Abstract
Non-alcoholic fatty liver disease (NAFLD) is emerging as the most common chronic liver disease worldwide. It refers to a range of liver conditions affecting people who drink little or no alcohol. NAFLD comprises non-alcoholic fatty liver and non-alcoholic steatohepatitis (NASH), the more aggressive form of NAFLD. NASH is featured by steatosis, lobular inflammation, hepatocyte injury, and various degrees of fibrosis. Although much progress has been made over the past decades, the pathogenic mechanism of NAFLD remains to be fully elucidated. Hepatocyte nuclear factor 4α (HNF4α) is a nuclear hormone receptor that is highly expressed in hepatocytes. Hepatic HNF4α expression is markedly reduced in NAFLD patients and mouse models of NASH. HNF4α has been shown to regulate bile acid, lipid, glucose, and drug metabolism. In this review, we summarize the recent advances in the understanding of the pathogenesis of NAFLD with a focus on the regulation of HNF4α and the role of hepatic HNF4α in NAFLD. Several lines of evidence have shown that hepatic HNF4α plays a key role in the initiation and progression of NAFLD. Recent data suggest that hepatic HNF4α may be a promising target for treatment of NAFLD.
Keywords: Nonalcoholic fatty liver disease, Hepatocyte nuclear factor 4α, Lipogenesis, Inflammation, Fibrosis, Liver, Lipotoxicity, Apoptosis
Introduction
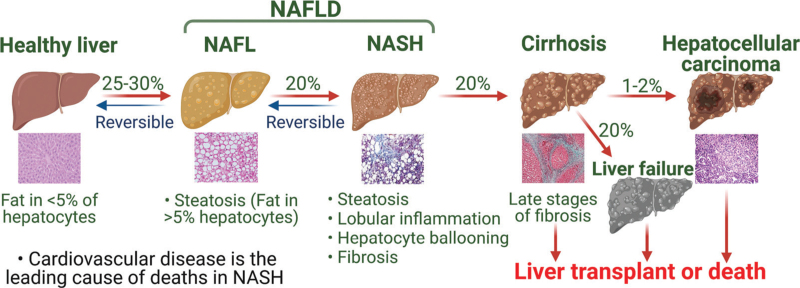
Non-alcoholic fatty liver disease (NAFLD) is emerging as the leading chronic liver disease due to the rising rates of obesity and diabetes. It refers to a range of liver conditions affecting people who drink little or no alcohol with the presence of steatosis in ≥5% hepatocytes. There are two subtypes of NAFLD, non-alcoholic fatty liver (NAFL) and non-alcoholic steatohepatitis (NASH). NASH is the more advanced subtype of NAFLD, which is characterized by liver steatosis, lobular inflammation, hepatocyte ballooning, and various degrees of fibrosis. NASH may further progress to cirrhosis, hepatocellular carcinoma (HCC), and liver failure [Figure 1]. NAFLD is often associated with diabetes, obesity, and dyslipidemia, and is considered as the hepatic manifestation of metabolic syndrome.[1]
Figure 1.
Progression of NAFLD. NAFLD encompasses NAFL and NASH. NASH may further progress to cirrhosis, HCC, and liver failure. Patients without cirrhosis may also develop HCC. Cardiovascular disease is the leading cause of deaths in NASH. HCC: Hepatocellular carcinoma; NAFL: Non-alcoholic fatty liver; NAFLD: Nonalcoholic fatty liver disease; NASH: Non-alcoholic steatohepatitis.
Hepatocyte nuclear factor 4α (HNF4α) is a nuclear hormone receptor that is highly abundant in the liver and highly conserved across the species. In the liver, HNF4α is best known for its role as a master regulator of liverspecific gene expression and its essential role in both fetal and adult liver functions. The expression of HNF4α is markedly reduced in NAFLD patients and mouse models of NASH[2,3] or fibrotic livers.[4–6] Dysregulation of HNF4α expression is associated with many human diseases, such as NAFLD, liver cirrhosis, HCC, ulcerative colitis, colon cancer, and maturity on-set diabetes of the young. In this review, we briefly overview the pathogenic mechanisms, diagnosis, and treatment of NAFLD, but focus on the regulation of hepatic HNF4α expression, the role of HNF4α in the pathogenesis of NAFLD, and the potential of HNF4α as a therapeutic target for NAFLD.
Pathogenic Mechanisms of NAFLD
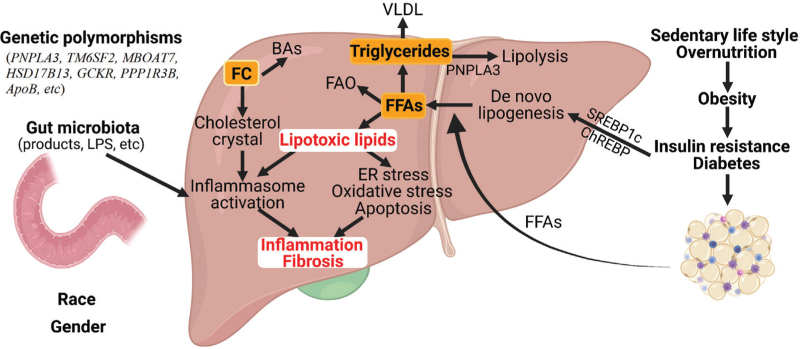
The pathogenic mechanisms of NAFLD are yet to be fully understand. Multiple lines of evidence have indicated that the pathogenesis of NAFLD is a complicated and multifactorial process involving interactions among nutrition, metabolism, genetic predisposition, and environment [Figure 2]. Historically, a “two-hit” hypothesis is first proposed, in which fats accumulate in the liver (first hit) followed by other insults (e.g., inflammatory cytokines, oxidative stress, mitochondrial dysfunction) leading to inflammation and fibrogenesis (second hit).[7,8] Due to the complexity of the pathogenesis, a “multiple-hit” hypothesis is brought forward, in which multiple insults act together on genetically predisposed subjects to induce NAFLD.[9,10]
Figure 2.
Molecular mechanisms of NAFLD. NAFLD is a complex and multifactorial disease. The development and progression of NAFLD is affected by insulin resistance, genetic polymorphisms, gut microbiota, race, gender, etc. Under insulin resistance or diabetes, the influx of FFAs from adipose tissue as well as DNL is increased. FFAs, particularly saturated FFAs, can cause ER stress, oxidative stress, apoptosis, and inflammasome activation via lipotoxic lipids (LPCs, ceramides, DAG, etc.). Cholesterol crystals also promote inflammasome activation. The change in the gut barrierallows LPS from gut microbiota to enter the portal circulation and activate toll-like receptors or inflammasome (pyroptosis) for induction of inflammation. The change in other gut microbiota products (ethanol, secondary bile acids, etc.) may also contribute to the development of NAFLD. ApoB: Apolipoprotein B; BAs: Bile acids; chREBP: Carbohydrate response element-binding protein; DNL: de novo lipogenesis; DAG: Diacylglycerols; ER: Endoplasmic reticulum; FC: Freecholesterol; FAO: Fatty acid oxidation; FFAs: Free fatty acids; GCKR: Glucokinase regulatory; HSD17B13: Hydroxysteroid 17-beta dehydrogenase 13; LPS: Lipopolysaccharides; LPC: Lysophosphatidylcholine; MBOAT7: Membrane-bound O- acyltransferase domain-containing 7; NAFLD: Nonalcoholic fatty liver disease; PPP1R3B: Protein phosphatase 1 regulatory subunit 3B; PNPLA3: Palatin-like phospholipase domain containing 3; SREBP-1c: Sterol regulatory element-binding protein 1c; TM6SF2: Transmembrane 6 superfamily 2.
