Abstract
Epilepsy is one of the most frequent neurological diseases, with focal epilepsy accounting for the largest number of cases. The genetic alterations involved in focal epilepsy are far from being fully elucidated.
Here, we show that defective lipid signalling caused by heterozygous ultra-rare variants in PIK3C2B, encoding for the class II phosphatidylinositol 3-kinase PI3K-C2β, underlie focal epilepsy in humans. We demonstrate that patients’ variants act as loss-of-function alleles, leading to impaired synthesis of the rare signalling lipid phosphatidylinositol 3,4-bisphosphate, resulting in mTORC1 hyperactivation. In vivo, mutant Pik3c2b alleles caused dose-dependent neuronal hyperexcitability and increased seizure susceptibility, indicating haploinsufficiency as a key driver of disease. Moreover, acute mTORC1 inhibition in mutant mice prevented experimentally induced seizures, providing a potential therapeutic option for a selective group of patients with focal epilepsy.
Our findings reveal an unexpected role for class II PI3K-mediated lipid signalling in regulating mTORC1-dependent neuronal excitability in mice and humans.
Keywords: class II PI3K, mTOR, epilepsy, variants, PI3K-C2B
Gozzelino et al. show that defective lipid signalling caused by heterozygous ultra-rare variants in PIK3C2B gives rise to focal epilepsy via mTOR hyperactivation. In vivo, mutant Pik3c2b alleles cause dose-dependent neuronal hyperexcitability and increased seizure susceptibility, identifying haploinsufficiency as a key driver of disease.
Introduction
With an estimated lifetime prevalence of 6.4‰, epilepsy is one of the most frequent neurological diseases world-wide.1,2 Epilepsy is under a clear genetic influence,3–5 and large-scale sequencing studies have established that a subset of the rare epilepsy syndromes are distinct monogenic entities.6,7 Focal epilepsy (FE), characterized by seizures originating within one brain hemisphere, represents the most common epilepsy type.8,9 Historically, genes associated with a FE phenotype encoded for proteins directly implicated in synaptic function: ion channels (e.g. KCNT1, KCNQ2, KCNQ3, SCN1A), ionotropic neurotransmitter receptors (e.g. CHRNA4, CHRNA2, CHRNB2) and synaptic proteins (LGI1, RELN, CNTNAP2).8–12 Recent work has paved the way towards novel mechanisms in epilepsy, implicating loss-of-function variants in genes encoding components of the Gap Activity TOward Rags 1 (GATOR1) complex, a negative regulator of the mechanistic Target of Rapamycin Complex 1 (mTORC1), a central signalling hub for cell growth and metabolism, and with specific roles in brain development and function.13–15 In spite of this progress, most patients suffering from FE remain without known genetic aetiology and, hence, without the perspective of targeted therapies.
In a concerted effort to close this knowledge gap we have identified ultra-rare variants in PIK3C2B (MIM*602838) in FE. The encoded PI3K-C2β protein is a ubiquitously expressed class II phosphatidylinositol 3-kinase (PI3K) that has been associated with various biological activities including the migration of cancer cells,16 insulin signalling via the growth-promoting kinase Akt17 and the local repression of mTORC1 activity in serum-starved cells.18,19 These putative physiological functions have been associated with the ability of PI3K-C2β to synthesize phosphatidylinositol 3-phosphate [PI(3)P] or phosphatidylinositol 3,4-bisphosphate [PI(3,4)P2], possibly in a cell-type and/or subcellular location-specific manner.20 Whether PI3K-C2β exhibits a specific function in the CNS is unknown. We show that Pik3c2b haploinsufficiency underlies FE in human patients and leads to increased seizure susceptibility in mouse models. We further demonstrate that the epileptic phenotype associated with impaired PI3K-C2β function is a consequence of impaired PI(3,4)P2 synthesis resulting in mTORC1 hyperactivation and elevated neuronal network excitability. Importantly, epilepsy in mice is potently rescued by mTORC1 inhibition. These findings establish an unprecedented link between class II PI3K-mediated lipid signalling and protection from neuronal hyperexcitability in mice and humans.
Materials and methods
Reagents
Antibodies for immunoblotting
PI3K-C2β (BD Biosciences, #611342, 1:1000), phospho-p70 S6 kinase (Thr389; Cell Signaling, #9205, 1:500), p70 S6 kinase (Cell Signaling, #9202, 1:1000), S6 ribosomal protein (Cell Signaling, #2217, 1:1000), phospho-S6 ribosomal protein (Ser235/236; Cell Signaling, #2211, 1/1000), phospho-Akt (Ser473) antibody (Cell Signaling, #9271, 1:1000), Akt (pan; Cell Signaling, #4691, 1:1000), GAPDH (Cell Signaling, #2118, 1/2500), PSD95 (Synaptic Systems, #124 011, 1:1000), vGAT (Synaptic Systems, #131011, 1:1000), vGLUT (Synaptic Systems, #135302, 1:1000), synaptotagmin 1 (1Synaptic Systems, #105 011, 1:5000), mTOR (Cell Signaling, #7C10, 1:1000), GluA1 (Millipore, # MAB2263, 1:1000), Homer1 (Synaptic Systems, #160003, 1:1000), Rab7 (Cell Signaling, #9367, 1:1000), p62 (Cell Signaling, #5114, 1:1000), LC3 (Abcam, #ab48394, 1:1000), GFP (Abcam, #ab290, 1:1000), Vinculin (Abcam, #ab129002, 1:5000).
Antibodies for mouse brain immunofluorescence
Gephyrin (1:200, Synaptic Systems, #147318), GFAP (1:500, Sigma, G3893), MAP2 (1:1000, Sigma-Aldrich, #M9942), phospho-S6 ribosomal protein (Ser235/236; 1:200, Cell Signaling, #2211), p62 (Progen, #GP62-C, 1:400), PSD95 (1:400, Synaptic Systems, #124 011), neurofilament M (1:200, Chemicon #AB1987), vGAT (1:400, Synaptic Systems, #131011C2), vGLUT (1:400, Synaptic Systems, #135302).
Plasmids and primers
The following plasmids were used: wild-type (WT) and kinase inactive eGFP-PI3K-C2β [28572395]; empty pEGFP-C1. PIK3C2B cDNA mutants were generated by site-directed mutagenesis (Agilent Quikchange Lightning Site-Directed Mutagenesis kit, # 210518) using the following pairs of primers: 5′-AACCCTGGGGAGGACAAGTATGAGAAGGCTG-3′ and 5′-CAGCCTTCTCATACTTGTCCTCCCCAGGGTT-3′ for p.E1169K; 5′-GGTCACATGTTCCACGTTGATTTTGGCCGCTTCC-3′ and 5′-GGAAGCGGCCAAAATCAACGTGGAACATGTGACC-3′ for p.I1212V; 5′-GATGTTTGGCAACATCAAGCCGCTCCGTGCCCCCTTTGTCTTC-3′, 5′-GAAGACAAAGGGGGCACGGAGCGGCTTGATGTTGCCAAACATC-3′ and 5′-CATCAAGCCGCTCCCTGCCCCCTTTGTCTTC-3′, 5′-GAAGACAAAGGGGGCAGGGAGCGGCTTGATG-3′ in a two-step reaction for the PI(3)P-only producing mutant (KRDR1228-1231KPLP);21 5′-GCATGGTCATCTTCCTCTGCTTCTCCACCGG-3′ and 5′-CCGGTGGAGAAGCAGAGGAAGATGACCATGC-3′ for p.R1118L; 5′-CCTGAACTCTCAGACCTGCAGGACCTCAAGTATGTG-3′ and 5′-CACATACTTGAGGTCCTGCAGGTCTGAGAGTTCAGG-3′ for p.E1294Q; 5′-GGGCTTGCAACTGCTCTAGGATGGAAATGACCC-3′ and 5′-GGGTCATTTCCATCCTAGAGCAGTTGCAAGCCC-3′ for p.Q1533X; 5′-GACCCCTATGTGAAAATGTACCTCCTTCCTGACCC-3′ and 5′-GGGTCAGGAAGGAGGTACATTTTCACATAGGGGTC-3′ for I1544M. All mutants were verified by standard Sanger sequencing.
Patient enrolment
Italian patients
Following the approval by the Human Research Ethics Committee of Bellaria Hospital in Bologna, Italy (Prot. N 945/CE; cod CE: 13084), between January 2014 and June 2017 we recruited 106 consenting Italian patients presenting with sporadic FE, based on a negative family history of epilepsy within the third degree of kinship. Parents of 37 patients also consented to participate in the study. Definition of the epilepsy type depended on anatomo-electroclinical data collected during clinical assessment and was reached by agreement among three clinical epileptologists.
A comprehensive clinical record was compiled, including epilepsy phenotype, age at seizure onset, seizure frequency at last assessment, antiepileptic treatment and therapeutic outcome, interictal epileptiform abnormalities, neuroimaging data and neuropsychiatric comorbidities. Targeted 1.5 T or 3 T brain MRI was acquired in all cases with refractory epilepsy. Most patients (n = 96/106) were referred to the Epilepsy and Sleep Centre of the Bellaria Hospital in Bologna (Italy), while the remaining ones (n = 10/106) were recruited through other Italian centres, in the context of the LICE (Italian League Against Epilepsy) collaborative network.
DNA was extracted from peripheral blood samples with the QIAamp DNA Blood Mini Kit (Qiagen), following the manufacturer’s protocol.
Australian patients
Following the guidelines of the Human Research Ethics Committee of Austin Health, Melbourne, Australia, informed consent was obtained from living subjects or their relatives. Patients with temporal or frontal lobe epilepsy without a known acquired cause were recruited from the first seizure and epilepsy clinics at Austin Health, private practices of the investigators and by referral for genetics research over a period of 25 years, regardless of reported family history of epilepsy. Clinical data and a detailed family history were obtained using a validated seizure questionnaire, personal evaluation and review of medical records, including EEG and neuroimaging investigations.22 MRI (generally 1.5 T) was performed when appropriate. Cases that did not receive MRI had normal CT scans and no history of an acquired insult. All available family members were included in the segregation analysis for variants of interest. For most samples, whole venous blood was obtained and genomic DNA extracted using a Qiagen QIAamp DNA Maxi Kit according to the manufacturer’s instructions. In some cases, only saliva samples were available, and DNA was extracted from these specimens using a prepIT•L2P kit (DNA Genotek Inc) as per the manufacturer’s instructions.
German patients
The sample recruitment site for the German patients was the Department of Epileptology at the University of Bonn. The collection of DNA samples from patients with epilepsy was conducted from 2007 to 2015 within the projects Epicure (Functional Genomics in Neurobiology of Epilepsy: A Basis for New Therapeutic Strategies) and NGEN-Plus (Genetic basis of Levetiracetam pharmacoresistance and side effects in human epilepsy) and has been approved by the Ethics Committee of University Bonn Medical Centre (approval code: 040/07). The genomic DNA was isolated from 10 ml aliquots of EDTA-anticoagulated blood by a salting-out technique and then used for exome sequencing.
Trio-whole-exome sequencing analysis
The 37 Italian patient-parents trios and German patients underwent whole-exome sequencing (WES). Exome library capture and enrichment was performed with BGI kit (BGI Genomics, n = 104) or the NimbleGen SeqCap EZ (Roche, n = 7) and sequenced as paired-end reads with an Illumina system. As previously described,23 sequencing reads were processed to obtain annotated variant calls and retain for downstream analyses only variants located in coding exons or canonical splice-site junctions. Guidelines for sequence variant interpretation of the American College of Medical Genetics (ACMG)24 were followed for clinical classification of variants in established epilepsy genes by using the online platform Varsome (https://varsome.com/).
We calculated the mean rate of de novo variants (DNVs) in the main variant classes (missense, nonsense, splicing affecting, frame-shift, in-frame indels and synonymous single nucleotide variants) and used for comparison data from a large control cohort of 1911 trios previously published.25 Although we did not formally account for differences in variant discovery workflows,25 they have similarities in both enrichment methods and bioinformatic pipelines.
To select candidate FE genes, we used the following criteria: (i) the DNV has allele count ≤1 across control samples in the genome aggregation database (gnomAD) data set v2.1.1 including counts on 60146 chromosomes; (ii) the DNV has CADD Phred score (v1.4) ≥ 15; (iii) the gene is expressed in brain according to the Human Protein Atlas and Human Allen Brain Atlas databases; (iv) the encoded protein is important for brain development and function based on NCBI Entrez Gene Summary and UniprotKB databases as well as bibliographic research; and (v) the gene is intolerant to variation according to gnomAD v2.1.1 measures for genetic constraints (missense Z score > 3 and/or pLI > 0.9) or already associated with epileptic seizures in humans.
Single-molecule molecular inversion probe analysis
Targeted sequencing of the candidate genes was performed by a custom single-molecule molecular inversion probes (smMIPs) assay, in 69 patients, 225 Australian patients and in 431 Italian control individuals with no apparent manifestation of epilepsy (males/females 221/210), whose DNA samples were collected as controls as previously described.23
MIPs were designed with MIPgen26 to cover the coding exons and the exon-flanking 5-nucleotide sequences of genes, and were molecularly collapsed on their 5-nucleotides single-molecule unique tags synthesized by an external manufacturer (Integrated DNA Technologies). Target capture and library preparation was performed according to a published protocol27 and variant calling and annotation as previously described.23
Following the observation that disease-causing variants in common epilepsies are characterized by allelic counts ≤1 in large-scale data sets of genomic variants representative of the general population,28 we defined ultra-rare variants (URVs) using criteria (i) and (ii) described above for the selection of candidate FE genes bearing deleterious protein-altering URVs.
We used the Depth of Coverage utility in the Genome Analysis Toolkit (GATK v3.8)29 to calculate the per-sample percentage of targeted bases covered by >5 unique reads (Depth of Coverage >5X) on each gene (coding exons and flanking canonical splice sites). Candidate variants were validated by Sanger sequencing.
Variant clustering analysis
We used CLUMP30 to assess whether the distribution of PIK3C2B singleton missense URVs observed in FE patients showed preferential clustering to key regions of the protein. To this end, we focused on PIK3C2B singleton missense URVs (smURVs) identified among the cases from this study and in the 3597 NAFE (non-acquired focal epilepsy) cases from Epi25 (https://epi25.broadinstitute.org/, accessed August 2020), and we compared their distribution with that of smURVs in Epi25 controls and in controls used in this study. As the gnomAD whole data set includes not only controls but also samples from neurological and oncological cohorts, in this analysis the variants were required to have allele count ≤1 in gnomAD controls to facilitate comparison between FE cases and controls. As samples may have been analysed by smMIPs and subsequently included in the Epi25 study, the variants that were identified in the same samples by both approaches were counted only once.
