Abstract
Culture enrichments and culture-independent molecular methods were employed to identify and confirm the presence of novel ammonia-oxidizing bacteria (AOB) in nitrifying freshwater aquaria. Reactors were seeded with biomass from freshwater nitrifying systems and enriched for AOB under various conditions of ammonia concentration. Surveys of cloned rRNA genes from the enrichments revealed four major strains of AOB which were phylogenetically related to the Nitrosomonas marina cluster, the Nitrosospira cluster, or the Nitrosomonas europaea-Nitrosococcus mobilis cluster of the β subdivision of the class Proteobacteria. Ammonia concentration in the reactors determined which AOB strain dominated in an enrichment. Oligonucleotide probes and PCR primer sets specific for the four AOB strains were developed and used to confirm the presence of the AOB strains in the enrichments. Enrichments of the AOB strains were added to newly established aquaria to determine their ability to accelerate the establishment of ammonia oxidation. Enrichments containing the Nitrosomonas marina-like AOB strain were most efficient at accelerating ammonia oxidation in newly established aquaria. Furthermore, if the Nitrosomonas marina-like AOB strain was present in the original enrichment, even one with other AOB, only the Nitrosomonas marina-like AOB strain was present in aquaria after nitrification was established. Nitrosomonas marina-like AOB were 2% or less of the cells detected by fluorescence in situ hybridization analysis in aquaria in which nitrification was well established.
Recent studies of many environments have demonstrated a large amount of diversity among ammonia-oxidizing bacteria (AOB) (4, 14, 21, 31, 39). AOB are responsible for the first step in nitrification, the oxidation of ammonia to nitrite, and are generally members of the β subdivision of the class Proteobacteria except for the marine genus Nitrosococcus, which belongs to the γ subdivision (37). Historically, Nitrosomonas europaea has generally been believed to be the bacterium responsible for ammonia oxidation, as it was commonly isolated from nitrifying systems by traditional culturing techniques (3). However, the application of cultivation-independent molecular techniques, including rRNA gene surveys (38), fluorescent in situ hybridization (FISH) (2), and denaturing gradient gel electrophoresis (DGGE) (24), has removed biases associated with cultivation, and additional AOB have been identified from a number of environments, including soils (34), sand dunes (16), biofilms (32), fluidized bed reactors (30, 31), lakes (11), wastewater (14), and seawater (26).
Purkhold et al. (29) recently summarized the results of the many phylogenetic studies on AOB and improved upon an existing AOB phylogenetic framework (27) by using nearly full-length sequences of 16S rRNA and partial sequences for amoA genes. This work resulted in the formation of seven general clusters of AOB which can serve as a template for constructing relationships of newly discovered AOB. However, molecular techniques alone may not provide detailed ecological information, unless they are combined with physiological and environmental data to establish the conditions under which particular microbial species will flourish. Furthermore, cultivation-independent molecular techniques alone may not always identify the microorganisms of interest, as evidenced by a previous study which failed to identify the bacteria responsible for ammonia oxidation in freshwater aquaria (12). However, in seawater aquaria, the same methods were able to detect AOB of the β subdivision of the class Proteobacteria (12). These findings implying a physiological difference between the species of AOB responsible for ammonia oxidation in the two systems, which differ only in terms of salinity. The effect of salinity on the growth and physiology of AOB has been demonstrated, and it has been shown that some AOB require salt for growth while salt is inhibitory for others (9).
The aim of this study was to combine cultivation-independent molecular techniques and cultivation methods to identify AOB responsible for ammonia oxidation in freshwater aquarium systems. Several enrichments of AOB were established from material collected from actively nitrifying freshwater systems. A range of molecular techniques were used to identify, target, and confirm the presence of putative AOB in the enrichments. Lastly, the establishment of ammonia oxidation in newly established aquaria seeded with enrichments of different AOB strains was compared to that in unseeded aquaria.
MATERIALS AND METHODS
Sources of nitrifying seed biomass.
Nitrifying biomass for enrichments was collected from biofarms (Table 1). Biofarms are proprietary self-contained units dedicated to producing nitrifying biofilms on BioWheels. BioWheels are a form of rotating biological contactor used as a biological filter in aquatic life support systems manufactured by The Aquaria Group (Moorpark, Calif.). Biofarms received a daily dose of ammonium chloride (2.86 mol) and sodium bicarbonate as a buffer to maintain the pH above 8.0.
TABLE 1.
Source and age of the nitrifying biomasses chosen for clone library analysis along with the range of ambient ammonia concentrations maintained in the culture system
| Clone library | Seed or source of biomass | Age of biomass when sampled | Ammonia concn (mg of NH3-N/liter) |
|---|---|---|---|
| BioFarm16 | BioFarm sump and BioWheel biomass | 3 yr | 40–60 |
| BC5 | BioFarm sump and BioWheel biomass | 1.5 yr | 5–10 |
| BC5(2) | BioFarm sump and BioWheel biomass | 1.5 yr | 5–10 |
| R3 | Biomass from an aquarium cultured for 335 days | 6 mo in reactor | 5–10a |
| R7 | BC5 biomass | 6 mo in reactor | 5–10 |
| R5 | Water filter from ammonia feed stock solution reservior | 6 mo in reactor | 40–60 |
| R7PostBA | BioWheel from aquarium inoculated with R7 material | 1 mo in aquarium | 5–10 |
Initially was a reactor at 40 to 60 mg of NH3-N per liter.
Operation of bioreactors.
Laboratory-scale bioreactors were used to produce the enriched nitrifying biomass. The bioreactors were circular columns of clear polyvinyl chloride pipe 198 mm in diameter and 600 mm tall for a maximum volume of 18.5 liters. Each reactor had a lid to minimize aerial contamination and aerosol production. A magnetic stirrer and air diffuser served to maximize mixing and maintain the dissolved oxygen concentration above 5 mg/liter. The bioreactors were kept in darkened cabinets at 26°C. The influent comprised a simple autotrophic medium, free of organic carbon, consisting of potassium phosphate (0.5 mg/liter) and ammonium chloride. The ammonia N concentration in the enrichment medium was kept in the range of 5 to 10 mg/liter for the low-concentration ammonia reactors and 40 to 60 mg/liter for the high-concentration ammonia reactors (Table 1). Bioreactors were monitored daily and maintained at their predefined ammonia N concentrations by feeding the autotrophic media when required. Ammonia, nitrite, and nitrate concentrations were routinely monitored by flow injection analysis and ion chromatography (12). The pH of the bacterial suspensions was kept at or above a pH of 8.0 through the addition of sodium bicarbonate. Fifty percent water changes were performed weekly by allowing the nitrifying biomass to settle, decanting and discarding the appropriate volume of supernatant, and replacing it with the equivalent volume of deionized water.
For analysis of the enriched bioreactor biomass, a sample was removed via the sampling port after the bioreactor's internal surfaces had been scrubbed and the biomass was evenly resuspended.
