ABSTRACT
Adverse pregnancy outcomes disproportionately affect non-Hispanic (NH) Black patients in the United States. Structural racism has been associated with increased psychosocial distress and inflammation and may trigger oxidative stress. Thus, the nitric oxide (NO) pathway (involved in the regulation of inflammation and oxidative stress) may partly explain the underlying disparities in obstetric outcomes.
Cohort study of 154 pregnant patients with high-risk obstetric histories; n = 212 mRNAs and n = 108 microRNAs (miRNAs) in the NO pathway were evaluated in circulating white blood cells. NO pathway mRNA and miRNA transcript counts were compared by self-reported race; NH Black patients were compared with women of other races/ethnicities. Finally, miRNA-mRNA expression levels were correlated.
Twenty-two genes (q < 0.10) were differentially expressed in self-identified NH Black individuals. Superoxide dismutase 1 (SOD1), interleukin-8 (IL-8), dynein light chain LC8-type 1 (DYNLL1), glutathione peroxidase 4 (GPX4), and glutathione peroxidase 1 (GPX1) were the five most differentially expressed genes among NH Black patients compared to other patients. There were 63 significantly correlated miRNA-mRNA pairs (q < 0.10) demonstrating potential miRNA regulation of associated target mRNA expression. Ten miRNAs that were identified as members of significant miRNA-mRNA pairs were also differentially expressed among NH Black patients (q < 0.10).
These findings support an association between NO pathway and inflammation and infection-related mRNA and miRNA expression in blood drawn during pregnancy and patient race/ethnicity. These findings may reflect key differences in the biology of inflammatory gene dysregulation that occurs in response to the stress of systemic racism and that underlies disparities in pregnancy outcomes.
KEYWORDS: Gene expression, microRNA, nitric oxide pathway, pregnancy complications, health disparities
Introduction
In the United States, the rates of nearly all adverse obstetric outcomes vary by patient race. Non-Hispanic/Latinx (NH) Black individuals carry the highest risks of several major pregnancy complications, including preterm birth and preeclampsia, at rates up to twofold higher than patients who identify with other racial/ethnic groups [1–3]. Preterm birth, defined as delivery <37 weeks’ gestation, is the leading cause of morbidity and mortality among non-anomalous neonates in the United States and formerly preterm children who survive carry a disproportionate share of lifelong complications compared with children born full term [4–8]. NH Black patients are also more likely to deliver at the earliest gestational ages, when the risk of neonatal morbidity and mortality is highest [9–12]. Similarly, pre-eclampsia is a serious pregnancy complication that is associated with an increased risk of medically indicated preterm birth, and can be accompanied by systemic dysfunction, seizures, and death of the patient and/or neonate [13–15]. For the remainder of this manuscript, we will use the term ‘NH Black’ patients to encompass both race and ethnicity. Further, we note that race as used here serves not as a ‘biologic variable’ but as a social construct and is an indicator of the lived experience of being Black in the United States.
Though the precise mechanisms remain elusive, stress related to longstanding personal, structural, and societal racism – rather than the social construct of race itself – likely best explains the racial disparities that are observed in pregnancy and birth outcomes [16–19]. Regardless of an individual’s personal experience with racism, structural and systemic racism are terms that are often used interchangeably and refer to a pervasive pattern in society. In the United States, structural and systemic racism have penetrated nearly all facets of life, including housing, education, employment, credit, healthcare, and criminal justice, and have perpetuated discriminatory beliefs, values, and distribution of resources – whereby ‘whiteness’ is associated with advantage and ‘colour’ is associated with disadvantage and the need to endure and adapt over time[20]. Structural racism differs slightly from systemic racism in that it focuses more on the historical, cultural, and social psychological aspects of society. Experiencing, being exposed to, and living in a society – such as that which is present in the United States – in which there is both structural racism and systemic racism is associated with poorer mental and physical health [21]. The negative effects of structural and systemic racism are not abrogated by age, sex, birthplace, or education, suggesting effects on entire populations [21].
The nitric oxide pathway has biologic plausibility in the pathophysiology of adverse obstetric outcomes including preterm birth and preeclampsia. Nitric oxide is a biologic messenger that is ubiquitous throughout the body; it has roles as a neurotransmitter, vasodilator, and bactericide[22]. During pregnancy, endogenous and exogenous progestogens stimulate nitric oxide synthesis via transcriptional and non-transcriptional pathways in human endothelial cells [23–26]. Nitric oxide also increases cyclic guanine monophosphate, which relaxes both peripheral vessels (contributing to the physiologic drop in blood pressure that occurs in pregnancy) and in the myometrium (helping to prevent the onset of uterine contractions and parturition). The nitric oxide pathway also mediates inflammation, a key driver of spontaneous preterm birth; tissue-derived inducible nitric oxide synthase is up-regulated in response to infectious stimuli (e.g., lipopolysaccharide)[27]. Previous studies have found differences in nitric oxide gene expression in the blood of individuals destined to deliver preterm vs. full term [28,29], differences in nitric oxide pathway CpG methylation and gene expression patterns in placentas from patients with recurrent preterm birth vs. term births[30], and differential nitric oxide pathway microRNA (miRNA) expression among placentas from patients with severe preeclampsia compared to those without preeclampsia[31]. miRNAs are small, single-stranded, non-coding RNA molecules that are known to play a critical role in post-transcription control of gene expression. Specifically, miRNAs induce messenger RNA (mRNA) degradation or repress translation by binding to the 3ʹ untranslated region of the target mRNA[32]. Therefore, an increase in miRNA expression commonly corresponds to a subsequent decrease in target mRNA gene expression, though in some cases miRNAs may activate gene expression and be positively associated with mRNA expression [32,33].
Structural and systemic racism have been associated with heightened psychosocial distress and elevated levels of inflammation and oxidative stress in pregnant and non-pregnant individuals [34–36]. Thus, the nitric oxide pathway – intimately involved in the regulation of inflammation and oxidative stress – provides a plausible mechanism linking patient race and adverse pregnancy outcomes. However, it is unknown whether epigenetic differences in the nitric oxide and related pathways may partly explain the underlying disparities in obstetric outcomes. Thus, the hypothesis of this study was that nitric oxide pathway gene expression during mid-pregnancy differs among self-reported NH Black patients compared to patients of other racial/ethnic backgrounds. Further, we also hypothesized that differentially expressed genes would be correlated with miRNAs known to regulate their expression and that these miRNAs would also demonstrate differences by patient race.
