Abstract
A culture-independent molecular analysis of archaeal communities in waters collected from deep South African gold mines was performed by performing a PCR-mediated terminal restriction fragment length polymorphism (T-RFLP) analysis of rRNA genes (rDNA) in conjunction with a sequencing analysis of archaeal rDNA clone libraries. The water samples used represented various environments, including deep fissure water, mine service water, and water from an overlying dolomite aquifer. T-RFLP analysis revealed that the ribotype distribution of archaea varied with the source of water. The archaeal communities in the deep gold mine environments exhibited great phylogenetic diversity; the majority of the members were most closely related to uncultivated species. Some archaeal rDNA clones obtained from mine service water and dolomite aquifer water samples were most closely related to environmental rDNA clones from surface soil (soil clones) and marine environments (marine group I [MGI]). Other clones exhibited intermediate phylogenetic affiliation between soil clones and MGI in the Crenarchaeota. Fissure water samples, derived from active or dormant geothermal environments, yielded archaeal sequences that exhibited novel phylogeny, including a novel lineage of Euryarchaeota. These results suggest that deep South African gold mines harbor novel archaeal communities distinct from those observed in other environments. Based on the phylogenetic analysis of archaeal strains and rDNA clones, including the newly discovered archaeal rDNA clones, the evolutionary relationship and the phylogenetic organization of the domain Archaea are reevaluated.
Recent molecular phylogenetic analyses based on small-subunit (SSU) rRNA gene (rDNA) sequencing have revealed that the phylogenetic diversity of Archaea in naturally occurring microbial communities is much greater than previously assumed on the basis of the results obtained with standard cultivation and isolation methods (3, 6, 14, 15, 20, 24, 43, 45). Initially, a small collection of isolates was referred to as archaebacteria, and now this varied assemblage is known to be both ubiquitous and cosmopolitan. Molecular phylogenetic approaches have revealed that environmental archaeal populations are both diverse and complex, often consisting of uncultivated and unidentified members. Because pure-culture phenotypic characterizations of many environmental Archaea are currently not possible, the physiological features and ecological significance of archaeal communities remain difficult to assess. The phylogenetic structure derived from archaeal rDNA clones from a given habitat, however, frequently corresponds to measurable environmental constraints (8, 42). When phylogenetic features intrinsic to archaeal communities are related to the environment, they may provide important insights into the physiological functions and ecological roles of the communities.
The gold mines of South Africa are the deepest accessible excavations in the world and provide a unique opportunity for direct exploration of the deep subsurface. The stratigraphic sequence and hydrogeological setting of the Transvaal region south of Johannesburg are well known (17). These mines harbor unique environments for microorganisms, both natural and anthropogenic, including high-temperature–high-pressure and saline groundwater systems, endolithic habitats, and diverse mine process and drainage waters ranging from acidic to strongly alkaline. The purpose of the research described here was to investigate the population structure and phylogenetic diversity of the Archaea in water samples from these environments and to relate the archaeal community to the geological setting and geochemical characteristics of the water. The isotopic composition of the water, its δ18O and δD, was utilized to distinguish between different types of water encountered at depth in the gold mines. The isotopic composition of groundwater usually lies on the global meteoric water line (GMWL) (12), and its exact position is determined by the history of meteorological processes (i.e., evaporation and precipitation) and the age of the water. In some extreme environments, the interaction between groundwater and the aquifer matrix or subsurface gases causes the isotopic composition to deviate from the GMWL. Archaeal communities in the water samples were analyzed by performing a PCR-mediated terminal restriction fragment length polymorphism (T-RFLP) analysis of rDNA in conjunction with a sequencing analysis of archaeal rDNA libraries. The phylogenetic features of the archaeal communities present in the deep gold mine environments were compared with those of communities from other extreme and nonextreme environments.
MATERIALS AND METHODS
Description of sites.
All samples used in this study were obtained from gold mines owned and operated by Gold Fields Ltd. of South Africa. Three of the mines, the East and West Driefontein (now Driefontein Consolidated) and Kloof mines, form a loose cluster of shaft complexes on the West Rand about 70 km west of Johannesburg near Carletonville. Beatrix Mine is located in the extreme southern Witwatersrand Basin, approximately 400 km south of the other mines, near Welkom.
The mines of the West Rand have a common stratigraphy. With increasing depth, the mines first penetrate Pretoria Group sandstones and then the Transvaal dolomites (2.2 gigaannum [Ga]), which range from nonexistent at Kloof Mine Shaft 4 to more than 2 km deep at Driefontein Consolidated. The dolomite is a partially dewatered karstic aquifer that overlies the Ventersdorp Supergroup volcanic strata (2.7 Ga) which, in turn, overlie the Witwatersrand Supergroup quartzites and carbonaceous gold-bearing reef horizons (2.9 Ga). At Beatrix Mine, the dolomite and Ventersdorp sequences are absent, and Karoo (200 to 300 megaannum [Ma]) sediments overlie the Witwatersrand Supergroup. Whereas the highly permeable dolomite aquifer is seasonally recharged by surface meteoric water, the low-permeability Ventersdorp and Witwatersrand strata are dominated by fluid flow along subvertical fractures with different connectivities to the surface. The Witwatersrand Basin is hydraulically compartmentalized by a swarm of vertical, cross-cutting, impermeable, 1.4-Ga syenite dykes radiating from the Pilanesburg Complex to the north. Similar compartmentalization occurs at Beatrix Mine due to 200-Ma Karoo basaltic dykes. Where dykes cross the hydraulic gradient, groundwater rises to the surface along the fractured dyke contacts, forming springs and pans at the surface.
The geothermal gradients were found to vary among the four mines and appeared to be correlated with the amounts of water moving through fractures or along dyke contacts. At East Driefontein only a few slowly flowing boreholes were noted at the time of this study and the geothermal gradient is 9°C km−1, whereas at West Driefontein, Kloof, and Beatrix fissure water emanating from boreholes is common and the geothermal gradients are 13, 16, and 29°C km−1, respectively (T. C. Onstott, D. P. Moser, M. F. DeFlaun, L. M. Pratt, and B. Sherwood Lollar, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., p. 515, 2001).
Ultradeep gold mines utilize large volumes of chilled service water to cool the ventilation air, for drilling, and for dust suppression. The mines utilized in this study employ closed-loop systems for their service water; process water is collected in settling ponds at the bottom of a shaft, pumped to the surface, disinfected by chlorination, chilled, and reused. The water is a mixture of all of the fluids intersected by the mines, water purchased from the regional water board, and dolomite aquifer water injected into the stream via intermediate pump compartments. During its migration from the stope face, along the access tunnels, and to the settling ponds, the service water may enter fractures at different levels, which are opened during mining or for pressure release purposes, and it may migrate into open boreholes.
Most water samples were collected from exploratory boreholes; the only exception was sample WDF1 (= sample A1) from Driefontein, which was collected from a roof fracture. The boreholes had different configurations and ages that ranged from 2 weeks to 4 years at the time of sampling. All holes were uncased except for the outermost 1 m, which was lined with a steel outlet pipe. High-flow-rate holes (thousands of liters per hour) were each managed with a steel pressure reduction valve, whereas boreholes with low flow rates (tens of liters per hour to less than 1 liter h−1) did not have valves and were allowed to flow freely.
