Abstract
Degradation of lignin-related aromatic compounds is an important ecological process in the highly productive salt marshes of the southeastern United States, yet little is known about the mediating organisms or their catabolic pathways. Here we report the diversity of a gene encoding a key ring-cleaving enzyme of the β-ketoadipate pathway, pcaH, amplified from bacterial communities associated with decaying Spartina alterniflora, the salt marsh grass that dominates these coastal systems, as well as from enrichment cultures with aromatic substrates (p-hydroxybenzoate, anthranilate, vanillate, and dehydroabietate). Sequence analysis of 149 pcaH clones revealed 85 unique sequences. Thirteen of the 53 amino acid residues compared were invariant in the PcaH proteins, suggesting that these residues have a required catalytic or structural function. Fifty-eight percent of the clones matched sequences amplified from a collection of 36 bacterial isolates obtained from seawater, marine sediments, or senescent Spartina. Fifty-two percent of the pcaH clones could be assigned to the roseobacter group, a marine lineage of the class α-Proteobacteria abundant in coastal ecosystems. Another 6% of the clones matched genes retrieved from isolates belonging to the genera Acinetobacter, Bacillus, and Stappia, and 42% of the clones could not be assigned to a cultured bacterium based on sequence identity. These results suggest that the diversity of the genes encoding a single step in aromatic compound degradation in the coastal marsh examined is high.
In southeastern United States salt marshes, lignin-related aromatic compounds comprise a significant fraction of the total organic carbon pool. These compounds arise primarily from Spartina alterniflora, a grass responsible for more than 80% of the total primary production (33), and from other vascular plants that decompose in the marsh sediments. While it is widely recognized that bacteria play a major role in transformation of vascular plant material (24–26), the bacteria responsible and the enzymatic pathways involved have yet to be properly characterized.
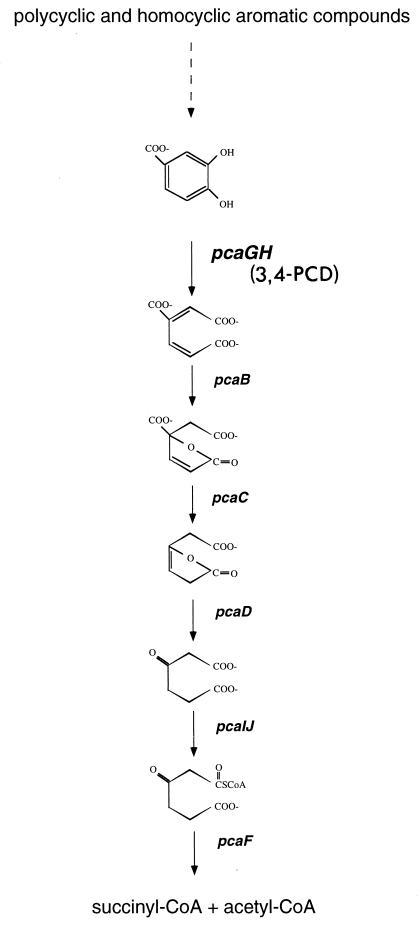
In terrestrial soils a major catabolic route for lignin-related aromatic compounds is the β-ketoadipate pathway (31). This primarily chromosomally encoded convergent pathway plays an integral role in the catabolism of a vast array of phenolic compounds and is widespread in phylogenetically diverse soil bacteria and fungi (18). In this pathway, polycyclic and homocyclic aromatic compounds are transformed into one of two dihydroxylated intermediates, catechol or protocatechuate. Each of these phenolic compounds is then cleaved between its two hydroxyl groups (ortho cleavage) by catechol 1,2 dioxygenase or protocatechuate 3,4-dioxygenase (3,4-PCD). Following ring cleavage the products are converted to β-ketoadipate, the intermediate for which the pathway is named. Two additional steps complete the conversion of β-ketoadipate to tricarboxylic acid cycle intermediates (Fig. 1). While this pathway has been identified in a number of bacterial genera, including Acinetobacter, Alicaligenes, Azotobacter, Bacillus, Pseudomonas, Rhodococcus, and Streptomyces (7, 18), it is not known whether it is prevalent in marine communities.
FIG. 1.
Protocatechuate branch of the β-ketoadipate pathway. Gene designations are in italics. CoA, coenzyme A.
The β-ketoadipate pathway is biochemically conserved and the structural genes encoding enzymes in this pathway are similar in the phylogenetically diverse organisms that possess it (18). Both 3,4-PCD and catechol 1,2-dioxygenase belong to a large class of non-heme-iron-containing dioxygenases. 3,4-PCD is composed of equimolar amounts of two nonidentical subunits, termed α and β, which are encoded by the usually cotranscribed pcaG and pcaH genes, respectively. The β-subunit contains all of the ligands required for formation of the catalytic site, which may explain the greater similarity of PcaH sequences than of PcaG sequences in various organisms (29). This conservation of PcaH facilitates the use of molecular tools to detect the corresponding gene in isolates and environmental samples.
Although the β-ketoadipate pathway is an important catabolic pathway in soil bacteria, alternative routes of aromatic compound degradation, including meta and para cleavage pathways, have been identified (18). However, since studies of these pathways have also focused primarily on soil organisms, their relevance in marine systems remains relatively unexplored. In this study, we investigated the potential ecological role of the β-ketoadipate pathway in coastal marine environments by assessing the presence and diversity of pcaH gene pools in natural bacterial communities associated with decaying Spartina. We also identified pcaH gene fragments in marine isolates cultured from seawater, marine sediments, and decomposing Spartina and used them for comparative studies with genes from uncultivated organisms. Our results suggest that the β-ketoadipate pathway is widespread in southeastern United States coastal bacteria and that members of the roseobacter lineage, an ecologically important marine clade, may be the dominant aromatic compound-degrading bacteria in these systems.
MATERIALS AND METHODS
Natural community DNA.
Spartina detritus was collected from a marsh at the Skidaway Institute of Oceanography (Savannah, Ga.) in April 2000. Spartina leaves were vigorously agitated in filter-sterilized (pore size, 0.2 μm) seawater to dislodge bacteria. The rinse water was passed through a series of Nitrex filters (140, 70, and 30 μm) to remove larger plant pieces and sediment. The bacterial community was captured by passing 100 ml of the screened rinse water through a 0.2-μm-pore-size filter, and DNA was extracted from the filter with a soil DNA extraction kit (Mega Size; MoBio, Solana Beach, Calif.). The remaining rinse water was used as the inoculum for enrichments as described below.
Amplification of pcaH from the natural community.