DNA extraction.
Sampled biomass was initially centrifuged at 12,000 × g for 10 min. The supernatant was discarded, and the pellet was resuspended in 200 μl of cell lysis buffer (40 mM EDTA, 50 mM Tris-HCl [pH 8.3]). Lysozyme was added to achieve a final concentration of 10 mg/ml and incubated at 37°C for 90 min. Sodium dodecyl sulfate (SDS; 20% ) was added to a final concentration of 1% and incubated at 37°C for 60 min. The sample was then subjected to four freeze-thaw cycles at −20 and 65°C, respectively. Proteinase K solution was added (stock concentration, 10 mg/ml) to a final concentration of 2 mg/ml and incubated at 50°C for 35 min. After cell lysis, the extracted DNA was recovered using the DNA Easy extraction kit (Qiagen Inc., Santa Clarita, Calif.) per the manufacturer's instructions. The DNA was eluted with 50 μl of supplied elution buffer and quantified by Hoechst type 33258 dye binding and fluorometry (DynaQuant 200; Hoefer Pharmacia Biotech Inc., San Francisco, Calif.).
Amplification and cloning of 16S rDNA.
Seven general 16S rDNA clone libraries and two AOB-specific clone libraries were constructed to examine the bacterial diversity of nitrifying biomass (Table 1).
General clone libraries were constructed using 16S rRNA genes (16S rDNA) from the total extracted DNA. The 16S rDNA was amplified by using the bacterial conserved primers 27f (GTTTGATCCTGGCTCAG) and 1492r (GGTTACCTTGTTACGACTT) (17). PCR conditions, cycle parameters, and reaction components were as described by DeLong (8). The nearly complete 16S rDNA PCR products were evaluated by agarose gel electrophoresis, purified, and concentrated with a Centricon concentrator (Amicon, Inc. Beverly, Mass.) and TE buffer (pH 8.0). PCR fragments were cloned by using TA cloning kits (Invitrogen, Carlsbad, Calif.). Inserts from the individual clones from each clone library were amplified and grouped according to restriction enzyme analysis (REA) using previously described methods (5, 7). At least two clone representatives from each REA group were reamplified and cleaned for subsequent sequencing using the PCR purification kit (catalog no. 28142; Qiagen). The representative clones from each REA group were initially screened by sequencing with the 1100r primer (GGGTTGCGCTCGTTG) (17) and then tentatively identified by BLAST analysis (1). From these results, clones were chosen for complete 16S rDNA sequencing using a range of conserved primers (27f, 357f, 519r, 530f, 1492r, 907r, and 926f) (17). Sequencing was performed with a LiCor 4000L automated DNA sequencer on template that had been cycle-sequenced with fluorescently labeled primers and SequiTherm ExcelII DNA sequencing kits (Epicentre Technologies, Madison, Wis.).
Clone libraries specific for AOB belonging to the β subdivision of the Proteobacteria were also constructed from two nitrifying biomasses, BioFarm16 and R7 (Table 1). The methods used in constructing and characterizing the specific clone libraries were as described above for the general clone libraries except that the 16S rRNA genes were amplified with the conserved bacterial primer 27f and the β proteobacterial AOB-specific primer NITROSO4E (12). Representative clones from each REA group were initially sequenced with the primer 519r. Then the full 640-nucleotide 16S rDNA fragment for each clone of interest was determined by using the sequencing primers 27f, 357f, and 519r.
Data analysis.
Partial and full-length 16S rDNA sequences were analyzed using the CHECK_CHIMERA program (19) to detect sequence chimeras. Nonchimeric 16S rDNA sequences were subjected to BLAST (1) followed by phylogenetic analysis using several programs (ARB, PHYLIP, and PAUP). ARB (O. Strunk and W. Ludwig, Department of Biology, Technische Universtität München, Munich, Germany) was used initially for sequence alignment and phylogenetic analysis. The topology of the phylogenetic tree generated in ARB was confirmed by using neighbor joining and parsimony analysis with bootstrap values (PAUP* version 4.0b2a; Sinauer Associates, Sunderland, Mass.). Similarity matrices between sequences and their closest bacterial relatives were done using the PHYLIP program (version 3.5c; J. Felsenstein, Department of Genetics, University of Washington, Seattle). Phylogenetic analyses were performed for the full-length and partial (specific AOB, 640 nucleotides) 16S rDNA clone sequences.
PCR primer and oligonucleotide probe design.
Primers and oligonucleotide probes were developed through the manual alignment of full-length 16S rDNA sequences and utilization of the ARB probe design tool and probe match programs (Table 2). The specificity of the oligonucleotide probes was verified with BLAST (1) and CHECK_PROBE (18). The probes were labeled at the 5′ end with indocarbocyanine dye (Cy-3) and/or with fluorescein isothiocyanate (FITC). Primers and probes were synthesized by Operon Tech Inc. (Alameda, Calif.).
TABLE 2.
The PCR sequencing primers and FISH oligonucleotides for general and specific detection of AOB
| Primer or probe | Sequence (5′–3′) | Target site | Annealing temp (°C) | NaCl concn (mM) | % Formamide | Specificity | Reference |
|---|---|---|---|---|---|---|---|
| CTO189f | GGA GRA AAG YAG GGG ATC G | 189–207 | 57 | β proteobacterial AOB | 16 | ||
| NITROSO4Er | CAC TCT AGC YTT GTA GTT TC | 639–658 | 57 | β proteobacterial AOB | 12 | ||
| NSMR71f | CGG AAC GTA TCC AGA AGA | 126–143 | 54 | Nitrosomonas marina-like AOB | This study | ||
| NSMR74r | ATC TCT AGA AAA TTC GCT | 1,000–1,017 | 54 | Nitrosomonas marina-like AOB | This study | ||
| NSMR32f | ATC GGA ACG TAT CTT CG | 125–141 | 56 | Nitrosospira tenuis-like AOB | This study | ||
| NSMR33r | CCA CCT CTC RGC GGG C | 1,006–1,021 | 56 | Nitrosospira tenuis-like AOB | This study | ||
| NSMR52f | TCA GAA AGA AAG AAT CAT G | 443–461 | 56 | Nitrosomonas europaea-like AOB | This study | ||
| NSMR53r | GTC TCC AYT AGA TTC CAA G | 999–1,017 | 56 | Nitrosomonas europaea-like AOB | This study | ||
| NMOB1f | GTT GGG AAG AAA CGA TTR CA | 442–461 | 56 | Nitrosococcus mobilis-like AOB | This study | ||
| NMOB1r | CAC TTT TAT GTC TCC GTA AAA | 1,006–1,026 | 56 | Nitrosococcus mobilis-like AOB | This study | ||
| EUB338 | GCT GCC TCC CGT AGG AGT | 338–355 | 225 | 20 | Bacteria | 2 | |
| NITROSO4E | CAC TCT AGC YTT GTA GTT TC | 639–658 | 225 | 20a | β proteobacterial AOB | This study | |
| Nso190 | CGA TCC CCT GCT TTT CTC C | 190–209 | 20 | 55 | β proteobacterial AOB | 23 | |
| NSMR76 | CCC CCC TCT TCT GGA TAC | 132–149 | 225 | 20 | Nitrosomonas marina-like AOB | This study | |
| NSMR34 | TCC CCC ACT CGA AGA TAC G | 131–149 | 225 | 20 | Nitrosospira tenuis-like AOB | This study | |
| S-G-Ntspa-0685-a-A-22 | CAC CGG GAA TTC CGC GCT CCT C | 664–685 | 225 | 20b | Nitrospira-like NOB | This study |
The specificity of the designed primers was determined by amplifying template from inserts of known AOB 16S rDNA clones or pure bacterial cultures. Stepwise increments of annealing temperatures from 46 to 60°C were used to determine the optimal primer annealing temperature.