Sample collection, processing, and physical measurements.
Service water samples were collected near underground work areas with mining hoses in East Driefontein Mine (sample S1) and Kloof Mine (sample S2). When possible, airtight connections were used to limit the contact of fissure water with air. Borehole B2-25-FW1 (sample F2) at Beatrix Mine was sealed with an inflatable packer to facilitate sampling. The high back pressures and flow rates of the boreholes at West Driefontein Mine, (borehole WDF2b; sample F1), East Driefontein Mine (borehole E4-IPC-DW1; sample D1), and Kloof Mine (borehole K4-41-FW1; sample F3) precluded management of the water flow with packers. Instead, an inlet hose was threaded a short distance into the outlet pipe downstream of the pressure reduction valve, and a portion of the flow was redirected into the sampling system (see below). Sample A1 from borehole WDF1 at West Driefontein Mine was collected in a sterile container from a flowing roof fracture.
For molecular analyses, water was collected in tightly sealed 11.6-liter stainless steel canisters (Cornelius, Inc., Anoka, Minn.). The canisters and associated hardware, such as quick connects and tubing, were autoclaved prior to use. The canisters were filled to overflowing and transported to the surface at the ambient temperature for processing. In the laboratory, a filtering manifold was attached to each tank outlet port, and a slight headspace overpressure was maintained with N2 gas to force the water through sterile 47- or 25-mm-diameter polysulfone membrane filters (pore size, 0.2 μm; Supor; Gelman, Ann Arbor, Mich.). The sample throughput varied from 1 to 12 liters per filter. The filters were stored in cryotubes with 1 ml of 5 M pH 5.0 guanidineisothiocyanate (9) and were frozen at −20°C on site and at −70°C after shipment to the United States.
Temperature and pH measurements were made at the time of sampling with a digital thermometer and a hand-held pH meter (Hanna Instruments Inc. Woonsocket, R.I.). Water samples used for chemical analyses either were collected unfiltered or were syringe filtered into collection vials. The unfiltered samples included those used to determine the contents of dissolved oxygen, δD, and δ18O, as well as the contents of total organic carbon (TOC) dissolved organic carbon (DOC). Samples used for dissolved O2 determinations were collected with hoses in 300-ml biological oxygen demand bottles and fixed on site with MnSO4 and KOH powder pillows (Hach, Loveland, Colo.). Samples used for δ18O and δD analyses were collected in 15-ml serum vials that were crimp sealed without a headspace under Teflon septa. Samples used for DOC and TOC analyses were collected in 45-ml screw-cap vials that had been preloaded with 2 ml of concentrated HCl under Teflon septa. Samples to be analyzed for anion and cation contents were filtered through 0.22-μm-pore-size nylon acrodisk membranes into acid-washed 50-ml Nalgene bottles (Gelman). Samples used for cation analyses were fixed with 2.5 ml of concentrated HNO3.
Chemical analyses.
Samples preserved for dissolved O2 analysis were titrated with 0.1 M Na2S3O4 by using an adaptation of the Winkler method (20) within 24 h of collection. Anion samples were analyzed by ion chromatography (Dionex). Cation concentrations, which were used to calculate total dissolved solids (TDS) contents, were determined by inductively coupled plasma mass spectrometry (Actlabs, Ancaster, Ontario, Canada). The δD content was determined by high-temperature reduction of water with zinc, followed by analysis of the resulting H2 gas by isotope ratio mass spectrometry (University of Waterloo, Waterloo, Ontario, Canada). The δ18O content of water was determined by equilibration with CO2(g) of known isotopic composition, followed by analysis by isotope ratio mass spectrometry (Rocky Mountain Mass Spectrometry, Boulder, Colo.). The TOC content was determined by converting the organic carbon to CO2 by catalytic combustion by Environmental Protection Agency method 415.1 (18). The CO2 was then measured directly with an infrared detector (Tekmar Dohrman DC-190). The DOC content was measured by using the procedure used to measure TOC content, except that particulate material was first removed by centrifugation at 10,000 × g for 10 min and only the supernatant was analyzed. TDS values were calculated from the sums of anion and cation concentrations. The particulate organic carbon content was calculated from the difference between the measured TOC and DOC values.
Extraction of nucleic acids.
DNA was extracted from preserved filters with a Soil DNA Mega Prep kit (Mo Bio Laboratories, Inc., Solana Beach, Calif.) used according to the manufacturer's suggested protocol. A blank (no filter) was routinely extracted in the same manner as a control (9). As a control for mine- and field laboratory-introduced contamination, an autoclaved 250-ml glass bottle containing 200 ml of deionized water was transported to the sampling site on several occasions and then recapped to simulate sample handling underground. The control was transported to the surface along with the samples and processed as described above.
Quantification of archaeal rDNA.
Quantification of archaeal rDNA in whole microbial DNA assemblages was performed by using a quantitative fluorescent PCR method as previously described (44).
T-RFLP analysis of archaeal rDNA.
In order to rapidly identify dominant sequences in the samples, a T-RFLP analysis of SSU rDNA was performed (31). Archaeal rDNA was amplified from DNA extracts by PCR by using LA Taq polymerase (TaKaRa, Kyoto, Japan) and oligonucleotide primers Arch21F-HEX (13) and Arch915R-TET (41). These primers were 5′ labeled with the phosphoramidite dyes 5-hexachlorofluorescein and 5-tetrachlorofluorescein, respectively. Reaction mixtures containing the oligonucleotide primers at concentrations of 0.1 μM and template DNA at a concentration of 1 ng μl−1 were prepared. Thermal cycling was performed with a GeneAmp 9600 (Perkin-Elmer, Foster City, Calif.) under the following conditions: denaturation at 96°C for 25 s, annealing at 50°C for 45 s, and extension at 72°C for 120 s for 30 cycles. When no apparent product was recovered with 30 reaction cycles, the number of cycles was increased to 40.
Amplified rDNA from five separate reaction mixtures was pooled and subjected to agarose gel electrophoresis. The products were cut from each gel lane and extracted sequentially with phenol, phenol-chloroform-isoamyl alcohol, and chloroform-isoamyl alcohol. DNA was precipitated with ethanol and centrifuged, and the pellet was suspended in double-distilled water. The purified rDNA was digested by using restriction enzymes with 4-bp recognition sites (HaeIII or HhaI or both). The terminal restriction fragments (T-RFs) were analyzed with a model 377 automated sequencer equipped with GeneScan software, version 3.0, according to the recommendations of the manufacturer (PE Applied Biosystems, Foster City, Calif.). The precise lengths of T-RFs were determined by comparison with an internal size standard (TAMRA 2500; PE Applied Biosystems) that was added to each digested sample. Apparent standard fragment sizes were adjusted downward by 18 bp as recommended by the manufacturer for denaturing gels.
Cloning and sequencing of archaeal rDNA.
