been recognized that certain members of the provisional family “Geobacteraceae” may contribute to hybridization with probe S-F-Dsv-0687-a-A-16 (27), the association of Desulfovibrio-like species with the anaerobic biotransformation of a halogenated aromatic compound has previously been observed. Two sodium-dependent Desulfovibrio species that are able to grow via reductive dehalogenation of a haloaromatic compound have recently been isolated (9, 47). The marine isolate Desulfovibrio dechloracetivorans utilizes 2-CP as a terminal electron acceptor (47). Desulfovibrio strain TBP-1 was isolated from estuarine sediments and couples reductive dehalogenation of 2,4,6-tribromophenol with the oxidation of lactate (9). Desulfovibrio-like populations have also been identified as members of anaerobic halobenzoate-degrading cocultures (17, 42) and a mixed community that reductively dechlorinated trichlorobenzenes (1), although the roles of the Desulfovibrio-like populations in these mixed cultures were not determined. Thus, it is possible that a Desulfovibrio-like population contributed to, and benefited from, the initial reductive dehalogenation of 2-CP in the sediment community.
Other modes of metabolism for a Desulfovibrio-like population in the amended reactor were most likely not favored. Sulfate was not added or detected in the anaerobic reactors (5). Thus, it is unlikely that members of the “Desulfovibrionaceae” were growing via sulfate reduction in the 2-CP-amended reactor. Growth of characterized Desulfovibrio isolates in the absence of an external electron acceptor is limited to fermentation of pyruvate, malate, or fumarate (49).
An important question regarding the role of the Desulfovibrio-like member of the amended-reactor community is as follows: if a Desulfovibrio-like population carried out reductive dehalogenation of 2-CP, why was the amount of SSU rRNA targeted by probe S-F-Dsv-0687-a-A-16 not higher in the amended reactor? “Desulfovibrionaceae” SSU rRNA constituted approximately 4 to 13% of the bacterial SSU rRNA in the amended reactor; however, the amount of “Desulfovibrionaceae” SSU rRNA in the amended reactor never exceeded that in the control reactor by more than approximately 1.8 ng/ml (Fig. 8). One possible explanation for these results is that additional, unidentified populations in the amended reactor community also contributed to the reductive dehalogenation of 2-CP. In addition, the possibility that the predominant Desulfovibrio populations in the amended and control reactors were different cannot be ruled out.
Under methanogenic conditions, the characterized members of the genus Syntrophus are able to ferment benzoate in the presence of H2-consuming populations (25, 35, 48). Therefore, after the onset of benzoate removal between days 42 and 49, a relationship between the level of SSU rRNA targeted by the Syntrophus-specific probe (S-G-Syn-0424-a-A-18) and benzoate metabolism was expected.
The Syntrophus-like SSU-rRNA and benzoate concentration data do suggest that members of the sediment community belonging to the genus Syntrophus were associated with fermentation of the benzoate produced during the biotransformation of 2-CP. Although fluctuations in the SSU rRNA targeted by the more general bacterial probe occurred prior to day 42, Syntrophus-like SSU-rRNA levels did not change significantly until day 42, when a dramatic increase in the abundance of Syntrophus-like SSU rRNA was observed (Fig. 7A). Subsequently, a rapid decline in the concentration of benzoate, which accumulated transiently through day 42, began, and a parallel decrease in Syntrophus-like SSU-rRNA levels was observed (Fig. 7B).
According to equation 1, fermentation of benzoate by Syntrophus strains produces H2 and acetate. This explains why on day 49, a significant increase in the inferred acetate concentration (Fig. 6A) and a H2 burst (Fig. 2) occurred concomitantly with the rapid removal of benzoate in the 2-CP-amended reactor. We expected that the onset of removal of 2-CP-derived benzoate and the associated release of electron donors would alter the impact of the chlorinated substrate on the metabolic processes and population structure of the sediment community in the amended reactor.
Several lines of evidence suggest that methanogenic populations were significantly influenced by the input of electrons derived from 2-CP in the amended reactor. First, whereas prior to day 42, CH4 levels were higher in the control than in the amended reactor, after day 49, the cumulative amounts of CH4 in the amended reactor were consistently greater than in the control reactor (Fig. 3). The stimulating effects of 2-CP-derived H2 and acetate on methanogenic populations are also clearly reflected in the archaeal SSU-rRNA data (Fig. 6). From day 49 on, the level of archaeal SSU rRNA in the amended reactor almost always exceeded that in the control. The first significant increase in the amended-reactor archaeal SSU rRNA (peak 1) occurred on day 49 and coincided with the H2 burst in the amended reactor. This suggests that the first increase in archaeal SSU rRNA reflects growth of the hydrogenotrophic methanogens due to the rapid production of H2 from the fermentation of accumulated benzoate. Peaks 2, 3, and 4 in the archaeal SSU-rRNA concentrations coincide with, or slightly trail, spikes in the inferred acetate concentration in the amended reactor and therefore probably reflect increases in the abundance of aceticlastic methanogens. Finally, the production of methanogenic electron donors via benzoate fermentation, presumably carried out by Syntrophus-like populations, is consistent with the observation that archaeal and Syntrophus-like SSU rRNA levels followed similar trends in the amended reactor (Fig. 7C).