A degenerate PCR primer set based on conserved regions in PcaH (P340IDf [5′ YTI GTI GAR RTI TGG CAR CGI AAY GC 3′] and P340IDr [5′ ICY IAI RTG IAY RTG IGC IGG ICK CCA 3′]), where Y = C or T, R = A or G, and K = T or G, was used to amplify a 212-bp fragment of pcaH (3). Each PCR mixture contained 1× buffer (10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl; pH 8.3), each deoxynucleoside triphosphate at a concentration of 2 mM, each primer at a concentration of 1.0 μM, 50 ng of DNA, and 1 U of Taq polymerase. The PCR was performed with a DNA Engine (MJ Research, Incline Village, Nev.) by using an initial cycle of 3 min at 95°C, followed by 30 cycles of 45 s at 95°C, 45 s at 60°C, and 45 s at 72°C. Products of the appropriate size were recovered from the gel with a QiaSpin gel extraction kit (Qiagen, Valencia, Calif.), and the PCR products were cloned by using a TA cloning kit (Invitrogen Corp., Carlsbad, Calif.).
Enrichment design.
Enrichment cultures consisting of 10 liters of filter-sterilized seawater (salinity, 27 practical salinity units) amended with a single substrate were established in 20-liter polycarbonate carboys. The natural community described above was used as the inoculum for the enrichments at a 1:40 dilution. The substrates (acetate, p-hydroxybenzoate, anthranilate, vanillate, and dehydroabietate) were added at zero time (final concentration, 10 μM) and again on days 2, 5, 8, and 11. A preparation that received no substrate was also included. The enrichments were prepared in duplicate and were incubated at room temperature in the dark; the carboys were manually shaken every other day. On day 14, bacterial cells were collected on 293-mm-diameter, 0.2-μm-pore-size polycarbonate filters. The filters were cut in half; one half of each filter was processed immediately, and the other half was stored at −70°C. DNA was extracted from the filter halves with a soil DNA extraction kit (Mega Size; MoBio). The bacterial abundance in each enrichment was determined by acridine orange direct counting (19) at zero time and on day 14. The dissolved organic carbon concentration was measured at zero time with a TOC-5000 (Shimadzu Corp., Norcross, Ga.).
Amplification of pcaH from enrichment communities.
pcaH clone libraries were established for the enrichment communities by using the protocol used for the natural community. Each clone sequence was named by using the substrate used in the enrichment (Table 1) and a number.
TABLE 1.
Bacterial cell growth during a 2-week enrichment period with aromatic substrates and pcaH clone recovery from the enrichments and the original salt marsh community
| Sample | Substrate | Increase in no. of cells (fold) | No. of pcaH clones sequenced | % Unique sequences |
|---|---|---|---|---|
| SMCa | NAb | NA | 21 | 76 |
| NocA | None | 1.6 | 10 | 40c |
| NocB | None | 1.1 | NA | |
| AcetA | Acetate | 5.1 | 12 | 52 |
| AcetB | Acetate | 3.2 | 11 | 52 |
| PhbA | p-Hydroxybenzoate | 3.4 | 15 | 42 |
| PhbB | p-Hydroxybenzoate | 2.7 | 11 | 42 |
| VanlA | Vanillate | 2.4 | 10 | 60 |
| VanlB | Vanillate | 3.2 | 10 | 60 |
| AnthA | Anthranilate | 1.6 | 10 | 55 |
| AnthB | Anthranilate | 1.7 | 10 | 55 |
| DhaA | Dehydroabietate | 2.8 | 10 | 57 |
| DhaB | Dehydroabietate | 1.9 | 11 | 57 |
SMC, salt marsh community.
NA, not applicable.
Calculated for the NocA sample only.
A nondegenerate version of the P340 primer set based on the pcaH sequence previously obtained from isolate Y3F (3) was also designed. Primers Y3Ffor (5′ CTG GTG GAG ATC TGG CAG GCC AAT GC 3′) and Y3Frev (5′ CGA AAC GTG GAT ATG CGC GGG CCG CCA 3′) were used to amplify a product from one replicate of the p-hydroxybenzoate enrichments and the natural community. The PCR and cloning procedures used were those described above. Clones obtained with this PCR primer set were designated by using the prefix Y and numbers.
T-RFLP analysis.
16S rRNA genes were amplified from enrichment DNA by using general bacterial primers 8F and 1522R (12). The primer 8F was fluorescently labeled at the 5′ end with either FAM or TET. The PCR was carried out with Ready-To-Go PCR beads (Amersham Pharmacia, Piscataway, N.J.) by using each primer at a concentration of 0.2 μM, and 50 ng of DNA. An initial incubation for 3 min at 95°C was followed by 25 cycles of 1 min at 95°C, 1 min at 60°C, and 1.5 min at 72°C. Products of the correct size (ca. 1,500 bp) were recovered from a 1.0% agarose gel with a QiaSpin gel extraction kit (Qiagen), followed by an additional purification step with a PCR purification kit (MoBio). Restriction digestion was carried out in a 10-μl (total volume) mixture containing 100 ng of purified PCR product and 10 U of either CfoI or RsaI (Roche, Indianapolis, Ind.). Digestion was carried out at 37°C for 3 h, after which samples were precipitated in ethanol and suspended in 12 μl of deionized formamide with 1 μl of the fluorescently labeled DNA fragment length standard Genescan-2500 (TAMRA; Applied Biosystems). The terminal restriction fragment lengths were determined with an ABI PRISM 310 (Applied Biosystems) in GeneScan mode. Typically, DNA extracted from replicate enrichments were analyzed simultaneously by using the FAM label for one replicate and the TET label for the other and coinjecting the samples. Similarities among the enrichment assemblage terminal restriction fragment length polymorphism (T-RFLP) profiles were determined by cluster analysis using KyPlot, version 2.0 (http://ftp.vector.co.jp/pack/Win95/business/calc/graph).
Bacterial isolation and 16S rDNA analysis.
Most isolates examined in this study were cultured from seawater, sediments, or decaying salt marsh grass collected in estuaries and coastal waters of the southeastern United States. Several of the strains had been described previously, having been isolated from lignin or aromatic monomer enrichment cultures (isolates Y3F, Y4I, and IC4, Sagittula stellata E-37, and Sulfitobacter sp. strain EE-36) (3, 17). Some strains were cultured directly from coastal seawater by using nonselective, low-nutrient seawater plates (all isolates with the prefix GAI) (15, 17). Some isolates were derived from a marine dimethylsulfoniopropionate enrichment (isolate DSS-3) (16). Additional strains were isolated for this study from Spartina detritus collected at the Skidaway Institute of Oceanography during October 1999 (all isolates with the prefix SE). The SE isolates (a total of 176 isolates) were obtained by grinding Spartina leaves in a blender with filter-sterilized seawater and spreading the liquid onto low-nutrient seawater plates containing, (per liter) 10 mg of peptone (Difco Laboratories, Detriot, Mich.), 5 mg of yeast extract (Difco), and 1.5% purified agar (Difco) in filter-sterilized diluted Sargasso Sea water that had been aged for more than 1 year in the dark (final salinity, 24 psu) (15). Finally, the following two isolates that were not obtained from the southeastern United States coast were examined: Sulfitobacter pontiacus ChLG 10, which was cultured from the Black Sea (40); and strain ISM, which was cultured from the Caribbean Sea (10).
