Abstract
Focal cortical dysplasia is a highly epileptogenic cortical malformation with few treatment options.
Here, we generated human cortical organoids from patients with focal cortical dysplasia type II. Using this human model, we mimicked some focal cortical dysplasia hallmarks, such as impaired cell proliferation, the presence of dysmorphic neurons and balloon cells, and neuronal network hyperexcitability. Furthermore, we observed alterations in the adherens junctions zonula occludens-1 and partitioning defective 3, reduced polarization of the actin cytoskeleton, and fewer synaptic puncta. Focal cortical dysplasia cortical organoids showed downregulation of the small GTPase RHOA, a finding that was confirmed in brain tissue resected from these patients. Functionally, both spontaneous and optogenetically-evoked electrical activity revealed hyperexcitability and enhanced network connectivity in focal cortical dysplasia organoids.
Taken together, our findings suggest a ventricular zone instability in tissue cohesion of neuroepithelial cells, leading to a maturational arrest of progenitors or newborn neurons, which may predispose to cellular and functional immaturity and compromise the formation of neural networks in focal cortical dysplasia.
Keywords: focal cortical dysplasia, cortical organoids, cell adhesion, cell proliferation, neuronal network
Avansini et al. generate a human cortical organoid model for focal cortical dysplasia that recapitulates dysmorphic neurons, balloon cells, and neuronal network hyperexcitability. Alterations observed in adherens junctions suggest a ventricular zone instability that can predispose to cellular and functional immaturity.
Introduction
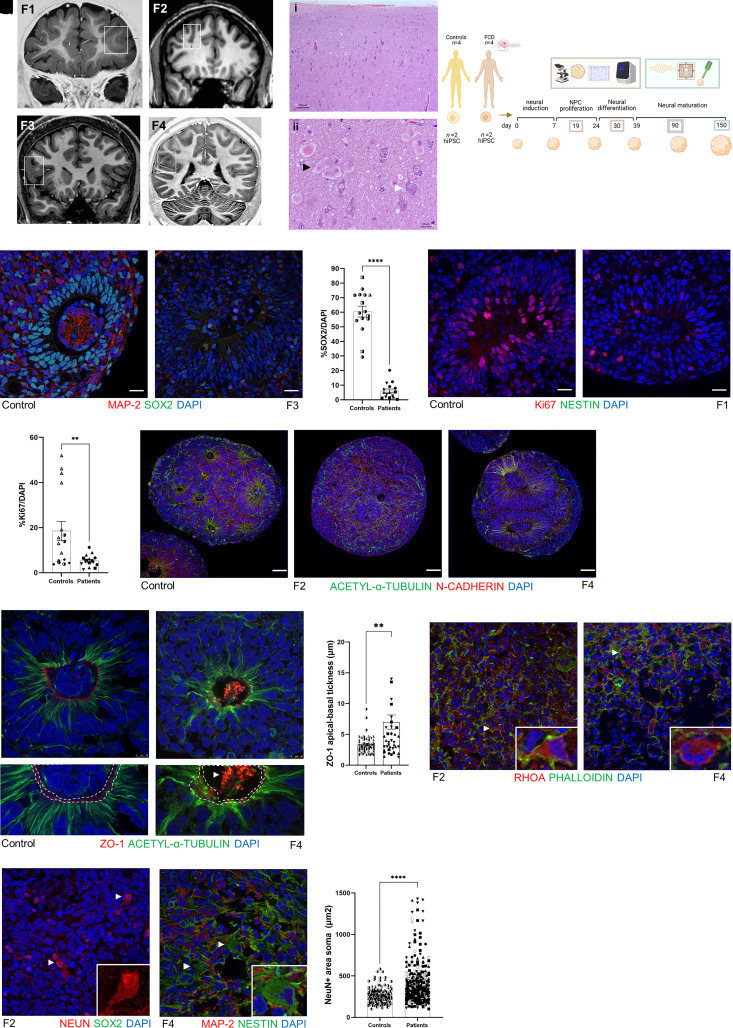
Focal cortical dysplasia (FCD) is one of the most frequent causes of pharmacoresistant focal epilepsy in children and adults.1 Treatment of these patients remains a challenge due to surgical inaccessibility in eloquent cortical areas and the absence of aetiology-specific antiepileptic drugs.2 FCD type II is characterized by focal lesions (Fig. 1A), cortical dyslamination [Fig. 1B(i)] associated with cellular abnormalities such as dysmorphic neurons [Fig. 1B(ii), white arrow], and balloon cells [Fig. 1B(ii), black arrow].3 These two abnormal cells seem to derive from radial glial progenitor cells at the ventricular zone.4
Figure 1.
Cellular characterization of FCD cortical organoids. (A) Coronal T1-weighted MRI from four patients with FCD type II. The square in each image indicates the abnormality suggestive of FCD. Patient F1, 5 years of age, the square shows increased cortical thickness and blurring of the cortical-subcortical transition in the anterior part of the left middle and inferior frontal gyrus and orbital gyri; Patient F2, 37 years of age, the square shows an abnormal deep sulcus in the transition of the right superior and middle frontal gyrus; Patient F3, 15 years of age, the square shows an increased cortical thickness of the right inferior frontal gyrus; and Patient F4, 19 years of age, the square shows increased cortical thickness and blurring of the cortical-subcortical transition in the parietal operculum and posterior insula. (B) Haematoxylin and eosin staining of a specimen from Patient F1 shown in: (i) with cortical dyslamination; and (ii) the presence of balloon cells (black arrow) and dysmorphic neurons (white arrow). (C) Schematic representation of the timeline of the cortical organoid generation and the experimental design using four patients with FCD and four healthy controls, with two clones of iPSCs per subject for posterior cortical organoid generation in four time points: 19, 30, 90 and 150 days. Cellular and molecular characterization was performed in 19-, 30- and 90-day-old organoids (represented by beige rectangle), and functional analyses were performed in 90- and 150-day-old organoids (represented by green rectangle). (D and E) Representative micrographs showing proliferating neural progenitor cells (NPC) at the neural rosette stained with SOX2+ (green) and mature MAP2+ neurons (red) from a 30-day-old cortical organoid from (D) WT83 control and (F) Patient F3. (F) Quantification of SOX2+ cells per DAPI+ cells in 30-day-old organoids (n = 4 organoids per experiment per each subject, P < 0.0001). (G and H) Representative micrographs Ki67+ cells (red) at neural rosette staining from 30-day-old cortical organoid from (G) WT83 control and (H) Patient F1. (I) Quantification of Ki67+ cells per DAPI+ cells in 30-day-old organoids (n = 4 organoids per experiment per each subject, P = 0.0044). (J–L) Nineteen-day-old cortical organoid stained with N-cadherin (red) and acetyl-α-tubulin (green), showing that stable microtubules were well organized from the apical to basal surface at neural rosettes in the 4C control (J). (K) In Patient F2 cortical organoids, we noticed fragmented microtubules surrounding the forme fruste neural rosette. (L) Organoids from Patient F4 presented enlarged rosettes with an increased lumen diameter. These two phenotypes were observed in the two different clones of the iPSCs from these two patients (Patients F2 and F4) and in consecutive batches of organoid generation. (M and N) Representative micrographs of ZO-1 (red) and acetyl-α-tubulin (green) immunostaining of 19-day-old cortical organoids, showing a continuous ring of ZO-1 in WT83 control (M; white dashed line; bottom figure represents an enlarged view of M), and a disrupted belt (N; white dashed line; bottom figure represents an enlarged view of N) with a concentration of ZO-1 protein in the middle of the lumen (white arrow) in the F4-derived organoids. (O) Quantification of ZO-1 apical-basal thickness, showing an irregular distribution of ZO-1 reflected in increased thickness in FCD organoids (n = 12 controls organoids, n = 12 patients organoids). (P and Q) Representative micrographs of RHOA (red) and phalloidin (green) immunostaining of 90-day-old cortical organoids. (P) In F2 organoids, the inset shows a neuron compatible with a dysmorphic neuron. (Q) Organoid derived from Patient F4, the inset indicates a round cell with four nuclei, features that indicates a balloon cell. (R) Representative micrographs of NeuN (red) immunostaining of a 90-day-old Patient F2-derived cortical organoid. The arrows and inset indicate neurons with enlarged NeuN+ soma staining that is more intense in the nucleus than the cytoplasm, features compatible with dysmorphic neurons. (S) Representative micrograph of MAP2 (red), and nestin (green) immunostaining of 90-day-old F4 derived cortical organoid. The arrows and inset indicate a nestin+ cell with abnormal soma morphology and a laterally displaced nucleus, features characteristic of a balloon cell. (T) Area of the soma in NeuN+ evidencing that neurons in FCD organoids were twice as large as controls (n = 145 controls, n = 316 neurons patients; mean: 264.2 µm2 controls and mean: 454.6 µm2 patients. Nuclei were stained with DAPI (blue). The results are presented as the mean ± SEM. A one-sample t-test was used to assess statistical significance, *P < 0.05, **P < 0.001, ****P < 0.0001. Scale bar = 100 μm [B(i) and J–L], 10 μm (M and N) or 20 μm (all others). Controls: WT83 clone 1◑, clone 2◐; 4C clone 1◨, clone 2 ◧; 969 clone 1◮, clone 2 ◭, 121 clone 1 ◆ clone 2 ◇; patients: F1 clone 1●, clone 2 ○; F2 clone 1■, clone 2 □, F3 clone 1▲, clone 2 △, F4 clone 1▼, clone 2▽.
Brain postzygotic mammalian target of rapamycin (mTOR) variants have been associated with FCD, but such genetic mosaicism cannot explain all cases.5–7 Moreover, the lack of a reliable human FCD model that recapitulates the emergence of the condition during human neurodevelopment is one of the major limitations to developing effective treatments. To better understand the mechanism leading to FCD, we generated patient-derived cortical organoids from induced pluripotent stem cells (iPSCs) of four patients with pathologically confirmed FCD type II.
Materials and methods
Human subjects
The four patients with FCD type II included in this study were characterized at the time of recruitment at the outpatient epilepsy clinic of the University of Campinas (UNICAMP) hospital. All patients had been diagnosed with pharmacoresistant epilepsy after extensive clinical assessment, routine EEG, prolonged video-EEG monitoring, and structural and functional neuroimaging. Because of the lack of response to clinical treatment, the patients underwent surgical resection of the epileptogenic zone. According to the Engel classification, three of these patients did not present a good postoperative seizure outcome, Engel class III, due to proximity of the epileptogenic zone to eloquent cortical areas. The adult patients signed a written informed consent approved by the Research Ethics Committee UNICAMP; the patients younger than age 18 years gave their assent, and their parents signed the consent on their behalf. All clinical information obtained from the patients’ records for this research was accessed only after informed consent from the patients and/or their legal guardians. Neuropathological diagnoses of FCD type II were confirmed according to the International League Against Epilepsy (ILAE) classification.3 Clinical, pathological and mTOR mutation findings from these patients are summarized in Table 1.
Table 1.