Methanogens were able to utilize benzoate-derived electron donors because the mineralization of 1 mol of benzoate generates more electrons than are needed to reductively dehalogenate 1 mol of 2-CP. The surplus electrons were available to the methanogens. Thus, unlike the period preceding the onset of benzoate degradation, after day 49, methanogenesis and reductive dechlorination did not have to directly compete for reducing equivalents. This is consistent with results obtained with benzoate-degrading cocultures (16). H2 levels were lower when both 3-CB and CO2 were present as terminal electron-accepting sinks for H2 generated by benzoate fermentation, compared to a coculture containing only a benzoate-degrading syntroph and a hydrogenotrophic methanogen. However, the lower H2 concentration resulted in increased H2 production, thus obviating competition between the two hydrogenotrophic populations. Similarly, in this study, H2 levels were lower in the amended reactor, in which both reductive dehalogenation and methanogenesis occurred, than in the methanogenic unamended control reactor.
In this study, we used an integrated experimental approach of chemical analyses and SSU-rRNA-based monitoring of key populations to quantify and interpret system-level processes and population-level changes in a complex chlorophenol-degrading community. The results we obtained are consistent with the established paradigms of competition, syntrophy, and electron flow in anaerobic food chains and therefore provide a foundation for additional high-resolution studies of structure-function relationships in other systems impacted by contaminants.
ACKNOWLEDGMENT
This research was supported by United States Environmental Protection Agency grant R823351 to D.A.S. and B.E.R.
REFERENCES
- 1.Adrian L, Manz W, Szewzyk U, Görisch H. Physiological characterization of a bacterial consortium reductively dechlorinating 1,2,3- and 1,2,4-trichlorobenzene. Appl Environ Microbiol. 1998;64:496–503. doi: 10.1128/aem.64.2.496-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basu S K, Oleszkiewicz J A, Sparling R. Dehalogenation of 2-chlorophenol (2-CP) in anaerobic batch cultures. Water Res. 1996;30:315–322. [Google Scholar]
- 5.Becker J G. Characterization of anaerobic microbial communities that adapt to 3-chlorobenzoate and 2-chlorophenol. Dissertation. Evanston, Ill: Northwestern University; 1998. [Google Scholar]
- 6.Becker J G, Berardesco G, Rittmann B E, Stahl D A. Molecular and metabolic characterization of a 3-chlorobenzoate degrading anaerobic microbial community. In: Wickramanayake G B, Hinchee R E, editors. Natural attenuation, chlorinated and recalcitrant compounds. Columbus, Ohio: Battelle Press; 1998. pp. 93–98. [Google Scholar]
- 7.Becker J G, Stahl D A, Rittmann B E. Reductive dehalogenation and conversion of 2-chlorophenol to 3-chlorobenzoate in a methanogenic sediment community: implications for predicting the environmental fate of chlorinated pollutants. Appl Environ Microbiol. 1999;65:5169–5172. doi: 10.1128/aem.65.11.5169-5172.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bisaillon J-G, Lépine F, Beaudet R, Sylvestre M. Potential for carboxylation-dehydroxylation of phenolic compounds by a methanogenic consortium. Can J Microbiol. 1993;39:642–648. doi: 10.1139/m93-093. [DOI] [PubMed] [Google Scholar]
- 9.Boyle A W, Phelps C D, Young L Y. Isolation from estuarine sediments of a Desulfovibrio strain which can grow on lactate coupled to the reductive dehalogenation of 2,4,6-tribromophenol. Appl Environ Microbiol. 1999;65:1133–1140. doi: 10.1128/aem.65.3.1133-1140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole J R, Cascarelli A L, Mohn W W, Tiedje J M. Isolation and characterization of a novel bacterium growing via reductive dehalogenation of 2-chlorophenol. Appl Environ Microbiol. 1994;60:3536–3542. doi: 10.1128/aem.60.10.3536-3542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies-Venn C, Young J C, Tabak H H. Impact of chlorophenols and chloroanilines on the kinetics of acetoclastic methanogenesis. Environ Sci Technol. 1992;26:1627–1635. [Google Scholar]
- 12.Devereux R, Kane M D, Winfrey J, Stahl D A. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst Appl Microbiol. 1992;15:601–609. [Google Scholar]
- 13.Dolfing J, Harrison B K. Gibbs free energy of formation of halogenated aromatic compounds and their potential role as electron acceptors in anaerobic environments. Environ Sci Technol. 1992;26:2213–2218. [Google Scholar]