Dysregulation of lipid metabolism and NAFL
About 25% of the population has NAFLD worldwide.[11] NAFLD is often associated with obesity and diabetes. Nonetheless, NAFLD is also found in non-obese or overweight children and adults, ranging from 3.3% to 21.2% of the population (with a body mass index <25 kg/m2).[12] The prevalence of NAFLD is higher in Hispanics and whites than in Black individuals[13,14] and is twice as much in men as in women.[15] Globally, about 55.5% people with type 2 diabetes and up to 90% of obese people have NAFLD[16,17] Among people with NAFLD, cardiovascular disease is the leading cause of death, followed by cancer and liver-related death.[1]
NAFLD often starts with lipid accumulation in the liver that is not the consequence of alcohol drinking, a condition called NAFL. Triglycerides (TG), free fatty acids (FFAs), free cholesterol (FC), and cholesterol esters (CEs) may accumulate in NAFL, albeit largely in the form of TG. The accumulation of TG in the liver may result from increased de novo lipogenesis (DNL) and impaired very low-density lipoprotein (VLDL) secretion or lipolysis. Impaired fatty acid oxidation (FAO) may also lead to FA and TG accumulation in the liver.[18]
Insulin resistance is a major risk factor for NAFLD. Under insulin resistance, more FFAs are released from adipose tissue and delivered to the liver. Hyperinsulinemia also transcriptionally induces genes that promote DNL. Sterol regulatory element-binding protein 1c (SREBP-1c) is a transcription factor that induces the lipogenic genes, such as fatty acid synthase, acetyl-CoA carboxylase (ACC), and stearoyl-CoA desaturase 1. Insulin activates SREBP-1c by inducing SREBP1c mRNA levels and SREBP-1c proteolytic processing, which can be blocked by wortmannin, an inhibitor of phosphatidylinositol 3-kinase, and low concentrations of rapamycin, an inhibitor of the mechanistic target of rapamycin complex 1 (mTORC1).[19] Furthermore, insulin-induced SREBP-1c proteolytic processing can be blocked by inhibition of p70 S6 kinase (S6K),[20] suggesting that activation of the mTORC1/S6K pathway is responsible for SREBP-1c processing. Under overnutrition, endoplasmic reticulum (ER) stress promotes insulin-induced SREBP-1c cleavage.[21] Unlike insulin, glucose promotes lipogenesis via activation of carbohydrate response element-binding protein (ChREBP). In response to increased glucose concentration, ChREBP is dephosphorylated and translocated to the nucleus, leading to induction of lipogenic genes and liver-type pyruvate kinase.[22] However, under insulin resistance or overnutrition, NAFLD is often accompanied by increased VLDL secretion and hyperlipidemia due to increased TG availability and microsomal triglyceride transfer protein (MTP) production.[23] By contrast, the contribution of FAO to steatosis in NAFLD has been less clear. It has been shown that NAFLD patients with insulin resistance have impaired ATP production[24,25] but increased hepatic FAO.[26] Consistent with the latter finding, high fat diet (HFD) feeding increases the function of tricarboxylic acid cycle in mice.[27] Additional studies with a larger sample size may be needed to clarify the role of FAO in fat deposit in NAFLD.
Lipolysis also plays a role in NAFLD. Adipose triglyceride lipase (ATGL; PNPLA2) is the major hepatic triglyceride lipase,[28,29] although some other lipases are also reported to display triglyceride hydrolase (TGH) in the liver, such as some of the carboxylesterase (CES) family, lysosomal acid lipase, etc.[30] Defective lipolysis contributes to hepatic TG accumulation. Multiple observations have uncovered that the common I148M missense mutation in palatin-like phospholipase domain containing 3 (PNPLA3; adiponutrin) is consistently associated with NAFLD.[31,32] In the presence of obesity or chronic alcohol intake, the variant is associated with hepatitis or cirrhosis.[32] PNpLA3 (I148M) promotes steatosis by inhibition of ATGL activity through interaction with comparative gene identification-58 (CGI- 58; ABHD5), a co-activator of ATGL.[33,34]
In addition to PNPLA3, other genetic variants are also found to play a role in hepatic fat accumulation and/or inflammation. The E167K variant in transmembrane six superfamily two (TM6SF2) causes fatty liver and elevates alanine aminotransferase (ALT) levels by impairing normal VLDL secretion.[35,36] The membrane-bound O- acyltransferase domain-containing 7 (MBOAT7; LPIATI) variant rs641738 increases risk of NAFLD,[37,38] which appears to be mediated by changes in hepatic phosphatidylinositol acyl-chain remodeling.[37,38] Further studies in mice show that ablation of Mboat7 causes accumulation of its substrate lysophosphatidylinositol (LPI) lipids, and that administration of LPI promotes hepatic inflammation and fibrogenesis.[39] In contrast, the rs72613567 variant with an adenine insertion in hydroxysteroid 17-beta dehydrogenase 13 (HSD17B13), an enzyme that is elevated in NAFLD and targets lipid droplets, is associated with a reduced risk for NASH.[40–42] The HSD17B13 rs72613567 variant is also shown to interact with PNPLA3 I148M and reduce the risk for liver disease conferred by PNPLA3 I148M.[42] Other studies have also shown that the variants in glucokinase regulatory gene or protein phosphatase 1 regulatory subunit 3B are also associated with NAFLD.[43]
Progression of NAFL to NASH
About 20% NAFL patients will develop NASH and 20% NASH patients will develop cirrhosis over time.[1,44] About 1% to 2% or 20% cirrhosis patients may develop HCC or liver failure over 1 or 2 years, respectively.[1] Inflammation is a key driver of NAFL progression to NASH. Under insulin resistance, excessive fatty acid influx from adipose tissue and increased DNL in the liver promote accumulation of lipotoxic lipids, which contribute to oxidative stress, ER stress, inflammasome activation, and apoptotic cell death, leading to inflammation and fibrogenesis[45] [Figure 2]. However, NAFLD is not always associated with insulin resistance. Other factors, such as genetic polymorphism, gut microbiota, etc., also contribute to the progression of NAFLD.
Lipotoxicity
Hepatic toxic lipid species accumulate when the liver cannot handle excessive carbohydrates and fatty acids. FFAs (saturated and trans fatty acids), diacylglycerols (DAG), lysophosphatidylcholine (LPC), ceramides, and FC are considered lipotoxic species, which can mediate inflammation in NAFLD by causing ER stress, oxidative stress, and inflammasome activation, leading to apoptosis, necroptosis, release of cytokines or chemokines (tumor necrosis factor [TNF] α, interleukin 1β [IL-1β], IL-6, IL- 18, tumor growth factor beta [TGF-β], etc.), and activation of stellate cells. [18,46] Inflammasome is a cytoplasmic protein complex that responds to danger-associated molecular patterns (saturated fatty acids, cholesterol crystals, etc.) and pathogen- associated molecular proteins (e.g., products of gut microbiota).[47] Activation of inflammasome leads to expression and release of IL-1β and IL-18, and promotes inflammation via activation of caspase-1[48,49] and induces a form of death called proptosis.[50]
Apoptosis
Apoptosis plays a key role in the progression of NABLD.[51,52] NASH patients have significant levels of apoptosis and caspase 3 activation.[53,54] Caspase 2 appears to be an initiator caspase in multiple apoptotic pathways. Caspase 2 expression is markedly upregulated in NAFL and NASH patients and animal models of NASH, and its deficiency reduces lipid-induced hepatocyte apoptosis (lipoapoptosis) and liver fibrosis.[55] Ablation of caspase 8 in hepatocytes inhibits methionine-choline deficient diet-induced inflammation, fibrosis, and liver injury[56] Saturated FFAs induce c-Jun N-terminal kinase (JNK)-dependent lipoapoptosis by activating the proa- poptotic B-cell lymphoma protein 2 (Bcl-2) proteins Bim and Bax.[57] Inhibition of apoptosis by the pan-caspase inhibitors VX-166 or Emricasan reduces inflammation or the development of fibrosis in mouse models with NASH.[58–60]
Extracellular vesicles (EVs)
EVs are non-nucleated, lipid-bound particles that include endosome-derived exosomes (30–150 nm in diameter) and plasma membrane-derived microvesicles (50–1000 nm). EVs can carry mRNAs, non-coding RNAs, lipids (cholesterol, ceramides, sphingomyelin, phosphatidylcholine, phosphatidylserine), proteins (heat shock proteins HSP70, HSP90, tubulin, actin, etc.), and mitochondrial DNA, and deliver them to other cell types[61,62] EVs are important for cell-cell communications and also act as drivers of inflammation in NAFLD.[63,64] Kakazu et al[65] show that lipotoxic hepatocytes induced by palmitate secrete EVs enriched in C16:0 ceramide, which in turn activate macrophage chemotaxis via formation of sphingosine-1-phosphate from 16:0 ceramide. Treatment of hepatocytes with palmitate or the palmitate metabolite LPC increases the release of EVs containing TNF-related apoptosis-inducing ligand, which are capable of inducing the expression of IL-1β and IL-6 in macrophages.[66]
Gut microbiome
