Abstract
BACKGROUND
Non-tuberculous mycobacteria (NTMs) cause diseases known as mycobacteriosis and are an important cause of morbidity and mortality. The diagnosis of pulmonary disease caused by NTM is hampered by its clinical similarity with tuberculosis (TB) and by the lack of an accurate and rapid laboratory diagnosis.
OBJECTIVES
Detect DNA from NTMs directly from lung samples using real-time polymerase chain reaction (qPCR) for amplification of 16S rRNA. Additionally, DNA sequencing (hsp65 and rpoB genes) was used to identify the species of MNTs.
METHODS
A total of 68 sputum samples (54 with suspected NTMs and 14 with TB) from patients treated at a referral hospital were used.
FINDINGS
Of these, 27/54 (50%) were qPCR positive for NTMs and 14/14 TB patients (controls) were qPCR negative with an almost perfect concordance (Kappa of 0.93) with the Mycobacterium spp. culture. Sequencing confirmed the presence of NTM in all positive samples. The most common species was Mycobacterium gordonae (33%), followed by Mycobacterium abscessus (26%), Mycobacterium fortuitum (22%), Mycobacterium avium (15%) and Mycobacterium peregrinum (4%).
MAIN CONCLUSIONS
The qPCR technique for detecting NTMs targeting 16S rRNA has the potential to detect NTMs and rapidly differentiate from Mycobacterium tuberculosis. However, it is necessary to identify the species to help in the differential diagnosis between disease and contamination, and to guide the choice of the therapeutic scheme.
Key words: non-tuberculous mycobacteria (NTMs), DNA, real-time PCR (qPCR)
Non-tuberculous mycobacteria (NTMs) consist of species of the genus Mycobacterium spp., which do not belong to the Mycobacterium tuberculosis complex (that causes tuberculosis - TB). They are present in nature and can eventually cause diseases, including pulmonary ones, with symptoms similar to those of TB, especially in immunosuppressed people, such as those with HIV. 1 , 2 Despite the high morbidity and mortality, the lack of compulsory notification of mycobacteriosis makes NTMs prevalence data scarce. 3 Therefore, an increase in prevalence has been reported in different regions of the world, including Brazil. 4 , 5
Lung infections are the most common disease triggered by NTMs, as they are often associated with structural changes in the lung, such as chronic obstructive pulmonary disease, sequelae of previous pulmonary TB, in addition to coinfection with HIV and in transplant patients. 6 The disease occurs predominantly in the population over 50 years of age and is related to the presence of additional comorbidities. 6
An accurate diagnosis that identifies pulmonary disease caused by NTMs is a challenge in clinical practice, 7 since, in addition to nonspecific symptoms, infections can originate from colonisation and transient contamination. 8 Thus, proper treatment and resolution of the disease depend on a diagnosis that associates clinical findings with laboratory tests. 4 , 8 According to guidelines established by the American Thoracic Society (ATS), European Respiratory Society (ERS), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), and the Infectious Diseases Society of America (IDSA), at least two cultures of NTMs are required for evidence of disease, and susceptibility testing is recommended due to the presence of resistance in relevant species, 9 which makes rapid identification of NTMs important for patient management and disease control. 4 , 8
The identification of NTMs is carried out mainly by phenotypic methods based on culture and biochemical characteristics, which makes the process time-consuming and laborious. 10 , 11
The molecular techniques for identifying the species of NTMs that are being used are mainly based on the polymerase chain reaction (PCR), such as GenoType ® Mycobacterium CM and PRA-hsp65, which have the advantage of quickly obtaining results, however, the high cost and/or complexity of the process limit its use in the routine. 12
This study was developed to detect DNA from NTMs by real-time PCR (qPCR) in lung samples, with the aim of differentiating from the M. tuberculosis Complex and, additionally, to identify the species through sequencing for comparison of results.
SUBJECTS AND METHODS
Clinical isolates (culture) and lung samples of NTMs - All samples used in the study (68 sputum samples) were from patients at the Phthisiology Outpatient Clinic of Hospital Sanatório Partenon (HSP), a state reference (Rio Grande do Sul, Brazil) for cases of NTMs, treated in the period from September 2018 to December 2019.
Clinical suspicion, as well as diagnosis, followed the criteria established in international guidelines. 9 The study was approved by the Ethics Committee on Health Research of the Escola de Saúde Pública (CEPS-ESP) in accordance with the Resolution 466/2012, under the number CAAE 96556418.4.0000.5312 of September 24, 2018.