CLUMP carries out an unsupervised clustering analysis of amino-acid residues hit by variants, with no prior assumption on annotated protein domains.
We also examined the proximity of smURVs relative to the known domains (‘domain-proximity’ analysis). To do this, we retrieved the protein sequence positions of the amino-acid residues representing the N-terminal (PN) and C-terminal (PC) boundaries of the domains annotated in the Uniprot database record for PI3K-C2β (O00750). We calculated the distance (D) between the position of amino-acid residue hit by each variant (PV) and each domain separately in the following way: (i) D = 0 if PN ≤ PV ≤ PC; (ii) D = PV–PC if PV > PC; and (iii) D = PN–PV if PV < PN.
PI3K-C2β kinase assay
HEK293T cells growing in 15-cm dishes were transfected 12 h prior to analysis with DNA mixtures containing 12 µg of empty-GFP, WT GFP-PI3K-C2β, or GFP-PI3K-C2β variants plasmids, using the calcium phosphate method. Cells were harvested and homogenized in lysis buffer (20 mM Tris–HCl, pH 8.0, 138 mM NaCl, 3 mM EDTA, 2.7 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 1% Nonidet P-40 (NP40), 5% glycerol) supplemented with protease inhibitor cocktail (Roche, #05892970001), 50 mM sodium fluoride, 10 mM sodium pyrophosphate and 1 mM sodium orthovanadate, as previously described.31 Lysates were cleared by centrifugation at 13 000 rpm at 4°C for 10 min. Protein concentration was determined by Bradford method. 4.5 mg of protein was incubated with 2 µg of anti-GFP antibody (Abcam, #ab290) on a rotating rack for 2 h at 4°C with Protein-G sepharose beads (GE Healthcare, #GE17-0618-01). Samples were collected by centrifugation and washed twice with lysis buffer, twice with washing buffer (0.1 M Tris–HCl pH7.4, 0.5 M LiCl) and twice with kinase buffer 1× (40 mM Tris–HCl pH 7.5, 20 mM MgCl2, 0.1 mg/ml BSA). Part of the immunoprecipitate was immunoblotted with GFP antibody as described below. The remaining part of the immunoprecipitate was resuspended in kinase buffer 2× and mixed 1:1 with substrate solution. Substrate solution was obtained drying 300 µl of 1 mg/ml phosphatidylserine and either 300 µl of 1 mg/ml phosphatidylinositol-4-phosphate (Echelon, #P-4016) or 300 µl of 1 mg/ml phosphatidylinositol (Echelon, #P-0016) under a stream of nitrogen gas, resuspending lipids in 300 µl of resuspension buffer (HEPES 10 mM pH 7.5, EDTA 1 mM) and sonicating the solution. The immunoprecipitate–lipid mixture was then incubated for 1.5 h at room temperature with 200 µM ATP. PI3K-C2β activity was measured with the ADP-Glo™ Kinase Assay kit (Promega, #V6930), according to the manufacturer’s instructions. One-way ANOVA was used for statistical testing.
Pik3c2b mouse model
All animal experiments were authorized by the Italian Ministry of Health (authorization number 548/2020-PR) or the Landesamt für Gesundheit und Soziales (LAGeSo, Berlin; authorization numbers T0243/08, S0313/17, G0162/18) and carried out according to the European Community guiding principles in the care and use of animals. Mice (C57BL/6 strain) were selected for experimental groups based solely on genotype. DNA was isolated from newborns and analysed by PCR and sequencing. Mice of each genotype were matched as closely as possible for age within constraints of availability.
Preparation of mouse brain lysates
Adult mouse brains were homogenized in 5 ml per gram of brain homogenization buffer. The brain and buffer were placed in a tube and homogenized 20 times with a EUROSTAR power basic stirrer (IKA) at 900 rpm. The samples were placed on ice for 10 min and then centrifuged at 17 000g in a Micro Star 17R microcentrifuge (VWR) at 4°C for 10 min. The supernatant was transferred to a new Eppendorf tube and centrifugation repeated to remove any remnant debris. The supernatant was again transferred to a new Eppendorf tube, a sample taken for protein quantification, then sodium dodecyl sulphate (SDS) sample buffer was added and the samples were stored at −20°C for analysis by SDS-PAGE and immunoblotting.
HEK293T cell culture
HEK293T cells were cultured in DMEM medium (Gibco®, #61965026) supplemented with 10% fetal bovine serum (Gibco®, 10270-106) and kept in culture for less than 20 passages. All media were supplemented with 1% penicillin–streptomycin (10 000 U/ml). Cells were grown at 37°C with 5% CO2. All cell lines were tested for Mycoplasma contamination every 2 weeks by PCR. Twelve hours before harvesting, cells were transfected with PIK3C2B cDNA mutants using Lipofectamine 2000 (Thermo Fisher Scientific, #11668019) according to the manufacturer’s guidelines, and serum starved for 4 h prior homogenization, as previously described.18
Immunoblot analysis of HEK293T cell lysates
HEK293T cells were harvested and homogenized in lysis buffer (120 mM NaCl, 50 mM Tris–HCl pH = 8, 1% Triton X-100) supplemented with protease inhibitor cocktail (Roche, #05892970001), 50 mM sodium fluoride, 10 mM sodium pyrophosphate and 1 mM sodium orthovanadate. Lysates were cleared by centrifugation at 13 000 rpm at 4°C for 10 min. Protein concentration was determined by the Bradford method. SDS-PAGE and immunoblotting followed standard procedures. Enhanced chemiluminescence (ECL) detection reagents were purchased from Bio-Rad Laboratories. All other chemicals were purchased from Sigma Aldrich. Signal quantification was performed using Image Lab (Bio-Rad Laboratories). One-way ANOVA was used for statistical testing.
Generation of the Pik3c2b+/G1501fs*1 mouse model
We modelled the loss of the terminal C2 domain of PI3K-C2β using the CRISPR/Cas9 technology. In mice, the terminal C2 domain starts at position 1504 (Uniprot Database), the residue encoded by exon 31. We designed a single guide RNAs (sgRNAs) targeting this region (TCCCCACCGACCTTCCCAACCGG), in order to generate frameshift-derived stop codons. We electroporated zygotes with sgRNAs and Cas9 protein and transferred them to the oviducts of pseudopregnant females.32,33 DNA was isolated from newborns and analysed by PCR and sequencing. We obtained a founder (Pik3c2b+/G1501fs*1) bearing a stop codon at position 1502, resulting from a frameshift involving a single amino acid. The selected mice were backcrossed with WT C57BL/6 mice for more than five generations.
Neuronal cell culture
Neuronal cultures were prepared by surgically removing the hippocampi from littermate animals (aged P1–4). Neurons were dissociated by trypsin digestion. Hippocampal neurons (100 000) were plated as 40 μl drops on poly-lysine-coated, acid washed coverslips. Two millilitres of plating medium [basic medium (MEM; 0.5% glucose; 0.02% NaHCO3; 0.01% transferrin) containing 10% FBS, 2 mM l-glutamine, insulin and penicillin/streptomycin] was added 1 h after plating. After 1 day in vitro (DIV 1) 1 ml of plating medium was replaced by 1 ml of growth medium (basic medium containing 5% FBS; 0.5 mM l-glutamine; 2% B27 supplement; penicillin/streptomycin). AraC (2 μM) was added to the culture medium at DIV 2 to limit glial proliferation.
Confocal imaging of immunostained hippocampal neurons
Cultured hippocampal neurons were fixed at DIV 14 using 4% (w/v) paraformaldehyde (PFA) and 4% sucrose in phosphate-buffered saline (PBS) for 15 min at room temperature. Fixed neurons were blocked and permeabilized with PBS containing 10% (v/v) normal goat serum and 0.3% Triton X-100 for 1 h, followed by incubation with primary antibodies in 0.3% Triton X-100 in PBS for 1 h. Corresponding secondary antibodies (Alexa Fluor 488, 568 and 647, Life technologies) were then incubated for 30 min in PBS–0.3% Triton X-100. Coverslips were mounted with DAPI-containing Immumount (Thermo-Fisher). Imaging was performed with a Zeiss Axiovert 200 M equipped with the Perkin-Elmer Ultra View ERS system and a Hamamatsu C9100 EM-CCD camera under the control of Volocity software (Perkin-Elmer). Fluorescent intensities were semi-automatically quantified using a custom Fiji macro.34 Manual regions of interest of the soma were defined based on the neuronal marker and mean intensities within these regions of interest were extracted based on the p-S6 channel (macro is available at doi:10.5281/zenodo.5652147). Camera background was subtracted by the intensity values of a blank image, and small artefacts/uneven background were controlled for using the rolling ball method with a 200-pixel radius. Segmentation was conducted with the MAP2 channel using a median filter of 2-pixel radius followed by the Huang2 automatic threshold.
Mice transcardial perfusion and brain dissection
Mice were weighed and terminally anaesthetized by intraperitoneal injection of 10 µl/g of 1.2% (v/v) ketamine (Medistar) and 0.16% (v/v) xylazine (Medistar) dissolved in PBS. Animals were perfused first with PBS pH 7.4 at a rate of 5 ml/min and then with 4% (w/v) PFA in PBS 4 for 20 min. The skull was severed at the neck, opened up the midline to separate the skull plates and the nasal bone severed between the eyes to enable brain recovery. The left and right hemispheres were separated and placed in PFA on a roller overnight at 4°C for post-fixation. Following post-fixation, PFA was exchanged for 15% sucrose (w/v) in PBS and samples were placed back on a roller for 6–12 h until tissue sank in the tube then the solution was exchanged for 30% sucrose (w/v) in PBS overnight. Samples were drained of excess liquid and stored at −80°C.
Cryosectioning of mouse brains
Isolated frozen brain hemispheres were put in Optimal Cutting Temperature compound (O.C.T.) and cut in sagittal sections at a Microm HM430 Sliding Microtome (Thermo Scientific). Stereological representative sampling principles were used while acquiring sections. Series of 40-μm sections were acquired.
Immunofluorescence of mouse brain sections
After thawing, one series of sections was transferred to a 12-well plate (one well per mouse), washed three times with 0.125 M phosphate buffer (PB) for 10 min each and then blocked and permeabilized in PB blocking buffer for 2–3 h at room temperature. Primary antibodies were dissolved in PB blocking buffer, 500 μl added to each well and incubated at 4°C for 2 days. Sections were washed three times with PB blocking buffer, 30 min each at room temperature. Fluorescently conjugated secondary antibodies were diluted in PB blocking buffer, 500 μl added to each well and incubated at 4°C overnight. Sections were washed three times with PB blocking buffer, 30 min each at room temperature and then once with 0.125 M PB for 30 min. Sections were then mounted in order in the same orientation on gelatinized slides, excess PB removed and 20 μl VECTASHIELD Antifade Mounting Medium (Vector laboratories) added to each section. Coverslips were placed on top, sealed with nail varnish and stored at 4°C.
To quantify p62 levels a z-series of 9–11 images (1024 × 1024 pixels/image with zoom factor 1.3, each averaged twice) chosen to cover the entire region of interest from top to bottom was acquired using a Zeiss LSM780 confocal microscope equipped with a 20× objective. The resulting z-stack was ‘flattened’ into a single image using maximum projection. All acquisition settings were set equally for sections of all groups within each immunostaining. Average p62 intensities were determined in rectangular areas of 100 × 50 μm in the stratum pyramidale or stratum radiatum of CA1. Images were quantified using Fiji/ImageJ software (NIH).
Stereological analysis of cryosectioned mouse brains
A Zeiss LSM 780 with xyz coordinate tracking was used for imaging DAPI-stained brain sections co-labelled for pS6. The border of the subiculum and the CA1 of a slice was placed at the border of the frame of the 40× oil objective (1024 × 1024 pixel image, 354.25 × 354.25 µm, pinhole set for 1 µm z-acquisition). A dice was rolled, with the result defining the start position of image acquisition (1–2 = 0 µm, 3–4 = 100 µm 5–6 = 200 µm away from subiculum), introducing random sampling. Two images were acquired with a z-separation of 10 µm. The field of view was repositioned 400 µm rostrally and a further z-stack taken. This was repeated until the border of CA1–CA2 was reached whereby no further images were acquired. The process was repeated across all sections to achieve representative sampling of the entire structure. Image analysis was conducted using Fiji. Three 40-µm square regions of interest were placed at equal intervals along the desired structure using only the DAPI channel for localization. The counting of nuclei was conducted across both z-planes; any nuclei present in both images were counted only once, which provides an estimate of overall cell density. Then within the same region of interest, the number of cells with the desired marker (i.e. pS6) were also counted. To only count cells with high expression of pS6 the brightness threshold of the image was adjusted to a level which would only show the top 10% intense pixels in the control group and the settings were then applied to all images. Each count was recorded, and a mean was generated from the six counts per image, leading to an estimate of the number of cells within a 40 × 40 × 10 µm cube that were converted into cells/mm3.
Electrophysiology
Slice preparation and instrumentation
Mice [2–3-month-old WT, knockout (KO) and heterozygous (HET)] were used for acute hippocampal slice electrophysiology. Mice were sacrificed by cervical dislocation followed by decapitation, and the brains quickly extracted into ice cold dissection artificial CSF containing: 2.5 mM KCl, 1.25 mM NaH2PO4, 24 mM NaHCO3, 1.5 mM MgSO4, 2 mM CaCl2, 25 mM glucose, 250 mM sucrose. Transversal slices 350-µm thick were prepared using Vibroslicer (Leica, VT 1200S) from both hemispheres in oxygenated (95% O2/5% CO2) dissection artificial CSF at 2–4°C. Three to four slices containing hippocampus were collected from each hemisphere in a resting chamber (Harvard apparatus, BSC-PC) filled with resting/recording artificial CSF supplemented with 120 mM NaCl instead of 250 mM sucrose. Resting solution was continuously oxygenated and kept at room temperature (22–24°C) letting slices recover for at least 1.5 h before recordings (pH 7.35–7.40). After recovery, slices were transferred to a submerged recording chamber (Warner instruments RC-27L), filled with oxygenated artificial CSF at room temperature with solution exchange of 3–5 ml per min. Slices were fixed to prevent floating and an upright microscope (Olympus, BX61WI) was used for positioning the stimulating and recording electrodes. Slices treated with rapamycin were incubated separately for 2 h with 1 µM rapamycin dissolved in DMSO. Rapamycin treatment was applied only during the incubation period and not during recordings. Control treatment contained just DMSO in artificial CSF (1:1000) at the same concentration as used to dissolve rapamycin. Incubation of slices with this low concentration of DMSO did not affect any electrophysiological parameters; thus, we merged data of control recordings with and without DMSO. A glass stimulating electrode (Hilgenberg) filled with artificial CSF (1–1.5 MΩ) was positioned to stimulate the Schaffer collaterals of the CA1 stratum radiatum. A similar glass electrode (1.5–2.5 MΩ) placed in the distal part of the CA1 region was used for field excitatory postsynaptic potential (fEPSP) and population spikes (PS) recordings. Electrodes were prepared using the micropipette puller Sutter P-1000 (Sutter Instruments). Data were recorded at a sampling rate of 10 kHz, low-pass filtered at 3 kHz and analysed using PatchMaster software (Heka Elektronics). No gender-specific tendency for any electrophysiological parameters was noticed. We therefore combined data from both genders for analyses. Data were collected and analysed blind to genotype of animals.