Archaeal rDNA was amplified by PCR by using the protocol used for the T-RFLP analysis except that nonlabeled primers (Arch21F and 1492R [30]) were used. When no apparent product was obtained after 30 reaction cycles under the conditions described above, the reverse primer was replaced with Arch915R and the number of cycles was increased to 40. Amplified rDNA from five separate reaction mixtures was purified as described above. Purified rDNA was cloned into vector pCR2.1 by using an Original TA cloning kit (Invitrogen, Carlsbad, Calif.).
Clones containing inserts from picked colonies were identified by direct PCR analysis by using M13 primers. Following PCR, the reaction mixtures were treated with exonuclease I and shrimp alkaline phosphatase (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) and directly sequenced by the dideoxynucleotide chain termination method using a Big Dye sequencing kit (PE Applied Biosystems). The 515F primer (45) was used in a partial sequencing analysis.
Sequence and phylogenetic analyses.
Single-strand sequences approximately 400 nucleotides long were analyzed. Sequence similarity was determined by the FASTA component program of DNASIS (Hitachi Software, Tokyo, Japan). rDNA sequences having ≥98% similarity as determined by FASTA were assigned to the same clone type. A representative sequence of each clone type was subjected to a sequence similarity analysis with the prokaryotic SSU rRNA database and the nonredundant nucleotide sequence databases of GenBank, EMBL, and DDBJ by using gapped-BLAST (1, 4).
Approximately 1.5 or 0.9 kb of sequence of each representative rDNA clone was determined for both strands. The data from rDNA clone library and T-RFLP analyses were linked by calculating the T-RF length for each sequenced clone. Clones with a predicted T-RF length identical (±1 bp) to a T-RF length found by T-RFLP analysis were assigned to a T-RFLP ribotype. The sequences were manually aligned with prokaryotic SSU rDNA data from Ribosomal Database Project II (33) based on primary- and secondary-structure considerations and were also subjected to analysis with the Chimera Check program of Ribosomal Database Project II. Phylogenetic analyses were restricted to nucleotide positions between the Arch21F and Arch915R primer sites that could be unambiguously aligned. Neighbor-joining analysis was performed by using the PHYLIP package (version 3.5; obtained from J. Felsenstein, University of Washington, Seattle). Bootstrap analysis (100 replicates) was used to obtain confidence estimates for phylogenetic tree topologies.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the DDBJ database under accession numbers AB050205 (SAGMA-A) to AB050214 (SAGMA-J), AB050215 (SAGMA-J2), AB050216 (SAGMA-K) to AB050231 (SAGMA-Z), AB050232 (SAGMA-1) to AB050235 (SAGMA-4), and AB050236 (SAGMA-6) to AB050247 (SAGMA-17).
RESULTS
Geochemistry of water samples.
Chemical analyses (Table 1) indicate that while chilled service water samples S1 and S2 were collected at depth, they were saturated with dissolved O2. This result is consistent with the cycling of the water through a treatment plant at the surface. Dolomite aquifer sample D1 and fissure water sample F3 were subsaturated with dissolved O2 (1.5 mg liter−1). The values obtained probably represent maximum values since when service waters were resampled with an inflatable packer later (data not shown), the levels of dissolved O2 in water samples from both sources were below the limit of detection. The levels of O2 in fissure water samples F1 and F2 were below the limit of detection.
TABLE 1.
Geochemical characteristics of water samples
| Sample
|
Source water | Mine (shaft) | Depth (km) | Vol filtered (liters) | Temp (°C) | pH | Concn of:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Designation | Type | Dissolved O2 (ppm) | TDS (%, wt/wt) | Cl− (ppm) | NO3− (ppm) | SO4− (ppm) | δ18O (‰, V-SMOWb) | δD (‰, V-SMOWb) | TOC (ppm) | DOC (ppm) | Particulate organic carbon (ppm) | ||||||
| S1 | Surface water | EDS-1 | Driefontein (5E) | 3.2 | 4 | 17.6 | 7.9 | 15.3 | 0.074 | 222.0 | 47.0 | 40.0 | −2.7 | −8.59 | 13 | 9 | 4 |
| S2 | Surface water | K4-41-SW1 | Kloof (4) | 3.08 | 1 | 19 | 6.8 | NAa | 0.134 | 287.0 | 12.7 | 635.0 | −2.75 | NA | NA | NA | NA |
| A1 | Acid mine water | WDF1 | Driefontein (2W) | 1.8 | 3 | 30 | 3.0 | 5 | >0.13 | 102.8 | 44.0 | 769.3 | −3.85 | NA | <1 | <1 | <1 |
| D1 | Dolomite aquifer | E4-IPC-DW2 | Driefontein (4E) | 0.7 | 6 | 25 | 7.0 | 1.5 | 0.022 | 15.5 | <1 | 80.0 | −4.55 | −20.28 | 6 | 5 | 1 |
| F1 | Fissure water | WDF2b | Driefontein (6W) | 2.7 | 6 | 45 | 10.0 | 0.0 | 0.118 | 610.0 | <1 | 112.0 | −6.58 | NA | 2 | 1 | 1 |
| F2 | Fissure water | B2-25-FW1 | Beatrix (2) | 0.87 | 7.4 | 35 | 9.5 | 0.0 | 0.214 | 1,295.0 | <1 | 0.7 | 1.92 | −36.86 | 1 | <1 | 1 |
| F3 | Fissure water | K4-41-FW1 | Kloof (4) | 3.08 | 11.6 | 60 | 9.6 | 1.1 | 0.323 | 1,898.5 | <1 | 0.1 | −5.71 | −27.69 | 2 | 1 | 1 |
NA, not analyzed.
V-SMOW, Vienna Standard Mean Ocean Water.
The pH values of the service and dolomite water samples were circumneutral, whereas the pH values of the fissure water samples ranged from 9.5 to 10. The sample obtained from a dripping roof fracture in West Driefontein Mine shaft 6 (sample A1) had a pH of 3.0. The levels of salinity, as determined by the TDS method, ranged from 0.02% (wt/wt) in the dolomite water (sample D1) to 0.32% in the fissure water from Kloof Mine (sample F3). The Cl concentrations ranged from 15.5 mg liter−1 for the dolomite sample (sample D1) to 1,898.5 mg liter−1 for sample F3. The nitrate levels were below the limit of detection (<1 mg liter−1) for all samples except mine service water and the acid mine water (sample A1). The sulfate concentrations varied from 0.1 mg liter−1 for sample F2 to 635 and 769 mg liter−1 for samples S2 and A1, respectively. There was considerable variation in the concentrations of TOC and DOC between samples. East Driefontein service water (sample S1) contained the highest TOC and DOC concentrations (13 and 9 mg liter−1, respectively). Dolomite aquifer water yielded TOC and DOC values of 6 and 5 mg liter−1, respectively. The fissure water samples yielded TOC concentrations of 1 or 2 mg liter−1, while the DOC values ranged from 1 to <1 mg liter−1.
