Abstract
An agar-degrading marine bacterium identified as a Microscilla species was isolated from coastal California marine sediment. This organism harbored a single 101-kb circular DNA plasmid designated pSD15. The complete nucleotide sequence of pSD15 was obtained, and sequence analysis indicated a number of genes putatively encoding a variety of enzymes involved in polysaccharide utilization. The most striking feature was the occurrence of five putative agarase genes. Loss of the plasmid, which occurred at a surprisingly high frequency, was associated with loss of agarase activity, supporting the sequence analysis results.
Bacteria have long been exploited as a source of natural products for use in medicine, agriculture, and industry. Until recently, most studies utilized microbes that had been isolated from clinical or terrestrial environments. It is now recognized that bacteria living in marine environments provide an abundant and as-yet-untapped source of metabolic properties (8). Marine sediments are rich in bacteria, with reports of 108 to 1010 microbes per g of sediment for the first few centimeters of sediment (21). Estimates of bacterial diversity in natural environments have indicated that, while a few organisms may predominate, such environments still represent a highly complex assemblage of microbes (45).
We have initiated a study to identify and characterize the genes present on plasmids isolated from bacteria present in coastal marine sediments. Extrachromosomal elements by definition encode functions that are not essential for cell growth but which provide an advantage to the host bacterium under certain growth conditions. It is therefore not surprising that a wide variety of traits in bacteria have been found to be plasmid encoded. In an earlier study (41), ca. 30% of more than 1,000 aerobic heterotrophic bacteria isolated from coastal California marine sediments contained at least one plasmid that ranged in size from 5 to >250 kb. These plasmids appeared to contain novel and generally uncharacterized replication regions since no homology was detected between ca. 300 plasmids of marine origin (41) and 15 replicon probes derived from plasmids found in bacteria isolated from mammalian or terrestrial sources (10). These findings suggested that plasmids in marine sediment microbial communities are a unique and diversified set of extrachromosomal elements.
We present here the characterization of an agar-degrading marine isolate, Microscilla sp. strain PRE1. This organism contains a 101-kb plasmid, designated pSD15, which potentially encodes five different agarases and is essential for the ability of the bacterium to degrade agar. The complete DNA sequence and analysis of pSD15 are reported.
MATERIALS AND METHODS
Isolation and characterization of marine bacterial isolate PRE1.
Marine sediment associated with the roots of pickleweed (Salicornia virginica) was collected from the Kendall-Frost Mission Bay Marsh Reserve in San Diego, Calif. The sediment was resuspended in artificial seawater (0.3 M NaCl, 0.1 M KCl, 0.01 M CaCl2, 0.05 M MgSO4), and serial dilutions were plated on M7 medium (0.1% tryptone in artificial seawater) solidified with 15 g of agar per liter. The plates were incubated for 2 to 4 days at 30°C. Isolated colonies, selected on the basis of differing morphology and color, were patched on to M7 medium and the bacteria were screened for total plasmid content by a modification of the Kieser method (22) as described previously (41). One isolate, PRE1, which appeared to degrade agar as detected by pitting around each colony and which contained a single covalently closed circular DNA plasmid ca. 100 kb in size, was selected for further study.
The morphology and cell size of isolate PRE1 were determined in an exponentially growing culture by measuring 100 cells after staining of the cell membrane with FM4-64 at 5 μg/ml (Molecular Probes, Eugene, Oreg.) and of the DNA with DAPI (4′,6′-diamidino-2-phenylindole) at 5 μg/ml. Cells were also Gram stained by the standard method. Growth at 25, 30, 37, 42, and 45°C was monitored in M10 broth (0.25% yeast extract–0.4% tryptone, prepared in artificial seawater). The ability to grow in different NaCl concentrations was determined by shaking cultures for 24 h at 30°C in M10 broth modified to contain NaCl concentrations from 0.5 to 6%. The growth requirement for K+, Mg2+, or Ca2+ was tested in liquid M10 medium prepared with artificial seawater missing the cation in question.
Sensitivity to antibiotics was tested for both isolate PRE1 and a derivative cured of plasmid pSD15. Antibiotic discs (Difco/BD Biosciences, Franklin Lakes, N.J.) containing gentamicin (10 μg), kanamycin (10 μg), neomycin (30 μg), streptomycin (10 μg), ampicillin (10 μg), trimethoprim (5 μg), rifampin (5 μg), tetracycline (30 μg), erythromycin (15 μg), carbenicillin (100 μg), chloramphenicol (30 μg), nalidixic acid (30 μg), and novobiocin (30 μg) were applied to lawns of the two strains spread on M10 plates. Plates were incubated for 24 to 36 h at 30°C, and resistance was determined by the method of Bauer et al. (5).
Identification of PRE1 by using 16S ribosomal DNA (rDNA) sequence.
For 16S rDNA analysis, genomic DNA was purified from 1 ml of an overnight culture of PRE1 grown at 30°C in M10 broth by using the QIAampTissue Kit (Qiagen, Chatsworth, Calif.). The 16S rDNA gene was amplified from 0.1 to 0.5 μg of genomic DNA by using the primers fD1 and rD1 (47) and Taq2000 DNA polymerase (Stratagene, La Jolla, Calif.) as described previously (42). The amplified products were electrophoresed on a 0.8% agarose gel, and a fragment of ca. 1.5 kb was purified by using GeneClean II (Q-Biogene, Carlsbad, Calif.). Partial DNA sequences were obtained for the 1.5-kb fragment by using three primers corresponding to the following positions of the Escherichia coli 16S rRNA gene sequence: primer A, positions 519 to 536; primer B, positions 907 to 926; and primer C, positions 1392 to 1406 (25). Sequence alignment was carried out by using the Ribosomal Database Project II online analysis tools with the SSU prokaryotic data set (26).
Plasmid pSD15 stability.
A single colony of Microscilla sp. strain PRE1 able to degrade agar (as evidenced by pitting of the solid medium) was picked from a fresh plate and resuspended in 5 ml of M10 broth. The culture was incubated at 30°C with shaking, and at 15 h (mid-log phase), 20 h (late log phase), and 26 h (stationary phase), a 10-μl aliquot was removed, serially diluted, and spread on M10 agar plates. After incubation at 30°C for ca. 36 h, by which time tiny colonies had appeared, single colonies were patched onto M10 plates, which were then incubated at 30°C. The number of patched colonies capable of degrading agar could be determined by direct visualization after 18 to 24 h. The assay was repeated twice, starting each time with a unique single colony isolate. The results reported are for a total of 500 colonies for each growth phase from the three repetitions.
Isolation of supercoiled plasmid DNA.
A 5-ml overnight culture of Microscilla sp. strain PRE1 grown in M10 broth was transferred to 1 liter of M10 broth and incubated with vigorous aeration at 30°C. To avoid excess polysaccharide formation, the culture was harvested after ca. 13 h of incubation (mid-exponential phase). Supercoiled plasmid DNA was prepared as described previously (42) by using the alkaline lysis method of Birnboim and Doly (9), except that RNase A was omitted from the first solution and that the sample was not extracted with phenol-chloroform prior to precipitation by isopropanol. Plasmid DNA was subsequently purified by two rounds of cesium chloride-ethidium bromide gradient centrifugation.