Field excitatory postsynaptic potential recordings
For fEPSP recordings, stimulating and recording electrodes were placed in a visually preselected area of stratum radiatum and slowly advanced until maximum responses were obtained (Supplementary Fig. 4A). Basal stimulation of 0.2 ms electrical stimuli were delivered at 0.05 Hz at the stimulation intensity, which induced approximately 30–50% of the maximum responses. After 10 min of stable baseline recordings input/output stimulus response curves were made as a measure of basal excitatory synaptic transmission. Slopes of the fEPSPs were plotted against fibre volley amplitudes as a function of increasing stimulation intensity. Stimulation intensity was increased by 20 µA steps until the maximal amplitude of fEPSP was obtained, defined as a response with superimposed population spike component on decaying fEPSP responses (Supplementary Fig. 4A). Paired pulse facilitation (PPF) was measured as a parameter for presynaptic short-term plasticity, by delivering two pulses at time intervals between 10 and 500 ms, at a stimulation intensity which induced one-third of the maximal responses. PPF was calculated as a percentage increase of the slope of the second response compared to the first. For short intervals (10 and 20 ms), the first responses were digitally subtracted before measurements of the second. Each value measured is an average of three consecutive stimulations repeated every 20 s for stimulus response and every 30 s for PPF measurements. To test long-term synaptic plasticity, we induced long-term potentiation by 5-theta burst stimulation which consisted of 8 burst stimuli delivered at 5 Hz, each burst containing four pulses at 100 Hz. For long-term potentiation experiments, the stimulation intensity was selected to elicit 50% of its maximum amplitude, and basal stimulation was monitored at 0.05 Hz for 10 min before and 1 h after long-term potentiation induction. The mean slope of the fEPSP recorded 0–10 min before theta burst stimulation application is taken as 100% and long-term potentiation is measured 50–60 min after its induction. Each value is an average of three consecutive time points recorded every 20 s.
Population spike recordings
We recorded output action potentials population spikes from pyramidal cells as a function of the excitability of neuronal networks. For these sets of experiments the recording electrode was placed ∼300 μm away from the stimulating electrode in the CA1 stratum pyramidale (Supplementary Fig. 4A). After stable baseline recordings, we recorded input output stimulus response curves with stimulation intensities from 0 to 300 µA, with 30 µA step increase. The ratios of population spikes over fibre volley amplitudes were plotted as a function of increasing stimulation intensity to measure neuronal excitability of hippocampal networks. To probe GABAergic feedback inhibition, we delivered two pulses of stimulation at the supramaximal intensity of 200 µA at different time intervals (10–500 ms). Paired pulse modulation (PPM) of the amplitudes of the second compared to the first population spike were calculated and expressed in %. Each value measured is an average of three consecutive stimulations repeated every 20 s for stimulus response and every 30 s for paired pulse modulation measurements. To probe if slices in vitro are prone to epileptic seizure-like activity, we induced activity-dependent disinhibition of neuronal networks by delivering 60 pulses at 1 Hz with the supramaximal intensity of 200 µA. Facilitation of the population spike and appearance of polyspikes, induced by activity-dependent disinhibition, were calculated as the sum area changes of population spikes and polyspikes using a custom-written algorithm implemented in IGOR Pro software. SigmaPlot (Systat Software, Inc.) and IGOR Pro (WaveMetrics, Inc.) were used for electrophysiological data analyses and presentations. Values are depicted as mean ± SEM. N indicates the number of animals tested, n refers to the number of slices analysed.
Patch-clamp recordings of inhibitory postsynaptic currents
Whole-cell voltage clamp experiments were performed in CA1 pyramidal neurons of 4–6 week-old WT and Pik3c2b KO mice. Mice were decapitated after short sedation with isoflurane and slices prepared as described above. CA1 pyramidal neurons were visually identified using a 40× objective. Differential interference contrast was used to visually control cell attachment of the patch electrode. Following establishment of the whole-cell configuration neurons were clamped at −60 mV. Cells with a series resistance >20 MΩ (compensated to 70%) and leak currents >200 pA were discarded from further analyses. Recordings were performed at room temperature (22–24°C) in the same ASCF as described for field recordings (pH 7.35–7.4; 310–320 mOsm). Inhibitory postsynaptic currents (IPSCs) were evoked using a glass stimulation electrode filled with artificial CSF (1–1.5 MΩ) placed at 50–60 μm distance to the recorded pyramidal cell. The patch electrodes had a resistance of 3–5 MΩ when filled with an internal solution containing (in mM) 140 CsCl, 10 HEPES, 10 EGTA, 2 Mg-ATP and 5 mM QX-314 (pH 7.35; 295–305 mOsm). IPSCs were pharmacologically isolated using 20 μM CNQX (AMPA/kainate receptor antagonist) and 50 μM APV (NMDA receptor antagonist) for at least 10 min before the recordings. After obtaining a stable whole-cell configuration (to allow intrapipette solution to diffuse into the cell) for 10 min we first recorded spontaneous IPSCs (sIPSCs) for 3 min. The frequency of sIPSCs was analysed using the Mini Analysis Program (Synaptosoft Inc.). Synaptic events above a threshold of 10 pA were used to calculate spontaneous synaptic activity. To measure evoked inhibitory transmission, we generated stimulus response curves by plotting increasing stimulation intensities from 10 to 80 μA in steps of 10 μA and compared IPSC amplitudes. Each value measured is an average of three consecutive stimulations repeated every 20 s. At the end of some experiments, the GABAA receptor antagonist picrotoxin (100 μM) was applied for 20 min to verify IPSCs (see Supplementary Fig. 5A and B).
Seizure behaviour
Two- to three-month-old male35 littermates (WT, Pik3c2b HET and Pik3c2b KO) were put into separate cages and injected intraperitoneally with 1 mg/ml atropine sulphate (Sigma Aldrich, #A0257) dissolved in PBS.36 After 30 min, mice were injected intraperitoneally with 350 mg/kg pilocarpine hydrochloride (Sigma Aldrich, #P6503) and scored for seizures for 90 min. For everolimus rescue experiments, mice were injected intraperitoneally with 10 mg/ml everolimus (Cayman Chemical, #11597) dissolved in PBS 5% Tween-20 60 min before atropine injection. Mice behaviours were scored on a scale ranging from 1 to 7 (1 = immobility; 2 = tail extension; 3 = repetitive movements; 4 = forelimb clonus, rearing and falling; 5 = repetitive stage 4; 6 = epileptic seizure; 7 = death).37 Observers were blinded to the genotype of animals.
Statistical analyses
CLUMP analysis incorporates a Wilcoxon signed-rank test.
A two-tailed Student’s t-test was used to compare the mean rate of DNVs in the main variant classes (missense, nonsense, splicing affecting, synonymous and frameshift/non-frameshift indels) between our cohort and a large control cohort of 1911 trios.25 In domain-proximity analysis, we used a two-tailed Student’s t-test to evaluate the difference in the mean distance (D) between the position of the variant amino acid residue and each domain separately in cases and controls. P-values were adjusted for multiple comparisons over the 6 PI3K-C2β Uniprot domains with Bonferroni correction. Biochemical data were analysed using the GraphPad Prism6 software: significance was calculated with Student’s t-test and one- or two-way ANOVA followed by Bonferroni’s post hoc analysis, or Mantel Cox log-rank test where appropriate. Data are reported as mean ± SEM. The number of independent experiments analysed is represented by ‘n’. Significance is denoted using asterisks *P < 0.05, **P < 0.01, ***P < 0.001 and P > 0.05 or not significant (ns). Confocal microscopy data from hippocampal slices and cultured neurons were analysed using GraphPad Prism6. Student’s t-test was used for experiments with two genotypes and one-way ANOVA followed by Bonferroni’s post hoc analysis for experiments with more than one variable. Data are reported as mean ± SEM. The number of independent experiments analysed is represented by ‘n’. Significance levels were *P < 0.05, **P < 0.01, ***P < 0.001 and P > 0.05 is not significant (ns). Electrophysiological data curves were statistically evaluated using two-way ANOVA with repeated measures (RM, significance depicted over a line encompassing the curve). For comparisons of two groups statistical significance was tested with a two-tailed unpaired Student’s t-test. When several experimental groups were compared, statistical significance was tested with a one-way ANOVA with RM. Statistics were calculated using SigmaPlot (Systat Software, Inc.) and are indicated as ***P < 0.001; **P < 0.01; *P < 0.05.
Data availability
Data supporting the findings of this study are available within the article and its Supplementary material. All supporting data in this study are available from the corresponding author on request.
Results
PIK3C2B ultra-rare variants in FE patients are loss-of-function or cluster in proximity to key protein domains
We assembled an initial cohort of 106 probands with sporadic FE through an Italian collaborative network. The mean age of patients at their last clinical assessment was 36.5 ± 13.6 years and their clinical features are summarized in Table 1. Reasoning that the identification of DNVs could facilitate the selection of candidate genes impacted by ultra-rare variants, we performed an unbiased trio WES analysis in 37 probands. According to the ACMG guidelines,24 we found five FE patients carrying pathogenic or likely pathogenic variants in established epilepsy genes, of which three were DNVs (Supplementary Table 1). We compared the mean number of DNVs detected in FE patients to that reported in a control cohort of 1911 trios.25 Although our trios and the previously published controls were analysed with slightly different strategies, this analysis suggested that FE patients had a higher mean number of protein-altering DNVs, with missense and loss-of-function single nucleotide variants holding significance on separate analyses (Supplementary Table 2). These findings suggested that protein-altering DNVs could point to genes contributing to FE in these trios, allowing us to make a shortlist of eight candidate genes based on protein-altering DNV ultra-rarity (allele count ≤ 1 in gnomAD v2.1.1), deleteriousness (CADD score v1.4 ≥ 15), documented gene expression and function in brain and intolerance of the gene to variation (Supplementary Table 3).
Table 1.
Clinical features of the initial cohort
| Total Patients n = 106 |
Valid % | Missing (%) | |
|---|---|---|---|
| Sex, male:female | 57:49 | ||
| Mean age at onset, years | 15.3 ± 10.8 | ||
| Brain MRI | |||
| Unremarkable | 86 | 81.1 | — |
| Abnormal | 20 | 18.9 | — |
| Hippocampal sclerosis | 3 | 2.8 | — |
| MCD | 14 | 13.3 | — |
| Other | 3 | 2.8 | — |
| Epilepsy type | |||
| Temporal | 57 | 53.8 | — |
| Extra-temporal | 49 | 46.2 | — |
| Focal to bilateral tonic–clonic seizures | 72 | 68.6 | 1 (1) |
| Epileptiform interictal EEG | 66 | 62.9 | 1 (1) |
| Drug resistance | 43 | 41.7 | 3 (2.8) |
| Personal history | |||
| Febrile seizures | 11 | 10.4 | — |
| ID/borderline IQ | 10 | 9.6 | 2 (1.9) |
| Psychiatric disorders | 17 | 16.2 | 1 (1) |
Valid: percentage of patients showing the feature. Missing: number and percentage of patients without information for the feature. MCD = malformations of cortical development [including focal cortical dysplasia (FCD); alterations suggestive of dysplasia of temporal pole and amygdala; transmantle columnar heterotopia] FCD was observed in 11 patients (10.4%) and was confirmed by histopathological analysis in six who underwent surgery, revealing FCD2b in five and FCD2a + encephalocele in one. Based on Engel Epilepsy Surgery Outcome scale, five were in Engel class I/Ia, one in Engel class IIb. ID = intellectual disability, defined as IQ < 70 or borderline IQ, between 70 and 80. Psychiatric disorders ranged from attention deficit hyperactivity disorder to anxiety/depression requiring pharmacologic treatment.
We then resequenced these genes by smMIPs in the remaining 69 cases and in controls ascertained as previously reported23 (Supplementary material and Supplementary Table 4). Consistent with the criteria used for protein-altering DNVs in the candidate gene shortlist, URVs were defined in this study as protein-altering variants with CADD Phred score v1.4 ≥ 15 and allele count in gnomAD v2.1.1 ≤ 1. Interestingly, two of the candidate genes, namely PDE2A and PIK3C2B, were reported with nominally significant P-values for the association with NAFE in the Epi25 collaborative, the largest WES study in epilepsy to date.38 While we did not find any additional URV in PDE2A, two heterozygous PIK3C2B URVs were identified in FE cases: a stop-gain p.Q1533* variant and a missense p.I1544 M variant in the C2 domain, which is known to autoregulate class II PI3K catalytic activity.20,39 As the initial PIK3C2B DNV was a heterozygous missense p.E1294Q variant occurring in the core PI3K catalytic domain, we hypothesized that URVs identified in FE patients could act via a loss-of-function mechanism resulting in decreased protein expression or in impaired lipid kinase activity. This hypothesis was confirmed by the finding of four additional URVs in cohorts of mostly sporadic FE from Australia (the frameshift variant p.E807fs45* and the missense variant p.I1098V within the catalytic domain) and Germany (the stop-gain variant p.Y1298* and the missense variant p.R1118L, also within the catalytic domain). In the PIK3C2B URV-positive patients, we could exclude the presence of a concomitant pathogenic/likely pathogenic variant in established FE genes or in FE-related genes along the mTOR pathway axis (details presented in Supplementary material), except in an Australian patient carrying the p.I1098V variant where a pathogenic variant p.R886* in PCDH19 (ClinVar variation ID 206341) was also identified (Supplementary Table 5) and was therefore not considered further in this study. The six remaining PIK3C2B variants (Table 2) were all heterozygous URVs including three protein-truncating variants and three missense changes affecting the catalytic or C2 domains (Fig. 1A). Conversely, URVs identified in controls were five missense variants situated mostly distant from domains involved in catalysis, with only one variant (p.E1168V) within the catalytic domain (Supplementary Table 4). None of the patients’ variants appeared in the whole data sets available in the gnomAD v2.1.1 and v3.1.1 browsers. In the Epi25 data set, the trend reported for this gene was towards an excess of loss-of-function variants in NAFE cases: 5/3597 samples (0.01%), including the p.Q1533* variant originally identified by smMIPs, versus 2/8364 controls (0.002%).
