Abstract
We previously showed that very thin filamentous bacteria affiliated with the division green non-sulfur bacteria were abundant in the outermost layer of thermophilic methanogenic sludge granules fed with sucrose and several low-molecular-weight fatty acids (Y. Sekiguchi, Y. Kamagata, K. Nakamura, A. Ohashi, H. Harada, Appl. Environ. Microbiol. 65:1280–1288, 1999). Further 16S ribosomal DNA (rDNA) cloning-based analysis revealed that the microbes were classified within a unique clade, green non-sulfur bacteria (GNSB) subdivision I, which contains a number of 16S rDNA clone sequences from various environmental samples but no cultured representatives. To investigate their function in the community and physiological traits, we attempted to isolate the yet-to-be-cultured microbes from the original granular sludge. The first attempt at isolation from the granules was, however, not successful. In the other thermophilic reactor that had been treating fried soybean curd-manufacturing wastewater, we found filamentous microorganisms to outgrow, resulting in the formation of projection-like structures on the surface of granules, making the granules look like sea urchins. 16S rDNA-cloning analysis combined with fluorescent in situ hybridization revealed that the projections were comprised of the uncultured filamentous cells affiliated with the GNSB subdivision I and Methanothermobacter-like cells and the very ends of the projections were comprised solely of the filamentous cells. By using the tip of the projection as the inoculum for primary enrichment, a thermophilic, strictly anaerobic, filamentous bacterium, designated strain UNI-1, was successfully isolated with a medium supplemented with sucrose and yeast extract. The strain was a very slow growing bacterium which is capable of utilizing only a limited range of carbohydrates in the presence of yeast extract and produced hydrogen from these substrates. The growth was found to be significantly stimulated when the strain was cocultured with a hydrogen-utilizing methanogen, Methanothermobacter thermautotrophicus, suggesting that the strain is a sugar-fermenting bacterium, the growth of which is dependent on hydrogen consumers in the granules.
Upflow anaerobic sludge blanket (UASB) processes have been used worldwide over the past decades mainly for the treatment of medium- and high-strength organic wastewaters (22, 31). Although thermophilic (50 to 60°C) UASB processes could be applied to high-temperature wastewaters discharged from certain manufacturing processes in some industries (22, 42, 51), almost all practical UASB processes constructed so far have been operated at mesophilic (30 to 40°C) or ambient temperatures. At present, there are only a few practical UASB processes running at thermophilic temperatures. One of the primary reasons for the lack of popularity of thermophilic UASB processes is the difficulty or lower reliability in accomplishing granulation of sludge under thermophilic conditions (40, 42).
For start-up of UASB processes, successful formation of stable granules from seed sludge is undoubtedly the major premise. Several factors are known to affect granulation, i.e., physicochemical properties of constituents in sludge such as surface hydrophobicity of cells (4, 11, 30), the presence of extracellular polymers (30, 46), and the composition of microorganisms (8, 16, 30, 52, 53). Among these, the microbial constituents are thought to be the decisive factor in many cases.
In general, filamentous and aggregating types of Methanosaeta cells are commonly observed other than Methanosarcina cells in mesophilic UASB granules (8, 12, 20, 23, 24, 32, 53). They are considered important for making cores of sludge granules by constructing web-like structures (53). In contrast, the long-filament type of Methanosaeta cells is seldom observed in thermophilic UASB granules, whereas the short-filament type (dispersing type) of Methanosaeta cells can frequently be found (18, 19, 23, 37, 39, 40, 45). Instead of Methanosaeta, very thin filamentous microorganisms, which are morphologically different from Methanosaeta, have been considered essential for the formation of well-settleable thermophilic granules. This type of microbe was frequently (or almost always) observed and found to entirely cover the thermophilic granules, forming a web-like coating on granules (32, 37, 39, 40). However, no detailed information on the microbes is available.
We previously analyzed the community structure of the thermophilic (55°C) methanogenic granular sludge in a UASB reactor treating an artificial wastewater (34) and determined the phylogenetic position and spatial distribution of the thin-filamentous organisms through the full-cycle rRNA approach (32). One of the interesting findings in the community structure analysis was that unidentifiable clones within the division green non-sulfur bacteria (GNSB) were frequently obtained from the sludge. Subsequent fluorescence in situ hybridization using an oligonucleotide probe targeting 16S rRNAs of the uncultured GNSB revealed that the GNSB in the reactors were thin-filamentous cells and predominated only in the outermost layers of thermophilic sludge granules as one of the significant populations (32).
To elucidate their physiology and metabolic function, we have attempted to cultivate the thin-filament microorganisms belonging to the GNSB group from several anaerobic wastewater sludges. In this paper, we report the isolation and partial characterization of a microorganism that is affiliated with the GNSB group and suggest its ecological importance in anaerobic wastewater treatment processes.
MATERIALS AND METHODS
Operation of UASB reactors.
Granules were collected from two lab-scale UASB reactors (reactor I and reactor II, 13-liter capacity), both of which had been operated at a thermophilic (55°C) temperature. Reactor I was fed with a synthetic substrate containing sucrose, acetate, propionate, and yeast extract (4.5:2.25:2.25:1, chemical oxygen demand [COD] ratio) over 3 years of operation (34). Reactor II had received the actual organic wastewater which had been discharged from a fried soy bean curd-manufacturing factory. Both reactors exhibited good performance for COD removal, with 90 to 95% removal efficiencies, and good methane formation throughout the experimental period.
Microorganisms, media, and cultivation.
The following organisms were used in this study. Thin-filamentous bacterium strain UNI-1 was enriched and isolated in this study. Methanothermobacter thermautotrophicus (formerly Methanobacterium thermoautotrophicum [49]) strain ΔH (DSM 1053) was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany). The culture medium used for enrichment and isolation of strain UNI-1 was prepared as described previously (17, 33). All cultivations were carried out at 55°C in 50-ml serum vials containing 20 ml of medium (pH25°C, 7.2) under an atmosphere of 80% N2–20% CO2 (vol/vol) without shaking. Neutralized substrates were added to the vials from stock solutions prior to inoculation. M. thermautotrophicus ΔH was cultivated at 55°C using the medium mentioned above except that hydrogen was added to the gas phase (80% N2–20% CO2, vol/vol) in the vials as the energy source. For cocultivation, M. thermautotrophicus ΔH and strain UNI-1 were inoculated into the medium supplemented with sucrose (20 mM) and yeast extract (0.1%) (5% inoculum of each).
