Abstract
To determine the potential of DNA array technology for assessing functional gene diversity and distribution, a prototype microarray was constructed with genes involved in nitrogen cycling: nitrite reductase (nirS and nirK) genes, ammonia mono-oxygenase (amoA) genes, and methane mono-oxygenase (pmoA) genes from pure cultures and those cloned from marine sediments. In experiments using glass slide microarrays, genes possessing less than 80 to 85% sequence identity were differentiated under hybridization conditions of high stringency (65°C). The detection limit for nirS genes was approximately 1 ng of pure genomic DNA and 25 ng of soil community DNA using our optimized protocol. A linear quantitative relationship (r2 = 0.89 to 0.94) was observed between signal intensity and target DNA concentration over a range of 1 to 100 ng for genomic DNA (or genomic DNA equivalent) from both pure cultures and mixed communities. However, the quantitative capacity of microarrays for measuring the relative abundance of targeted genes in complex environmental samples is less clear due to divergent target sequences. Sequence divergence and probe length affected hybridization signal intensity within a certain range of sequence identity and size, respectively. This prototype functional gene array did reveal differences in the apparent distribution of nir and amoA and pmoA gene families in sediment and soil samples. Our results indicate that glass-based microarray hybridization has potential as a tool for revealing functional gene composition in natural microbial communities; however, more work is needed to improve sensitivity and quantitation and to understand the associated issue of specificity.
Microorganisms play an integral and unique role in ecosystem function and sustainability. Understanding the structure and composition of microbial communities and their responses and adaptations to environmental perturbations such as toxic contaminants, climate change, and agricultural and industrial practices is critical in maintaining or restoring desirable ecosystem functions (5, 10, 16). However, because less than 1% of microorganisms have been cultivated (1), characterization and detection of microbial populations in natural environments present a great challenge to microbial ecologists. Current methods for analyzing microbial communities, especially their key functions, are too cumbersome. Rapid, simple, reliable, quantitative, and cost-effective tools that can be operated in real-time and in heterogeneous field-scale environments are needed.
The DNA microarray (or microchip) technology is a powerful tool for studying gene expression and regulation on a genomic scale (7, 9, 13, 14, 24, 27, 32, 34) and detecting genetic polymorphisms (4, 12, 31) in both eukaryotes and prokaryotes. Compared to conventional membrane-based hybridization, glass slide-based microarrays offer the additional advantages of rapid detection, lower cost, automation, and low background levels (25). Although microarray-based genomic technology is potentially an extremely powerful tool for characterizing microbial communities and their biological functions (11), the concept and performance of microarray hybridization have not been tested for environmental applications.
In contrast to studies using pure cultures, microarray-based analysis of environmental nucleic acids presents a number of technical challenges. In environmental studies, the target and probe sequences can be very diverse (29), and it is not clear whether the performance of microarrays with diverse environmental samples is similar to that with pure culture samples and how sequence divergence affects microarray hybridization. Also, environmental samples generally contain humic acid and other organic materials, which may inhibit DNA hybridization on microarrays (19, 21). In contrast to pure cultures, the biomass that can be retrieved from environmental samples is generally low (18). Not surprisingly, information on the performance of microarrays with complex environmental samples is lacking. It is not clear whether microarray hybridization is sensitive enough for detecting microorganisms in environmental samples and whether microarray-based detection can be quantitative. Environmental studies require experimental tools that not only detect the presence or absence of particular groups of microorganisms but also provide quantitative data to help evaluate their biological activities.
To examine the potential of microarray-based genomic technology for environmental studies, a prototype microarray was constructed for understanding the specificity, sensitivity, and quantification of microarray hybridization within the context of complex environmental samples. The prototype microarrays contained approximately 100 functional genes encoding dissimilatory heme- and copper-containing nitrite reductases (NirS and NirK, respectively), ammonia mono-oxygenase (AmoA), and the evolutionarily related methane mono-oxygenase (PmoA). These genes code for key enzymes in the ecosystem processes of denitrification, nitrification, and methane oxidation, respectively. Glass slide microarrays containing DNA fragments from different functional genes for monitoring various environmental processes are referred to in this paper as functional gene arrays (FGAs). Our results evaluate the conditions and performance of glass slide-based microarrays as tools for assessing functional gene distribution in natural samples.
MATERIALS AND METHODS
Bacterial strains, environmental clones, and samples.
The majority of the denitrifying bacteria used in this study were isolated from continental margin sediments off the coast of the state of Washington (2). The other denitrifying and nitrifying bacteria were obtained from our culture collection at Michigan State University (8, 26). The amoA genes from pure cultures of four nitrifying bacteria were also cloned into plasmids as described previously (37). Plasmid clones containing nirS, nirK, amoA, and pmoA genes from marine sediments were kindly provided by Gesche Braker (3) and Stephen Nold (17). For simplicity, the probe group consisting of amoA and pmoA genes is collectively referred to as amoA unless otherwise noted.
To evaluate the performance of microarray hybridization, marine sediment samples from the Washington margin and soil samples from Michigan State University were used. Marine sediment samples (W303 and W307) (3) were provided by Allan Devol at The University of Washington. Most of the denitrifiers and environmental clones used in this study were isolated from these marine sediment samples (3). The surface soil samples O22, J19, H17, and M24 were from the Cannelton site (J.-Z. Zhou, B. C. Xia, D. S. Treves, T. L. Marsh, R. V. O'Neill, L.-Y. Wu, A. V. Palumbo, and J. M. Tiedje, submitted for publication), a wetland located on the shore of St. Mary's River in northern Michigan. These samples, which contain different levels of organic matter and chromium, were supplied by Terry Marsh at Michigan State University. Yeast genomic DNA was prepared from Saccharomyces cerevisiae ATCC 18824.
DNA purification, quantitation, and PCR amplification.
Genomic DNA was extracted from isolates as previously described (35). Community DNA from marine sediments and soils was isolated according to the method described by Zhou et al. (36). DNA concentration was determined in the presence of ethidium bromide by fluorometric measurement of the excitation at 360 nm and emission at 595 nm using an HTS700 BioAssay Reader (Perkin-Elmer, Norwalk, Conn.).
To construct functional gene arrays, nirS, nirK, and amoA genes were amplified from the genomic DNA of pure cultures with primers described previously (3, 17). The desired inserts of the environmental clones were amplified with vector-specific primers (37). 16S rRNA genes were amplified from genomic DNA from pure cultures of denitrifying bacteria using PCR primers (TA-F and TA-R) described elsewhere (22). To determine the effects of probe size on microarray hybridization signal intensity, DNA fragments of varying sizes from different gene regions in Shewanella oneidensis MR-1 were selected. The primers for these DNA fragments were designed based on unpublished genome sequences provided as a courtesy from The Institute for Genomic Research (Rockville, Md.). The size and annotation of the amplified DNA products were 2.3 kb from a putative sigma 54 factor, 1.4 kb from putative RNA polymerase sigma factor N, 0.8 kb from a putative two-component sensor kinase, 0.65 kb from a putative alcohol dehydrogenase homolog, and 0.33 kb from putative heat shock protein A.