Clinical, neuropathological and variants in the mTOR pathway findings in four FCD type II patients and four controls
| ID | Symbols | Age | Sex | Typea | Site of surgery | Seizure frequencyb | Seizure onset, years | Number of surgeriesc | Engeld | Variants in the mTOR brain tissue/leucocytes | Variants in the mTOR cortical organoids | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mosaic braine | Germlinef | Mosaic | Germline | ||||||||||
| F1 | ● ○ | 5 | Female | IIb | FrE; TE | 60 | 1 | 2 | III | MTOR; p.Ala1459Pro; 3.98% FrL; 0.403% TL; VUS; heterozygous | Not found | Not found | Not found |
| F2 | ▪■□ | 37 | Female | IIb or TSC | FrR; FrR; FrTR | 150 | 10 | 3 | III | Not found | TSC1; p.Trp676Arg; VUS; heterozygous TSC2; p.Arg367Gln; VUS; homozygous | Not found | TSC1; p.Trp676Arg; VUS; heterozygous. TSC2; p.Arg367Gln; VUS; homozygous |
| F3 | ▲ △ | 15 | Female | IIa | FrR; FrR | 90 | 9 | 2 | III | Not found | Not found | Not found | Not found |
| F4 | ▼ ▽ | 19 | Female | IIb | PR | 90 | 5 | 1 | I | Not found | Not found | Not found | Not found |
| 4C1 | ◨ ◧ | 41 | Female | Control | – | – | – | – | – | – | – | Not found | Not found |
| WT83 | ◑ ◐ | 32 | Male | Control | – | – | – | – | – | – | – | Not found | Not found |
| 121 | ◆ ◇ | 14 | Female | Control | – | – | – | – | – | – | – | NP | NP |
| 969 | ◮ ◭ | 2 | Female | Control | – | – | – | – | – | – | – | NP | NP |
Fr = frontal; NP = the analysis was not performed; P = parietal; R = right; T = temporal; TSC = tuberous sclerosis complex; VUS = variant of uncertain significance. (–) = not applicable.
Subtype of FCD according to the ILAE.
Monthly seizure frequency before first surgery.
Number of surgical treatments to control seizures.
Classification scale for surgical outcome.
Percentage of mosaic variants found in brain tissue using mTOR pathway panel.
Germline variant in mTOR pathway using whole exome sequencing from genomic DNA.
For the molecular characterization, we also used fresh-frozen brain tissues from additional patients with FCD type II controls individuals. The clinical characteristics of these subjects used in the molecular characterization are summarized in Table 2.
Table 2.
Clinical findings of fresh-frozen brain tissue from additional FCD patients and controls used in molecular characterization
| ID | Sex | Age | Site of surgery | Subject; cause of death: surgery |
|---|---|---|---|---|
| P1 | Male | 40 | TPO/R | Patient; FCD type IIb |
| P2 | Male | 26 | T/L | Patient; FCD type IIa |
| P3 | Male | 6 | TO/R | Patient; FCD type IIb |
| P13 | Male | 29 | Fr/R | Patient; FCD type IIa |
| P16 | Male | 18 | Fr/L | Patient; FCD type IIb |
| P20 | Male | 32 | Fr/L | Patient; FCD type IIb |
| P21 | Female | 21 | Fr/L | Patient; FCD type IIb |
| C2 | Female | 33 | Fr/R | Control; autopsy: respiratory failure, diffuse lymphoma |
| C20 | Male | 55 | Fr/L | Control; autopsy: septic shock |
| C21 | Female | 37 | Fr/R | Control; autopsy: septic shock/bronchopneumonia |
| C24 | Male | 62 | Fr/R | Control; autopsy: hepatic impairment |
| C23 | Male | 48 | Fr/R | Control; autopsy: pulmonary embolism |
| C25 | Male | 47 | Fr/R | Control; autopsy: septic shock |
| C26 | Male | 50 | T | Control; biopsy: anterior temporal lobectomy |
| C27 | Male | 38 | T | Control; biopsy: anterior temporal lobectomy |
| C28 | Female | 44 | T | Control; biopsy: anterior temporal lobectomy |
| C29 | Male | 40 | T | Control; biopsy: anterior temporal lobectomy |
| C30 | Female | 49 | T | Control; biopsy: anterior temporal lobectomy |
| C31 | Female | 45 | T | Control; biopsy: anterior temporal lobectomy |
| C32 | Male | 51 | T | Control; biopsy: anterior temporal lobectomy |
| C33 | Female | 28 | T | Control; biopsy: anterior temporal lobectomy |
| C34 | Female | 54 | T | Control; biopsy: anterior temporal lobectomy |
Control samples were obtained from autopsies of individuals whose cause of death was non-neurological, and from the biopsy of normal temporal cortex obtained during anterior temporal lobectomy performed in patients with mesial temporal sclerosis. All control samples used in this work were evaluated and considered normal cytoarchitecture of the cerebral cortex by histopathological examination. Fr = frontal; L = left; O = occipital; P = parietal; R = right; T = temporal.
Detection and validation of germline and somatic variants of the mTOR pathway
Genomic DNA was extracted from surgically resected brain tissue and blood leucocytes by using a phenol-chloroform protocol. First, in order to detect mTOR germline and mosaic variants, we used deep whole-exome sequencing. Capture and enrichment were performed by using the Agilent SureSelect V6-Post, following the manufacturer’s instructions. Samples were sequenced with HiSeq 2500 and NovaSeq Illumina sequencer, using a 150 bp paired-end run to achieve 1000× coverage (∼90 Gb/sample). Alignment, BAM processing, and variant detection were performed by using the Genome Analysis Toolkit Best Practices Workflows (https://gatk.broadinstitute.org/hc/en-us/sections/360007226651-Best-Practices-Workflows, version 4.1.4.1) and the human reference genome based on GRCh37. Germline and mosaic variants were filtered, prioritizing: (i) stop codon, frameshift, missense, nonsense and splicing site mutations; (ii) variants whose minor allele frequency is <0.01; (iii) variants absent in gnomAD (Exome Aggregation Consortium) and in a publicly available genomic database of Brazilian samples (BipMed, Brazilian Initiative on Precision Medicine, http://bipmed.iqm.unicamp.br/genes); and (iv) variants were classified as mosaic mutations when <10% of reads were not aligned to the human genome reference. The deleterious effect on the protein functions of the variants was evaluated by using SIFT, PolyPhen-2 and the Franklin database.
To validate our results, the same brain tissue samples and cortical organoids (patients and controls) were submitted to deep sequencing using a customized enrichment panel comprising genes from the mTOR and GATOR pathways. Capturing and enrichment were performed with the Agilent SureSelectXT, following the manufacturer’s instructions. The list of the 68 genes included in the panel is available in Supplementary Table 1. Samples were sequenced in a Miseq (Illumina) to achieve 600–2000× of coverage. Mosaic variants were called according to the Genome Analysis Toolkit Best Practices Workflows and were evaluated using the following somatic variant caller tools: Mutect2, VarScan and Strelka. Mosaic variants were filtered considering: (i) only variants that were found in at least two somatic variant caller algorithms; (ii) when <10% of reads were not aligned to the reference human genome; (iii) present only in brain tissue resected from patients with FCD; (iv) absent in GenomAD and BipMed; and (v) variants whose minor allele frequency is <0.01. Variants were also filtered by prioritizing stop codon, frameshift, missense, nonsense, and splicing site mutations.
Validation of mTOR mosaic variants was performed by using droplet digital PCR (QX200TM Droplet Digital™ PCR System; Bio-Rad). Briefly, the droplet digital PCR reaction consisted of droplet digital PCR Supermix for Probes (no dUTP; Bio-Rad), mutant and reference probes (0.25 µmol/l each; acquired from Bio-Rad and IDT), forward and reverse primers (0.9 µmol/l each), and 60 ng of the DNA sample, which was previously digested by restriction enzyme (10 U/µg DNA). The reaction was emulsified into at least 10 000 droplets by using a QX200 Droplet Generator (Bio-Rad). PCR amplification was performed with the following parameters: 10 min at 95°C, 40 cycles of 30 s at 94°C and 70 s at 58°C, and 10 min at 98°C, using a ramp rate of 2°C/s between the steps. The analysis was performed using the QuantaSoft software, version 1.7.4.0917.
Reprogramming of skin fibroblasts into iPSCs
The skin biopsies from FCD patients were performed under sterile conditions by a dermatologist at the UNICAMP hospital. Fibroblasts derived from mechanical dissection of skin were cultured in Dulbecco’s modified Eagle medium (DMEM/F12) containing 10% foetal bovine serum and penicillin/streptomycin. The cellular reprogramming of fibroblasts into iPSCs was performed with a Cytotune iPS 2.0 Sendai reprogramming kit (Thermo Fisher Scientific), following manufacturer instructions (Supplementary Fig. 1F and G). Briefly, fibroblast cultures were transduced with transcription factors OCT4, SOX2, KLF4 and MYC. Seven days after transduction, cells were replated onto a feeder layer composed of murine embryonic fibroblasts in DMEM/F12 containing 20% Knockout Serum Replacement (Thermo Fisher Scientific), 1% non-essential amino acids (NEAA; Thermo Fisher Scientific) and 100 μM 2-mercaptoethanol. IPSC colonies were identified after 2 weeks and transferred to 6-cm plates coated with Matrigel (BD Biosciences). The iPSCs were cultured in StemFlex medium (Thermo Fisher Scientific) and passaged manually. Ten iPSC clones were selected from each subject from this study and three of these were chosen for further validation of stem cell phenotypes using a combination of immunostaining, expression analysis and single nucleotide polymorphism mapping to rule out the presence of unwanted chromosomal abnormalities and mutations. Four iPSC controls were included in this study, which are part of a bank of iPSCs from the Muotri lab.
Validation of iPSCs was performed by immunostaining for SOX2, OCT4 and LIN28 (Supplementary Fig. 1A–C). Briefly, iPSCs were grown inside the wells of LabTek II 8-well chambered slides (Thermo Fisher Scientific). Colonies were then fixed with 4% paraformaldehyde (PFA) solution for 10 min, washed once with 1× PBS, permeabilized with 1% Triton X-100 for 5 min, washed in 1× PBS containing 10% bovine serum albumin (BSA) and 1% Triton X-100. Incubation of primary antibodies was performed in the same blocking solution overnight at 4°C. The primary antibodies used were anti-SOX2 (Abcam; ab97959; 1:1000), anti-OCT4 (Abcam; ab19857; 1:300) and anti-LIN28 (Cell Signaling; 3978; 1:1000). After three washes in 1× PBS, colonies were incubated with fluorescently labelled secondary antibodies for 3 h, and nuclei were counterstained with 1 μg/ml 4′6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific) for 30 min. Slides were mounted with ProLong Gold anti-fading mountant (Thermo Fisher Scientific).
Quantitative reverse transcription PCR (RT-qPCR) using TaqMan assays (Thermo Fisher Scientific) NANOG (Hs02387400_g1), LIN28 (Hs00702808_s1), SOX2 (Hs04234836_s1), MYC (Hs00153408_m1) and KLF4 (Hs00358836_m1) were used to quantify the expression of stem cell markers (Supplementary Fig. 1D). Single nucleotide polymorphism mapping-based karyotyping using genome-wide profiling for amplifications, deletions, copy number variations and rearrangements was performed on genomic DNA extracted from the iPSC clones using the iScan system (Illumina) and the Infinium HumanCytoSNP-12 BeadChip (Illumina; Supplementary Fig. 1E). After confirmation of the pluripotent state of these cells, we chose two different iPSC clones from each subject for cortical organoid generation. All cellular cultures were routinely tested for mycoplasma.