Gut microbiota is a complex ecosystem whose composition and relative abundance of species are comparable between healthy people but are affected by environmental and host-related factors, such as diets, drugs, physical activity, geographic locations, etc.[67] A less diverse microbiota population is observed in NASH patients in comparison with that of healthy subjects[18] Some studies have suggested a link between gut dysbiosis and the progression of NAFLD. In one study, Bacteroides and Ruminococcus have been identified as independently associated with steatohepatitis and fibrosis, respective- ly.[68] The change in gut microbiota composition may regulate the development and progression of NAFLD via their metabolites (short-chain fatty acids, ethanol, etc.), endotoxemia due to increased gut permeability, and changes in hormones and bile acid signalling.[67] Lipopolysaccharides (LPS) activate Toll-like receptor (TLR) 4 and TLR9 on Kupffer cells to induce production of proinflammatory cytokines and chemokines. PAMPs derived from gut microbial products activate inflamma- somes (NRLP3 and NLRP6) to release IL-18 and IL-1β.[18] The contribution of gut microbiota to NASH progression is also validated by the use of germ-free animal models.[69]
Diagnosis and Treatment of NAFLD
Diagnosis of NAFLD
NAFL is histologically defined by the presence of macro- vesicular steatosis in >5% of hepatocytes whereas NASH is histologically characterized by hepatic steatosis and hepatocellular injury, including hepatocyte ballooning, lobular inflammation, and various degrees of pericellular fibrosis. The majority of NAFLD patients are asymptomatic until NAFLD progresses to cirrhosis. Serum ALT and aspartate aminotransferase (AST) levels are often elevated with ALT levels higher than AST levels.[70] Hepatic steatosis can be identified non-invasively by ultrasound, computed tomography (CT), or magnetic resonance imaging (MRI). MRI can detect as little as 5% steatosis whereas ultrasound or CT can detect ≥20% steatosis.[18] Magnetic resonance elastography measures the stiffness of liver tissue, and offers a high accuracy in detection of liver fibrosis.[71,72] So far, no practically useful surrogate makers can be used for diagnosis of NASH, and liver biopsy remains the gold standard for diagnostic evaluation of liver inflammation and fibrosis.[18]
Treatment of NAFLD
Lifestyle change, including a low-calorie diet (a daily reduction of 500–1000 kcal calorie intake) and 30 min of daily moderate exercise, is highly recommended. Lifestyle change-induced weight loss by ≥10% is associated with NASH resolution and fibrosis regression.[73] For patients with NASH and obesity, bariatric surgery is associated with a significant lower risk of major adverse liver outcomes (progression to cirrhosis, HCC, liver transplantation, liver-related mortality) and major adverse cardiovascular events (coronary artery or cerebrovascular events, heart failure, cardiovascular death). [74] No drugs have been approved for NASH treatment, although some pharmacological therapies at various phases of clinical trials show promising outcomes. Ongoing major clinical trials of pharmacotherapies for NASH treatment mainly target metabolism, inflammation, and/or apoptosis. Peroxisome proliferation-activated receptors α/β/γ (PPARα/β/γ) and farnesoid X receptor (FXR) are nuclear hormone receptors that play an important role in regulating metabolic pathways and inflammatory response. PPARγ ligands (such as pioglita zone) have been shown to improve steatohepatitis but also induce weight gain, fluid retention, osteopenia, and fracture risk.[75,76] PPARα/γ or PPARα/δ dual agonists are also being tested in clinical trials.[18] FXR ligands improve insulin sensitivity and NASH in mice and humans.[77,78] Obeticholic acid (OCA) is a well-characterized FXR agonist which also causes pruritus and a moderate increase in low-density lipoprotein cholesterol (LDL-C) levels at 25 mg/day.[77] Apoptosis signaling kinase 1 (ASK-1) activates the P38/JNK pathway to induce cell death.[79] Inhibition of ASK-1 by selonsertib ameliorates NASH and fibrosis in humans.[80] Other potential therapies are also being evaluated for NASH treatment, such as glucagon-like peptide-1 receptor agonists (e.g., Liraglutide), ACC inhibitors, a thyroid hormone receptor β-selective agonist, CCR2–CCR5 inhibitors, etc. (see recent reviews).[18,81]
Overview and Regulation of HNF4α
HNF4α (NR2A1) is a nuclear hormone receptor that is highly expressed in the liver, and to a lesser extent in pancreas, intestine, and kidney.[82] In hepatocytes, HNF4α is a master regulator of many genes involved in hepatocyte differentiation and morphogenesis, drug metabolism, gluconeogenesis, lipid homeostasis, bile acid synthesis and conjugation, ureagenesis, cell proliferation and inflammation.[83–92] Global Hnf4α−/− mice are embryonically lethal,[93] highlighting the importance of HNF4α in development. Loss-of-function mutation of HNF4α causes maturity onset diabetes of the young type 1.[94] Crystallization studies show that HNF4α has long-chain fatty acids in its ligand-binding domain.[95] HNF4α is constitutively active as fatty acids constantly bind to the binding pocket of the ligand binding domain.[96] HNF4α binds as a homodimer to the direct repeat 1 or DR2 sequences in the target genes to regulate gene transcription.
HNF4α is regulated at the transcriptional and post- transcriptional levels. Fasting is known to induce HNF4α mRNA expression,[97] but the underlying mechanism is not clear. More studies have been focused on post- transcriptional regulation of HNF4α expression. Studies by liquid chromatography with tandem mass spectrometry (LC-MS/MS) have identified several phosphorylation sites (S142, T166, S167, T432, S436),[98–100] ubiquitylation sites (K234, K307) and one acetylation site (K458).[98] Sun et al[101] show that protein kinase C phosphorylates a highly conserved serine (S78) to increase HNF4α cytoplasmic localization and degradation. Phosphorylation by protein inase A,[99] AMP-activate protein kinase,[102] proto-oncogene tyrosine-protein kinase Src (c-Src),[103] or ERK1/2 signaling[104] has also been shown to reduce the DNA binding activity and/or stability of HNF4α. Interestingly, inhibition of p38 mitogen-activated protein kinase (MAPK) activity reduces the phosphorylation and nuclear rotein levels of HNF4α[105] suggesting that phosphorylation by p38 MAPK is important for the nuclear retention of HNF4α. Acetylation at lysine residues by CREB-binding protein is reported to be crucial for the proper nuclear retention of HNF4α.[106]
HNF4α may physically interact with forkhead box O1[107] or tribbles homolog 1[108] to reduce HNF4α stability and transcriptional activity. HNF4α may also interact with the co-activator PPARγ coactivator 1α (PGC1α) to induce gluconeogenesis during fasting[97,109] or steroid receptor co-activators (SRC-1, -3) to enhance the transcriptional activity of HNF4α,[110,111] whereas interaction with the co-repressor Hes family basic helix-loop-helix transcription factor 6 [112] represses HNF4α transcription activity. HNF4α may also physically interact with FXR,[113,114] p53,[115] sterol regulatory-binding protein element 1 (SREBP1),[116] Smad3/Smad4,[117,118] specificity protein 1 (SP1),[119] cyclin D1,[120] or small heterodimer partner (SHP)[121] to regulate HNF4α activity.
TGF-β1 is shown to induce HNF4α degradation in the proteosome while nitric oxide incites nitrosylation to inhibit HNF4α activity.[122] The protein arginine N-methyltransferase PRMT1 is shown to bind to and methylate the DNA binding domain of HNF4α, therefore enhancing the binding affinity of HNF4α to target genes.[123]
Epigenetic regulation of HNF4α expression by micro- RNAs has been extensively studied. MicroRNAs are small, non-coding RNA molecules that regulate gene expression often by binding to the 3′UTR of target genes. Several microRNAs, including miR-34a, [2,124–127] miR-24, miR-21,[127] miR-449,[125,126] miR-103a,[128] miR-483-5p,[129] let-7b,[130] and miR-122,[131] have been reported to regulate HNF4α mRNA and/or protein levels.
HNF4α in the Pathogenesis of NAFLD
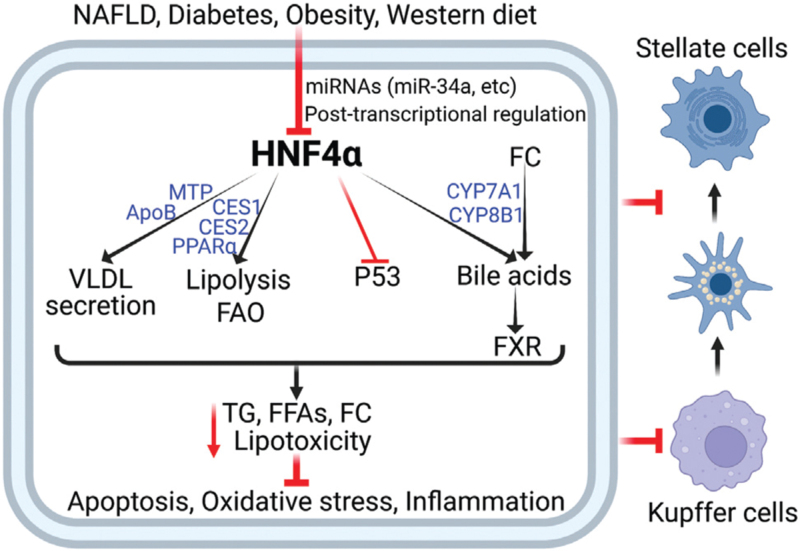
Hepatic HNF4α expression is markedly reduced in NAFLD patients and diabetic or HFD-fed mice.[2,3] The reduction in hepatic HNF4α expression may be partly due to the induction of miR-34a as hepatic miR-34a expression is induced in NAFLD patients and diabetic or HFD-fed mice,[2,132] and overexpression of miR-34a markedly represses HNF4α expression in the liver[2] [Figure 3]. FFAs, FC, and p53 are shown to induce miR-34a expression and repress HNF4α expression.[2,133] During the development and progression of NAFLD, Kupffer cells may secrete pro-inflammatory cytokines. Treatment of HepG2 cells with IL-1β [134] or TNFα [135] represses HNF4α expression, but it remains unclear whether and how IL-1β or TNFa inhibits HNF4α expression in vivo.
Figure 3.
Hepatic HNF4α regulates the development and progression of NAFLD via multiple pathways. Hepatic HNF4α expression is reduced in NAFLD, diabetes and obesity, and by western diet feeding. HNF4α reduces hepatic lipotoxicity by regulating several pathways, including the induction of lipolysis, FAO, VLDL secretion, and bile acid synthesis. HNF4α also inhibits P53 activity. As a result, hepatic apoptosis, oxidative stress, inflammation, and fibrogenesis are inhibited. FC: Free cholesterol; FAO: Fatty acid oxidation; FFAs: Free fatty acids; FXR: Farnesoid X receptor; HNF4α: Hepatocyte nuclear factor 4α; NAFLD: Nonalcoholic fatty liver disease; TG: Triglycerides; VLDL: Very low- density lipoprotein.