The purity of the strain isolated in this study was routinely examined by microscopy and incubation of the cultures with the medium containing 0.1% yeast extract and a mixture of carbohydrates (sucrose, glucose, arabinose, and fructose, each at 2 mM) at 35 or 55°C.
In situ hybridization.
Fixation of granules was done as described previously (32). Whole-cell in situ hybridization was performed by the method described elsewhere (32, 34). For in situ hybridization with spine-like projections on sea urchin-like granules, we carefully cut the fixed projections with a surgical knife under a binocular microscope and immobilized the projections on glass slides coated with Vectabond (Vector Laboratories).
The following 16S rRNA-targeted oligonucleotide probes were used in this study: GNSB633, specific for clones MUG9 and TUG8, -9, and -10, which were classified in the GNSB group as described previously (32); EUB338 for the domain Bacteria (2); ARC915 for the domain Archaea (36); MX825 for the genus Methanosaeta (27); and MB1174 for the family Methanobacteriaceae (27). For in situ hybridization, we adjusted the stringency of hybridization by adding formamide to the hybridization buffer (20% [vol/vol] for EUB338, MX825, and GNSB633, 35% for MB1174 and ARC915). For double staining of the granule sections, indodicarbocyanine (Cy5)- and rhodamine-labeled probes were used simultaneously.
Construction of 16S rDNA clone library from spine-like structures formed on granules.
DNA extraction, PCR amplification, cloning, and sequencing procedures for constructing a 16S ribosomal DNA (rDNA) clone library were performed as previously reported (34) with slight modifications. For DNA extraction from spine-like structures on granules, we washed the granules with phosphate-buffered saline (PBS; 0.13 M NaCl, 10 mM sodium phosphate buffer, pH 7.2) several times and carefully cut and collected the projections with a surgical knife under a binocular microscope, dispersed the projections by weak sonication (50 W, 5 to 10 s), and subjected them to DNA extraction.
For construction of the 16S rDNA clone library from the projections, we used the following primer set for the PCR amplification of bacterial 16S rRNA genes: Bacteria-specific primer EUB341F (5′-GGTTACCTTGTTACGACTT-3′, positions 341 to 357 in Escherichia coli) (25) and prokaryote-specific primer 1490R (5′-GGTTACCTTGTTACGACTT-3′, 1491 to 1509 in E. coli) (50). The PCR products were purified with a MicroSpin column (Amersham Pharmacia Biotech), followed by cloning into plasmids using the TA cloning kit (Novagen). Twenty clonal rDNAs were randomly picked and subjected to sequencing.
DNA extraction and amplification of 16S rDNA from a pure culture.
DNA extraction from pure culture of several isolates was performed by the method of Hiraishi (13), and 16S rDNA was amplified by PCR as described above. In this PCR, the following primers were used: the Bacteria-specific primer 8F (5′-AGAGTTTGATCCTGGCTCAG-3′, 8 to 27 in E. coli) and universal primer 1490R (50). The PCR products were purified with a MicroSpin column (Amersham Pharmacia Biotech) and subjected to further analysis.
Sequencing and phylogenetic analysis.
Sequences of representative rDNA clones as well as the 16S rDNA of pure cultures were determined by Thermo Sequenase fluorescent-labeled primer cycle sequencing kit (Amersham Pharmacia Biotech) and an automated sequence analyzer (DSQ-1000L; Shimadzu) as described previously (17). Sequence data were aligned with the Clustal X package (38) and corrected by manual inspection. Phylogenetic trees were constructed by the neighbor-joining method (28) with the MEGA version 2 package (Kumar et al., submitted for publication). Bootstrap resampling analysis (10) for 100 replicates was performed for the estimation of confidence of tree topologies.
Microscopy and analytical methods.
Cells immobilized and hybridized on glass slides were viewed with a fluorescent microscope (Olympus BX50F), and the sections and spine-like projections hybridized with the probes were examined under a confocal laser scanning microscope (Olympus Fluoview BX50). Scanning electron microscopy (SEM) was performed as described previously (40). Short-chain fatty acids were determined with a gas chromatograph (Shimadzu GC-14A; flame ionization detector; packing material, FAL-M (G. L. Science); column temperature, 125°C). Alcohols and other compounds were determined by high-pressure liquid chromatography (HPLC) using an RSpak KC-811 column (Shodex; eluent, 3 mM HClO4; column temperature, 50°C) and a UV detector (210 nm; Shimadzu SPD-10A). Carbohydrates such as sucrose and glucose were determined by HPLC using an SCR101-H column (Shimadzu; eluent, H2O; column temperature, 60°C) and a refractive index detector (Shimadzu RID-10A). Methane, hydrogen, and carbon dioxide were determined by gas chromatography (GL Science model 370; detector type, thermal conductivity detection; packing material, Unibeads C; column temperature, 145°C).
Nucleotide sequence accession number.
The 16S rDNA sequence of strain UNI-1 was deposited in the EMBL/GenBank/DDBJ databases under accession no. AB046413.
RESULTS
Detailed analysis of phylogenetic positions of uncultured clones recovered from UASB processes.
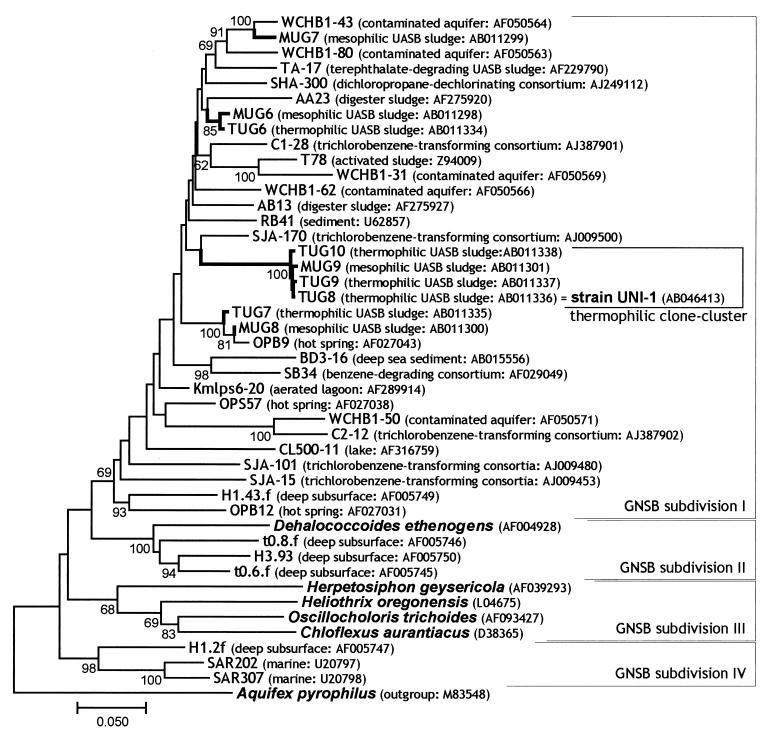
In our previous study, we recovered several types of uncultured clones affiliated with GNSB from mesophilic (35°C) and thermophilic (55°C) UASB granular sludges (34). The rDNA cloning-based analysis indicated that these clones accounted for approximately 20% of the total clones in libraries of both mesophilic (110 clones) and thermophilic (115 clones) sludges (34). In addition, we had designed a fluorescently labeled oligonucleotide probe (GNSB633) specific to those clones (MUG9, TUG8, TUG9, and TUG10 clones, referred to as thermophilic UASB cluster in Fig. 1) to elucidate their in situ morphology and spatial distribution within granular sludges, resulting in the detection of only filamentous cells in the outermost layer of thermophilic sludge granules (32).