Table 2.
Molecular findings and clinical features of PIK3C2B URV-positive patients in this study
| Patient ID | Molecular findings | Clinical findings | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Transcript annotation (NM_002646) | Protein change | CADD v1.4 | AC | Sex | AAO, years | Drug resistance | Psychiatric comorbidities/ID | EEG | Brain MRI | |
| AU-1 | c.2424_2426delTGG;c.2419_insGAGGCCCCAAG | p.E807fs45* | — | 0 | M | 31 | No | Yes/NA | Normal | Normal |
| GE-1 | c.3353G>T | p.R1118L | 28.7 | 0 | F | 11 | No | No/NA | Normal | Normal |
| IT-10 | c.3880G>C | p.E1294Q | 21.5 | 0 | M | 5 | No | No/No | Interictal epileptiform abnormalities, L T ictal recording of HM seizure | Normal |
| GE-2 | c.3894T>G | p.Y1298* | 36 | 0 | M | 17 | No | Yes/NA | Ictal recording of seizure, R T (mesial onset) interictal theta abnormalities, bil F-T | Cavernoma, T mesialright side |
| IT-14 | c.4597C>T | p.Q1533* | 43 | 0 | F | 4 | Yes | No/No | Interictal epileptiform abnormalities, R F ictal recording of HM seizure | Normal |
| IT-15 | c.4632T>G | p.I1544M | 22.6 | 0 | F | 2 | Yes | Yes/No | Interictal epileptiform abnormalities, L T | Suggestive of HS, L T |
AAO = age at onset; protein change = in bold font, loss-of-function variants; CADD = CADD Phred score v1.4; AC = minor allele counts in gnomAD v2.1.1; bil = bilateral; F = frontal; F-T = fronto-temporal; HM = hypermotor; HS = hippocampal sclerosis; ID = intellectual disability/borderline IQ; L = left; R = right; T = temporal; NA = not applicable.
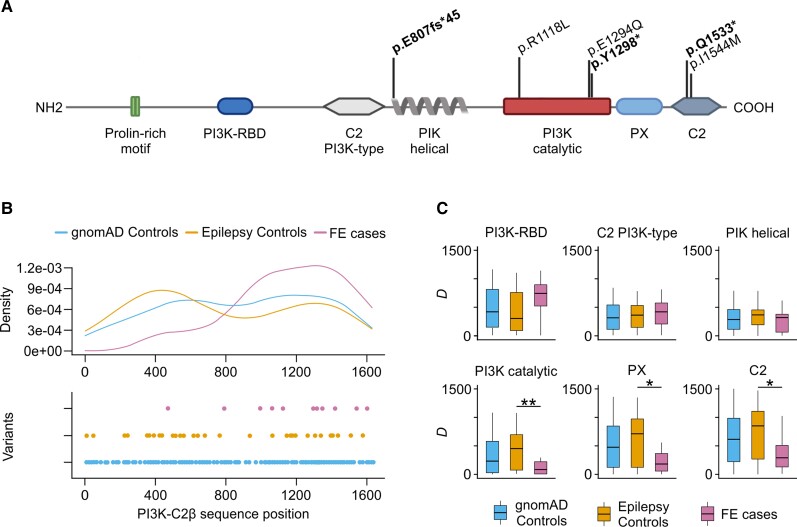
Figure 1.
URVs in PIK3C2B are loss-of-function. (A) Protein-altering, deleterious URVs in PIK3C2B identified in FE patients from this study, and their location with respect to protein length and protein domains according to the Uniprot 000750 record. ‘PI3K Catalytic’ refers to the ‘PI3K/PI4K’ domain intervals in the Uniprot reference. Loss-of-function URVs are shown in bold. (B) Density (top) and point (bottom) distribution of smURVs with respect to the protein sequence positions. Combined smURVs in FE cases from this study and the Epi25 NAFE group are compared to controls from this study and the Epi25 control group. Skewed distribution of FE-associated smURVs towards the C-terminal end of the protein results in a significant difference of CLUMP scores between cases and controls (P = 0.007). (C) Domain-proximity analysis comparing D, the mean distance of smURVs from protein domain boundaries, in cases and controls. The most significant difference in D between cases and controls is found for the PI3K catalytic domain (P = 0.006), followed by the PX (P = 0.017) and C2 (P = 0.024) C-terminal domains. For all analyses, singleton missense variants in gnomAD v2.1.1 controls (allele count ≤1) are also shown.
As clustering of singleton missense variants provides insights into the molecular mechanisms underlying epileptic disorders,40 we further explored whether PIK3C2B singleton missense URVs (smURVs) identified in FE patients were preferentially clustered in key regions of the protein. We compared the distribution of combined smURVs identified in FE cases from this study and the Epi25 NAFE group (12 smURVs) with that of the Epi25 control group and controls in this study (42 smURVs; Supplementary Table 6). The distribution of smURVs in NAFE cases was skewed towards the C-terminal end, while smURVs of controls were sparser and their density distribution closely resembled that of variants with allele count ≤1 in gnomAD v2.1.1 controls (Fig. 1B). Unsupervised clustering analysis with CLUMP detected a significant difference (2.93) between the average scores of controls (4.70) and cases (1.76), thus indicating a non-random localization of patients’ smURVs (Fig. 1B). To determine whether the clustering of smURVs observed in FE cases involved key functional domains, we carried out a domain-proximity analysis by calculating the mean distance of smURV amino-acid residues from each domain separately in cases and controls. Significant clustering was detected in cases within or near the catalytic domain as well as the C-terminal lipid-binding autoregulatory domains C2 and PX, although to a lesser extent (Fig. 1C and Supplementary Table 7), suggesting that URVs located within or near the main known functional domains were more likely to be involved in FE. Interestingly, one of the smURVs in Epi25 (p.N1311D) recurred in two genomically unrelated German NAFE cases: this smURV was the closest to the p.E1294Q DNV on the protein sequence, that was present only once in the whole gnomAD data set and was the only variant with population allele frequency <5% shared by these two subjects.
To assess familial segregation of other variants than the p.E1294Q DNV, we could access parental samples of two Epi25 NAFE cases carrying URVs of interest. The first was a familial case in which the proband inherited the missense p.G1345S variant from the affected father. A paternal uncle was also reported to have experienced seizures, but no further information was available. This URV was not present in the whole gnomAD data sets and was situated immediately downstream to the catalytic domain. Another Epi25 NAFE case inherited a loss-of-function URV (p.K584*) from the unaffected father, suggesting incomplete penetrance.
None of the NAFE cases with smURVs was found to carry a concomitant pathogenic/likely pathogenic variant in established FE genes or in FE-related genes along the mTOR pathway axis (details presented in Supplementary material).
URV-positive patients did not display specific clinicopathological features (Table 2) compared to the characteristics of the whole initial cohort (Table 1) except for an earlier mean age at epilepsy onset (11.28 ± 11.0 versus 15.5 ± 10.8) and a higher prevalence of psychiatric comorbidities (43% versus 16.2%).
These results support the proposition that heterozygous URVs in PIK3C2B underlie epilepsy in patients by preventing proper protein expression due to nonsense-mediated decay or, possibly, impairing the lipid kinase activity of the encoded PI3K-C2β enzyme.
Patients’ mutations impair PI3K-C2β lipid-synthesizing activity and lead to mTORC1 hyperactivation
We next asked whether protein-truncating and missense PI3K-C2β URVs result in a loss-of-function mechanism. We therefore used site-directed mutagenesis to generate three missense mutants (p.R1118L, p.E1294Q and p.I1544M) and one mutant inserting a premature stop codon (p.Q1533*) predicted to cause the truncation of the C2 domain, which is known to be a critical determinant for the kinase activity of class II enzymes.39 We also generated control cDNAs of p.I1212V and p.E1169K variants that have been observed in FE patients but present in the general population. These variants, located within the PI3K catalytic domain are listed in the gnomAD database and reported with allele frequencies 1.19e-4 and 1.00e-4 in gnomaAD v2.1.1 controls, respectively. Furthermore, we generated two mutant forms of PI3K-C2β that either eliminate or alter enzymatic activity: first, we took advantage of a kinase-dead protein lacking PI3K activity whose expression has been shown to result in elevated mTORC1 signalling.18 Second, we used information derived from sequence comparison of the substrate-binding loops of different classes of PI3Ks21,41 to engineer a substrate-shifted variant of PI3K-C2β, which selectively lacks the ability to generate PI(3,4)P2 but retains capability to synthesize PI(3)P (p.KRDR1228-1231KPLP, thereafter named KPLP; Supplementary Fig. 1A and B).
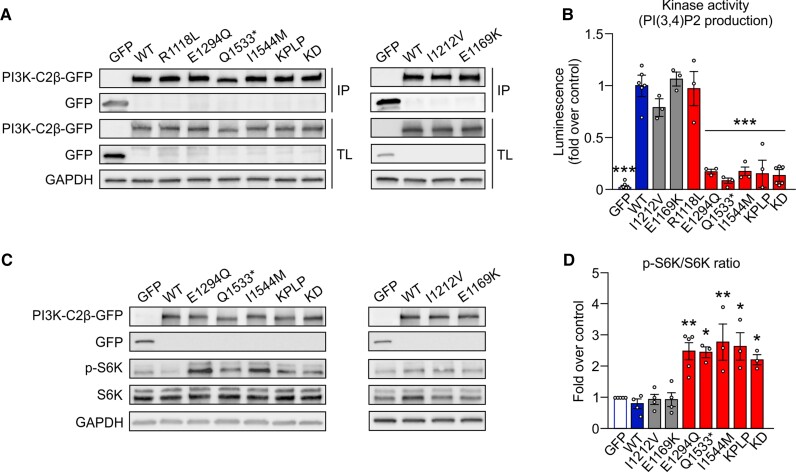
We next determined the effect of missense URVs on the kinase activity of PI3K-C2β. To this aim, we immunoprecipitated WT or mutant PI3K-C2β from transfected HEK293T cells31 (Fig. 2A and Supplementary Fig. 1A) and analysed their ability to synthesize PI(3,4)P2 from PI(4)P in vitro (Fig. 2B). The majority of patients’ variants (p.E1294Q, p.Q1533* and p.I1544M), but not the p.R1118L URV, were found to have little PI(3,4)P2-synthesizing activity, akin to the KPLP or kinase-dead PI3K-C2β mutants. The production of PI(3)P was also impaired, excluding a differential substrate preference upon mutation of these residues (Supplementary Fig. 1B). In contrast, both control variants selected from the general population displayed normal kinase activity (Fig. 2B, grey bars).
Figure 2.
Loss of PIK3-C2β activity causes mTORC1 hyperactivation in vitro. (A) Representative immunoblot of PI3K-C2β variants transfected in HEK293T cells immunoprecipitated with GFP antibody (IP), as used for the kinase assay, and total lysates (TL). (B) Quantification of the kinase activity on PI(4)P of PI3K-C2β URVs, gnomAD variants (grey bars) and controls compared to WT PI3K-C2β. One-way ANOVA, *P < 0.05, **P < 0.01. n > 3 independent experiments. Error bars represent SEM. (C) Representative immunoblot of GFP, PI3K-C2β-GFP, p-S6K, S6K and GAPDH content in HEK293T cells transfected with GFP, WT PI3K-C2β, PI3K-C2β URVs (left), or gnomAD variants (right). (D) Quantification of the p-S6K/total S6K of data shown in C. One-way ANOVA, *P < 0.05, **P < 0.01. n > 3 independent experiments. Error bars represent SEM.
We then assessed the effect of over-expression of either WT or mutant PI3K-C2β in HEK293T cells using mTORC1 signalling monitored by the phosphorylation status of ribosomal protein S6 kinase (S6K) as a readout. Overexpression of WT PI3K-C2β did not modify S6K phosphorylation, indicating that endogenous expression has already reached a saturating inhibitory activity on the pathway. Conversely, overexpression of loss-of-function p.E1294Q, p.Q1533*, or p.I1544M alleles, as well as kinase-dead or KPLP mutant forms of PI3K-C2β, significantly hyperactivated the mTORC1 pathway (Fig. 2C and D), indicating that the mutant alleles compete with the WT protein. In contrast, expression of any of the control variants or the p.R1118L URV had no significant effect on the phosphorylation status of S6K (Fig. 2C and D and Supplementary Fig. 1C and D). Within human cells and tissues, mTOR kinase is part of mTORC1 and mTORC2, two distinct protein complexes with different signalling outputs. We found that in contrast to its effect on mTORC1 activity, expression of the FE-associated PI3K-C2β variants did not alter mTORC2 signalling, monitored by phosphorylation of AKT1/2 (Supplementary Fig. 1E).
These data demonstrate that the majority of PIK3C2B URVS identified in FE patients are mutations that act through a common loss-of-function mechanism impairing PI(3,4)P2 synthesis, and that overexpression of these mutants leads to mTORC1 hyperactivity. They are further consistent with previous data suggesting that PI3K-C2β can act as a negative regulator of mTORC1 activity18 rather than a driver of EGF16 or Akt signalling,17 at least in brain.
Pik3c2b KO and HET mice display mTORC1 hyperactivity
As patients’ URVs in PIK3C2B resulted in loss-of-function, mice lacking PI3K-C2β could serve as an experimental model to study the mechanisms underlying PIK3C2B-related FE. We therefore took advantage of a mouse model with complete genetic inactivation of Pik3c2b (KO)42 to evaluate the effects of defective PI3K-C2β function in brain.