The stable isotope compositions of the water (δD and δ18O) fell on or near the GMWL (Fig. 1). The ratio of δD to δ18O for mine service water (sample SW1) was near the observed mean annual value for modern precipitation and was also near the value reported for the Vaal River (35), one of the local service water sources. The value for the dolomite water sample, sample D1, fell on the local meteoric water line, and this sample had an isotopic composition that overlapped the lowest recorded precipitation values. Fissure water samples from the Kloof and West Driefontein mines (samples F1 and F3) had isotope concentrations that placed them close to the GMWL but were significantly less than the modern precipitation and D1 values and were near the value obtained for the Florisbad Hot Springs (35). The value for sample F2 was significantly removed from the GMWL.
FIG. 1.
δ18O and δD values of the water samples. The cross-hatched region represents International Atomic Energy Agency values for local precipitation, and the values for the Vaal River and Florisbad Hot Springs are from reference 35. Most of the water samples fall on the global meteoric water line; the only exception is water from Beatrix Mine (sample F2). V-SMOW, Vienna Standard Mean Ocean Water. MW, meteoric water.
Archaeal rDNA abundance.
Relative archaeal rDNA abundance values for the whole microbial DNA assemblages extracted from the various groundwater samples were determined by quantitative fluorogenic PCR. The proportions of archaeal rDNA in the whole microbial rDNA populations were determined to be 17.7% for sample F1, 26.1% for sample F2, 74.4% for sample A1, 0.1% for sample S1, 0.1% for sample D1, and below the detection limit for samples F3 and S2. In most of the groundwater samples, the proportion of bacterial rDNA was higher than that of archaeal rDNA in the whole microbial DNA assemblages, whereas the archaeal rDNA population dominated the bacterial population in the acid mine water sample (sample A1). In addition, the fissure water samples from West Driefontain (sample F1) and Beatrix Mine (sample F2) contained relatively high proportions of archaeal rDNA. These results suggest that the proportions of the archaeal communities are different in different microhabitats in the South African gold mines but that these communities can be significant or even dominant components in some environments.
T-RFLP analysis.
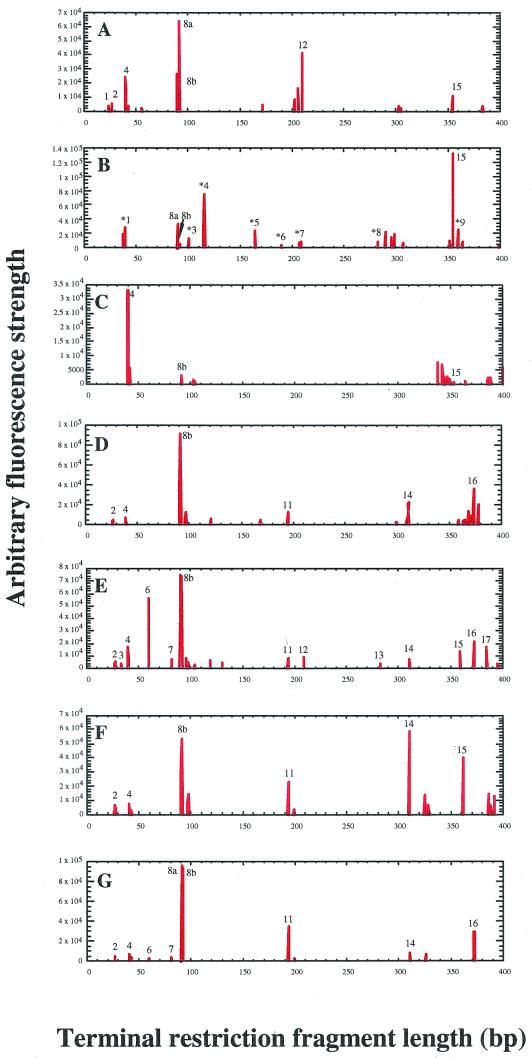
Figure 2 shows typical electropherograms generated with a labeled reverse primer and HhaI-HaeIII double digests. Approximately 17 major T-RFs that were 400 bp or less long (ribotypes 1 to 17) were generally found in all samples, whereas sample F2 contained a number of novel T-RFs (ribotypes *1 to *9). Although many of the major T-RFs were present across the range of samples, the patterns that grouped most closely were the patterns for water samples obtained from similar sources. For example, ribotypes 8b, 11, and 14 were prominent in all of the T-RFLP profiles for service water, dolomite water, and acid mine water. Within this set, however, diagnostic patterns were also observed that allowed differentiation. For example, in service water sample S1, ribotypes 6 and 17 were uniquely prominent. Service water sample S2, however, was dominated by ribotype 14, a minor peak in the other members of this set. Likewise, dolomite water sample D1 was dominated by ribotype 8a (Fig. 2).
FIG. 2.
Typical electropherograms of archaeal T-RFLPs generated from rDNAs with a labeled reverse primer and HhaI-HaeIII digests obtained from representative water samples. The numbers above or next to the peaks indicate major ribotypes commonly observed in various samples, and the numbers with asterisks indicate novel ribotypes unique to a sample. (A to G) Typical archaeal patterns for the fissure water from West Driefontein Mine (sample F1), fissure water from Beatrix Mine (sample F2), fissure water from Kloof Mine (sample F3), acid mine water from West Driefontein (sample A1), service water from East Driefontein (sample S1), service water from Kloof Mine (sample S2) and dolomite aquifer water (sample D1), respectively.
In contrast, the fissure water samples displayed unique T-RFLP patterns. Except for ribotypes 15, 8a, and 8b, the profile for Beatrix Mine fissure water (sample F2) was composed entirely of ribotypes not represented in any other sample. Sample F1 was dominated by the otherwise absent or minor ribotype 12, while in sample F3 ribotype 4 was the most prominent ribotype.
Phylogenetic analyses of archaeal rDNA clone libraries.
Archaeal rDNA clones from representative water samples were characterized by partial sequencing (ca. 550 to 600 nucleotides) and sequence similarity analysis. Approximately 1.5-kb rDNAs were amplified from samples F2, A1, S1, S2, and D1 by using 30 reaction cycles and primers Arch21F and 1492R. rDNA fragments that were 0.9 kb long were obtained from F1 and F3 by using 40 reaction cycles and the Arch21F and Arch915R primers. The numbers of archaeal rDNA clones characterized for the samples were 50 for sample F1, 64 for sample F2, 48 for sample F3, 45 for sample A1, 46 for sample S1, 40 for sample S2, and 43 for sample D1. The major archaeal ribotypes observed in the T-RFLP analyses were also found among the rDNA clones characterized (Fig. 3).