The 16S ribosomal DNA (rDNA) sequences of the following isolates have been reported previously: DSS-3 (accession no. AF09491), EE-36 (AF007254), GAI-05 (AF007256), GAI-37 (AF007260), GAI-111 (AF098494), IC4 (AF254098), ISM (AF098495), and Y3F (AF253467). If not already available, 16S rDNA sequences for the isolates were obtained by PCR amplification using the general bacterial primers 27F and 1522R (12). Genomic DNA was prepared from each isolate by a colony boil method as previously described (3). Approximately 500 bp of the PCR product was directly sequenced by using primer 27F and an ABI PRISM 310 genetic analyzer (Applied Biosystems) following purification with an Ultra Clean PCR clean-up kit (MoBio). Sequences were analyzed by using Genetics Computer Group program package 10.0 (Wisconsin Package version; Madison, Wis.).
pcaH genes were amplified from isolates by using the degenerate primer set as described above, except that 3 μl of cell lysate was used in the PCR mixture. Both strands of pcaH gene fragments were sequenced.
Sequence and phylogenetic analyses.
Sequence analysis was performed with an ABI PRISM 310 genetic analyzer by using a BigDye terminator cycle sequencing kit (Applied Biosystems). DNA sequences were determined with M13 primers that recognized the cloning vectors. Phylogenetic trees were constructed with the PHYLIP package by using evolutionary distances (Jukes-Cantor) and the neighbor-joining method.
Nucleotide sequence accession numbers.
Sequences determined in this study have been deposited in the GenBank database under the following accession numbers: AF388307, AF388308, AY038900 to AY038926, and AY040248 to AY040273.
RESULTS
pcaH diversity in the salt marsh community.
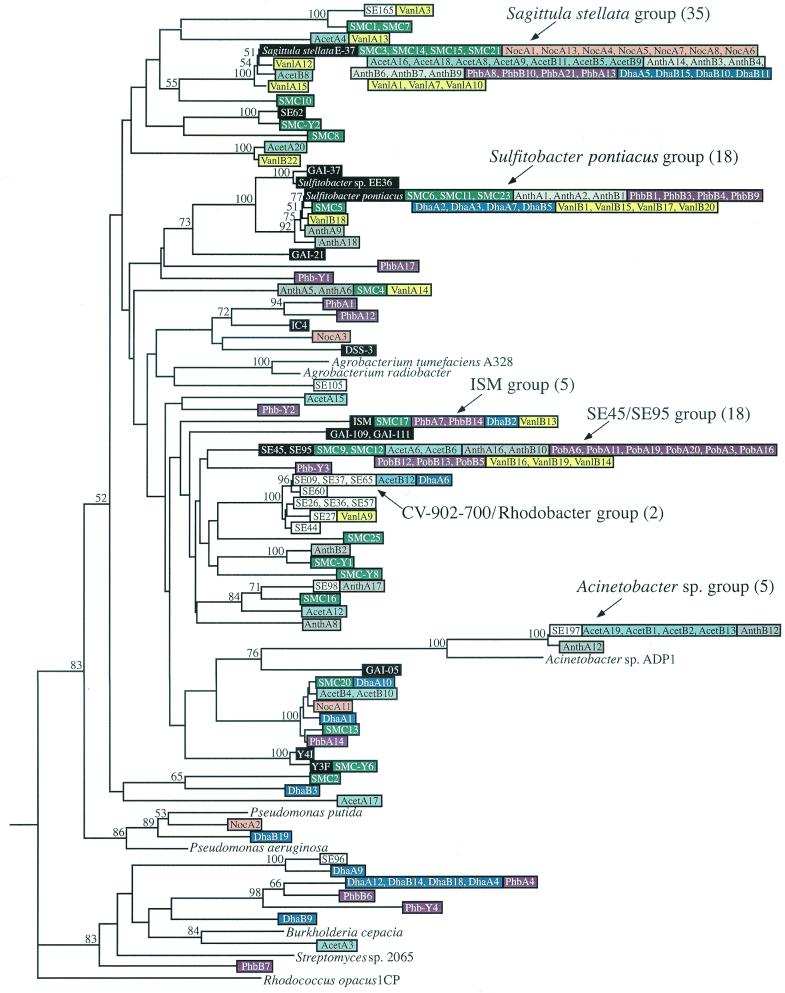
A pcaH clone library was established for the natural bacterial community associated with decaying Spartina (referred to below as the salt marsh community) by amplifying DNA with the degenerate primer set. Twenty-one clones were sequenced, which yielded 14 unique sequences (Fig. 2). Homology searches with sequences from GenBank confirmed that amino acid sequences deduced from the PCR products of the clones had the highest levels of similarity with the approximately 240-residue PcaH molecules from members of various bacterial genera. The deduced levels of amino acid similarity ranged from 82 to 100%, and the levels of identity ranged from 73 to 100%. Furthermore, two residues demonstrated to be involved in Fe2+ binding, Tyr408 and Tyr447, and a residue involved in substrate specificity, Trp449 (29, 43), were conserved in all sequences.
FIG. 2.
Phylogenetic tree of pcaH sequences from isolates, the natural salt marsh community, and the enrichment communities. The tree is based on the 159 nucleotides located in between the degenerate primer binding sites and is unrooted; pcaH from Rhodoccocus opacus 1CP is the outgroup. Major clone groups are indicated, and the numbers in parentheses are the numbers of clones. Isolate sequences are color coded with either black type (sequences from roseobacter group isolates) or white type (sequences from members of other phylogenetic groups). Sequences from the salt marsh community and enrichment communities are color coded by treatment and are identified by the designations shown in Table 1. Bootstrap values greater than 50% are indicated at branch nodes.
Aromatic substrate enrichments.
Enrichments were established to monitor the responses of the bacterial community from decaying Spartina to specific aromatic substrates representing compounds associated with vascular plant decay. Anthranilate, p-hydroxybenzoate, and vanillate are aromatic monomers that have been shown to be degraded through the β-ketoadipate pathway in soil microorganisms (18). Studies of soil microbes have indicated that p-hydroxybenzoate and vanillate are converted to protocatechuate, whereas anthranilate is typically converted to catechol prior to intradiol ring cleavage (18). Dehydroabietate is a plant diterpenoid that is commonly associated with pulp and paper mill effluent, and an extradiol cleavage pathway has only recently been elucidated for this compound (22). Enrichments with acetate, a nonaromatic compound, and no-carbon controls were established for comparison with the aromatic compound enrichments.
The natural dissolved organic carbon concentration in the filter-sterilized coastal seawater was 365 μM, and the four additions of substrate (10 μM each) during the enrichments increased this value by <11%. Direct counts obtained at zero time and on day 14 showed that there were increases in the numbers of bacterial cells in all enrichments; the average increases were 2.8-fold during the 2-week enrichment period for the enrichments to which substrates were added and 1.4-fold for the no-carbon controls (Table 1). The larger increases in numbers of cells in the presence of added substrates suggest that bacteria capable of metabolizing the compounds became established in the enrichment cultures.