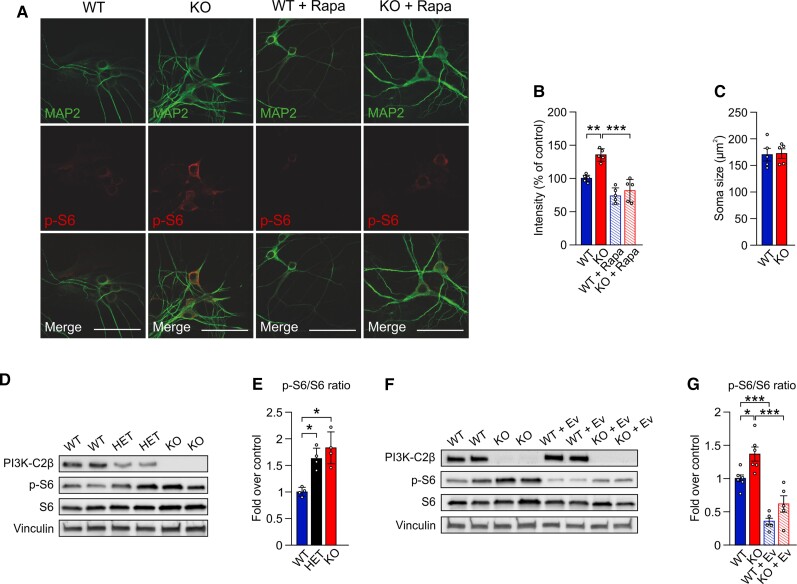
Pik3c2b KO mice were born at normal Mendelian ratios and did not display any overt phenotypic abnormalities.42 Moreover, no gross alterations in hippocampal lamination (not shown) or in the levels of key synaptic proteins (e.g. postsynaptic AMPA-type glutamate receptor or the synaptic vesicle protein synaptotagmin 1) and of mTOR kinase were detected (Supplementary Fig. 2A and B). Differently from loss of either Tsc243 or the GATOR1 component Depdc5,44Pik3c2b KO mice did not show signs of neuroinflammation such as astrogliosis (Supplementary Fig. 2C and D). We next assessed mTORC1 signalling by analysing the levels of phosphorylated S6 (p-S6), a downstream target of the mTORC1 substrate S6K, in primary hippocampal neurons. Confocal imaging combined with semi-automated image analysis revealed elevated levels of p-S6 in MAP2-positive neurons from Pik3c2b KO mice, while pre-fixation treatment with the mTORC1 inhibitor rapamycin greatly reduced p-S6 staining in neurons from either WT or KO animals to a similar level (Fig. 3A and B). However, elevated mTORC1 activity in hippocampal neurons from Pik3c2b KO mice was not accompanied by alterations in average soma size (Fig. 3C).
Figure 3.
Loss of PI3K-C2β causes mTORC1 hyperactivity in cultured neurons and in mouse brain. (A) Immunohistochemical detection of p-S6-positive cells in hippocampal neurons (DIV 14) from WT and Pik3c2b KO mice left untreated or treated with the mTORC1 inhibitor rapamycin (200 nM). Scale bar = 50 μm. Rapamycin treatment essentially eliminates p-S6 staining. (B) Semi-automated intensity quantification of representative data shown in A. One-way ANOVA, **P < 0.005, ***P < 0.001, n = 5 independent cultures. Error bars represent SEM. (C) Quantification of neuronal soma size. Unpaired t-test; n = 5 independent cultures. Soma area is unaffected (P = 0.5813). (D and E) Dose-dependent loss of PI3K-C2β activates mTORC1 signalling. (D) Representative immunoblot of PI3K-C2β, p-S6, S6 and vinculin content in whole brain lysates from 8-week-old WT, HET and KO mice. (E) Quantification of the p-S6/S6 ratio as shown in D. One-way ANOVA, *P < 0.05. n = 4 independent experiments. Error bars represent SEM. (F and G) mTOR hyperactivation in mouse brain is rescued by acute everolimus treatment. (F) Representative immunoblot of PI3K-C2β, p-S6, S6 and vinculin content in whole brain lysates from 8-week-old WT and KO mice, untreated or pretreated with 10 mg/kg of everolimus. (G) Quantification of the p-S6/S6 ratio as shown in F. Everolimus treatment potently downmodulates mTORC1 signalling in both WT and KO mice. One-way ANOVA, *P < 0.05, ***P < 0.001. n = 6 independent experiments. Error bars represent SEM.
To corroborate these data, we monitored hippocampal mTORC1 signalling in brain sections from perfused WT or Pik3c2b KO mice using stereological principles. Loss of PI3K-C2β expression was accompanied by a significant elevation in the fraction of cells that display robust p-S6 signals within the hippocampal pyramidal cell layer (Supplementary Fig. 2E and F), while total cell numbers (Supplementary Fig. 2G) were unaltered. Elevated mTORC1 signalling was further confirmed by immunoblot analysis of whole brain lysates from 8-week-old WT and KO mice (Fig. 3D and E). Interestingly, mTORC1 signalling was also upregulated in Pik3c2b HET mice, suggesting a haploinsufficiency effect of PI3K-C2β in the repression of mTORC1 activity (Fig. 3D and E). Importantly, a single treatment with 10 mg/kg of the rapamycin derivative everolimus 90 min before mice were sacrificed was sufficient to normalize mTORC1 activity in KO adult brains (Fig. 3F and G).
As activated mTORC1 may repress autophagy, we monitored the steady-state levels of p62 and LC3, two common markers of autophagy. Immunofluorescence analysis of adult mouse brains did not reveal any significant differences in the levels of p62 in either stratum radiatum or stratum pyramidale between WT and Pik3c2b KO mice (Supplementary Fig. 3A and B). This result was confirmed by immunoblot analysis of total brain lysates of 8-week-old WT and KO mice (Supplementary Fig. 3C and D). Moreover, we did not detect variation in the intensities of either the non-lipidated or lipidated forms of LC3 (respectively, LC3-I and LC3-II, Supplementary Fig. 3C and D). Hence, brain autophagy does not appear to be affected by Pik3c2b loss.
Collectively, our data demonstrate that decreased PI3K-C2β expression in brain results in mTORC1 hyperactivation. This phenotype supports haploinsufficiency as a molecular mechanism underlying the pathogenic effect of heterozygous mutations in patients.
Neuronal activity and network excitability are increased and dose-dependent in Pik3c2b KO and HET mouse brains
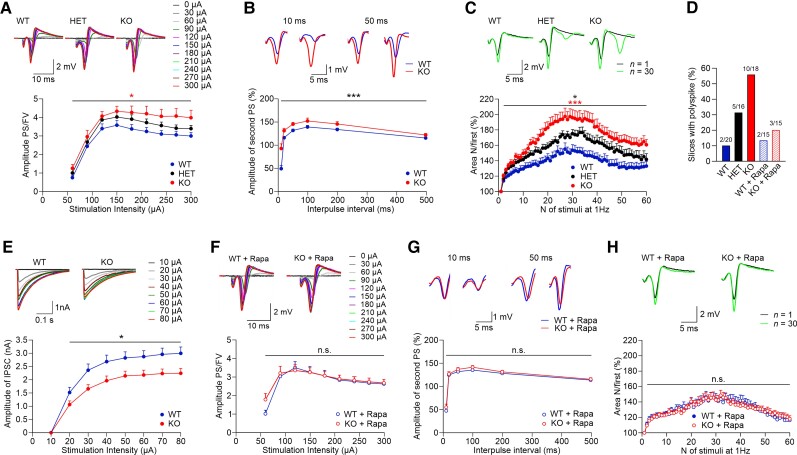
Because the degree of mTORC1 hyperactivation was dependent on Pik3c2b dosage, we tested both KO and HET mice to profile the electrophysiological phenotypes that may arise as a consequence of defective PI3K-C2β function in the brain. Previous work has shown that mTORC1 hyperactivity can result in increased neuronal network excitability.37,45 We therefore asked whether abolished or halved PI3K-C2β expression affects synaptic transmission and excitability in our Pik3c2b mouse model. To this aim, we recorded fEPSPs of CA3–CA1 synapses in acute hippocampal slices (Supplementary Fig. 4A). Analyses of stimulus response curves revealed elevated basal synaptic transmission in Pik3c2b KO mice as slopes of fEPSPs over fibre volley amplitudes were significantly increased in slices from Pik3c2b KO mice (Supplementary Fig. 4B). The fact that presynaptic PPF was unaltered in Pik3c2b KO mice (Supplementary Fig. 4C) further suggests that the observed increase in fEPSPs slopes is unlikely to be due to an increased presynaptic release probability. Instead, it may result from altered neuronal network excitability, a hypothesis further corroborated below. Hippocampal long-term potentiation in the CA1 area was significantly reduced in slices from Pik3c2b KO animals (Supplementary Fig. 4D and E), consistent with findings from other models of mTORC1 hyperactivity characterized by increased neuronal network excitability.46,47 These data suggest that loss of PI3K-C2β expression leads to increased excitatory neurotransmission and reduced plasticity.
As elevated excitatory fEPSP slopes may result from increased neuronal excitability, a hallmark of the epileptic phenotype, we recorded population spikes in CA1 stratum pyramidale as a measure of neuronal network excitability (Supplementary Fig. 4A). Analysis of the input–output relationships between stimulus intensities and the ratios of population spikes over fibre volley amplitudes showed significantly elevated neuronal excitability in Pik3c2b KO and, importantly, in Pik3c2b HET mice (Fig. 4A). Next, we functionally tested the modulation of neuronal excitability,48,49 by assessing feedback inhibition following paired stimulation delivered within a short time interval. Pik3c2b KO mice displayed larger responses to the second stimulus (Fig. 4B), suggesting that action potential-generated GABAergic feedback inhibition is reduced in KO animals. Moreover, in Pik3c2b HET and, more pronouncedly, in Pik3c2b KO mice we observed increased activity-dependent disinhibition of population spikes (Fig. 4C), a phenotype that was accompanied by marked polyspiking activity. Indeed, 55% of slices from Pik3c2b KO mice exhibited distinct polyspiking activity (0.5 mV and higher), while only 10% of slices from Pik3c2b WT mice displayed polyspiking (Fig. 4D). Importantly, 31% of slices from Pik3c2b HET animals showed distinct polyspiking (Fig. 4D). To further study possible alterations in action potential-induced inhibitory neurotransmission in Pik3c2b KO mice, we pharmacologically isolated IPSCs from patch-clamped CA1 pyramidal neurons. Induced IPSCs evoked by electrical stimulation were significantly lower in Pik3c2b KO neurons compared to WT controls (Fig. 4E and Supplementary Fig. 5A and B), while spontaneous activity of non-stimulated neurons was significantly upregulated (Supplementary Fig. 5C and D). These results indicate that loss of Pi3kc2b weakens evoked inhibitory input possibly as a consequence of increased spontaneous inhibitory activity, thereby resembling other models of mTORC1 hyperactivation, i.e. KO of Tsc1.37
Figure 4.
Dose-dependent PI3K-C2β expression causes mTORC1-dependent hippocampal hyperexcitability. (A) Input–output relationships between stimulus intensities and the ratios of population spikes (PS) over fibre volley (FV) amplitudes show increased neuronal excitability in Pik3c2b KO mice. Two-way repeated measures (RM) ANOVA detected a significant difference between WT and KO mice [P= 0.013; WT n = 20 (slices) from N = 6 (mice); HET n = 16, N = 5; KO n = 18, N = 6]. Inset samples (above) show representative responses evoked by increasing stimulation intensities from 0 to 300 µA for each genotype. (B) Reduced paired-pulse modulation of population spikes in Pik3c2b KO mice. Representative traces of population spike paired-pulse modulation at 10 and 50 ms intervals show that the amplitudes of the second population spikes are greater in Pik3c2b KO mice, indicating reduced perisomatic feedback inhibition. Two-way RM ANOVA detected a significant difference between WT and KO mice (P < 0.001; WT n = 20 N = 6; KO n = 18, N = 6). (C) Activity-dependent facilitation of population spikes and appearance of polyspikes were significantly increased in KO (n = 18, N = 6) and HET (n = 16, N = 5) compared to WT (n = 20, N = 6) mice. Two-way RM ANOVA with P < 0.001 and P = 0.042, respectively. KO slices tended to facilitate more pronouncedly compared with HET (P = 0.065), indicating an effect of PI3K-C2β gene dosage on neuronal excitability. The areas of population spikes and polyspikes were calculated and given as ratios over the area of the first response during 1 Hz stimulation. Inset samples (above) show representative responses for each genotype evoked with 200 µA at first (black) and 30th stimuli (green). Note the appearance of polyspikes in HET and, more pronouncedly, in KO mice. (D) Fraction of slices (%) that display prominent (≥0.5 mV) polyspiking. The elevated fraction of slices with prominent polyspiking in KO mice is rescued by rapamycin (1 µM; N and n are as in C and H). (E) Representative IPSCs of both genotypes recorded from CA1 pyramidal neurons at stimulation intensities from 10 to 80 µA, showing reduced inhibitory synaptic transmission in Pik3c2b KO mice (top). Stimulation-evoked IPSCs (from 20 to 80 µA) are significantly reduced in Pik3c2b KO mice compared to WT littermates (bottom, two-way RM ANOVA P = 0.019; WT n = 13, N = 7; KO n = 14, N = 7). (F) The increased excitability of Pik3c2b KO mice is rescued by pretreatment with rapamycin (1 µM). Two-way RM ANOVA detected no significant difference between WT and KO mice (P = 0.705; WT n = 15, N = 5; KO n = 15, N = 5). Inset samples (above) show representative responses evoked by increasing stimulation intensities from 0 to 300 µA for each genotype. (G) Reduced paired-pulse modulation of population spikes in Pik3c2b KO mice is rescued by pretreatment of slices with rapamycin (1 µM). Representative traces of population spikes at 10 and 50 ms intervals show no difference in perisomatic feed-back inhibition. Two-way RM ANOVA detected no significant difference between WT and KO mice (P = 0.260; for N and n see F). (H) Increased activity-dependent facilitation of population spikes and polyspikes in Pik3c2b KO mice are rescued by pretreatment of slices with rapamycin (1 µM). Two-way RM ANOVA detected no significant difference between WT and KO mice (P = 0.968; for N and n see F). Inset samples (above) show representative responses for each genotype evoked with 200 µA at first (black) and 30th stimuli (green).