Materials and methods
Cohort description
This cross-sectional study was derived from the prospective University of North Carolina Preterm Birth Biobank (UNC Preterm Birth Biobank) cohort study. The UNC Preterm Birth Biobank enrolled 271 patients at high risk for spontaneous preterm birth from outpatient obstetric clinics and inpatient obstetric units at the University of North Carolina-Chapel Hill (Chapel Hill, NC), 2015–2017. Patients were considered ‘high-risk’ for spontaneous preterm birth if they met at one or more of the following criteria: (a) at least one spontaneous preterm birth between 16°/[7] and 366/[7] weeks’ gestation in a previous pregnancy; (b) a short transvaginal cervical length (<25 mm prior to 24°/[7] weeks’ gestation) in the current pregnancy; (c) a twin or triplet gestation; and/or (d) admission to the high-risk antepartum service due to either preterm pre-labour rupture of membranes or threatened preterm labour (defined as cervical change and/or cervical dilation in the setting of at least 6 symptomatic uterine contractions per hour). Patients carrying foetuses with major structural anomalies or aneuploidy were ineligible. Each patient’s pregnancy due date was determined using a combination of the last menstrual period (if available) and earliest ultrasound using standard American College of Obstetricians and Gynaecologists criteria[37]. At enrolment, participants met with trained research staff and were asked a set of standardized clinical interview questions to obtain demographic, medical, and prior and current pregnancy history data, including an open-ended question regarding what race they best identified with, and whether they identified as Hispanic/Latinx. Interview data was verified and supplemented by information from the electronic medical record. Antenatal characteristics, labour and delivery course, delivery indication, and neonatal outcomes were also collected from the electronic medical record. All clinical management decisions were made at the discretion of each patient’s obstetric provider. Blood was collected by standard venipuncture at enrolment into a PaxGene RNA tube (Qiagen, Valencia, CA), which was then frozen at −80°C until analysis. Study data were collected and managed using REDCap (Research Electronic Data Capture) tools, a secure, web-based application designed to support data capture for research studies, hosted at UNC-Chapel Hill[38]. All participants provided written, informed consent and this study was approved by the Institutional Review Board of the UNC-Chapel Hill, Chapel Hill.
For the purposes of the current analysis, we included 155 participants after excluding 115 patients with evidence of active preterm labour (defined as at least 6 uterine contractions per hour in the presence of cervical dilation) or preterm prelabour rupture of membranes prior to 37 weeks’ gestation at the time of study enrolment and one patient with missing race/ethnicity data.
mRNA and miRNA isolation and quantification
Total RNA was isolated from stored maternal blood using the PaxGene Blood miRNA/RNA kit (Qiagen, Valencia, CA) according to manufacturer’s instructions. The Zymo Clean and Concentrate kit (Zymo Research, Irvine, CA) was used, according to manufacturer’s instructions, to clean and concentrate RNA. Standard 260:230 and 260:280 spectrophotometer analysis was used to quantify RNA and assess RNA sample purity. A custom 212-gene NanoString (NanoString Technologies, Seattle, WA) mRNA panel (containing n = 202 candidate genes and n = 10 housekeeping genes) was then used to evaluate RNA transcript counts for each subject. Genes in the nitric oxide pathway (per the Kyoto Encyclopaedia of Genes and Genomes), those associated with inflammation and infection (e.g., cytokines), and those previously associated with spontaneous preterm birth were selected for inclusion on the panel [39–45]. A custom 108-gene NanoString (NanoString Technologies, Seattle, WA) miRNA panel (containing n = 99 candidate miRNA and n = 9 housekeeping miRNA) was similarly designed. The miRSystem (ver. 20,160,513) database was then used to predict miRNA-mRNA target pairs; this database integrates multiple miRNA target gene prediction programs (DIANA, miRanda, miRBridge, PicTar, PITA, rna22, and TargetScan)[46]. We selected miRNAs for inclusion that specifically target the genes included on the custom mRNA panel in addition to individual miRNAs previously associated with adverse pregnancy outcomes (NanoString Technologies, Seattle, WA). When we considered transcripts included on both the mRNA and miRNA panels, there were 162 known mRNA-miRNA pairings, 36 miRNAs not known to target any of the analysed mRNA, and 20 mRNAs not known to be a target of the included miRNAs.
Filtering, normalization, and QA/QC of nanostring mRNA transcripts
Universally low expressed transcripts were excluded, as per our previous genome-wide mRNA analyses, in order to remove transcripts that may bias results [47–49]. Specifically, after conducting a sensitivity analysis of various filtering approaches, genes with a median signal intensity above 5, which was the median signal intensity of all 10 mRNA housekeeping genes, remained in the analysis. Based on these parameters, n = 159 genes remained in the final analysis (n = 53 genes were excluded due to low transcript counts).
Raw, filtered transcript counts were evaluated using the DeSeq2 package (v 1.28.0) in the ‘R’ statistical software [50,51]. First, counts were normalized in order to adjust for RNA composition bias and library size. Specifically, the DeSeq2 normalization was used in which counts are divided by a sample-specific size factor, resulting in to ‘log transformed normalized counts’ that approximate the ratio of transcript count divided by size factor. Comprehensive QA/QC was also then conducted to determine if there were any substantial outliers in the data warranting removal. Specifically, hierarchical clustering, using the hclust function, and principal component analysis, via the prcomp function were used. No sample was deemed a significant outlier to require removal from the analysis. The Remove Unwanted Variation (RUVSeq) (v 1.122.0) package in R was used to account for cell type heterogeneity, such as variation in gene expression associated with white blood cell counts that may otherwise bias results. RUV functions through empirically calculating a surrogate variable representative of background variation in the data, based on the expression of housekeeping genes. This surrogate variable was then added to the multivariable model, detailed below. This package is designed for use with NanoString data [52,53].
Statistical analysis
Patients were grouped by their self-reported race/ethnicity for the purposes of analysis; at the time of the original study interview, individuals who self-identified with more than one race and/or ethnicity were asked to identify with a primary race/ethnicity. Because the greatest disparity in obstetric outcomes occurs among NH Black patients, those individuals who self-identified as NH Black were compared to patients who self-identified as belonging to other racial/ethnic groups. Demographic characteristics of the cohort were evaluated using bivariate tests of clinical characteristics, comparing NH Black patients to those of other racial/ethnic groups using two-tailed t-test, chi-square, and Wilcoxon rank-sum as appropriate using STATA statistical software (version 15.1, College Station, TX). Medians were compared for continuous data that were not normally distributed.
Negative binomial generalized linear models within DESeq2 were used to identify mRNAs that were differentially expressed among NH Black patients (dependent variable), controlling for covariates. Model covariates included the gestational age (considered continuously, in weeks and days) at the time of blood sample collection, and the RUV surrogate variable to account for cell type heterogeneity. The gestational age at sample collection was included based on prior knowledge of variation in gene expression by gestational age [54–57]. There were no subjects missing data with regards to these key covariates. In brief, negative binomial generalized linear models calculate shrunken logarithmic fold changes in expression, which are then divided by their standard error values to produce z-statistics. These z-statistics are compared to standard normal distribution curves to generate Wald test p-values. To account for multiple testing, the Benjamini–Hochberg (BH) procedure was used to control for the false discovery rate (FDR). Statistical significance for mRNA gene expression was set at q-value of <0.10.
Identification of miRNA regulators and correlation between miRNA and mRNA expression
We next evaluated the correlation between miRNA and mRNA expression utilizing in silico prediction and correlation analysis. First, miRNA targets of significantly differentially expressed mRNA were identified utilizing the publicly available miRSystem database [46,58]. Multiple predicted miRNA-mRNA pairs were generated; these pairs were then filtered to miRNAs that were measured and were evaluated for significant correlation of expression levels[46]. Specifically, the correlation between normalized mRNA and normalized miRNA counts was evaluated utilizing the two-tailed Spearman rank test, which accounts for non-normal distributions. miRNA count data were normalized using similar methodology to the normalization of mRNA data, also utilizing the DeSeq2 package. The miRNA data, however, were not filtered to remove universally lowly expressed counts because the miRNA data were not used for a differential expression analysis. miRNA-mRNA pairs with a correlation with p < 0.05 were considered statistically significant due to the exploratory nature of this analysis; however, FDR corrected p values were also calculated utilizing the BH procedure described above for reference. In sum, this analytical step identified miRNA-mRNA expression pairs wherein a) the mRNA was differentially expressed by patient race/ethnicity; b) the miRNA is predicted to target the mRNA; c) the miRNA and mRNA normalized counts were significantly correlated in this cohort.