A general AOB PCR was used to screen DNA extracted from nitrifying biomass for the presence of AOB using two published oligonucleotides, CTO189f (16) and NITROSO4Er (12), at an annealing temperature of 57°C for 30 cycles. Samples were also analyzed for the presence of specific strains of AOB using primers developed in this study. Nitrosomonas marina-like AOB were detected with NSMR71f and NSMR74r (54°C annealing temperature, 35 cycles), Nitrosospira tenuis-like AOB were detected with NSMR32f and NSMR33r (56°C, 35 cycles), Nitrosomonas europaea-like AOB were detected with NSMR52f and NSMR53r (56°C, 35 cycles), and Nitrosococcus mobilis-like AOB were detected with NMOB1f and NMOB1r (56°C, 30 cycles) (Table 2).
Bioreactor biomass and pure cultures of Nitrosomonas europaea, Nitrosospira multiformis, and Nitrosomonas cryotolerans were used to evaluate the specific probes designed in this study. Two new probes (NSMR76 and NSMR34) and one previously reported probe (NITROSO4E) (12) were tested. To determine the stringency for optimal probe specificity and signal intensity, stepwise 5% increments in formamide concentrations from 0 to 40% were used. The specificity of the NITROSO4E probe was tested in a series of single hybridizations on a range of bioreactor biomass and pure cultures and also in a series of dual hybridizations with the Nso190 probe (23) at increasing formamide stringencies. The two specific AOB probes were then confirmed for specificity through a series of dual hybridizations with the NITROSO4E probe.
FISH.
After sampling, the biomass was immediately processed and fixed in 4% paraformaldehyde for 3 h at 4°C. Then the biomass was washed with phosphate-buffered saline (pH 7.4) and stored at a 1:1 ratio of phosphate-buffered saline and 100% ethanol at −20°C.
In situ hybridizations were performed as described by Manz et al. (20). Fixed cell biomass was spotted onto clean microscope slides, dehydrated (3 min in 50, 80, and 98% ethanol), and air dried. After the addition of the selected probe (5 ng/μl), slides were incubated at 46°C for 120 min. The probe was added to a hybridization buffer containing 20 mM Tris-HCl (pH 7.4), 0.9 M NaCl, 0.01% SDS, and the appropriate formamide concentration. A stringent wash step followed, using a wash buffer at 48°C for 15 min. The wash solution contained 20 mM Tris-HCl (pH 7.4), 0.01% SDS, and the appropriate NaCl concentration. After washing, the slides were removed and rinsed with distilled water and air dried. The slides were mounted with Citifluor (Ted Pella Inc., Redding, Calif.) to avoid bleaching and examined with a Axioskop 2 epifluorescent microscope (Carl Zeiss, Jena, Germany).
The presence of nitrite-oxidizing bacteria (NOB) of the genus Nitrospira in the flocs was accessed by FISH using a probe previously designed for slot blot analysis (13) (Table 2).
A semiquantitative method was established by visualizing the flocs and expressing the proportion of cells that hybridized to a particular probe in relation to another, more general oligonucleotide probe. Generally, the percentage of cells which hybridized to the AOB strain-specific probes was expressed as the proportion of cells that hybridized to the NITROSO4E or EUB probe.
Biomasses were dually stained for AOB-AOB or AOB-NOB analysis and photographed with a Spot SP100 cooled digital color charge-coupled-device camera (Diagnostic Instruments, Inc., Sterling Heights, Mich.). Captured images were overlaid in Adobe Photoshop 6.0.
DGGE analysis and profiling.
General DGGE was performed to describe the microbial diversity and complexity of the nitrifying biomass. For DGGE analysis, rDNA fragments were amplified with the 357f primer (Table 2) with a 40-nucleotide GC clamp on the 5′ end and the primer 519r (Table 2). The PCR procedure and subsequent analysis were as described by Hovanec et al. (13).
A specific AOB DGGE was performed to examine the diversity of AOB in nitrifying biomasses. The method was the same as that described above except that the NITROSO4E primer replaced the 519r primer.
Representative clone AOB were run alongside the seven biomass samples as standards for the detection of candidate AOB in both the general and AOB-specific DGGE. Putative AOB DGGE bands were excised and sequenced with the 357f and either the 519r (general DGGE) or NITROSO4E (AOB-specific DGGE) primers. Confirmation of the sequence's identity was performed by BLAST and phylogenetic analysis.
Nucleotide sequence accession numbers.
The sequences reported in this study are available in GenBank under accession no. AF386746 to AF386757.
RESULTS
General clone library analysis.
A total of 643 clones were screened from the seven libraries, with 92 clones fully sequenced and another 348 clones partially sequenced (Table 3). Sequencing revealed that the general clone libraries contained bacteria belonging to a number of bacterial phyla, including the Proteobacteria, Cytophagales, Actinomycetales, low-G+C gram-positive bacteria, Acidobacteria, Nitrospira, OP11, green nonsulfur bacteria, and Planctomycetales. The most common clones in each library were affiliated with either Nitrospira or Proteobacteria. In the libraries BioFarm16, R7, and R5, a large number of proteobacterial clones were shown to be related to known AOB belonging to the β subdivision of the Proteobacteria by BLAST analysis. No clones in any library were related to AOB belonging to the γ subdivision of the Proteobacteria.
TABLE 3.