Hot-start PCR amplification (6) was carried out using 1 × Taq polymerase buffer (10 mM Tris-Cl [pH 9.0], 1.5 mM MgCl2, 50 mM KCl, and 0.1% Triton X-100), 200 μM deoxynucleoside triphosphates, 100 pmol of each primer, 1 ng of plasmid DNA or 100 ng of genomic DNA, and 0.5 U of Taq DNA polymerase in a total reaction volume of 100 μl. Samples were initially denatured at 94°C for 2 min, followed by 30 sequential cycles at 94°C (30 s), 60°C (1 min) for nir and amoA primer sets or 58°C (1 min) for 16S ribosomal DNA and yeast gene primers, 72°C (1 min), and a final extension at 72°C for 7 min. PCR products were analyzed for correct size and the presence of a single product by agarose gel electrophoresis and ethidium bromide staining. The PCR products were then purified using the QIAquick PCR Purification kit (Qiagen, Chatsworth, Calif.) according to the manufacturer's instructions.
Microarray construction and postprocessing.
The prototype DNA microarrays for monitoring bacteria involved in nitrogen cycling contained the following PCR-amplified gene products (for details, see our website: http://www.esd.ornl.gov/facilities/genomics/index.html): (i) 22 nirS, 9 nirK, and 4 amoA genes from pure bacterial cultures; (ii) 27 nirS, 9 nirK, 11 pmoA, and 7 amoA genes cloned from marine sediment samples; (iii) 16S rRNA genes from pure cultures of 10 denitrifying bacteria as positive controls; and (iv) five yeast genes encoding mating pheromone α-factors (mfα1, mfα2), mating-type α-factor pheromone receptor (ste3), actin (act1), and GTP-binding protein involved in the regulation of the cyclic AMP pathway (ras1) as negative controls. A group of seven nirS genes from pure cultures were represented on the microarray as 1.4-kb fragments. All other nirS genes were 0.76 kb in size. To avoid confusion, the DNA deposited on the array is referred to as the probe, whereas the fluorescently labeled DNA is designated as the target. The size, G+C content, and GenBank accession number for each gene probe are listed in a table on our website.
Purified PCR products were diluted to a final concentration of 200 ng μl−1 in 50% dimethyl sulfoxide (DMSO; Sigma, St. Louis, Mo.). Ten microliters of each sample was then transferred to a 384-well microplate for printing. DNA samples were arrayed with a single pin (ChipMaker 3; TeleChem International, Sunnyvale, Calif.) at a spacing distance of 250 μm on silane-coated 25- by 75-mm glass slides using a PixSys 5500 robotic printer (Cartesian Technologies, Inc., Irvine, Calif.) under conditions of 62% relative humidity. Based on performance in preliminary studies and cost, the silane-modified slides from Cel Associates (Houston, Tex.) were selected for microarray fabrication in this study. All of the 104 probes were arranged as a matrix of 16 rows × 7 columns. The exact location of each gene in the matrix is listed in the table on our website. Each glass slide contained three replicates of the gene probe array. For optimizing hybridization conditions, smaller arrays consisting of nirS genes were also constructed.
DNA microarrays were rehydrated over a 60°C water bath for 20 s and dried on a heating block at 80°C for 5 s. The DNA on the microarrays was fixed by UV cross-linking at 65 mJ in a UV Stratalinker 1800 (Stratagene, La Jolla, Calif.). The glass slides were then treated with 0.17 M succinic anhydride (Sigma) dissolved in 240 ml of 1-methyl-2-pyrrolidinone (EM Science, Gibbstown, N.J.) and 10.7 ml of 1 M boric acid (pH 8.0) (J. T. Baker, Phillipsburg, N.J.). Immediately following blocking, the DNA was denatured by immersing the slides in deionized water (dH2O) at 95°C for 2 min. The microarrays were then rinsed briefly in 95% ethanol, air dried at room temperature, and stored dry in the dark at room temperature.
To evaluate the quality of printing and the retention of arrayed DNA elements, a single slide from the same printed set of slides was stained for 30 min in a solution of PicoGreen (Molecular Probes, Eugene, Oreg.), diluted 1:200 in 1× Tris-EDTA (TE) buffer (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA). The slides were then washed sequentially in 1× TE, 0.5× TE, and sterile dH2O for 1 min each prior to being scanned with the ScanArray 5000 Microarray Analysis System (GSI Lumonics, Watertown, Mass.).
Preparation of fluorescently labeled DNA.
Two methods were employed to fluorescently label DNA. In a direct labeling procedure, 1 pg to 2.5 μg of genomic DNA was denatured by boiling for 2 min and immediately chilled on ice for labeling. Each 40-μl labeling reaction mixture contained denatured genomic DNA; 1.5 μg of random hexamers (Gibco BRL, Gaithersburg, Md.); 1× EcoPol buffer (New England Biolabs, Beverly, Mass.); 50 μM dATP, dTTP, and dGTP; 20 μM dCTP; 10 μM Cy3-dCTP or Cy5-dCTP (Amersham Pharmacia Biotech, Piscataway, N.J.); 2.5 mM dithiothreitol; and 10 U of the large Klenow fragment of DNA polymerase I (New England Biolabs). The reaction mixture was incubated at 37°C for 2 h, heat treated in a 100°C heating block for 3 min, and chilled on ice. Labeled target DNA was purified with a QIAquick PCR purification column according to the manufacturer's instructions, concentrated in a Savant SC110 Speedvac (Savant Instruments, Inc., Holbrook, N.Y.) at 40°C for 1.5 h, and resuspended in 10 μl of dH2O for hybridization, except for sensitivity experiments, in which the labeled target DNA was resuspended in 2 μl of dH2O.
For labeling by PCR amplification, each reaction mixture contained 1× Taq polymerase buffer; 20 ng of genomic DNA template or 10 pg of plasmid containing the desired target gene; 100 pmol each of nirS, nirK, amoA, 16S rDNA, or vector-specific primers; 200 μM dATP, dTTP, and dGTP; 80 μM dCTP; 50 μM Cy3- or Cy5-dCTP (Amersham Pharmacia Biotech); and 0.5 U of Taq DNA polymerase in a 100-μl reaction volume. PCRs were programmed for 30 sequential cycles, and the labeled PCR products were purified as described above.
Microarray hybridization.
Microarray experiments for testing specificity were carried out in duplicate (a total of six replicates per gene probe), while all other microarray hybridizations were performed in triplicate (a total of nine replicates per gene probe). Hybridization solutions contained 3× SSC (1× SSC contained 150 mM NaCl and 15 mM trisodium citrate), 1 μg of unlabeled herring sperm DNA (Promega, Madison, Wis.), and 0.3% sodium dodecyl sulfate in a total standard volume of 15 μl. A reduced hybridization solution volume of 2 μl was used for testing detection sensitivity and monitoring target genes in environmental samples. In this case, the hybridization solution was deposited directly onto the immobilized DNA prior to placing a coverslip (6.25 by 8 mm) over the array. Fluorescently labeled DNA was denatured in the hybridization solution at 100°C for 2 min, cooled to ambient temperature, and deposited onto glass coverslips. The microarray (array side down) was placed on the coverslip and then into a waterproof slide chamber (TeleChem International). Fifteen microliters of 3× SSC was dispensed into the hydration wells on either side of the microarray slide, and hybridization was carried out for 12 to 15 h at 45 or 65°C. For experiments determining the effect of temperature on signal intensity, hybridization was carried out at 40, 45, 50, 55, 60, 65, or 75°C. Following hybridization, the arrays were washed with 1× SSC, 0.2% SDS and 0.1× SSC, and 0.2% SDS for 5 min each at ambient temperature and then with 0.1× SSC for 30 s (ambient temperature) prior to being air dried in the dark. For experiments testing the effect of washing conditions on microarray hybridization signals, slides were washed in four separate wash treatments: (i) 1× SSC + 0.2% SDS (twice, 5 min) and dH2O (30 s); (ii) 1× SSC + 0.2% SDS (once, 5 min), 0.5× SSC + 0.2% SDS (once, 5 min), and dH2O (30 s); (iii) 1× SSC + 0.2% SDS (once, 5 min), 0.1× SSC + 0.2% SDS (once, 5 min), and dH2O (30 s); and (iv) 1× SSC + 0.2% SDS (once, 5 min), 0.01× SSC + 0.2% SDS (once, 5 min), and dH2O (30 s).