Plasmid library construction.
A small-insert random-fragment library was constructed from purified pSD15 DNA as follows. First, 5 to 10 μg of pSD15 DNA was sheared by sonication, treated with Bal 31 nuclease followed by T4 DNA polymerase to repair the ends, and then size fractionated by agarose gel electrophoresis. Fragments 2 to 4 kb in size were recovered from the gel and ligated into the SmaI site of commercially available bacterial alkaline phosphatase (BAP)-treated pUC18 (Amersham Pharmacia Biotech, Piscataway, N.J.). The ligation mixture was subjected to StrataClean Resin (Stratagene, La Jolla, Calif.) treatment to remove ligase and then transformed into E. coli XL1-Blue by electroporation. An aliquot of the transformants was plated on a Luria-Bertani (LB) agar containing ampicillin (100 μg/ml), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 32 μg/ml), and IPTG (isopropyl-β-d-thiogalactopyranoside; 32 μg/ml) to determine the percentage of recombinants based on blue and white screening. The range of insert sizes was determined by a convenient whole-cell cracking procedure (Promega Protocols and Application Guide, 2nd ed., 1991 [Promega, Madison, Wis.]).
DNA sequencing and data annotation.
Sanger sequencing reactions were performed on 1,824 randomly chosen subclones by using BigDye Terminators (Applied Biosystems) and then analyzed on ABI377 automated sequencing machines (Applied Biosystems). This generated seven- to eightfold sequence coverage across the 100-kb plasmid. The sequences were evaluated for quality and error probability by using the program phred (15), assembled by using the phrap assembler (14), and viewed by using consed (16). All contigs were ordered and oriented, and all gaps were closed by using a directed primer walking strategy. A final quality value of phred40 (1-base error in 10,000 bases) with no gap regions, double coverage, or two chemistries across single-stranded areas was achieved.
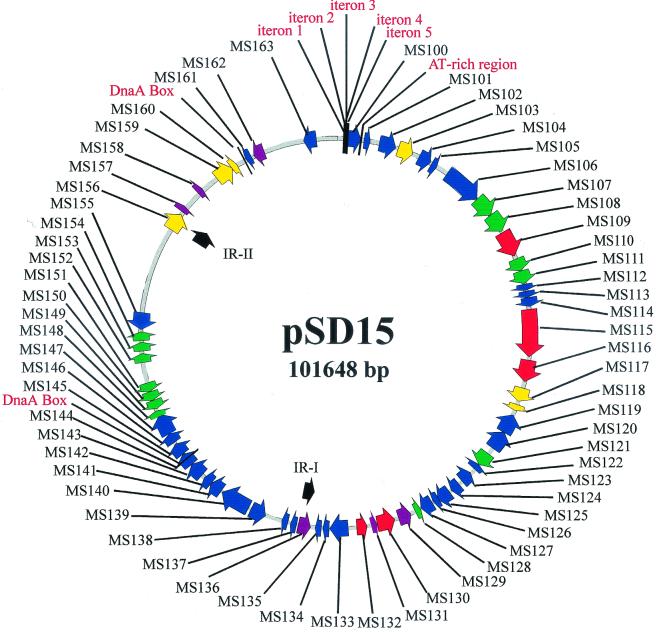
The pSD15 DNA sequence was analyzed by using the computer programs Vector NTI (version 5.5; InforMax, North Bethesda, Md.) and the GCG Wisconsin Package (Oxford Molecular Group). The initial assignments of putative open reading frames (ORFs) were made according to the criteria that (i) the start codon was ATG or GTG; (ii) the stop codon was TAA, TGA, and TAG; and (iii) the ORF was more than 100 amino acids in length. TestCode analysis, in the GCG Wisconsin package, was used to identify those ORFs most likely to express a gene. ORFs likely to code for a protein were given the designations MS100 to MS163, starting with MS100 and proceeding clockwise as shown on the map in Fig. 1. Putative ORFs were compared to the protein databases by using BLASTX (1). Only searches that returned E values of <10−5 were considered sufficiently significant to be reported in Table 1.
FIG. 1.
Graphical map of plasmid pSD15. The putative ORFs identified on pSD15 are indicated by arrows. ORFs related to agarases are red, those related to mobile elements are yellow, ORFs similar to noncharacterized proteins are purple, ORFs with no homologs in GenBank are green, and all other ORFs are dark blue. The ORFs are more fully described in Table 1. Also indicated on the map are the large inverted repeats, IR-I and IR-II, putative DnaA boxes, and the five iterons and an AT-rich region that may play a role in replication.
TABLE 1.
Putative pSD15 ORFs and their functions
| pSD15 ORF | Gene position
(nucleotide)
|
Strand | Size of putative protein (aa) | Function, closest match | GenBank accession no. of closest match | pSD15 ORF alignment region (aa, range) | Homolog alignment region [aa range (total aa)] | % Identity | |
|---|---|---|---|---|---|---|---|---|---|
| Start | Stop | ||||||||
| MS100 | 871 | 1911 | + | 347 | Replication protein (RepA), Riemerella anatipestifer | AAD33095 | 115–298 | 95–263 (309) | 25 |
| MS101 | 2210 | 3049 | + | 280 | Partition protein (SOJ homolog), Pyrococcus abyssi | B75038 | 2–248 | 4–251 (257) | 31 |
| MS102 | 3502 | 4929 | + | 478 | Replicative DNA helicase (DnaB), Bacillus stearothermophilus | AAD20314 | 34–473 | 14–438 (454) | 36 |
| MS103 | 5139 | 6377 | + | 413 | Transposase (similar to ISRm3) from plasmid pMT1, Yersinia pestis | AAC13227 | 49–402 | 46–398 (402) | 46 |
| MS104 | 6997 | 8046 | + | 350 | 2-Keto-3-deoxygluconate kinase (KdgK), Thermotoga maritima | AAD35161 | 5–350 | 2–339 (339) | 47 |
| MS105 | 8173 | 8838 | + | 222 | 2-Dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase (KdpG), Thermotoga maritima | AAD35160 | 14–209 | 8–199 (205) | 38 |
| MS106 | 9720 | 12986 | + | 1089 | Outer membrane protein, Porphyromonas gingivalis | CAA10226 | 48–1089 | 2–1017 (1017) | 25 |
| MS107 | 13001 | 14716 | + | 572 | ORF, no homolog | ||||
| MS108 | 14742 | 16355 | + | 538 | ORF, no homolog | ||||
| MS109 | 16469 | 18676 | + | 736 | β-Agarase, Pseudoalteromonas atlantica | P13734 | 29–485 | 26–482 (505) | 38 |
| MS110 | 18732 | 19955 | + | 408 | Hypothetical protein from Streptomyces coelicolor A3(2) | CAB61805 | 54–407 | 7–362 (370) | 50 |
| MS111 | 19967 | 21235 | + | 423 | ORF, no homolog | ||||
| MS112 | 21352 | 21711 | + | 120 | Na+/glucose symporter, Vibrio parahaemolyticus | BAA11215 | 9–117 | 1–109 (530) | 78 |
| MS113 | 21813 | 22124 | + | 104 | Na+/glucose symporter, Vibrio parahaemolyticus | BAA11215 | 1–83 | 147–229 (530) | 60 |
| MS114 | 22054 | 22953 | + | 300 | Na+/glucose symporter, Vibrio parahaemolyticus | BAA11215 | 2–300 | 231–530 (530) | 56 |
| MS115 | 23245 | 27234 | + | 1330 | β-Agarase precursor, Pseudoalteromonas atlantica | P13734 | 25–524 | 26–482 (505) | 33 |
