Abstract
We have developed a new, effective procedure for detecting Vibrio parahaemolyticus in seafoods using enrichment and plating onto a chromogenic agar medium. Samples were cultured in salt Trypticase soy broth, which is a nonselective medium, and then a portion of the culture was cultured with salt polymyxin broth, which is a selective medium for V. parahaemolyticus. This two-step enrichment was more effective than the one-step enrichment in salt polymyxin broth alone. The enrichment cultures were then plated onto a new chromogenic agar containing substrates for beta-galactosidase. The V. parahaemolyticus colonies developed a purple color on this growth medium that distinguished them from other related bacterial strains. V. parahaemolyticus was isolated more frequently from naturally contaminated seafood samples using the chromogenic agar than thiosulfate citrate bile salts sucrose agar medium, which is currently used for the isolation of V. parahaemolyticus. Our findings suggest that this new enrichment and isolation scheme is more sensitive and accurate for identifying V. parahaemolyticus in seafood samples than previously used methods.
Vibrio parahaemolyticus is a major food-borne pathogen that causes worldwide health problems. Prevention of V. parahaemolyticus contamination of foods, especially contamination with serotype O3:K6, which has been associated with recent outbreaks (3, 5, 6, 7), is an important public health concern. It has been observed that the new O3:K6 clone of V. parahaemolyticus spread to Taiwan, Laos, Japan, Thailand, Korea, and the United States between 1997 and 1998 (10).
To establish effective control measures to reduce the risk of V. parahaemolyticus infection and to ensure the safety of foods, efficient analytical methods for the detection of V. parahaemolyticus in foods and the environment must be available. Selective enrichment with alkaline peptone water (APW) or salt polymyxin broth (SPB) and plating of the enrichment culture onto thiosulfate citrate bile salts sucrose (TCBS) agar have been widely used for selective isolation of V. parahaemolyticus from foods. Although this existing method for detecting this organism in foods is inadequate, V. parahaemolyticus isolation schemes have not been carefully studied. It has been observed for other bacteria that nonselective enrichment prior to selective enrichment is effective for the detection of bacteria injured by various environmental stresses (4, 13). Therefore, we studied the effectiveness of nonselective enrichment prior to selective enrichment for V. parahaemolyticus. Furthermore, we have noted in a survey of seafoods that V. parahaemolyticus colonies on TCBS agar are difficult to distinguish visually from other bacterial colonies, since they can be covered by a yellow color produced by sucrose-fermenting bacteria. To develop a more efficient method, we studied the effectiveness of a method involving a two-step enrichment with nonselective and selective media and plating onto a chromogenic agar medium. This agar medium containing substrates for beta-galactosidase was developed specifically to differentiate V. parahaemolyticus from other bacteria by using a chromogenic substrate, instead of sugar fermentation, in traditional growth media such as TCBS.