Cortical organoid generation
To generate cortical organoids, we used our previously published protocol.8 Briefly, iPSC colonies were dissociated using Accutase (Thermo Fisher Scientific; diluted with an equal volume of 1× PBS) for 12 min at 37°C. After centrifugation for 3 min at 150g, the individualized cells were resuspended in StemFlex Medium (Thermo Fisher Scientific) supplemented with 10 µM SB431542 (Stemgent), 1 µM dorsomorphin (R&D Systems) and 5 µM Rho kinase inhibitor (Y-27632; Calbiochem, Sigma-Aldrich). Approximately 4 × 10 iPSCs were seeded into each well of a low-binding six-well plate and placed on a shaker inside a CO2 incubator at 95 rpm and 37°C. Formed spheres were fed at 24 and 48 h with StemFlex supplemented with 10 µM SB431542 and 1 µM dorsomorphin. After 3 days, the medium was replaced to neural induction medium consisting of Neurobasal Medium (Thermo Fisher Scientific) containing GlutaMAX (Thermo Fisher Scientific), 1% Gem21 NeuroPlex supplement (Gemini Bio-Products), 1% N2 NeuroPlex supplement (Gemini Bio-Products), 1% of NEAA (Thermo Fisher Scientific), 1% penicillin/streptomycin (Thermo Fisher Scientific), 10 µM SB431542 and 1 µM dorsomorphin, for 7 days. Next, the medium was replaced to proliferation medium, consisting of Neurobasal medium containing GlutaMAX, 1% Gem21, 1% NEAA and 20 ng/ml basic fibroblast growth factor (FGF-2; Thermo Fisher Scientific) for 10 days, followed by an additional 7 days in the same medium further supplemented with 20 ng/ml epidermal growth factor (PeproTech). Neuronal differentiation and organoid maturation were achieved by switching to Neurobasal medium containing 1% GlutaMAX, 1% Gem21, 1% NEAA, 10 ng/ml of brain-derived neurotrophic factor, 10 ng/ml of glial cell-derived neurotrophic factor, 10 ng/ml of neurotrophin-3 (all from PeproTech), 200 mM l-ascorbic acid, and 1 mM dibutyryl-cAMP (Sigma-Aldrich) for 15 days. After this period, cortical organoids were maintained in Neurobasal medium containing GlutaMAX, 1% Gem21, and 1% NEAA for as long as needed, with media changes every 3–4 days and kept in culture until 150 days (Supplementary Fig. 1H). For every subject and their respective clones, we conducted two independent batches at four time points: 19, 30, 90, and 150 days of culture.
Immunofluorescence staining
Cortical organoids were fixed with 4% PFA overnight at 4°C and cryoprotected in 30% sucrose for at least 2 days. They were then embedded in TissueTek (Leica Microsystems) and sectioned on a Leica VT1000S cryostat to produce 20 µm sections. For staining, slides were air dried for 10 min, permeabilized in 1× PBS containing 1% Triton X-100 for 2 min and blocked with 1× PBS containing 0.1% Triton X-100 and 3% BSA for 1 h at 25°C, followed by incubation with primary antibodies in the same solutions. Primary antibodies used were: rat anti-CTIP2 (Abcam; ab18465; 1:500); rabbit anti-SATB2 (Abcam; ab34735; 1:200); chicken anti-MAP2 (Abcam; ab5392; 1:1000); rabbit anti-SOX2 (Abcam; ab97959; 1:1000); rabbit anti-CUX1 (CUTL1 or CASP, Abcam; ab54583; 1:200); rabbit anti-vesicular glutamate transporter 1 (VGLUT1; Synaptic Systems; 135311; 1:500); rabbit anti-cleaved caspase 3 (CC3 Cell Signaling; 9664S; 1:500); mouse anti-RHOA (Abcam; ab54835;1:100); mouse anti-postsynaptic density protein 95 (PSD-95; NeuroMab, UC Davis, 1:1000); rabbit anti-synapsin I (Sigma-Aldrich; AB1543P, 1:1000); mouse anti-zona occludens 1 (ZO-1;Thermo Fisher Scientific; ZO1-1A12; 1:500); rabbit anti-partitioning defective 3 (PARD3; Thermo Fisher Scientific; 11085-1-AP; 1:300); mouse anti-N-Cadherin (BD; Clone 32; 1:150); rabbit anti-NeuN (Abcam; ab128886; 1:1000); mouse anti-NESTIN (Abcam; ab22035; 1:200); and rabbit anti-Ki67 (Abcam; ab15580; 1:100). The primary antibodies were incubated overnight at 4°C. After, slides were washed three times in 1× PBS, for 5 min each, and incubated with fluorescent secondary antibodies (Thermo Fisher Scientific; conjugated to Alexa Fluor 488 or 555; 1:500 dilution) and Alexa Fluor 488 Phalloidin (Thermo Fisher Scientific; A12379; 1:40) for 3 h at 25°C. After further washes in 1× PBS, slides were counterstained with DAPI solution (1 µg/ml) for 5 min and mounted with ProLong Gold. All images were taken using a Zeiss fluorescence microscope equipped with Apotome (Axio Observer Apotome, Zeiss). For cell quantification in cortical organoids, we used the Analyze Particles plugin on the ImageJ software (National Institutes of Health, Bethesda, MD, USA) and three random 70 × 70 µm regions of interest for each selected organoid slice analysed. For quantification of the ZO-1 apical-basal thickness, four measurements (at 90°, 180°, 270° and 360°) for each ventricular structure were performed in cortical organoids, according to Iefremova et al.9 For quantification of neuronal soma area, we also used ImageJ software after staining with NeuN antibody. Five to ten neurons were quantified per organoid slice (500 μm × 500 μm). For quantification of percentage of dysmorphic versus normal neurons, neurons with area of soma >500 µm2 were considered dysmorphic. This value was standardized based on the average of area of soma from neurons analysed in controls organoids, which was approximately 300 µm2.
Three-dimensional imaging of cleared organoids for synaptic puncta quantification
Three-dimensional imaging of solvent cleared cortical organoids was performed using an adapted method of 3DISCO/FDISCO10,11 with the aim to quantify synaptic puncta. This process was divided into four phases: staining, clearing, imaging, and quantification.
Staining
Organoids were fixed in 4% PFA and stored at 4°C in 1× PBS containing 0.02% NaN3. After washing in 1× PBS, organoids were permeabilized in PTx.2 solution (1% 1× PBS, 0.2% Triton X-100, ddH2O) with 2.3% glycine and 2% dimethyl sulphoxide (DMSO) at room temperature overnight, incubated in PTx.2 with 3% serum and 2% DMSO to block non-specific protein binding, and incubated for 72–96 h with primary antibody at 25°C under agitation. Cortical organoids were labelled with chicken anti-MAP2 (Abcam; ab53921; 1:1000), mouse anti-VGLUT1 (Synaptic systems; 135311; 1:250), and rabbit anti-Homer1, a postsynaptic density scaffold protein (Synaptic systems; 160003; 1:500). They were then washed five times for 15 min each in PTwH solution (1% 1× PBS, 0.2% Tween-20, 1 mg/100 ml heparin, ddH2O). Next, they incubated in fluorescent secondary antibodies (1:1000; goat anti-chicken conjugated to Alexa Fluor 555; goat anti-mouse conjugated to Alexa Fluor 488, and goat anti-rabbit conjugated to Alexa Fluor 647; Thermo Fisher Scientific) diluted in PTwH with 3% serum for 72–96 h at 25°C on a shaker plate.
Clearing
Organoids were dehydrated in a methanol/H2O series as follows: 20%, 50%, 70%, 80%, 100% and100%, 15 min each. They were then transferred to sealed glass vials: the first containing 33% dichloromethane (Sigma; 34856) and 66% methanol, and the second containing 100% dichloromethane. Finally, they were cleared in 100% dibenzyl ether (ACROS Organic; 148400010) directly in the chamber.
Imaging
Because of the small size of the organoids, each sample was directly imaged after clearing using an inverted SP5 confocal microscope with a 63× water objectives lens. For imaging 35–65 µm z-stack images were captured with an interval of 0.3 µm.
Synaptic puncta quantification
Co-localized synaptic puncta were quantified by using Imaris (Bitplane), the microscopy image analysis software (Oxford Instruments). In the Imaris analysis, pre- and postsynaptic markers in the 3D images were identified as ‘spots’ with dimensions established considering the x/y and z axes. It was set to detect identically sized objects presenting a proximity based on the minimal distance of their centers of mass. A semi-automatic threshold was determined to avoid ‘false spots’; the same parameters were used for all images. The Imaris co-localization function was used to identify the co-localized spots, and the number of co-localized spots was normalized by the total number of spots found in the whole 3D image.
RT-qPCR
Total RNA from cortical organoids and human fresh-frozen brain tissue was extracted by using TRIzol (Thermo Fisher Scientific) following the manufacturer’s recommendations. Subsequently, 1.8 µg of total RNA was reverse transcribed into cDNA using the SuperScript® IV Reverse Transcriptase with DNase treatment included in the reaction (Thermo Fisher Scientific). RT-qPCR was performed using TaqMan probes (Thermo Fisher Scientific) and the TaqMan universal master mix II without UNG (Thermo Fisher Scientific) on a CFX Connect Real Time PCR detection system (Bio-Rad), with the following cycling parameters: 94°C for 3 min, followed by 40 cycles of 94°C for 30 s and 68°C for 1 min. To quantify mRNA expression, we used TaqMan gene expression assays. The following TaqMan probes were used: endogenous controls, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Hs02758991_g1) and β-actin (ACTB) (Hs01060665_g1); target mRNAs, RHOA (Hs00357608_m1), PARD3 (Hs00969077_m1), ZO-1 (TJP1, Hs01551871_m1), RAC1 (Hs01902432_s1), RND2 (Hs00183269_m1), RND3 (Hs01003594_m1), and CDC42 (Hs00918044_g1). PCR reactions were performed in triplicate. Relative expression was calculated using the 2−ΔΔCt method.
Western blotting analysis
Cell lysates from cortical organoids and human fresh-frozen brain tissue were collected by lysing cells with radioimmunoprecipitation assay buffer [50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulphate (SDS)] and mixed with protease inhibitor cocktail (Sigma-Aldrich) and phosphatase inhibitor cocktail (Roche). After removing the cell debris by centrifuging at 15 000 rpm for 30 min at 4°C, the protein quantification was determined with a bicinchoninic acid protein assay (Pierce). Total protein extracts (15–20 μg) were separated on Bolt 4–12% Bis-Tris Plus Gels (Life Technologies) and transferred onto a nitrocellulose membrane using iBlot2 dry blotting system (Thermo Fisher). After blocking non-specific protein binding (Rockland Immunochemicals, VWR International), membranes were incubated with primary antibodies overnight at 4°C. The primary antibodies used were mouse anti-RHOA (Abcam; ab54835; 1:100), mouse anti-PSD-95 (NeuroMab, 1:1000), rabbit anti-synapsin I (Sigma-Aldrich; AB1543P, 1:1000), and rabbit anti-MMP2 (Abcam, ab97779 1:500). Then, the membrane was incubated with secondary antibodies for 3 h at room temperature. Proteins were detected by using the Odyssey CLx infrared imaging system (LI-COR Biosciences).