| MS116 | 27486 | 29327 | + | 614 | β-Agarase B precursor, Cytophaga drobachiensis | AF098955 | 37–377 | 59–351 (353) | 38 |
| MS117 | 29826 | 31211 | + | 462 | Maturase, Pseudomonas alcaligenes | AAB68949 | 45–452 | 55–464 (490) | 43 |
| MS118 | 31698 | 32120 | + | 141 | Transposase of ISW1, Wolbachia sp. | BAA73610 | 1–137 | 1–137 (144) | 49 |
| MS119 | 33875 | 32256 | − | 540 | Fucosidase, Thermotoga maritima | AAD35394 | 53–476 | 5–392 (449) | 31 |
| MS120 | 35465 | 33879 | − | 529 | Fucosidase, Thermotoga maritima | AAD35394 | 45–483 | 8–415 (449) | 30 |
| MS121 | 35878 | 37314 | + | 479 | ORF, no homolog | ||||
| MS122 | 38281 | 37469 | − | 271 | Esterase, Caldicellulosiruptor saccharolyticus | P23553 | 43–266 | 34–264 (266) | 27 |
| MS123 | 39417 | 38284 | − | 378 | Alcohol dehydrogenase, Streptomyces coelicolor A3(2) | CAB46803 | 13–377 | 8–365 (365) | 31 |
| MS124 | 39589 | 40437 | + | 283 | Transcriptional activator (MtlR), Pseudomonas fluorescens | AAC34292 | 18–276 | 22–290 (301) | 24 |
| MS125 | 40616 | 41773 | + | 386 | Cytochrome P-450, Bacillus subtilis | P27632 | 75–364 | 86–385 (405) | 31 |
| MS126 | 41809 | 42114 | + | 102 | Ferredoxin, Caulobacter crescentus | P37098 | 1–100 | 1–101 (106) | 45 |
| MS127 | 42170 | 43420 | + | 417 | Ferredoxin reductase, Pseudomonas putida | P16640 | 9–415 | 8–417 (422) | 41 |
| MS128 | 44151 | 43609 | − | 181 | ORF, no homolog | ||||
| MS129 | 45397 | 44336 | − | 354 | Hypothetical protein, Mycobacterium tuberculosis | CAB06436 | 48–351 | 8–295 (299) | 41 |
| MS130 | 47228 | 45738 | − | 497 | β-Agarase, Pseudoalteromonas atlantica | P13734 | 1–237 | 241–467 (505) | 35 |
| MS131 | 47791 | 47267 | − | 175 | Hypothetical protein (YwoF), Bacillus subtilis | CAB05379 | 51–127 | 31–107 (468) | 41 |
| MS132 | 48975 | 48112 | − | 288 | β-Agarase, Streptomyces coelicolor | P07883 | 31–286 | 44–309 (309) | 24 |
| MS133 | 49678 | 51213 | + | 512 | Arylsulfatase A, Mus musculus | P50428 | 28–428 | 15–372 (506) | 32 |
| MS134 | 51265 | 51870 | + | 202 | Arylsulfatase E, Homo sapiens | NP_000038 | 31–185 | 30–189 (589) | 39 |
| MS135 | 51955 | 52701 | + | 249 | Arylsulfatase B, Mus musculus | ARSB_MOUSE_2 | 1–153 | 99–252 (252) | 36 |
| MS136 | 54108 | 52855 | − | 418 | Hypothetical protein, Staphylococcus aureus | BAA86649 | 1–418 | 30–464 (507) | 35 |
| MS137 | 54073 | 54624 | + | 184 | Dehydrogenase, Haemophilus influenzae | Q57517 | 15–183 | 169–342 (342) | 26 |
| MS138 | 54975 | 55412 | + | 146 | Dehydrogenase, Synechocystis sp. | BAA18061 | 4–137 | 106–243 (249) | 36 |
| MS139 | 58035 | 56464 | − | 524 | GGDEF family protein, Deinococcus radiodurans | F75583 | 37–154 | 74–180 (533) | 24 |
| MS140 | 58243 | 60804 | + | 854 | Beta-galactosidase, Thermoanaerobacter ethanolicus | P77989 | 37–436 | 7–384 (743) | 28 |
| MS141 | 60926 | 61942 | + | 339 | Dehydrogenase, Haemophilus influenzae | Q57517 | 29–328 | 33–332 (342) | 28 |
| MS142 | 61972 | 62739 | + | 256 | Gluconate dehydrogenase, Bacillus subtilis | P50842 | 5–256 | 6–254 (254) | 51 |
| MS143 | 62770 | 64227 | + | 486 | Aldehyde dehydrogenase, E. coli | P25553 | 3–483 | 1–478 (479) | 42 |
| MS144 | 64230 | 65357 | + | 376 | Racemase, Pseudomonas putida | 443131 | 113–361 | 110–355 (357) | 33 |
| MS145 | 65362 | 66414 | + | 351 | Aldose epimerase, Acinetobacter calcoaceticus | P05149 | 6–343 | 26–377 (381) | 38 |
| MS146 | 66414 | 67415 | + | 334 | l-Rhamnose transporter, E. coli | P27125 | 4–330 | 8–341 (344) | 19 |
| MS147 | 67545 | 69335 | + | 597 | Chemotaxis protein, Bacillus subtilis | P39217 | 195–553 | 295–653 (662) | 26 |
| MS148 | 69402 | 69731 | + | 110 | ORF, no homolog | ||||
| MS149 | 70199 | 69765 | − | 145 | ORF, no homolog | ||||
| MS150 | 70934 | 70287 | − | 216 | ORF, no homolog | ||||
| MS151 | 71423 | 70947 | − | 159 | ORF, rather weak similarity to HigB, proteic killer active protein from plasmid Rts1 (JC4694, 29% over 77 amino acids) | ||||
| MS152 | 74351 | 75130 | + | 260 | ORF, no homolog | ||||
| MS153 | 75148 | 75906 | + | 253 | ORF, no homolog | ||||
| MS154 | 75920 | 76501 | + | 194 | ORF, no homolog | ||||
| MS155 | 77927 | 76518 | − | 470 | Mobilization protein (MocB), Bacteroides fragilis | B48487 | 71–206 | 61–213 (318) | 29 |
| MS156 | 85243 | 86772 | + | 510 | Transposase, Staphylococcus aureus prophage phiPV83 | NP_061635 | 34–510 | 27–520 (548) | 36 |
| MS157 | 87182 | 87874 | + | 231 | Hypothetical protein, Lactobacillus bacteriophage phi-adh | CAB52516 | 118–227 | 25–131 (170) | 23 |
| MS158 | 89336 | 90091 | + | 252 | Hypothetical protein, Pseudomonas aeruginosa | AAG03933 | 1–235 | 1–232 (247) | 29 |
| MS159 | 91015 | 92565 | + | 517 | Transposase, Pseudomonas alcaligenes | AAB48471 | 10–516 | 9–508 (509) | 34 |
| MS160 | 92534 | 93319 | + | 262 | Transposase, Rhizobium sp. strain NGR234 | P55617 | 25–258 | 6–239 (245) | 41 |
| MS161 | 94522 | 93842 | − | 227 | HsdS of type I restriction-modification system, E. coli | CAA10700 | 1–197 | 8–205 (463) | 22 |
| MS162 | 95628 | 94525 | − | 368 | Hypothetical protein, Xylella fastidiosa | AAF84517 | 15–342 | 10–336 (371) | 26 |
| MS163 | 100016 | 98931 | − | 362 | HsdM of type I restriction-modification system, Xylella fastidiosa | AAF85527 | 6–362 | 16–351 (519) | 44 |
Direct sequence repeats (iterons) were detected by using a modified version of the program Internal Repeat Finder, which was created and modified by Matteo Pellegrini (http://www.doe-mbi.ucla.edu/people/matteo/repeats.html). The G+C content was determined by using MacVector, while the GC profile was obtained with the BioPlot module of Vector NTI with a window size of 500. SignalP V1.1 (http://www.cbs.dtu.dk/services/SignalP/) was used to identify possible signal sequences in the putative agarases encoded by pSD15.