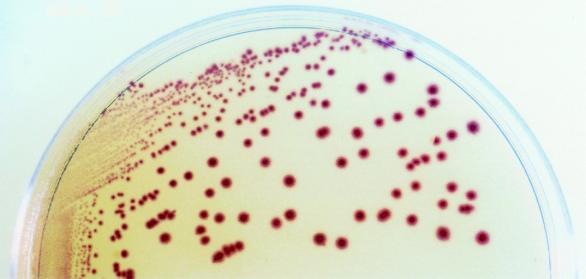
First, we tested strains of various bacteria (Table 1) which could contaminate seafoods during harvesting and processing for colony formation on the new chromogenic agar medium CHROMagar Vibrio (CV) agar medium (CHROMagar Microbiology, Paris, France). Photobacterium damselae (IFO15633) and Vibrio alginolyticus (IFO15630) were provided by the Institute for Fermentation (Osaka, Japan). Pseudomonas aeruginosa (ATCC 9027), Vibrio hollisae (ATCC 33564), and Vibrio mimicus (ATCC 33656) were from the American Type Culture Collection (Manassas, Va.). Vibrio vulnificus (JCM3725) was from the Japan Collection of Microorganisms (Saitama, Japan). The others were isolates obtained from clinical and environmental samples. Vibrio spp. and the other bacteria were cultured at 37°C for 18 h in tryptic soy broth (TSB; Difco, Detroit, Mich.) supplemented with 2% NaCl (NTSB) and TSB alone, respectively. A loopful of each culture was streaked onto CV medium. After incubation for 20 h at 35 to 37°C, the colonies were examined for size and color. All 68 strains of V. parahaemolyticus, including 24 strains of serotype O3:K6, formed violet colonies on CV agar (Fig. 1; Table 1). The other strains did not form colonies or formed colonies of other colors. V. mimicus and V. vulnificus, both of which cannot easily be distinguished from V. parahaemolyticus on TCBS agar, formed green colonies distinguishable from those of V. parahaemolyticus on CV agar. Vibrio cholerae and V. alginolyticus formed pale-blue and milk-white colonies, respectively.
TABLE 1.
Colony morphologies of various bacteria grown on CV medium and TCBS medium
| Species | No. of strains tested | CV medium
|
TCBS medium
|
||
|---|---|---|---|---|---|
| Size of colonya | Color of colony | Size of colony | Color of colony | ||
| Citorobacter freundii | 1 | NG | NG | ||
| Edwardsiella tarda | 1 | NG | NG | ||
| Enterobacter cloacae | 2 | NG | NG | ||
| Escherichia coli O157:H7 | 2 | NG | NG | ||
| Klebsiella ornithinolytica | 1 | NG | NG | ||
| Klebsiella oxytoca | 1 | NG | NG | ||
| Photobacterium damselae | 1 | NG | NG | ||
| Proteus mirabilis | 2 | Minute | Milk white | Minute | Blue-green |
| Providencia rettgeri | 1 | 1 | Milk white | Minute | Blue-green |
| Pseudomonas aeruginosa | 1 | NG | NG | ||
| Salmonella enteritidis | 2 | NG | NG | ||
| Serratia marcescens | 1 | NG | NG | ||
| Shigella sonnei | 2 | NG | NG | ||
| Vibrio alginolyticus | 4 | 5–6 | Milk white | 3–4 | Yellow |
| Vibrio cholerae O1 | 2 | 3 | Pale blue | 3 | Yellow |
| Vibrio hollisae | 1 | 4 | Milk white | 3 | Green |
| Vibrio mimicus | 2 | 3–4 | Pale blue | 1–2 | Green |
| Vibrio parahaemolyticus | 68 | 3–5 | Violet | 2–4 | Green |
| Vibrio vulnificus | 1 | 5 | Pale blue | 1 | Green |
NG, no growth. Values are in millimeters.
FIG. 1.
Appearance of V. parahaemolyticus on CV agar.
Subsequently, we compared various enrichment and plating combinations for the ability to isolate V. parahaemolyticus from naturally contaminated seafoods. Whole bodies of grunt (Parapristipoma trilineatum), horse mackerel (Trachurus japonicus), mackerel (Scomber japonicus), right-eyed flounder (Pleuronectidae), sardine (Sardinops melanostictus), saury (Cololabis saina), octopus (Octopus vulgaris), squid (Sepioteuthis lessoniana), scallop (Patinopecten yessoenisis), spiny top shell (Turbo cornutus), and short-necked clam (Tapes japonica) meat removed from shells and cut meat of codfish (Gadus macrocephalus) were purchased from retail shops in Tokyo in the autumn of 2000. Each seafood specimen (200 to 500 g) had been packed separately in plastic packages when purchased. The seafood specimens were transferred with ice from retail shops to our laboratories within 1 h and kept at 4°C for less than 6 h until they were used in the experiments. For scallop and spiny top shells, whole bodies removed from shells were used as test samples. Fish, octopus, and squid purchased as whole bodies were cut into several portions. Portions including the gills and skin from fish and those including legs from octopus and squid were used as test samples. Total viable bacterial counts and coliform counts were determined for each sample (16).