FIG. 1.
Phylogenetic tree of GNSB, including environmental clones. The 16S rDNA sequences obtained in the previous study (MUG and TUG clones) and reference sequences were aligned, and phylogenetic trees were constructed by the neighbor-joining method (pairwise analysis). Bar represents 5 nucleotide substitutions per 100 nucleotides in 16S rDNA sequences. The sequence of Aquifex pyrophilus was used to root the tree. Bootstrap values above 50% are indicated at the branch points. The accession number of each reference sequence and the origin of environmental clones are also shown in parentheses. Names of cultivated organisms are shown in bold and italic. Our UASB clones, referred to as thermophilic UASB cluster and strain UNI-1, are shown in a bracket.
To precisely determine the phylogenetic positions of those GNSB clones, we performed phylogenetic analysis of the clonal sequences with all known 16S rDNA/RNAs of cultured and uncultured organisms belonging to GNSB (Fig. 1). Through this analysis, we found our GNSB clones to be classified within GNSB subdivision I (14), which comprises a number of environmental 16S rDNA sequences but contains no cultured microorganisms. Since the specific DNA probe (GNSB633) reacted with the filamentous microorganisms at the outermost layer of thermophilic granules, we focused on these organisms for further studies.
Attempts to cultivate GNSB filaments from UASB granules in reactor I.
To elucidate the physiology and metabolic function of the probe GNSB633-positive uncultured filaments, we attempted to isolate these microorganisms from granules in reactor I, which was the same thermophilic reactor used in the previous studies (17, 32, 34), and had been fed with sucrose, acetate, and propionate. Since the filamentous organisms were observed only in the outermost layer of the thermophilic methanogenic granules (32), it was very likely that these cells could utilize primary substrates such as carbohydrates. We therefore used sucrose, glucose, and several other carbohydrates as carbon and energy sources for primary enrichment. The surface layer of the granules was carefully scraped using a micromanipulator under binocular microscopy, sonicated briefly, and then inoculated into several media containing carbohydrate and yeast extract.
To judge whether targeted cells were grown in the media, fluorescence in situ hybridization with GNSB633 probe was applied to the cells in the cultures. We also tried to enrich the filaments in both liquid and solid media in which serially diluted samples were inoculated. However, no cultures contained the targeted cells. In many cases, irrelevant fast-growing microbes such as Thermoanaerobacterium and Thermoanaerobacter species (data not shown) outcompeted the target organisms.
Morphological changes in sludge granules in a thermophilic UASB reactor (reactor II).
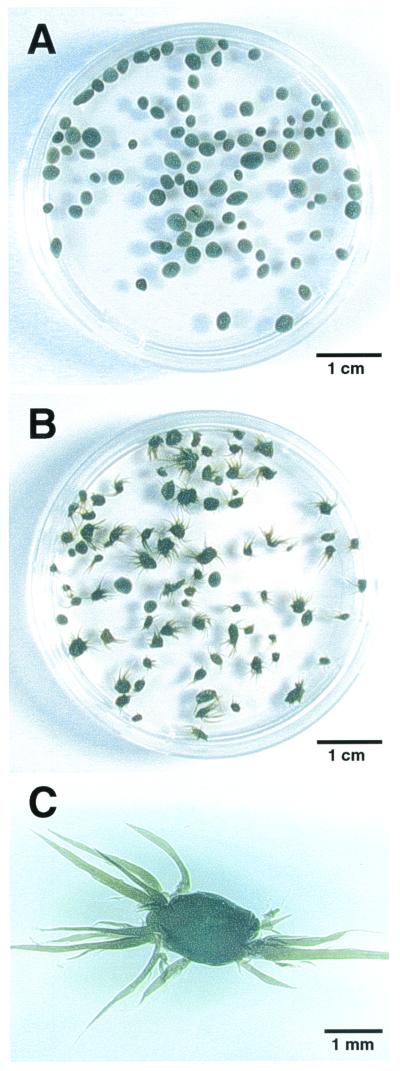
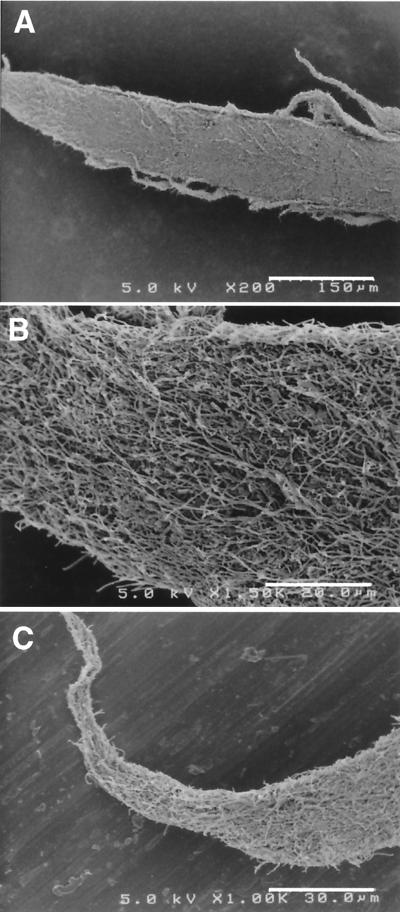
During the attempts, we unexpectedly found a unique phenomenon in the other thermophilic UASB reactor (reactor II). Reactor II had been treating actual organic wastewater from a fried soybean curd-manufacturing factory under thermophilic (55°C) conditions, exhibiting sufficient COD removal throughout the operation of the reactor. During the first operational period (days 0 to 100), the reactor contained normal granular sludge which had a good settleability (Fig. 2A). However, after 100 days of operation, we found changes in the configuration of sludge granules, i.e., the sludge granules became fluffy (Fig. 2B). The sludge still retained the granular structure, but a number of filaments were found to be sticking out of the surface of the granules, just like sea urchins (Fig. 2C). Over time, the number of sea urchin-like granules gradually increased, and finally they became predominant in the reactor. The formation of the fluffy granules did not significantly affect COD removal and methane production in the reactor, but the fluffy granules had far less settling ability than the normal granules, indicating anaerobic bulking of granular sludge. SEM observations of the spine-like projections on the granules showed that the projections comprised mainly filamentous organisms (Fig. 3).