Additional considerations
This was an exploratory analysis derived from an existing cohort. For this reason, sample size calculations were not performed. Further, because post-hoc power analysis is often considered by epidemiologists and statisticians to be misleading, no post-hoc power analysis was performed[59]. All laboratory and analytic personnel were blinded to the clinical data through the mRNA QC steps, and patient race/ethnicity data were available for each subject only in the final steps of the analysis. De-identified datasets are available to external investigators upon request.
Results
Cohort description
In total, 155 patients met inclusion criteria and were included. Of these, 51 (32.9%) self-identified as NH Black, 59 (38.1%) as NH White, 38 (24.5%) as Hispanic/Latinx ethnicity with unspecified race, and 7 (4.5%) as belonging to ‘other’ racial/ethnic groups. Demographic and obstetric characteristics of the study population are shown in Table 1. Changes in maternal circulating blood gene expression were assessed at a median 19.1 [interquartile range (IQR) 12.9–24.9] weeks’ gestation (range: 66/[7] through 406/[7] weeks’ gestation); the gestational age at the time of the sample collection did not differ by NH Black patients vs. other patients (p = 0.31). The vast majority of patients (141/155, 90.6%), had their blood drawn early (prior to 28 weeks’ gestation), and the distribution of individuals with an early blood draw was also similar between NH Black patients and other patients.
Table 1.
Demographic, prior pregnancy, and antenatal characteristics of the study cohort. Data are n(%) unless otherwise specified; the denominator is provided, as applicable, if there are missing values for a variable.
| Characteristic |
Non-Hispanic Black patients n = 51 |
Patients of other race and/or ethnic groups n = 104 |
p-value |
|---|---|---|---|
| Maternal age, mean years (± SD) | 31.4 ± 6.2 | 32.1 ± 6.6 | 0.546 |
| Married | 23 (45.1) | 77 (75.5) | <0.001 |
| Low socioeconomic status* | 14 (27.5) | 35 (34.3) | 0.391 |
| Smoked cigarettes during pregnancy | 5 (9.8) | 14 (13.9) | 0.475 |
| Pre-pregnancy body mass index, median kg/m2 (IQR) | 32.7 (26.0, 39.1) | 28.3 (23.2, 33.3) | 0.003 |
| No prior pregnancy to reach at least 14 weeks’ gestation | 11 (21.6) | 22 (21.2) | 0.953 |
| Prior preterm birth <37 weeks’ gestation** | 38 (95.0) | 79 (96.3) | 0.726 |
| Gestational age of earliest prior preterm birth, median weeks (IQR)† | 22.1 (19.4, 30.7) | 26.0 (20.9, 32.6) | 0.257 |
| Shortest endovaginal cervical length, median mm (IQR)‡ | 28 (9, 37) | 31.5 (16, 40) | 0.568 |
| Endovaginal cervical length less than 25 mm‡ | 21/47 (44.7) | 39/94 (41.5) | 0.718 |
| Cervical cerclage placed | 26 (51.0) | 41 (39.4) | 0.172 |
| Received 17-alpha hydroxyprogesterone caproate** | 33/38 (86.8) | 67/79 (84.8) | 0.770 |
| Received vaginal progesterone | 17 (33.3) | 24 (23.1) | 0.174 |
* defined as one or more of the following: no healthcare insurance, annual household income <$24,000 annually, or less than high school education
**among 122 patients with at least one prior pregnancy reaching 14 weeks’ gestation
†among patients with ≥1 prior preterm birth and prior history available
‡among 141 patients with ≥1 endovaginal cervical length 16.0–23.9 weeks’ gestation
mRNA transcripts associated with NH black race
In multivariable models controlling for the gestational age at sample collection and the RUV surrogate variable, a total of 22 genes (q < 0.10) were differentially expressed among NH Black patients compared to other patients (Table 2; Figure 1). A nearly equal number of differentially expressed genes were down-regulated (negative log-fold change; 10/22, 45.4%) and up-regulated in NH Black patients (12/22, 54.5%) compared to other patients. The five most differentially expressed genes between NH Black compared to other patients were SOD1, log-fold change 0.674, q = 4.21 × 10−[7]), interleukin-8 IL-8 (also known C-X-C motif chemokine ligand 8 (CXCL8); log-fold change −1.110, q = 5.78 × 10−[7]), dynein light chain LC8-type 1 (DYNLL1; log fold change 0.330, q = 5.27 × 10−[4]), glutathione peroxidase 4 (GPX4; log fold change 0.435, q = 8.33 × 10−[4]), and glutathione peroxidase 1 (GPX1; log fold change 0.513, q = 9.32 × 10−[4]).
Table 2.
Genes differentially expressed among non-Hispanic Black patients compared to other patients after adjusting for gestational age at sample collection.
| Gene | Gene Symbol | Log fold change | Standard Error | q-value |
|---|---|---|---|---|
| Superoxide dismutase | SOD1 | 0.674 | 0.114 | 4.2e-07 |
| Interleukin-8 | IL8 | −1.11 | 0.194 | 5.8e-07 |
| Dynein light chain 1, cytoplasmic | DYNLL1 | 0.336 | 0.074 | 2.0e-04 |
| Glutathione peroxidase 4 | GPX4 | 0.439 | 0.104 | 5.8e-04 |
| Glutathione peroxidase 1 | GPX1 | 0.513 | 0.126 | 9.3e-03 |
| Cyclin dependent kinase inhibitor 1A | CDKN1A | 0.301 | 0.078 | 1.4e-03 |
| Neutrophil cytosolic factor 1 | NCF1 | −0.258 | 0.069 | 2.7e-03 |
| CREB binding protein | CREBBP | −0.213 | 0.076 | 0.058 |
| RAP2C, member of RAS oncogene family | RAP2C | −0.126 | 0.046 | 0.065 |
| Signal transducer and activator of transcription 1 | STAT1 | −0.247 | 0.091 | 0.066 |
| Peroxiredoxin 2 | PRDX2 | 0.387 | 0.144 | 0.066 |
| Major histocompatibility complex, class II, DQ alpha 1 | HLADQA1 | 1.121 | 0.422 | 0.068 |
| AKT serine/threonine kinase 1 | AKT1 | −0.165 | 0.063 | 0.070 |
| Fos proto-oncogene, AP-1 transcription factor subunit | FOS | −0.311 | 0.124 | 0.080 |
| Phosphatidylinositol-4,5-bisphosphate 3-Kinase catalytic subunit delta | PIK3CD | −0.195 | 0.078 | 0.080 |
| Galactosidase alpha | GLA | 0.128 | 0.051 | 0.078 |
| Thioredoxin reductase 2 | TXNRD2 | 0.225 | 0.092 | 0.086 |
| Methionine sulphoxide reductase A | MSRA | −0.170 | 0.070 | 0.087 |
| Interleukin-6 receptor | IL6R | −0.194 | 0.081 | 0.087 |
| Hypoxanthine phospho-ribosyltransferase 1 | HPRT1 | 0.133 | 0.056 | 0.087 |
| Erythropoietin receptor | EPOR | 0.160 | 0.068 | 0.096 |
| Cold shock domain containing E1 | CSDE1 | 0.168 | 0.073 | 0.096 |
Figure 1.