Number of clones screened and sequenced for each clone library and numbers of clones for each AOB strain from this study found in the libraries
| Group | No. of clones for:
|
||||||
|---|---|---|---|---|---|---|---|
| Bio-Farm16 | BC5 | BC5(2) | R3 | R7 | R5 | R7PostBA | |
| Clones screeneda | 54 | 76 | 104 | 36 | 185 | 105 | 83 |
| Clones fully sequenced | 20 | 21 | 2 | 12 | 18 | 15 | 4 |
| Clones partially sequenced | 34 | 47 | 86 | 6 | 110 | 21 | 44 |
| β proteobacteria AOB | |||||||
| Nitrosomonas marina-like AOB | 4 | 2 | 2 | 0 | 13 | 0 | 0 |
| Nitrosospira tenuis-like AOB | 16 | 0 | 0 | 7 | 0 | 62 | 0 |
| Nitrosomonas europaea-like AOB | 0 | 0 | 0 | 1 | 0 | 19 | 0 |
| Nitrosococcus mobilis-like AOB | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Nitrospira-like NOB | 0 | 2 | 14 | 24 | 51 | 0 | 2 |
Screened by REA.
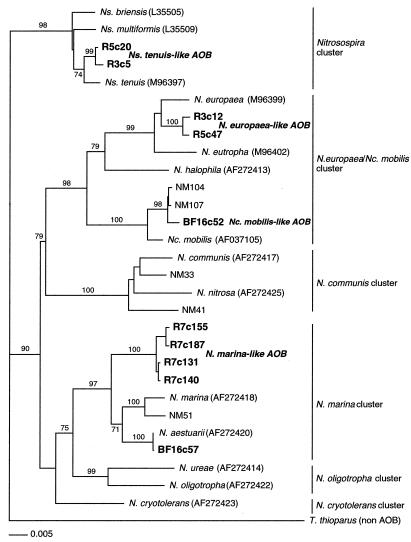
Full-length sequencing and subsequent phylogenetic and BLAST analyses of the putative AOB clones showed that there were four strains of AOB in the enrichments which could be grouped in three clusters, using the terminology of Purkhold et al. (29), of the β subdivision of the Proteobacteria (Fig. 1; Table 3). The three clusters were the Nitrosospira cluster (Nitrosospira tenuis-like AOB), the Nitrosomonas europaea-Nitrosococcus mobilis cluster (Nitrosomonas europaea-like or Nitrosococcus mobilis-like AOB), and the Nitrosomonas marina cluster (Nitrosomonas marina-like AOB) (Fig. 1).
FIG. 1.
Phylogenetic relationships of the four strains of AOB (in bold) found in this study inferred from comparative analysis of 16S rDNA sequences. The tree is based on neighbor-joining distance analysis of nearly full-length sequences (minimum of 1,370 nucleotides). Parsimony bootstrap values of 70% or greater are presented at the nodes (from 100 replicates). AOB cluster designations, at right, are adapted from the work of Purkhold et al. (29).
A single AOB clone (BF16c57) most similar to Nitrosomonas aestuarii was found but not studied any further (Fig. 1). In many instances, multiple clones from each AOB strain were identified in a number of the clone libraries (Table 3). All putative AOB clones were partially sequenced to confirm their identity, but only a subset were fully sequenced for phylogenetic and oligonucleotide design purposes. For phylogenetic studies, several fully sequenced clones of three of the AOB strains were randomly chosen. For the Nitrosococcus mobilis-like AOB, the only clone found was used. Tree topologies generated by PAUP and ARB (not shown) analyses were identical (Fig. 1).
Nitrosomonas marina-like AOB clones, represented by clone R7c140, were found in four of the general clone libraries (Fig. 1; Table 3). Similarity analysis showed this clone sequence to be most similar to Nitrosomonas marina (98.8%). Nitrosomonas marina-like AOB clones represented 7% of all R7 clones, 2% in the two BC5 clone libraries, and 7% of the BioFarm16 library clones (Table 3). A majority of the Nitrosomonas marina-like AOB clones found in the R7 clone library were of a second Nitrosomonas marina-like AOB clone sequence (R7c155) (Fig. 1). This sequence differed by 5 bases, of over 1,450 bases, from the Nitrosomonas marina-like AOB sequence and was not found in the other three clone libraries. Similarity analysis showed the two sequences to be 99.6% similar to each other and most likely represent multiple 16S rDNA operons of the Nitrosomonas marina-like AOB.
Nitrosospira tenuis-like AOB clones (98.8% similar to Nitrosospira tenuis) were found in three general clone libraries and represented 30, 19, and 59% of all of the clones in the BioFarm16, R3, and R5 clone libraries, respectively (Table 3). Two of these enrichments were high-ammonia-concentration reactors, while the third enrichment (R3) had been initially maintained as a high-ammonia reactor and then switched to a low-ammonia reactor.
Nitrosomonas europaea-like AOB clones were found in only two general clone libraries, R3 (3% of all clones) and R5 (18% of all clones). This clone sequence was determined to be 98.4% similar to the sequence of Nitrosomonas europaea.
A single Nitrosococcus mobilis-like AOB clone was found in the high ammonia concentration reactor of BioFarm16 and was determined to be most similar to Nitrococcus mobilis (97.6%).
The two clone libraries created from the same BC5 biomass (BC5 and BC5) (2) to test the reproducibility of the clone library technique had similar general bacterial diversity (Table 3), and each contained only two Nitrosomonas marina-like AOB clones.
Specific AOB clone library analysis.
The BioFarm16 and R7 specific clone libraries provided a greater resolution in identifying AOB present in the biomass than the general clone libraries. Phylogenetic analysis of clones from both the specific and general Biofarm16 clone libraries using 640 nucleotides indicated that the clones were nearly identical. Both clone libraries contained Nitrosomonas marina-like AOB, Nitrosospira tenuis-like AOB, and Nitrosococcus mobilis-like AOB. However, REA analysis of the specific BioFarm16 clone library produced five patterns, whereas only three patterns were found in the general BioFarm16 clone library. Sequencing determined that the fourth and fifth patterns were due to the presence of Nitrosomonas europaea-like AOB and the second sequence for the Nitrosomonas marina-like AOB. Neither of these strains of AOB were detected in the general BioFarm16 clone library. In addition, the AOB diversity of the specific R7 clone library was greater than that of the general R7 clone library. Nitrosomonas marina-like AOB were the only AOB identified in the general R7 clone library, whereas the specific R7 clone library identified clones belonging to all four strains of AOB found in this study.
FISH.
The NITROSO4E probe, used as a general AOB FISH probe, specifically hybridized to pure cultures of Nitrosomonas europaea, Nitrosospira multiformis, and Nitrosomonas cryotolerans in both single- and dual-hybridization experiments with the AOB-specific Nso190 probe. The NITROSO4E probe yielded an optimal signal at 20% formamide.
Neither the NSMR76 probe, designed for the detection of Nitrosomonas marina-like AOB, nor the NSMR34 probe, designed for the detection of Nitrosospira tenuis-like AOB, hybridized to pure cultures of Nitrosomonas europaea, Nitrosospira multiformis, or Nitrosomonas cryotolerans at the tested formamide stringencies, demonstrating specificity for their target AOB. The optimal signal for NSMR76 and NSMR34 was determined to be at 20% formamide. The NSMR34 probe did not hybridize to “Nitrosomonas marina”-like, Nitrosomonas europaea-like, or Nitrosococcus mobilis-like AOB cells. The NSMR76 probe did not hybridize to Nitrosospira tenuis-like, Nitrosomonas europaea-like, or Nitrosococcus mobilis-like AOB cells at the tested formamide stringencies (Fig. 2).