The smear slides, culture and GeneXpert/RIF tests were performed in the diagnostic routine of the HSP, as recommended. 11 , 13 Patients with an insufficient sample (less than 500 µL) for all tests were excluded from the study, in addition to those being monitored for TB control and responding to treatment. The NTMs strains used as controls in this study came from the Institute of Biological Sciences of Universidade Federal do Rio Grande (Rio Grande, RS, Brazil).
DNA extraction - The extraction of DNA directly from culture samples and lung samples was performed by the method known as “sonication”. 14 The concentration of DNA extracted from the cultures was determined by spectrophotometry using the Eppendorf BioSpectrometer ® basic equipment (Hamburg, Germany).
The study was divided into three stages: standardisation of the qPCR technique with control strains; detection of NTM DNAs in lung samples using the standardised technique and sequencing of the positive samples.
qPCR standardisation from clinical isolates - A qPCR (TaqMan® hydrolysis linear probes) methodology was standardised for amplification of the 16S rRNA region 15 in the 7500 equipment (Applied Biosystems). For this, 11 DNAs from NTMs extracted from culture and confirmed by sequencing were used, as described in item C (Table I).
TABLE I. Comparison of results between culture, real-time polymerase chain reaction (qPCR) and sequencing tests for Non-tuberculous mycobacteria (NTMs).
| Sample nº | NTM culture | qPCR | Sequencing |
|---|---|---|---|
| 1 | M. kansasii | + | M. kansasii |
| 2 | M. fortuitum | + | M. fortuitum |
| 3 | M. szulgai | + | M. szulgai |
| 4 | M. malmoense | + | M. malmoense |
| 5 | M. abscessus | + | M. abscessus |
| 6 | M. marinum | + | M. marinum |
| 7 | M. peregrinum | + | M. peregrinum |
| 8 | M. chelonae | + | M. chelonae |
| 9 | M. gordonae | + | M. gordonae |
| 10 | M. chitae | + | M. chitae |
| 11 | M. avium | + | M. avium |
+: positive (16S target detection).
Primer efficiency was evaluated from a serial dilution using known amounts (100 ng) of M. avium DNA. The detection limit was defined using known concentrations of DNA from cultured M. avium and serially diluted, in ultrapure water, in factor 10 (16 ng to 0.016 fg) (Figure).
Graph of the analysis of the detection limit and evaluation of the technical efficiency (This figure has not been previously published in any journal).
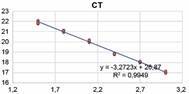
Assay validation was performed in triplicate with six mixtures of different concentrations, starting with primers and probes at 10 ng (oligonucleotide sequences are based on the study by Kim et al. 15 ). Analysing the amplification profile from the curve formed and based on the level of detectable fluorescence (threshold). The smallest amount of primers and probe with the best performance was considered as the standard for the reaction. 16
Detection of NTMs by qPCR in a lung sample - The qPCR for amplification of 16S rRNA from NTMs was performed as standardised, using DNAs extracted directly from sputum samples, by sonication, as described above. The positive control (PC) was a DNA from M. avium and the negative control (NC) was ultrapure water. After carrying out the assay using 16S rRNA as a target, the extracted DNAs were amplified by PCR using primers that amplify regions of the IS6110 to identify the presence of M. tuberculosis complex DNA or mixed infections. 17 The reactions used M. tuberculosis DNA from the reference strain H37Rv as PC and ultrapure water as NC. 18
The analysis of the amplification curve followed the predefined parameters (threshold and baseline). An evaluation of the methodology was performed by the amplification of DNAs from previously genotyped mycobacteria.