FIG. 2.
Photographs of granules formed in reactor II. (A) Sludge granules at day 91 of operation. (B) Fluffy granules formed at day 133. (C) Magnified view of a fluffy granule.
FIG. 3.
Scanning electron micrographs of spine-like projections sticking out of the fluffy granules (sea urchin granules) in reactor II. (A) Whole view of a projection (bar, 150 μm). (B) Higher magnification of the bottom part of the projection (bar, 20 μm). (C) Higher magnification of the tip of the projection (bar, 30 μm)
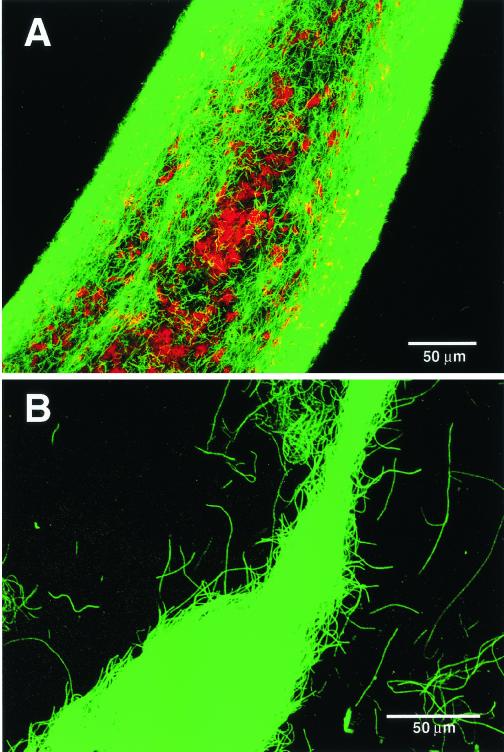
To identify the populations within the projections, we performed 16S rRNA-based analysis. Whole cell in situ hybridization analysis with a universal bacterial probe (EUB338) (2) showed that only a few cells within the projections reacted with the probe; instead, almost all filamentous populations hybridized with the GNSB633 probe (Fig. 4). Inside the projections, some archaeal cells could be detected using a universal archaeal probe (ARC915) (36) (Fig. 4) and were stained with the Methanosaeta-specific probe MX825 or the Methanobacteriaceae-specific probe MB1174 (27) (data not shown).
FIG. 4.
In situ hybridization of a projection from the fluffy thermophilic granules viewed by confocal laser scanning microscopy (digitally produced images with pseudocolors). The projection was simultaneously hybridized with Cy5-labeled archaeal probe ARC915 (indicated as red) and rhodamine-labeled probe GNSB633 for thermophilic UASB cluster of GNSB subdivision I (indicated as green). (A) Magnified view of the bottom part of the projection (bar, 50 μm). (B) Magnified view of the tip portion of the projection (bar, 50 μm).
To determine the phylogenetic position of the GNSB-like organisms present in the projections, an rDNA clone library was constructed from DNAs extracted from the projection part using a bacterial universal primer set. Through this analysis, we found that half of the total bacterial clones were identical, and the identical sequence was assigned to the thermophilic UASB cluster of GNSB subdivision I. Moreover, the clonal sequence was identical to that of clone TUG8, which was obtained in our previous cloning analysis (Fig. 1). Remaining clones were clustered within other clades, such as the gram-positive bacteria Clostridium/Bacillus group and the OP9 division (15) (data not shown).
Cultivation of GNSB-like filaments from spine-like projections of sea urchin granules in reactor II.
Since the probe GNSB633-positive filamentous cells were found to be highly abundant in the spine-like projections, particularly at the tips of the projections (Fig. 4B), we again attempted to cultivate the GNSB subdivision I filaments using the tips as the inoculum. We washed the fluffy granules and carefully cut the tip of the projections with a surgical knife under a binocular microscope, dispersed the cells by sonication, serially diluted them, and placed them into a liquid medium supplemented with several substrates. Of a number of substrates tested, we found that medium containing sucrose (20 mM) plus yeast extract (0.1%) or glucose (20 mM) plus yeast extract (0.1%) supported the growth of the filamentous cells after 2 weeks of incubation at 55°C.
Although the fast-growing anaerobes outgrew the filamentous cells at lower dilutions, the filamentous cells grew slowly at higher dilutions. Cotton-like dense flocs appeared in the liquid cultures receiving the highest dilution, and the floc-like aggregates contained a number of thin-filamentous cells and F420-autofluorescent rods resembling Methanothermobacter, associating together (data not shown). The thin-filamentous microbes were reactive with the GNSB633 probe.
By using the highest dilutions, we attempted to isolate the filamentous cells by the roll-tube method with the addition of bromoethane sulfonate (final concentration, 1 mM in culture) to inhibit the growth of the concomitant methanogens. Colonies that were only 0.1 to 0.2 mm in diameter appeared in solid medium after 1 month of incubation, and the same isolation procedure was repeated several times. The filamentous strain designated UNI-1 was eventually obtained in pure culture.
Physiological and genetic properties of strain UNI-1.