Quantification of F-actin versus G-actin
Quantification of the F-actin versus G-actin content in the cortical organoids was realized using a G-Actin/F-Actin In Vivo Assay Biochem Kit (Cytoskeleton Inc., BK037) according to the manufacturer’s instructions. Briefly, we homogenized cortical organoids in F-actin stabilization buffer, followed by ultracentrifugation at 100 000g to separate F-actin from G-actin. The fractions are then separated by SDS-PAGE and actin was quantitated by western blotting. The actin antibody was provided by the company. β-Tubulin III was used as a housekeeping protein.
Gene expression using the NanoString Human Neuropathology Panel
The expression of 770 neuropathology-related genes in 90-day-old cortical organoids from four patients with FCD and four controls was analysed by using the nCounter Human Neuropathology Panel (NanoString Technologies) according to the manufacturer’s protocol. The list of genes included in the panel is available in Supplementary Table 1. Total RNA from cortical organoids was extracted by using the RNeasy Micro Kit (Qiagen) following the manufacturer’s recommendations. One hundred nanograms of RNA per sample were prepared for analysis. The assay was performed on the nCounter MAX Analysis System (Sanford Consortium for Regenerative Medicine Stem Cell Genomics Core) according to the manufacturer’s instructions. The nSolver software (v4.0) by NanoString was used to normalize gene expression data. ROSALIND software (OnRamp Bioinformatics, https://rosalind.onramp.bio/) was then used to interpret targeted gene expression data and to create heat maps. The data were then analysed by ROSALIND® (https://rosalind.onramp.bio/), with a HyperScale architecture developed by OnRamp BioInformatics, Inc. (San Diego, CA, USA) to interpret targeted gene expression data and to create heat maps. The read distribution percentages, violin plots, identity heat maps, and sample MDS plots were generated as part of the QC step. The Limma R library12 was used to calculate fold changes and P-values and to perform optional covariate correction. Clustering of genes for the final heat map of differentially expressed genes was done using the Partitioning around medoids method. This process employed the fpc R library that takes into consideration the direction and type of all signals on a pathway, the position, role, and type of every gene, etc. A hypergeometric distribution was used to analyse the enrichment of pathways, gene ontology, domain structure, and other ontologies. The topGO R library,13 was used to determine local similarities and dependencies between GO terms in order to perform Elim pruning correction. Several database sources were referenced for enrichment analysis, including Interpro,14 NCBI,15 MSigDB,16,17 REACTOME,18 and WikiPathways.19 Enrichment was calculated relative to a set of background genes relevant for the experiment. Functional enrichment analysis of pathways, gene ontology, domain structure and other ontologies was performed by using HOMER.20
Electrophysiology in cortical organoids
We measured the electrical activity in cortical organoids by using extracellular electrophysiology. We recorded spontaneous neural activity in organoids plated on multi electrode arrays, and we recorded light-evoked response in organoids expressing the channelrhodopsin (CheRiff) by means of high-density silicon microelectrodes.
Extracellular recording using multi-electrode array analysis
Multi-electrode array (MEA) electrophysiological recordings were performed once per week by using the Maestro MEA system. Briefly, 45-day-old cortical organoids were plated on 12-well MEA plates (Axion Biosystems). Each well contains 64 low-impedance (0.04 MΩ) platinum microelectrodes of 30 µm in diameter, with a 200 µm space between the electrodes. The plates were previously coated with 100 µg/ml poly-l-ornithine and 10 µg/ml laminin. The seeded organoids were fed once a week with BrainPhys medium (STEMCELL Technologies). The recordings were performed with the AxIS Software Spontaneous Neural Configuration (Axion Biosystems) with a band-pass filter of 10 Hz and 2.5 kHz cut-off frequencies. Spike detection was computed with an adaptive threshold of 5.5 times the standard deviation of the estimated noise for each electrode. Three minutes of data were recorded. Two independent experiments were conducted for each subject, with three independent replicates per subject in each experiment. The data were analysed by using the Axion Biosystems’ Neural Metrics Tool. Wells with fewer than five active electrodes were eliminated from the analysis. Bursts were identified using an inter-spike interval threshold of five spikes with a maximum interspike interval of 100 ms. Network bursts were detected when a minimum of 15 spikes were found under the same interspike interval with a minimum of 25% active electrodes. The synchrony index was calculated using a cross-correlogram synchrony window of 20 ms. Custom software was used for inference of effectivity connectivity from MEA recordings. Briefly, spike file recordings were converted to .mat files using Axion Biosystem provided functions and analysed offline with custom MATLAB functions and scripts developed by Puppo et al.21 We used spike sorting based on principal component analysis and a k-means clustering to group spikes with similar shape into different clusters, each corresponding to a different unit (neuron). The number of cluster k was inferred via the gap statistic.22 This approach allowed us to isolate the activity of a few units per electrode, which resulted in the reconstruction of the activity of multiple detected neurons in the MEA well. Then, we used custom MATLAB routines (described by Puppo et al.21 and available online at https://github.com/fpuppo/ECRtools.git) for the following steps: (i) to detect functional interactions (statistical dependencies) between neurons via computations of pairwise correlations; (ii) to detect correlation peaks associated with indirect and apparent connections; and (iii) to reconstruct the directed causal connectivity between recorded neurons in the well. Connectivity maps were generated from the reconstructed networks to evaluate visually differences in connectivity between control and FCD organoids plated on MEAs.
Gene delivery
The optogenetic probe CheRiff was delivered to organoids via lentiviral transduction. Lentivirus was produced by VectorBuilder (Cyagen) from the CamKIIa-CheRiff plasmid.23 DRH313:FCK-CheRiff-eGFP was a gift from Adam Cohen (Addgene plasmid #51693; http://n2t.net/addgene:51693; RRID: Addgene_51693). Transduction of CheRiff was carried out following the procedures already described24 with some modifications. Briefly, 2 days before the viral transfection, we eliminated penicillin/streptomycin from the cortical organoid maturation medium. The organoids were then transferred to an ultra-low-attachment 96-well plate. Thirty-five microlitres of 1.4 × 107 transducing units (TU/ml) of DRH313-FCK-CheRiff-eGFP lentivirus in 1× PBS were added to each well. After 4 h of incubation at 37°C and 5%CO2, the organoids were transferred to a well containing 3 ml of organoid maturation medium in a six-well plate and incubated at 37°C and 5% CO2 under agitation for at least 10 days. The medium was changed every 4 days.
Extracellular recording using high-density silicon microelectrodes
Extracellular neuronal activity of mature cortical organoids (90 and 150 days old) was recorded by using a high-density penetrating silicon probe with 128 channels distributed onto four shanks each including a 2-column, 16-row configuration of 177 µm2 iridium recording sites of 46 µm pitch (Neuronexus; a 4 × 32-poly2-5mm-23s-200-177; Supplementary Fig. 5F). Signals were processed and digitized on a headstage proximal to the silicon probe (Intan; RHD2164), then acquired by an OpenEphys Acquisition Board.25 Recordings of CheRiff-expressing cortical organoids were performed in glass bottom MatTek dishes containing artificial CSF (ACSF; Ecocyte Bioscience) bubbled with carbogen (95% O2, 5% CO2) at room temperature. The dish was loaded onto an inverted epi-fluorescence microscope with a custom illumination scheme. During electrophysiological recordings, the organoids were maintained in the dark. The illumination was provided by a diode laser at 473-nm (100 mW, Cobolt 06-MLD) for CheRiff actuation. The laser beam was directed towards a lens (AC508-400-A-ML, f = 400 mm), focusing the light onto the rear focal plane of a high numerical aperture objective lens [Olympus UPlanFL N 20×, numerical aperture = 0.5 (air)]. The lens was translated in the plane perpendicular to the optical axis to illuminate a wider area of the organoid surface (∼300 µm beam diameter). The light hit the surface of the organoid in contact with the bottom of the dish. The extracellular probe was inserted into the organoid from the top by means of a micromanipulator (MP285; Sutter). In this configuration, we probed the internal activity of the organoid resulting from the electrical response elicited in CheRiff-expressing cells at the organoid surface and propagating towards the more internal layers where the probe was positioned (Supplementary Fig. 5G).
Multi-unit activity recordings were made for 60 s to measure the response to optogenetic stimulation, in at least three different recording sites per organoid. Optogenetic actuation was provided by delivering sequences of 50-ms long laser pulses at different frequency (0.01 Hz and 2 Hz; Supplementary Fig. 5H). The intensity of the 473-nm beam at the sample was 10 mW/cm2. Light stimulation was controlled by in-house MATLAB scripts through a National Instruments Data Acquisition board used to deliver TTL pulses to the laser control box.
Raw data were stored for analysis. Then, post-processing, multi-unit spike detection and all descriptive analysis were carried out via custom MATLAB routines. Spike detection was performed following a previously published protocol.8 Each channel was first referenced to the well mean for every time point, like a common average reference (128 channels). Next, the re-referenced signal was bandpass filtered for 300–3000 Hz with a third-order Butterworth filter. The adaptative spike threshold was set to be 5.5 times the standard deviations, where the standard deviation was estimated from the mean as previously described by Quiroga et al.26 Spike timestamps were taken as the peak time after the absolute value of the signal crossed the threshold, but at least 1 ms from another spike. The response rate was calculated as the total number of successful recordings (recordings with at least two responding channels) out of the total number of recordings performed. Spike events in response to optogenetic stimulation were searched in the post-processed signal by considering the 50-ms window following the optogenetic stimulus.
Statistical analyses
The data are presented as the mean ± standard error of the mean (SEM). No statistical method was used for a power analysis to predetermine the sample size and adjustments were made for multiple comparisons (Bonferroni test). The statistical analyses were performed by using Prism software (GraphPad). Student’s t-test or ANOVA with post hoc tests were used as indicated. Significance was defined as *P < 0.05, *P < 0.01 or *P < 0.001.
Data availability
Data from whole exome sequencing and mTOR/GATOR panel that support the findings of this study have been deposited at NCBI BioProject ID PRJNA730529. For gene expression using NanoString Human Neuropathology Panel, GEO accession number is GSE174605.
Results
All patients had been diagnosed with pharmacoresistant epilepsy, secondary to FCD type II, and underwent epilepsy surgery. The clinical and pathological characterization of the selected patients and their respective genetic mTOR analyses are summarized in Table 1. Four iPSCs, derived from healthy individuals, were included as controls. We generated forebrain cortical organoids using an optimized functional protocol8 and subsequently characterized the cortical organoids at three levels: morphological, molecular and functional (Fig. 1C).
The most notorious histopathological hallmarks of FCD are disruption of cell proliferation, characterized by localized cortical lesions associated with the presence of disorganized lamination and misshapen cells, such as dysmorphic neurons and balloon cells.3 These guided us to first investigate cell proliferation in FCD cortical organoids in the early stages of development, specifically in the neural rosette, the structure in the organoid that most closely resembles the embryonic neural tube.
We used immunofluorescence to quantify the proliferation markers NESTIN, SOX2 and Ki67, and to investigate eventual cellular alterations. The neural rosettes displayed radially organized columnar neuroepithelium cells surrounding an apical lumen whose continuous radial glial basal processes were stained by NESTIN (Supplementary Fig. 2F1–J1). We observed that FCD organoids harbored a substantially decreased number of SOX2+ progenitor cells compared with controls (Fig. 1D–F and Supplementary Figs 2A1–E4 and 6I), and those few cells localized mainly to the edges of the organoids (Supplementary Fig. 2A1–E1). Compared with controls organoids, Ki67+ proliferative cells in FCD organoids were also reduced (Fig. 1G–I and Supplementary Figs 2F1–J5 and 6J). We observed no differences in cell apoptosis between FCD and control organoids, as measured by cleaved caspase-3 (Supplementary Fig. 3A and B).