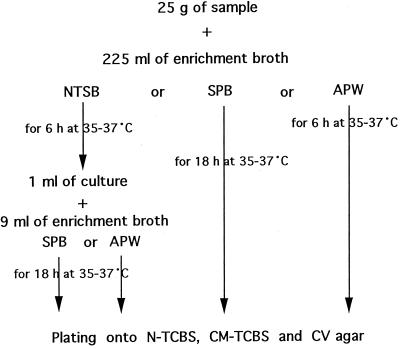
In the first series of experiments, the presence of V. parahaemolyticus in short-necked clam meat and horse mackerel was examined using CV agar, TCBS agar (Nissui Co., Tokyo, Japan) (N-TCBS), and cholera medium TCBS agar (Oxoid, Unipath Ltd., Basingstoke, Hampshire, United Kingdom) (CM-TCBS) in combination with different enrichment procedures. N-TCBS and CM-TCBS have been widely used as selective agar media for Vibrio spp., including V. cholera and V. parahaemolyticus. The formulae of both media are similar except for the concentrations of sucrose, ox bile, and agar. The total numbers of viable bacteria and coliform bacteria in short-necked clams were 5.41 and 3.49 log CFU/g, respectively, and those in horse mackerel were 5.89 and 4.13 log CFU/g, respectively. Duplicate portions (25 g) from each package were cultured with 225 ml of NTSB, APW (Nissui Co.), or SPB (Nissui Co.) at 37°C for 6, 6, or 18 h, respectively (Fig. 2). One milliliter of the NTSB culture was further cultured in 9 ml of APW or SPB for 18 h at 37°C as NTSB-APW or NTSB-SPB, respectively. A loopful of each enrichment culture was streaked onto N-TCBS, CM-TCBS, and CV agars. After the agars were incubated at 37°C for 18 h, colonies suspected of being V. parahaemolyticus were incubated in triple-sugar iron medium (Eiken Co., Tokyo, Japan), nutrient broth (NB; Difco), and NB supplemented with 8% NaCl for confirmation of glucose fermentation, sucrose nonfermentation, nongrowth in NB, and growth in NB supplemented with 8% NaCl. The levels of efficiency of the plating media for identification of V. parahaemolyticus colonies were evaluated by classifying each plate as negative or positive when no colonies or more than one colony was identified as V. parahaemolyticus, respectively.
FIG. 2.
Protocol of the method of enrichment and plating for isolation of V. parahaemolyticus from seafood samples.
V. parahaemolyticus was isolated from all short-necked clam samples using CV agar in combination with all enrichment conditions (Table 2). In contrast, the organism was detected in 50 to 70% of the samples when CM-TCBS agar was used in combination with any of the enrichment conditions and in 80 to 90% of the samples when N-TCBS agar was used in combination with enrichment with either APW, NTSB-APW, or NTSB-SPB (Table 2). In addition, V. parahaemolyticus was isolated from horse mackerel samples more frequently with CV agar than with the other agars (Table 2). The numbers of colonies identified as V. parahaemolyticus tended to be greater on CV agar plates than on the other plates for both food items (data not shown). The presence of the toxR gene, which is specific for V. parahaemolyticus, was confirmed in 33 colonies randomly selected from the colonies identified as V. parahaemolyticus using the PCR method (9). Thus, the methods using CV agar in combination with SPB or NTSB-SPW were superior for both food items when the numbers of identified colonies were compared. NTSB-SPB was superior to SPB for horse mackerel but not for short-necked clam (data not shown).
TABLE 2.