The GenBank nucleotide sequence accession number for pSD15 is AF339846.
RESULTS AND DISCUSSION
Morphology and growth characteristics of isolate PRE1.
Isolate PRE1 is a gram-negative rod-shaped bacterium ca. 6 μm in length. Colonies growing on M10 or M7 solid medium are pigmented orange and exhibit a strong agar-degrading activity. The strain grows optimally at 30°C, more slowly at 25°C, very poorly at 37 or 42°C, and not at all at 45°C. Cultures grown on solid medium at 30°C lose their viability very rapidly upon storage at room temperature.
Salt-dependency testing indicates that isolate PRE1 is truly a marine bacterium, requiring NaCl for growth. Growth occurs over a range of 0.5 to 6% NaCl, but significant growth only occurs at between 1.5 and 3% NaCl. K+ can be omitted from artificial seawater but Mg2+ and Ca2+ are indispensable for growth.
The 16S rRNA gene is a useful target for establishing prokaryotic phylogeny based on sequence similarity (25). Analysis of the sequence obtained for isolate PRE1 by using three conserved primers indicated that it is a Microscilla sp. in the Flexibacter group belonging to the Flexibacter-Cytophaga-Bacteroides phylum. The characteristics of PRE1, as described above, also fit the description of Microscilla species given in Bergey's Manual of Determinative Bacteriology (7).
Microscilla sp. strain PRE1 was resistant to gentamicin, kanamycin, neomycin, streptomycin, ampicillin, and trimethoprim and sensitive to rifampin, tetracycline, erythromycin, carbenicillin, chloramphenicol, nalidixic acid, and novobiocin at the concentrations tested. An identical pattern of resistance and sensitivity was observed for the strain cured of the pSD15 plasmid. This suggested the absence of any common antibiotic resistance determinants on pSD15, which was later confirmed by the sequencing results.
Agarase activity and plasmid stability.
The ability to degrade agar is a striking feature of Microscilla sp. strain PRE1 grown on solid medium. During the initial culturing of the organism, colonies with the same morphology but unable to degrade agar appeared at a relatively high frequency. Analysis of cells from these colonies by Gram staining, microscopic observation, antibiotic susceptibility, and 16S rDNA analysis indicated that they were identical to Microscilla sp. strain PRE1. The possibility that the loss of agar-degrading ability was related to loss of the native plasmid, pSD15, was therefore examined.
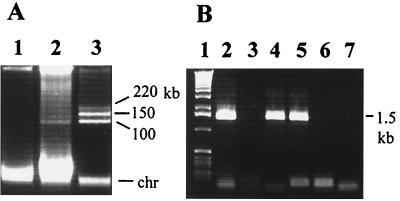
Several colonies which were no longer able to degrade agar were screened for plasmid content. While the native 101-kb plasmid was detected in the original parent, plasmid DNA was not detected in any of the strains which lost the ability to degrade agar (Fig. 2A; strains unable to degrade agar were designated PRE1-0). To confirm that plasmid pSD15 was indeed missing from these strains and did not integrate into the chromosome, we screened for its presence by PCR of total DNA by using a pair of primers specific to pSD15. To do so, a clone was selected from the library prepared for sequencing pSD15, and both ends of the ca. 2-kb insert were sequenced. On the basis of this sequence, two primers were designed and used to PCR amplify DNA from cell lysates prepared by boiling a suspension of cells from several colonies of Microscilla PRE1 or PRE1-0. As shown in Fig. 2B, a PCR fragment of the expected size was obtained from colonies able to degrade agar, while no product appeared from colonies unable to degrade agar. This confirmed that the nondegrading isolates arose from the parent Microscilla sp. strain PRE1 after loss of the native 101-kb plasmid, pSD15.
FIG. 2.
Plasmid pSD15 is not present in non-agar-degrading isolates. (A) Total DNA isolated from Microscilla colonies that had lost the ability to degrade agar (lane 1; PRE1-0) and from colonies capable of degrading agar (lane 2; PRE1). Lane 3 is DNA isolated from Rhizobium meliloti, harboring three plasmids, used as a molecular mass marker with sizes as indicated. The position of chromosomal DNA on the gel is also indicated (chr). (B) PCR amplification to identify the presence of a 1.5-kb pSD15 fragment in PRE1 colonies (lane 2), PRE1-0 colonies (lane 3), supercoiled pSD15 DNA (lane 4), a library clone carrying this fragment (from which primers were designed) (lane 5), and a library clone selected at random (lane 6). Lane 7 is a no-DNA template control. Lane 1 has a molecular mass marker (1-kb ladder; Gibco).
The stability of pSD15 in Microscilla sp. strain PRE1 was then tested by using the ability to degrade agar as a phenotypic marker for the presence of the plasmid. Colonies from mid-exponential-, late-exponential-, and stationary-phase cultures were checked for the ability to degrade agar on M10 medium as described in Materials and Methods. On average 2% of the cells at each stage of growth had lost the plasmid. This is a surprisingly high loss rate even for a low-copy-number plasmid. Most naturally occurring plasmids are lost at frequencies of 10−5 to 10−4 per generation. As discussed below, sequence analysis of pSD15 did not provide any evidence for a stabilization mechanism (such as partitioning, postsegregational killing, or conjugal transfer), which would help to limit its loss from the host. It is possible that this high loss rate is a result of culturing the strain in the lab as opposed to growth in the natural environment, where the ability to degrade agar (or any other traits conferred on the host by the plasmid) may provide enough of a selective pressure to maintain the plasmid. Another possibility is that Microscilla sp. strain PRE1 is not the “natural” host for pSD15. However, it should be noted that the putative replication initiation protein of pSD15, MS100, is very similar to replication initiation proteins of plasmids isolated from Riemerella anatipestifer, another member of the Flexibacter-Cytophaga-Bacteroides group of bacteria (see Table 1).