HNFα and NAFL
Hepatocyte-specific Hnf4α−/− (Hnf4αΔHep) mice have reduced plasma TG and cholesterol levels and increased hepatic neutral lipid accumulation.[86] Acute ablation of hepatic HNF4α by shRNA also markedly decreases plasma TG and cholesterol levels and increases hepatic TG levels by four-fold.[136] The drastic changes in plasma and hepatic lipid levels likely result from a profound reduction in VLDL secretion as hepatic expression of apolipoprotein B and MTP are markedly reduced.[86,136] In contrast adeno-associated virus serotype 8 (AAV8)-mediated overexpression of human HNF4α in hepatocytes prevents the development of hepatosteatosis induced by a diet enriched im high fat/cholesterol/fructose (HFCF).[56]
In addition to regulating VLDL secretion, hepatocyte HNF4α is an essential regulator of hepatic lipolysis and FAO.[137] Hepatic CES1 and CES2 are shown to have TGH activity and their overexpression increases hepatic triglyceride hydrolysis and FAO, leading to reduced hepatic TG levels.[138,139] Both CES1 and CES2 are direct target genes of HNF4α.[139,140] Overexpression of hepatocyte HNF4α promotes lipolysis and FAO, whereas loss of hepatocyte HNF4α has opposite effects.[137] Thus, CES1 and CES2 may be partly involved in the regulation of lipolysis and FAO and hepatic TG levels by HNF4α.
HNF4α and NASH
AAV8-mediated overexpression of human HNF4α in hepatocytes protects against HFCF diet-induced steatohepatitis, whereas loss of hepatocyte HNF4α has an opposite effect.[137] P53 is a tumor suppressor and a primary stress sensor that is induced in the liver of NAFLD patients and experimental NASH.[141–143] Ablation or inhibition of p53 attenuates diet-induced apoptosis and steatohepatitis.[141,144] Overexpression of HNF4α inhibits p53 expression and apoptosis in a p53-dependent manner.[137] HNF4α plays an important role in regulating bile acid synthesis. Cholesterol 7a-hydroxylase (CYP7A1) and sterol 12a-hydroxylase (CYP8B1) are two of the key enzymes in the classic pathway of bile acid biosynthesis. Both Cyp7a1 and Cyp8b1 are reduced in Hnf4αΔHep mice. Recapitulation of hepatic Cyp7a1 and Cyp8b1 expression in Hnf4αΔHep mice prevents HFCF diet-induced NASH, which likely results from activation of FXR[137] as bile acids are endogenous ligands for FXR. FXR activation by OCA is shown to inhibit p53 activation and apoptosis.[145] Overexpression of hepatocyte HNF4α also reduces hepatic FC and FFA levels whereas loss of hepatocyte HNF4α has opposite effects. The changes in hepatic FC and FFA levels may also contribute to hepatic lipotoxicity and NASH development. In addition, HNF4α is shown to inhibit the expression and nuclear translocation of RelA (p65) and NF-κB activation via induction of miR-7 and miR-124.[146] NASH is a risk factor for HCC. Overexpression of HNF4α inhibits the development of HCC likely by inhibiting β-catenin activation.[147,148]
HNF4α as a therapeutic target
Since hepatic HNF4α is markedly repressed in NASH and liver fibrosis.[2–6] HNF4α may be a therapeutic target for treatment of NAFLD. Adenovirus-mediated overexpression of HNF4α is shown to attenuate liver fibrosis induced by dimethylnitrosamine or bile duct ligation.[5] AAV8- mediated overexpression of HNF4α under the control of an albumin promoter is shown to attenuate HFCF diet- induced NAFL and NASH.[137] Yang et al[6] show that delivery of HNF4α mRNA in lipid nanoparticles to four different mouse models protects against hepatoxin- and cholestasis-induced liver fibrosis. Compounds that can induce HNF4α expression or activation have also been investigated. Lee et al[149] show that N-trans caffeoyltyr- amine (NCT) is an HNF4α activator, and administration of this compound can prevent HFD-induced hepatostea- tosis, although its role in NASH needs to be evaluated. These promising findings suggest that HNF4α may be a good candidate for treatment of NASH.
Conclusion and Future Perspectives
NAFLD is the most common chronic liver disease in developed countries. So far, the pathogenic mechanisms of NAFLD remain to be fully elucidated. No drugs have been approved for NASH treatment. As one of the most abundantly expressed genes in the liver, HNF4α appears to be a key player in the pathogenesis of NAFLD, which is supported by several lines of evidence. First, the expression of hepatic HNF4α is markedly reduced in NAFLD patients, diabetic or HFD-fed mice, and fibrotic livers. Second, ablation of hepatocyte HNF4α promotes the development and progression of NAFLD in a mouse model of NASH. Third, AAV8-mediated overexpression of HNF4α in hepatocytes attenuates steatohepatitis in mice. Delivery of HNF4α by adenovirus or lipid nanoparticles-embedded mRNA inhibits liver fibrogenesis. Administration of a compound that induces HNF4α expression prevents HFD from inducing hepatosteatosis. These findings highlight the importance of HNF4α in the pathogenesis of NAFLD and suggest that hepatic HNF4α may be targeted for treatment of NAFLD.
Hepatic HNF4α inhibits the development and progression of NAFLD via regulation of multiple pathways, including VLDL secretion, lipolysis, FAO, apoptosis, lipotoxicity, and inflammation. P53 and bile acid signaling pathways play an important role in the progression of NAFL to NASH mediated by HNF4α. Although increased hepatic HNF4α expression may cause hyperlipidemia via increased VLDL secretion, Huang et al[150] report that delivery of small activating RNA specific for upregulating HNF4α to rats improves FAO and liver steatosis, and lowers plasma TG levels, suggesting that raising hepatic HNF4α expression may even improve dyslipidemia. Hepatic HNF4α can increase TG hydrolysis, FAO, and the conversion of cholesterol to bile acids via inducing CYP7A1 and CYP8B1 expression, which may help to reduce VLDL-TG or VLDL- cholesterol levels. Considering the factors discussed above, it is plausible to summarize that hepatic HNF4α is a promising therapeutic target for NASH.
Acknowledgements
The figures were created via BioRenders.com.
Funding
This work is supported by the grants from National Institutes of Health (R01DK102619, R01DK118941, R01DK118805, and R0DK121548).
Conflicts of interest
None.