Comparison of enrichment procedures and agar media for isolation of V. parahaemolyticusa
| Enrichment culture | Agar medium | No. of short-necked clam samples
|
No. of horse mackerel samples
|
||
|---|---|---|---|---|---|
| − | + | − | + | ||
| APW | CV | 0 (0)a* | 10 (100) | 1 (10) | 9 (90) |
| N-TCBS | 1 (10) | 9 (90) | 4 (40) | 6 (60) | |
| CM-TCBS | 5 (50)* | 5 (50) | 3 (30) | 7 (70) | |
| SPB | CV | 0 (0) | 10 (100) | 0 (0)** | 10 (100) |
| N-TCBS | 0 (0) | 10 (100) | 3 (30) | 7 (70) | |
| CM-TCBS | 3 (30) | 7 (70) | 6 (60)** | 4 (40) | |
| NTSB-APW | CV | 0 (0)* | 10 (100) | 2 (20) | 8 (80) |
| N-TCBS | 2 (20) | 8 (80) | 5 (50) | 5 (50) | |
| CM-TCBS | 5 (50)* | 5 (50) | 4 (40) | 6 (60) | |
| NTSB-SPB | CV | 0 (0) | 10 (100) | 0 (0) | 10 (100) |
| N-TCBS | 1 (10) | 9 (90) | 2 (20) | 8 (80) | |
| CM-TCBS | 4 (40) | 6 (60) | 4 (40) | 6 (60) | |
Values are numbers of samples showing growth (+) or no growth (−) of V. parahaemolyticus out of 10 samples tested (percentages). * and **, significant differences (P < 0.05 and P < 0.01, respectively) between results with CV agar and results with other agars.
In the second series of experiments, the presence of V. parahaemolyticus was tested in 12 different seafoods using CV and N-TCBS agars in combination with enrichment with SPB and NTSB-SPW. A 25-g portion of each seafood sample was cultured with 225 ml of NTSB or SPB at 37°C for 6 or 18 h, respectively, so that each portion from the same package was assigned to the two enrichment media. One milliliter of the NTSB culture was transferred to 9 ml of SPB and incubated for 18 h at 37°C. A loopful of each enrichment culture was streaked onto N-TCBS and CV agars. After incubation at 37°C for 18 h, colonies were confirmed as V. parahaemolyticus by the methods mentioned above. The levels of efficiency of the plating media were evaluated by the methods mentioned above. Significant differences between the numbers of samples detected as V. parahaemolyticus were analyzed by Fisher's exact probability test (15).
V. parahaemolyticus was isolated more frequently with CV agar than with N-TCBS agar by either of the two enrichment protocols; V. parahaemolyticus was isolated from 65 and 80% of samples using CV agar with SPB and NTSB-SPB, respectively, and from 50 and 70% of samples using N-TCBS with SPB and NTSB-SPB, respectively (Table 3). CV agar was also superior to N-TCBS agar in terms of the number of identified colonies (Table 3). The total numbers of bacteria in these seafood samples were 3.06 to 6.90 log CFU/g for viable bacteria and a nondetectable number to 5.70 log CFU/g for coliform bacteria. However, the total numbers of viable and coliform bacteria were not related to contamination of V. parahaemolyticus.
TABLE 3.