Sequence analysis.
pSD15, the single plasmid found in Microscilla sp. strain PRE1, is a circular plasmid of 101,648 bp (Fig. 1). It has a total of 98 ORFs larger than 100 amino acids, of which 64 are predicted to code for a protein as determined by TestCode analysis. Of these 64 putative proteins, 46 are coded for on one strand and 18 are coded on the other. Possible functions of these putative proteins, based on amino acid sequence similarities, are given in Table 1.
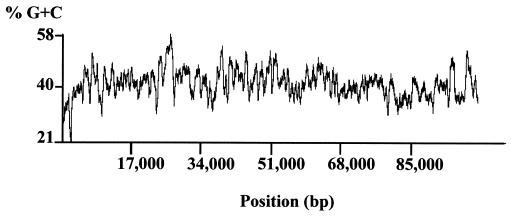
Analysis of the base composition of pSD15 revealed an overall G+C content of 42%. The chromosomal G+C content of Microscilla furvescens, Microscilla sp. strain PRE1's closest match in the RDP database, is reported to be 44% (7). As shown in Fig. 3, except for an AT-rich region located in the putative replication origin, the G+C content is distributed evenly throughout pSD15.
FIG. 3.
Percent G+C profile of pSD15. The GC profile was obtained by the BioPlot module of Vector NTI (version 5.5) with a window size of 500.
In general, pSD15 is a catabolic plasmid, conferring the ability to degrade and utilize polysaccharide polymers such as agar. A small part of the plasmid is dedicated to autonomous plasmid replication functions, and there are some additional putative functions, such as a DNA modification system and several transposases. In addition, ca. 25% of the plasmid, from about position 78000 to position 101648, has little coding sequence. Of the eight ORFs in this region (designated MS156 to MS163), three (MS156, MS159, and MS160) are similar to transposases, two appear to encode a restriction modification system (MS161 and MS163), and three are similar to proteins of unknown function from other organisms.
Analysis of putative agarase genes.
The most remarkable feature of pSD15 is the presence of five different genes potentially encoding agarases. Agar-degrading bacteria are ubiquitous in marine environments but are found as well in freshwater, sewage, and soil. These bacteria belong to diverse genera, including Alterococcus (38), Bacillus (23), Pseudomonas (17), and Microscilla (31), as well as those cited in Table 2. Bacteria capable of fixing nitrogen while growing on agar (39) or growing anaerobically with agar as the sole carbon source (38) have been isolated. Agarases have been purified and characterized from several different bacteria (2, 3, 23, 31, 36, 44, 46, 49), and in several cases the proteins have been overexpressed (24, 34, 35). Several of the agarase-encoding genes have also been cloned but no organism has been found to encode more then two agarases (Table 2).
TABLE 2.
Previously sequenced agarase genes
| Agarase gene | Organism | Putative protein (no. of
aa)a
|
Type of agarase | Accession no. | |
|---|---|---|---|---|---|
| Signal peptide | Mature peptide | ||||
| aagA | Aeromonas sp. | –b | 290 | β-Agarase | AAF03246 |
| agaA | Alteromonas agarilytica | 26 | 1,403 | α-Agarase | AAF26838 |
| agaA | Cytophaga drobachiensis | 19 | 520 | β-Agarase | AAF21820 |
| agaB | Cytophaga drobachiensis | 18 | 335 | β-Agarase | AAF21821 |
| agrA | Pseudoalteromonas atlantica | 23 | 482 | β-Agarase | P13734 |
| dagA | Pseudoalteromonas atlantica | 21 | 269 | β-Agarase | M73783 |
| dagA | Streptomyces coelicolor | 30 | 279 | β-Agarase | CAA29257 |
| agaA | Vibrio sp. | – | 995 | β-Agarase | BAA03541 |
| agaB | Vibrio sp. | – | 955 | β-Agarase | BAA04744 |
aa, amino acids.
–, no signal peptide reported.
The agarase-encoding genes of pSD15, MS109, MS115, MS116, MS130, and MS132 are dispersed in a 32.5-kb region from bp 16469 to bp 48975. An alignment of the deduced amino acid sequence for the five genes was performed by using Vector NTI after the introduction of gaps to compensate for size differences between the proteins. The analysis did not identify any region of homology common to all five putative agarases. However, the C-terminal two-thirds of MS109 (from amino acids 241 to 736) showed 32.7% identity to MS130, while the N-terminal half of MS116 (amino acids 1 to 311) showed 18.4% identity to MS132. The method of Belas (6) was also used to compare the predicted protein sequences for the five genes. The amino acid sequences were first simplified by categorizing each residue according to the general functional properties: (i) neutral, weakly hydrophobic (P, A, G, S, or T residues); (ii) hydrophilic, acid amine (Q, N, E, and D); (iii) hydrophilic, basic (H, K, and R); (iv) hydrophobic (L, I, V, and M); (v) hydrophobic, aromatic (F, Y, and W); or (vi) cross-link forming (C). Again, there was no single region common to all five proteins.
The five putative agarases were then compared to the deduced amino acid sequence data for nine different agarase genes available in public databases (described in Table 2). As shown in Table 1, three putative agarases from pSD15, MS109, MS115, and MS130 match with the agrA-encoded protein from Pseudoalteromonas atlantica. MS116 is most similar to the β-agarase B precursor of Cytophaga drobachiensis, while MS132 is most similar to β-agarase from Streptomyces coelicolor. When the deduced amino acid sequences for all 14 agarase proteins were aligned (five from pSD15 plus the nine previously reported), only a few randomly distributed residues were found to be conserved in all 14 proteins (data not shown). This result agrees with a prior conclusion that microorganisms appear to degrade agar by using a series of enzymes with narrow specificities rather than a single enzyme with a broad specificity (43).
A signal peptide is common to most secreted proteins. Six of the nine previously reported agarases contain a signal peptide of 18 to 30 amino acids in length (Table 2). Analysis of the five putative agarases encoded by pSD15 by using the SignalP program resulted in the identification of a signal peptide within the first 40 amino acids for four of the proteins (Table 3) (32).
TABLE 3.
Signal peptides identified in the N-terminal 40 amino acids of putative agarases encoded by pSD15a
| ORF | Sequence |
|---|---|
| MS109 | MRIKIWISYGLCFILMIFLGSDILA↓QKVEVDVQFNVKHVV |
| MS115 | MYGKVVLFTVLFLGNIFCLYSQ↓GVQ↓VDVNLNVKHSVGGVS |
| MS116 | MKSPIVIYYFTKKPSNMKTHLTLLITWIAFLGAKA↓QDWSG |
| MS132 | MKKTYLGLALLFIAYQSTLAQS↓QPTVNEGEPVAQLEWELV |
No signal peptide was identified in MS130. The downward arrow indicates the position(s) of the predicted cleavage site(s).