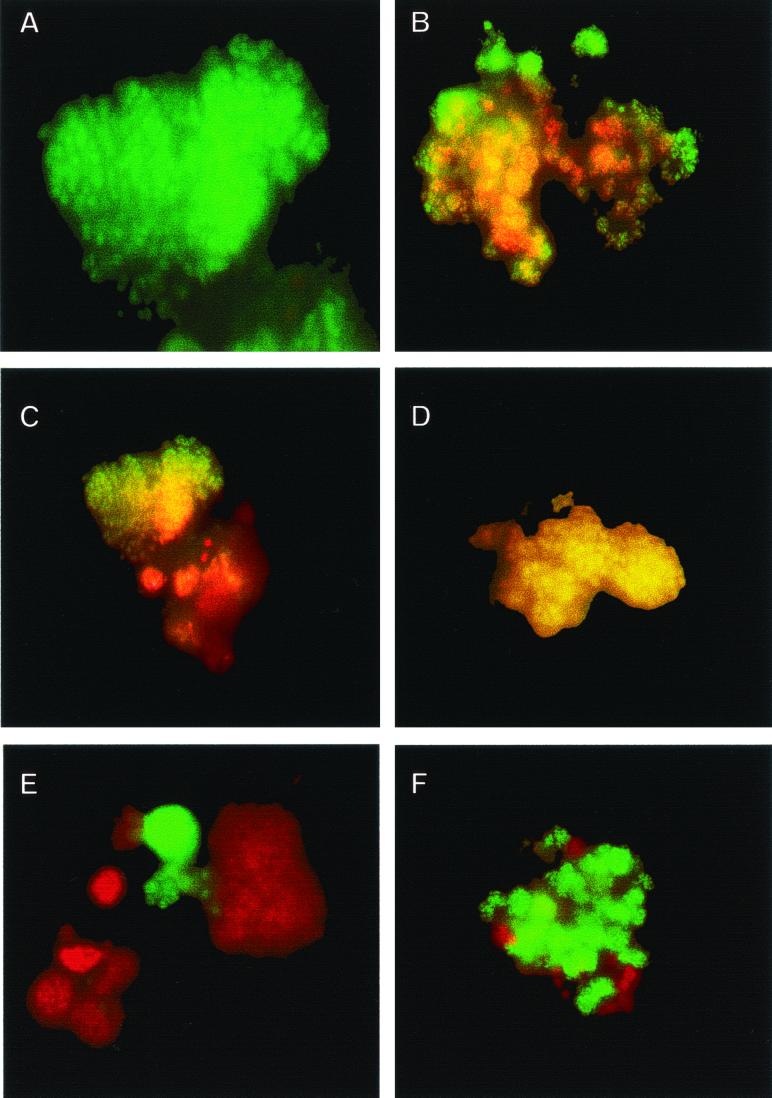
FIG. 2.
Whole-cell FISH of nitrifying biomasses. (A) FITC stain of Nitrosomonas marina-like AOB enrichment to be added to aquaria. (B) Simultaneous hybridization of AOB enrichment with Cy-3 stain for all AOB (red) and FITC stain for Nitrosomonas marina-like AOB, resulting in a yellow color for this AOB strain. (C) Simultaneous hybridization of biomass enrichment before addition to a newly established aquarium with Cy-3 stain for all AOB (red) and FITC stain for Nitrosomonas marina-like AOB, resulting in a yellow color for Nitrosomonas marina-like AOB and showing some non Nitrosomonas marina-like AOB. (D) Biomass material harvested from an aquarium with active ammonia oxidation after inoculation with the enrichment from panel C showing the presence of only Nitrosomonas marina-like AOB, which are yellow from the simultaneous hybridization with Cy-3 stain for all AOB and FITC stain for Nitrosomonas marina-like AOB. (E) Dual staining of nitrifying enrichment with Cy-3 for all AOB and FITC for Nitrospira spp. showing the proximity of AOB to NOB. (F) Dual staining of nitrifying biomass with FITC for Nitrosomonas marina-like AOB and Cy-3 for Nitrospira sp. NOB, elucidating the structure of the nitrifying consortium.
FISH analysis of biofilms.
FISH analysis of the BioFarm16 biomass revealed that about 50% of the EUB-positive cells cohybridized with the general AOB probe. However, only about 2% of these AOB-positive cells were estimated to be Nitrosomonas marina-like AOB. The majority (>90%) of the AOB-positive cells were Nitrosospira tenuis-like, with the remaining 8% or so being unidentifiable with regard to strain type.
The BC5 biomass sample was highly autofluorescent. AOB detected with the AOB general probe comprised less than 5% of the EUB-positive cells. Virtually 100% of these cells hybridized with the Nitrosomonas marina-like AOB probe. No cells were positive with the specific probes for the other AOB strains found in this study.
General AOB probing revealed two distinct AOB strains in the R3 biomass. By using the specific AOB probes in dual hybridization experiments, it was estimated that Nitrosospira tenuis-like AOB comprised 90% of the NITROSO4E-positive cells while the remaining 10% of the cells were unidentifiable as to the strain of AOB. No Nitrosomonas marina-like AOB-positive cells were detected in the R3 biomass.
In the R5 reactor sample, over 90% of the EUB-positive cells hybridized to the general AOB FISH probe, indicating a large concentration of AOB cells in this reactor biomass. FISH analysis with the AOB-specific probes demonstrated that 90% of the general AOB-positive cells hybridized with the Nitrosospira tenuis-like AOB probe, with the remaining 10% being unidentifiable as to strain type. No Nitrosomonas marina-like AOB were detected in the R5 biomass by FISH.
Only about 10% of the EUB-positive cells in the R7 biomass hybridized to the general AOB probe. More than 95% of these AOB positive cells hybridized with the Nitrosomonas marina-like AOB-specific probe.
Nitrospira spp. were found to be in close association with AOB in the biomass from each reactor (Fig. 2).
FISH analysis of aquaria inoculated with AOB biomass.
The R7PostBA biomass, which was collected from an aquarium inoculated with a biomass dominated by Nitrosomonas marina-like AOB, was microbiologically complex, with many bacterial morphotypes hybridizing with the EUB probe. The only AOB strain detected was Nitrosomonas marina-like cells. Probing of samples from aquaria inoculated with biomass from reactors BC5 (Nitrosomonas marina-like AOB) and R3 (Nitrosospira tenuis-like AOB) were also dominated by Nitrosomonas marina-like AOB, although the R3 sample did contain some Nitrosospira tenuis-like AOB. However, the total percentage of AOB in each sample was 2% or less of the total bacterial community. The biofilm collected from an aquarium inoculated with R5 biomass (Nitrosospira tenuis-like AOB) was the only one in which FISH analysis did not detect Nitrosomonas marina-like AOB. Nitrospira spp. were detected in the biomass from each aquarium by FISH (Fig. 2).