Detection of V. parahaemolyticus from various foods
| Seafood samplea | Total no. ofb:
|
Test resultc on indicated agar after enrichment with:
|
||||
|---|---|---|---|---|---|---|
| NTSB-SPB
|
SPB
|
|||||
| Viable bacteria | Coliform bacteria | N-TCBS | CV | N-TCBS | CV | |
| Short-necked clam 1 | 5.09 | 4.09 | ++ | +++ | ++ | +++ |
| Short-necked clam 2 | 6.90 | 4.14 | ++ | +++ | ++ | +++ |
| Short-necked clam 3 | 6.81 | 3.98 | ++ | +++ | +++ | +++ |
| Short-necked clam 4 | 4.18 | 2.21 | + | ++ | ++ | +++ |
| Codfish | 3.86 | 2.93 | + | ++ | + | + |
| Grunt | 5.62 | 3.65 | + | + | − | + |
| Horse mackerel 1 | 3.24 | ND | − | − | − | − |
| Horse mackerel 2 | 5.48 | 3.99 | + | ++ | − | + |
| Horse mackerel 3 | 6.03 | 4.12 | − | − | − | − |
| Mackerel | 4.73 | 1.53 | + | +++ | ++ | +++ |
| Octopus | 6.08 | 5.70 | + | ++ | + | ++ |
| Right-eyed flounder | 5.83 | 4.51 | ++ | + | − | − |
| Saury | 3.13 | 1.78 | + | ++ | ++ | ++ |
| Sardine 1 | 4.46 | 2.51 | − | + | + | ++ |
| Sardine 2 | 4.62 | 3.16 | + | + | + | ++ |
| Scallop 1 | 3.06 | 2.44 | + | + | − | + |
| Scallop 2 | 3.83 | 3.02 | + | + | − | − |
| Squid 1 | 3.68 | 1.90 | − | − | − | − |
| Squid 2 | 4.48 | 3.62 | − | − | − | − |
| Spiny top shell | 3.99 | 0.9 | − | + | − | − |
Different numbers indicate different packages.
Values are numbers of log CFU per gram. ND, not detected.
Appearance of V. parahaemolyticus on agar medium. −, no colonies of V. parahaemolyticus; +, less than 10 colonies of V. parahaemolyticus; ++, more than 11 colonies of V. parahaemolyticus; +++, most colonies were V. parahaemolyticus.
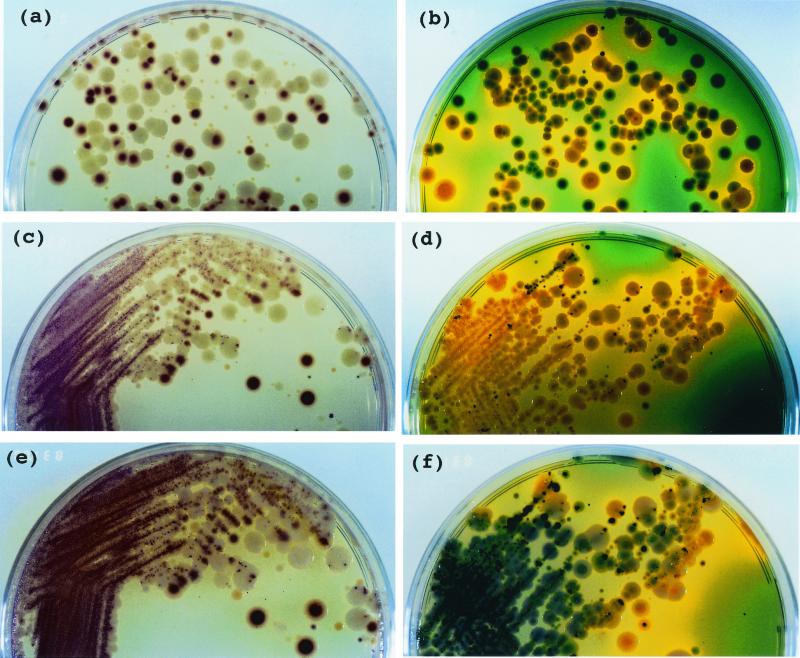
A portion (0.1 ml) of diluted enrichment cultures from the octopus samples was spread on CV and N-TCBS agars, which were then cultured at 37°C for 18 h. Violet (Fig. 3a) and green (Fig. 3b) colonies were observed on CV and N-TCBS agars, respectively, as being typical of V. parahaemolyticus colonies. However, the typical colonies were clearly distinguished only on CV agar (Fig. 3c) when a loopful of the same enrichment culture was streaked on both agars, which were then incubated at 37°C for 18 h. Furthermore, the color of N-TCBS agar changed to green and black after being left for 24 h at ambient room temperature (Fig. 3f), whereas the color of CV agar remained unchanged under the same conditions (Fig. 3e).