It is possible that increased excitatory transmission and neuronal network activity in Pik3c2b KO mice are a consequence of altered numbers of inhibitory and/or excitatory neurons. However, quantitative analysis of the density of parvalbumin-positive interneurons in the hippocampus of Pik3c2b KO mice failed to reveal any difference (Supplementary Fig. 6A and B). Also, elevated excitatory neurotransmission was not the consequence of altered numbers of excitatory (Supplementary Fig. 6C) or inhibitory synapses (Supplementary Fig. 6D). These data strongly support the hypothesis that increased excitatory transmission and neuronal network excitability in Pik3c2b KO mice are caused by elevated mTORC1 signalling, which can modulate excitatory, e.g. downstream of muscarinic acetylcholine receptors,50–52 and inhibitory neurotransmission.37 To test this hypothesis further, we analysed the effects of mTORC1 inhibition in the presence of rapamycin. Pre-incubation of hippocampal slices with the mTORC1 inhibitor rapamycin fully rescued elevated basal synaptic transmission (Supplementary Fig. 4F) and defective hippocampal long-term potentiation (Supplementary Fig. 4E and H) in slices from Pik3c2b KO mice along with normal presynaptic PPF (Supplementary Fig. 4G). Most importantly, rapamycin treatment rescued the increased neuronal excitability of Pik3c2b KO mice. No difference between rapamycin-treated WT or Pik3c2b KO slices was observed with respect to the input–output ratios of population spikes over fibre volley amplitudes (Fig. 4F), feedback GABAergic inhibition following paired stimulation (Fig. 4G), activity-dependent disinhibition of population spikes (Fig. 4H) and the fraction of slices exhibiting pronounced polyspiking (Fig. 4D).
Collectively, these analyses show that the increase in neuronal activity and network excitability, arising as a consequence of decreased PI3K-C2β expression, is dose-dependent, suggesting that haploinsufficiency might lead to the critical electrophysiological phenotypes. These defects likely result from excitatory/inhibitory imbalance downstream to excessive mTORC1 signalling and are rescued by rapamycin administration.
Seizure susceptibility is increased and dose-dependent in Pik3c2b HET and KO mouse models
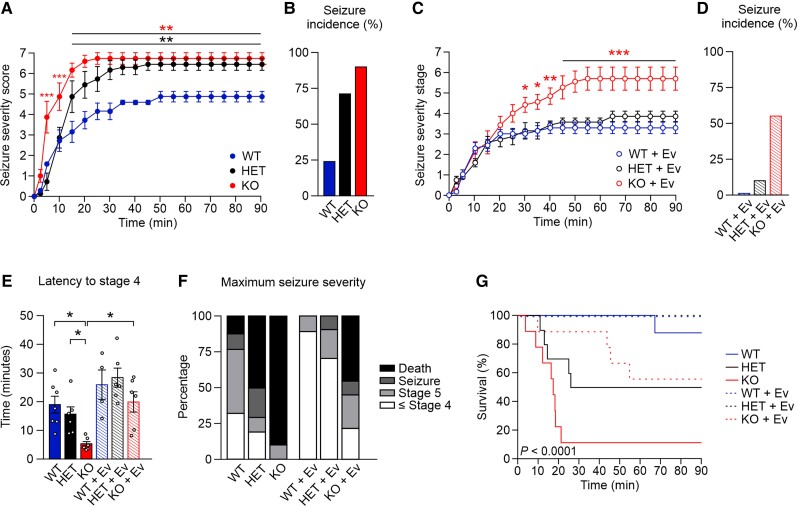
To probe the hypothesis that elevated neuronal network excitability caused by mTORC1 hyperactivity underlies epilepsy, we analysed the response of Pik3c2b HET and Pik3c2b KO mice to epileptogenic drugs. To this aim, we tested the ability of pilocarpine, a muscarinic receptor agonist known to cause sustained increases in extracellular glutamate levels and to stimulate mTORC1 signalling,50 to elicit epileptic seizures in Pik3c2b HET and Pik3c2b KO mice. Seizure severity was scored for a period of 90 min using a modified Racine scale,37,53 in which high values correspond to more severe seizures (Supplementary Videos 1–4). To avoid side effects induced by peripheral cholinergic activation, mice were treated with 1 mg/kg of atropine 30 min before pilocarpine injection.36 Administration of pilocarpine at 350 mg/kg had a severe effect on Pik3c2b KO mice; already at 10 min post-injection, 66% (n = 6/9) of Pik3c2b KOs displayed forelimb clonus (stage 4 in the seizure scale), while at 15 min 78% (n = 7/9) of Pik3c2b KO mice showed severe pilocarpine-induced tonic–clonic seizures (status epilepticus, stage 6 on the seizure scale; Fig. 5A and B and Supplementary Fig. 7A). Progression of status epilepticus in WT mice was significantly slower with only 22% (n = 2/9) of WT controls reaching status epilepticus stage and at later time points (about 30 min; Fig. 5A, B, E, Supplementary Fig. 7A and Supplementary Video 5). Moreover, a mortality rate of 89% was observed for Pik3c2b KO mice, compared to 11% for WT littermates (Fig. 5F and G). The response of Pik3c2b HET mice was comparable to WT during the first stages (Fig. 5A, B and E). However, 70% (n = 7/10) of HET animals developed seizures at the end of the observation, suggesting a mechanism of haploinsufficiency for this pilocarpine susceptibility model (Fig. 5A, B, E–G and Supplementary Fig. 7A).
Figure 5.
Dose-dependent PI3K-C2β expression predisposes to pilocarpine-induced epileptic seizure s. (A) Severity of seizure behaviour over time following intraperitoneal injection of 350 mg/kg pilocarpine in 2–3-month-old male Pik3c2b KO, HET and littermate WT mice. Higher scores correspond to more severe seizure status; a score of 6 indicates status epilepticus, a score of 7 indicates mortality. Two-way ANOVA, **P < 0.01. n = 9 WT, n = 10 HET, n = 9 KO. Error bars represent SEM. (B) Incidence of seizures in KO, HET and WT mice. n is as in A. (C) Severity of seizure behaviour over time following intraperitoneal injection of 350 mg/kg pilocarpine in 2–3-month-old Pik3c2b KO, HET and littermate WT mice pretreated with 10 mg/kg everolimus (Ev) 90 min before pilocarpine injection. Higher scores correspond to more severe seizure status; a score of 6 indicates status epilepticus, a score of 7 indicates mortality. Two-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001. n = 9 WT, n = 10 HET, n = 9 KO. Error bars represent SEM. (D) Incidence of seizures in KO, HET and WT mice treated with everolimus. n is as in C. (E) Time needed for experimental groups to reach seizure stage 4 (forelimb clonus, rearing and falling). One-way ANOVA, *P < 0.05 n > 9 for each experimental group. Error bars represent SEM. (F) Maximal severity stage reached by experimental groups at the end of the behavioural observation. n as in A and C. (G) Kaplan–Meier survival curve for untreated (continuous line) or everolimus-treated (dotted line) WT, HET and KO littermates injected with 350 mg/kg pilocarpine. P < 0.0001. n as in A and C.
If mTORC1 hyperactivity was the cause of evoked seizures, acute inhibition of mTORC1 activity should at least partially rescue this phenotype. We therefore assessed the effect of the mTORC1 inhibitor everolimus in preventing seizure onset. We treated mice with a single dose of everolimus (10 mg/kg) 90 min before pilocarpine treatment and scored mouse behaviour. Everolimus treatment rescued the phenotype of Pik3c2b HET mice and greatly improved the response of the Pik3c2b KO mice (Fig. 5C and D and Supplementary Fig. 7A). In Pik3c2b HET mice, seizure onset was considerably delayed and 90% of the mice did not reach the status epilepticus stage. The latency to early stages was also increased in Pik3c2b KO mice (like untreated WT controls; Fig. 5E). Moreover, only 55% (n = 5/9) of the Pik3c2b KO mice displayed seizures at later time points (30 min) and the mortality rate was reduced to 45% (Fig. 5F and G).
Collectively, these in vivo studies demonstrate that the severity of the epileptic phenotype due to defective PI3K-C2β is dose-dependent and significantly rescued by mTORC1 inhibition, thus matching the above results on neuronal activity and network excitability.
A Q1533* mouse model shows increased mTORC1 signalling and susceptibility to pilocarpine
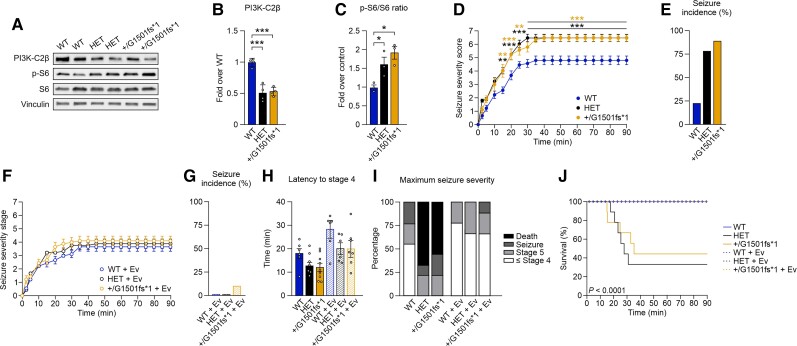
Our genetic and biochemical data indicate that defective activity of mutant PI3K-C2β is associated with the severity of the epileptic phenotype in a dose-dependent manner, supporting haploinsufficiency as a molecular mechanism underlying the pathogenic effect of heterozygous mutations in patients. To model the predicted functional effect of the patient’s p.Q1533* mutation in the C2 terminal domain of PI3K-C2β in vivo, we used CRISPR/Cas9 technology to generate a mutant mouse line heterozygous for a premature stop codon (Pik3c2b+/G1501fs*1). Immunoblot analysis of Pik3c2b+/G1501fs*1 mouse brains confirmed the expression of only the WT allele at a level similar to Pik3c2b HET mice, but did not detect a lower molecular weight band (Fig. 6A and B). Consistent with the absence of the truncated protein, Pik3c2b+/G1501fs*1 mice showed hyperactive mTORC1 signalling at a level comparable to that of Pik3c2b HET mice (Fig. 6A and C). These data suggest that despite the decreased enzymatic activity observed in vitro, in a physiological context the mutant protein is likely to be unstable, resulting in an effect on mTORC1 activity superimposable to the null allele.
Figure 6.
A Q1533* mouse model displays a haploinsufficiency phenotype in response to pilocarpine treatment. (A–C) The Pik3c2b+/G1501fs*1 mutation does not result in the expression of the truncated allele, but activates mTORC1 at levels comparable to HET mice. (A) Representative immunoblot of PI3K-C2β, p-S6, S6 and vinculin content in whole brain lysates from 8-week-old WT, HET and Pik3c2b+/G1501fs*1 mice. (B) Quantification of the PI3K-C2β content as shown in A. No PI3K-C2β bands at lower molecular weight, corresponding to the Pik3c2bG1501fs*1 allele, were detected. One-way ANOVA, ***P < 0.001. n = 4 independent experiments. Error bars represent SEM. (C) Quantification of the p-S6/S6 ratio as shown in A. One-way ANOVA, *P < 0.05. n = 3 independent experiments. Error bars represent SEM. (D) Severity of seizure behaviour over time following intraperitoneal injection of 350 mg/kg pilocarpine in 2–3-month-old male WT, HET and Pik3c2b+/G1501fs*1 mice. Higher scores correspond to more severe seizure status; a score of 6 indicates status epilepticus, a score of 7 indicates mortality. Two-way ANOVA, **P < 0.01, ***P < 0.001. n = 6 WT, n = 6 Pik3c2b+/G1501fs*1, n = 6 KO. Error bars represent SEM. (E) Incidence of seizures in WT, HET and Pik3c2b+/G1501fs*1 mice. n is as in D. (F) Severity of seizure behaviour over time following intraperitoneal injection of 350 mg/kg pilocarpine in 2–3-month-old HET, Pik3c2b+/G1501fs*1 and littermate WT mice pretreated with 10 mg/kg everolimus (Ev) 90 min before pilocarpine injection. Higher scores correspond to more severe seizure status; a score of 6 indicates status epilepticus, a score of 7 indicates mortality. Two-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001. n = 9 WT, n = 10 HET, n = 9 KO. Error bars represent SEM. (G) Incidence of seizures in WT, HET and Pik3c2b+/G1501fs*1 mice treated with everolimus. n is as in D. (H) Time needed for experimental groups to reach seizure stage 4 (forelimb clonus, rearing and falling). One-way ANOVA. n is as in D and F. Error bars represent SEM. (I) Maximal severity stage reached by experimental groups at the end of the behavioural observation. n is as in D and F. (J) Kaplan–Meier survival curve for untreated (continuous line) or everolimus-treated (dotted line) WT, HET and Pik3c2b+/G1501fs*1 mice injected with 350 mg/kg pilocarpine. P < 0.0001. n as in D and F.
We next assessed the response of Pik3c2b+/G1501fs*1 mice to pilocarpine treatment. Similar to Pik3c2b HET mice, Pik3c2b+/G1501fs*1 behaviour was comparable to WT controls through the first stages (Fig. 6D and H and Supplementary Fig. 7B). However, towards the end of the observation 88.9% (n = 8/9) of the Pik3c2b+/G1501fs*1 animals developed seizures, while only 2/9 WT mice did (Fig. 6D and E). Moreover, mortality was significantly higher (55.6% of Pik3c2b+/G1501fs*1 mice compared to 11.1% of controls; Fig. 6I and J).
Next, we tested the ability of everolimus in preventing seizure onset in our mouse model mimicking the Q1533* patient’s mutation. Treatment with a single dose of everolimus (10 mg/kg) was sufficient to completely rescue the phenotype of Pik3c2b+/G1501fs*1 mice, akin to Pik3c2b HET mice (Fig. 6F–I and Supplementary Fig. 7B). Seizure incidence of Pik3c2b+/G1501fs*1 mice dropped to 11.1% (n = 1/9) and all animals survived until the end of the experiment. The striking similarity of Pik3c2b HET and Pik3c2b+/G1501fs*1 mice thus strongly supports the hypothesis that haploinsufficiency is responsible for the clinical phenotype observed in patients.
Discussion
Our data unravel defective lipid signalling caused by mutations in PIK3C2B, encoding class II PI 3-kinase C2β, as a novel mechanism to underlie FE in humans. Mutations in proteins that participate in phosphoinositide signalling pathways (e.g. PIK3CA, PTEN, SYNJ1) have so far been implicated in rare severe infantile epilepsies or developmental multisystemic syndromes,54 but not in FE. We show that disturbance of PI3K-C2β-mediated synthesis of PI(3,4)P2, i.e. a lipid hitherto not associated with brain or synaptic function, is a key factor responsible for a common and relatively mild form of focal epilepsy.