Previous studies have indicated that there are two pathways for the degradation of agar, which is a linear sulfated galactan composed of two regularly repeated galactose units alternatively linked by β-d-(1,4) and α-l-(1,3) linkages. The β-agarases cleave the β-d-(1,4) linkages, while the α-agarases cleave α-l-(1,3) linkages. As shown in Table 2, most purified agarases have been identified as β-agarases, with only the Alteromonas agarilytica enzyme being an α-agarase. The putative agarases encoded by pSD15 on the basis of amino acid sequence are most similar to β-agarases; however, confirmation of this must await purification and biochemical analysis of the activity of these proteins.
Replication and plasmid maintenance.
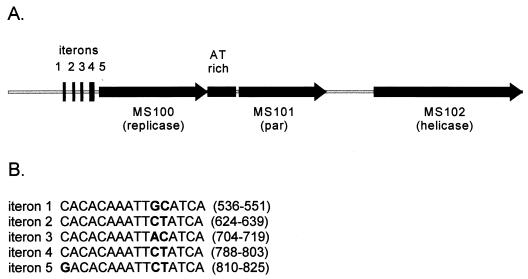
The region from position 1 to bp 4929 of pSD15 has an organization typical of many prokaryotic replicons (19). It includes five direct repeats (putative iterons), an ORF (MS100) encoding a putative 374-amino-acid replication initiation protein, and a 316-bp AT-rich region (86% AT; positions 1916 to 2231). The five iterons share a 16-bp consensus sequence and are separated by 73, 65, 69, and 7 bp, respectively (Fig. 4). Immediately downstream of the AT-rich region are the coding regions for two proteins, MS101 and MS102, which share similarities, respectively, to a component of the partitioning system from Borrelia burgdorferi and the replicative helicase, DnaB, of Bacillus stearothermophilus. Two putative DnaA box sequences, TTATCCACA and TGTGGATAA, are distantly separated from each other, and both boxes were located a considerable distance from the origin of replication, making them unlikely to be involved in replication (see Fig. 1).
FIG. 4.
Putative replication region of pSD15. (A) A 5-kb region of pSD15 contains a putative replicase (MS100), helicase (MS102), partition protein (MS101), five iterons, and a 316-bp AT-rich region. (B) The sequence of the five iterons is shown, with the base-pair position on pSD15 indicated in parentheses. Mismatched bases are indicated in boldface.
In an attempt to clone a fragment of pSD15 capable of autonomous replication, a ca. 5-kb fragment containing the five iterons, the AT-rich region, and ORFs MS100 to MS102 was obtained by PCR and cloned into pBR325 (which requires PolA for its replication). Attempts to introduce this construct, designated pZZ3, into a polA E. coli strain by electroporation failed, suggesting that the cloned replicon could not function in E. coli. Since pBR325 can be mobilized (4), attempts were then made to conjugally transfer pZZ3 into PRE1-0 (the Microscilla isolate cured of the native plasmid) by using E. coli SM10 (40) (which has the Tra+ plasmid RP4 integrated in the chromosome) carrying pZZ3 as the donor. We also failed to obtain any exconjugates, which suggests either that the cloned fragment doesn't contain all regions required for autonomous replication or that the selective marker on pZZ3, chloramphenicol, is not expressed and/or functional in Microscilla sp. strain PRE1.
No evidence for tra-type genes, which would encode the elements necessary for conjugal transfer, was found on pSD15. Furthermore, attempts to transfer pSD15 from the parent to a spontaneous rifampin-resistant derivative of the Microscilla PRE1-0 strain failed. The only ORF encoded by pSD15 that has homology to a protein involved in plasmid transfer is MS155. This protein has significant homology with MocB (for mobilization cassette) of Tn4399 isolated from Bacteroides fragilis. Derivatives of nonconjugal plasmids that carry Tn4399 can be mobilized in trans by the broad-host-range IncP plasmid pRK231 or R751 in E. coli (29). This mobilization requires a 2.8-kb region of Tn4399 that encodes both the MocA and MocB proteins. A homolog of MocA was not found on pSD15.
MS161 and MS163 are similar to HsdS of E. coli and HsdM of Xylella fastidiosa, respectively. These are two components of the type I family of restriction-modification systems (see reference 30 for a review). HsdS is the specificity subunit, and HsdM is the modification subunit. An often-found third component, HsdR, which is the restriction subunit, was not identified on pSD15. However, hsdS and hsdM usually form an operon unlinked to hsdR. While HsdR is necessary for restriction, HsdM and HsdS by themselves are sufficient for modification methylation. Type II restriction-modification systems have been shown to function as a postsegregational killing system for plasmid stabilization (18), but there is no report of the type I system having similar properties.
Polysaccharide utilization.
A number of genes dedicated to polysaccharide or oligosaccharide catabolism were found on pSD15. MS106 is most similar to an outer membrane protein from Porphyromonas gingivalis, and it shows significant identity to an outer membrane protein from Bacteroides thetaiotaomicron (accession number JC6027) that is essential for the utilization of malto-oligosaccharides and starch (data not shown). An attractive hypothesis is that MS106 enables Microscilla sp. strain PRE1 to import the oligosaccharides that are generated by the activities of its extracellular agarases.
MS104 is similar to 2-keto-3-deoxygluconate kinase, which is a component of the glucuronic acid catabolism pathway. Glucuronic acid is one of the building blocks of glycosaminoglycans, a very abundant heteropolysaccharide. Glucuronate catabolism genes are usually part of catabolic pathways of polysaccharide degradation. MS105 putatively encodes 2-dehydro-3-deoxyphosphogluconate aldolase (KdpG). This is a key enzyme in the Entner-Doudoroff pathway and participates in the regulation of the intracellular level of glyoxylate. MS119 and MS120 are predicted to encode proteins with sequence similarity to a fucosidase. These two proteins themselves are 64.4% identical. MS122 is similar to an esterase that is involved in xylan degradation. MS140 is predicted to be a β-galactosidase. MS145 is similar to aldose epimerase, a mutarotase active in galactose metabolism. In addition, six putative dehydrogenase genes were found. MS123, MS137, MS138, and MS141 appear to be alcohol dehydrogenases, MS142 putatively is a gluconate dehydrogenase, and MS143 putatively is an aldehyde dehydrogenase.
Transposases and insertion sequences.
Five ORFs with homology to transposases—MS103, MS118, MS156, MS159, and MS160—were found in pSD15. Inverted repeats (with mismatches underlined) could be identified near each of these ORFs. MS159 and MS160 are flanked by inverted repeats, which start at positions 90875 (5′-GTGCCAGTGATTACGG) and 93368 (5′-CCGAAATCACTGGCAC). MS156 is flanked by 5′-TCTTTTTTT (starting at position 85165) and 5′-AAAAAAAGA (starting at position 88860). The inverted repeats of MS118, 5′-TTTGCGATATTTC, start at position 31682 (just upstream of the ATG start of MS118) and at position 31775 (within the coding sequence). There were two sets of inverted repeats flanking MS103, each located within another ORF (positions 3813 [5′-TGGAGCTGC] and 9813 [5′-GCAGCTCCA] and positions 3490 [5′-CTTTCTTCTTTTCCTGAA] and 10204 [5′-TTCAGAAAAAGAAGGAAG]).