16S rDNA T-RFLP analysis of enrichment communities.
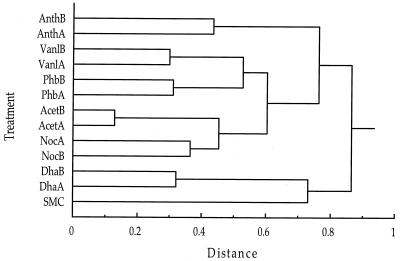
The enriched bacterial communities were characterized by using the 16S rDNA T-RFLP procedure (20a). Independent PCR amplification and GeneScan analysis of each sample on at least two occasions confirmed the reproducibility of T-RFLP profiles. Replicate enrichments with the same substrate typically developed very similar bacterial communities. A cluster analysis performed by using the relative peak area of each of the major peaks in the T-RFLP chromatograms digested with CfoI (31 peaks) and RsaI (32 peaks) confirmed that replicates supplemented with the same aromatic compound were most similar in terms of the amplifiable 16S rRNA genes (Fig. 3). The vanillate and p-hydroxybenzoate enrichment culture communities formed a subgroup in this analysis, perhaps due to the structural similarity of these two compounds. The two preparations that were not supplemented with an aromatic compound (the acetate and no-carbon-addition preparations) also formed a distinct cluster.
FIG. 3.
Cluster analysis of 16S rRNA T-RFLP profiles from the salt marsh and enrichment communities based on the relative areas of the major peaks. A similarity matrix was constructed by using Euclidean distances, and clustering was performed by using Ward's method. The enrichment designations are described in Table 1.
pcaH in enrichment communities.
To characterize the ring cleavage genes harbored by the enriched bacterial communities, pcaH clone libraries were established for 11 of the 12 enrichments by amplification with the degenerate primer set. The remaining sample (no-carbon replicate NocB) did not yield a PCR product when it was amplified with the degenerate primer set (although it did produce a product when it was amplified with 16S rDNA primers). Repeated attempts to obtain a PCR product from this replicate (including carrying out a second DNA extraction with the unused filter half) were not successful, and thus this sample was not characterized further. From all of the other samples, a total of 120 pcaH clones were sequenced; at least 10 clones were sequenced from each library (Table 1).
Seventy-six unique sequences were identified, and five of these sequences matched sequences retrieved from the natural community (Fig. 2). The pcaH sequences did not segregate according to enrichment substrate. For example, the 20 pcaH sequences retrieved from the replicate vanillate enrichments were distributed throughout the pcaH tree, and 19 clustered with sequences from other types of enrichments. Similarly, 14 of the 20 sequences obtained from the anthranilate enrichments clustered with sequences from other enrichments (Fig. 2).
Phylogeny of Spartina-associated isolates.
For comparative purposes, a collection of culturable marine bacteria harboring the pcaH gene was assembled. An initial screening of the 176 SE isolates obtained from decaying Spartina was carried out with the degenerate PCR primers targeting pcaH. For all 28 isolates (16%) that gave a PCR product of the correct size, a phylogenetic analysis of 16S rDNA sequences was carried out. Twenty-three of these isolates showed close phylogenetic affinities to members of the class α-Proteobacteria previously isolated from marine environments. Eighteen of these 23 isolates fell into the Rhizobium-Agrobacterium group, exhibiting ≥96.7% similarity to a symbiont isolated from the eastern oyster, Crassostrea virginica (isolate CV902-700) (1). The closest previously described relative of these CV902-700-like isolates is Stappia stellulata (originally described as an Agrobacterium), an organism isolated from marine sediments and seawater (37). The remaining α-proteobacterial isolates were affiliated with the rhodobacter group or the roseobacter group. Two of the isolates showed affiliations with γ-proteobacteria, and three were closely related to Bacillus spp. (Table 2).
TABLE 2.
Phylogenetic affiliations of marine isolates with amplifiable pcaH genes
| Isolate(s) | Major taxa | Group | Closest relative (accession no.) | % 16S rDNA similarity |
|---|---|---|---|---|
| Sagittula stellata E-37 | α-Proteobacteria | Roseobacter | NAa | NA |
| Sulfitobacter pontiacus | α-Proteobacteria | Roseobacter | NA | NA |
| DSS-3 | α-Proteobacteria | Roseobacter | Ruegeria sp. strain AS-36 (AJ391197) | 97 |
| EE-36 | α-Proteobacteria | Roseobacter | Sulfitobacter pontiacus (Y13155) | 99 |
| GAI-05 | α-Proteobacteria | Roseobacter | Marine isolate JP88.1 (AY007684) | 98 |
| GAI-21 | α-Proteobacteria | Roseobacter | Sulfitobacter sp. strain GAI-37 (AF007260) | 98 |
| GAI-37 | α-Proteobacteria | Roseobacter | Sulfitobacter sp. strain GAI-21 (AF007257) | 98 |
| GAI-111, GAI-109 | α-Proteobacteria | Roseobacter | Roseobacter clone NAC11-6 (AF245634) | 94 |
| IC4 | α-Proteobacteria | Roseobacter | Hydrothermal vent strain TB66 (AF254109) | 98 |
| ISM | α-Proteobacteria | Roseobacter | C. virginica symbiont CV919-312 (AF114484) | 96 |
| Y3F, Y4I | α-Proteobacteria | Roseobacter | Marine bacterium PP-154 (AJ296158) | 97 |
| SE03b | α-Proteobacteria | Rhizobium-Agrobacterium | C. virginica symbiont CV902-700 (AF246615) | 97 |
| SE09 | α-Proteobacteria | Rhizobium-Agrobacterium | C. virginica symbiont CV902-700 (AF246615) | 98 |
| SE11 | α-Proteobacteria | Rhizobium-Agrobacterium | C. virginica symbiont CV902-700 (AF246615) | 97 |
| SE65 | α-Proteobacteria | Rhizobium-Agrobacterium | C. virginica symbiont CV902-700 (AF246615) | 99 |
| SE45, SE95 | α-Proteobacteria | Roseobacter | Hydrothermal vent strain AG33 (AF254108) | 98 |
| SE62 | α-Proteobacteria | Roseobacter | Isolate GAI-37 (AF007260) | 96 |
| SE37 | α-Proteobacteria | Rhodobacter | Marine isolate Sippewissett 2-21 (AF055822) | 99 |
| SE197 | γ-Proteobacteria | Moraxellaceae | Acinetobacter calcoaceticus (AF159045) | 99 |
| SE96 | γ-Proteobacteria | Halomonadaceae | Noctiluca scintillans endocyte (AF262750) | 97 |
| SE98 | Firmicutes | Bacillus-Clostridium | Bacillus cereus (AF274244) | 99 |
| SE105 | Firmicutes | Bacillus-Clostridium | Bacillus sp. strain OS-5 (BSP296095) | 99 |
| SE165 | Firmicutes | Bacillus-Clostridium | Bacillus subtilis (BAC180K) | 91 |
NA, not applicable.