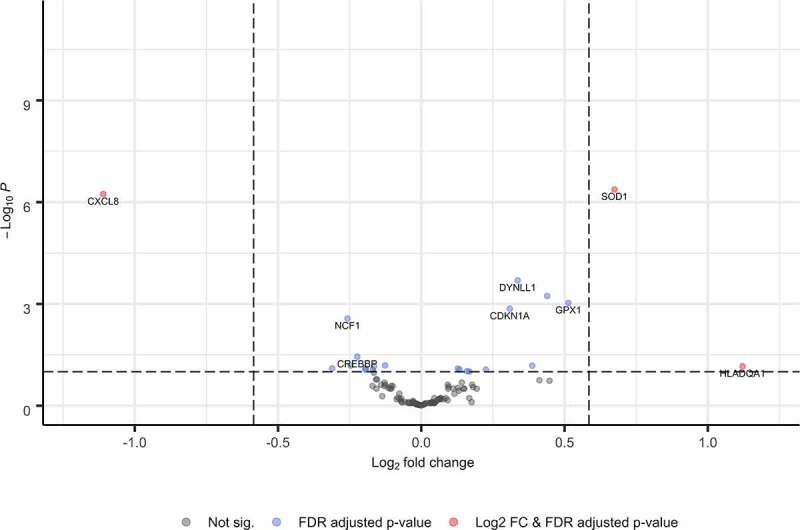
Volcano plot of genes differentially expressed by race and ethnicity.
Identification of miRNAs potentially targeting differentially expressed mRNA
Because miRNAs are known to regulate mRNA expression, we next evaluated whether miRNA expression was correlated with the differentially expressed mRNA. We found 162 miRNA-mRNA pairs that included 36 unique miRNAs. Overall, 63 of the 162 miRNA-mRNA pairs (44%) were significantly correlated (q < 0.10), Table 3. Of the 63 significant miRNA-mRNA pairs, a nearly equal number (n = 30, 47.6%) were negatively associated (e.g., increased miRNA expression was associated with decreased mRNA expression) as were positively associated. Correlation between the most significantly correlated miRNA-mRNA pairs is shown in Figure 2.
Table 3.
Significantly correlated miRNA-mRNA pairs.
| Gene name | Gene symbol | miR | Coefficient | p-value |
|---|---|---|---|---|
| AKT serine/threonine kinase 1 | AKT1 | miR-495-3p | −0.467 | 9.18E-06 |
| miR-222-3p | 0.406 | 1.83E-04 | ||
| miR-143-3p | −0.35 | 1.60E-03 | ||
| miR-539-5p | −0.338 | 2.41E-03 | ||
| Cyclin dependent kinase inhibitor 1A | CDKN1A | miR-15a-5p | 0.296 | 9.10E-03 |
| CREB binding protein | CREBBP | miR-181a-5p | 0.409 | 1.62E-04 |
| miR-495-3p | −0.366 | 8.72E-04 | ||
| miR-140-5p | 0.357 | 1.24E-03 | ||
| miR-494-3p | −0.312 | 5.51E-03 | ||
| miR-145-5p | 0.293 | 1.00E-02 | ||
| miR-340-5p | −0.267 | 2.09E-02 | ||
| miR-106b-5p | 0.253 | 2.98E-02 | ||
| miR-539-5p | −0.239 | 4.09E-02 | ||
| Cold shock domain containing E1 | CSDE1 | miR-15a-5p | 0.377 | 5.88E-04 |
| let-7 c-5p | −0.246 | 3.44E-02 | ||
| miR-539-5p | −0.228 | 5.28E-02 | ||
| miR-222-3p | 0.221 | 6.11E-02 | ||
| miR-940 | −0.216 | 6.81E-02 | ||
| miR-106b-5p | 0.214 | 7.08E-02 | ||
| miR-140-5p | 0.203 | 8.88E-02 | ||
| C-X-C motif chemokine ligand 8 | CXCL8 | miR-876-5p | 0.438 | 4.33E-05 |
| miR-539-5p | 0.388 | 3.70E-04 | ||
| miR-106b-5p | −0.376 | 6.17E-04 | ||
| miR-340-5p | 0.268 | 2.03E-02 | ||
| miR-214-3p | 0.225 | 5.67E-02 | ||
| Dynein light chain 1, cytoplasmic | DYNLL1 | miR-143-3p | 0.423 | 8.25E-05 |
| miR-204-5p | 0.242 | 3.86E-02 | ||
| miR-876-5p | 0.198 | 9.82E-02 | ||
| Fos proto-oncogene, AP-1 transcription factor subunit | FOS | miR-29b-3p | 0.273 | 1.74E-02 |
| miR-222-3p | −0.232 | 4.85E-02 | ||
| Hypoxanthine phospho-ribosyltransferase 1 | HPRT1 | miR-146b-5p | 0.445 | 3.05E-05 |
| miR-494-3p | 0.374 | 6.46E-04 | ||
| miR-432-5p | 0.243 | 3.73E-02 | ||
| Interleukin 6 receptor | IL6R | miR-34 c-5p | −0.427 | 7.02E-05 |
| miR-15a-5p | 0.396 | 2.75E-04 | ||
| miR-143-3p | −0.392 | 3.20E-04 | ||
| miR-495-3p | −0.381 | 5.09E-04 | ||
| miR-145-5p | 0.368 | 8.24E-04 | ||
| miR-146b-5p | −0.35 | 1.60E-03 | ||
| miR-140-5p | 0.312 | 5.50E-03 | ||
| miR-106b-5p | 0.286 | 1.21E-02 | ||
| miR-103a-3p | −0.281 | 1.39E-02 | ||
| miR-432-5p | −0.239 | 4.08E-02 | ||
| miR-150-5p | 0.221 | 6.15E-02 | ||
| miR-21-5p | 0.217 | 6.67E-02 | ||
| miR-214-3p | −0.211 | 7.62E-02 | ||
| Methionine sulphoxide reductase A | MSRA | miR-876-5p | −0.357 | 1.21E-03 |
| miR-495-3p | −0.234 | 4.57E-02 | ||
| Phosphatidylinositol-4,5-bisphosphate 3-Kinase catalytic subunit delta | PIK3CD | miR-222-3p | 0.475 | 6.08E-06 |
| miR-495-3p | −0.442 | 3.46E-05 | ||
| miR-92a-3p | 0.425 | 7.68E-05 | ||
| miR-34 c-5p | −0.416 | 1.17E-04 | ||
| miR-494-3p | −0.408 | 1.68E-04 | ||
| miR-539-5p | −0.377 | 5.87E-04 | ||
| miR-663a | −0.368 | 8.39E-04 | ||
| miR-342-3p | 0.322 | 4.10E-03 | ||
| miR-940 | −0.259 | 2.53E-02 | ||
| miR-214-3p | −0.235 | 4.52E-02 | ||
| RAP2C, member of RAS oncogene family | RAP2C | miR-301a-3p | −0.214 | 7.11E-02 |
| miR-340-5p | 0.213 | 7.34E-02 | ||
| miR-146b-5p | 0.206 | 8.36E-02 | ||
| Signal transducer and activator of transcription 1 | STAT1 | miR-214-3p | 0.223 | 5.91E-02 |
| Thioredoxin reductase 2 | TXNRD2 | miR-539-5p | −0.267 | 2.04E-02 |
Figure 2.