There are two additional large inverted repeats in pSD15. These repeats, termed IR-I and IR-II, are 99.6% identical. IR-I is 1,381 bp long, starts at position 54124, and ends at position 52744. IR-II starts at position 85503, ends at position 86886, and is 1,384 bp long. These inverted repeats overlap MS136 and MS156.
Additional possible functions.
There are three ORFs—MS112, MS113, and MS114—that show similarities to different regions of the 531-amino-acid Na+/glucose symporter encoded by the sglS gene of Vibrio parahaemolyticus, a slightly halophilic marine bacterium (37). MS112 (120 amino acids) matches amino acids 1 to 109 of the V. parahaemolyticus symporter with 78% identity. MS113 (104 amino acids) matches residues 147 to 229 with 60% identity and MS114 (300 amino acids) matches residues 231 to 530 with 56% identity. It is possible that pSD15 encodes the symporter in three parts, which are assembled together to form a functional unit.
Na+/glucose transporters have been shown to utilize a Na+ electrochemical gradient to drive sugar transport in mammalian cells (33), but except for the SglS transporter from V. parahaemolyticus, this is unusual in bacteria (37). A conserved amino acid sequence (G… AXXXXLXXXGR) has been proposed to be involved in Na+ recognition or binding and has been found in several Na+-coupled symporter proteins (11, 48). This motif was found in MS114 from residues 110 to 158 (G110… A148XXXXLXXXGR158) with the motif of SglS from V. parahaemolyticus (G339… S377XXXXLXXXGR387) being very similar (37). Since the first 83 residues of MS113 show 60% identity (78% similarity) with residues 147 to 229 of SglS, which have been shown to be involved in sugar recognition or transport (37), it is likely that MS113 contributes to sugar recognition or transport.
Upstream of the putative transposase MS118, an ORF (MS117) showed 43% identity to a maturase-related protein of a group II intron from Pseudomonas alcaligenes (50). Group II introns found in bacteria are almost always associated with or inserted in another mobile genetic element (see references 13 and 27 for reviews). Maturase is an enzyme that promotes RNA splicing, which is commonly found in various introns, particularly in group II introns (12, 20, 28). Previous studies indicate that all known maturases are expressed from intronic ORFs fused to the upstream exon (20). Our sequence analysis, however, did not identify the presence of any potential exon(s) in proximity to this maturase-encoding ORF.
Concluding remarks.
Analysis of plasmid pSD15 has revealed a rich array of genes whose putative products appear to be involved in polysaccharide degradation and sugar transport and modification. This plasmid, while dispensable for growth of the bacterium under laboratory culturing, undoubtedly plays an important role in the adaptation of its host organism to its marine environment. This is reflected in the presence of five genes related to known agarases. While attempts to demonstrate conjugal transfer of the plasmid under laboratory conditions failed, the possibility of a conjugal transfer system unrelated to known systems cannot be ruled out, particularly in view of the surprisingly high frequency of spontaneous loss of the plasmid during culturing in the lab. Much remains to be done to understand the many novel features contributed by this plasmid to its host.
ACKNOWLEDGMENTS
We thank Lynn Zuo for advice on the construction of a random small-insert library and Matteo Pellegrini for modifying his computer program to identify repeats in DNA sequences.
This work was supported by BioSTAR Project Award S97-03 funded by the University of California and Calgene-Monsanto, Davis, Calif.
REFERENCES
- 1.Altschul S F, Madden T L, Scheaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki T, Araki T, Kitamikado M. Purification and characterization of novel β-agarase from Vibriosp. AP-2. Eur J Biochem. 1990;187:461–465. doi: 10.1111/j.1432-1033.1990.tb15326.x. [DOI] [PubMed] [Google Scholar]
- 3.Araki T, Hayakawa M, Lu Z, Karita S, Morishita T. Purification and characterization of agarases from a marine bacterium, Vibriosp. PO-303. J Mar Biotechnol. 1998;6:260–265. [PubMed] [Google Scholar]
- 4.Balbas P, Soberon X, Merino E, Zurita M, Lomeli H, Valle F, Flores N, Bolivar F. Plasmid vector pBR322 and its special-purpose derivatives: a review. Gene. 1986;50:3–40. doi: 10.1016/0378-1119(86)90307-0. [DOI] [PubMed] [Google Scholar]
- 5.Bauer A W, Kirby W M, Sherris J C, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- 6.Belas R. Sequence analysis of the agrA gene encoding β-agarase from Pseudomonas atlantica. J Bacteriol. 1989;171:602–605. doi: 10.1128/jb.171.1.602-605.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergey D H, Holt J G. Bergey's manual of determinative bacteriology. 9th ed. Baltimore, Md: The Williams & Wilkins Co.; 1994. [Google Scholar]
- 8.Bernan V S, Greenstein M, Maiese W M. Marine microorganisms as a source of new natural products. Adv Appl Microbiol. 1997;43:57–90. doi: 10.1016/s0065-2164(08)70223-5. [DOI] [PubMed] [Google Scholar]
- 9.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couturier M F, Bex F, Bergquist P L, Maas W K. Identification and classification of bacterial plasmids. Microb Rev. 1988;52:375–395. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deguchi Y, Yamato I, Anraku Y. Nucleotide sequence of gltS, the Na+/glutamate symport carrier gene of Escherichia coliB. J Biol Chem. 1990;265:21704–21708. . (Erratum, 266:11404, 1991.) [PubMed] [Google Scholar]
- 12.Doetsch N A, Thompson M D, Hallick R B. A maturase-encoding group III twintron is conserved in deeply rooted euglenoid species: are group III introns the chicken or the egg? Mol Biol Evol. 1998;15:76–86. doi: 10.1093/oxfordjournals.molbev.a025850. [DOI] [PubMed] [Google Scholar]
- 13.Edgell D R, Belfort M, Shub D A. Barriers to intron promiscuity in bacteria. J Bacteriol. 2000;182:5281–5289. doi: 10.1128/jb.182.19.5281-5289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 15.Ewing B, Hillier L, Wendl M C, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 16.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 17.Ha J-C, Kim G-T, Kim S-K, Oh T-K, Yu J-H, Kong I-S. Beta-agarase from Pseudomonas sp. W7: purification of the recombinant enzyme from Escherichia coliand the effects of salt on its activity. Biotechnol Appl Biochem. 1997;26:1–6. [PubMed] [Google Scholar]