We next examined the essential components involved in neural rosette formation, namely adherens junctions and cytoskeletal elements (actin and tubulin). Some adherens junctions, such as N-cadherin, β-catenin, PARD3, CDC42, and the tight junction, ZO-1, are located preferentially at the ventricular zone of neuroepithelium cells. They function to anchor the radial glia to the ventricular surface and maintain apical radial glia polarity while regulating the division of neural progenitors.27,28
Using acetylated α-tubulin antibody, which labels stable microtubules (Fig 1J and Supplementary Fig. 3C1–4), we identified two distinct phenotypes in 19-day-old organoids. We detected forme fruste rosettes, characterized by the absence of an organized column of neuroepithelium cells (Fig. 1K and Supplementary Fig. 3D1–4), in 20% of all FCD organoids generated (Supplementary Fig. 3F), and we also observed enlargement of rosettes with an increased lumen diameter (Fig. 1L and Supplementary Fig 3E). Abnormal F-actin filaments (stained by phalloidin) appeared sparsely as thick dense actin cables at the apical side of the rosette (Supplementary Fig. 3H1–4). Upon examining adherens junctions in control organoids, we noticed a well-delimited and continuous belt of ZO-1 at the apical surface of the rosette (Fig. 1M and Supplementary Fig. 3J1–4). By contrast, the FCD organoids portrayed a fragmented ring with an accumulation of ZO-1 as bright droplets in the middle of the lumen (Fig. 1N and Supplementary Fig. 3K1–L4). Furthermore, the ZO-1 apical-basal thickness in FCD organoids was two times higher than the controls (Figs 10 and 6K). We also noticed that the integrity of the neuroepithelial barrier between tight junctions and cytoskeletal tubulin seemed disrupted in FCD organoids (Supplementary Fig. 3K5–L5).
Based on these initial findings, we further extended our morphological evaluation to later-stage organoids. Considering that the key regulator of adherens junctions and actin is the small Rho GTPase family,29 we decided to investigate the role of the small GTPase Rho A (RHOA). We observed some abnormalities in cell morphology in FCD compared with control organoids. We noticed that FCD organoids exhibited morphologies absent in control organoids, including cells with enlarged cell bodies (Fig. 1P), and sparse emergence of round multi-nucleated cells (Fig. 1Q). To confirm these cell abnormalities as evidence of FCD hallmarks, we used markers of mature neurons and noticed that NeuN+ cells had somas two times enlarged compared with cells found in controls (Fig. 1R and T and Supplementary Figs 4B1–C3 and 6R). The abnormal soma size was found in 25% of all neurons analysed in FCD organoids (Supplementary Fig. 6S). Large MAP2+ pyramidal-shaped cells were also observed (Supplementary Videos 1 and 2). Together, these cellular abnormalities are morphologically compatible with those observed in dysmorphic neurons. Using nestin and MAP2, we discerned several cells with aberrant soma morphology, including lateral displacement of the nucleus and phenotypic features suggestive of a mixed progenitor cell and neural lineage, histologically similar to balloon cells (Fig. 1S and Supplementary Fig. 4E1–F4). None of these abnormal cells were present in the control organoids (Supplementary Figs 4A1–3 and D1–3). Both FCD and control organoids expressed the cortical laminar markers CUX1, SATB2 and TBR1, recapitulating some characteristics of human cortical development (Supplementary Fig. 4G1–J3).
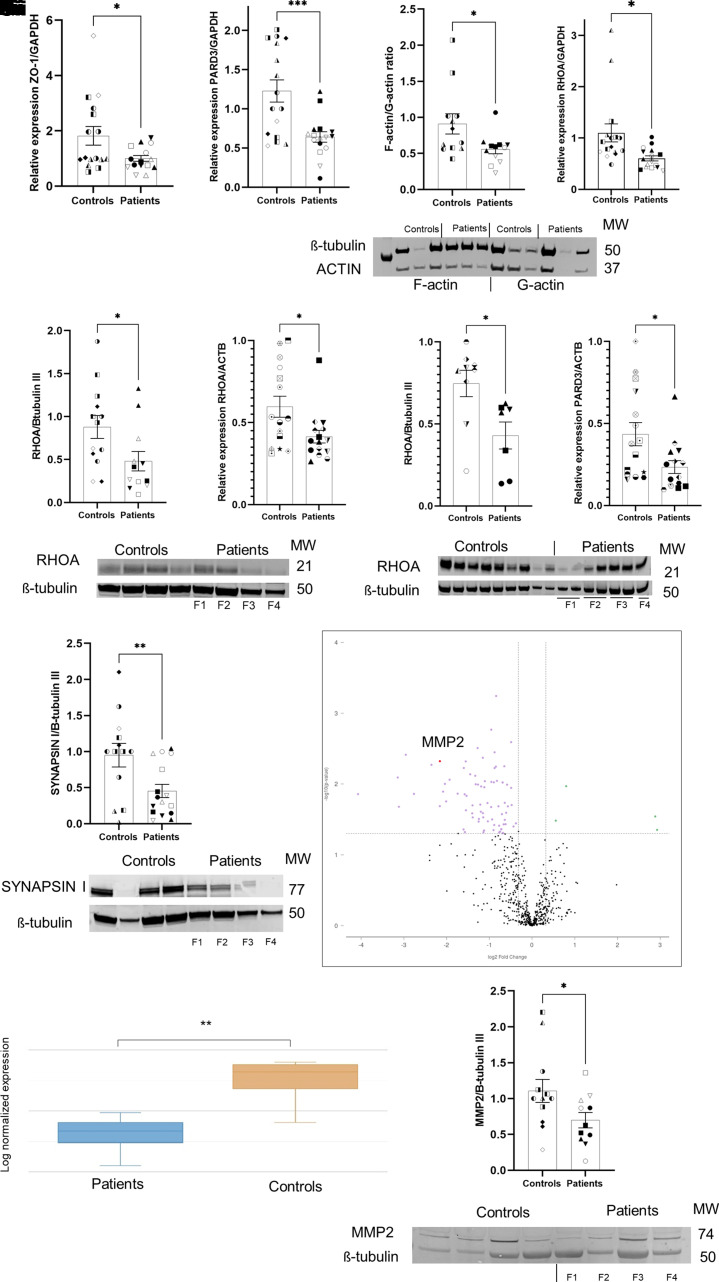
Using RT-qPCR, we observed the downregulation of ZO-1 (encoded by TJP1) in FCD compared with controls organoids (Fig. 2A and Supplementary Fig. 6A). Similarly, we noticed the downregulation of another adherens junction gene, PARD3, in FCD organoids (Fig. 2B and Supplementary Figs 4K and 6B). Moreover, the balance between actin polymerization and depolymerization (F-actin/G-actin ratio) is essential during corticogenesis,30 and its instability has been associated with the disassembly of the radial glial scaffold.31 Using ultrafractionated actin samples, FCD organoids were found to have a 38% decreased F-actin/G-actin ratio compared with controls organoids (Fig. 2C and Supplementary Fig. 6C).
Figure 2.
Molecular characterization of FCD cortical organoids. (A) Relative expression (RT-qPCR) of ZO-1 in 19-day-old-organoid from four independent batches of organoids per subject (n = 16 control and n = 16 patients). Each subject comprised two different clones of iPSCs, with three technical replicates per sample, expression levels were normalized to GAPDH;
P = 0.0387. (B) RT-qPCR of PARD3 in 90-day-old-organoids from four independent batches of organoids per subject (n = 16 controls and n = 16 patients). Each subject comprised two different clones of iPSCs, with three technical replicates per sample, expression levels were normalized to GAPDH;
P = 0.0008. (C) Quantification and western blot of ultrafractionated actin samples (F-actin and G- actin) from 90-day-old organoids; four independent batches of organoids per subject (n = 12 controls and n = 12 patients). Each subject comprised two different clones of iPSCs. The expression levels were normalized to β-tubulin III; P = 0.0312. (D) RT-qPCR of RHOA in 90-day-old-organoids from four independent batches of organoids per subject (n = 16 controls and n = 16 patients). Each subject was composed of two different clones of iPSCs, with three technical replicates per sample, expression levels were normalized to GAPDH;
P = 0.0110. (E) Quantification and western blot of RHOA protein from 90 day-old-organoids; four independent batches of organoids per subject (n = 11 controls and n = 10 patients; without F1 organoids). Each subject comprised two different clones of iPSCs. The expression levels were normalized to β-tubulin III; P = 0.0241. (F) RT-qPCR of RHOA from fresh-frozen brain tissue of patients with FCD type II (n = 15 controls and n = 15; Table 2), with three technical replicates per sample. The expression levels were normalized to ACTB; P = 0.0214. (G) Quantification and western blot of RHOA protein from fresh-frozen brain tissue of patients with FCD type II (n = 7) and controls (n = 9). The expression levels were normalized to β-tubulin III; P = 0.0167. (H) RT-qPCR of PARD3 from fresh-frozen brain tissue of patients with FCD type II (n = 15) and controls (n = 15), with three technical replicates per sample. The expression levels were normalized to ACTB; P = 0.0199. (I) Quantification and western blot of SYNAPSIN I protein from 90-day-old-organoids; four independent batches of organoids per subject (n = 13 controls and n = 16 patients). Each subject comprised two different clones of iPSCs. The expression levels were normalized to β-tubulin III; P = 0.0098. (J–
L) Gene expression analysis based on Nanostring neuropathology panel from 90-day-old organoids, FCD versus control, (n = 4 controls and n = 4 patients; Patients F1, F2, F3 and F4). (J) Volcano plot evidencing the differentially expressed genes (purple dots) found when FCD organoids and controls organoids were compared. (K) Box plot evidencing the differential expression of MMP2 when comparing patients and controls; P = 0.00476 and a fold change of −4.45. J and K were obtained from the ROSALIND™ platform. (L) Quantification and western blotting of MMP2 from 90-day-old organoids; four independent batches of organoids per subject (n = 12 control and n = 11 patients). Each subject comprised two different clones of iPSCs. The expression levels were normalized to β-tubulin III; P = 0.0489. The results are presented as the mean ± SEM. A one-sample t-test was used to assess statistical significance; *P < 0.05, **P < 0.001, ****P < 0.0001. Controls: WT83 clone 1◑, clone 2◐; 4C clone 1◨, clone 2 ◧; 969 clone 1◮, clone 2 ◭, 121 clone 1 ◆ clone 2 ◇; patients: F1 clone 1●, clone 2 ○; F2 clone 1■, clone 2 □, F3 clone 1▲, clone 2 △, F4 clone 1▼, clone 2▽.
We also noticed that FCD organoids exhibited decreased RHOA transcript (Fig. 2D and Supplementary Fig. 6D), and protein (Fig. 2E and Supplementary Fig. 6E) expression compared with controls organoids. This observation was also orthogonally validated in fresh-frozen brain tissue resected from these same patients (Fig. 2F, G and Table 2). Other members of the small Rho GTPase family were evaluated, but no statistical significance was found (Supplementary Fig. 6T). We similarly observed PARD3 downregulation in FCD brain tissue compared to controls (Fig. 2H). Collectively, these results suggest that FCD organoids have cell proliferation defects that are likely caused by abnormal regulation of RHOA, indicating that at certain points on the ventricular surface, compromised actin-anchored adhesion might affect the integrity of the neuroepithelium.