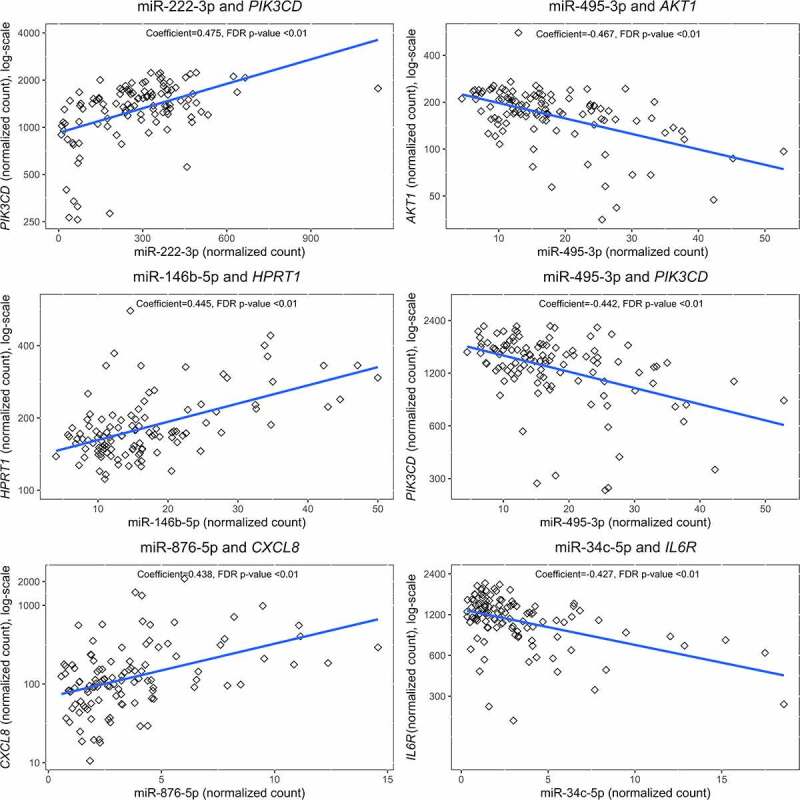
Scatterplots of top 6 correlated miRNA-mRNA pairs.
miRNA transcripts associated with NH Black race
Finally, we evaluated the direct association between miRNA expression and NH Black race and found that 10 miRNA that were identified as members of significant miRNA-mRNA pairs were also independently associated with NH Black race (q < 0.10). The miRNAs that differed most significantly by race included miR-539-5p, miR-15a-5p, and miR-21-5p, Table 4.
Table 4.
Shown are median expression levels (IQR) of microRNAs differentially expressed among non-Hispanic Black patients compared to other patients.
| miRNA |
non-Hispanic Black patients n = 51 |
Patients of other racial and/or ethnic groups n = 104 |
P-value |
|---|---|---|---|
| miR-539-5p | 5.71 (3.52, 7.89) |
8.43 (4.21, 12.66) |
0.0029 |
| miR-15a-5p | 369.1 (147.8, 590.5) |
230.1 (67.7, 392.5) |
0.0029 |
| miR-21-5p | 25.0 (17.4, 32.7) |
18.3 (11.2, 25.3) |
0.0079 |
| miR-204-5p | 6.04 (3.58, 8.51) |
8.88 (5.28, 12.48) |
0.0118 |
| miR-940 | 3.40 (2.08, 4.73) |
4.26 (2.23, 6.28) |
0.0374 |
| miR-146b-5p | 12.3 (9.5, 15.2) |
15.8 (10.6, 21.1) |
0.0495 |
| miR-876-5p | 2.17 (1.20, 3.14) |
3.24 (1.87, 4.60) |
0.0703 |
| miR-663a | 1.26 (0.10, 2.42) |
1.98 (0.87, 3.09) |
0.0888 |
| miR-200a-3p | 13.6 (9.3, 17.9) |
12.3 (8.5, 16.1) |
0.0929 |
| miR-340-5p | 4.36 (3.03, 5.68) |
5.91 (2.21, 9.61) |
0.0997 |
Discussion
We found that the nitric oxide pathway and related gene expression in inflammatory pathways in maternal blood obtained during pregnancy differs in NH Black patients compared to those of other races/ethnicities. Furthermore, we found evidence for miRNA regulation of this gene expression. Disparate pregnancy outcomes in NH Black individuals in the US occurs – at least in part – as a result of systemic and/or personal perceived racism and discrimination [16–19,21]. Though this study was not designed to evaluate the aetiologies underlying the biologic differences observed here, it is possible that the acute and chronic stress associated with structural, institutional, and systemic racism may result in epigenetic changes in genes such as those studied here. These epigenetic changes result in biologic effects that subsequently increase an individual’s risk for adverse obstetric outcomes.
Despite the prevalence of disparities in obstetrics, a paucity of other research studies have evaluated epigenetic or gene expression differences by self-reported race and/or ethnicity in pregnant patients. In one study that included 148 Latinx patients residing in the US and evaluated DNA methylation from maternal blood in two regions associated with inflammation [(FPXP3 Treg-cell-specific demethylated region and the promoter of tumour necrosis factor-alpha (TNF-α)], investigators found that those reporting high levels of perceived discrimination during pregnancy had differential regulation depending on whether or not they ultimately developed postnatal emotional distress or depression and anxiety symptoms[60]. The researchers concluded that epigenetic regulation of inflammatory regions of the genome may provide an important mechanism by which individuals ultimately exhibit resilience or sensitivity to discrimination-related prenatal stress. In another study, Carroll, et al. studied 103 pregnant individuals (33 classified by their group as ‘African-American’ and 70 classified only as ‘White’) and found upregulation of proinflammatory transcription factors NF-κB, activator protein 1 (AP1), and cyclic adenosine monophosphate response element-binding protein (CREB) and down-regulation of anti-inflammatory glucocorticoid responses and anti-viral response factors (IRF) among African-American individuals with sleep disturbances during pregnancy; these differences were less pronounced in White individuals[61]. In another study, Fortunato, et al. evaluated inflammatory related gene expression of amniochorionic membranes ex vivo following stimulation with endotoxins, and racially disparate immune responses in membranes derived from African-American and White patients[62]. Specifically, an increase in protective soluble tumour necrosis alpha (TNF-α) gene expression was observed in the membranes of White patients but not African-American patients. Further, the gene expression of matrix metalloproteinase 9 (MMP-9) was increased in the membranes of African-American patients in response to endotoxin, but there was no change in the membranes of White patients[62].
The findings of increased expression of some genes and decreased expression of others when evaluating changes in gene expression by race reflects the complexity of the conditions being studied. In addition, though increased miRNA expression is traditionally associated with decreased mRNA expression, we found both positive and negative miRNA-mRNA correlations. This is not unexpected and reflects the complex bidirectional regulation of miRNA targets as has been observed in previous studies [63–65]. Furthermore, algorithms identifying predicted miRNA-mRNA pairs are not cell-type specific and are unable to account for post-transcriptional modifications (e.g., histone acetylation and DNA methylation).