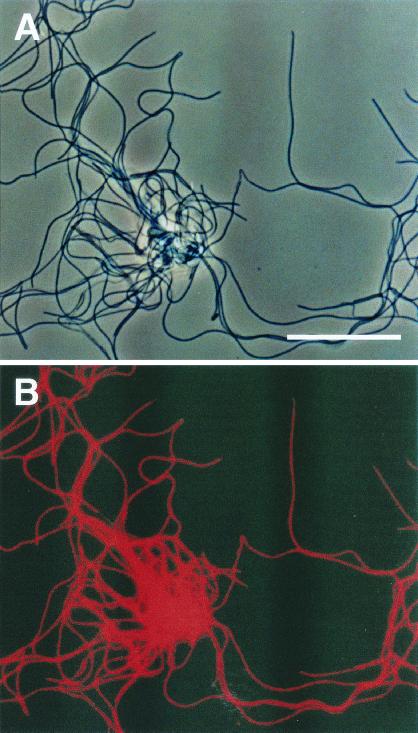
The strain was a strictly anaerobic, thin (0.2 to 0.3 mm)-filamentous bacterium, as shown in Fig. 5. The strain was moderately thermophilic (optimum temperature, 55°C). It required yeast extract for growth, and only a limited range of substrates, such as sucrose, glucose and arabinose, could be utilized in the presence of yeast extract. The strain was a slow-growing, fermentative bacterium which produced hydrogen, acetate, and carbon dioxide from these substrates (the approximate doubling time estimated from the production of hydrogen in the medium with glucose and yeast extract was 3 days).
FIG. 5.
In situ hybridization of strain UNI-1 cells isolated in this study. The cells were hybridized with the rhodamine-labeled GNSB633 probe. (A) Phase-contrast micrograph of strain UNI-1 cells (bar, 10 μm). (B) Fluorescent micrograph of the same field as panel A, showing that all cells were stained with the thermophilic UASB cluster-specific probe GNSB633.
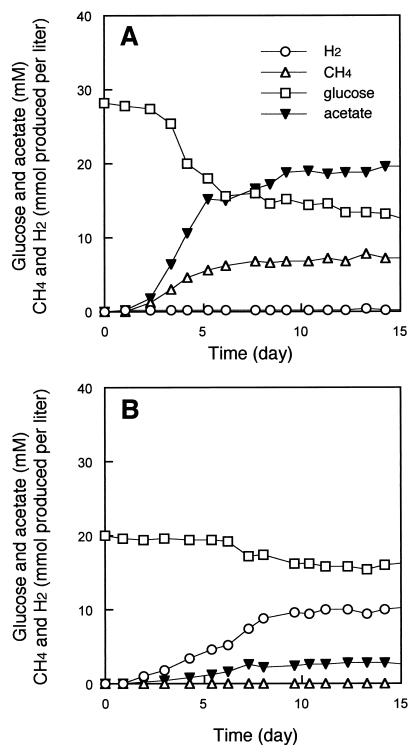
Growth was found to stagnate after a certain amount of hydrogen had accumulated. However, significant improvement of growth was observed when the strain was cocultivated with a hydrogen-utilizing methanogen, Methanothermobacter thermautotrophicus (Fig. 6). This coculture converted 223 μmol of glucose to 150 μmol of methane, 408 μmol of carbon dioxide, and 355 μmol of acetate in the presence of 0.1% yeast extract (94% carbon recovery and 75% electron recovery on the basis of glucose degraded).
FIG. 6.
Fermentation of glucose by strain UNI-1 in the presence (A) and the absence (B) of the hydrogenotrophic methanogen Methanothermobacter thermautotrophicus strain ΔH. The amounts of gases are shown as millimoles of gas produced per liter of culture.
Phylogenetic analysis of strain UNI-1 based on 16S rDNA sequences revealed that the strain was clustered with subdivision I of GNSB, which had the same sequence as the clone TUG8 (see Fig. 1). Through detailed analysis, the 16S rDNA sequence of strain UNI-1 was found to have two mismatches with the EUB338 probe sequence (Fig. 7). In fact, this strain could not be stained by in situ hybridization with the EUB338 probe at the standard hybridization stringency (data not shown). Moreover, the bacterial 16S rDNA-specific primer EUB341F (25), which was used in the construction of the 16S rDNA clone library for the projections in this study, also contained two mismatches with the 16S rDNA sequence of strain UNI-1. This was probably why only half of the total clones analyzed from the projections of the granules were recovered as GNSB clones, although the projections comprised primarily this type of organism, as shown in in situ hybridization (see above).
FIG. 7.
Alignment of target sequences of the EUB338 probe suite with the corresponding regions of 16S rRNA of members of the GNSB group, showing mismatches with the Bacteria universal probe EUB338 and its sequence variations (EUB338-II and EUB338-III). Mismatches with the EUB338 probe sequence are shaded.
DISCUSSION
Contribution of UNI-1-type filamentous cells to granulation of anaerobic sludge.
Our data presented in this study show that at least one of the filamentous populations on the surface of the thermophilic granules was a slow-growing bacterium affiliated with GNSB subdivision I, which is able to utilize only limited types of carbohydrates in the presence of yeast extract. It has been reported that the granulation of thermophilic sludges was difficult to achieve when only volatile fatty-acid mixtures were used as the sole substrate, while the addition of sucrose or glucose to the influent wastewater resulted in the formation of granular sludge with good settleability (40, 42, 43, 45, 54). In thermophilic UASB processes having well-settleable sludge granules, it was always observed that UNI-1 type of thin-filamentous microbes predominated on the surface of the sludge granules (32, 37, 39, 40). Considering these findings together with the physiological properties of strain UNI-1, the strain UNI-1 type of filamentous organism is one of the indispensable organisms for the granulation of thermophilic UASB sludges.
In liquid culture, strain UNI-1 cells grew and formed well-settleable dense, cotton-like flocs. This trait seems to contribute to granule formation and preservation of the structure, as mesophilic Methanosaeta cells do in mesophilic UASB granules. In the mesophilic UASB processes, UNI-1-like thin-filamentous cells were occasionally observed in addition to filamentous Methanosaeta cells (32, 57). The detailed phylogenetic positions and physiological properties of the mesophilic, thin-filamentous populations are not known. However, considering the facts that GNSB subdivision I-type clones were frequently recovered from several mesophilic UASB granules (34, 56) and that mesophilic filamentous cells probably require carbohydrates, it is strongly suggested that such microorganisms are also affiliated with the GNSB subdivision I group. In terms of sludge granule formation under mesophilic conditions, the thin-filamentous microorganisms seem to be less significant than the filamentous bacteria in thermophilic processes, since well-settleable granules could be made in the absence of the thinner filamentous cells (57).