The heterozygous PIK3C2B mutations identified in patients from this study were either predicted to be protein-truncating or corresponded to single amino-acid substitutions situated within protein domains that exert or control lipid-synthesizing activity (i.e. the PI3K catalytic domain and the C2 autoregulatory domain). Although spatial clustering is commonly taken to indicate a mechanism different from loss-of-function,55 the preferential location of smURVs from this study and from Epi25 NAFE cases around key protein regions conceivably indicated that both missense and protein-truncating URVs could result in null functional alleles. Consistently, the majority of tested patients’ URVs resulted in impaired enzyme activity and mTOR pathway hyperactivation when expressed in human HEK293T cells. In this cellular background, the decrease in PI(3,4)P2 synthesis and the increase in mTORC1 activity resulting from patients’ mutations was as dramatic as that observed for the kinase-dead PI3K-C2β isoform, thus supporting their loss-of-function effect. On the other hand, one tested URV, the p.R1118L, retained its catalytic activity and had no significant effect on mTORC1 when transfected in HEK293T cells. The clinical phenotype of the patient carrying the p.R1118L is remarkably mild, which would be consistent either with a less pronounced effect on kinase activity and mTORC1 activation at the molecular level, or with a pathogenic mechanism that is triggered through different molecular pathways. PIK3C2B is a gene highly intolerant to loss-of-function variation in the general population according to gnomAD data, where only 15 loss-of-function variants are observed against an expected number of 82.8. Moreover, in the Database of Genomic Variants, deletions of PIK3C2B coding regions are rarely reported, i.e. appearing in only two healthy individuals. However, the presence of gene-disrupting variants at low allele frequencies in individuals without epilepsy (including the unaffected father of a Epi25 NAFE case) suggests reduced penetrance, a common feature of other mTOR pathway genes previously implicated in FE including the major GATOR1 complex gene DEPDC5 (displaying 24 observed loss-of-function variants in gnomAD).
Motivated by the loss-of-function nature of patients’ mutations, we chose Pik3c2b KO mice as an experimental model to test the impact of PI3K-C2β alterations on the disease. Functional analysis of hippocampal neurotransmission by electrophysiological recordings showed increased neuronal excitability and network activity, i.e. functional signatures typically associated with epileptic phenotypes, in KO mice. This was coupled to the absence of prominent morphological alterations of brain architecture, hippocampal layering, cell number or size, or alterations in excitatory versus inhibitory synapse numbers between Pik3c2b KO mice and WT animals, thus arguing against major developmental alterations. Accordingly, the PIK3C2B URV-positive patients reported here do not present structural anomalies identified previously in a subset of GATOR1-mutated FE patients (such as focal cortical dysplasia). However, we cannot exclude the presence of subtler developmental defects or alterations in brain regions not investigated in our study, as reported in bona fide mTORopathy models.56 Finally, the presence of brain abnormalities in patients may require a second somatic hit occurring in the developing brain, as reported for other mTORC1 inhibitors TSC1/2 and DEPDC5; additional studies will be needed to further explore this possibility.57 Moreover, it is possible that somatic events that add up to germline mutations explain the focal nature of PIK3C2B-related disease, as brain mosaic mutations inside and outside the mTOR pathway have been associated with lesional and non-lesional focal epilepsies.57,58
Our data favour a model in which acute alterations in PI3K-C2β-mediated lipid signalling via PI(3,4)P2 causes mTORC1 hyperactivity in neurons, which in turn leads to excitatory/inhibitory imbalance and epileptic seizures. Although not addressed in subcellular detail in this study, the most likely mechanism by which defective lipid signalling results in elevated mTORC1 activity in neurons is by reduced local production of the signalling lipid PI(3,4)P2 at late endosomes or lysosomes, i.e. organelles where active mTORC1 is located. Localized PI(3,4)P2 represses mTORC1 activity via triggering the association of Raptor with inhibitory 14-3-3 proteins18 and potentially additional factors. Although we cannot formally rule out an effect of PI3K-C2β-mediated production of other PI 3-phosphate lipids, most notably PI(3)P which can be synthesized by PI3K-C2β in various non-neuronal cell types,17,59,60 these are unlikely to explain mTORC1-mediated epilepsy in patients and mice. Indeed, PI(3)P is known to activate mTORC1,61 thus loss-of-function variants would be predicted to result in impaired mTORC1 signalling and protection from seizures.
Interestingly, the phenotypic consequences of impaired PI3K-C2β-mediated lipid synthesis appeared to be dose-dependent. Abnormal mTORC1 signalling and neuronal activity, determined as increased population spikes and polyspiking activity, were indeed most prominent in the brains of Pik3c2b KO mice, reflecting the extreme vulnerability to developing seizures after acute treatment with pilocarpine. Notably, these molecular, cellular and seizure phenotypes were already remarkable in Pik3c2b HET animals, suggesting that the critical threshold for epileptogenesis is exceeded with the loss of only one Pik3c2b allele. Most importantly, in the brains of Pik3c2b+/G1501fs*1 mice, which model the premature translational stop codon occurring in patients with the p.Q1533* mutation, lowered PI3K-C2β expression levels were sufficient to hyperactivate mTORC1 which, in turn, increased susceptibility to epileptic seizures. We therefore propose haploinsufficiency as the main molecular mechanism underlying epilepsy related to PIK3C2B mutations.
A further important finding of our study is the fact that increased neuronal network activity and seizure susceptibility can be acutely rescued by pharmacological inhibition of mTORC1 activity. These data open the possibility that the use of everolimus, an FDA-approved rapamycin analogue for the treatment of tuberous sclerosis, could be beneficial in the treatment of patients selectively stratified for PIK3C2B mutations, as already proposed for other genetic FEs.62
Disentangling the genetic determinants of individual disease risks remains a core challenge in epilepsy research. A central question is whether, and to what extent, URVs of large effect drive disease susceptibility, and if defects in specific pathways are shared or unique between rare syndromes and common forms. Focal epilepsies, in spite of being the most commonly encountered in the clinic,63 remain largely elusive to genetic dissection and are thought to be characterized by oligogenic/polygenic architecture as other neuropsychiatric conditions.64,65 Combining multiple layers of genetic, functional and animal data, we provide here compelling arguments that defects in an mTORC1 inhibitor, previously unlinked to human disease, represent a piece in the heterogeneous genetic puzzle of FE.
In conclusion, we implicate mutations in PIK3C2B as a genetic aetiology in FEs, mostly of sporadic origin, and provide insights into the variety of cellular pathways involved in epileptogenesis. These findings highlight the genetic complexity of FE and pave the way to unravel novel molecular mechanisms underlying neuronal hyperexcitability that are amenable to therapeutic intervention.
Web resources
Genome Aggregation Database: https://gnomad.broadinstitute.org/
Database of Genomic Variants: http://dgv.tcag.ca/dgv/app/home
Varsome: https://varsome.com/
Epi25: https://epi25.broadinstitute.org/
CADD score: https://cadd.gs.washington.edu/
Supplementary Material
Acknowledgements
We are thankful to the patients and their families for their participation in this study. We acknowledge Elena Bacchelli (Dipartimento di Farmacia e Biotecnologie, University of Bologna, Italy) for providing access to Italian control samples. Whole Exome Sequencing data were generated at Beijing Genomic Institute, Shenzhen, China. Pik3c2b+/G1501fs*1 genetically modified mice were generated by the MBC Core Facility for Gene Transfer and Mutagenesis (University of Torino, Italy). Finally, we thank Dr Chistopher Schmied [Cellular Imaging Facility, Leibniz Forschungsinstitut für Molekulare Pharmakologie (FMP), Berlin, Germany] for generating a Fiji-based macro for semi-automated neuron segmentation (available at: DOI 10.5281/zenodo.5652147).
Abbreviations
- DNV
de novo variant
- FE
focal epilepsy
- fEPSP
field excitatory postsynaptic potential
- HET
heterozygous
- IPSC
inhibitory postsynaptic current
- KO
knockout
- mTORC1
mechanistic Target of Rapamycin Complex 1
- NAFE
non-acquired focal epilepsy
- PI(3)P
phosphatidylinositol 3-phosphate
- PI(3,4)P2
phosphatidylinositol 3,4-bisphosphate
- PI3K
phosphatidylinositol 3-kinase
- (sm)URV
(singleton missense) ultra-rare variant
- WT
wild-type
Appendix 1
Collaborative Group of Italian League Against Epilepsy (LICE) Genetic Commission
Full details are available in the Supplementary material. Amedeo Bianchi, Pasquale Striano Antonio Gambardella, Giuseppe d'Orsi, Giovanni Crichiutti, Stefano Meletti, Roberto Dilena, Margherita Santucci, Carla Marini, Aglaia Vignoli, Giuseppe Gobbi, Eleonora Briatore, Massimo Mastrangelo, Barbara Mostacci.
Contributor Information
Luca Gozzelino, Department of Molecular Biotechnology and Health Sciences, University of Torino, Torino, Italy.
Gaga Kochlamazashvili, Department of Molecular Pharmacology and Cell Biology, Leibniz Forschungsinstitut für Molekulare Pharmakologie (FMP), Robert-Roessle-Strasse 10, 13125 Berlin, Germany.
Sara Baldassari, Department of Medical and Surgical Sciences (DIMEC), University of Bologna, Bologna, Italy; Sorbonne Université, Institut du Cerveau—Paris Brain Institute—ICM, Inserm, CNRS, F-75013 Paris, France.
Albert Ian Mackintosh, Department of Molecular Pharmacology and Cell Biology, Leibniz Forschungsinstitut für Molekulare Pharmakologie (FMP), Robert-Roessle-Strasse 10, 13125 Berlin, Germany.
Laura Licchetta, IRCCS Istituto delle Scienze Neurologiche di Bologna, Epilepsy Center (Reference Center for Rare and Complex Epilepsies—EpiCARE), Bologna, Italy.
Emanuela Iovino, Department of Medical and Surgical Sciences (DIMEC), University of Bologna, Bologna, Italy.
Yu Chi Liu, Epilepsy Research Centre, Department of Medicine, University of Melbourne, Austin Health, Melbourne, Victoria, Australia; Population Health and Immunity Division, The Walter and Eliza Hall Institute of Medical Research, Melbourne, Victoria, Australia; Department of Medical Biology, University of Melbourne, Melbourne, Victoria, Australia.
Caitlin A Bennett, Epilepsy Research Centre, Department of Medicine, University of Melbourne, Austin Health, Melbourne, Victoria, Australia.
Mark F Bennett, Epilepsy Research Centre, Department of Medicine, University of Melbourne, Austin Health, Melbourne, Victoria, Australia; Population Health and Immunity Division, The Walter and Eliza Hall Institute of Medical Research, Melbourne, Victoria, Australia; Department of Medical Biology, University of Melbourne, Melbourne, Victoria, Australia.
John A Damiano, Epilepsy Research Centre, Department of Medicine, University of Melbourne, Austin Health, Melbourne, Victoria, Australia.
Gábor Zsurka, Department of Experimental Epileptology and Cognition Research and Department of Epileptology, University Bonn Medical Center, Venusberg Campus 1, D-53105 Bonn, Germany.
Caterina Marconi, Department of Medical and Surgical Sciences (DIMEC), University of Bologna, Bologna, Italy.
Tania Giangregorio, Department of Medical and Surgical Sciences (DIMEC), University of Bologna, Bologna, Italy.
Pamela Magini, U.O. Genetica Medica, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Bologna, Italy.
Marijn Kuijpers, Department of Molecular Pharmacology and Cell Biology, Leibniz Forschungsinstitut für Molekulare Pharmakologie (FMP), Robert-Roessle-Strasse 10, 13125 Berlin, Germany.
Tanja Maritzen, Department of Molecular Pharmacology and Cell Biology, Leibniz Forschungsinstitut für Molekulare Pharmakologie (FMP), Robert-Roessle-Strasse 10, 13125 Berlin, Germany; Department of Nanophysiology, Technische Universität Kaiserslautern, 67663 Kaiserslautern, Germany.
Giuseppe Danilo Norata, Department of Excellence in Pharmacological and Biomolecular Sciences, Università degli Studi di Milano, Milan and Center for the Study of Atherosclerosis, SISA Bassini Hospital Cinisello B, Italy.
Stéphanie Baulac, Sorbonne Université, Institut du Cerveau—Paris Brain Institute—ICM, Inserm, CNRS, F-75013 Paris, France.
Laura Canafoglia, Unit of Integrated Diagnostics for Epilepsy, Fondazione IRCCS Istituto Neurologico Carlo Besta, Milan, Italy.
Marco Seri, Department of Medical and Surgical Sciences (DIMEC), University of Bologna, Bologna, Italy; U.O. Genetica Medica, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Bologna, Italy.
Paolo Tinuper, IRCCS Istituto delle Scienze Neurologiche di Bologna, Epilepsy Center (Reference Center for Rare and Complex Epilepsies—EpiCARE), Bologna, Italy; Department of Biomedical and NeuroMotor Sciences (DIBINEM), University of Bologna, Bologna, Italy.
Ingrid E Scheffer, Epilepsy Research Centre, Department of Medicine, University of Melbourne, Austin Health, Melbourne, Victoria, Australia; Murdoch Children’s Research Institute, Royal Children’s Hospital, Parkville, Melbourne, Victoria, Australia; Florey Institute for Neuroscience and Mental Health, University of Melbourne, Melbourne, Victoria, Australia; Department of Pediatrics, University of Melbourne, Royal Children's Hospital, Melbourne, Victoria, Australia.
Melanie Bahlo, Spedali Civili, Neuropsychiatric Department, Brescia, Italy; Faculty of Biology, Chemistry, Pharmacy, Freie Universität Berlin, 14195 Berlin, Germany.
Samuel F Berkovic, Epilepsy Research Centre, Department of Medicine, University of Melbourne, Austin Health, Melbourne, Victoria, Australia.
Michael S Hildebrand, Epilepsy Research Centre, Department of Medicine, University of Melbourne, Austin Health, Melbourne, Victoria, Australia; Murdoch Children’s Research Institute, Royal Children’s Hospital, Parkville, Melbourne, Victoria, Australia.
Wolfram S Kunz, Department of Experimental Epileptology and Cognition Research and Department of Epileptology, University Bonn Medical Center, Venusberg Campus 1, D-53105 Bonn, Germany.
Lucio Giordano, Spedali Civili, Neuropsychiatric Department, Brescia, Italy.
Francesca Bisulli, IRCCS Istituto delle Scienze Neurologiche di Bologna, Epilepsy Center (Reference Center for Rare and Complex Epilepsies—EpiCARE), Bologna, Italy; Department of Biomedical and NeuroMotor Sciences (DIBINEM), University of Bologna, Bologna, Italy.
Miriam Martini, Department of Molecular Biotechnology and Health Sciences, University of Torino, Torino, Italy.