In this study, the IL-8 gene was among the top differentially expressed genes; expression was reduced among NH Black patients compared to other patients (log fold change −1.11, standard error 0.201, q = 2.7e−06). IL-8 is a pro-inflammatory cytokine; previous studies have found elevated IL-8 cytokine levels in those with spontaneous preterm deliveries, other studies have found that IL-8 cytokine levels are not associated with spontaneous preterm birth. Though elevated levels of IL-8 in NH Black women would be expected compared to those of other races, these results are consistent with prior studies evaluating IL-8 in pregnancy by race. One study found high-risk Black birthing people had lower levels of IL-8 in blood compared to high-risk White birthing people (median 12.1 vs. 18.9, p = 0.02)[66]. Another study found IL-8 amniotic fluid concentrations were similar between Black women who delivered preterm vs at term (742 pg/mL vs 731 pg/mL, p = 0.90) but higher in White women who delivered preterm vs. at term (1362 pg/mL vs. 533.5 pg/mL, p = 0.005)[67]. Finally, in a study evaluating the concentration of IL-8 in foetal membranes, similar IL-8 concentrations were found in unstimulated control membranes, but higher IL-8 concentrations were detected in membranes from Caucasian vs. African American women after stimulation with the proinflammatory infectious endotoxin. One possible mechanism for these findings of reduced IL-8 gene expression (this present study) and lower cytokine levels in various tissues (literature) in NH Black patients may be mediated through progesterone[68]. In in vitro human decidual cell models, addition of progesterone to an inflammatory cell model reduces IL-8 expression compared to models without progesterone[69]. Though a similar proportion of NH Black and NH White individuals received progestogen supplementation, factors including compliance, absorption, and drug levels from participants in this study are unknown, and progesterone use may have affected IL-8 levels through this mechanism.
The STAT1 gene also exhibited reduced expression among NH Black patients (log fold change −2.47, standard error 0.091, q = 0.064). In contrast to IL-8, STAT1 provides defence against pathogens by mediating response to pro-inflammatory cytokines[70]. In one study evaluating genetic pathways associated with spontaneous preterm birth and preterm pre-labour rupture of membranes, STAT1 was the predicted upstream transcription factor for multiple genes implicated in preterm pre-labour rupture of membranes, including cluster of differentiation 14 (CD14), FAS cell surface death receptor (FAS), and insulin-like growth factor 1 receptor precursor (IGF1R)[70]. STAT1 is also associated with the renin-angiotensin system and genes that regulate blood pressure[71].
It is important to note that other explanations – beyond the aforementioned hypotheses related to racism and discrimination – may be responsible for the observed differences in mRNA and miRNA gene expression by patient race in this study. For example, some medical conditions are more common among individuals of certain ancestry groups due to different carrier rates for minor alleles associated with autosomal recessive disorders (e.g., it is estimated that the carriage rate for a panel of germline mutations known to cause cystic fibrosis – is 1:25 among individuals of European ancestry whereas it is 1:65 among individuals of African ancestry)[72]. Prior studies have also demonstrated that individuals with darker skin pigmentation are more likely to experience vitamin D deficiency, due to a reduction in the absorption of ultraviolet radiation in the presence of increasing amounts of melanin pigment and subsequently, an increased dietary requirement[73]. Indeed, vitamin D has roles as an antioxidant and antimicrobial; in prior studies, it has been found to directly impact nitric oxide pathway function; vitamin D deficiencies have been proposed to partly explain disparities in other areas of medicine [74,75].
Study strengths and limitations
There are several strengths to the current study. Use of peripheral blood samples during pregnancy offers clear advantages as a non-invasive approach to evaluate differences in biology during pregnancy. Inflammation, preterm birth, and white blood cells are closely linked, lending biologic plausibility for using maternal blood as a matrix to evaluate gene expression during pregnancy [76–78]. These data add to the current paucity of transcriptomic data from NH Black patients during pregnancy. The study of paired mRNA and miRNA expression at the same time during pregnancy permitted assessment of the association between miRNA-mediated gene expression changes by patient race/ethnicity. Evaluation of self-identified NH Black patients may capture and reflect the effects of structural, institutional, and systemic racism (pervasive throughout society) and other potential biologic factors on samples collected during pregnancy [21]. Further, these findings support the previously observed clinical findings of different clinical preterm birth phenotypes, specifically, higher rates of inflammation and histologic chorioamnionitis among NH Black compared to NH White patients delivering preterm [79].
These results should be interpreted in the context of the study limitations. Our study was not designed to evaluate an individual’s perceived stress or personal experiences with racism or discrimination, nor did we have additional biologic variables such as vitamin D variables; all factors which may provide key insight into the mechanisms underlying the observed differences. Multiple miRNAs target individual genes and each miRNA typically targets more than one gene; thus, our focused custom mRNA and miRNA panels were not all-inclusive. In addition, it is possible that covariates or population characteristics other than the ones considered in this analysis contributed to our findings. The findings from this high-risk pregnancy cohort may not be applicable to the general obstetric population. Our sample size did not allow evaluation of gene expression among patients of other races and ethnicities, nor did it allow for direct comparison between NH Black patients and patients of only one other racial and ethnic group.
Conclusions
These findings provide evidence of an association between NO pathway and inflammation and infection-related mRNA and miRNA expression in blood drawn during pregnancy among NH Black patients. These findings may reflect key differences in the biology of inflammatory gene dysregulation that underlies differences in pregnancy outcomes, using self-reported patient race/ethnicity as a surrogate marker for the lived experiences of individuals of colour in the United States – which are all encompassing and may also include underlying differences in other biologic processes. Further study – e.g., through direct assessment of one’s lived experience of structural, institutional, and systemic racism – is required to determine whether these biologic changes in gene expression are directly related to acute and/or chronic stress or other factors unique to the lived experience of persons of colour in the United States, whether downstream protein expression is also affected, and how the observed findings may contribute to disparate pregnancy outcomes.
Funding Statement
This work was supported by the National Institutes of Health [R01-MD011609]; National Institutes of Health [K24-ES031131].
Disclosure statement
No potential conflict of interest was reported by the authors.
Financial disclosure
Funded by R01-MD011609 and K24-ES031131.
Ethical disclosure
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, written, informed consent has been obtained from the participants involved.