Molecular identification of NTMs species by sequencing - Molecular identification of NTMs in qPCR-positive samples was performed by sequencing the rpoB and hsp65 genes after PCR amplification with primers described by Telenti and coworkers. 19 PCR for the rpoB gene was performed with an initial denaturation step at 94ºC (5 min), proceeding with 40 cycles of denaturation at 94ºC (90 s), hybridisation at 65ºC (2 min), extension 72ºC (3 min) and a final extension cycle at 72ºC (10 min). The PCR for the hsp65 gene follows the model described above, changing to 45 cycles of denaturation at 94ºC (1 min), hybridisation at 60ºC (1 min) and 72ºC extension (1 min). The reactions were performed on the Applied Biosystems® Veriti® 96-Well ThermalCycler StepOne equipment. As controls, DNAs from NTMs previously identified by culture and confirmed in the sequencing were used (Table I) and as NC, ultrapure water. After checking the DNA amplifications on an agarose gel, the PCR amplicons were subjected to purification by polyethylene glycol (PEG) described by Rosenthal et al. 20 Cycling with the Big Dye® Kit version 3.1 (Applied Biosystems) was performed according to the manufacturer’s instructions. 21
The PCR products were labeled with 5 pmol of the TB11 primer (5’-ACCAACGATGGTGTGTGTCAT-3’, for the hsp65 gene) or with 5 pmol of the MycoF primer (5’-GGCAAGGTCACCCCGAAGGG0-3’, for the rpoB gene), together with 1 µL of reagent (BigDye Terminator v3.1 Cycle Sequencing Kit; Applied Biosystems), to 4.5 µL of purified PCR product in a final volume of 10 µL. Labeling reactions were performed in a 96-well thermal cycler (Veriti; AppliedBiosystems). The sequencing was performed in a genetic analyser from Applied Biosystems® 3130/3130xl Genetic Analysers (Data Collection Software 4).
The sequences obtained were analysed using the Laser gene SeqMan software (DNASTAR, Madison, USA), and aligned through the basic local alignment search tool (BLAST) with other sequences deposited in GenBank (National Centre for Biotechnology Information - NCBI - http://blast.ncbi.nlm.nih.gov). 8 The sequences were analysed considering the greatest coverage and identity of the species, with a mean coverage of 98.8%.
Statistical analysis - Statistical analysis was carried out using the statistical program Statistical Package for Social Sciences - SPSS v.21 (SPSS Ins. Chicago, IL, USA). The agreement between the tests, when performed with clinical isolates, was analysed by the Kappa test. 22
RESULTS
qPCR standardisation - The primers used in the assay for detection of NTMs (16S rRNA) showed an efficiency of 98% (0.988), as shown in Figure. The efficiency was calculated from the reaction slope of -3.35, which is the linear coefficient of the standard curve, indicating the regression coefficient of the curve, showing the efficiency of the reaction amplification. Reproducibility of duplicates (R2) was 0.998. 23
The qPCR detection limit was 160 pg of DNA of the NTM (M. avium) [corresponding to cycle threshold (CT) 30]. The standardisation of the primer concentrations in the validation was 0.4 ng and 0.2 ng for the probe, which showed the best performance.
The test reproducibility was confirmed by the DNA amplification pattern of the 11 NTMs used (Table I).
Detection of NTM DNA in lung samples - The 68 lung samples analysed consisted of: 54 from patients with suspected mycobacteriosis (NTM) and 14 from patients with TB (tested for acid-fast bacilli - AFB, M. tuberculosis culture and GeneXpert/RIF positive) used as negative controls to assess specificity.
In 28/54 (52%), the culture was positive for Mycobacterium (NTMs) that do not belong to the M. tuberculosis complex. Of these, 27/28 (96.4%) were positive in the qPCR test for NTMs with CT ranging from 12-17 (Table II). All 26 samples negative for NTMs culture and the 14 samples that were from patients with tuberculosis did not have qPCR amplification for NTM (sensitivity and specificity were 100% and 96%, respectively).
TABLE II. Results of positive clinical samples in one of the tests culture, acid-fast bacilli (AFB), and real-time polymerase chain reaction (qPCR) and sequencing.
| Sample nº | AFB | Culture | Threshold cycles | Sequencing | |
| NTM | M. tuberculosis | CT - PCR 16S | |||
| 1 | + | + | - | 14 | M. fortuitum |
| 2 | + | + | - | 16 | M. abscessus |
| 3 | + | + | - | 16 | M. abscessus |
| 4 | - | + | - | 13 | M. gordonae |
| 5 | + | + | - | 14 | M. abscessus |
| 6 | - | + | - | 14 | M. abscessus |
| 7 | - | + | - | 17 | M. gordonae |
| 8 | + | + | - | 16 | M. gordonae |
| 9 | + | + | - | 12 | M. avium |
| 10 | - | + | - | 16 | M. fortuitum |
| 11 | - | + | - | 18 | M. gordonae |
| 12 | - | + | - | 14 | M. fortuitum |
| 13 | + | + | - | 15 | M. fortuitum |
| 14 | + | + | - | 13 | M. abscessus |
| 15 | + | + | - | 14 | M. fortuitum |
| 16 | + | + | - | 12 | M. gordonae |
| 17 | - | + | - | 13 | M. avium |
| 18 | + | + | - | 15 | M. gordonae |
| 19 | + | + | - | 14 | M. avium |
| 20 | + | + | - | 15 | M. avium |
| 21 | + | + | - | 13 | M. gordonae |
| 22 | + | + | - | 17 | M. gordonae |
| 23 | + | + | - | 17 | M. fortuitum |
| 24 | - | + | - | 16 | M. abscessus |
| 25 | - | + | - | 13 | M.peregrinum |
| 26 | - | + | - | 17 | M. gordonae |
| 27 | - | + | - | 16 | M. abscessus |
| 28 | + | + | - | - | - |
+: positive; -: negative; CT: cycle threshold; NTM: non-tuberculous mycobacteria; PCR: polymerase chain reaction.