Volker Haucke, Department of Molecular Pharmacology and Cell Biology, Leibniz Forschungsinstitut für Molekulare Pharmakologie (FMP), Robert-Roessle-Strasse 10, 13125 Berlin, Germany; Faculty of Biology, Chemistry, Pharmacy, Freie Universität Berlin, 14195 Berlin, Germany.
Emilio Hirsch, Department of Molecular Biotechnology and Health Sciences, University of Torino, Torino, Italy.
Tommaso Pippucci, U.O. Genetica Medica, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Bologna, Italy.
Funding
We acknowledge the Epi25 collaborative for the use of Epi25 data: this work was part of the Centers for Common Disease Genomics (CCDG) program, funded by the National Human Genome Research Institute (NHGRI) and the National Heart, Lung, and Blood Institute (NHLBI). CCDG-funded Epi25 research activities at the Broad Institute, including genomic data generation in the Broad Genomics Platform, are supported by NHGRI grant UM1 HG008895 (PIs: Eric Lander, Stacey Gabriel, Mark Daly, Sekar Kathiresan). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This study has been partially supported by Fondazione Telethon Project GGP13200 to P.T. and T.P., Fondazione Telethon Project GGP19146 to G.D.N., Ministero dell'Università e della Ricerca, PRIN 2017 K55HLC to G.D.N., Ministero della Salute Project GR-2009-1574072 to T.P. and F.B. and the Deutsche Forschungsgemeinschaft (TRR186/A08) to V.H.
Competing interests
E.H. is a founder of Kither Biotech, a company involved in the development of PI3K inhibitors, which has no involvement in this study. The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Fiest KM, Sauro KM, Wiebe S, et al. Prevalence and incidence of epilepsy: A systematic review and meta-analysis of international studies. Neurology. 2017;88(3):296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fisher RS, van Emde Boas W, Blume W, et al. Epileptic seizures and epilepsy: Definitions proposed by the International League Against Epilepsy (ILAE) and the International Bureau for Epilepsy (IBE). Epilepsia. 2005;46(4):470–472. [DOI] [PubMed] [Google Scholar]
- 3. Berkovic SF, Howell RA, Hay DA, Hopper JL. Epilepsies in twins: Genetics of the major epilepsy syndromes. Ann Neurol. 1998;43(4):435–445. [DOI] [PubMed] [Google Scholar]
- 4. Helbig I, Scheffer IE, Mulley JC, Berkovic SF. Navigating the channels and beyond: Unravelling the genetics of the epilepsies. Lancet Neurol. 2008;7(3):231–245. [DOI] [PubMed] [Google Scholar]
- 5. Hildebrand MS, Dahl HH, Damiano JA, Smith RJ, Scheffer IE, Berkovic SF. Recent advances in the molecular genetics of epilepsy. J Med Genet. 2013;50(5):271–279. [DOI] [PubMed] [Google Scholar]
- 6. Epi4K Consortium, Epilepsy Phenome/Genome Project, Allen AS, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501(7466):217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. EuroEPINOMICS-RES Consortium, Epilepsy Phenome/Genome Project, Epi4K Consortium . De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am J Hum Genet. 2014;95(4):360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Scheffer IE, French J, Hirsch E, et al. Classification of the epilepsies: New concepts for discussion and debate-Special report of the ILAE Classification Task Force of the Commission for Classification and Terminology. Epilepsia Open. 2016;1(1-2):37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Perucca P. Genetics of focal epilepsies: What do we know and where are we heading? Epilepsy Curr. 2018;18(6):356–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pippucci T, Licchetta L, Baldassari S, et al. Epilepsy with auditory features: A heterogeneous clinico-molecular disease. Neurol Genet. 2015;1(1):e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bisulli F, Rinaldi C, Pippucci T, et al. Epilepsy with auditory features: Contribution of known genes in 112 patients. Seizure. 2021;85:115–118. [DOI] [PubMed] [Google Scholar]
- 12. Licchetta L, Pippucci T, Baldassari S, et al. Sleep-related hypermotor epilepsy (SHE): Contribution of known genes in 103 patients. Seizure. 2020;74:60–64. [DOI] [PubMed] [Google Scholar]
- 13. Ricos MG, Hodgson BL, Pippucci T, et al. Mutations in the mammalian target of rapamycin pathway regulators NPRL2 and NPRL3 cause focal epilepsy. Ann Neurol. 2016;79(1):120–131. [DOI] [PubMed] [Google Scholar]
- 14. Baldassari S, Licchetta L, Tinuper P, Bisulli F, Pippucci T. GATOR1 complex: The common genetic actor in focal epilepsies. J Med Genet. 2016;53(8):503–510. [DOI] [PubMed] [Google Scholar]
- 15. Baldassari S, Picard F, Verbeek NE, et al. The landscape of epilepsy-related GATOR1 variants. Genet Med. 2019;21(2):398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Domin J, Harper L, Aubyn D, et al. The class II phosphoinositide 3-kinase PI3K-C2beta regulates cell migration by a PtdIns3P dependent mechanism. J Cell Physiol. 2005;205(3):452–462. [DOI] [PubMed] [Google Scholar]
- 17. Alliouachene S, Bilanges B, Chicanne G, et al. Inactivation of the class II PI3K-C2beta potentiates insulin signaling and sensitivity. Cell Rep. 2015;13(9):1881–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marat AL, Wallroth A, Lo WT, et al. mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate. Science. 2017;356(6341):968–972. [DOI] [PubMed] [Google Scholar]
- 19. Wallroth A, Koch PA, Marat AL, Krause E, Haucke V. Protein kinase N controls a lysosomal lipid switch to facilitate nutrient signalling via mTORC1. Nat Cell Biol. 2019;21(9):1093–1101. [DOI] [PubMed] [Google Scholar]
- 20. Margaria JP, Ratto E, Gozzelino L, Li H, Hirsch E. Class II PI3Ks at the intersection between signal transduction and membrane trafficking. Biomolecules. 2019;9(3):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Posor Y, Eichhorn-Gruenig M, Puchkov D, et al. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 2013;499(7457):233–237. [DOI] [PubMed] [Google Scholar]
- 22. Reutens DC, Howell RA, Gebert KE, Berkovic SF. Validation of a questionnaire for clinical seizure diagnosis. Epilepsia. 1992;33(6):1065–1071. [DOI] [PubMed] [Google Scholar]
- 23. Pippucci T, Licchetta L, Baldassari S, et al. Contribution of ultrarare variants in mTOR pathway genes to sporadic focal epilepsies. Ann Clin Transl Neurol. 2019;6(3):475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iossifov I, O’Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boyle EA, O’Roak BJ, Martin BK, Kumar A, Shendure J. MIPgen: Optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics. 2014;30(18):2670–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. O’Roak BJ, Vives L, Fu W, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338(6114):1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bennett CA, Petrovski S, Oliver KL, Berkovic SF. ExACtly zero or once: A clinically helpful guide to assessing genetic variants in mild epilepsies. Neurol Genet. 2017;3(4):e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1––11.10.33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Turner TN, Douville C, Kim D, et al. Proteins linked to autosomal dominant and autosomal recessive disorders harbor characteristic rare missense mutation distribution patterns. Hum Mol Genet. 2015;24(21):5995–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ciraolo E, Perino A, Hirsch E. Measuring PI3K lipid kinase activity. Methods Mol Biol. 2012;795:55–67. [DOI] [PubMed] [Google Scholar]
- 32. Hashimoto M, Yamashita Y, Takemoto T. Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse. Dev Biol. 2016;418(1):1–9. [DOI] [PubMed] [Google Scholar]
- 33. Wang W, Kutny PM, Byers SL, et al. Delivery of Cas9 protein into mouse zygotes through a series of electroporation dramatically increases the efficiency of model creation. J Genet Genomics. 2016;43(5):319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Scharfman HE, MacLusky NJ. Sex differences in the neurobiology of epilepsy: A preclinical perspective. Neurobiol Dis. 2014;72(Pt B):180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lenz M, Ben Shimon M, Deller T, Vlachos A, Maggio N. Pilocarpine-induced status epilepticus is associated with changes in the actin-modulating protein synaptopodin and alterations in long-term potentiation in the mouse hippocampus. Neural Plast. 2017;2017:2652560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bateup HS, Johnson CA, Denefrio CL, Saulnier JL, Kornacker K, Sabatini BL. Excitatory/inhibitory synaptic imbalance leads to hippocampal hyperexcitability in mouse models of tuberous sclerosis. Neuron. 2013;78(3):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Epi25 Collaborative . Ultra-rare genetic variation in the epilepsies: A whole-exome sequencing study of 17,606 individuals. Am J Hum Genet. 2019;105(2):267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang H, Lo WT, Vujičić Žagar A, et al. Autoregulation of class II alpha PI3K activity by its lipid-binding PX-C2 domain module. Mol Cell. 2018;71(2):343–351.e4. [DOI] [PubMed] [Google Scholar]
- 40. Traynelis J, Silk M, Wang Q, et al. Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation. Genome Res. 2017;27(10):1715–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Campa CC, Margaria JP, Derle A, et al. Rab11 activity and PtdIns(3)P turnover removes recycling cargo from endosomes. Nat Chem Biol. 2018;14(8):801–810. [DOI] [PubMed] [Google Scholar]
- 42. Harada K, Truong AB, Cai T, Khavari PA. The class II phosphoinositide 3-kinase C2beta is not essential for epidermal differentiation. Mol Cell Biol. 2005;25(24):11122–11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zeng LH, Rensing NR, Zhang B, Gutmann DH, Gambello MJ, Wong M. Tsc2 gene inactivation causes a more severe epilepsy phenotype than Tsc1 inactivation in a mouse model of tuberous sclerosis complex. Hum Mol Genet. 2011;20(3):445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yuskaitis CJ, Jones BM, Wolfson RL, et al. A mouse model of DEPDC5-related epilepsy: Neuronal loss of Depdc5 causes dysplastic and ectopic neurons, increased mTOR signaling, and seizure susceptibility. Neurobiol Dis. 2018;111:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. LaSarge CL, Pun RY, Muntifering MB, Danzer SC. Disrupted hippocampal network physiology following PTEN deletion from newborn dentate granule cells. Neurobiol Dis. 2016;96:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chevere-Torres I, Kaphzan H, Bhattacharya A, et al. Metabotropic glutamate receptor-dependent long-term depression is impaired due to elevated ERK signaling in the DeltaRG mouse model of tuberous sclerosis complex. Neurobiol Dis. 2012;45(3):1101–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Richter JD, Klann E. Making synaptic plasticity and memory last: Mechanisms of translational regulation. Genes Dev. 2009;23(1):1–11. [DOI] [PubMed] [Google Scholar]
- 48. Freund TF. Interneuron Diversity series: Rhythm and mood in perisomatic inhibition. Trends Neurosci. 2003;26(9):489–495. [DOI] [PubMed] [Google Scholar]
- 49. Freund TF, Buzsaki G. Interneurons of the hippocampus. Hippocampus. 1996;6(4):347–470. [DOI] [PubMed] [Google Scholar]
- 50. Smolders I, Khan GM, Manil J, Ebinger G, Michotte Y. NMDA receptor-mediated pilocarpine-induced seizures: Characterization in freely moving rats by microdialysis. Br J Pharmacol. 1997;121(6):1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao LX, Ge YH, Li JB, et al. M1 muscarinic receptors regulate the phosphorylation of AMPA receptor subunit GluA1 via a signaling pathway linking cAMP-PKA and PI3K-Akt. FASEB J. 2019;33(5):6622–6631. [DOI] [PubMed] [Google Scholar]
- 52. Huang X, Zhang H, Yang J, et al. Pharmacological inhibition of the mammalian target of rapamycin pathway suppresses acquired epilepsy. Neurobiol Dis. 2010;40(1):193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Morrison RS, Wenzel HJ, Kinoshita Y, Robbins CA, Donehower LA, Schwartzkroin PA. Loss of the p53 tumor suppressor gene protects neurons from kainate-induced cell death. J Neurosci. 1996;16(4):1337–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Volpatti JR, Al-Maawali A, Smith L, Al-Hashim A, Brill JA, Dowling JJ. The expanding spectrum of neurological disorders of phosphoinositide metabolism. Dis Model Mech. 2019;12(8):dmm038174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lelieveld SH, Wiel L, Venselaar H, et al. Spatial clustering of de novo missense mutations identifies candidate neurodevelopmental disorder-associated genes. Am J Hum Genet. 2017;101(3):478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marsan E, Baulac S. Review: Mechanistic target of rapamycin (mTOR) pathway, focal cortical dysplasia and epilepsy. Neuropathol Appl Neurobiol. 2018;44(1):6–17. [DOI] [PubMed] [Google Scholar]
- 57. Baldassari S, Ribierre T, Marsan E, et al. Dissecting the genetic basis of focal cortical dysplasia: A large cohort study. Acta Neuropathol. 2019;138(6):885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Winawer MR, Griffin NG, Samanamud J, et al. Somatic SLC35A2 variants in the brain are associated with intractable neocortical epilepsy. Ann Neurol. 2018;83(6):1133–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chikh A, Ferro R, Abbott JJ, et al. Class II phosphoinositide 3-kinase C2beta regulates a novel signaling pathway involved in breast cancer progression. Oncotarget. 2016;7(14):18325–18345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sabha N, Volpatti JR, Gonorazky H, et al. PIK3C2B inhibition improves function and prolongs survival in myotubular myopathy animal models. J Clin Invest. 2016;126(9):3613–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hong Z, Pedersen NM, Wang L, Torgersen ML, Stenmark H, Raiborg C. PtdIns3P controls mTORC1 signaling through lysosomal positioning. J Cell Biol. 2017;216(12):4217–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Myers KA, Scheffer IE. DEPDC5 as a potential therapeutic target for epilepsy. Expert Opin Ther Targets. 2017;21(6):591–600. [DOI] [PubMed] [Google Scholar]
- 63. Petrovski S, Kwan P. Unraveling the genetics of common epilepsies: Approaches, platforms, and caveats. Epilepsy Behav. 2013;26(3):229–233. [DOI] [PubMed] [Google Scholar]
- 64. Gramm M, Leu C, Perez-Palma E, et al. Polygenic risk heterogeneity among focal epilepsies. Epilepsia. 2020;61(11):e179–e185. [DOI] [PubMed] [Google Scholar]
- 65. Geschwind DH, Flint J. Genetics and genomics of psychiatric disease. Science. 2015;349(6255):1489–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available within the article and its Supplementary material. All supporting data in this study are available from the corresponding author on request.