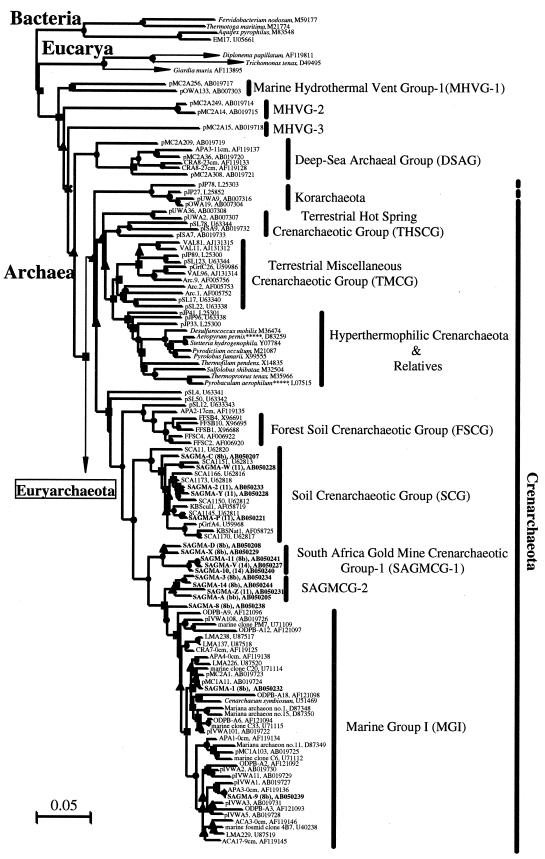
FIG. 3.
Phylogenetic trees based on SSU rDNA sequences, including various rDNA clones obtained from deep subsurface South African gold mine environments. Each tree was inferred by a neighbor-joining analysis of 637 homologous positions of the rDNA sequence. (Left) Tree showing the phylogenetic relationships among the domains Bacteria and Eucarya, the deep branches of uncultivated Archaea, and Crenarchaeota; (Right) tree showing the phylogenetic organization in the Euryarchaeota. The symbols on the branches indicate bootstrap confidence values, as follows: ●, >80%; ▪, 60 to 80%; ▴, 40 to 60%; × <40%. The bars indicate the expected number of changes per sequence position. Boldface type indicates rDNA clones obtained in this study. The numbers in parentheses after the SAGMA clone designations indicate the T-RFLP types shown in Fig. 2. The beginnings of designations indicate rDNA clones corresponding to uncultivated organisms derived from the following environments: pMC1A, pMC2A, pISA, and pIVWA, deep-sea hydrothermal vent environments (42); pOWA and pUWA, shallow marine hydrothermal ventwater and terrestrial acidic hot spring water, respectively (45); pJP and pSL, sediments in Yellowstone National Park hot springs (2, 3); CRA, APA, ACA, and LMA, deep-sea sediments (49); VAL, boreal forest lake water (27); SCA, soil clone from agricultural soil (6); KBScul and KBSnat, soils (8); pGrfA and LMA, freshwater lake sediments (23, 32); Arc, a subsurface paleosol (10); marine clones C and PM, ocean water samples (36); ODPB-A, subseafloor sediments and water (5); Mariana archaeon, sediments in Mariana Trench (28); TA and BA, marine anoxic methane-oxidizing layers (24); ARR and ABS, rice roots and paddy soils (11); KTK, deep-sea brine sediments (19); MSP, hypersaline lake sediments (21); SBAR, OARB, and WHARN, coastal water (13); 2MT, salt marsh sediment (37), WCHD, a subsurface soil (16); SAGMA, South African gold mine environments. The rDNA clones with asterisks have putative archaeal rRNA introns.
As observed in the T-RFLP analyses, the archaeal rDNA phylotypes were different in the samples from different water sources and even in samples from the same water source. In general, the fissure water contained rDNA clones phylogenetically affiliated with the Euryarchaeota, whereas crenarchaeotic rDNA sequences were associated with service water samples S1 and S2, acid mine water sample A1, and dolomite water sample D1. The crenarchaeotic rDNA clones from samples S1, S2, A1, and D1 were placed into three groups based on sequence similarity. One common group bore a strong similarity (>97%) to previously described crenarchaeotic soil rDNA clones (soil group) (6, 8) and contained representative clone types SAGMA-C (five clones in sample A1, three clones in sample S2, two clones in sample D1), SAGMA-W (two clones in sample S2), SAGMA-2 (two clones in sample S2), SAGMA-Y (five clones in sample S2, 20 clones in sample D1), and SAGMA-P (two clones in sample S1, three clones in sample S2). Another group, obtained from the service water, was similar (>97%) to marine group I (MGI), and its representative clone types were SAGMA-8 (two clones in sample S1, four clones in sample S2), SAGMA-1 (one clone in sample S2), and SAGMA-9 (six clones in sample F1, one clone in sample S1, four clones in sample in S2), whereas the final group was moderately related (levels of similarity, between 88 and 94%) to both the soil group and MGI and was represented by clone types SAGMA-D, -X, -11, -V, -10, -3, -14, -Z, and -A.
These crenarchaeotic phylotypes dominated the archaeal rDNA assemblages of service water sample S2 (one, one, four, two, two, three, and three clones of SAGMA-X, -11, -V, -10, -3, -Z, and -A, respectively), acid mine water sample A1 (40 clones of SAGMA-D), and dolomite water sample D1 (16 clones of SAGMA-14) and were also recovered from fissure water sample F1 (one clone of SAGMA-14). Service water from East Driefontein (sample S1) also contained euryarchaeotic rDNAs. In this sample, most of the euryarchaeotic rDNA clones were closely related to cultivated strains of methanogens. The most abundant clone types were phylogenetically associated with Methanospirillum hungatei (six clones of SAGMA-4, five clones of SAGMA-7, and 13 clones of SAGMA-J2 were recovered and had >94% similarity) in the order Methanomicrobiales and with Methanosaeta concilli (eight clones of SAGMA-N and one clone of SAGMA-L had >96% similarity) and Methanosarcina barkeri (one clone of SAGMA-M and one clone of SAGMA-O had >95% similarity, and one clone of SAGMA-6 had >92% similarity) in the order Methanosarcinales. In addition to rDNA clones closely related to methanogens, two clones of a novel phylotype (SAGMA-B) from sample S1 were not similar to any other archaeal rDNA sequences and possessed three putative archaeal rRNA introns.
The putative introns (83, 24, and 149 bp) formed bulge-helix-bulge type secondary-structure signatures at the junctions between possible exons and introns (the exon-intron junction positions and the sequences of the introns of SAGMA-B are described in the DDBJ database [accession number AB050206]).
Service water samples S1 and S2 contained genetically diverse rDNA clones related to clones in samples A1 and D1. The archaeal rDNA compositions of the fissure water differed markedly from those of the service water, acid mine water, and dolomite water. In the fissure water sample obtained from Kloof Mine, sample F3, only two clone types were observed (43 clones of SAGMA-16 and five clones of SAGMA-17). The most abundant clone type (SAGMA-16) was strongly associated with species of the hyperthermophilic Archaea genus Pyrococcus, particularly Pyrococcus abyssi (99.4%). The other clone type (SAGMA-17) was only distantly related to known archaeal rDNAs and also contained an archaeal rRNA intron-like intervening sequence (71 bp) similar to that found in the sequence of clone type SAGMA-B. The euryarchaeotic phylotypes obtained from the fissure water samples from the West Driefontein and Beatrix mines, samples F1 and F2, were relatively similar to one another and exhibited no apparent phylogenetic association with other archaeal rDNAs in available databases. Fissure water sample F1 contained 37, 3, and 3 clones of representative clone types SAGMA-12, -13, and -15, respectively, and fissure water sample F2 contained 21, 21, 3, 2, 1, 1, 1, 1, 1, 1, and 1 clones of SAGMA-E, -F, -G, -Q, -H, -I, -J, -R, -S, -T, and -U respectively. Thus, these clones may represent novel deep subsurface archaeal phylotypes.