Footnotes
How to cite this article: Pan X, Zhang Y. Hepatocyte nuclear factor 4α in the pathogenesis of non-alcoholic fatty liver disease. Chin Med J 2022;135:1172–1181. doi: 10.1097/CM9.0000000000002092
References
- 1.Loomba R, Friedman SL, Shulman GI. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 2021; 184:2537–2564. doi: 10.1016/j.cell.2021.04.015. [DOI] [PubMed] [Google Scholar]
- 2.Xu Y, Zalzala M, Xu J, Li Y, Yin L, Zhang Y. A metabolic stressinducible miR-34a-HNF4alpha pathway regulates lipid and lipoprotein metabolism. Nat Commun 2015; 6:7466.doi: 10.1038/ncomms8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Y, Hu S, Jadhav K, Zhu Y, Pan X, Bawa FC, et al. Hepatocytic activating transcription factor 3 protects against steatohepatitis via hepatocyte nuclear factor 4α. Diabetes 2021; 70:2506–2517. doi: 10.2337/db21-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishikawa T, Bell A, Brooks JM, Setoyama K, Melis M, Han B, et al. Resetting the transcription factor network reverses terminal chronic hepatic failure. J Clin Invest 2015; 125:1533–1544. doi: 10.1172/JCI73137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yue HY, Yin C, Hou JL, Zeng X, Chen YX, Zhong W, et al. Hepatocyte nuclear factor 4alpha attenuates hepatic fibrosis in rats. Gut 2010; 59:236–246. doi: 10.1136/gut.2008.174904. [DOI] [PubMed] [Google Scholar]
- 6.Yang T, Poenisch M, Khanal R, Hu Q, Dai Z, Li R, et al. Therapeutic HNF4α mRNA attenuates liver fibrosis in a preclinical model. J Hepatol 2021; 75:1420–1433. doi: 10.1016/j.jhep.2021.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Day CP, James OF. Steatohepatitis: a tale oftwo “hits”? Gastroenterology 1998; 114:842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- 8.Day CP. From fat to inflammation. Gastroenterology 2006; 130:207–210. doi: 10.1053/j.gastro.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 9.Buzzetti E, Pinzani M, Tsochatzis EA. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism 2016; 65:1038–1048. doi: 10.1016/j.metabol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Fang YL, Chen H, Wang CL, Liang L. Pathogenesis of nonalcoholic fatty liver disease in children and adolescence: from “two hit theory” to “multiple hit model”. World J Gastroenterol 2018; 24:2974–2983. doi: 10.3748/wjg.v24.i27.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Younossi Z, Tacke F, Arrese M, Sharma BC, Mostafa I, Bugianesi E, et al. Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology 2019; 69:2672–2682. doi: 10.1002/hep.30251. [DOI] [PubMed] [Google Scholar]
- 12.Kim D, Kim WR. Nonobese fatty liver disease. Clin Gastroenterol Hepatol 2017; 15:474–485. doi: 10.1016/j.cgh.2016.08.028. [DOI] [PubMed] [Google Scholar]
- 13.Rich NE, Oji S, Mufti AR, Browning JD, Parikh ND, Odewole M, et al. Racial and ethnic disparities in nonalcoholic fatty liver disease prevalence, severity, and outcomes in the United States: a systematic review and meta-analysis. Clin Gastroenterol Hepatol 2018; 16:198–210.e2.doi: 10.1016/j. cgh.2017.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 2018; 15:11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 15.Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018; 67:328–357. doi: 10.1002/hep.29367. [DOI] [PubMed] [Google Scholar]
- 16.Polyzos SA, Kountouras J, Mantzoros CS. Obesity and nonalcoholic fatty liver disease: from pathophysiology to therapeutics. Metabolism 2019; 92:82–97. doi: 10.1016/j.metabol.2018. 11.014. [DOI] [PubMed] [Google Scholar]
- 17.Younossi ZM, Golabi P, de Avila L, Paik JM, Srishord M, Fukui N, et al. The global epidemiology of NAFLD and NASH in patients with type 2 diabetes: a systematic review and meta-analysis. J Hepatol 2019; 71:793–801. doi: 10.1016/j.jhep.2019.06.021. [DOI] [PubMed] [Google Scholar]
- 18.Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med 2018; 24:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, Brown MS, Goldstein JL. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci USA 2010; 107:3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Owen JL, Zhang Y, Bae SH, Farooqi MS, Liang G, Hammer RE, et al. Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc Natl Acad Sci USA 2012; 109:16184–16189. doi: 10.1073/pnas.1213343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kammoun HL, Chabanon H, Hainault I, Luquet S, Magnan C, Koike T, et al. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Invest 2009; 119:1201–1215. doi: 10.1172/JCI37007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdul-Wahed A, Guilmeau S, Postic C. Sweet sixteenth for ChREBP: established roles and future goals. Cell Metab 2017; 26:324–341. doi: 10.1016/j.cmet.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Kawano Y, Cohen DE. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J Gastroenterol 2013; 48:434–441. doi: 10.1007/s00535-013-0758-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmid AI, Szendroedi J, Chmelik M, Krssak M, Moser E, Roden M. Liver ATP synthesis is lower and relates to insulin sensitivity in patients with type 2 diabetes. Diabetes Care 2011; 34:448–453. doi: 10.2337/dc10-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cortez-Pinto H, Chatham J, Chacko VP, Arnold C, Rashid A, Diehl AM. Alterations in liver ATP homeostasis in human nonalcoholic steatohepatitis: a pilot study. JAMA 1999; 282:1659–1664. doi: 10.1001/jama.282.17.1659. [DOI] [PubMed] [Google Scholar]
- 26.Sanyal AJ, Campbell-Sargent C, Mirshahi F, Rizzo WB, Contos MJ, Sterling RK, et al. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology 2001; 120:1183–1192. doi: 10.1053/gast.2001.23256. [DOI] [PubMed] [Google Scholar]
- 27.Satapati S, Sunny NE, Kucejova B, Fu X, He TT, Mendez-Lucas A, et al. Elevated TCA cycle function in the pathology of diet-induced hepatic insulin resistance and fatty liver. J Lipid Res 2012; 53:1080–1092. doi: 10.1194/jlr.M023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ong KT, Mashek MT, Bu SY, Greenberg AS, Mashek DG. Adipose triglyceride lipase is a major hepatic lipase that regulates triacylglycerol turnover and fatty acid signaling and partitioning. Hepatology 2011; 53:116–126. doi: 10.1002/hep.24006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turpin SM, Hoy AJ, Brown RD, Rudaz CG, Honeyman J, Matzaris M, et al. Adipose triacylglycerol lipase is a major regulator of hepatic lipid metabolism but not insulin sensitivity in mice. Diabetologia 2011; 54:146–156. doi: 10.1007/s00125-010-1895-5. [DOI] [PubMed] [Google Scholar]
- 30.Quiroga AD, Lehner R. Pharmacological intervention of liver triacylglycerol lipolysis: the good, the bad and the ugly. Biochem Pharmacol 2018; 155:233–241. doi: 10.1016/j.bcp.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2008; 40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romeo S, Huang-Doran I, Baroni MG, Kotronen A. Unravelling the pathogenesis of fatty liver disease: patatin-like phospholipase domain-containing 3 protein. Curr Opin Lipidol 2010; 21:247–252. doi: 10.1097/mol.0b013e328338ca61. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Kory N, BasuRay S, Cohen JC, Hobbs HH. PNPLA3, CGI-58, and inhibition of hepatic triglyceride hydrolysis in mice. Hepatology 2019; 69:2427–2441. doi: 10.1002/hep.30583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang A, Mottillo EP, Mladenovic-Lucas L, Zhou L, Granneman JG. Dynamic interactions of ABHD5 with PNPLA3 regulate triacylglycerol metabolism in brown adipocytes. Nat Metab 2019; 1:560–569. doi: 10.1038/s42255-019-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjaerg-Hansen A, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2014; 46:352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahdessian H, Taxiarchis A, Popov S, Silveira A, Franco-Cereceda A, Hamsten A, et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci USA 2014; 111:8913–8918. doi: 10.1073/pnas.1323785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mancina RM, Dongiovanni P, Petta S, Pingitore P, Meroni M, Rametta R, et al. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European Descent. Gastroenterology 2016; 150:1219–1230.e6.doi: 10.1053/j.gastro.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luukkonen PK, Zhou Y, Hyotylainen T, Leivonen M, Arola J, Orho-Melander M, et al. The MBOAT7 variant rs641738 alters hepatic phosphatidylinositols and increases severity of nonalcoholic fatty liver disease in humans. J Hepatol 2016; 65:1263–1265. doi: 10.1016/j.jhep.2016.07.045. [DOI] [PubMed] [Google Scholar]