The following isolates had 16S rRNA gene sequences identical to that of SE03: SE22, SE26, SE27, SE32, SE35, SE36, SE39, SE44, SE49, SE55, SE57, SE60, SE83, SE97, and SE114.
Many pcaH-containing SE isolates were related to organisms in which 3,4-PCD activity has been reported previously. These organisms include members of the roseobacter group (3), Agrobacterium species (5, 30, 32), Acinetobacter species (11, 31, 42), and Bacillus species (23). 3,4-PCD activity has not been found in members of the rhodobacter group or the Halomonadaceae group (in which isolates SE37 and SE96 cluster), although both of these groups contain members capable of metabolizing aromatic compounds (9, 35).
pcaH in marine isolates.
In addition to the 28 SE isolates described above, strains previously isolated from seawater or sediments and belonging to the roseobacter clade were also screened for pcaH. Nine members of the roseobacter group yielded a PCR product of the correct size when the pcaH primers were used (ISM, Y4I, DSS-3, GAI-05, GAI-21, GAI-109, GAI-111, GAI-37, and S. pontiacus). We previously identified this gene in four other roseobacter group isolates (S. stellata E-37, EE-36, Y3F, and IC4) (3) and included these organisms in all of the analyses described here. The PCR products from most SE isolates and roseobacter group isolates were sequenced; the only exceptions were the PCR products from the SE isolates exhibiting very high levels of similarity as determined by 16S rDNA analysis to the C. virginica symbiont CV902-700. Due to the strain level 16S rDNA sequence identity of the 19 isolates examined, only 8 were selected for pcaH sequence analysis. Altogether, 26 pcaH sequences were obtained from marine isolates.
Similarity of pcaH sequences was typically observed for closely related isolates. S. pontiacus, EE-36, GAI-37, and GAI-21 formed a cluster in the roseobacter lineage based on 16S rRNA analysis, and pcaH genes from these organisms also clustered with a high bootstrap value (Fig. 2), exhibiting ≥81.8% sequence similarity at the nucleotide level. Isolates Y3F and Y4I had a level of 16S rDNA sequence similarity of 100% and a level of pcaH sequence similarity of 97.5%. Isolate SE197 and Acinetobacter calcoaceticus exhibited 99.7% 16S rRNA gene sequence similarity, and their pcaH sequences formed a distinct cluster that was supported by a high bootstrap value (Fig. 2). All of the C. virginica symbiont CV902-700-like isolates had pcaH sequences that were ≥97.5% similar and deduced amino acid sequences that were ≥94.3% identical. Finally, two pairs of isolates, isolates GAI-109 and GAI-111 and isolates SE45 and SE95, had identical 16S rDNA sequences and identical pcaH sequences.
The 16S rDNA and pcaH phylogenies were not always congruent, however. Comparisons of isolate SE37 and the CV902-700-like strains revealed only ca. 84% sequence similarity based on 16S rDNA analysis but as little as 1-bp difference when the pcaH sequences were compared. Furthermore, the pcaH gene sequences available for two agrobacterial strains related to the CV902-700-like isolates did not appear to cluster with the pcaH gene sequences for these isolates obtained in our analysis (Fig. 2). Finally, the obvious lack of similarity among pcaH sequences retrieved from Bacillus isolates SE98, SE105, and SE165 suggests that this gene may be highly divergent in these organisms, although no other Bacillus pcaH genes are available for comparision (i.e., this is the first report of pcaH sequences for Bacillus isolates).
Comparisons of the PcaH sequences for all of the isolates examined in this study and sequences previously deposited in GenBank revealed levels of sequence similarity of ≥52.2% at the nucleotide level and levels of similarity and identity of ≥52.8 and ≥43.4%, respectively, at the deduced amino acid level. Furthermore, the conservation of 13 residues in all clone and isolate sequences suggests that these residues have a required catalytic or structural function.
Comparison of pcaH in clones and isolates.
Of the 21 pcaH clones obtained from the salt marsh community with the degenerate primer set, 10 (44%) were considered matches (i.e., ≤1-bp difference) with genes from roseobacter group isolates. One additional pcaH clone, SMC5, had a level of nucleotide similarity of >98% with the sequence of the roseobacter group isolate S. pontiacus. Finally, clones SMC1 and SMC7 exhibited >94% sequence similarity with Bacillus isolate SE165, which brought the total number of clones that clustered with pcaH sequences from isolates to 13 (56%).
Of the 120 clones obtained from the enrichments, 67 (54%) were considered matches (i.e., ≤1-bp difference) with genes from roseobacter group isolates. A number of the remaining clones differed from isolate pcaH sequences at more than one position but nonetheless exhibited notable sequence similarity with isolates (Fig. 2). Three clones (Van1A12, AcetB8, and Van1A15) grouped with the S. stellata E-37 pcaH sequence and exhibited ≥96.9% sequence similarity. Four clones clustered with S. pontiacus and exhibited within-group levels of nucleotide sequence similarity of ≥96.9%, which brought the total number of clones that grouped with roseobacter group isolates to 74 (60%). Two other clones had sequences which were identical to the sequences of two Bacillus isolates (SE165 and SE98), five clones had sequences identical to the Acinetobacter isolate SE197 sequence, and three clones had sequences identical to the sequences of CV902-700-like isolates and rhodobacter group isolate SE37. Clone DhaA9 was 93.1% identical at the nucleotide level and 98.1% identical at the amino acid level to the pcaH fragment of γ-proteobacterial isolate SE96.
Only a few pcaH clone sequences grouped with sequences from isolates not identified in this study. NocA2 and DhaB19 exhibited >85% sequence identity and clustered with the Pseudomonas putida and Pseudomonas aeruginosa pcaH sequences in GenBank. AcetA3 was 84% identical to the pcaH sequence from the β-proteobacterium Burkholderia cepacia.
Clustering of clone pcaH sequences from the same enrichment type was generally not observed. One of the few exceptions to this was the finding that clones resembling Acinetobacter sp. were recovered only the from the acetate- and anthranilate-amended enrichments. In addition, seven clones from the dehydroabietate and p-hydroxybenzoate enrichments formed a cluster with pcaH genes from isolates with different phylogenetic affinities (γ- and β-proteobacteria and Streptomyces sp.). Finally, 7 of the 10 clones obtained from the NocA library exhibited sequence similarity to pcaH from S. stellata E-37. This relatively low level of clonal diversity may suggest that the pcaH-containing community in this preparation was composed of only a few organisms. Indeed, a low abundance of pcaH genes in the absence of aromatic substrates may also explain our inability to obtain a PCR product from the second no-substrate replicate (NocB).