In order to determine the phylogenetic positions of the archaeal rDNA clones from the South African gold mines and to clarify the phylogenetic relationships between these clones and other archaeal strains and an increasing number of environmental clones from other sources, a phylogenetic tree was constructed. Representative rDNA sequences from major archaeal groups were incorporated into a single tree by using the neighbor-joining method (Fig. 3).
In this study, the crenarchaeotic rDNA clones were obtained mainly from service, dolomite, and acid mine water samples and were affiliated with the soil crenarchaeotic group, MGI, and a newly described group intermediate between the soil crenarchaeotic group and MGI. This intermediate group was separated into two subgroups, South Africa gold mine crenarchaeotic group 1 (SAGMCG 1) and SAGMCG 2 (Fig. 3A). The topology of these branches was supported by relatively high bootstrap confidence values (Fig. 3A). As demonstrated by Buckley et al. (8), nonthermophilic Crenarchaeota rDNA clones tend to form distinct phylogenetic clusters strongly associated with their habitats, such as freshwater lake water and sediments, forest soils, agricultural soils, and marine water and sediments. Hence, the distinct phylogenetic assemblage of SAGMCG 1 and SAGMCG 2 within the Crenarchaeota suggests that the crenarchaeotic rDNA sequences were derived from novel archaeal members that specifically inhabit deep terrestrial subsurface environments.
The Euryarchaeota rDNA clones obtained from the service water of East Driefontein (sample S1) plot close to cultivated strains in the Methanosarcinales and Methanomicrobiales, as expected from sequence similarity analysis (Fig. 3B). The novel euryarchaeotic rDNA clones recovered from fissure water samples from the West Driefontein and Beatrix mines, however, constitute a new phylogenetic group that branches more deeply than the divergence between the order Thermoplasmales and a diverse clade of uncultivated environmental clones (Fig. 3B).
The new lineage of rDNA clones formed two subgroups (Fig. 3B). One, South Africa gold mine euryarchaeotic group 1 (SAGMEG 1), consisted mainly of rDNA clones from the fissure water from Beatrix Mine, while the other, SAGMEG 2, consisted mainly of clones from the fissure water from West Driefontein Mine. The rDNA sequences belonging to both SAGMEG 1 and SAGMEG 2 had relatively high G+C contents (ranging from 57.3% for SAGMA-U to 60.2% for SAGMA-12) and relatively short branch lengths.
Of the rDNA clones that did not fall within these two subgroups, SAGMA-T from sample F2 plotted near subsurface archaeal clone WCHD3-16 (16), SAGMA-B from sample S2 was affiliated with the deep-sea euryarchaeotic group, and SAGMA-17 from sample F3 plotted in the biogeographically and phylogenetically diverse miscellaneous euryarchaeotic group. The last two clones possessed putative intron-containing rDNA. In general, the archaeal clones obtained from South African gold mine environments were distantly associated with the archaeal strains and clones obtained from other environments (Fig. 3).
DISCUSSION
The presence of novel archaeal communities was evident after culture-independent molecular analyses of rDNAs recovered from deep South African gold mine environments. To date, a great number of novel archaeal phylotypes have been identified from a variety of microbial habitats, including open ocean waters (20, 36), coastal waters (13, 34, 39), a polar sea (15), sediments from continental shelf (50) and pelagic (28, 49) environments, a salt marsh (37), a freshwater lake (23, 32, 40), agricultural and forest soils, including the rhizosphere (6–8, 48) and paddy field soil (11, 22, 29), alkaline hypersaline lakes (21, 25), hot springs (2, 3, 45), deep-sea hydrothermal vents (42), and a deep subsurface geothermal pool (43). The phylogenetic diversity of Archaea has been significantly extended by these investigations. In addition, comparative phylogenetic analyses of environmental archaeal clones have revealed that the phylogenetic features of a given archaeal community are relatively conserved in its environment.
The geochemical, biological, and hydrological processes at work in the groundwater habitats in the South African gold mines most likely influenced archaeal community structure. All of the water was originally derived from precipitation, and there was no apparent marine input. Nevertheless, the three classes of water samples (fissure water, dolomite aquifer water, and service water) were readily distinguished by their geochemical and isotopic signatures, as well as by their molecular fingerprints.
The plot of δD versus δ18O (Fig. 1) indicates that service water sample S1 is isotopically similar to modern precipitation (Pretoria annual mean precipitation) and Vaal River water (35) from near the mines. The service water often contains nitrate, probably as a result of the explosives used in the mining process, and sulfate, which originates from oxidation of sulfide minerals. Dolomite aquifer water sample D1 is isotopically lighter than either the service water samples or modern precipitation. This result is not consistent with rapid recharge of the dolomite aquifer by modern surface meteoric water as the sole source and suggests that a portion of the dolomite water originates from an isotopically lighter source. The δ18O of acidic mine water sample A1 was similar to that of the dolomite water, suggesting that either there is substantial infiltration of water from the overlying dolomite aquifer into the West Driefontein Mine or a large portion of the service water at West Driefontein is comprised of dolomite water. Fissure water samples F1 and F3 were isotopically similar to each other, and their values clustered with borehole water values reported for other South African gold mines by Duane et al. (17). The most likely explanation for the isotopic similarity is that the recharge age coincides with a time interval during which the climate in South Africa was much cooler (at least as old as the Pleistocene). The value for fissure water sample F2 was horizontally displaced from the GMWL towards a heavier δ18O value. Such displacement is normally interpreted as being caused by high-temperature isotopic exchange between the oxygen of the meteoric water and the isotopically heavier oxygen of quartz (the most common oxygen-bearing mineral in the Witwatersrand Supergroup). This signature is frequently observed in hot spring waters (47). Although the Beatrix Mine fissure water was collected at a depth of only 0.8 km and had a temperature of 37°C, the mining district occurs in a region of the Kaapvaal Craton where the geothermal gradient is greater (26) than that observed in the mines of the West Rand and where the presence of hot springs, such as those at Florisbad, suggests that deep circulation of meteoric water occurs. Sample F2, therefore, may have originated from a much deeper and, therefore, higher-temperature source.
The presence of archaeal rDNA clones related to the soil crenarchaeotic group in dolomite water is consistent with infiltration of precipitation from the surface to the dolomite layer. The MGI crenarchaeota are ubiquitous archaea in global marine environments, and several clones from a freshwater environment have been described (32). Since the dolomite was deposited during the last marine incursion in this part of the Kaapvaal Craton, the possibility that the MGI members in the dolomite aquifer water are descendants of ancient marine archaea cannot be excluded. Given that the dolomite was deposited 2.3 billion years ago, however, and given that the MGI rDNA sequences obtained from the dolomite aquifer are closely related to those of extant MGI members, a more likely explanation is that freshwater MGI organisms were introduced via infiltration of relatively modern surface water. The novel SAGMCG sequences, also retrieved from the dolomite aquifer, may reflect an unusual microbial habitat within the dolomite aquifer or may be associated with the isotopically lighter, second source of the dolomite water.