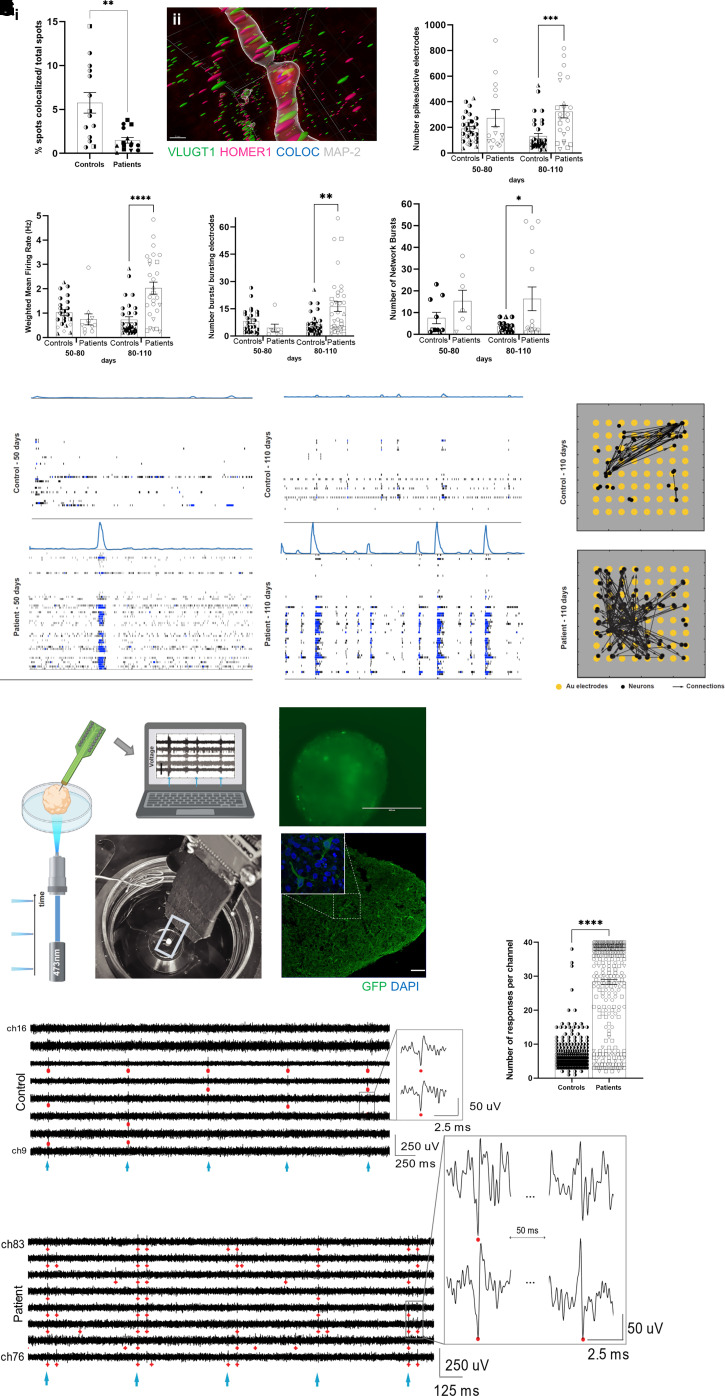
Because of the known role of F-actin and cell adhesion in the early stages of synaptogenesis assembly,32 we hypothesized that reduced F-actin in FCD organoids could affect synapse formation. Using immunoblotting, we found a 52% decrease in the presynaptic protein synapsin 1 in FCD compared with controls organoids (Fig. 2I and Supplementary Fig. 6F). Differences in PSD-95 were not detected (Supplementary Fig. 5A). We next evaluated the complexity of synaptic connections by using high-resolution 3D imaging of cleared cortical organoids with the whole mount immunofluorescence for presynaptic (VGLUT1) and postsynaptic (HOMER1) proteins. Both 90- and 150-day-old FCD organoids showed a reduction of co-localized synaptic puncta (Fig. 3A and Supplementary Figs 5B, C and 6L).
Figure 3.
Functional characterization of FCD cortical organoids. [A(i)] Reduction of co-localized synaptic puncta, using VGLUT1 (presynaptic antibody) and HOMER1 (postsynaptic), in 90- and 150-day-old organoids (n = 13 controls and n = 15 patients); 70 × 70 µm regions of interest per organoid; P = 0.0014. [A(ii)] Representative image of spot co-localization (blue sphere) using VGLUT1 (green sphere), HOMER1 (magenta sphere), and MAP2+ neurite (grey) from a 90-day-old control organoid. The image was obtained by using 3D imaging of cleared organoids for whole-mount immunofluorescence staining and Imaris software. (B–
E) Extracellular recordings using MEAs of 3-min intervals of spontaneous activity at different developmental stages (50–80 and 80–110 days) in control and FCD organoids. MEA analyses of FCD organoids with 80–110-day-old organoids revealed an increase in: (B) the number of spikes (P = 0.0002); (C) the mean firing rate (P < 0.0001); (D) the number of bursts (P = 0.0017); and (E) the number of spontaneous network bursts (P = 0.024). Each dot represents the result of a weekly measurement of a single well of an MEA plate (two to three organoids plated per well). Two independent experiments were conducted for each subject, with three technical replicates per subject in each experiment. (F) Raster plots of network spiking activity relative to 50-day-old (left) and 110-day-old (right) in WT83 control and F1 cortical organoids. (G) Effective connectivity inferred from MEA recordings of 110-old-day control (top) and F1 (bottom) cortical organoids reflect higher connectivity in the FCD organoids. Each visual map consists of a 1.2 × 1.2 mm MEA plate (grey area), an 8-by-8 array of micro-electrodes (yellow circles), and the estimated causal and direct connections (black arrows) between spike sorted neurons. The active neurons are represented by black dots; they are randomly distributed around their corresponding sensing electrode within a radius of 50 µm. The MEA results are reported as the mean ± SEM; statistical significance is based on two-way ANOVA and Bonferroni’s multiple comparisons test. (H–
K) Extracellular recordings using silicon high-density penetrating probes and stimulation via optogenetic actuation of CheRiff-expressing organoids. (H) Schematics and photograph of our setup for optogenetic stimulation with a 473-nm laser light and 3D extracellular recordings in organoids using penetrating high-density silicon microelectrodes. (I) Representative image of an FCD organoid transduced with DRH313-FCK-CheRiff-eGFP (top) and immunostaining (bottom) of 150-day-old organoid with GFP (green; indicating expression of the EGFP-CheRiff; scale bar = 100 μm) and DAPI (blue). (J) Representative recordings of optogenetic-evoked activity in control (top) and FCD (bottom) 150-day-old-organoids showing response in most of the considered channels. The cyan markers indicate pulses of 473-nm light (frequency of stimulation of 2 Hz). The red markers indicate detected spikes in the recorded activity (threshold of spike detection 5.5 × standard deviation). The insets show an enlarged view of neuronal spiking in response to optogenetic stimulation. Predominantly in FCD organoids, we observed double-spike event behaviour: the first spike followed by a second event of different polarity at a latency of ∼40–50 ms. FCD organoids presented (K) more optogenetic-elicited responses per channel than the controls (n = 365 controls and n = 758 patients) channels; all responding channels from all successful experiments were considered to be as independent variables. Extracellular recordings using silicon high-density probes results are presented as the mean ± SEM. Statistical significance based on one-sample t-test; *P < 0.05, **P < 0.001 and ****P < 0.0001. Controls: WT83 clone 1◑, clone 2◐; 4C clone 1◨, clone 2 ◧; 969 clone 1◮, clone 2 ◭, 121 clone 1 ◆ clone 2 ◇; patients: F1 clone 1●, clone 2 ○; F2 clone 1■, clone 2 □, F3 clone 1▲, clone 2 △, F4 clone 1▼, clone 2▽.
Next, to gain further insights into FCD mechanism, we performed targeted RNA-sequencing. Differential expression analysis comparing FCD and control organoids revealed 82 dysregulated genes (Fig. 2J), some of which were enriched for matrix remodelling, chromatin modification and neuronal cytoskeleton (Supplementary Table 2). Among those with higher fold-changes, we identified MMP2, which encodes a matrix metalloproteinase with several cellular functions and is also known to mediate synaptic remodelling.33 This gene was decreased 4.4-fold compared with control organoids (Fig. 2K and Supplementary Fig. 6G), and this downregulation was further confirmed in the protein assay (Fig. 2L and Supplementary Fig. 6H). Taken together, these data indicate that FCD organoids have compromised and late synapse formation.
Finally, we interrogated whether FCD organoids showed any alteration at the functional level. We performed extracellular recordings of spontaneous electrical activity in organoids plated on top of MEAs (Supplementary Fig. 5E), as optimized previously.8 We noticed that over time, the cortical organoids exhibited an increase in electrical activity demonstrating the development of the neuronal network. FCD cortical organoids 80–110 days old displayed enhanced neuronal excitability, evidenced by increases in the number of spikes, the mean firing rate, and the number of bursts (Fig. 3B, D and Supplementary Fig. 6M–O, respectively), when compared with control organoids. None of these alterations were present in 50-80-day-old organoids. FCD organoids also displayed an increased number of neuronal network bursts over time (Fig. 3E and Supplementary Fig. 6P), with evidence of more structured network activity already at the earlier development stages (Fig. 3F). Although we did not detect differences in synchrony between FCD and control organoids (Supplementary Fig. 5D), inferred effective (direct and causal) connectivity from MEA recordings demonstrated increased network connectivity in FCD (Fig. 3G), corroborating the hyperactive and coordinated electrical behaviour of neuronal networks found in FCD organoids.
The abnormal neuronal excitability and enhanced network connectivity prompted us to investigate network functionality by means of 3D extracellular recordings and optogenetic stimulation. Measurements using high-density silicon microelectrodes (Fig. 3H and Supplementary Fig. 5F–H) in 150-day-old organoids expressing CheRiff (Fig. 3I) demonstrated a successful response in both control and FCD organoids upon stimulation with 473 nm laser pulses (Fig. 3J). Compared with control organoids, we found that FCD organoids presented a higher global response rate to the stimulus and approximately a four-times more optogenetic-elicited responses per channel (Fig. 3K and Supplementary Fig. 6Q). We further observed that these optogenetic stimulations triggered a first spike followed by a second event of different polarity at a latency of ∼40–50 ms (Fig. 3J and Supplementary Fig. 5I and J). Collectively, these observations support the presence of increased neuronal hyperexcitability, detected both spontaneously and via optogenetic-evoked activity, associated with heightened connectivity in FCD organoids.
Discussion
Using four patient-derived cortical organoids, none harboring mosaic mTOR variants (Table 1 and Supplementary Fig. 1O–R), we recapitulated several aspects of FCD type II, including impaired cell proliferation, the presence of dysmorphic neurons, and balloon cells, and neuronal network hyperexcitability. We suggest that disruption of RHOA expression likely contributes to the perturbation of cell-cell contacts (ZO-1 and PARD3) as well as, impaired polarization of the actin cytoskeleton and it might cause disruption in the ventricular zone. Furthermore, this subtle instability in the cohesion of the neuroepithelium apical anchoring could lead to delayed detachment of radial glia cells, affecting cell fate commitment and promoting maturational arrest of both progenitor cells and newborn neurons.