- 39.Helsley RN, Varadharajan V, Brown AL, Gromovsky AD, Schugar RC, Ramachandiran I, et al. Obesity-linked suppression of membranebound O-acyltransferase 7 (MBOAT7) drives non-alcoholic fatty liver disease. Elife 2019; 8:e49882.doi: 10.7554/eLife.49882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma Y, Belyaeva OV, Brown PM, Fujita K, Valles K, Karki S, et al. 17-Beta hydroxysteroid dehydrogenase 13 is a hepatic retinol dehydrogenase associated with histological features of nonalcoholic fatty liver disease. Hepatology 2019; 69:1504–1519. doi: 10.1002/hep.30350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pirola CJ, Garaycoechea M, Flichman D, Arrese M, San Martino J, Gazzi C, et al. Splice variant rs72613567 prevents worst histologic outcomes in patients with nonalcoholic fatty liver disease. J Lipid Res 2019; 60:176–185. doi: 10.1194/jlr.P089953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abul-Husn NS, Cheng X, Li AH, Xin Y, Schurmann C, Stevis P, et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med 2018; 378:1096–1106. doi: 10.1056/NEJMoa1712191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romeo S, Sanyal A, Valenti L. Leveraging human genetics to identify potential new treatments for fatty liver disease. Cell Metab 2020; 31:35–45. doi: 10.1016/j.cmet.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 44.Sheka AC, Adeyi O, Thompson J, Hameed B, Crawford PA, Ikramuddin S. Nonalcoholic steatohepatitis: a review. JAMA 2020; 323:1175–1183. doi: 10.1001/jama.2020.2298. [DOI] [PubMed] [Google Scholar]
- 45.Powell EE, Wong VW, Rinella M. Non-alcoholic fatty liver disease. Lancet 2021; 397:2212–2224. doi: 10.1016/S0140-6736 (20)32511-3. [DOI] [PubMed] [Google Scholar]
- 46.Neuschwander-Tetri BA. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: the central role of nontriglyceride fatty acid metabolites. Hepatology 2010; 52:774–788. doi: 10.1002/hep.23719. [DOI] [PubMed] [Google Scholar]
- 47.Xiao J, Tipoe GL. Inflammasomes in non-alcoholic fatty liver disease. Front Biosci (Landmark Ed) 2016; 21:683–695. doi: 10.2741/4414. [DOI] [PubMed] [Google Scholar]
- 48.Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 2010; 464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rajamaki K, Lappalainen J, Oorni K, Valimaki E, Matikainen S, Kovanen PT, et al. Cholesterol crystals activate the NLRP3 inflammasome in human macrophages: a novel link between cholesterol metabolism and inflammation. PLoS One 2010; 5:e11765.doi: 10.1371/journal.pone.0011765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Carvalho Ribeiro M, Szabo G. Role of the inflammasome in liver disease. Annu Rev Pathol 2022; 17:345–365. doi: 10.1146/annurev-pathmechdis-032521-102529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Afonso MB, Castro RE, Rodrigues CMP. Processes exacerbating apoptosis in non-alcoholic steatohepatitis. Clin Sci (Lond) 2019; 133:2245–2264. doi: 10.1042/CS20190068. [DOI] [PubMed] [Google Scholar]
- 52.Alkhouri N, Carter-Kent C, Feldstein AE. Apoptosis in nonalcoholic fatty liver disease: diagnostic and therapeutic implications. Expert Rev Gastroenterol Hepatol 2011; 5:201–212. doi: 10.1586/egh.11.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feldstein AE, Canbay A, Angulo P, Taniai M, Burgart LJ, Lindor KD, et al. Hepatocyte apoptosis and fas expression are prominent features of human nonalcoholic steatohepatitis. Gastroenterology 2003; 125:437–443. doi: 10.1016/s0016-5085(03)00907-7. [DOI] [PubMed] [Google Scholar]
- 54.Ferreira DMS, Castro RE, Machado MV, Evangelista T, Silvestre A, Costa A, et al. Apoptosis and insulin resistance in liver and peripheral tissues of morbidly obese patients is associated with different stages of non-alcoholic fatty liver disease. Diabetologia 2011; 54:1788–1798. doi: 10.1007/s00125-011-2130-8. [DOI] [PubMed] [Google Scholar]
- 55.Machado MV, Michelotti GA, de Almeida Pereira T, Boursier J, Kruger L, Swiderska-Syn M, et al. Reduced lipoapoptosis, hedgehog pathway activation and fibrosis in caspase-2 deficient mice with non-alcoholic steatohepatitis. Gut 2015; 64:1148–1157. doi: 10.1136/gutjnl-2014-307362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). UK Prospective Diabetes Study (UKPDS) Group. Lancet 1998; 352:837–853. [PubMed] [Google Scholar]
- 57.Malhi H, Bronk SF, Werneburg NW, Gores GJ. Free fatty acids induce JNK-dependent hepatocyte lipoapoptosis. J Biol Chem 2006; 281:12093–12101. doi: 10.1074/jbc.M510660200. [DOI] [PubMed] [Google Scholar]
- 58.Witek RP, Stone WC, Karaca FG, Syn WK, Pereira TA, Agboola KM, et al. Pan-caspase inhibitor VX-166 reduces fibrosis in an animal model of nonalcoholic steatohepatitis. Hepatology 2009; 50:1421–1430. doi: 10.1002/hep.23167. [DOI] [PubMed] [Google Scholar]
- 59.Anstee QM, Concas D, Kudo H, Levene A, Pollard J, Charlton P, et al. Impact of pan-caspase inhibition in animal models of established steatosis and non-alcoholic steatohepatitis. J Hepatol 2010; 53:542–550. doi: 10.1016/j.jhep.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 60.Barreyro FJ, Holod S, Finocchietto PV, Camino AM, Aquino JB, Avagnina A, et al. The pan-caspase inhibitor Emricasan (IDN- 6556) decreases liver injury and fibrosis in a murine model of nonalcoholic steatohepatitis. Liver Int 2015; 35:953–966. doi: 10.1111/liv.12570. [DOI] [PubMed] [Google Scholar]
- 61.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002; 2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 62.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013; 200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Srinivas AN, Suresh D, Santhekadur PK, Suvarna D, Kumar DP. Extracellular vesicles as inflammatory drivers in NAFLD. Front Immunol 2020; 11:627424.doi: 10.3389/fimmu.2020.627424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cai S, Cheng X, Pan X, Li J. Emerging role of exosomes in liver physiology and pathology. Hepatol Res 2017; 47:194–203. doi: 10.1111/hepr.12794. [DOI] [PubMed] [Google Scholar]
- 65.Kakazu E, Mauer AS, Yin M, Malhi H. Hepatocytes release ceramide-enriched pro-inflammatory extracellular vesicles in an IRE1a-dependent manner. J Lipid Res 2016; 57:233–245. doi: 10.1194/jlr.M063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hirsova P, Ibrahim SH, Krishnan A, Verma VK, Bronk SF, Werneburg NW, et al. Lipid-induced signaling causes release of inflammatory extracellular vesicles from hepatocytes. Gastroenterology 2016; 150:956–967. doi: 10.1053/j.gastro.2015.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gil-Gomez A, Brescia P, Rescigno M, Romero-Gomez M. Gutliver axis in nonalcoholic fatty liver disease: the impact of the metagenome, end products, and the epithelial and vascular barriers. Semin Liver Dis 2021; 41:191–205. doi: 10.1055/s-0041-1723752. [DOI] [PubMed] [Google Scholar]
- 68.Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016; 63:764–775. doi: 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bashiardes S, Shapiro H, Rozin S, Shibolet O, Elinav E. Nonalcoholic fatty liver and the gut microbiota. Mol Metab 2016; 5:782–794. doi: 10.1016/j.molmet.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Torres DM, Williams CD, Harrison SA. Features, diagnosis, and treatment of nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol 2012; 10:837–858. doi: 10.1016/j.cgh.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 71.Park CC, Nguyen P, Hernandez C, Bettencourt R, Ramirez K, Fortney L, et al. Magnetic resonance elastography vs transient elastography in detection of fibrosis and noninvasive measurement of steatosis in patients with biopsy-proven nonalcoholic fatty liver disease. Gastroenterology 2017; 152:598–607.e2.doi: 10.1053/j. gastro.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiao G, Zhu S, Xiao X, Yan L, Yang J, Wu G. Comparison of laboratory tests, ultrasound, or magnetic resonance elastography to detect fibrosis in patients with nonalcoholic fatty liver disease: a meta-analysis. Hepatology 2017; 66:1486–1501. doi: 10.1002/hep.29302. [DOI] [PubMed] [Google Scholar]
- 73.Vilar-Gomez E, Martinez-Perez Y, Calzadilla-Bertot L, Torres-Gonzalez A, Gra-Oramas B, Gonzalez-Fabian L, et al. Weight loss through lifestyle modification significantly reduces features of nonalcoholic steatohepatitis. Gastroenterology 2015; 149:367–378.e5.doi: 10.1053/j.gastro.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 74.Aminian A, Al-Kurd A, Wilson R, Bena J, Fayazzadeh H, Singh T, et al. Association of bariatric surgery with major adverse liver and cardiovascular outcomes in patients with biopsy-proven nonalcoholic steatohepatitis. JAMA 2021; 326:2031–2042. doi: 10.1001/jama.2021.19569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mahady SE, Webster AC, Walker S, Sanyal A, George J. The role of thiazolidinediones in non-alcoholic steatohepatitis - a systematic review and meta analysis. J Hepatol 2011; 55:1383–1390. doi: 10.1016/j.jhep.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 76.Sanyal AJ, Chalasani N, Kowdley KV, McCullough A, Diehl AM, Bass NM, et al. Pioglitazone, vitamin E, or placebo for nonalcoholic steatohepatitis. N Engl J Med 2010; 362:1675–1685. doi: 10.1056/NEJMoa0907929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mudaliar S, Henry RR, Sanyal AJ, Morrow L, Marschall HU, Kipnes M, et al. Efficacy and safety of the farnesoid X receptor agonist obeticholic acid in patients with type 2 diabetes and nonalcoholic fatty liver disease. Gastroenterology 2013; 145:574–582.e1.doi: 10.1053/j.gastro.2013.05.042. [DOI] [PubMed] [Google Scholar]