Use of AOB-specific PCR primers.
The four primer sets developed in this study for specific strains of AOB amplified only their target templates at the optimal annealing temperature, producing PCR products of the correct size (Table 2). Analysis of the bioreactor biomass with the AOB-specific primer sets showed Nitrosomonas marina-like AOB to be present in all bioreactor samples except R5. Nitrosospira tenuis-like AOB were detected in the BioFarm16, R3, R7, and R5 biomasses but could not be detected in the BC5 sample. No Nitrosomonas europaea-like AOB could be detected in biomasses harvested from aquaria inoculated with the various bioreactor enrichments. Nitrosococcus mobilis-like AOB were detected only in the BioFarm16 biomass.
DGGE.
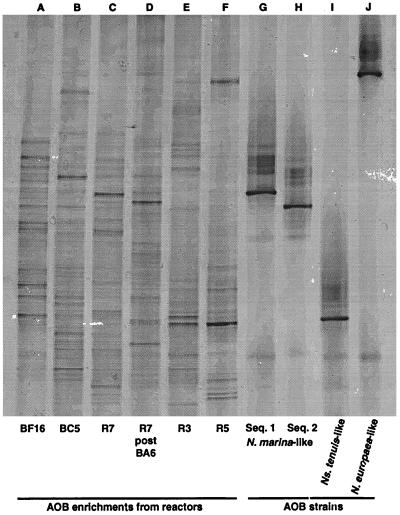
General DGGE analysis revealed a pattern of multiple bands of various intensities which reflects the complex microbial community in each sample (Fig. 3). The sequencing of selected DGGE bands that had migrated the same distance in the gel as the clonal representatives of the AOB strains identified in this study confirmed the presence of the various AOB strains in the bioreactor biomass.
FIG. 3.
DGGE of AOB enrichments, amplified with a universal and specific eubacterial primer set, from which clone libraries were constructed. Lane D is material collected from an aquarium which was seeded with the R7 enrichment (lane C). Lanes G through J are from clones representing Nitrosomonas marina-like AOB (sequence type 1), Nitrosomonas marina-like AOB (sequence type 2), Nitrosospira tenuis-like AOB, and Nitrosomonas europaea-like AOB, respectively.
The two sequences for Nitrosomonas marina-like AOB which differ by only a single base pair in the amplified fragment were easily differentiated in the general DGGE (Fig. 3).
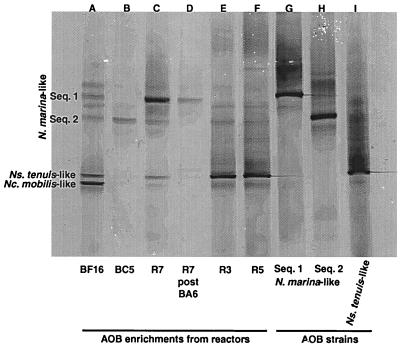
The AOB-specific DGGE was able to detect three of the four AOB strains found in this study with good spatial resolution (Fig. 4). Only the Nitrosomonas europaea-like AOB strain could not be reliably detected in the AOB-specific DGGE. Agarose gel analysis of cloned Nitrosomonas europaea-like AOB DNA amplified with the specific PCR primers showed a positive reaction (data not shown). However, when the material was run on DGGE, it remained at or near the top of the gel, which may represent a problem with the percentage of denaturant used in the DGGE gel.
FIG. 4.
DGGE of AOB enrichments (same samples as in Fig. 3) except that the samples were amplified with an AOB-specific primer set. Nitrosomonas europaea-like AOB could not be successfully visualized under the gel conditions of this DGGE.
DISCUSSION
A suite of culture-independent molecular techniques were combined with long-term enrichments to identify novel AOB in freshwater aquarium systems. These techniques were in close, but not absolute, agreement with each other in terms of the presence or absence in our samples of the four strains of AOB found in this study.
16S rDNA clone library analysis revealed that enriched nitrifying biomass were microbially diverse which is consistent with other observations of autotrophic microbial communities from bioreactors fed a simple nutrient solution (7, 22). However, clone library analysis also showed that the most common clones in many of the enrichments were from either the Nitrospira phylum, which contains NOB commonly found in a number of aquatic environments, including aquaria (13) and wastewater facilities (7, 14, 31), or associated with the class Proteobacteria.
All putative AOB identified in the clone libraries were members of the β subdivision of the class Proteobacteria. The phylogeny of the four strains of AOB recovered from biomass in this study, with the phylogenetic 16S rRNA tree of Purkhold et al. (29) superimposed, is shown in Fig. 1. The four strains of this study fall into three of the clusters described by Purkhold et al. (29): the Nitrosomonas marina cluster, the Nitrosospira cluster, and the Nitrosomonas europaea-Nitrosococcus mobilis cluster. However, only the Nitrosospira tenuis-like AOB, Nitrosomonas europaea-like AOB, and Nitrosococcus mobilis-like AOB showed a high similarity with previously published AOB sequences (Fig. 1).
The Nitrosomonas marina-like AOB strain from this study likely represents a new species of freshwater AOB, since full-length 16S rDNA sequences are only 95% similar to Nitrosomonas marina. It has been shown that at 16S rRNA similarity values below 97%, the DNA similarity between two organisms is likely to be less than 70%, and thus, the organisms are probably distinct species (33). That these criteria apply to AOB belonging to the β subdivision of the class Proteobacteria was confirmed by Purkhold et al. (29).
Nitrosomonas marina-like AOB are the bacteria most likely responsible for ammonia oxidation in aquaria, as they were found by multiple molecular techniques in all but one of the bioreactors maintained at low ammonia concentrations (5 to 10 mg of NH3 N per liter) and enrichments containing Nitrosomonas marina-like AOB successfully accelerated nitrification in aquaria. These criteria were not matched by any of the other AOB strains found in this study. In addition, Nitrosomonas marina-like AOB were detected by FISH in all the biomasses extracted from nitrifying aquaria accelerated with an enrichment except for one (R5). However, Nitrosomonas marina-like AOB seem to represent only a small percentage of the microbial community in an aquaria, as neither they nor any other AOB strain was found in the clone library developed from biomass extracted from a nitrifying aquarium. The relatively low number of Nitrosomonas marina-like AOB cells in the microbial community of aquaria may make detection by various molecular methods difficult and could explain why these microorganisms were not previously detected as the putative AOB in freshwater aquaria (12).