Besides the importance of the UNI-1-type organisms in thermophilic UASB reactors, our study also shows the other aspect of this type of microorganism as a potential causative agent of the bulking of granular sludges. Endo and Tohya previously found anaerobic bulking in an anaerobic mesophilic (30°C) contact process, which was also caused by the outbreak of thin filamentous cells (9). These filamentous microbes were thought to be heterotrophic bacteria, because they could be eliminated by introducing acidified wastewater, similar to the case of Wu et al. (57). In addition, Alphenaar observed similar fluffy anaerobic granules with hairy surface structures as the result of the outgrowth of filamentous cells in mesophilic UASB reactors (1). A similar observation was also reported in a high-rate thermophilic UASB reactor (44). These observations, together with our data, suggest that it is important to control the growth of these filamentous cells not only to enhance granule formation but also to prevent the bulking of sludge granules.
In our experiment, from day 100 of the operation of the soybean wastewater-treating UASB process, we used diluted wastewater to reduce the COD loading for the reactor from 70 kg of COD/m3/day to 30 kg of COD/m3/day. In addition, the influent wastewater has been changed from day 100; in the earlier stage of operation, we used wastewater directly discharged from the manufacturer, which contained large amounts of suspended organic solids, but we used the supernatant of wastewater as the feed to eliminate the suspended solids from day 100 of the operation, since the severe formation of scum inside the reactor was found to occur due to the suspended solids in the wastewater. The major difference between the two wastewaters was that the latter wastewater contained relatively high concentrations of carbohydrates compared with the previous one (data not shown).
It is not clear which factors caused the outbreak of UNI-1-like filaments during the process. However, considering the physiological properties of strain UNI-1, the decrease in COD loading together with the increase in the carbohydrate content in the wastewater might have triggered the outbreak, since (i) the low COD loading might have enhanced the autolysis of microbial cells in the sludge, resulting in providing a yeast extract-like nutrient that is essential for the growth of strain UNI-1, and (ii) relatively high concentrations of carbohydrates, which can be substrates for UNI-1 cells, might have enhanced the growth of the strain. As suggested by Alphenaar (1), two-phase separation, in which a controlled acidifying phase was installed prior to the methanogenic UASB process, will be helpful to maintain an appropriate concentration of carbohydrates in the influent wastewater to prevent the anaerobic bulking of granular sludge as well as to enhance granulation.
Partial characterization of strain UNI-1.
Based on 16S rRNA/DNA sequences, the GNSB group has been divided into four subdivisions, I, II, III, and IV (Fig. 1) (14). Subdivision I contains the most diverse environmental clones among the four subdivisions; those clones were derived from sediments (55), subsurface (3), hot spring (15), aquifer samples (7), anaerobic dechlorinating consortia (29, 47, 48), a sulfate-reducing consortium (26), and anaerobic sludges (6, 34, 56). Some of these clones came from virtually aerobic environments like fresh water (ultraoligotrophic lake) (41), activated sludge (35), and an aerated lagoon (58). Apparently, strain UNI-1 belongs to subdivision I, but much more information on the tangible microbes within this subdivision must be accumulated to address the question of why these microorganisms can be so elusive, refusing isolation in the laboratory.
Interestingly, the strain was found to be sensitive to some extent to hydrogen, and growth could be greatly improved in coculture with a hydrogenotrophic methanogen similar to “Syntrophococcus sucromutans,” which is known as a syntroph which utilizes various carbohydrates in the presence of hydrogenotrophic methanogens (21). As a matter of fact, we found that Methanothermobacter-like hydrogen-consuming methanogens were associating with this microorganism in the projections of the granules. In addition to this characteristic feature, the strain is a very slow-growing organism and hence is easily outcompeted by fast-growing heterotrophs. This was probably the primary reason why our first attempts to isolate the organism from the surface layer of healthy granules in reactor I failed.
The other interesting aspect of strain UNI-1 is that its 16S rRNA sequence had two mismatches with the EUB338 probe, which is a 16S rRNA-targeted oligonucleotide probe and has been used widely to detect many members of the domain Bacteria by in situ hybridization (2). Since several other known microbes and environmental clones were found to have those mismatches, two supplementary versions of the EUB338 probe (EUB338-II and EUB338-III probes) have recently been developed by Daims et al. (5). One of the modified probes (EUB338-III) was fully complementary to the 16S rRNA of strain UNI-1 (see Fig. 7), and hence it was possible to detect the strain with the modified universal probe suite (data not shown). In addition, bacterial 16S rDNA-specific primer EUB341F, which is a widely used forward primer for 16S rDNA-based microbial community analysis, particularly for denaturing gradient gel electrophoresis (25), also contained two mismatches with the corresponding sequence of strain UNI-1. These findings indicate that those microbes have not been recognizable by conventional universal probes or primers commonly used in hybridization and PCR detection.
In summary, it is not appropriate to hasten the conclusion that subdivision I of GNSB comprises solely heterotrophic, anaerobic microbes, but our results suggest that unseen microbes in the GNSB subdivision I group may be relatively slower growers than commonly cultivatable microbes and/or need to be associated with other microbes (syntrophy) for growth. Our results also indicate that it would be still feasible to apply traditional techniques to the isolation of unseen microbes if the investigators use them thoughtfully in combination with molecular tools and carefully select good inocula which contain sufficient amounts of targeted cells.
ACKNOWLEDGMENTS
We thank Takeshi Yamada and Hiroyuki Imachi at Nagaoka University of Technology for help with cultivation of strain UNI-1.
This study was financially supported by research grant 11794002 of the Grant-in-aid for University and Society Collaboration subsidized by the Ministry of Education, Culture, Sports, Science and Technology, Japan.