- 14.Dolfing J, Tiedje J M. Acetate as a source of reducing equivalents in the reductive dechlorination of 2,5-dichlorobenzoate. Arch Microbiol. 1991;156:356–361. [Google Scholar]
- 15.Dolfing J, Tiedje J M. Hydrogen cycling in a three-tiered food web growing on the methanogenic conversion of 3-chlorobenzoate. FEMS Microbiol Ecol. 1986;38:293–298. [Google Scholar]
- 16.Dolfing J, Tiedje J M. Kinetics of two complementary hydrogen sink reactions in a defined 3-chlorobenzoate degrading methanogenic coculture. FEMS Microbiol Ecol. 1991;86:25–32. [Google Scholar]
- 17.Drzyzga O, Jannsen S, Blotevogel K-H. Mineralization of monofluorobenzoate by a diculture under sulfate-reducing conditions. FEMS Microbiol Lett. 1994;116:215–220. doi: 10.1111/j.1574-6968.1994.tb06703.x. [DOI] [PubMed] [Google Scholar]
- 18.Fennell D E, Gossett J M, Zinder S H. Comparison of butyric acid, ethanol, lactic acid, and propionic acid as hydrogen donors for the reductive dechlorination of tetrachloroethene. Environ Sci Technol. 1997;31:918–926. [Google Scholar]
- 19.Fitzgerald S A. The biogeochemistry of amino acids in sediments from the Great Lakes. Ph.D. dissertation. Milwaukee: University of Wisconsin-Milwaukee; 1989. [Google Scholar]
- 20.Freedman D L, Gossett J M. Biological reductive dechlorination of tetrachloroethylene and trichloroethylene to ethylene under methanogenic conditions. Appl Environ Microbiol. 1989;55:2144–2151. doi: 10.1128/aem.55.9.2144-2151.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerritse J, Renard V, Gomes T M P, Lawson P A, Collins M D, Gottschal J C. Desulfitobacterium sp. strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols. Arch Microbiol. 1996;165:132–140. doi: 10.1007/s002030050308. [DOI] [PubMed] [Google Scholar]
- 22.Hach Company. Hach water analysis handbook. 3rd ed. Loveland, Colo: Hach Company; 1997. [Google Scholar]
- 23.Holliger C, Wohlfarth G, Diekert G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev. 1999;22:383–398. [Google Scholar]
- 24.Horowitz A, Suflita J M, Tiedje J M. Reductive dehalogenations of halobenzoates by anaerobic lake sediment microorganisms. Appl Environ Microbiol. 1983;45:1459–1465. doi: 10.1128/aem.45.5.1459-1465.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson B E, Bhupathiraju V K, Tanner R S, Woese C R, McInerney M J. Syntrophus aciditrophicus sp. nov., a new anaerobic bacterium that degrades fatty acids and benzoate in syntrophic association with hydrogen-using microorganisms. Arch Microbiol. 1999;171:107–114. doi: 10.1007/s002030050685. [DOI] [PubMed] [Google Scholar]
- 26.Lin C, Stahl D A. Taxon-specific probes for the cellulolytic genus Fibrobacter reveal abundant and novel equine-associated populations. Appl Environ Microbiol. 1995;61:1348–1351. doi: 10.1128/aem.61.4.1348-1351.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lonergan D J, Jenter H L, Coates J D, Phillips E J P, Schmidt T M, Lovley D R. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacGregor B J, Moser D P, Alm E W, Nealson K H, Stahl D A. Crenarchaeota in Lake Michigan sediment. Appl Environ Microbiol. 1997;63:1178–1181. doi: 10.1128/aem.63.3.1178-1181.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackiewicz M, Wiegel J. Comparison of energy and growth yields for Desulfitobacterium dehalogenans during utilization of chlorophenol and various traditional electron acceptors. Appl Environ Microbiol. 1998;64:352–355. doi: 10.1128/aem.64.1.352-355.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madigan M T, Martinko J M, Parker J. Brock biology of microorganisms. 9th ed. Upper Saddle River, N.J: Prentice Hall; 2000. [Google Scholar]
- 31.McCarty P L. Stoichiometry of biological reactions. Prog Water Technol. 1975;7:157–172. [Google Scholar]
- 32.McMahon K D, Stahl D A, Raskin L. A comparison of the use of in vitro-transcribed and native rRNA for the quantification of microorganisms in the environment. Microbiol Ecol. 1998;36:362–371. doi: 10.1007/s002489900122. [DOI] [PubMed] [Google Scholar]