Nondegenerate pcaH primers.
Due to the phylogenetic differences among the organisms whose pcaH genes had previously been sequenced, the design of universal pcaH primers required a high degree of DNA sequence degeneracy. In an attempt to investigate the potential bias of the degenerate primers, a nondegenerate primer set was designed based on the pcaH sequence from roseobacter group isolate Y3F, an isolate for which no similar sequences were found among the 141 pcaH clones obtained with the degenerate primer set. This second primer set was used to amplify pcaH gene fragments from both the salt marsh community and one replicate of the p-hydroxybenzoate enrichments (PhbA). Four representatives of the cloned PCR products were sequenced for each sample. One of the clones analyzed, SMCY6, had a pcaH sequence identical to that of isolate Y3F. In addition, SMCY1 was 96.2% similar at the nucleotide level and identical at the deduced amino acid level to the pcaH fragment of SE62, another roseobacter group isolate. Finally, clone PobY3 exhibited 87.4% nucleotide similarity and 98.1% amino acid identity to roseobacter group isolates SE45 and SE95. The remaining five clones had no identifiable sequence similarity with either an isolate or a clone.
A total of 86 (58%) of the 149 pcaH clones obtained from the salt marsh community and enrichments were considered matches (≤1-bp mismatch) with the gene sequences from isolates examined in this study, and 78 (52%) matched one of five roseobacter group isolates. Sixty-three (42%) of the cloned pcaH sequences did not closely match the sequence of any isolated bacterium. In almost all cases, the branch topologies of trees based on nucleotide sequences were maintained when deduced amino acids were analyzed (data not shown).
DISCUSSION
The ecological significance of the β-ketoadipate pathway for degradation of naturally occurring aromatic compounds has been inferred from studies of a select group of soil microorganisms. While these studies were instrumental in characterizing structural and sequence similarities, as well as the regulation and function of the pathway in certain bacteria, they did not establish that this catabolic route is widely distributed in many natural systems. With the development of a degenerate primer set targeting all known pcaH sequences, we can now begin to investigate the importance and diversity of this key aromatic ring cleavage gene in a variety of natural bacterial communities.
pcaH diversity in salt marsh and enrichment communities.
The pcaH gene diversity in the bacterial communities associated with decaying Spartina was high. Fourteen of the 21 clones derived by using the degenerate primer set were unique sequences. Enrichment cultures were established to assess the diversity of pcaH genes harbored in marine bacterial assemblages by varying the amount and type of aromatic substrates available, and T-RFLP analysis of 16S rRNA genes indicated that distinct bacterial communities indeed developed in each preparation (Fig. 3). Analysis of these communities revealed an additional 76 gene sequences out of 120 partial pcaH sequences. Five of these sequences matched sequences found after direct amplification from the salt marsh community, but 71 were novel sequences. The Y3F-specific primers yielded even more novel pcaH sequences from the Spartina-associated bacterial community; eight new sequences were obtained from eight clones, and only one of these sequences was identical to the sequence from the isolate for which this primer set was specifically designed. The possibility that some fraction of the pcaH diversity found in this study resulted from chimeric artifacts generated during PCR amplification does not change the overall conclusion that there is significant diversity of this key gene in aromatic compound degradation. Furthermore, the small size of the amplified product reduced the likelihood of heteroduplex formation (44).
High levels of functional gene diversity in environmental samples are not unprecedented and have been noted previously for genes involved in denitrification (2, 38), bisulfite reduction (6), and nitrogen fixation (21, 28). However, it is not typical that functional genes retrieved directly from environmental samples have such high levels of sequence similarity to genes from cultured bacteria. For example, Scala and Kerkof (38) identified 37 unique nosZ genes in marine sediments, none of which resembled the nosZ genes of cultivated organisms. Similarly, Lovell et al. (21) found 43 unique nifH sequences in the 59 clones which they analyzed, none of which matched sequences of known nitrogen fixers. Yet for pcaH genes retrieved from decaying Spartina, 58% of the clones matched (i.e., ≤1-bp difference) sequences found in a companion collection of marine isolates. Nearly one-half of the 25 genes amplified from the salt marsh community (44%) matched the gene of one of five roseobacter group isolates cultured from decaying Spartina detritus or seawater. Similarly, more than one-half the 124 clones from the enrichment communities (54%) matched the sequence of a roseobacter group isolate (Table 3). A more conservative definition requiring no mismatches between sequences still resulted in 32 and 28% of the salt marsh and enrichment community pcaH sequences, respectively, matching sequences of cultured roseobacter group species. It is unlikely that this predominance of roseobacter group-like pcaH sequences is due to a particular bias in the degenerate primers, since the primers were designed to target PcaH in 14 phylogenetically diverse organisms representing several bacterial lineages (e.g., α-, β-, and γ-proteobacteria, gram-positive organisms). Moreover, the pcaH gene was previously found to be quite common in culturable members of the roseobacter clade (3).
TABLE 3.
Phylogenetic affiliations of pcaH sequences from cultured members (SE isolate collection) and uncultured members (salt marsh community clones) of the bacterial community associated with decaying Spartina and from enrichments of that community with a variety of aromatic substrates (enrichment clones)a
| Organisms | % of sequences affiliated with:
|
|||||
|---|---|---|---|---|---|---|
| Roseobacter group | Rhodobacter group | Acinetobacter | Halomonas | Bacillus | Unidentified organisms | |
| Salt marsh community clones (n = 25) | 44 | 0 | 0 | 0 | 0 | 56 |
| Enrichment clones (n = 124) | 54 | 2 | 4 | 0 | 2 | 38 |
| SE isolate collection (n = 28) | 11 | 71 | 3.5 | 3.5 | 11 | NAb |
Clone affiliations were inferred based on ≤1-bp difference with pcaH sequences from isolates.
NA, not applicable.
Members of the roseobacter clade are abundant in many coastal and open-ocean environments (16, 27, 41) and have been found to contribute up to 30% of the bacterioplankton 16S rRNA genes in southeastern United States coastal systems (15). Unlike other dominant marine bacterial lineages that have no close relatives in culture, roseobacter group members are readily isolated from coastal and open-ocean systems (10, 13, 15, 20). Roseobacter group members have also been shown to be primary colonists on surfaces in coastal salt marshes (8). Both surface colonization and plant degradation typically involve the production of exopolysaccharide, holdfast structures, or fibrils which can assist in cellular attachment (34). The largest number of pcaH clones clustered with the gene from roseobacter group isolate S. stellata E-37, a bacterium able to attach selectively to the surfaces of lignocellulose particles and to mineralize cellulose and synthetic lignin (14).