- 78.Radun R, Trauner M. Role of FXR in bile acid and metabolic homeostasis in NASH: pathogenetic concepts and therapeutic opportunities. Semin Liver Dis 2021; 41:461–475. doi: 10.1055/s- 0041-1731707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ogier JM, Nayagam BA, Lockhart PJ. ASK1 inhibition: a therapeutic strategy with multi-system benefits. J Mol Med (Berl) 2020; 98:335–348. doi: 10.1007/s00109-020-01878-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loomba R, Lawitz E, Mantry PS, Jayakumar S, Caldwell SH, Arnold H, et al. The ASK1 inhibitor selonsertib in patients with nonalcoholic steatohepatitis: a randomized, phase 2 trial. Hepatology 2018; 67:549–559. doi: 10.1002/hep.29514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parlati L, Regnier M, Guillou H, Postic C. New targets for NAFLD. JHEP Rep 2021; 3:100346.doi: 10.1016/j.jhepr.2021.100346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Drewes T, Senkel S, Holewa B, Ryffel GU. Human hepatocyte nuclear factor 4 isoforms are encoded by distinct and differentially expressed genes. Mol Cell Biol 1996; 16:925–931. doi: 10.1128/MCB.16.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Babeu JP, Boudreau F. Hepatocyte nuclear factor 4-alpha involvement in liver and intestinal inflammatory networks. World J Gastroenterol 2014; 20:22–30. doi: 10.3748/wjg.v20.i1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hwang-Verslues WW, Sladek FM. HNF4α - role in drug metabolism and potential drug target? Curr Opin Pharmacol 2010; 10:698–705. doi: 10.1016/j.coph.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Inoue Y, Yu AM, Yim SH, Ma X, Krausz KW, Inoue J, et al. Regulation of bile acid biosynthesis by hepatocyte nuclear factor 4alpha. J Lipid Res 2006; 47:215–227. doi: 10.1194/jlr.M500430-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 2001; 21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Inoue Y, Hayhurst GP, Inoue J, Mori M, Gonzalez FJ. Defective ureagenesis in mice carrying a liver-specific disruption of hepatocyte nuclear factor 4alpha (HNF4alpha). HNF4alpha regulates ornithine transcarbamylase in vivo. J Biol Chem 2002; 277:25257–25265. doi: 10.1074/jbc.M203126200. [DOI] [PubMed] [Google Scholar]
- 88.Lu H, Gonzalez FJ, Klaassen C. Alterations in hepatic mRNA expression of phase II enzymes and xenobiotic transporters after targeted disruption of hepatocyte nuclear factor 4 alpha. Toxicol Sci 2010; 118:380–390. doi: 10.1093/toxsci/kfq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bonzo JA, Ferry CH, Matsubara T, Kim JH, Gonzalez FJ. Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4a in adult mice. J Biol Chem 2012; 287:7345–7356. doi: 10.1074/jbc.M111.334599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Inoue Y, Yu AM, Inoue J, Gonzalez FJ. Hepatocyte nuclear factor 4alpha is a central regulator of bile acid conjugation. J Biol Chem 2004; 279:2480–2489. doi: 10.1074/jbc.M311015200. [DOI] [PubMed] [Google Scholar]
- 91.Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev 2000; 14:464–474. doi: 10.1101/gad.14.4.464. [PMC free article] [PubMed] [Google Scholar]
- 92.Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev 2006; 20:2293–2305. doi: 10.1101/gad.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, et al. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev 1994; 8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- 94.Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, et al. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1). Nature 1996; 384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 95.Dhe-Paganon S, Duda K, Iwamoto M, Chi YI, Shoelson SE. Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand. J Biol Chem 2002; 277:37973–37976. doi: 10.1074/jbc.C200420200. [DOI] [PubMed] [Google Scholar]
- 96.Wisely GB, Miller AB, Davis RG, Thornquest AD, Jr, Johnson R, Spitzer T, et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure 2002; 10:1225–1234. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- 97.Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 2001; 413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 98.Yokoyama A, Katsura S, Ito R, Hashiba W, Sekine H, Fujiki R, et al. Multiple post-translational modifications in hepatocyte nuclear factor 4a. Biochem Biophys Res Commun 2011; 410:749–753. doi: 10.1016/j.bbrc.2011.06.033. [DOI] [PubMed] [Google Scholar]
- 99.Viollet B, Kahn A, Raymondjean M. Protein kinase A-dependent phosphorylation modulates DNA-binding activity of hepatocyte nuclear factor 4. Mol Cell Biol 1997; 17:4208–4219. doi: 10.1128/MCB.17.8.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Daigo K, Kawamura T, Ohta Y, Ohashi R, Katayose S, Tanaka T, et al. Proteomic analysis of native hepatocyte nuclear factor-4a (HNF4α) isoforms, phosphorylation status, and interactive cofactors. J Biol Chem 2011; 286:674–686. doi: 10.1074/jbc.M110.154732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun K, Montana V, Chellappa K, Brelivet Y, Moras D, Maeda Y, et al. Phosphorylation of a conserved serine in the deoxyribonucleic acid binding domain of nuclear receptors alters intracellular localization. Mol Endocrinol 2007; 21:1297–1311. doi: 10.1210/me.2006-0300. [DOI] [PubMed] [Google Scholar]
- 102.Hong YH, Varanasi US, Yang W, Leff T. AMP-activated protein kinase regulates HNF4alpha transcriptional activity by inhibiting dimer formation and decreasing protein stability. J Biol Chem 2003; 278:27495–27501. doi: 10.1074/jbc.M304112200. [DOI] [PubMed] [Google Scholar]
- 103.Chellappa K, Jankova L, Schnabl JM, Pan S, Brelivet Y, Fung CLS, et al. Src tyrosine kinase phosphorylation of nuclear receptor HNF4α correlates with isoform-specific loss of HNF4α in human colon cancer. Proc Natl Acad Sci USA 2012; 109:2302–2307. doi: 10.1073/pnas.1106799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Veto B, Bojcsuk D, Bacquet C, Kiss J, Sipeki S, Martin L, et al. The transcriptional activity of hepatocyte nuclear factor 4 alpha is inhibited via phosphorylation by ERK1/2. PLoS One 2017; 12:e0172020.doi: 10.1371/journal.pone.0172020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xu Z, Tavares-Sanchez OL, Li Q, Fernando J, Rodriguez CM, Studer EJ, et al. Activation of bile acid biosynthesis by the p38 mitogen-activated protein kinase (MAPK): hepatocyte nuclear factor-4alpha phosphorylation by the p38 MAPK is required for cholesterol 7alpha-hydroxylase expression. J Biol Chem 2007; 282:24607–24614. doi: 10.1074/jbc.M611481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol Cell 2000; 5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 107.Ganjam GK, Dimova EY, Unterman TG, Kietzmann T. FoxO1 and HNF-4 are involved in regulation of hepatic glucokinase gene expression by resveratrol. J Biol Chem 2009; 284:30783–30797. doi: 10.1074/jbc.M109.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Soubeyrand S, Martinuk A, McPherson R. TRIB1 is a positive regulator of hepatocyte nuclear factor 4-alpha. Sci Rep 2017; 7:5574.doi: 10.1038/s41598-017-05768-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rhee J, Inoue Y, Yoon JC, Puigserver P, Fan M, Gonzalez FJ, et al. Regulation of hepatic fasting response by PPARgamma coactiva-tor-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proc Natl Acad Sci USA 2003; 100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang JC, Stafford JM, Granner DK. SRC-1 and GRIP1 coactivate transcription with hepatocyte nuclear factor 4. J Biol Chem 1998; 273:30847–30850. doi: 10.1074/jbc.273.47.30847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee YK, Dell H, Dowhan DH, Hadzopoulou-Cladaras M, Moore DD. The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: two mechanisms for repression. Mol Cell Biol 2000; 20:187–195. doi: 10.1128/MCB.20.1.187-195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Martinez-Jimenez CP, Kyrmizi I, Cardot P, Gonzalez FJ, Talianidis I. Hepatocyte nuclear factor 4alpha coordinates a transcription factor network regulating hepatic fatty acid metabolism. Mol Cell Biol 2010; 30:565–577. doi: 10.1128/MCB.00927-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Thomas AM, Hart SN, Li G, Lu H, Fang Y, Fang J, et al. Hepatocyte nuclear factor 4 alpha and farnesoid X receptor co-regulates gene transcription in mouse livers on a genome-wide scale. Pharm Res 2013; 30:2188–2198. doi: 10.1007/s11095-013-1006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Caron S, Samanez CH, Dehondt H, Ploton M, Briand O, Lien F, et al. Farnesoid X receptor inhibits the transcriptional activity of carbohydrate response element binding protein in human hepatocytes. Mol Cell Biol 2013; 33:2202–2211. doi: 10.1128/MCB.01004-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Maeda Y, Seidel SD, Wei G, Liu X, Sladek FM. Repression of hepatocyte nuclear factor 4alpha tumor suppressor p53: involvement of the ligand-binding domain and histone deacetylase activity. Mol Endocrinol 2002; 16:402–410. doi: 10.1210/mend.16.2.0769. [DOI] [PubMed] [Google Scholar]
- 116.Yamamoto T, Shimano H, Nakagawa Y, Ide T, Yahagi N, Matsuzaka T, et al. SREBP-1 interacts with hepatocyte nuclear factor-4 alpha and interferes with PGC-1 recruitment to suppress hepatic gluconeogenic genes. J Biol Chem 2004; 279:12027–12035. doi: 10.1074/jbc.M310333200. [DOI] [PubMed] [Google Scholar]