Array scanning and quantitative analysis of hybridization signals.
Microarrays were scanned initially at a resolution of 50 μm to obtain a quick display image and then at 5 μm with the scanning laser confocal fluorescence microscope of the ScanArray 5000 System. The emitted fluorescent signal was detected by a photomultiplier tube (PMT) at 570 nm (Cy3) or 670 nm (Cy5). For sensitivity experiments and analysis of environmental samples, the laser power and PMT gain were both 100%. For all other microarray experiments, the laser power was 95% and the PMT gain was 90%.
The scanned image displays were saved as 16-bit TIFF files and analyzed by quantifying the pixel density (intensity) of each hybridization spot using the software of ImaGene version 3.0 (Biodiscovery, Inc., Los Angeles, Calif.). A grid of individual circles defining the location of each DNA spot on the array was superimposed on the image to designate each fluorescent spot to be quantified. Mean signal intensity was determined for each spot. The data sheet from ImaGene was then exported to Excel for further processing. The local background signal was subtracted automatically from the hybridization signal of each separate spot. Fluorescence intensity values for the five yeast genes (negative controls) were averaged and then subtracted from the final quantitation values for each hybridization signal. Statistical analysis was performed using SigmaPlot 5.0 (Jandel Scientific, San Rafael, Calif.).
RESULTS
Optimization of parameters for microarray fabrication.
The following buffers for DNA deposition were also evaluated: 3× SSC, 1.5× SSC + 50% DMSO, 1× TE, 0.5× TE + 50% DMSO, 10 to 50% DMSO (at increments of 10%), and dH2O. DMSO at a concentration of 50% was the most appropriate buffer for microarray fabrication in terms of hybridization intensity, spot homogeneity, and evaporation loss during printing (data not shown). Arrayed DNA that was rehydrated at 60°C for 20 s, in contrast to 0 and 10 s, gave the highest and most reproducible signal intensities following microarray hybridization (data not shown). When 50% DMSO and a 20-s rehydration time were used, differences in the denaturation time (ranging from 2 to 4 min at 1-min increments) for arrayed genes did not have a significant effect on the fluorescence intensity (data not shown). Hence, a denaturation time of 2 min was selected as part of the standard protocol.
The effect of DNA probe concentration on hybridization intensity was also examined. PCR-generated nirS DNA was prepared for array deposition in 50% DMSO at various concentrations and rehydrated for 20 s at 60°C prior to hybridization. No substantial difference in hybridization signal intensities for nirS was observed when the DNA concentration was greater than 100 ng μl−1 (data not shown). A DNA probe concentration of 200 ng μl−1 was therefore used for microarray construction in later experiments.
Specificity of DNA microarray hybridization.
To determine the specificity of microarray hybridization, functional gene arrays consisting of nitrite reductase genes (nirS and nirK), ammonia mono-oxygenase genes (amoA), and methane mono-oxygenase genes (pmoA) were used. PicoGreen staining indicated that the shapes (diameters) of the DNA spots on the glass slides were uniform (data not shown).
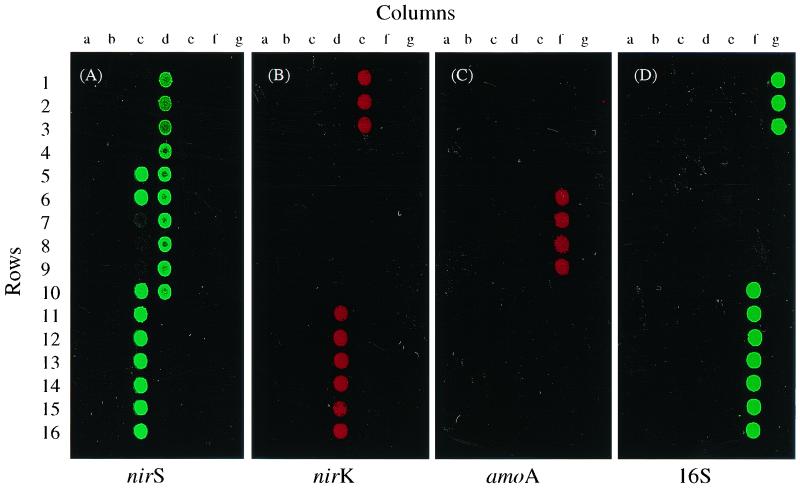
Microarray hybridization was performed in duplicate at 45°C (low stringency) and at 65°C (high stringency) for 12 h with genomic DNA from pure cultures, and the arrays were washed and scanned as described above (see Materials and Methods). At low stringency, all of the labeled target DNAs hybridized to their cognate genes from both pure cultures and environmental samples (data not shown). By contrast, probes hybridized strongly to their complementary sequences from pure cultures at 65°C, with no or only weak cross-hybridization with genes cloned from environmental samples (Fig. 1A to D). At high stringency, Cy3-labeled nirS from the γ-proteobacterium Pseudomonas stutzeri E4-2 hybridized strongly with nirS genes from other P. stutzeri strains that were 92 to 100% identical (Fig. 1A, 5c, 6c, 10c, and 10d). All of the other nirS gene sequences that were less than 75% identical (Fig. 1A, 1a to 15a, and 1b to 4c) did not hybridize, except for three nirS genes (7c to 9c), which showed weak hybridization. The nirK genes showing more than 88% identity hybridized strongly with the labeled nirK gene C3-2 (Fig. 1B, 11d to 16d), while the nirK genes displaying less than 79% identity showed no visible hybridization with the labeled target DNA (Fig. 1B, 16a and 7b to 14b). The Cy5-labeled amoA target gene from Nitrosospira sp. NP39-19 showed hybridization to the sequences from pure cultures of Nitrosolobus and Nitrosospira species with 87 to 100% sequence identity (Fig. 1C, 6f to 9f) but not to the genes from environmental clones with sequence identities of less than 80% (Fig. 1C, 4e to 5f). The 16S genes from different bacteria hybridized very well with the labeled target 16S gene (Fig. 1D, 10f to 3g). Cross-hybridization among different gene groups was not observed at either temperature, regardless of whether the probe was prepared by random primer labeling or PCR labeling with gene-specific primers (Fig. 1). In addition, no hybridization was observed with any of the five yeast genes on the array. These results indicate that different levels of hybridization specificity can be achieved using glass slide microarrays.
FIG. 1.
Fluorescence images showing the specificity of nirS, nirK, amoA, and 16S rRNA target genes in DNA microarray hybridization. Target DNA was labeled with either Cy3 (green pseudocolor; nirS and 16S rRNA genes from pure cultures) or Cy5 (red pseudocolor; nirK and amoA genes from pure cultures) using the method of PCR amplification and hybridized separately at high stringency (65°C) to functional gene arrays containing nirS, nirK, and amoA gene probes from both pure bacterial cultures and environmental clones. 16S rRNA and yeast genes served as positive and negative controls, respectively. Shown are nirS (A), nirK (B), amoA (C), and 16S rDNA (D).