The effect of heterotrophic bacterial growth on the percentage of AOB and NOB in aquarium biofilm samples is evident upon examination of the NOB numbers in biomass samples before and after inoculation. In reactor 7, Nitrospira-like bacteria were nearly 28% of the clones screened. However, after 1 month in an aquarium only 2.4% of clones screened in aquarium biomass were Nitrospira-like NOB. This value compares well with previous results obtained with oligonucleotide probes for Nitrospira-like NOB in aquaria (13). In that study, there was 1.5 to 3.4% hybridization of the Nitrospira-like NOB probe relative to the eubacterial probe for biomass extracted from aquaria after 50 days.
Figure 2E and F show the close association of AOB and NOB cells with each other in the nitrifying biomass. The association of these two groups of bacteria in the nitrifying flocs points out the difficulty in obtaining pure cultures of AOB. This association has been demonstrated previously in the nutrient-rich environment of wastewater systems (14, 31) and is now extended to the comparatively nutrient-poor aquarium systems. Furthermore, the structure of the nitrifying consortium is reminiscent of the consortium of archaea and sulfate-reducing bacteria responsible for anaerobic oxidation of methane on the ocean floor (6, 25) and would be a good candidate for the further application of coupled FISH and secondary ion mass spectrometry (25).
The topology of the phylogenetic tree, parsimony analysis bootstrap analysis, and similarity matrix analysis suggest that the Nitrosospira tenuis-like AOB represent a unique clade which is distinct from Nitrosospira briensis, Nitrosospira multiformis, and Nitrosospira tenuis (Fig. 1). Nitrosospira tenuis-like AOB grew best in the reactors maintained with a high concentration of ammonia. Nevertheless, enrichments of Nitrosospira tenuis-like AOB were able to accelerate nitrification when added to new aquaria. However, Nitrosospira tenuis AOB could not be detected by PCR or FISH in the majority of aquarium biomass samples several weeks after being added. Thus, it appears that Nitrosomonas marina-like AOB may outcompete Nitrosospira tenuis-like AOB in the low-ammonia-concentration environment of an aquarium.
The Nitrosomonas europaea-like AOB are phylogenetically most closely related to Nitrosomonas europaea and were found only in reactors with a history of high ammonia concentration. This strain of AOB was also absent in clone libraries and FISH analysis of biomass grown at consistently low ammonia concentrations, suggesting their affinity for high-ammonia-concentration environments.
PCR and FISH analyses did, at times, produce conflicting results. In the process of identifying the active AOB in the nitrifying biomass by FISH analysis, it became apparent that some of the results contradicted results of PCR analysis. Two biomass samples which were PCR positive for a specific strain of AOB were negative in the AOB-specific FISH studies (R3 and R7). The occurrence of a positive PCR but negative FISH result can be due to the presence of active but scarce AOB cells, or the PCR result could be a false positive caused by the amplification of DNA from inactive cells or dead cells. The latter can be expected due to the presence of extracellular DNA, which is stable long-term, and the passive dispersal of cells (29). Under these circumstances, AOB implicated with a positive PCR-negative FISH result in a particular sample could not be absolutely associated with ammonia oxidation. Our results suggest that when there was a conflict, the PCR tests provided a false indication of the presence of an active AOB strain.
It was apparent from the results of this study that by altering the ammonia concentrations in the bioreactors, different populations of AOB were generated. The ability to phylogenetically differentiate AOB on the basis of the ambient ammonia concentration has been previously demonstrated under a variety of conditions (10, 15, 28, 35, 36). Koops et al. (15) used maximum ammonia tolerance as one criterion to classify eight new species of AOB. Princic et al. (28) examined shifts in the AOB community at ammonium concentrations ranging from 50 to 3,000 mg of N per liter, which overlaps the higher ranges of this study. At these ammonia concentrations, Princic et al. (28) found AOB that fell into the Nitrosomonas europaea-Nitrococcus mobilis cluster of Purkhold et al. (29), which correlates with our results for the Nitrosomonas europaea-like and Nitrosococcus mobilis-like AOB. Gieseke et al. (10) found a spatial separation of Nitrosomonas europaea-Nitrosococcus mobilis cluster AOB and Nitrosomonas oligotropha cluster AOB in a phosphate-removing biofilm, with only Nitrosomonas oligotropha being present in the deeper (lower-ammonia-concentration) layers of the biofilm, which further supports the possibility of there being physiological differences between the Nitrosomonas europaea-like and Nitrosococcus mobilis-like AOB found in high-ammonia environments and the low-ammonia Nitrosomonas marina-like AOB.
That AOB can be phylogenetically differentiated on the basis of the ambient ammonia concentration was also demonstrated by Suwa et al. through isolation studies (35) and 16S rDNA sequence analysis for detecting two general groups of AOB based on their degree of sensitivity to (NH4)2SO4 (36). The (NH4)2SO4-insensitive strains found by these researchers, which could tolerate (NH4)2SO4 concentrations above 30 mM, would be grouped in the Nitrosomonas europaea-Nitrosococcus mobilis cluster of Purkhold et al. (29), and this compares favorably with our finding that AOB strains from high-ammonia reactors also fall into this cluster. The (NH4)2SO4-sensitive AOB of Suwa et al. (35, 36), which grew at 3.57 mM (NH4)2SO4 but were inhibited at 10.7 mM (NH4)2SO4, would be grouped in the Nitrosomonas oligotropha cluster, which is on the same main branch leading to the Nitrosomonas marina cluster containing the Nitrosomonas marina-like AOB (low-ammonia-concentration AOB) found in this study.
When ammonia concentrations were varied, AOB population shifts did occur, thereby altering the presence and activity of important AOB. Low-ammonia environments will likely produce Nitrosomonas marina-like AOB, while as the ammonia concentration increases, Nitrosospira tenuis-like and Nitrosomonas europaea-like AOB will become important until at the highest ammonia concentration Nitrosococcus mobilis-like AOB may be predominant. Our results suggest that the AOB found in fish culture environments, such as public aquaria, aquaculture facilities, and home aquaria, where the ambient ammonia concentration rarely exceeds 5 mg of N per liter, are different from the traditional Nitrosomonas europaea-Nitrosococcus mobilis cluster type AOB, which are prevalent in the high-ammonia concentrations typically found in environment such as wastewater and sewage treatment facilities. This, and our results with enrichments of the various strains of AOB in newly set-up aquaria, strongly suggest that start-up inocula for the establishment of nitrification in aquatic culture systems should optimally consist of Nitrosomonas marina-like AOB rather than Nitrosomonas europaea-Nitrosococcus mobilis cluster AOB.