The aromatic compounds used in the enrichment experiments represent fused-ring and hydroxy-, methyl-, and amino-substituted structures. The presence of roseobacter group-like pcaH genes in all enrichments with aromatic substrates suggests that these bacteria are capable of converting a variety of ring structures and therefore may contain multiple sets of catabolic genes. Anthranilate, p-hydroxybenzoate, and vanillate have been shown to require a unique set of upper pathway genes for conversion to a dihydroxylated intermediate, such as protocatechuate or catechol (4, 18, 36, 39).
While T-RFLP profiles of 16S rDNA amplicon pools from enrichment community DNA indicated that distinctive bacterial communities developed in response to each enrichment substrate (Fig. 3), there was surprisingly little evidence that pcaH gene sequences likewise segregated according to enrichment type (Fig. 2). This absence of pcaH clustering by enrichment type suggests that the marine bacteria responsible for aromatic ring cleavage are nutritional generalists that are able to funnel a variety of different aromatic structures through the protocatechuate branch of the β-ketoadipate pathway. Two alternative explanations for the lack of apparent pcaH clustering by enrichment substrate are that the diversity of pcaH clones was high relative to the sample size of the clone libraries (i.e., clustering may have been evident if more clones had been sequenced per enrichment treatment) and that the distinct T-RFLP patterns obtained for enrichment preparations reflected compositional differences of the component of the bacterial community that was not involved in aromatic ring cleavage.
Ecological significance.
Despite the significance of the β-ketoadipate pathway in the processing and degradation of aromatic compounds in a variety of systems, the ecological role of this pathway has not been demonstrated yet outside soil ecosystems. Here we describe the importance and diversity of a gene encoding a key enzyme of the pathway, 3,4-PCD, in both natural and enriched bacterial communities from a southeastern United States salt marsh. If we presume that successful amplification of a portion of pcaH indicates the presence of a functional 3,4-PCD enzyme (i.e., pcaH and pcaG), these results suggest that taxonomically diverse marine bacteria, some of which have yet to be identified, are involved in the processing of aromatic compounds via a mechanism that has been well described for soil bacteria. In the environment from which these genes were amplified, lignin and lignin degradation products are the most likely sources of naturally occurring aromatic substrates.
The pcaH primer set used here was based on previously retrieved pcaH genes and therefore may target only a subset of the ring cleavage dioxygenases present in the system studied. Furthermore, at least six different ring cleavage dioxygenases in addition to 3,4-PCD have been identified in soil bacteria, and these or other novel dioxygenases may also be present in coastal marine marshes. Nonetheless, the pcaH gene is present in the decomposer community of this coastal marsh, and at least 85 different versions are present, as indicated by sequence differences in the 159-bp fragment amplified from pcaH.
The radiation of similar pcaH sequences raises questions about phenotypic microheterogeneity in the salt marsh bacterial community and potentially has interesting implications for population dynamics and ecological function. The sequence microheterogeneity observed in the pcaH fragment may reflect genetic divergence within the phylogenetically broad clade that has little ecological significance. Alternatively, it may serve as the basis for slight differences in enzyme activity or stability under different environmental conditions. Because members of the roseobacter lineage are amenable to culturing, it should be possible to perform laboratory-based physiological and genetic studies of aromatic compound degradation by members of this bacterial clade having distinctive pcaGH sequences. Access to the physiology of ecologically relevant bacteria via culturing is uncommon in microbial ecology, and such access may lead to unique insights into the role of functional gene microheterogeneity in natural environments.
ACKNOWLEDGMENTS
We are grateful to José González for advice concerning experimental design.
This work was supported by the NSF through grants from the Microbial Observatory Program (grant MCB-0084164 to M.A.M.) and the Molecular and Cellular Biosciences Program (grant MCB-9808784 to E.L.N.) and a traineeship (to A.B.) provided through a research training grant in prokaryotic diversity (grant BIR-9413235).
REFERENCES
- 1.Boettcher K J, Barber B J, Singer J T. Additional evidence that juvenile oyster disease is caused by a member of the Roseobacter group and colonization of nonaffected animals by Stappia stellulata-like strains. Appl Environ Microbiol. 2000;66:3924–3930. doi: 10.1128/aem.66.9.3924-3930.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braker G, Zhou J, Wu L, Devol A H, Tiedje J M. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl Environ Microbiol. 2000;66:2096–2104. doi: 10.1128/aem.66.5.2096-2104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchan A, Collier L S, Neidle E L, Moran M A. Key aromatic-ring-cleaving enzyme, protocatechuate 3,4-dioxygenase, in the ecologically important marine Roseobacter lineage. Appl Environ Microbiol. 2000;66:4662–4672. doi: 10.1128/aem.66.11.4662-4672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bundy B M, Campbell A L, Neidle E L. Similarities between the antABC-encoded anthranilate dioxygenase and the benABC-encoded benzoate dioxygenase of Acinetobacter sp. strain ADP1. J Bacteriol. 1998;180:4466–4474. doi: 10.1128/jb.180.17.4466-4474.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contzen M, Stolz A. Characterization of the genes for two protocatechuate 3,4-dioxygenases from the 4-sulfocatechol-degrading bacterium Agrobacterium radiobacter strain S2. J Bacteriol. 2000;182:6123–6129. doi: 10.1128/jb.182.21.6123-6129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cottrell M, Cary S C. Diversity of dissimilatory bisulfite reductase genes of bacteria associated with the deep-sea hydrothermal vent polychaete annelid Alvinella pompejana. Appl Environ Microbiol. 1999;65:1127–1132. doi: 10.1128/aem.65.3.1127-1132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dagley S. Biochemistry of aromatic hydrocarbon degradation in pseudomonads. In: Sokatch J R, editor. The bacteria. Vol. 10. New York, N.Y: Academic Press Inc.; 1986. pp. 527–555. [Google Scholar]
- 8.Dang H, Lovell C R. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl Environ Microbiol. 2000;66:467–475. doi: 10.1128/aem.66.2.467-475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esham E C, Ye W Y, Moran M A. Identification and characterization of humic substances-degrading bacterial isolates from an estuarine environment. FEMS Microbiol Ecol. 2000;34:103–111. doi: 10.1111/j.1574-6941.2000.tb00759.x. [DOI] [PubMed] [Google Scholar]
- 10.Fuhrman J A, Lee S H, Masuchi Y, Davis A A, Wilcox R M. Characterization of marine prokaryotic communities via DNA and RNA. Microb Ecol. 1994;28:133–145. doi: 10.1007/BF00166801. [DOI] [PubMed] [Google Scholar]