REFERENCES
- 1.Alphenaar P A. Anaerobic granular sludge: characterization and factors affecting its functioning. Ph.D. thesis. Wageningen, The Netherlands: Wageningen Agricultural University; 1994. [Google Scholar]
- 2.Amann R I, Gish W, Miller W, Myers E W, Lipman D J. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandler D P, Brockman F J, Bailey T J, Fredrickson J K. Phylogenetic diversity of Archaea and Bacteria in a deep subsurface pleosol. Microb Ecol. 1998;36:37–50. doi: 10.1007/s002489900091. [DOI] [PubMed] [Google Scholar]
- 4.Daffonchio D D, Thaveesri J, Verstraete W. Contact angle measurement and cell hydrophobicity of granular sludge from upflow anaerobic sludge bed reactors. Appl Environ Microbiol. 1995;61:3676–3680. doi: 10.1128/aem.61.10.3676-3680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daims H, Breul A, Amann R, Schleifer K-H, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 6.Delbes C, Moletta R, Godon J J. Monitoring of activity dynamics of an anaerobic digester bacterial community using 16S rRNA polymerase chain reaction-single-strand conformation polymorphism analysis. Environ Microbiol. 2000;2:506–515. doi: 10.1046/j.1462-2920.2000.00132.x. [DOI] [PubMed] [Google Scholar]
- 7.Dojka M A, Hugenholtz P, Haack S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubourguier H C, Prensier G, Albagnac G. Structure and microbial activities of granular anaerobic sludge. In: Lettinga G, Zehnder A J B, Grotenhuis J T C, Hulshoff Pol L W, editors. Granular anaerobic sludge; microbiology and technology. Wageningen, The Netherlands: Centre for Agricultural Publishing and Documentation; 1987. pp. 18–41. [Google Scholar]
- 9.Endo G, Tohya Y. Ecological study on anaerobic sludge bulking caused by filamentous bacterial growth in an anaerobic contact process. Water Sci Technol. 1988;20:205–211. [Google Scholar]
- 10.Felsenstein J. Confidence limits of phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 11.Grotenhuis J T C, Plugge C M, Stams A J M, Zehnder A J B. Hydrophobicities and electrophoretic mobilities of anaerobic bacterial isolates from methanogenic granular sludge. Appl Environ Microbiol. 1992;58:1054–1056. doi: 10.1128/aem.58.3.1054-1056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grotenhuis J T C, Smit M, Plugge C M, Yuansheng X, van Lammeren A A M, Stams A J M, Zehnder A J B. Bacteriological composition and structure of granular sludge adapted to different substrate. Appl Environ Microbiol. 1991;57:1942–1949. doi: 10.1128/aem.57.7.1942-1949.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiraishi A. Direct automated sequencing of 16S rDNA amplified by polymerase chain reaction from bacterial cultures without DNA purification. Lett Appl Microbiol. 1992;15:210–213. doi: 10.1111/j.1472-765x.1992.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 14.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hulshoff Pol L W, Heijnekamp K, Lettinga G. The selection pressure as a driving force behind the granulation of anaerobic sludge. In: Lettinga G, Zehnder A J B, Grotenhuis J T C, Hulshoff Pol L W, editors. Granular anaerobic sludge; microbiology and technology. Wageningen, The Netherlands: Centre for Agricultural Publishing and Documentation; 1988. p. 153. [Google Scholar]
- 17.Imachi H, Sekiguchi Y, Kamagata Y, Ohashi A, Harada H. Cultivation and in situ detection of a thermophilic bacterium capable of oxidizing propionate in syntrophic association with hydrogenotrophic methanogens in a thermophilic methanogenic granular sludge. Appl Environ Microbiol. 2000;66:3608–3615. doi: 10.1128/aem.66.8.3608-3615.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamagata Y, Kawasaki H, Oyaizu H, Nakamura K, Mikami E, Endo G, Koga Y, Yamasato K. Characterization of three thermophilic strains of Methanothrix (“Methanosaeta”) thermophila sp. nov. and rejection of Methanothrix (“Methanosaeta”) thermoacetophila. Int J Syst Bacteriol. 1992;42:463–468. doi: 10.1099/00207713-42-3-463. [DOI] [PubMed] [Google Scholar]
- 19.Kamagata Y, Mikami E. Isolation and characterization of a novel thermophilic Methanosaeta strain. Int J Syst Bacteriol. 1991;41:191–196. doi: 10.1099/00207713-42-3-463. [DOI] [PubMed] [Google Scholar]
- 20.Kamagata Y, Mikami E. Some characteristics of 2 morphotypes of Methanothrix soehngenii from mesophilic anaerobic digesters. J Ferment Bioeng. 1990;70:272–274. [Google Scholar]
- 21.Krumholz L R, Bryant M P. Syntrophococcus sucromutans sp. nov. gen. nov. uses carbohydrates as electron donors and formate, methoxymonobenzoids or Methanobrevibacter as electron acceptor systems. Arch Microbiol. 1986;143:313–318. [Google Scholar]
- 22.Lettinga G. Anaerobic digestion and wastewater treatment systems. Antonie van Leeuwenhoek. 1995;67:3–28. doi: 10.1007/BF00872193. [DOI] [PubMed] [Google Scholar]
- 23.Macario A J L, Visser F A, van Lier J B, Conway de Macario E. Topography of methanogenic subpopulations in a microbial consortium adapting to thermophilic conditions. J Gen Microbiol. 1991;137:2179–2189. [Google Scholar]
- 24.MacLeod F, Guiot S R, Costerton J W. Layered structure of bacterial aggregates produced in an upflow anaerobic sludge bed and filter reactor. Appl Environ Microbiol. 1990;56:1598–1607. doi: 10.1128/aem.56.6.1598-1607.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denatured gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phelps C D, Kerkhof L J, Young L Y. Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microbiol Ecol. 1998;27:269–279. [Google Scholar]
- 27.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito N, Nei M. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 29.Schlötelburg C, von Wintzingerode F, Hauck R, Hegemann W, Göbel U B. Bacteria of an anaerobic 1,2-dichloropropane-dechlorinating mixed culture are phylogenetically related to those of other anaerobic dechlorinating consortia. Int J Syst Evol Microbiol. 2000;50:1505–1511. doi: 10.1099/00207713-50-4-1505. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt J E, Ahring B K. Granular sludge formation in upflow anaerobic sludge blanket (UASB) reactors. Biotech Bioeng. 1996;49:229–246. doi: 10.1002/(SICI)1097-0290(19960205)49:3<229::AID-BIT1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 31.Sekiguchi Y, Kamagata Y, Harada H. Recent advances in methane fermentation technology. Curr Opin Biotechnol. 2001;12:277–282. doi: 10.1016/s0958-1669(00)00210-x. [DOI] [PubMed] [Google Scholar]