ACKNOWLEDGMENTS
We thank Julia Sears-Hartley, Jennifer Coshland, Michele Barlow, Scott Wirtz, Jason Niemans, and Les Wilson for their assistance in the construction, operation, and water analysis of the bioreactors and biofarms. We also thank Edward DeLong, MBARI, for use of the microscope and digital imaging system.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argaman Y. Biological nutrient removal. In: Martin A M, editor. Biological degradation of wastes. Amsterdam, The Netherlands: Elsevier Applied Science; 1991. pp. 85–101. [Google Scholar]
- 4.Bano N, Hollibaugh J T. Diversity and distribution of DNA sequences with affinity to ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in the Arctic Ocean. Appl Environ Microbiol. 2000;66:1960–1963. doi: 10.1128/aem.66.5.1960-1969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackall L L, Burrell P C, William H G, Bradford D, Bond P L, Hugenholtz P. The use of 16S rDNA clone libraries to describe the microbial diversity of activated sludge communities. Water Sci Technol. 1998;37:451–454. [Google Scholar]
- 6.Boetius A, Ravenschlag K, Schubert C J, Rickert D, Widdel F, Gieseke A, Amann R, Barker J B, Witte U, Pfannkuche O. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 7.Burrell P C, Keller J, Blackall L L. Microbiology of a nitrite-oxidizing bioreactor. Appl Environ Microbiol. 1998;64:1878–1883. doi: 10.1128/aem.64.5.1878-1883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finstein M S, Bitzky M R. Relationships of autotrophic ammonium-oxidizing bacteria to marine salts. Water Res. 1972;6:31–40. [Google Scholar]
- 10.Gieseke A, Purkhold U, Wagner M, Amann R, Schramm A. Community structure and activity dynamics of nitrifying bacteria in a phosphate-removing biofilm. Appl Environ Microbiol. 2001;67:1351–1362. doi: 10.1128/AEM.67.3.1351-1362.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiorns W D, Hastings R C, Head I M, McCarthy A J, Saunders J R, Pickup R W, Hall G H. Amplification of 16S ribosomal RNA genes of autotrophic ammonia-oxidizing bacteria demonstrates the ubiquity of nitrosospiras in the environment. Microbiology. 1995;141:2793–2800. doi: 10.1099/13500872-141-11-2793. [DOI] [PubMed] [Google Scholar]
- 12.Hovanec T A, DeLong E F. Comparative analysis of nitrifying bacteria associated with freshwater and marine aquaria. Appl Environ Microbiol. 1996;62:2888–2896. doi: 10.1128/aem.62.8.2888-2896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hovanec T A, Taylor L T, Blakis A, DeLong E F. Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl Environ Microbiol. 1998;64:258–264. doi: 10.1128/aem.64.1.258-264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juretschko S, Timmermann G, Schmid M, Schleifer K-H, Pommerening-Roser A, Koops H-P, Wagner M. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3191. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koops H-P, Böttcher U C, Möller A, Pommerening-Röser A, Stehr G. Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov., Nitrosomonas halophila sp. nov. J Gen Microbiol. 1991;137:1689–1699. [Google Scholar]
- 16.Kowalchuk G A, Stephen J R, de Boer W, Prosser J I, Embley T M, Wolderdorp J W. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: Academic Press; 1991. pp. 115–175. [Google Scholar]
- 18.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manz W, Amann R, Ludwig W, Wagner M, Scheifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 21.McCaig A E, Phillips C J, Stephen J R, Kowalchuk G A, Harvey S M, Herbert R A, Embley T M, Prosser J I. Nitrogen cycling and community structure of proteobacterial beta-subgroup ammonia-oxidizing bacteria within polluted marine fish farm sediments. Appl Environ Microbiol. 1999;65:213–220. doi: 10.1128/aem.65.1.213-220.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDevitt C, Burrell P, Blackall L L, McEwan A G. Aerobic nitrate respiration in a nitrite oxidising bioreactor. FEMS Microbiol Lett. 2000;184:113–118. doi: 10.1111/j.1574-6968.2000.tb09000.x. [DOI] [PubMed] [Google Scholar]
- 23.Mobarry B K, Wagner M, Urbain V, Rittman B E, Stahl D A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplifies genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orphan V J, House C H, Hinrichs K-U, McKeegan K D, DeLong E F. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science. 2001;293:484–487. doi: 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- 26.Phillips C J, Smith Z, Embley T M, Prosser J I. Phylogenetic differences between particle-associated and planktonic ammonia-oxidizing bacteria of the β-subdivision of the class Proteobacteria in the northwestern Mediterranean Sea. Appl Environ Microbiol. 1999;65:779–786. doi: 10.1128/aem.65.2.779-786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pommerening-Röser A, Rath G, Koops H-P. Phylogenetic diversity within the genus Nitrosomonas. Syst Appl Microbiol. 1996;19:344–351. [Google Scholar]
- 28.Princic A, Mahne I, Megusar F, Paul E A, Tiedje J M. Effects of pH and oxygen and ammonium concentrations on the community structure of nitrifying bacteria from wastewater. Appl Environ Microbiol. 1998;64:3584–3590. doi: 10.1128/aem.64.10.3584-3590.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purkhold U, Pommerening-Roser A, Juretschko S, Schmid M C, Koops H-P, Wagner M. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol. 2000;66:5368–5382. doi: 10.1128/aem.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schramm A, de Beer D, van den Heuvel J C, Ottengraf S, Amann R. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl Environ Microbiol. 1999;65:3690–3696. doi: 10.1128/aem.65.8.3690-3696.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schramm A, de Beer D, Wagner M, Amann R. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol. 1998;64:3480–3485. doi: 10.1128/aem.64.9.3480-3485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schramm A, Larsen L H, Revsbech N P, Ramsing N B, Amann R, Schleifer K-H. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol. 1996;62:4641–4647. doi: 10.1128/aem.62.12.4641-4647.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stackebrandt E, Goebel M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 34.Stephen J R, Kowalchuk G A, Bruns M V, McCaig A E, Phillips C J, Embley T M, Prosser J I. Analysis of β-subgroup proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl Environ Microbiol. 1998;64:2958–2965. doi: 10.1128/aem.64.8.2958-2965.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suwa Y, Imamura Y, Suzuki T, Tashiro T, Urushigawa Y. Ammonia-oxidizing bacteria with different sensitivities to (NH4)2SO4 in activated sludges. Water Res. 1994;28:1523–1532. [Google Scholar]
- 36.Suwa Y, Sumino T, Noto K. Phylogenetic relationships of activated sludge isolates of ammonia oxidizers with different sensitivities in ammonium sulfate. J Gen Appl Microbiol. 1997;43:373–379. doi: 10.2323/jgam.43.373. [DOI] [PubMed] [Google Scholar]
- 37.Teske A, Alm E, Regan J, Toze S, Rittmann B, Stahl D. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J Bacteriol. 1994;176:6623–6630. doi: 10.1128/jb.176.21.6623-6630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward D M, Weller R, Bateson M M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 39.Whitby C B, Saunders J R, Rodriguez J, Pickup R W, McCarthy A. Phylogenetic differentiation of two closely related Nitrosomonas spp. that inhabit different sediment environments in an oligotrophic freshwater lake. Appl Environ Microbiol. 1999;65:4855–4862. doi: 10.1128/aem.65.11.4855-4862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]