References
- [1].Manuck TA. Racial and ethnic differences in preterm birth: a complex, multifactorial problem. Semin Perinatol. 2017;41(8):511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Martin JA, Hamilton BE, Osterman MJK, et al. Births: final data for 2018. November 27, 2019. [PubMed]
- [3].Martin JA, Hamilton BE, Osterman MJK, et al. Births: final data for 2018. Natl Vital Stat Rep. 2019;68(13):1–47. [PubMed] [Google Scholar]
- [4].Manuck TA, Rice MM, Bailit JL, et al. Preterm neonatal morbidity and mortality by gestational age: a contemporary cohort. Am J Obstet Gynecol. 2016;215(1):103e101–103 e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Manuck TA, Sheng X, Yoder BA, et al. Correlation between initial neonatal and early childhood outcomes following preterm birth. Am J Obstet Gynecol. 2014;210(5):426 e421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Natarajan G, Shankaran S.. Short- and Long-Term outcomes of moderate and late preterm infants. Am J Perinatol. 2016;33(3):305–317. [DOI] [PubMed] [Google Scholar]
- [7].Bodeau-Livinec F, Marlow N, Ancel PY, et al. Impact of intensive care practices on short-term and long-term outcomes for extremely preterm infants: comparison between the British Isles and France. Pediatrics. 2008;122(5):e1014–1021. [DOI] [PubMed] [Google Scholar]
- [8].Vohr B. Long-term outcomes of moderately preterm, late preterm, and early term infants. Clin Perinatol. 2013;40(4):739–751. [DOI] [PubMed] [Google Scholar]
- [9].Singh GK, Siahpush M, Liu L, et al. Racial/Ethnic, nativity, and sociodemographic disparities in maternal hypertension in the United States, 2014-2015. Int J Hypertens. 2018;2018:7897189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Thoma ME, Drew LB, Hirai AH, et al. Black-White disparities in preterm birth: geographic, social, and health determinants. Am J Prev Med. 2019;57(5):675–686. [DOI] [PubMed] [Google Scholar]
- [11].Burris HH, Lorch SA, Kirpalani H, et al. Racial disparities in preterm birth in USA: a biosensor of physical and social environmental exposures. Arch Dis Child. 2019;104(10):931–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].DeFranco EA, Hall ES, Muglia LJ. Racial disparity in previable birth. Am J Obstet Gynecol. 2016;214(3):394 e391–397. [DOI] [PubMed] [Google Scholar]
- [13].Lo J, Mission JF, Caughey AB. Hypertensive disease of pregnancy and maternal mortality. Curr Opin Obstet Gynecol. 2013;25(2):124–132. [DOI] [PubMed] [Google Scholar]
- [14].Lawn J, Blencowe H, Waiswa P, et al. Lancet ending preventable stillbirths series study group; lancet stillbirth epidemiology investigator group. stillbirths: rates, risk factors, and acceleration towards 2030. Lancet. 2016;387(10018):587–603. [DOI] [PubMed] [Google Scholar]
- [15].Bujold E, Roberge S, Lacasse Y, et al. Prevention of preeclampsia and intrauterine growth restriction with aspirin started in early pregnancy: a meta-analysis. Obstet Gynecol. 2010;116(2.1):402–414. [DOI] [PubMed] [Google Scholar]
- [16].Dole N, Savitz DA, Hertz-Picciotto I, et al. Maternal stress and preterm birth. Am J Epidemiol. 2003;157(1):14–24. [DOI] [PubMed] [Google Scholar]
- [17].Dominguez TP, Dunkel-Schetter C, Glynn LM, et al. Racial differences in birth outcomes: the role of general, pregnancy, and racism stress. Health Psychol. 2008;27(2):194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dole N, Savitz DA, Siega-Riz AM, et al. Psychosocial factors and preterm birth among African American and white women in central North Carolina. Am J Public Health. 2004;94(8):1358–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chambers BD, Baer RJ, McLemore MR, et al. Using Index of concentration at the extremes as indicators of structural racism to evaluate the association with preterm birth and infant mortality-California, 2011-2012. J Urban Health. 2019;96(2):159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bailey ZD, Krieger N, Agenor M, et al. Structural racism and health inequities in the USA: evidence and interventions. Lancet. 2017;389(10077):1453–1463. [DOI] [PubMed] [Google Scholar]
- [21].Paradies Y, Ben J, Denson N, et al. Racism as a determinant of health: a systematic review and meta-Analysis. PLoS One. 2015;10(9):e0138511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sladek SM, Magness RR, Conrad KP. Nitric oxide and pregnancy. Am J Physiol. 1997;272(2 Pt 2):R441–463. [DOI] [PubMed] [Google Scholar]
- [23].Selles J, Polini N, Alvarez C, et al. Nongenomic action of progesterone in rat aorta: role of nitric oxide and prostaglandins. Cell Signal. 2002;14(5):431–436. [DOI] [PubMed] [Google Scholar]
- [24].Simoncini T, Mannella P, Fornari L, et al. Differential signal transduction of progesterone and medroxyprogesterone acetate in human endothelial cells. Endocrinology. 2004;145(12):5745–5756. [DOI] [PubMed] [Google Scholar]
- [25].Simoncini T, Caruso A, Garibaldi S, et al. Activation of nitric oxide synthesis in human endothelial cells using nomegestrol acetate. Obstet Gynecol. 2006;108(4):969–978. [DOI] [PubMed] [Google Scholar]
- [26].Pang Y, Dong J, Thomas P. Progesterone increases nitric oxide synthesis in human vascular endothelial cells through activation of membrane progesterone receptor-alpha. Am J Physiol Endocrinol Metab. 2015;308(10):E899–911. [DOI] [PubMed] [Google Scholar]
- [27].Forstermann U, Sessa WC. Nitric oxide synthases: regulation and function. Eur Heart J. 2012;33(7):829–837d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Manuck TA, Watkins WS, Moore B, et al. Pharmacogenomics of 17-alpha hydroxyprogesterone caproate for recurrent preterm birth prevention. Am J Obstet Gynecol. 2014;210(4):321e321–321 e321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Manuck TA, Watkins WS, Esplin MS, et al. Pharmacogenomics of 17-alpha hydroxyprogesterone caproate for recurrent preterm birth: a case-control study. BJOG. 2018;125(3):343–350. [DOI] [PubMed] [Google Scholar]
- [30].Manuck TA, Smeester L, Martin EM, et al. Epigenetic regulation of the nitric oxide pathway, 17-alpha hydroxyprogesterone caproate, and recurrent preterm birth. Am J Perinatol. 2018;35(8):721–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jiang F, Li J, Wu G, et al. Upregulation of microRNA335 and microRNA584 contributes to the pathogenesis of severe preeclampsia through downregulation of endothelial nitric oxide synthase. Mol Med Rep. 2015;12(4):5383–5390. [DOI] [PubMed] [Google Scholar]
- [32].O’Brien J, Hayder H, Zayed Y, et al. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). 2018;9:402. doi: 10.3389/fendo.2018.00402.eCollection 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA. 2012;3(3):311–330. [DOI] [PubMed] [Google Scholar]
- [34].Giurgescu C, Engeland CG, Templin TN, et al. Racial discrimination predicts greater systemic inflammation in pregnant African American women. Appl Nurs Res. 2016;32:98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Giurgescu C, Zenk SN, Engeland CG, et al. Racial discrimination and psychological wellbeing of pregnant women. MCN Am J Matern Child Nurs. 2017;42(1):8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Szanton SL, Rifkind JM, Mohanty JG, et al. Racial discrimination is associated with a measure of red blood cell oxidative stress: a potential pathway for racial health disparities. Int J Behav Med. 2012;19(4):489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Committee opinion no 611: method for estimating due date. Obstet Gynecol. 2014;124(4):863–866. [DOI] [PubMed] [Google Scholar]