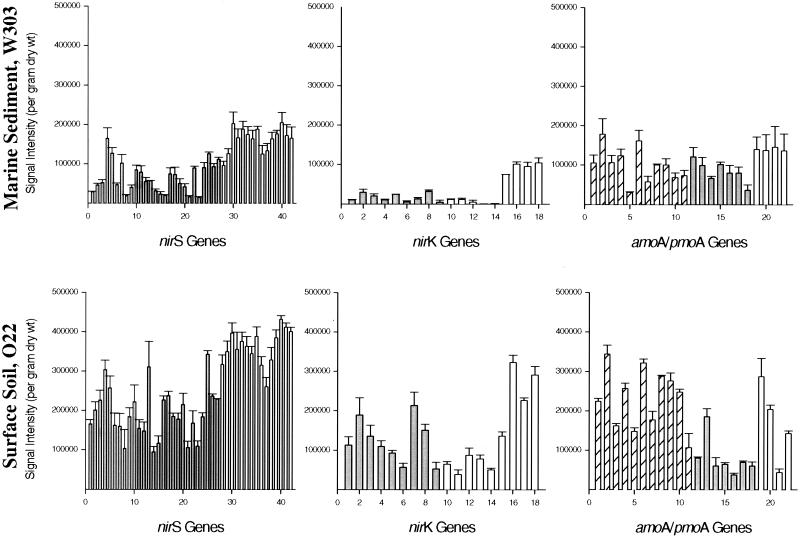
FIG. 5.
The normalized distribution of signal intensity levels for nirS, nirK, and amoA genes as determined by microarray-based analysis of community DNA from marine sediment (W303) and surface soil (O22) environments. Bulk community DNA isolated from two different environmental samples was directly labeled with Cy5 using Klenow fragment with random hexamer primers and hybridized with the microarrays at 65°C in separate experiments. The hybridization signal intensity for each gene is presented. Shaded bars represent probes from environmental clones, and striped boxes represent pmoA probes from environmental clones. Open bars designate probes from pure cultures. The data represent mean values obtained from nine replicates after subtracting background hybridization to yeast genes. For the nirS graphs, bars 1 to 42 correspond to individual genes S1-S42 (see assigned designation in Table 1 on the web site). Similarly, bars 1 to 18 for nirK correspond to genes K1 to K18, and bars 1 to 22 for amoA and pmoA represent genes M1 to M22 (see our table on the website cited in Materials and Methods). The standard deviation of signal intensity is indicated on the top of each bar.
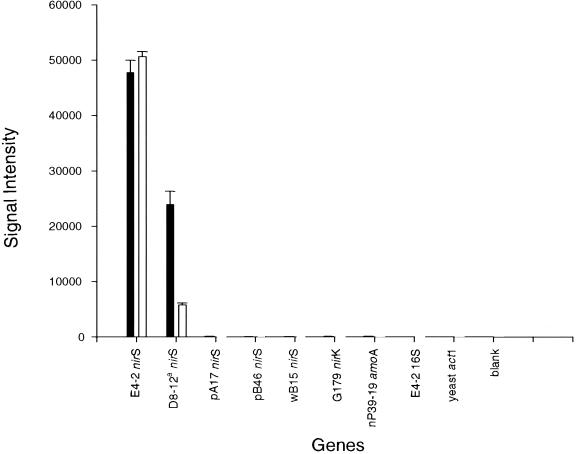
FIG. 6.
Effects of mixed templates on signal intensity. The nirS gene from P. stutzeri E4-2 was labeled with Cy3 using PCR amplification and nirS-specific primers. The signal intensity data presented in the filled bars were from hybridization experiments with the Cy5-labeled E4-2 nirS gene, while the data in open bars were from hybridization experiments with Cy5-labeled E4-2 nirS gene plus an equal amount of the D8-12 nirS gene. The nirS genes of E4-2 and D8-12 are 92% identical.
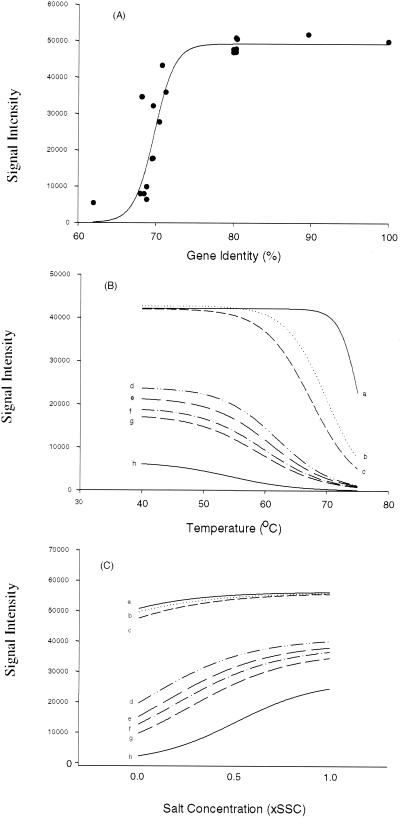
FIG. 4.
Effects of sequence divergence and hybridization stringency on signal intensity. The amoA gene from a pure culture was labeled with Cy5 by PCR amplification and hybridized in triplicate with the functional gene array under different stringencies for 12 to 15 h. (A) Relationship of sequence identity to signal intensity. Hybridizations were carried out at 45°C. (B) Effect of temperature on signal intensity for amoA genes with different sequence identities to the labeled target DNA. Hybridizations were carried out at 40, 45, 50, 55, 60, 65, 70, or 75°C, followed by washing in 0.1× SSC. The calculated intensity (y axis) based on fitted nonlinear models was plotted against temperature (x axis) (n = 72). The sequence identity of the amoA genes on the arrays to the labeled target DNA are as follows: curve a, 100% identity, determination efficient for the fitted model, r2 = 0.91; curve b, 89.7% identity, r2 = 0.95; curve c, 80.3% identity, r2 = 0.96; curve d, 72.0% identity, r2 = 0.92; curve e, 70.8% identity, r2 = 0.83; curve f, 69.7% identity, r2 = 0.83; curve g, 68.9% identity, r2 = 0.95; curve h, 62.0% identity, r2 = 0.78. (C) Effects of washing conditions on signal intensity. Hybridizations were carried out at 45°C. Slides were subjected to four different wash treatments that varied in salt (SSC) concentration (see Materials and Methods for details). The calculated intensity (y axis) based on fitted nonlinear models was plotted against salt concentrations (x axis) (n = 36). The same amoA genes as described for panel B were examined here. The determinant coefficient for the fitted models are as follows: curve a, r2 = 0.87; curve b, r2 = 0.89; curve c, r2 = 0.86; curve d, r2 = 0.84; curve e, r2 = 0.96; curve f, r2 = 0.91; curve g, r2 = 0.93; curve h, r2 = 0.92.
FIG. 3.