Indeed, knockout29 and knockdown31 of RhoA, respectively, during mouse brain development has been shown to generate massive dysplasia of neural progenitor cells and loss of the apical scaffold of radial glia with disruption of adherens junctions at the ventricular surface but with different effects in cell proliferation. Specifically, knockdown of ZO-1 has been observed to inhibit the apical polarization of β-catenin, Pard3, and tubulin,34 as well as to induce multi-lumen formation.35 Likewise, downregulation of Pard3 during mouse brain development was associated with abnormal symmetric division and overproduction of progenitors that were ectopically localized in the cortical plate.36 These findings indicate that dysregulation in adherens junctions’ coupling could potentially lead to the formation of dysmorphic neurons, detected in all patient-derived FCD organoids studied here, and balloon cells, seen in the organoids derived from Patient F4 (Supplementary Table 3).
The combined effects of ZO-1 and depolarized F-actin generating a punctual destabilization of the ventricular junctional barrier could also mimic a transmantle sign,37 as observed in the thick actin cables in cortical organoids and confirmed using magnetic resonance imaging obtained from the same patient (Supplementary Fig. 3I). The cytoskeletal disassembly also results in impaired neuronal polarity38 and dendritic spine formation,3 as observed in disoriented, heterotopic neurons and the intrinsic hypertrophy displayed in dysmorphic neurons,39 respectively. Dysmorphic neurons were also identified by other investigators using Tuberous Sclerosis Complex derived organoids,40,41 with four times larger neuronal soma size than found in control neurons.41
Interestingly, in contrast to the reduced synaptic puncta, we observed an increase in excitability in the FCD organoids. These seemingly conflicting results indicate a dysmaturity of the neuronal networks39 in FCD. The immature synaptic circuit is not only generated by delayed synaptogenesis but also by the presence of cells that exhibited signs of immature development. Balloon cells do not have voltage-gated currents,42 and some neurons displayed electrophysiological membrane properties42 and markers43 of developing neurons. Furthermore, in addition to the fact the developing networks are naturally more excitable,44 dysmorphic neurons also display inherent hyperexcitability.42
During the formation of early neuronal networks, neurons develop primary forms of excitability regulated by synaptic connections orchestrated by many genetic determinants.45 Our results suggest that this emerging excitability exceeds the normal threshold in FCD organoids, where neurons also appear hyperconnected and signal propagation is enhanced, as reflected by the higher response rate and the type of activity elicited by optogenetic stimulation found in the FCD organoids. These findings not only corroborate the presence of hyperexcitability in the FCD organoid, but also reflect a faster and increased spreading of spikes as typically observed in patients with epilepsy.46 This abnormal connectivity in FCD can be affected by neuronal morphological abnormalities,47 revealed by our model, and a still unknown genetic context, producing circuits that are more susceptible to becoming an epileptic network.
Also, the relationship between fewer synapses and hyperconnectivity in neuronal network could be explained by synaptic reorganization38,48 via a compensatory mechanism. In this way, the few synaptic sites with better locations along the dendrites could be compensated by redistributed synaptic strength resulting in a more connected network.48 This structural reconfiguration of the neuronal circuity seems to be reasonable given the reduction of dendritic spine density,38 passive membrane properties,49 and atypical dendritic processes38,50 noticed in dysmorphic neurons, all of which will impact the excitability of FCD circuitries. Therefore, it is plausible that the formation of neuronal networks in FCD appears to show aberrant wiring from the early stages of cortical development, which may be attributable to maturation, morphological and functional abnormalities.
The correlation between the genetic and clinical presentation of patients and organoid phenotypes is also noteworthy. At the genetic level, in brain tissue from these four patients with FCD, we identified a mosaic variant in the MTOR gene in Patient F1 and germline variants in TSC1 and TSC2 genes in Patient F2. Both variants are rare and considered to be of uncertain significance. In patient-derived organoids, we confirmed both germline mutations in Patient F2 and, we were not able to detect mTOR mosaic mutations in FCD organoids, include in F1-derived organoids (Table 1). At the clinical level, since carrying TSC1/TSC2 variants, presenting epilepsy and, mild intellectual disability, patient F2 does not present the clinical manifestation to be considered tuberous sclerosis complex, due to absence of multiple cortical tubers, and any alterations in the skin, heart, lung, and kidney. In cortical organoids from this patient, we noticed the absence of an organized column of neuroepithelium cells and an undefined apical lumen (Fig. 1K and Supplementary Fig. 3D1–4). Similarly, Costa et al.51 observed that biallelic deletion of TSC2 affected the structural organization of neural rosettes with aberrant proliferation and differentiation in a human 2D model. Therefore, our organoid model demonstrated that it is partially possible to mirror patient genetic status with faithful recapitulation of the germline mutations, a finding that provides some insights into faulty proliferation.
It is recognized that cell abnormalities5 and seizure generation52 in FCD type II are often associated with presence of brain de novo somatic variants in the mTOR pathway. From our data, we observed that regardless of the mosaic genetic background, we are able to recapitulate both the histological and functional hallmarks of FCD. The present data provide an understanding about the mechanisms leading to FCD, showing that mTOR mosaic variants might contribute to the clinical phenotypes, but they are not essential to trigger the process.
There are experimental caveats on our work. First, our organoid system is limited to the early phases of human corticogenesis. However, this limitation could be an opportunity to access the latent period to investigate the underlying epileptogenic mechanisms in FCD tissue. From this point of development, our interpretation is that increased excitability could be consequence of recurrent glutamatergic connectivity, when GABAergic synaptic transmission may not be present or functional enough to contribute to it. Second, our results should be considered preliminary due to the small number of patients. Moreover, despite the unavoidable differences in the genetic background among the selected patients with FCD and the variation among the iPSC clones (Supplementary Table 3), our model was able to capture key FCD hallmarks with robustness and reproducibility across subjects and clones. Future work should validate our initial observations in a larger cohort.
Overall, the molecular, cellular, and functional analyses of FCD cortical organoids presented here offer new insights into elucidating the mechanism underlying FCD. Successful establishment of this human FCD model opens new opportunities to investigate how abnormal early cortical development can contribute to epileptogenesis and may expand the search for novel forms of epilepsy treatment for this devastating disorder.
Supplementary Material
Acknowledgements
We kindly thank all patients and their families for their helpful cooperation. We are grateful to Dr F. Papes, Dr M. Shtrahman, Dr O. Platoshyn, Dr C. Trujillo, Dr P. Negraes, Dr I. Fernandes, C. Snethlage, Dr R. Herai, Dr F. Cugola, Dr R. Gao, Dr S. Benassi, Dr J. Sena, Dr W. de Souza, M. Lima Santos, L. Luz, L. Henry, and M. Patne for technical support and helpful discussions, and to Wirla Pontes, Johnnie Parks, and Sonia Neves Romeu for administrative help. We acknowledge the expertise and collaboration of the UNICAMP university hospital. We also greatly appreciate the help of the Neurology and Dermatology staff. We also thank the staffs at the Human Embryonic Stem Cell Core Facility, University of California San Diego (UCSD) School of Medicine Microscopy Core (NINDS P30 NS047101), the Institute for Genomic Medicine facility, Stem Cell Genomics Core (CIRM Major Facilities grant FA1-00607), the UCSD Stem Cell Program, and the Viral Vector Core at the Salk Institute for Biological Studies.
Abbreviations
- CheRiff
channelrhodopsin
- FCD
focal cortical dysplasia
- iPSCs
induced pluripotent stem cells
- MEA
multi-electrode array
- mTOR
mammalian target of rapamycin
Contributor Information
Simoni H Avansini, Department of Pediatrics/Rady Children’s Hospital-San Diego, Department of Cellular & Molecular Medicine, School of Medicine, University of California San Diego, La Jolla, CA 92037, USA; Department of Translational Medicine, School of Medical Sciences, University of Campinas, Campinas, Sao Paulo 13083-887, Brazil; Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil.
Francesca Puppo, Department of Pediatrics/Rady Children’s Hospital-San Diego, Department of Cellular & Molecular Medicine, School of Medicine, University of California San Diego, La Jolla, CA 92037, USA.
Jason W Adams, Department of Pediatrics/Rady Children’s Hospital-San Diego, Department of Cellular & Molecular Medicine, School of Medicine, University of California San Diego, La Jolla, CA 92037, USA.
Andre S Vieira, Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil; Department of Structural and Functional Biology, Institute of Biology, University of Campinas, Campinas Sao Paulo 13083-887, Brazil.
Ana C Coan, Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil; Department of Neurology, School of Medical Sciences, University of Campinas, Campinas Sao Paulo 13083-887, Brazil.
Fabio Rogerio, Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil; Department of Pathology, School of Medical Sciences, University of Campinas, Campinas, Sao Paulo 13083-887, Brazil.
Fabio R Torres, Department of Translational Medicine, School of Medical Sciences, University of Campinas, Campinas, Sao Paulo 13083-887, Brazil; Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil.
Patricia A O R Araújo, Department of Translational Medicine, School of Medical Sciences, University of Campinas, Campinas, Sao Paulo 13083-887, Brazil; Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil.
Mariana Martin, Department of Translational Medicine, School of Medical Sciences, University of Campinas, Campinas, Sao Paulo 13083-887, Brazil; Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil.
Maria A Montenegro, Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil; Department of Neurology, School of Medical Sciences, University of Campinas, Campinas Sao Paulo 13083-887, Brazil.
Clarissa L Yasuda, Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil; Department of Neurology, School of Medical Sciences, University of Campinas, Campinas Sao Paulo 13083-887, Brazil.
Helder Tedeschi, Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil; Department of Neurology, School of Medical Sciences, University of Campinas, Campinas Sao Paulo 13083-887, Brazil.
Enrico Ghizoni, Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil; Department of Neurology, School of Medical Sciences, University of Campinas, Campinas Sao Paulo 13083-887, Brazil.
Andréa F E C França, Department of Clinical Medicine, School of Medical Sciences, University of Campinas, Campinas, Sao Paulo 13083-887, Brazil.
Marina K M Alvim, Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil; Department of Neurology, School of Medical Sciences, University of Campinas, Campinas Sao Paulo 13083-887, Brazil.
Maria C Athié, Department of Translational Medicine, School of Medical Sciences, University of Campinas, Campinas, Sao Paulo 13083-887, Brazil; Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil.
Cristiane S Rocha, Department of Translational Medicine, School of Medical Sciences, University of Campinas, Campinas, Sao Paulo 13083-887, Brazil; Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil.
Vanessa S Almeida, Department of Translational Medicine, School of Medical Sciences, University of Campinas, Campinas, Sao Paulo 13083-887, Brazil; Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil.
Elayne V Dias, Department of Anesthesiology/Medical Center Hillcrest, School of Medicine, University of California San Diego, Hillcrest, CA 92103, USA.
Lauriane Delay, Department of Anesthesiology/Medical Center Hillcrest, School of Medicine, University of California San Diego, Hillcrest, CA 92103, USA.