- [38].Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Leone A, Roca MS, Ciardiello C, et al. Oxidative stress gene expression profile correlates with cancer patient poor prognosis: identification of crucial pathways might select novel therapeutic approaches. Oxid Med Cell Longev. 2017;2017:2597581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sheikh IA, Ahmad E, Jamal MS, et al. Spontaneous preterm birth and single nucleotide gene polymorphisms: a recent update. BMC Genomics. 2016;17(Suppl 9):759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shetty GA, Hattiangady B, Upadhya D, et al. Chronic oxidative stress, mitochondrial dysfunction, Nrf2 activation and inflammation in the hippocampus accompany heightened systemic inflammation and oxidative stress in an animal model of gulf war illness. Front Mol Neurosci. 2017;10:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Birben E, Sahiner UM, Sackesen C, et al. Oxidative stress and antioxidant defense. World Allergy Organ J. 2012;5(1):9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Musolino C, Allegra A, Innao V, et al. Inflammatory and Anti-Inflammatory equilibrium, proliferative and antiproliferative balance: the role of cytokines in multiple Myeloma. Mediators Inflamm. 2017;2017:1852517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Carey AJ, Tan CK, Ulett GC. Infection-induced IL-10 and JAK-STAT: a review of the molecular circuitry controlling immune hyperactivity in response to pathogenic microbes. JAKSTAT. 2012;1(3):159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Agarwal A, Aponte-Mellado A, Premkumar BJ, et al. The effects of oxidative stress on female reproduction: a review. Reprod Biol Endocrinol. 2012;10(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lu TP, Lee CY, Tsai MH, et al. miRSystem: an integrated system for characterizing enriched functions and pathways of microRNA targets. PLoS One. 2012;7(8):e42390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Rager JE, Auerbach SS, Chappell GA, et al. Benchmark dose modeling estimates of the concentrations of inorganic arsenic that induce changes to the neonatal transcriptome, proteome, and epigenome in a pregnancy cohort. Chem Res Toxicol. 2017;30(10):1911–1920. [DOI] [PubMed] [Google Scholar]
- [48].Rager JE, Bailey KA, Smeester L, et al. Prenatal arsenic exposure and the epigenome: altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environ Mol Mutagen. 2014;55(3):196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rager JE, Moeller BC, Miller SK, et al. Formaldehyde-associated changes in microRNAs: tissue and temporal specificity in the rat nose, white blood cells, and bone marrow. Toxicol Sci. 2014;138(1):36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Costa-Silva J, Domingues D, Lopes FM. RNA-Seq differential expression analysis: an extended review and a software tool. PLoS One. 2017;12(12):e0190152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Molania R, Gagnon-Bartsch JA, Dobrovic A, et al. A new normalization for Nanostring nCounter gene expression data. Nucleic Acids Res. 2019;47(12):6073–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gagnon-Bartsch JA, Speed TP. Using control genes to correct for unwanted variation in microarray data. Biostatistics. 2012;13(3):539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].De Magalhaes JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25(7):875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lian IA, Langaas M, Moses E, et al. Differential gene expression at the maternal-fetal interface in preeclampsia is influenced by gestational age. PLoS One. 2013;8(7):e69848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Weissenbacher T, Laubender RP, Witkin SS, et al. Influence of maternal age, gestational age and fetal gender on expression of immune mediators in amniotic fluid. BMC Res Notes. 2012;5(1):375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Menon R, Debnath C, Lai A, et al. Circulating exosomal miRNA profile during term and preterm birth pregnancies: a longitudinal study. Endocrinology. 2019;160(2):249–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].miRSystem - An integrated miRNA analytical system for characterizing enriched functions and pathways. 2020. cited 2020 April 27. http://mirsystem.cgm.ntu.edu.tw/index.php.
- [59].Levine M, Ensom MH. Post hoc power analysis: an idea whose time has passed? Pharmacotherapy. 2001;21(4):405–409. [DOI] [PubMed] [Google Scholar]
- [60].Sluiter F, Incollingo Rodriguez AC, Nephew BC, et al. Pregnancy associated epigenetic markers of inflammation predict depression and anxiety symptoms in response to discrimination. Neurobiol Stress. 2020;13:100273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Carroll JE, Rentscher KE, Cole SW, et al. Sleep disturbances and inflammatory gene expression among pregnant women: differential responses by race. Brain Behav Immun. 2020;88:654–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Fortunato SJ, Lombardi SJ, Menon R. Racial disparity in membrane response to infectious stimuli: a possible explanation for observed differences in the incidence of prematurity. Community award paper. Am J Obstet Gynecol. 2004;190(6):1557–1562. discussion 1562-1553. [DOI] [PubMed] [Google Scholar]
- [63].Diaz G, Zamboni F, Tice A, et al. Integrated ordination of miRNA and mRNA expression profiles. BMC Genomics. 2015;16(1):767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Cascione L, Gasparini P, Lovat F, et al. Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer. PLoS One. 2013;8(2):e55910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Nunez-Iglesias J, Liu CC, Morgan TE, et al. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PLoS One. 2010;5(2):e8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ekeke P, Mendez DD, Yanowitz TD, et al. Racial Differences in the Biochemical Effects of Stress in Pregnancy. Int J Environ Res Public Health. 2020;17(19):6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Menon R, Williams SM, Fortunato SJ. Amniotic fluid interleukin-1beta and interleukin-8 concentrations: racial disparity in preterm birth. Reprod Sci. 2007;14(3):253–259. [DOI] [PubMed] [Google Scholar]
- [68].Menon R, Dunlop AL, Kramer MR, et al. An overview of racial disparities in preterm birth rates: caused by infection or inflammatory response? Acta Obstet Gynecol Scand. 2011;90(12):1325–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Okabe H, Makino S, Kato K, et al. The effect of progesterone on genes involved in preterm labor. J Reprod Immunol. 2014;104-105:80–91. [DOI] [PubMed] [Google Scholar]
- [70].Capece A, Vasieva O, Meher S, et al. Pathway analysis of genetic factors associated with spontaneous preterm birth and pre-labor preterm rupture of membranes. PLoS One. 2014;9(9):e108578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Najjar I, Fagard R. STAT1 and pathogens, not a friendly relationship. Biochimie. 2010;92(5):425–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Strom CM, Crossley B, Buller-Buerkle A, et al. Cystic fibrosis testing 8 years on: lessons learned from carrier screening and sequencing analysis. Genet Med. 2011;13(2):166–172. [DOI] [PubMed] [Google Scholar]
- [73].Webb AR, Kazantzidis A, Kift RC, et al. Colour Counts: sunlight and skin type as drivers of vitamin D Deficiency at UK Latitudes. Nutrients. 2018;10(4):457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wolf ST, Jablonski NG, Ferguson SB, et al. Four weeks of vitamin D supplementation improves nitric oxide-mediated microvascular function in college-aged African Americans. Am J Physiol Heart Circ Physiol. 2020;319(4):H906–H914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Liu PT, Stenger S, Li H, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006;311(5768):1770–1773. [DOI] [PubMed] [Google Scholar]
- [76].Rafaeli-Yehudai T, Imterat M, Douvdevani A, et al. Maternal total cell-free DNA in preeclampsia and fetal growth restriction: evidence of differences in maternal response to abnormal implantation. PLoS One. 2018;13(7):e0200360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ribeiro VR, Romao-Veiga M, Romagnoli GG, et al. Association between cytokine profile and transcription factors produced by T-cell subsets in early- and late-onset pre-eclampsia. Immunology. 2017;152(1):163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Giorgi VS, Witkin SS, Bannwart-Castro CF, et al. Elevated circulating adenosine deaminase activity in women with preeclampsia: association with pro-inflammatory cytokine production and uric acid levels. Pregnancy Hypertens. 2016;6(4):400–405. [DOI] [PubMed] [Google Scholar]
- [79].Savitz DA, Blackmore CA, Thorp JM. Epidemiologic characteristics of preterm delivery: etiologic heterogeneity. Am J Obstet Gynecol. 1991;164(2):467–471. [DOI] [PubMed] [Google Scholar]