All 27 samples positive for the presence of NTM DNA in the qPCR were sequenced. The identified NTM species were M. gordonae in 9/27 (33%), M. abscessus in 7/27 (26%), M. fortuitum in 6/27 (22%), M. avium in 4/27 (15%) and M. peregrinum in 1/27 (4%) of the samples.
PCR results were also compared with smear slides tests. Eleven (40.7%) samples that were negative by sputum smear microscopy were positive by qPCR. All were positive in culture for NTM. Only one sample, with positive smear slide, did not show qPCR amplification.
The performance of qPCR, performed from DNA from lung samples, compared to culture, showed an almost perfect agreement (Kappa = 0.93), and with the AFB test the agreement was substantial (Kappa = 0.62).
DISCUSSION
Molecular methods for identifying mycobacterial species are more accurate and faster when compared to conventional methods. 8
The purpose of this study was to detect NTMs directly from lung samples from patients, using a qPCR with specific primers (16S rRNA) only for NTMs, thus being able to rapidly separate these from the M. tuberculosis complex. The 16S rRNA gene contains highly conserved and variable regions, being universally used to identify micobactéria. 18 , 24 In our study, these primers amplified DNA from several NTMs with an efficiency of 98% (90-100%), with a test detection limit of 160 pg for amplification of 100% of triplicates. Similar analytical sensitivity analyses were used in the study by Peixoto and his coworkers. 25
The standardised qPCR technique showed an almost perfect agreement with the culture (Kappa 0.93), which was confirmed by sequencing. Only one sample was positive for culture, but negative for qPCR and sequencing. In all the others, it was possible to detect DNA from NTMs. This negative sample had culture and positive AFB testing. In the hypothesis that it really is an NTM, it is believed that, possibly, this lack of amplification could be related to PCR inhibitors present in the sample, which were not totally removed in the DNA extraction process, 26 or loss of DNA during extraction, 27 since the targets (rpoB and hsp65) used in the sequencing also did not amplify.
Agreement with AFB testing was substantial (Kappa = 0.62), that is, 11 AFB negative samples were detected by PCR. Greater detection by qPCR was already expected, due to the need for a high bacterial load (around 10,000 bacilli per mL of sample) for sputum smear positivity, 28 unlike qPCR, where low DNA concentrations of NTMs could already be amplified. 16 , 29
When testing the 14 samples from TB positive patients, no amplification was detected, which may allow the use of qPCR to differentiate between TB and NTM. The presence of TB/NTM coinfection has been reported, 30 , 31 , 32 but it was not detected in this study.
These results suggest that the technique may be promising for detecting NTMs directly from lung samples, ruling out a diagnosis of TB and indicating the possibility of NTM infection/disease in the patient. The target used in this study (16S rRNA), together with the hsp65 gene, has been described for the identification and differentiation of NTMs from the M. tuberculosis complex. 16 , 33 , 34
Sequencing was used to identify species detected as NTMs. This is described by many authors as the molecular gold standard for the identification of mycobacteria, due to its high discriminatory capacity, allowing the characterisation of bacterial species. 35 , 36 The hsp65 and rpoB genes have been described for this purpose. 8 , 25 , 34
The most frequently identified NTM species in this study was M. gordonae (33%), being a common mycobacterial species. 37 M. gordonae is considered to be a low virulence strain that is also very associated with laboratory contamination; hence the importance of other factors to make the differential diagnosis of disease caused by NTM, thus reinforcing the need for careful procedures in order to differentiate contamination from infection, 4 , 38 since M. gordonae hardly causes disease. 37 The results of the study by Shin et al. 39 suggested that the clinical sensitivity of a test may be strains dependent.