- 117.Chou WC, Prokova V, Shiraishi K, Valcourt U, Moustakas A, Hadzopoulou-Cladaras M, et al. Mechanism of a transcriptional cross talk between transforming growth factor-beta-regulated Smad3 and Smad4 proteins and orphan nuclear receptor hepatocyte nuclear factor-4. Mol Biol Cell 2003; 14:1279–1294. doi: 10.1091/mbc.e02-07-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kardassis D, Pardali K, Zannis VI. SMAD proteins transactivate the human ApoCIII promoter by interacting physically and functionally with hepatocyte nuclear factor 4. J Biol Chem 2000; 275:41405–41414. doi: 10.1074/jbc.M007896200. [DOI] [PubMed] [Google Scholar]
- 119.Hwang-Verslues WW, Sladek FM. Nuclear receptor hepatocyte nuclear factor 4alpha1 competes with oncoprotein c-Myc for control of the p21/WAF1 promoter. Mol Endocrinol 2008; 22:78–90. doi: 10.1210/me.2007-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hanse EA, Mashek DG, Becker JR, Solmonson AD, Mullany LK, Mashek MT, et al. Cyclin D1 inhibits hepatic lipogenesis via repression of carbohydrate response element binding protein and hepatocyte nuclear factor 4alpha. Cell Cycle 2012; 11:2681–2690. doi: 10.4161/cc.21019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang M, Chiang JY. Transcriptional regulation of the human sterol 12alpha-hydroxylase gene (CYP8B1): roles of heaptocyte nuclear factor 4alpha in mediating bile acid repression. J Biol Chem 2001; 276:41690–41699. doi: 10.1074/jbc.M105117200. [DOI] [PubMed] [Google Scholar]
- 122.de Lucas S, Lopez-Alcorocho JM, Bartolome J, Carreno V. Nitric oxide and TGF-beta1 inhibit HNF-4alpha function in HEPG2 cells. Biochem Biophys Res Commun 2004; 321:688–694. doi: 10.1016/j.bbrc.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 123.Barrero MJ, Malik S. Two functional modes of a nuclear receptor- recruited arginine methyltransferase in transcriptional activation. Mol Cell 2006; 24:233–243. doi: 10.1016/j.molcel.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Takagi S, Nakajima M, Kida K, Yamaura Y, Fukami T, Yokoi T. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem 2010; 285:4415–4422. doi: 10.1074/jbc. M109.085431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang Z, Burke PA. The role of microRNAs in hepatocyte nuclear factor-4alpha expression and transactivation. Biochim Biophys Acta 2013; 1829:436–442. doi: 10.1016/j.bbagrm.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ramamoorthy A, Li L, Gaedigk A, Bradford LD, Benson EA, Flockhart DA, et al. In silico and in vitro identification of microRNAs that regulate hepatic nuclear factor 4a expression. Drug Metab Dispos 2012; 40:726–733. doi: 10.1124/dmd.111.040329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wirsing A, Senkel S, Klein-Hitpass L, Ryffel GU. A systematic analysis of the 3′UTR of HNF4α mRNA reveals an interplay of regulatory elements including miRNA target sites. PLoS One 2011; 6:e27438.doi: 10.1371/journal.pone.0027438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen M, Li MH, Zhang N, Sun WW, Wang H, Wang YA, et al. Pro-angiogenic effect of exosomal microRNA-103a in mice with rheumatoid arthritis via the downregulation of hepatocyte nuclear factor 4 alpha and activation of the JAK/STAT3 signaling pathway. J Biol Regul Homeost Agents 2021; 35:629–640. doi: 10.23812/20-537-A. [DOI] [PubMed] [Google Scholar]
- 129.Sun J, Li X, Wang W, Li W, Gao S, Yan J. Mir-483-5p promotes the malignant transformation of immortalized human esophageal epithelial cells by targeting HNF4α. Int J Clin Exp Pathol 2017; 10:9391–9399. [PMC free article] [PubMed] [Google Scholar]
- 130.Alizadeh E, Akbarzadeh A, Eslaminejad MB, Barzegar A, Hashemzadeh S, Nejati-Koshki K, et al. Up regulation of liver-enriched transcription factors HNF4α and HNF6 and liver-specific microRNA (miR-122) by inhibition of let-7b in mesenchymal stem cells. Chem Biol Drug Des 2015; 85:268–279. doi: 10.1111/cbdd.12398. [DOI] [PubMed] [Google Scholar]
- 131.Deng XG, Qiu RL, Wu YH, Li ZX, Xie P, Zhang J, et al. Overexpression of miR-122 promotes the hepatic differentiation and maturation of mouse ESCs through a miR-122/FoxA1/HNF4α-positive feedback loop. Liver Int 2014; 34:281–295. doi: 10.1111/liv.12239. [DOI] [PubMed] [Google Scholar]
- 132.Fu T, Choi SE, Kim DH, Seok S, Suino-Powell KM, Xu HE, et al. Aberrantly elevated microRNA-34a in obesity attenuates hepatic responses to FGF19 by targeting a membrane coreceptor (-Klotho. Proc Natl Acad Sci USA 2012; 109:16137–16142. doi: 10.1073/pnas.1205951109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hermeking H. p53 enters the microRNA world. Cancer Cell 2007; 12:414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 134.Simo R, Barbosa-Desongles A, Hernandez C, Selva DM. IL1α down-regulation of sex hormone-binding globulin production by decreasing HNF-4α via MEK-1/2 and JNK MAPK pathways. Mol Endocrinol 2012; 26:1917–1927. doi: 10.1210/me.2012-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mogilenko DA, Dizhe EB, Shavva VS, Lapikov IA, Orlov SV, Perevozchikov AP. Role of the nuclear receptors HNF4 alpha, PPAR alpha, and LXRs in the TNF alpha-mediated inhibition of human apolipoprotein A-I gene expression in HepG2 cells. Biochemistry 2009; 48:11950–11960. doi: 10.1021/bi9015742. [DOI] [PubMed] [Google Scholar]
- 136.Yin L, Ma H, Ge X, Edwards PA, Zhang Y. Hepatic hepatocyte nuclear factor 4a is essential for maintaining triglyceride and cholesterol homeostasis. Arterioscler Thromb Vasc Biol 2011; 31:328–336. doi: 10.1161/ATVBAHA.110.217828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xu Y, Zhu Y, Hu S, Xu Y, Stroup D, Pan X, et al. Hepatocyte nuclear factor 4a prevents the steatosis-to-NASH progression by regulating p53 and bile acid signaling (in mice). Hepatology 2021; 73:2251–2265. doi: 10.1002/hep.31604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xu J, Li Y, Chen WD, Xu Y, Yin L, Ge X, et al. Hepatic carboxylesterase 1 is essential for both normal and farnesoid X receptor-controlled lipid homeostasis. Hepatology 2014; 59:1761–1771. doi: 10.1002/hep.26714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li Y, Zalzala M, Jadhav K, Xu Y, Kasumov T, Yin L, et al. Carboxylesterase 2 prevents liver steatosis by modulating lipolysis, endoplasmic reticulum stress, and lipogenesis and is regulated by hepatocyte nuclear factor 4 alpha in mice. Hepatology 2016; 63:1860–1874. doi: 10.1002/hep.28472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xu J, Xu Y, Li Y, Jadhav K, You M, Yin L, et al. Carboxylesterase 1 is regulated by hepatocyte nuclear factor 4a and protects against alcohol- and MCD diet-induced liver injury. Sci Rep 2016; 6:24277.doi: 10.1038/srep24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tomita K, Teratani T, Suzuki T, Oshikawa T, Yokoyama H, Shimamura K, et al. p53/p66Shc-mediated signaling contributes to the progression of non-alcoholic steatohepatitis in humans and mice. J Hepatol 2012; 57:837–843. doi: 10.1016/j. jhep.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 142.Farrell GC, Larter CZ, Hou JY, Zhang RH, Yeh MM, Williams J, et al. Apoptosis in experimental NASH is associated with p53 activation and TRAIL receptor expression. J Gastroenterol Hepatol 2009; 24:443–452. doi: 10.1111/j.1440-1746.2009.05785.x. [DOI] [PubMed] [Google Scholar]
- 143.Panasiuk A, Dzieciol J, Panasiuk B, Prokopowicz D. Expression of p53, Bax and Bcl-2 proteins in hepatocytes in non-alcoholic fatty liver disease. World J Gastroenterol 2006; 12:6198–6202. doi: 10.3748/wjg.v12.i38.6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Derdak Z, Villegas KA, Harb R, Wu AM, Sousa A, Wands JR. Inhibition of p53 attenuates steatosis and liver injury in a mouse model of non-alcoholic fatty liver disease. J Hepatol 2013; 58:785–791. doi: 10.1016/j.jhep.2012.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Goto T, Itoh M, Suganami T, Kanai S, Shirakawa I, Sakai T, et al. Obeticholic acid protects against hepatocyte death and liver fibrosis in a murine model of nonalcoholic steatohepatitis. Sci Rep 2018; 8:8157.doi: 10.1038/s41598-018-26383-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ning BF, Ding J, Liu J, Yin C, Xu WP, Cong WM, et al. Hepatocyte nuclear factor 4α-nuclear factor-kB feedback circuit modulates liver cancer progression. Hepatology 2014; 60:1607–1619. doi: 10.1002/hep.27177. [DOI] [PubMed] [Google Scholar]
- 147.Ning BF, Ding J, Yin C, Zhong W, Wu K, Zeng X, et al. Hepatocyte nuclear factor 4 alpha suppresses the development of hepatocellular carcinoma. Cancer Res 2010; 70:7640–7651. doi: 10.1158/0008-5472.CAN-10-0824. [DOI] [PubMed] [Google Scholar]
- 148.Yin C, Lin Y, Zhang X, Chen YX, Zeng X, Yue HY, et al. Differentiation therapy of hepatocellular carcinoma in mice with recombinant adenovirus carrying hepatocyte nuclear factor-4alpha gene. Hepatology 2008; 48:1528–1539. doi: 10.1002/hep.22510. [DOI] [PubMed] [Google Scholar]
- 149.Lee SH, Veeriah V, Levine F. Liver fat storage is controlled by HNF4α through induction of lipophagy and is reversed by a potent HNF4α agonist. Cell Death Dis 2021; 12:603.doi: 10.1038/s41419-021-03862-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Huang KW, Reebye V, Czysz K, Ciriello S, Dorman S, Reccia I, et al. Liver activation of hepatocellular nuclear factor-4α by small activating RNA rescues dyslipidemia and improves metabolic profile. Mol Ther Nucleic Acids 2020; 19:361–370. doi: 10.1016/j.omtn.2019.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]