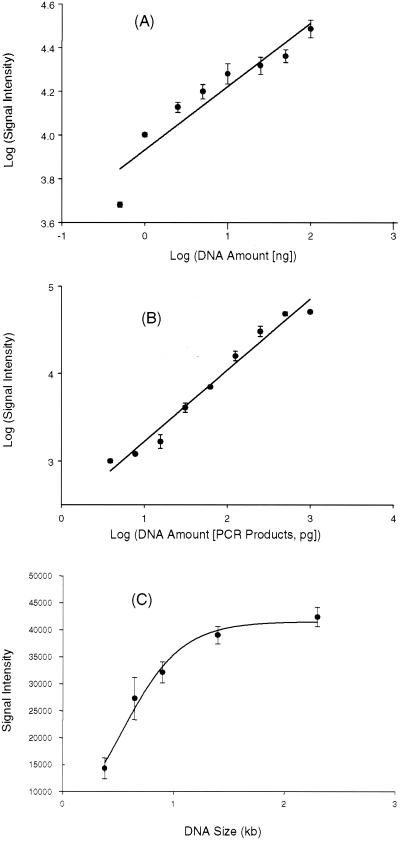
Quantitative analysis of functional gene arrays. (A) Relationship of hybridization signal intensity to DNA target concentration from a single pure culture. Genomic DNA from nirS-containing P. stutzeri E4-2 was labeled with Cy5 and hybridized to the microarrays at the following target concentrations: 0.5, 1, 2.5, 5, 10, 25, 50, and 100 ng. The plot shows the log-transformed average hybridization intensity versus the log-transformed target DNA concentration. (B) Relationship of hybridization signal intensity to DNA target concentration using a mixture of target DNAs. The PCR products from the following nine strains were mixed together in different quantities (in picograms): E4-2 (nirS), 1,000; G179 (nirK), 500; wc301–37 (amoA), 250; ps-47 (amoA), 125; pB49 (nirS), 62.5; Y32K (nirK), 31.3; wA15 (nirS), 15.6; ps-80 (amoA), 7.8; wB54 (nirK), 3.9. All of these genes are less than 80% identical. The mixed templates were labeled with Cy5. The plot shows the log-transformed average hybridization intensity versus the log-transformed target DNA concentration for each strain. (C) Effects of probe DNA size and composition on hybridization signal intensity. Microarrays contained DNA fragments (200 ng μl−1) of different sizes amplified from different regions in the S. oneidensis MR-1 genome. The target DNA was prepared by labeling MR-1 genomic DNA with Cy5 using the Klenow fragment with random hexamer primers. For panels A and B, the data points are mean values derived from three independent microarray slides, with three replicates on each slide (a total of nine data points). Error bars showing the standard deviations are presented.
Detection sensitivity of FGA hybridization.
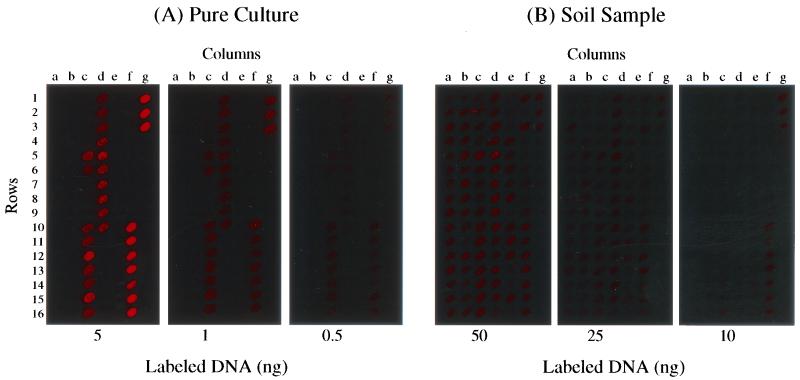
The detection sensitivity of hybridization with the gene array (104 genes) was determined using genomic DNA from both pure cultures and soil community samples (Fig. 2). Genomic DNA (at concentrations of 0.5, 1, and 5 ng) from a nirS-containing denitrifier, P. stutzeri E4-2, was randomly labeled with Cy5 as described. At high stringency, strong hybridization signals were observed with 5 ng of DNA for both nirS and 16S rRNA genes, whereas hybridization signals were weaker but detectable with 1 ng of DNA (Fig. 2A). The hybridization signal at low DNA concentrations was stronger for 16S rRNA genes than for nirS genes. Hybridization signals were measurable with 0.5 ng of genomic DNA, but the fluorescence intensity was poor (Fig. 2A). Therefore, the detection limit with randomly labeled pure genomic DNA under these hybridization conditions was estimated to be approximately 1 ng.
FIG. 2.
Array hybridization images showing the detection sensitivity with labeled pure genomic DNA and bulk community DNA from soil. (A) Genomic DNA from a pure culture of nirS-containing P. stutzeri E4-2 was labeled with Cy5 using the random primer labeling method. The target DNA was hybridized to the microarrays at total concentrations of 0.5, 1, and 5 ng. (B) Genomic DNA from surface soil O22 (10, 25, and 50 ng) was labeled with Cy5 as described for panel A and hybridized with the microarrays.
The detection sensitivity of microarray hybridization was also evaluated with community genomic DNA isolated from soil sample O22, which contained a large amount of organic matter. Community DNA was randomly labeled with Cy5 as described. All of the arrayed genes, with the exception of the five yeast genes, showed hybridization with 50 and 25 ng of labeled community DNA (Fig. 2B). Only the 16S rRNA genes could be detected when as little as 10 ng of soil community DNA was used in the hybridization reaction (Fig. 2B). Thus, with this microarray hybridization system, the detection sensitivities of nirS and 16S genes in this soil sample were considered to be approximately 25 and 10 ng of total environmental DNA, respectively.
Microarray hybridization-based quantitation.
To evaluate whether microarray hybridization can be used as a quantitative tool, the relationship between target DNA concentration and hybridization signal was examined. Genomic DNA from a pure culture of the nirS-containing denitrifier P. stutzeri E4-2 was fluorescently labeled with Cy5 as described and hybridized in triplicate with the microarray at total concentrations ranging from 0.5 to 100 ng. The fluorescence intensities obtained at each DNA concentration for nine data points (three independent microarrays with three replicates on each slide) were averaged, and the log of the concentration was compared to the corresponding log value of the mean fluorescence intensity (Fig. 3A). Within a DNA range of 0.5 to 100 ng, a linear relationship (r2 = 0.89) was observed between signal intensity and target DNA concentration; however, this relationship was stronger (r2 = 0.96) within a range of 1 to 100 ng, suggesting that microarray hybridization is quantitative for pure bacterial cultures within a limited range of DNA concentration.
Since environmental samples contain a mixture of target and nontarget templates, the presence of other nontarget templates could affect microarray-based quantification. To determine whether microarray hybridization is quantitative for targeted templates within the context of environmental samples, 11 nirS, nirK, and amoA genes (plasmid DNA) were mixed together in a series of twofold differences in concentrations ranging from 1 pg to 1 ng. The pairwise similarities of these genes are less than 80%. The mixed plasmid DNA was randomly labeled with Cy5 in triplicate and hybridized with the microarrays. Similarly, a linear relationship (r2 = 0.94) was observed between signal intensity and target DNA concentration (Fig. 3B), suggesting that microarray hybridization is also quantitative for mixed DNA templates.
The accuracy of quantitative measurements obtained from hybridization-based methods relies on hybridization signal intensity, which in turn depends on the fragment size and G+C content of the probes on the glass slides. The effects of size and G+C content of arrayed DNA elements on hybridization were therefore investigated. DNA fragments of different sizes, ranging from 0.33 to 2.3 kb, were amplified by PCR from different open reading frames distributed throughout the sequenced S. oneidensis MR-1 genome. The G+C content of the DNA fragments varied from 41.8 to 49.6%. Within this narrow percentage range, no substantial difference in hybridization intensity was observed due to G+C bias; however, a pronounced impact on signal intensity may be evident across a broader range of percentage of G+C values. The signal intensity increased linearly as the size of the probe increased up to about 1 kb (Fig. 3C), whereas no substantial difference in signal intensity was observed when the probe size was greater than 1.4 kb. This result suggests that the effect of probe size on signal intensity is negligible when the probe is larger than about 1.5 kb.
Effect of sequence divergence and hybridization stringency on signal intensity.