FIG. 3.
Colonies plated out from enrichment culture of seafoods on CV and TCBS agars. (a, c, and e) CV agar; (b, d, and f) TCBS agar; (a to d) incubation at 37°C for 18 h; (e and f) incubation at room temperature for 24 h followed by incubation at 37°C for 18 h; (a and b) spread plating; (c to f) streak plating. Violet colonies on CV agar and green colonies on TCBS agar were identified as V. parahaemolyticus.
Acids produced by sucrose-fermenting bacteria diffuse into and spread though the agar with time, changing the color of the agar from green to yellow in the area surrounding the colonies of sucrose-fermenting bacteria or even across the entire area of the agar plate. When V. parahaemolyticus colonies near sucrose-fermenting bacterial colonies are covered by the yellow color, the color of V. parahaemolyticus colonies also changes from green to yellow or yellow-green, leading to difficulties in the differentiation of V. parahaemolyticus from other bacteria. In contrast, the violet color of V. parahaemolyticus on CV agar was not affected by the presence of colonies of other bacteria on the same plate, because generation of the color depends on the reaction of the bacterial beta-galactosidase with the substrate contained in the media. Furthermore, the color of V. parahaemolyticus colonies did not change with time for up to 18 h at room temperature. Violet colonies of V. parahaemolyticus remained violet even when they were covered by other colors produced by other bacteria, differing from what occurred with TCBS agar, on which V. parahaemolyticus colonies were occasionally hidden by the yellow color produced by sucrose-fermenting bacteria such as V. alginolyticus. Perhaps because of these characteristics of CV agar, the rate of detection of V. parahaemolyticus in naturally contaminated seafood samples was higher when CV agar was used than when TCBS was used.
The present experiments, using various seafoods as test samples, demonstrated that the detection rate was higher when NTSB-SPB was used than when SPB alone was used for enrichment. It has been observed that V. parahaemolyticus, like many other bacteria, is injured by various environmental stresses such as low temperature (8, 11). Stressed bacteria are difficult to grow in selective enrichment broth (4, 13). These findings suggest that a nonselective medium such as TSB prior to enrichment with selective media might be effective in recovering injured cells. Furthermore, based upon the present findings, CV agar is recommended as the selective agar in combination with enrichment with NTSB-SPB for the isolation of V. parahaemolyticus from seafood samples.
In the present study, the total number of V. parahaemolyticus cells was targeted to assess the isolation procedure involving CV agar and two-step enrichment with NTSB-SPB. This procedure would also be effective in isolating thermostable direct hemolysin (TDH)- or TDH-related hemolysin-producing V. parahaemolyticus strains, because these pathogenic strains may behave similarly to total V. parahaemolyticus cells in the process of the isolation procedure, especially on CV agar. Thus, pathogenic V. parahaemolyticus, which has not been as frequently detected in foods as it has been in clinical specimens (2, 12, 14), might also be isolated more successfully by the use of the present procedure with CV agar.
The acceptable level for the total number of V. parahaemolyticus cells in fish fillets and shocked shellfishes for raw consumption is below 100 CFU/g of sample in Japan. Although the two-step enrichment procedure would not be suitable for routine tests for inspection in terms of the time required, CV medium would be very useful for routine tests, including those with the most-provable-number method (1).