The service water samples from the East Driefontein and Kloof mines had archaeal rDNA community structures similar to that of the dolomite water except for the methanogenic Archaea-like rDNA observed in the East Driefontein service water. The conserved community structure in the service water from the Kloof Mine shows that anthropogenic treatments, such as oxygenation, pH buffering, and chlorination, have little effect on the archaeal community structure. Prior to circulation, the service water for the East Driefontein Mine was pooled in an artificial reservoir at the surface. The methanogenic archaeal population may have formed in an anaerobic portion of the reservoir. The complex circulation patterns and the potential for contact or mixing with various sources make it difficult to draw firm conclusions regarding the source(s) of the sequences.
Acid mine water is commonly observed in metalliferous ore mines and is formed by both chemical and microbiological processes (38). In the South African gold mines, water dripping from wall and roof cracks was often acidic. The acidic water sample obtained from West Driefontein Mine was leaking from a crack covered with a crusty yellow microbial mat. This acid mine water contained a low-diversity Archaea community that was closely related to SAGMCG 1, a major archaeal component in the dolomite aquifer. Quantitative fluorogenic PCR analysis (14) of the acid mine water sample revealed that archaeal rDNA comprised 83% of the microbial rDNA assemblage (results not shown). These results suggest that an archaeal member originally derived from the dolomite aquifer predominates in the microbial community of the acid mine water. Although the microbial community structure of the yellow mats was not determined, the uncultivated archaeon represented by rDNA clone type SAGMA-D may inhabit the microbial mats and may also be involved in acidification of the water as a result of sulfur oxidation.
The archaeal community structures of the fissure water samples obtained from the Beatrix and West Driefontein mines were similar. The archaeal phylotypes in these fissure water samples represent entirely new groups (SAGMEG 1 and 2) at the division or order level in the domain Archaea. The rDNA sequences belonging to both SAGMEG 1 and SAGMEG 2 have relatively high G+C contents and relatively short branches. These phylogenetic features were found in most of the archaeal communities and suggest that the archaeal organisms may be thermophilic. The ambient temperatures of fissure water samples collected at Beatrix and West Driefontein, however, fall within the temperature range established for mesophiles. This implies that either the inference described above is incorrect or the fissure water originated from a greater depth (and hence a hotter environment) and migrated upwards, carrying its archaeal inhabitants with it. In the case of Beatrix Mine, the isotopic signatures are consistent with the latter hypothesis.
The most abundant rDNA clone in the fissure water sample obtained from Kloof Mine was closely related to Pyrococcus. Pyrococcus species are hyperthermophilic Archaea that previously have been found only in marine hydrothermal vent systems (46). The rDNA clones obtained from the fissure water of Kloof Mine may have been derived from Pyrococcus species living in anaerobic, saline groundwater at a depth of 5 to 6 km, where temperatures are in the hyperthermophile range, that migrated upward into Kloof Mine and mixed with meteoric freshwater. These are the first results that we are aware of that suggest the presence of Pyrococcus-like hyperthermophiles in the deep terrestrial subsurface.
The presence of diverse, novel Archaea sequences suggests that the South African gold mines and underlying strata contain unique thermophilic and perhaps hyperthermophilic habitats. The water moving through the fractures intersected during mining might convey microbial communities from deeper, more isolated habitats in some cases or from the surface in other cases. Thus, rDNA may provide important clues for elucidating details of hydrogeological fluid transport in this environment. Additional molecular, culture-based, and geochemical analyses are necessary, however, in order to understand the distribution, function, and interactions of deep subsurface Archaea.
ACKNOWLEDGMENTS
We gratefully acknowledge Goldfields Ltd. and Driefontein Consolidated for access to their mines and logistical support during sampling. We are also grateful for the assistance of Dawie Nel, Mel Haupf, and the other members of the East Driefontein Geology Departments who interpreted the mine stratigraphy and provided helpful comments during preparation of the manuscript. We also thank Jennifer Alexander, Margaret Grant, and others in the Microbiology Department of the University of Witwatersrand for laboratory support. We are grateful to M. Borscik of Princeton University for assistance with the geochemical analyses.
This research was supported by grant EAR-9714214 from the National Science Foundation LExEn program to Princeton University (to T. C. Onstott) and by National Geographic Society grant 6339-98 to T. C. Onstott for travel.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson D A, Karsch-Mizrachi I, Lipman D J, Ostell J, Rapp B A, Wheeler D L. GenBank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bidle K A, Kastner M, Bartlett D H. A phylogenetic analysis of microbial communities associated with methane hydrate containing marine fluids and sediments in the Cascadia margin (ODP site 892B) FEMS Microbiol Lett. 1999;177:101–108. doi: 10.1111/j.1574-6968.1999.tb13719.x. [DOI] [PubMed] [Google Scholar]
- 6.Bintrim S B, Donohue T J, Handelsman J, Roberts G P, Goodman R M. Molecular phylogeny of Archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckley D H, Graber J R, Schmidt T M. Phylogenetic analysis of nonthermophilic members of the kingdom Crenarchaeota and their diversity and their diversity and abundance in soils. Appl Environ Microbiol. 1998;64:4333–4339. doi: 10.1128/aem.64.11.4333-4339.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cary S C, Cottrell M T, Stein J L, Camacho F, Desbruyeres D. Molecular identification and localization of filamentous symbiotic bacteria associated with the hydrothermal vent annelid Alvinella pompejana. Appl Environ Microbiol. 1997;63:1124–1130. doi: 10.1128/aem.63.3.1124-1130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandler D P, Brockman F J, Fredrickson J K. Phylogenetic diversity of Archaea and Bacteria in a deep subsurface paleosol. Microb Ecol. 1998;36:37–50. doi: 10.1007/s002489900091. [DOI] [PubMed] [Google Scholar]
- 11.Chin K J, Lukow T, Conrad R. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl Environ Microbiol. 1999;65:2341–2349. doi: 10.1128/aem.65.6.2341-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig H. Isotopic variations in meteoric waters. Science. 1961;133:1702–1703. doi: 10.1126/science.133.3465.1702. [DOI] [PubMed] [Google Scholar]