The target functional genes in environmental samples may be highly divergent. Therefore, it is important to understand how sequence divergence affects microarray hybridization. Since, unlike amoA and pmoA genes, the sequence identity of the partial nirS and nirK genes on the arrays is not well distributed within the sequence range of 60 to 100%, only amoA genes were selected for examining the effect of sequence divergence and hybridization stringency on signal intensity. The relationship of sequence homology among amoA genes to hybridization signal intensity at low stringency (45°C) was investigated. A nonlinear S-shaped curve (r2 = 0.83) between the hybridization intensity and gene identity for the amoA genes was observed (Fig. 4A). At 45°C, little hybridization was observed for probes showing 60 to 70% sequence identity to the labeled target DNA, whereas the hybridization signals increased exponentially within the range of 70 to 75% identity. No substantial difference in fluorescence intensities was detected for amoA and pmoA probe genes exhibiting 80 to 100% identity to the labeled target DNA (Fig. 4A). Microarray hybridization was also carried out at 40, 50, 55, 60, 65, 70, or 75°C to test the effect of temperature on signal intensity. Nonlinear S-shaped curves (r2 = 0.84 to 0.96) showing saturation at sequence identities of about 80% were observed when hybridization temperatures were below 65°C (data not shown), whereas nonlinear hyperbolic curves with no saturation (r2 = 0.83 to 0.84) were obtained at hybridization temperatures of 70 and 75°C (data not shown). This suggests that the effect of sequence divergence on signal intensity may be negligible at certain temperatures for genes displaying sequence identities above a certain limit.
In order to understand the effect of hybridization temperature on signal intensity, eight amoA genes exhibiting a wide range of sequence identities to the labeled target gene were examined. The amoA gene on plasmid NP39-19 was labeled with Cy5 with a vector-specific primer. The relationship of signal intensity to temperature was fitted with nonlinear statistical models, and the calculated hybridization intensities based on the fitted nonlinear models were then plotted against hybridization temperatures (Fig. 4B). All of the experimental data fit the nonlinear models, as evident from the determination coefficient values (r2 = 0.78 to 0.96) (Fig. 4B). As expected, signal intensity decreased as the hybridization temperature increased and gene sequence identity decreased (Fig. 4B). Temperature melting profiles were found to be sequence-dependent. For example, at temperatures below 50°C, signal intensity decreased very slowly for genes with sequence identities ≥62%, and genes with >80% identity showed only a small decrease in signal intensity at temperatures below 60 or 70°C (Fig. 4B). The signal intensity for most of the genes decreased dramatically when the hybridization temperature was above 60°C. In addition, at temperatures below 50 or 60°C, no obvious differences in signal intensity were observed among genes with identities greater than 80%. When the hybridization temperature was above 70°C, little or no hybridization was evident even for genes showing about 90% identity to the labeled target DNA. These results suggest that hybridization specificity can be achieved by adjusting the hybridization temperature.
The effect of washing conditions on microarray hybridization at 45°C was also examined by varying the concentration of SSC from 0.01× to 1×. The hybridization data under different washing conditions were fitted with nonlinear models (r2 = 0.84 to 0.96), and the calculated hybridization intensity was plotted against salt concentration (Fig. 4C). As expected, signal intensity generally increased as the salt concentration increased, and the effect of washing stringency on signal intensity was found to be sequence dependent. Significant differences were observed under different salt concentrations for gene sequences that were about 70% identical to the labeled target DNA, whereas little or no difference was obtained for genes with identities greater than 80%.
Microarray-based detection of target genes in marine sediment and soil samples.
To evaluate the potential applicability of DNA microarrays for microbial community analysis, community DNA from two marine sediment samples and four surface soil samples was directly labeled with Cy5 using the random primer labeling method and hybridized with the FGAs in triplicate. Strong hybridization signals above the background to the yeast genes were obtained with both marine sediment and soil samples (only the results for the marine sediment sample W303 and the soil sample O22 are shown in Fig. 5). No hybridization with the five yeast control genes was observed. The average variation in signal intensity for all of the gene replicates in the samples was 12.8% with a standard deviation of 7.7%. The signal variation was lower for the soil sample (10.7% ± 6.9%) than for the marine sediment sample (14.9% ± 8.0%).
As shown in Fig. 5, microarray analysis of the composition of marine sediment sample W303 indicated that the most abundant genes appeared to be those genes showing sequence similarity to nirS from cultured denitrifiers (bars 28 to 42). P. stutzeri (bar 30) and A. tolulyticus (bar 40) nirS genes showed the highest hybridization signal intensity with labeled DNA from the marine sediment sample. Genes similar to nirS from environmental clone pB20 (bar 4) displayed high abundance relative to the other environmental clones (Fig. 5). Similar to what is observed in the distribution of nirS, gene sequences hybridizing to the nirK genes of cultured denitrifiers (Pseudomonas sp. G179 [bar 15], Bacillus azotoformans [bar 16], and Corynebacterium nephridii [bars 17 and 18]) were generally more abundant than genes hybridizing to the nirK genes from environmental clones. By contrast, genes similar to amoA and pmoA genes appeared to be evenly distributed among environmental clones and cultured nitrifiers.
Although most of the functional gene probes were derived from marine sediment environments, these probes also hybridized well with community DNA from soil samples, suggesting that these types of nitrifier and denitrifier genes are present in both environments. The distribution patterns for nirS- and nirK-like genes in the surface soil sample O22 generally resembled those patterns obtained for the marine sediment sample (Fig. 5). Genes showing the highest abundance based on hybridization signal intensity more closely resembled nirS and nirK genes from cultured denitrifiers than the environmental clones. For example, nirS genes similar to those from P. stutzeri (bar 30) and A. tolulyticus (bar 40) appear to be abundant in both the marine sediment and surface soil samples as indicated by hybridization with nirS probes (Fig. 5). However, genes similar to amoA and pmoA sequences from both environmental clones and cultured nitrifiers were less evenly distributed in the surface soil sample.
DISCUSSION
Specificity is one of the most critical parameters for any technique used to detect and monitor microorganisms in natural environments. The specificity of probe-target association depends on the degree of sequence divergence, which can be very high among target genes in natural microbial communities. However, different levels of specificity can be achieved by adjusting microarray hybridization conditions, depending on the experimental objectives. While microarray hybridization under conditions of low stringency allows the detection of microbial populations with a broad range of sequence divergence, high-stringency conditions permit the detection of more specific microbial populations. For example, at low stringency (45°C), all of the amoA and pmoA genes on the array hybridized with Cy5-labeled amoA target DNA from Nitrosospira sp. NP39-19 (Fig. 4A), and no cross-hybridization was observed for nontarget genes. By contrast, at high stringency (65°C) only the sequences from pure cultures of Nitrosolobus and Nitrosospira species with 87 to 100% identity showed strong hybridization, while genes from environmental clones with sequence identities of less than 80% did not show any detectable hybridization (Fig. 1C). At extremely high stringency (75°C), only the sequence displaying 100% identity showed strong hybridization, while the gene from the environmental clone with 90% identity showed very little hybridization (Fig. 4B). Since there were no amoA gene sequences with identities between the ranges of 80 to 90% and 90 to 100%, it is uncertain what minimum sequence divergence is required for differentiation at high or extremely high stringency. We estimate that our microarray hybridization conditions can differentiate sequences exhibiting a dissimilarity of approximately 15 to 20% at 65°C and 10% at 75°C. The level of specificity could be further improved by introducing formamide into the hybridization buffer.