REFERENCES
- 1.American Public Health Association. Recommended procedure for the examination of seawater and shellfish. 4th ed. Washington, D.C.: American Public Health Association; 1970. [Google Scholar]
- 2.Ayres P A, Barrow G I. The distribution of Vibrio parahaemolyticus in British coastal water: report of a collaborative study 1975–6. J Hyg. 1978;95:299–307. doi: 10.1017/s002217240005364x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bag P K, Nandi S, Bhadra R K, Ramamurthy T, Bhattacharya S K, Nishibuchi M, Hamabata T, Yamasaki S, Takeda Y, Nair G B. Clonal diversity among recently emerged strains of Vibrio parahaemolyticus O3:K6 associated with pandemic spread. J Clin Microbiol. 1999;37:2354–2357. doi: 10.1128/jcm.37.7.2354-2357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beuchat L R. Suitability of some enrichment broths and dilutions for enumerating cold- and heat-stressed Vibrio parahaemolyticus. Can J Microbiol. 1977;23:630–633. doi: 10.1139/m77-092. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Outbreak of Vibrio parahaemolyticus infections association with eating raw oysters—Pacific Northwest, 1997. Morb Mortal Wkly Rep. 1998;47:457–462. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Outbreak of Vibrio parahaemolyticus infection association with eating raw oysters and clams harvested from Long Island Sound—Connecticut, New Jersey, and New York, 1998. Morb Mortal Wkly Rep. 1999;48:48–51. [PubMed] [Google Scholar]
- 7.Infectious Diseases Surveillance Center; National Institute of Infectious Diseases. Vibrio parahaemolyticus, Japan, 1996–1998. Infect Agents Surv Rep. 1999;20:159. ′–160′. [Google Scholar]
- 8.Jiang X, Chai T-J. Survival of Vibrio parahaemolyticus at low temperatures under starvation conditions and subsequent resuscitation of viable, nonculturable cells. Appl Environ Microbiol. 1996;62:1300–1305. doi: 10.1128/aem.62.4.1300-1305.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y B, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol. 1999;37:1173–1177. doi: 10.1128/jcm.37.4.1173-1177.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumoto C, Okuda J, Ishibashi M, Iwanaga M, Garg P, Rammamurthy T, Wong H-C, Depaola A, Kim Y B, Albert M J, Nishibuchi M. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J Clin Microbiol. 2000;38:578–585. doi: 10.1128/jcm.38.2.578-585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muntada-Garriga J M, Rodrigues-Jerez J J, Lopez-Sabater E I, More-Ventura M T. Effect of chill and freezing temperatures on survival of Vibrio parahaemolyticus inoculated in homogenates of oyster meat. Lett Appl Microbiol. 2000;20:225–227. doi: 10.1111/j.1472-765x.1995.tb00433.x. [DOI] [PubMed] [Google Scholar]
- 12.Oliver J D, Kaper J B. Vibrio species. In: Doyle M P, Beuchat L R, Montville T J, editors. Food microbiology: fundamentals and frontiers. Washington, D.C.: American Society for Microbiology; 1997. pp. 228–264. [Google Scholar]
- 13.Ray B, Hawkins S M, Hackney C R. Method for the detection of injured Vibrio parahaemolyticus in seafoods. Appl Environ Microbiol. 1978;35:1121–1127. doi: 10.1128/aem.35.6.1121-1127.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakazaki R, Tamura K, Kato T, Obara Y, Yamai S, Hobo K. Studies on the enteropathogenic, facultatively halophilic bacteria, Vibrio parahaemolyticus III. Enteropathogenicity. Jpn J Med Sci Biol. 1968;21:325–331. doi: 10.7883/yoken1952.21.325. [DOI] [PubMed] [Google Scholar]
- 15.Siegel S, Castellan J N., Jr . Two independent samples. In: Siegel S, John Castellan N Jr, editors. Nonparametric statistic for the behavioral sciences. Singapore: McGraw-Hill Book Company; 1988. pp. 102–197. [Google Scholar]
- 16.Zipkes M R, Gilchrist J E, Peeler J T. Comparison of yeast and mold counts by spiral, pour, and streak plate methods. J Assoc Off Anal Chem. 1981;64:1465–1469. [PubMed] [Google Scholar]