- 33.Miller T L, Wolin M J. A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl Environ Microbiol. 1974;27:985–987. doi: 10.1128/am.27.5.985-987.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohn W W, Tiedje J M. Microbial reductive dehalogenation. Microbiol Rev. 1992;56:482–507. doi: 10.1128/mr.56.3.482-507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mountfort D O, Brulla W J, Krumholz L R, Bryant M P. Syntrophus buswellii gen. nov., sp. nov.: a benzoate catabolizer from methanogenic ecosystems. Int J Syst Bacteriol. 1984;34:216–217. [Google Scholar]
- 36.Muyzer G, Waal E C D, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes encoding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raskin L, Poulsen L K, Noguera D R, Rittmann B E, Stahl D A. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl Environ Microbiol. 1994;60:1241–1248. doi: 10.1128/aem.60.4.1241-1248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanford R A, Cole J R, Löffler F E, Tiedje J M. Characterization of Desulfitobacterium chlororespirans sp. nov., which grows by coupling oxidation of lactate to the reductive dechlorination of 3-chloro-4-hydroxybenzoate. Appl Environ Microbiol. 1996;62:3800–3808. doi: 10.1128/aem.62.10.3800-3808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharak Genthner B R. Preliminary characterization of four 2-chlorobenzoate-degrading anaerobic bacterial consortia. Biodegradation. 1999;10:27–33. doi: 10.1023/a:1008348123672. [DOI] [PubMed] [Google Scholar]
- 41.Sharak Genthner B R, Price II W A, Pritchard P H. Characterization of anaerobic dechlorinating consortia derived from aquatic sediments. Appl Environ Microbiol. 1989;55:1472–1476. doi: 10.1128/aem.55.6.1472-1476.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharak Genthner B R, Townsend G T, Blattmann B O. Reduction of 3-chlorobenzoate, 3-bromobenzoate, and benzoate to corresponding alcohols by Desulfomicrobium escambiense, isolated from a 3-chlorobenzoate-dechlorinating coculture. Appl Environ Microbiol. 1997;63:4698–4703. doi: 10.1128/aem.63.12.4698-4703.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharak Genthner B R, Townsend G T, Chapman P J. Anaerobic transformation of phenol to benzoate via para-carboxylation: use of fluorinated analogues to elucidate the mechanism of transformation. Biochem Biophys Res Commun. 1989;162:945–951. doi: 10.1016/0006-291x(89)90764-x. [DOI] [PubMed] [Google Scholar]
- 44.Sharak Genthner B R, Townsend G T, Chapman P J. para-Hydroxybenzoate as an intermediate in the anaerobic transformation of phenol to benzoate. FEMS Microbiol Lett. 1991;78:265–270. doi: 10.1016/0378-1097(91)90168-a. [DOI] [PubMed] [Google Scholar]
- 45.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suflita J M, Horowitz A, Shelton D R, Tiedje J M. Dehalogenation: a novel pathway for the anaerobic biodegradation of haloaromatic compounds. Science. 1982;218:1115–1117. doi: 10.1126/science.218.4577.1115. [DOI] [PubMed] [Google Scholar]
- 47.Sun B, Cole J R, Sanford R A, Tiedje J M. Isolation and characterization of Desulfovibrio dechloracetivorans sp. nov., a marine dechlorinating bacterium growing by coupling the oxidation of acetate to the reductive dechlorination of 2-chlorophenol. Appl Environ Microbiol. 2000;66:2408–2413. doi: 10.1128/aem.66.6.2408-2413.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wallrabenstein C, Gorny N, Springer N, Ludwig W, Schink B. Pure culture of Syntrophus buswellii, definition of its phylogenetic status, and description of Syntrophus gentianae sp. nov. Syst Appl Microbiol. 1995;18:62–66. [Google Scholar]
- 49.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. IV. New York, N.Y: Springer-Verlag; 1992. pp. 3352–3378. [Google Scholar]
- 50.Yang Y, McCarty P L. Competition for hydrogen within a chlorinated solvent dehalogenating anaerobic mixed culture. Environ Sci Technol. 1998;32:3591–3597. [Google Scholar]
- 51.Zhang X, Morgan T V, Wiegel J. Conversion of 13C-1 phenol to 13C-4 benzoate, an intermediate step in the anaerobic degradation of chlorophenols. FEMS Microbiol Lett. 1990;67:63–66. [Google Scholar]
- 52.Zhang X, Wiegel J. The anaerobic degradation of 3-chloro-4-hydroxybenzoate in freshwater sediment proceeds via either chlorophenol or hydroxybenzoate to phenol and subsequently to benzoate. Appl Environ Microbiol. 1992;58:3580–3585. doi: 10.1128/aem.58.11.3580-3585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X, Wiegel J. Sequential anaerobic degradation of 2,4-dichlorophenol in freshwater sediments. Appl Environ Microbiol. 1990;56:1119–1127. doi: 10.1128/aem.56.4.1119-1127.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]