- 32.Sekiguchi Y, Kamagata Y, Nakamura K, Ohashi A, Harada H. Fluorescence in situ hybridization using 16S rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules. Appl Environ Microbiol. 1999;65:1280–1288. doi: 10.1128/aem.65.3.1280-1288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekiguchi Y, Kamagata Y, Nakamura K, Ohashi A, Harada H. Syntrophothermus lipocalidus gen. nov., sp. nov., a novel thermophilic, syntrophic, fatty-acid-oxidizing anaerobe which utilizes isobutyrate. Int J Syst Evol Microbiol. 2000;50:771–779. doi: 10.1099/00207713-50-2-771. [DOI] [PubMed] [Google Scholar]
- 34.Sekiguchi Y, Kamagata Y, Syutsubo K, Ohashi A, Harada H, Nakamura K. Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. Microbiology. 1998;144:2655–2665. doi: 10.1099/00221287-144-9-2655. [DOI] [PubMed] [Google Scholar]
- 35.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Syutsubo K, Harada H, Ohashi A, Suzuki H. An effective start-up of thermophilic UASB reactor by seeding mesophilically-grown granular sludge. Water Sci Technol. 1997;36:391–398. [Google Scholar]
- 38.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uemura S, Harada H. Inorganic composition and microbial characteristics of methanogenic granular sludge grown in a thermophilic upflow anaerobic sludge blanket reactor. Appl Microbiol Biotechnol. 1995;43:358–364. [Google Scholar]
- 40.Uemura S, Harada H. Microbial characteristics of methanogenic sludge consortia developed in thermophilic UASB reactors. Appl Microbiol Biotechnol. 1993;39:654–660. [Google Scholar]
- 41.Urbach E, Vergin K L, Young L, Morse A, Larson G L, Giovannoni S J. Unusual bacterioplankton community structure in ultra-oligotrophic Crater Lake. Limnol Oceanogr. 2001;46:557–572. [Google Scholar]
- 42.van Lier J B. Limitations of thermophilic anaerobic wastewater treatment and the consequences for process design. Antonie van Leeuwenhoek. 1996;69:1–14. doi: 10.1007/BF00641606. [DOI] [PubMed] [Google Scholar]
- 43.van Lier J B, Boersma F, Debets M M W H, Lettinga G. High-rate thermophilic anaerobic wastewater treatment in compartmentalized upflow reactors. Water Sci Technol. 1994;30:251–261. [Google Scholar]
- 44.van Lier J B, Groeneveld N, Lettinga G. Development of thermophilic methanogenic sludge in compartmentalized upflow reactors. Biotech Bioeng. 1996;50:115–124. doi: 10.1002/(SICI)1097-0290(19960420)50:2<115::AID-BIT1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 45.van Lier J B, Grolle K C F, Stams A J M, de Macario E C, Lettinga G. Start-up a thermophilic upflow anaerobic sludge bed (UASB) reactor with mesophilic granular sludge. Appl Microbiol Biotechnol. 1992;37:130–135. doi: 10.1007/BF00174217. [DOI] [PubMed] [Google Scholar]
- 46.Veiga M C, Jain M K, Wu W-M, Hollingsworth R I, Zeikus J G. Composition and role of extracellular polymers in methanogenic granules. Appl Environ Microbiol. 1997;63:403–407. doi: 10.1128/aem.63.2.403-407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Wintzingerode F, Landt O, Ehrlich A, Göbel U B. Peptide nucleic acid-mediated PCR clamping as a useful supplement in the determination of microbial diversity. Appl Environ Microbiol. 2000;66:549–557. doi: 10.1128/aem.66.2.549-557.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Wintzingerode F, Selent B, Hegemann W, Göbel U B. Phylogenetic analysis of an anaerobic, trichlorobenzene-transforming microbial consortium. Appl Environ Microbiol. 1999;65:283–286. doi: 10.1128/aem.65.1.283-286.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wasserfallen A, Nöling J, Pfister P, Reeve J, Coway de Macario E. Phylogenetic analysis of 18 thermophilic Methanobacterium isolates supports the proposals to create a new genus, Methanothermobacter gen., nov., and to reclassify several isolates in three species, Methanothermobacter thermoautotrophicus comb. nov., Methanothermobacter wolfeii comb. nov., and Methanothermobacter marburgensis sp. nov. Int J Syst Evol Microbiol. 2000;50:43–53. doi: 10.1099/00207713-50-1-43. [DOI] [PubMed] [Google Scholar]
- 50.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiegant W M. Thermophilic anaerobic digestion for waste and wastewater treatment. Ph.D. thesis. Wageningen, The Netherlands: Wageningen Agricultural University; 1986. [Google Scholar]
- 52.Wiegant W M, de Man A W A. Granulation of biomass in thermophilic upflow anaerobic sludge blanket reactors treating acidified wastewaters. Biotechnol Bioeng. 1986;28:718–727. doi: 10.1002/bit.260280511. [DOI] [PubMed] [Google Scholar]
- 53.Wiegant W W. The “spaghetti theory” on anaerobic granular sludge formation, or the inevitability of granulation. In: Lettinga G, Zehnder A J B, Grotenhuis J T C, Hulshoff Pol L W, editors. Granular anaerobic sludge; microbiology and technology. Wageningen, The Netherlands: Centre for Agricultural Publishing and Documentation; 1987. pp. 146–152. [Google Scholar]
- 54.Wiegant W W, Lettinga G. Thermophilic anaerobic digestion of sugers in upflow anaerobic sludge blanket reactors. Biotechnol Bioeng. 1985;28:718–727. doi: 10.1002/bit.260271115. [DOI] [PubMed] [Google Scholar]
- 55.Wise M G, McArthur J V, Shimkets L J. Bacterial diversity of a Carolina bay as determined by 16S rRNA gene analysis: confirmation of novel taxa. Appl Environ Microbiol. 1997;63:1505–1514. doi: 10.1128/aem.63.4.1505-1514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu J-H, Liu W-T, Tseng I-C, Cheng S-S. Characterization of microbial consortia in a terephthalate-degrading anaerobic granular sludge system. Microbiology. 2001;147:373–382. doi: 10.1099/00221287-147-2-373. [DOI] [PubMed] [Google Scholar]
- 57.Wu W-M, Thiele J H, Pankratz H S, Hickey R F, Zeikus J G. Comparison of rod- versus filament-type methanogenic granules: microbial population and reactor performance. Appl Microbiol Biotechnol. 1993;39:795–803. [Google Scholar]
- 58.Yu Z, Mohn W W. Bacterial diversity and community structure in an aerated lagoon revealed by ribosomal intergenic spacer analyses and 16S ribosomal DNA sequencing. Appl Environ Microbiol. 2001;67:1565–1574. doi: 10.1128/AEM.67.4.1565-1574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]