- 13.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLong E F, Taylor L T, Marsh T L, Preston C M. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol. 1999;65:5554–5563. doi: 10.1128/aem.65.12.5554-5563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLong E F, Wu K Y, Prézolin B B, Jovine R V M. High abundance of Archaea in Antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 16.Dojka M A, Hugenholz P, Haack S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duane M J, Pigozzi G, Harris C. Geochemistry of some deep gold mine waters from the western portion of the Witwatersrand Basin, South Africa. J Afr Earth Sci. 1997;24:105–123. [Google Scholar]
- 18.Eaton A D, Clesceri L S, Greenburg A E. Standard methods for the examination of water and wastewater. 19th ed. Philadelphia, Pa: American Public Health Association, American Water Works Association, Water Pollution Control Federation; 1995. [Google Scholar]
- 19.Eder W, Ludwig W, Huber R. Novel 16S rRNA gene sequences retrieved from highly saline brine sediments of Kebrit Deep, Red Sea. Arch Microbiol. 1999;172:213–218. doi: 10.1007/s002030050762. [DOI] [PubMed] [Google Scholar]
- 20.Fuhrman J A, McCallum K, Davis A A. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 21.Grant S, Grant W D, Jones B E, Kato C, Li L. Novel archaeal phylotypes from an East African alkaline saltern. Extremophiles. 1999;3:139–145. doi: 10.1007/s007920050109. [DOI] [PubMed] [Google Scholar]
- 22.Grosskopf R, Stubner S, Liesack W. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl Environ Microbiol. 1998;64:4983–4989. doi: 10.1128/aem.64.12.4983-4989.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hershberger K L, Barns S M, Reysenbach A-L, Dawson S C, Pace N R. Crenarchaeota in low-temperature terrestrial environments. Nature. 1996;384:420. doi: 10.1038/384420a0. [DOI] [PubMed] [Google Scholar]
- 24.Hinrichs K U, Hayes J M, Sylva S P, Brewer P G, DeLong E F. Methane-consuming archaebacteria in marine sediments. Nature. 1999;398:802–805. doi: 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- 25.Jones B E, Grant W D, Duckworth A W, Owenson G G. Microbial diversity of soda lakes. Extremophiles. 1998;2:191–200. doi: 10.1007/s007920050060. [DOI] [PubMed] [Google Scholar]
- 26.Jones M Q W. Heat flow in the Witswaterrand Basin and environs and its significance for the South African shield geotherm and thickness. J Geophys Res. 1980;93:3243–3260. [Google Scholar]
- 27.Jurgens G, Lindström K, Saano A. Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl Environ Microbiol. 1997;63:803–805. doi: 10.1128/aem.63.2.803-805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato C, Li L N, Tamaoka J, Horikoshi K. Molecular analyses of the sediment of the 11000-m deep Mariana Trench. Extremophiles. 1997;1:117–123. doi: 10.1007/s007920050024. [DOI] [PubMed] [Google Scholar]
- 29.Kudo Y, Shibata S, Miyaki T, Aono T, Oyaizu H. Peculiar archaea found in Japanese paddy soils. Biosci Biotechnol Biochem. 1997;61:917–920. [PubMed] [Google Scholar]
- 30.Laue D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons Ltd.; 1991. pp. 115–147. [Google Scholar]
- 31.Liu W T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacGregor B J, Moser D P, Wheeler Alm E, Nealson K H, Stahl D A. Crenarchaeota in Lake Michigan sediment. Appl Environ Microbiol. 1997;63:1178–1181. doi: 10.1128/aem.63.3.1178-1181.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maidak B L, Cole J R, Lilburn T G, Parker C T, Saxman P R, Stredwick J M, Garrity G M, Li B, Olsen G J, Pramanik S, Schmidt T M, Tiedje J M. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 2000;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massana R, Murray A E, Preston C M, DeLong E F. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazor E, Verhagen B T. Dissolved ions, stable and radioactive isotopes and noble gases in thermal waters of South Africa. J Hydrol. 1983;63:315–329. [Google Scholar]
- 36.McInerney J O, Mullarkey M, Wernecke M E, Powell R. Phylogenetic analysis of Group I marine archaeal rRNA sequences emphasizes the hidden diversity within the primary group Archaea. Proc R Soc Lond Ser B Biol Sci. 1997;264:1663–1669. [Google Scholar]
- 37.Munson M A, Nedwell D B, Embley T M. Phylogenetic diversity of Archaea in sediment samples from a coastal salt marsh. Appl Environ Microbiol. 1997;63:4729–4733. doi: 10.1128/aem.63.12.4729-4733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nordstrom D K, Southam G. Geomicrobiology of sulfide mineral oxidation. In: Banfield J F, Nealson K H, editors. Geomicrobiology: interactions between microbes and minerals. Washington, D.C.: The Mineralogical Society of America; 1997. pp. 361–390. [Google Scholar]
- 39.Preston C M, Wu K Y, Molinski T F, DeLong E F. A psychrophilic crenarchaeon inhabits a marine sponge: Crenarchaeum symbiosum gen. nov. sp. nov. Proc Natl Acad Sci USA. 1996;93:6241–6246. doi: 10.1073/pnas.93.13.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schleper C, Holben W, Klenk H-P. Recovery of crenarchaeotal ribosomal DNA sequences from freshwater lake sediments. Appl Environ Microbiol. 1997;63:321–323. doi: 10.1128/aem.63.1.321-323.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stahl D A, Amann R. Development and application of nucleic acid probes in bacterial systematics. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley & Sons, Ltd.; 1991. pp. 205–248. [Google Scholar]
- 42.Takai K, Horikoshi K. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics. 1999;152:1285–1297. doi: 10.1093/genetics/152.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takai K, Horikoshi K. Molecular phylogenetic analysis of archaeal intron-containing genes coding for rRNA obtained from a deep-subsurface geothermal pool. Appl Environ Microbiol. 1999;65:5586–5589. doi: 10.1128/aem.65.12.5586-5589.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takai K, Horikoshi K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol. 2000;66:5066–5072. doi: 10.1128/aem.66.11.5066-5072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takai K, Sako Y. A molecular view of archaeal diversity in marine and terrestrial hot water environments. FEMS Microbiol Ecol. 1999;28:177–188. [Google Scholar]
- 46.Takai K, Sugai A, Itoh T, Horikoshi K. Palaeococcus ferrophilus gen. nov., sp. nov., a barophilic hyperthermophilic archaeon from a deep-sea hydrothermal vent chimney. Int J Syst Evol Microbiol. 2000;50:489–500. doi: 10.1099/00207713-50-2-489. [DOI] [PubMed] [Google Scholar]
- 47.Truesdell A H, Hulston J R. Isotopic evidence of environments of geothermal systems. In: Fritz P, Fontes J-C, editors. Handbook of environmental isotope geochemistry, vol. 1, The terrestrial environment. Amsterdam, The Netherlands: Elsevier; 1980. pp. 179–226. [Google Scholar]
- 48.Ueda T, Suga Y, Matsuguchi T. Molecular phylogenetic analysis of a soil microbial community in a soybean field. Eur J Soil Sci. 1995;46:415–421. [Google Scholar]
- 49.Vetriani C, Jannasch H W, MacGregor B J, Stahl D A, Reysenbach A L. Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl Environ Microbiol. 1999;65:4375–4384. doi: 10.1128/aem.65.10.4375-4384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vetriani C, Reysenbach A L, Dore J. Recovery and phylogenetic analysis of archaeal rRNA sequences from continental shelf sediments. FEMS Microbiol Lett. 1998;161:83–88. doi: 10.1111/j.1574-6968.1998.tb12932.x. [DOI] [PubMed] [Google Scholar]