- 11.Gaines G L, Smith L, Neidle E L. Novel nuclear magnetic resonance spectroscopy methods demonstrate preferential carbon source utilization by Acinetobacter calcoaceticus. J Bacteriol. 1996;178:6833–6841. doi: 10.1128/jb.178.23.6833-6841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giovannoni S J. The polymerase chain reaction. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons; 1991. pp. 177–201. [Google Scholar]
- 13.González J M, Kiene R P, Moran M A. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl Environ Microbiol. 1999;65:3810–3819. doi: 10.1128/aem.65.9.3810-3819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González J M, Mayer F, Moran M A, Hodson R E, Whitman W B. Sagittula stellata gen. nov., sp. nov., a lignin transforming bacterium from a coastal environment. Int J Syst Bacteriol. 1997;47:773–780. doi: 10.1099/00207713-47-3-773. [DOI] [PubMed] [Google Scholar]
- 15.González J M, Moran M A. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González J M, Simó R, Massana R, Covert J S, Casamayor E O, Pedrós-Alió C, Moran M A. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl Environ Microbiol. 2000;66:4237–4246. doi: 10.1128/aem.66.10.4237-4246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González J M, Whitman W B, Hodson R E, Moran M A. Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl Environ Microbiol. 1996;62:4433–4440. doi: 10.1128/aem.62.12.4433-4440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harwood C S, Parales R E. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 19.Hobbie J E, Daley R J, Jasper S. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ledyard K M, Delong E F, Dacey J W H. Characterization of a DMSP-degrading bacterial isolate from the Sargasso Sea. Arch Microbiol. 1993;160:312–318. [Google Scholar]
- 20a.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovell C R, Piceno Y M, Quattro J M, Bagwell C E. Molecular analysis of diazotroph diversity in the rhizosphere of the smooth cordgrass, Spartina alterniflora. Appl Environ Microbiol. 2000;66:3814–3822. doi: 10.1128/aem.66.9.3814-3822.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin V J J, Mohn W W. Genetic investigation of the catabolic pathway for degradation of abietane diterpenoids by Pseudomonas abietaniphila BKME-9. J Bacteriol. 2000;182:3784–3793. doi: 10.1128/jb.182.13.3784-3793.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mashetty S B, Manohar S, Karegoudar T B. Degradation of 3-hydroxybenzoic acid by a Bacillus species. Indian J Biochem Biophys. 1996;33:145–148. [PubMed] [Google Scholar]
- 24.Moran M A, Hodson R E. Formation and bacterial utilization of dissolved organic carbon derived from detrital lignocellulose. Limnol Oceanogr. 1989;34:1034–1047. [Google Scholar]
- 25.Moran M A, Hodson R E. Bacterial production on humic and nonhumic components of dissolved organic carbon. Limnol Oceanogr. 1990;35:1744–1756. [Google Scholar]
- 26.Moran M A, Hodson R E. Dissolved humic substances of vascular plant origin in a coastal marine environment. Limnol Oceanogr. 1994;39:762–771. [Google Scholar]
- 27.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisions reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 28.Ohkuma M, Noda S, Kudo T. Phylogenetic diversity of the nitrogen fixation genes in the symbiotic community in the gut of diverse termites. Appl Environ Microbiol. 1999;65:4926–4934. doi: 10.1128/aem.65.11.4926-4934.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohlendorf D H, Orville A M, Lipscomb J D. Structure of protocatechuate 3,4-dioxygenase from Pseudomonas aeruginosa at 2.15 Å resolution. J Mol Biol. 1994;244:586–608. doi: 10.1006/jmbi.1994.1754. [DOI] [PubMed] [Google Scholar]
- 30.Parke D. Acquistion, reorganization, and merger of genes: novel management of the β-ketoadipate pathway in Agrobacterium tumefaciens. FEMS Microbiol Lett. 1997;146:3–12. [Google Scholar]
- 31.Parke D, D'Argenio D A, Ornston L N. Bacteria are not what they eat: that is why they are so diverse. J Bacteriol. 2000;182:257–263. doi: 10.1128/jb.182.2.257-263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parke D, Ornston L N. Enzymes of the β-ketoadipate pathway are inducible in Rhizobium and Agrobacterium spp. and constitutive in Bradyrhizobium spp. J Bacteriol. 1986;165:288–292. doi: 10.1128/jb.165.1.288-292.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pomeroy L R, Darley W M, Dunn E L, Gallagher J L, Haines E B, Whitney D M. Primary production. In: Pomeroy L R, Wiegert R G, editors. The ecology of a salt marsh. New York, N.Y: Springer-Verlag; 1981. pp. 39–68. [Google Scholar]
- 34.Rogers G M, Baecker A A W. Theories on the degradation in wood associated with glycocalyx-producing bacteria. J Inst Wood Sci. 1987;11:78–84. [Google Scholar]
- 35.Rolden M D, Blasco R, Caballero F J, Castillo F. Degradation of p-nitrophenol by the phototrophic bacterium Rhodobacter capsulatus. Arch Microbiol. 1998;169:36–42. doi: 10.1007/s002030050538. [DOI] [PubMed] [Google Scholar]
- 36.Romero-Steiner S, Parales R E, Harwood C S, Houghton J E. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J Bacteriol. 1994;176:5771–5779. doi: 10.1128/jb.176.18.5771-5779.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rüger H-J, Holfe M G. Marine star-shaped-aggregate-forming bacteria: Agrobacterium atlantic sp. nov.; Agrobacterium meteor sp. nov; Agrobacterium ferruginous sp. nov., nom. rev.; Agrobacterium gelatinovorum sp. nov., nom. rev.; and Agrobacterium stellulatum sp. nov., nom. rev. Int J Syst Bacteriol. 1992;42:133–143. doi: 10.1099/00207713-42-1-133. [DOI] [PubMed] [Google Scholar]
- 38.Scala D J, Kerkhof L J. Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl Environ Microbiol. 1999;65:1681–1687. doi: 10.1128/aem.65.4.1681-1687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segura A, Bunz P V, D'Argenio D A, Ornston L N. Genetic analysis of a chromosomal region containing vanA and vanB, genes required for conversion of either ferulate or vanillate to protocatechuate in Acinetobacter. J Bacteriol. 1999;181:3494–3504. doi: 10.1128/jb.181.11.3494-3504.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sorokin D Y, Lysenko A M. Heterotrophic bacteria from the Black Sea oxidizing reduced sulfur compounds to sulfate. Microbiology (Engl Transl Mikrobiologiya) 1993;62:1018–1031. [Google Scholar]
- 41.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Ströbel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasydevan N, Mahadevan A. Degradation of nonphenolic β-ortho-4-lignin substructure model compounds by Acinetobacter sp. Res Microbiol. 1992;143:333–339. doi: 10.1016/0923-2508(92)90025-j. [DOI] [PubMed] [Google Scholar]
- 43.Vetting M W, D'Argenio D A, Ornston L N, Ohlendorf D H. A structure of Acinetobacter sp. ADP1 protocatechuate 3,4-dioxygenase at 2.2 Å resolution: implications for the mechanism of an intradiol dioxygenase. Biochemistry. 2000;39:7943–7955. doi: 10.1021/bi000151e. [DOI] [PubMed] [Google Scholar]
- 44.Wang G C-Y, Wang Y. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology. 1996;142:1107–1117. doi: 10.1099/13500872-142-5-1107. [DOI] [PubMed] [Google Scholar]