At high stringency (65°C), all of the sequences with >80 to 85% identity hybridized well with their labeled cognate target gene when a single target template was present (Fig. 1). However, in the case of mixed target templates, it is expected that the labeled target templates will preferentially hybridize with their perfect-match probes rather than with their mismatch probes, because the rate of hybridization will occur more rapidly and the probe-target association will be more stable between the perfectly matched sequences than between the mismatched sequences. As a result, the mismatch sequences will have less effect on the hybridization signal intensity of the target genes when the perfect-match sequences are present. To illustrate this, the nirS gene from P. stutzeri E4-2 was labeled with Cy3 in triplicate using PCR amplification and nirS-specific primers. The Cy3-labeled E4-2 DNA was divided into two aliquots, and one aliquot (∼100 ng) was mixed with an equal amount of PCR-amplified D8-12 products. The nirS gene sequences of these species are 92% identical. The labeled target DNA was hybridized with the microarrays at 65°C. Equivalent hybridization signals were obtained for E4-2 in both treatments (Fig. 6). However, the hybridization signal for D8-12 decreased by ∼75% when the perfect-match sequences from D8-12 were present (Fig. 6). These results support the expectation that mismatch sequences will have less effect on the hybridization signal intensity of target genes when perfect-match sequences are present.
The amoA and pmoA sequences having less than 80% identity showed some hybridization even at 65°C (Fig. 4B). This weak hybridization could be due to the amoA sequences that are less than 80% similar or to the primer sequences used for cloning and PCR amplification. Since all of the amoA and pmoA gene fragments on the arrays were first cloned with the amoA-specific primers, followed by amplification from plasmid clones with vector-specific primers, they have 80 bp of identical sequence at each end. Weak signals were also observed in some other experiments for the nirS gene fragments cloned from environmental samples but not for the nirS gene fragments from pure cultures when the target genes were labeled with vector-specific primers (data not shown). The arrayed nirS fragments cloned from environmental samples have ∼40 bp of identical sequence at each end, while those amplified from pure genomic DNA had 18 to 22 bp of identical sequence at each end. The presence of these vector-specific sequences in the nirS gene fragments could contribute to the weak hybridization signals observed in the microarray analysis. In addition, a good linear relationship was obtained for the mixed nine sequences that are all less than 80% similar (Fig. 3B). This also implies that the microarray hybridization is not influenced by sequences with less than 80% identity. Otherwise, no linear relationship between signal intensity and DNA concentration could be obtained.
To investigate the potential contribution of vector-specific sequences to hybridization intensity, the hybridization signals observed for the other target and nontarget probes in the above experiments were also analyzed (Fig. 6). Since genomic DNAs were used as templates, the vector primer sequence portion of the arrayed probes should not affect microarray hybridization. No substantial differences in hybridization signal intensity were observed among nirS genes with less than 80% sequence identity, or between the nontarget nirS genes and nirK, amoA, 16S, or yeast genes (Fig. 6). The signal intensities of the nontarget nirS genes and blank controls were extremely low and comparable, suggesting that no hybridization occurred among the nirS sequences that were less than 80% identical. These results implied that the weak hybridization signals observed in Fig. 4B for the amoA genes of less than 80% sequence identity are most likely due to the continuous stretch of primer sequences used for cloning and amplification.
The potential applicability and usefulness of DNA microarray hybridization to environmental samples will depend not only on the arrayed probe number, specificity, and representation, but also on the sequence diversity of the target genes in natural environments. Analysis based on published sequences and our own unpublished sequences indicates that the genes involved in many important biogeochemical processes such as nitrogen fixation (e.g., nifH), denitrification (e.g., nirS, nirK), sulfite reduction (e.g., dsrA/B) are diverse in natural environments. Many cognate sequences are less than 80 to 85% similar and hence should be resolvable. Collaborative efforts are needed to recover more specific, representative probe sequences from a variety of environments to build a more generally useful array.
If it is useful to differentiate sequences with more than 80 to 85% identity, oligonucleotide microarrays containing short oligonucleotide sequences could be used. Oligonucleotide-based microarrays have the advantage of minimizing the potentially confounding effects of cross-hybridization (33) and are uniquely suited for detecting genetic mutations and polymorphisms. Previous studies have indicated that single-nucleotide differences can be distinguished by oligonucleotide microarray hybridization (reviewed in reference 23). However, since only a very small portion of a gene is used for detection in oligonucleotide microarray hybridization, the signal intensity is generally low and thus detection sensitivity will be lower. In order to fully characterize microbial communities, both types of microarrays may be needed.
Sensitivity is another critical parameter for environmental application and may be the most difficult challenge. Using our optimized hybridization protocol with the reduced hybridization solution volume (2 μl), nirS genes were detected in only 1 ng of labeled pure genomic DNA and 25 ng of bulk community DNA from the surface soil samples (Fig. 2). We found that the detection sensitivity was approximately 500-fold lower when 15 μl of hybridization solution was used than when the reduced volume was used (data not shown). Also, we found that fresh reagents were very important in achieving highly sensitive detection and the detection sensitivity often varied with the batch of fluorescent dyes used. The level of detection sensitivity obtained with the reduced volume should be sufficient for many studies in microbial ecology. However, the sensitivity obtained with the glass-based microarray hybridization may still be 1,000 to 10,000-fold less than with PCR amplification (37), and probably 1,000-fold less than the one obtained with membrane-based hybridization (30). One of the main reasons that the sensitivity of glass-based microarray hybridization is lower than that of the membrane-based hybridization is probably that the probe-binding capacity on glass surfaces is much lower than that on porous membranes. We are currently exploring ways to enhance the level of detection sensitivity.
The quantitative aspects of microarray hybridization have not been well established. The accuracy of microarray-based quantitative assessments is uncertain due to inherent variations associated with array fabrication, labeling, target concentration, and scanning. Comparison of microarray hybridization results with previously known results suggested that microarray hybridization appears to be quantitative enough for detecting differences in gene expression patterns (7, 15, 28). DNA microarrays were also used to measure differences in DNA copy number in breast tumors (20). Single-copy deletions or additions can be detected, suggesting that microarray-based detection can be potentially quantitative. In this study, a linear relationship was observed between hybridization signal intensity and target DNA concentration for a pure culture and a population of mixed DNAs (Fig. 3A and B), suggesting that DNA microarrays may potentially be used for quantitative analysis of environmental samples. With our optimized protocol, experimental variation between array slides was reduced to below 15% (data not shown). This is consistent with the findings of microarray studies on gene expression (2).
The challenge in quantifying the abundance of microbial populations in natural environments based on hybridization signal intensity is how to distinguish differences in hybridization intensity due to population abundance from those due to sequence divergence. One possible solution is to carry out microarray hybridization under conditions of various levels of stringency. Based on the relationships among signal intensity, sequence divergence, hybridization temperature, and washing conditions determined in this study, it should be possible to distinguish, to some extent, the contributions of population abundance and sequence divergence to hybridization intensity. For instance, our results showed that at about 55 to 60°C, sequence divergence had little or no effect on signal intensity for amoA genes with greater than 80% identity to the labeled target DNA (Fig. 4B). This suggests that under such hybridization conditions the effect of sequence divergence on signal intensity is negligible for genes with >80% sequence identity; therefore, any significant differences in signal intensity are most likely due to differences in population abundance. Another possible solution to this problem is to use microarrays containing probes extremely specific to the target populations, such as those used in oligonucleotide microarrays.
This work evaluates specificity, sensitivity, sequence divergence and quantitation of DNA microarrays for environmental application. While this tool holds much potential for environmental studies, more development is needed, especially for improved sensitivity, quantitation, and the biological meaning of a detectable specificity before it can be used broadly and interpreted meaningfully.