- 18.Handa N, Ichige A, Kusano K, Kobayashi I. Cellular responses to postsegregational killing by restriction-modification genes. J Bacteriol. 2000;182:2218–2229. doi: 10.1128/jb.182.8.2218-2229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helinski D R, Toukdarian A E, Novick R P. Replication control and other stable maintenance mechanisms of plasmids. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 2295–2324. [Google Scholar]
- 20.Ho Y, Waring R B. The maturase encoded by a group I intron from Aspergillus nidulansstabilizes RNA tertiary structure and promotes rapid splicing. J Mol Biol. 1999;292:987–1001. doi: 10.1006/jmbi.1999.3070. [DOI] [PubMed] [Google Scholar]
- 21.Karl D M. Distribution, abundance, and metabolic state of microorganisms in the water column and sediments of the Black Sea. Limnol Oceanogr. 1978;23:936–949. [Google Scholar]
- 22.Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984;12:19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- 23.Kim B J, Kim H J, Ha S D, Hwang S H, Byun D S, Lee T H, Kong J Y. Purification and characterization of beta-agarase from marine bacterium Bacillus cereusASK202. Biotechnol Lett. 1999;21:1011–1015. [Google Scholar]
- 24.Kong J-Y, Hwang S-H, Kim B-J, Bae S-K, Kim J-D. Cloning and expression of an agarase gene from a marine bacterium Pseudomonassp. w7. Biotechnol Lett. 1997;19:23–26. [Google Scholar]
- 25.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maidak B L, Cole J R, Lilburn T G, Parker C T, Saxman P R, Stredwick J M, Garrity G M, Li B, Olsen G J, Pramanik S, Schmidt T M, Tiedje J M. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 2000;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martínez-Abarca F, García-Rodríguez F M, Toro N. Homing of a bacterial group II intron with an intron-encoded protein lacking a recognizable endonuclease domain. Mol Microbiol. 2000;35:1405–1412. doi: 10.1046/j.1365-2958.2000.01804.x. [DOI] [PubMed] [Google Scholar]
- 28.Michel F, Ferat J L. Structure and activities of group II introns. Annu Rev Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 29.Murphy C G, Malamy M H. Characterization of a “mobilization cassette” in transposon Tn4399 from Bacteroides fragilis. J Bacteriol. 1993;175:5814–5823. doi: 10.1128/jb.175.18.5814-5823.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray N E. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiol Mol Biol Rev. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naganuma T, Coury D A, Polne-Fuller M, Gibor A, Horikoshi K. Characterization of agarolytic Microscillaisolates and their extracellular agarases. Syst Appl Microbiol. 1993;16:183–190. [Google Scholar]
- 32.Nielsen H, Engelbrecht J, Brunak S, Von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 33.Panayotova-Heiermann M, Leung D W, Hirayama B A, Wright E M. Purification and functional reconstitution of a truncated human Na+/glucose cotransporter (SGLT1) expressed in E. coli. FEBS Lett. 1999;459:386–390. doi: 10.1016/s0014-5793(99)01292-2. [DOI] [PubMed] [Google Scholar]
- 34.Parro V, Mellado R P, Harwood C R. Effects of phosphate limitation on agarase production by Streptomyces lividansTK21. FEMS Microbiol Lett. 1998;158:107–113. [Google Scholar]
- 35.Parro V, Vives C, Godia F, Mellado R P. Overproduction and purification of an agarase of bacterial origin. J Biotechnol. 1997;58:59–66. doi: 10.1016/s0168-1656(97)00128-4. [DOI] [PubMed] [Google Scholar]
- 36.Potin P, Richard C, Rochas C, Kloareg B. Purification and characterization of the alpha-agarase from Alteromonas agarlyticus(Cataldi) comb. nov., strain GJ1B. Eur J Biochem. 1993;214:599–607. doi: 10.1111/j.1432-1033.1993.tb17959.x. [DOI] [PubMed] [Google Scholar]
- 37.Sarker R I, Ogawa W, Shimamoto T, Shimamoto T, Tsuchiya T. Primary structure and properties of the Na+/glucose symporter (SglS) of Vibrio parahaemolyticus. J Bacteriol. 1997;179:1805–1808. doi: 10.1128/jb.179.5.1805-1808.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shieh W Y, Jean W D. Alterococcus agarolyticus, gen. nov., sp. nov., a halophilic thermophilic bacterium capable of agar degradation. Can J Microbiol. 1998;44:637–645. doi: 10.1139/cjm-44-7-637. [DOI] [PubMed] [Google Scholar]
- 39.Shieh W Y, Simidu U, Maruyama Y. Nitrogen fixation by marine agar-degrading bacteria. J Gen Microbiol. 1988;134:1821–1826. [Google Scholar]
- 40.Simon R, Priefer U, Puhler A. Vector plasmids for in-vivo and in-vitro manipulations of gram-negative bacteria. In: Puhler A, editor. Molecular genetics of the bacteria-plant interaction. Berlin, Germany: Springer-Verlag; 1983. pp. 98–106. [Google Scholar]
- 41.Sobecky P A, Mincer T J, Chang M C, Helinski D R. Plasmids isolated from marine sediment microbial communities contain replication and incompatibility regions unrelated to those of known plasmid groups. Appl Environ Microbiol. 1997;63:888–895. doi: 10.1128/aem.63.3.888-895.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sobecky P A, Mincer T J, Chang M C, Toukdarian A, Helinski D R. Isolation of broad-host-range replicons from marine sediment bacteria. Appl Environ Microbiol. 1998;64:2822–2830. doi: 10.1128/aem.64.8.2822-2830.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugano Y, Matsumoto T, Noma M. Sequence analysis of the agaB gene encoding a new beta-agarase from Vibriosp. strain JT0107. Biochim Biophys Acta. 1994;1218:105–108. doi: 10.1016/0167-4781(94)90109-0. [DOI] [PubMed] [Google Scholar]
- 44.Sugano Y, Terada I, Arita M, Noma M, Matsumoto T. Purification and characterization of a new agarase from a marine bacterium, Vibriosp. strain JT0107. Appl Environ Microbiol. 1993;59:1549–1554. doi: 10.1128/aem.59.5.1549-1554.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torsvik V, Goksoyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vera J, Alvarez R, Murano E, Slebe J C, Leon O. Identification of a marine agarolytic Pseudoalteromonasisolate and characterization of its extracellular agarase. Appl Environ Microbiol. 1998;64:4378–4383. doi: 10.1128/aem.64.11.4378-4383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamato I, Anraku Y. Na+/substrate symport in prokaryotes. In: Bakker E P, editor. Alkaline cation transport system in prokaryotes. Boca Raton, Fla: CRC Press; 1992. pp. 53–76. [Google Scholar]
- 49.Yamaura I, Matsumoto T, Funatsu M, Shigeiri H, Shibata T. Purification and some properties of agarase from Pseudomonassp. PT-5. Agric Biol Chem. 1991;55:2531–2536. [PubMed] [Google Scholar]
- 50.Yeo C C, Tham J M, Yap M W-C, Poh C L. Group II intron from Pseudomonas alcaligenesNCIB 9867 (P25X): entrapment in plasmid RP4 and sequence analysis. Microbiology. 1997;143:2833–2840. doi: 10.1099/00221287-143-8-2833. [DOI] [PubMed] [Google Scholar]