Elsa Molina, Stem Cell Genomics and Microscopy Core, Sanford Consortium for Regenerative Medicine, University of California San Diego, La Jolla, CA 92037, USA.
Tony L Yaksh, Department of Anesthesiology/Medical Center Hillcrest, School of Medicine, University of California San Diego, Hillcrest, CA 92103, USA.
Fernando Cendes, Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil; Department of Neurology, School of Medical Sciences, University of Campinas, Campinas Sao Paulo 13083-887, Brazil.
Iscia Lopes Cendes, Department of Translational Medicine, School of Medical Sciences, University of Campinas, Campinas, Sao Paulo 13083-887, Brazil; Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), University of Campinas, Campinas, Sao Paulo 13083-888, Brazil.
Alysson R Muotri, Department of Pediatrics/Rady Children’s Hospital-San Diego, Department of Cellular & Molecular Medicine, School of Medicine, University of California San Diego, La Jolla, CA 92037, USA; Kavli Institute for Brain and Mind, Archealization Center (ArchC), Center for Academic Research and Training in Anthropogeny (CARTA), University of California San Diego, La Jolla, CA 92093, USA.
Funding
This work was supported by a grant to A.R.M. from the American Epilepsy Society (AES2017SD1), the UCSD Stem Cell Program, and by grants from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (2013/07559-3, 2017/50404-1, and 2019/09090-9), and by FAPESP fellowships to S.H.A. (2017/13220-0 and 2018/02967-0).
Competing interests
A.R.M. is a co-founder and has an equity interest in TISMOO, a company dedicated to genetic analysis and human brain organogenesis, focusing on therapeutic applications customized to autism spectrum disorders and other neurological diseases. The other authors declare no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1.Blumcke I, Spreafico R, Haaker G, et al. Histopathological findings in brain tissue obtained during epilepsy surgery. N Engl J Med. 2017;377(17):1648–1656. [DOI] [PubMed] [Google Scholar]
- 2.Guerrini R, Duchowny M, Jayakar P, et al. Diagnostic methods and treatment options for focal cortical dysplasia. Epilepsia. 2015;56(11):1669–1686. [DOI] [PubMed] [Google Scholar]
- 3.Blümcke I, Thom M, Aronica E, et al. The clinicopathologic spectrum of focal cortical dysplasias: A consensus classification proposed by an ad hoc Task Force of the ILAE Diagnostic Methods Commission. Epilepsia. 2011;52(1):158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamparello P, Baybis M, Pollard J, et al. Developmental lineage of cell types in cortical dysplasia with balloon cells. Brain. 2007;130(9):2267–2276. [DOI] [PubMed] [Google Scholar]
- 5.Lim JS, Kim WI, Kang HC, et al. Brain somatic mutations in MTOR cause focal cortical dysplasia type II leading to intractable epilepsy. Nat Med. 2015;21(4):395–400. [DOI] [PubMed] [Google Scholar]
- 6.D’Gama AM, Woodworth MB, Hossain AA, et al. Somatic mutations activating the mTOR pathway in dorsal telencephalic progenitors cause a continuum of cortical dysplasias. Cell Rep. 2017;21(13):3754–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldassari S, Ribierre T, Marsan E, et al. Dissecting the genetic basis of focal cortical dysplasia: A large cohort study. Acta Neuropathol. 2019;138(6):885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trujillo CA, Gao R, Negraes PD, et al. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell. 2019;25(4):558–569.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iefremova V, Manikakis G, Krefft O, et al. An organoid-based model of cortical development identifies non-cell-autonomous defects in Wnt signaling contributing to Miller-Dieker syndrome. Cell Rep. 2017;19(1):50–59. [DOI] [PubMed] [Google Scholar]
- 10.Ertürk A, Becker K, Jährling N, et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc. 2012;7(11):1983–1995. [DOI] [PubMed] [Google Scholar]
- 11.Qi Y, Yu T, Xu J, et al. FDISCO: Advanced solvent-based clearing method for imaging whole organs. Sci Adv. 2019;5(1):eaau8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexa A, Rahnenfuhrer J. topGO: Enrichment Analysis for Gene Ontology. R package version 2.37.0. Rahnenfuhrer. [Google Scholar]
- 14.Mitchell AL, Attwood TK, Babbitt PC, et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019;47(D1):D351–D360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geer LY, Marchler-Bauer A, Geer RC, et al. The NCBI BioSystems database. Nucleic Acids Res. 2010;38(SUPPL.1):D492–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabregat A, Jupe S, Matthews L, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46(D1):D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slenter DN, Kutmon M, Hanspers K, et al. WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018;46(D1):D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puppo F, Pré D, Bang AG, Silva GA. Super-selective reconstruction of causal and direct connectivity with application to in vitro iPSC neuronal networks. Front Neurosci. 2021;15:647877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. J R Stat Soc Series B: Stat Methodol. 2001;63(2):411–423. [Google Scholar]
- 23.Hochbaum DR, Zhao Y, Farhi SL, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014;11(8):825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Depla JA, Sogorb-Gonzalez M, Mulder LA, et al. Cerebral organoids: A human model for AAV capsid selection and therapeutic transgene efficacy in the brain. Mol Ther. 2020;18:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegle JH, López AC, Patel YA, Abramov K, Ohayon S, Voigts J. Open Ephys: An open-source, plugin-based platform for multichannel electrophysiology. J Neural Eng. 2017;14(4):045003. [DOI] [PubMed] [Google Scholar]
- 26.Quiroga RQ, Reddy L, Kreiman G, Koch C, Fried I. Invariant visual representation by single neurons in the human brain. Nature 2005;435(7045):1102–1107. [DOI] [PubMed] [Google Scholar]
- 27.Veeraval L, O’Leary CJ, Cooper HM. Adherens junctions: Guardians of cortical development. Front Cell Dev Biol. 2020;8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hakanen J, Ruiz-Reig N, Tissir F. Linking cell polarity to cortical development and malformations. Front Cellular Neurosci. 2019;13:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katayama KI, Melendez J, Baumann JM, et al. Loss of RhoA in neural progenitor cells causes the disruption of adherens junctions and hyperproliferation. Proc Natl Acad Sci U S A. 2011;108(18):7607–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heng JIT, Chariot A, Nguyen L. Molecular layers underlying cytoskeletal remodelling during cortical development. Trends Neurosci. 2010;33(1):38–47. [DOI] [PubMed] [Google Scholar]
- 31.Cappello S, Böhringer CRJ, Bergami M, et al. A radial Glia-specific role of RhoA in double cortex formation. Neuron. 2012;73(5):911–924. [DOI] [PubMed] [Google Scholar]
- 32.Zhang W, Benson DL. Stages of Synapse Development Defined by Dependence on F-Actin. 2001.
- 33.Dear ML, Dani N, Parkinson W, Zhou S, Broadie K. Two classes of matrix metalloproteinases reciprocally regulate synaptogenesis. Development. 2016;143(1):75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hříbková H, Grabiec M, Klemová D, Slaninová I, Sun YM. Calcium signaling mediates five types of cell morphological changes to form neural rosettes. J Cell Sci. 2018;131(3):jcs206896. [DOI] [PubMed] [Google Scholar]
- 35.Odenwald MA, Choi W, Buckley A, et al. ZO-1 interactions with F-actin and occludin direct epithelial polarization and single lumen specification in 3D culture. J Cell Sci. 2017;130(1):243––259.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu WA, Chen S, Li Z, et al. PARD3 dysfunction in conjunction with dynamic HIPPO signaling drives cortical enlargement with massive heterotopia. Genes Dev. 2018;32(11-12):763–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang DD, Deans AE, James Barkovich A, et al. Transmantle sign in focal cortical dysplasia: A unique radiological entity with excellent prognosis for seizure control . J Neurosurg. 2013;118(2):337–344. [DOI] [PubMed] [Google Scholar]
- 38.Rossini L, de Santis D, Mauceri RR, et al. Dendritic pathology, spine loss and synaptic reorganization in human cortex from epilepsy patients. Brain. 2021;144:251–265. [DOI] [PubMed] [Google Scholar]
- 39.Cepeda C, André VM, Levine MS, et al. Epileptogenesis in pediatric cortical dysplasia: The dysmature cerebral developmental hypothesis. Epilepsy Behav. 2006;9(2):219–235. [DOI] [PubMed] [Google Scholar]
- 40.Blair JD, Hockemeyer D, Bateup HS. Genetically engineered human cortical spheroid models of tuberous sclerosis. Nat Med. 2018;24(10):1568–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eichmüller OL, Corsini NS, Vértesy Á, et al. Amplification of human interneuron progenitors promotes brain tumors and neurological defects. Science. 2022;375(6579):eabf5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cepeda C, André VM, Vinters Hv, Levine MS, Mathern GW. Are cytomegalic neurons and balloon cells generators of epileptic activity in pediatric cortical dysplasia? Epilepsia. 2005;46:82–88. [DOI] [PubMed] [Google Scholar]
- 43.Englund C, Folkerth RD, Born D, Lacy JM, Hevner RF. Aberrant neuronal-glial differentiation in Taylor-type focal cortical dysplasia (type IIA/B). Acta Neuropathol. 2005;109(5):519–533. [DOI] [PubMed] [Google Scholar]
- 44.Rakhade SN, Jensen FE. Epileptogenesis in the immature brain: Emerging mechanisms. Nat Rev Neurol. 2009;5(7):380–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khazipov R, Luhmann HJ. Early patterns of electrical activity in the developing cerebral cortex of humans and rodents. Trends Neurosci. 2006;29(7):414–418. [DOI] [PubMed] [Google Scholar]
- 46.van Mierlo P, Höller Y, Focke NK, Vulliemoz S. Network perspectives on epilepsy using EEG/MEG source connectivity. Front Neurol. 2019;10:721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeong W, Jin SH, Kim M, Kim JS, Chung CK. Abnormal functional brain network in epilepsy patients with focal cortical dysplasia. Epilepsy Res. 2014;108(9):1618–1626. [DOI] [PubMed] [Google Scholar]
- 48.Wilson NR, Ty MT, Ingber DE, Sur M, Liu G. Synaptic reorganization in scaled networks of controlled size. J Neurosci. 2007;27(50):13581–13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.André VM, Wu N, Yamazaki I, et al. Cytomegalic interneurons: A new abnormal cell type in severe pediatric cortical dysplasia. J Neuropathol Exp Neurol. 2007;66(6):491–504. [DOI] [PubMed] [Google Scholar]
- 50.Cepeda C, Hurst RS, Flores-Hernández J, et al. Morphological and electrophysiological characterization of abnormal cell types in pediatric cortical dysplasia. J Neurosci Res. 2003;72(4):472–486. [DOI] [PubMed] [Google Scholar]
- 51.Costa V, Aigner S, Vukcevic M, et al. mTORC1 Inhibition corrects neurodevelopmental and synaptic alterations in a human stem cell model of tuberous sclerosis. Cell Rep. 2016;15(1):86–95. [DOI] [PubMed] [Google Scholar]
- 52.Hsieh LS, Wen JH, Claycomb K, et al. Convulsive seizures from experimental focal cortical dysplasia occur independently of cell misplacement. Nat. Commun. 2016;7:11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from whole exome sequencing and mTOR/GATOR panel that support the findings of this study have been deposited at NCBI BioProject ID PRJNA730529. For gene expression using NanoString Human Neuropathology Panel, GEO accession number is GSE174605.