ACKNOWLEDGMENTS
This research was supported by the United States Department of Energy under the Natural and Accelerated Bioremediation Research, the Biotechnology Investigations—Ocean Margins, and Microbial Genome Programs of the Office of Biological and Environmental Research. Oak Ridge National Laboratory is managed by University of Tennessee—Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartosiewicz M, Trounstine M, Barker D, Johnston R, Buckpitt A. Development of a toxicological gene array and quantitative assessment of this technology. Arch Biochem Biophys. 2000;376:66–73. doi: 10.1006/abbi.2000.1700. [DOI] [PubMed] [Google Scholar]
- 3.Braker G, Zhou J, Wu L, Devol A H, Tiedje J M. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl Environ Microbiol. 2000;66:2096–2104. doi: 10.1128/aem.66.5.2096-2104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chee M, Yang R, Hubbell E, Berno A, Huang X C, Stern D, Winkler J, Lockhart D J, Morris M S, Fodor S P A. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 5.Curtis P S, Zak D R, Pregitzer K S, Teeri J A. Above- and belowground response of Populus grandidentata to elevated atmospheric CO2 and soil N availability. Plant Soil. 1994;165:45–51. [Google Scholar]
- 6.D'Aquila R T, Bechtel L J, Videler J A, Eron J J, Gorczyca P, Kaplan J C. Maximizing sensitivity and specificity of PCR by preamplification heating. Nucleic Acids Res. 1991;19:3749. doi: 10.1093/nar/19.13.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 8.Fries M R, Zhou J-Z, Chee-Sanford J C, Tiedje J M. Isolation, characterization and distribution of denitrifying toluene degraders from a variety of habitats. Appl Environ Microbiol. 1994;60:2802–2810. doi: 10.1128/aem.60.8.2802-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Futcher B. Microarrays and cell cycle transcription in yeast. Curr Opin Cell Biol. 2000;12:710–715. doi: 10.1016/s0955-0674(00)00156-3. [DOI] [PubMed] [Google Scholar]
- 10.Gibson D T, Sayler G S. Scientific foundations of bioremediation: current status and future needs. Washington, D.C.: American Academy of Microbiology; 1992. [Google Scholar]
- 11.Guschin D Y, Mobarry B K, Proudnikov D, Stahl D A, Rittmann B E, Mirzabekov A D. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl Environ Microbiol. 1997;63:2397–2402. doi: 10.1128/aem.63.6.2397-2402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hacia J G. Resequencing and mutational analysis using oligonucleotide microarrays. Nat Genet. 1999;21:42–47. doi: 10.1038/4469. [DOI] [PubMed] [Google Scholar]
- 13.Khodursky A B, Peter B J, Cozzarelli N R, Botstein D, Brown P O, Yanofsky C. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:12170–12175. doi: 10.1073/pnas.220414297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lashkari D A, DeRisi J L, McCusker J H, Namath A F, Genetile C, Hwang S Y, Brown P O, Davis R W. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc Natl Acad Sci USA. 1997;94:13057–13062. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 16.National Research Council (NRC) In situ bioremediation: when does it work? Washington, D.C.: National Academy Press; 1993. [Google Scholar]
- 17.Nold S C, Zhou J, Devol A H, Tiedje J M. Marine sediment nitrifying communities of the Pacific Northwest contain β-proteobacteria. Appl Environ Microbiol. 2000;66:4532–4535. doi: 10.1128/aem.66.10.4532-4535.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogram A, Sun W H, Brocman F J, Fredrickson J K. Isolation and characterization of RNA from low-biomass deep-subsurface sediments. Appl Environ Microbiol. 1995;61:763–768. doi: 10.1128/aem.61.2.763-768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogram A V, Sayler G S, Barkay T. The extraction and purification of microbial DNA from sediments. J Microbiol Methods. 1987;7:57–66. [Google Scholar]
- 20.Pinkel D, Seagraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo W L, Chen C, Zhai Y, Dairkee S H, Ljung B M, Gray J W, Albertson D G. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 21.Porteous L A, Armstrong J L. Recovery of bulk DNA from soil by a rapid, small-scale extraction method. Curr Microbiol. 1991;22:345–348. [Google Scholar]
- 22.Qiu X Y, Wu L-Y, Huang H, McDonel P E, Palumbo A V, Tiedje J M, Zhou J-Z. An improved 16S rRNA-based cloning approach for reducing PCR-generated artifacts. Appl Environ Microbiol. 2001;67:880–887. doi: 10.1128/AEM.67.2.880-887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramsay G. DNA chips: state-of-the art. Nat Biotechnol. 1998;16:40–44. doi: 10.1038/nbt0198-40. [DOI] [PubMed] [Google Scholar]
- 24.Richmond C S, Glasner J D, Mau R, Jin H, Blattner F R. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shalon D, Smith S J, Brown P O. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 26.Smith G B, Tiedje J M. Isolation and characterization of a nitrite reductase gene and its use as a probe for denitrifying bacteria. Appl Environ Microbiol. 1992;58:376–384. doi: 10.1128/aem.58.1.376-384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sudarsanam P, Iyer V R, Brown P O, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taniguchi M, Miura K, Iwao H, Yamanaka S. Quantitative assessment of DNA microarrays—comparison with Northern blot analyses. Genomics. 2001;71:34–39. doi: 10.1006/geno.2000.6427. [DOI] [PubMed] [Google Scholar]
- 29.Tiedje, J. M., Jae-Chang Cho', A. Murray, D. Treves, B.-C. Xia, and J.-Z. Zhou. 2000. Soil teaming with life: new frontiers for soil science, p. 393–412. In R. M. Rees, B. C. Ball, C. D. Campbell, and C. A. Watson (ed.), Sustainable management of soil organic matter CABI International, New York, N.Y.
- 30.Voordouw G, Shen Y, Harrington C S, Telang A J, Jack T R, Westlake D W S. Quantitative reverse sample genome probing of microbial communities and its application to oil field production waters. Appl Environ Microbiol. 1993;59:4101–4114. doi: 10.1128/aem.59.12.4101-4114.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D G, Fan J B, Siao C J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, Kruglyak L, Stein L, Hsie L, Topaloglou T, Hubbell E, Robinson E, Mittmann M, Morris M S, Shen N, Kilburn D, Rioux J, Nusbaum C, Rozen S, Hudson T J, Lipshutz R, Chee M, Lander E S. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 32.White K P, Rifkin S A, Hurban P, Hogness D S. Microarray analysis of Drosophila development during metamorphosis. Science. 1999;286:2179–2184. doi: 10.1126/science.286.5447.2179. [DOI] [PubMed] [Google Scholar]
- 33.Wodicka L, Dong H, Mittmann M, Ho M-H, Lockhart D J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nature Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 34.Ye R W, Tao W, Bedzyk L, Young T, Chen M, Li L. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J Bacteriol. 2000;182:4458–4465. doi: 10.1128/jb.182.16.4458-4465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J-Z, Fries M R, Chee-Sanford J C, Tiedje J M. Phylogenetic analyses of a new group of denitrifiers capable of anaerobic growth on toluene: description of Azoarcus tolulyticus sp. nov. Int J Syst Bacterol. 1995;45:500–506. doi: 10.1099/00207713-45-3-500. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J-Z, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou J-Z, Davey M E, Figueras J B, Rivkina E, Glichinsky D, Tiedje J M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology. 1997;143:3913–3919. doi: 10.1099/00221287-143-12-3913. [DOI] [PubMed] [Google Scholar]