Mycobacterium abscessus was the second (26%) most frequently identified bacteria, followed by M. fortuitum (22%) and M. avium (15%). These potentially pathogenic NTM species are among the most frequent related to lung diseases, both in southern Brazil and in other regions of the country. 4 , 8 , 40
The limitation found in the present study was the small number of samples analysed. Despite this, the results showed a possibility for the rapid identification of NTM, excluding a diagnosis of TB, especially in patients with symptoms and suspicion of TB, facilitating the beginning of early treatment and contributing to reduce the transmission of this disease.
In conclusion - The results suggest that the application of the qPCR technique to detect NTMs using 16S rRNA as a target has the potential to detect NTMs and differentiate them, quickly and efficiently, from M. tuberculosis. However, it is still necessary to identify the species to assist in the differential diagnosis between disease and contamination, guiding the choice of the therapeutic scheme.
ACKNOWLEDGEMENTS
To the Postgraduate Program in Cellular and Molecular Biology Applied to Health - Lutheran University of Brazil (ULBRA), Institute of Biological Sciences of Universidade Federal do Rio Grande (Rio Grande, RS, Brazil) and Centre for Scientific and Technological Development (CDCT-RS).
REFERENCES
- 1.Cowman S, Van JI, Griffith D, Loebinger MR. Non-tuberculous mycobacterial pulmonary disease. Eur Respir J. 2019;54(1):1–23. doi: 10.1183/13993003.00250-2019. [DOI] [PubMed] [Google Scholar]
- 2.Falkinham JO. Nontuberculous mycobacteria in the environment. Clin Chest Med. 2002;23(3):529–551. doi: 10.1016/s0272-5231(02)00014-x. [DOI] [PubMed] [Google Scholar]
- 3.Fernandes C, Zárate-Bladés CR. Infecções por micobactérias não tuberculosas no Hospital Nereu Ramos, centro de referência em Santa Catarina. Bol Curso Med UFSC. 2019;5(5):88–90. [Google Scholar]
- 4.Carneiro MDS, Nunes LS, David SMM, Dias CF, Barth AL, Unis G. Nontuberculous mycobacterial lung disease in a high tuberculosis incidence setting in Brazil. J Bras Pneumol. 2018;44(2):106–111. doi: 10.1590/S1806-37562017000000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nunes-Costa D, Alarico S, Dalcolmo MP, Correia-Neves M, Empadinhas N. The looming tide of nontuberculous mycobacterial infections in Portugal and Brazil. Tuberculosis (Edinb) 2016;96:107–119. doi: 10.1016/j.tube.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 6.de Mello KG, Mello FC, Borga L, Rolla V, Duarte RS, Sampaio EP. Clinical and therapeutic features of pulmonary nontuberculous mycobacterial disease, Brazil, 1993-2011. Emerg Infect Dis. 2013;19(3):393–399. doi: 10.3201/eid1903.120735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO Global tuberculosis report 2019. World Health Organization. 2020 https://apps.who.int/iris/bitstream/handle/10665/329368/9789241565714-eng.pdf?ua=1 [Google Scholar]
- 8.Busatto C, Vianna JS, Silva ABS, Basso R, Silveira J, Groll AV. Nontuberculous mycobacteria in patients with suspected tuberculosis and the genetic diversity of Mycobacterium avium in the extreme south of Brazil. J Bras Pneumol. 2020;46(2):e20190184. doi: 10.36416/1806-3756/e20190184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daley CL, Iaccarino JM, Lange C, Cambau E, Wallace RJ, Jr, Andrejak C. Treatment of nontuberculous mycobacterial pulmonary disease an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Eur Respir J. 2020;56(1):2000535–2000535. doi: 10.1183/13993003.00535-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasperbauer SH, De Groote MA. The treatment of rapidly growing mycobacterial infections. Clin Chest Med. 2015;36(1):67–78. doi: 10.1016/j.ccm.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 11.MS - Ministério da Saúde . Manual nacional de vigilância laboratorial da tuberculose e outras micobactérias. Brasília: Ministério da Saúde; 2008. [Google Scholar]
- 12.Hain Lifesciences . GenoType(r) Mycobacterium CM. 2015. [Google Scholar]
- 13.MS Teste rápido molecular para tuberculose (TRM-TB). Nova tecnologia para o diagnóstico da tuberculose. Ministério da Saúde. 2016 http://www.saude.gov.br/images/pdf/2016/maio/18/folder-TRM-TB-grafica-reduzido.pdf [Google Scholar]
- 14.Bello GL, Morais FCL, Wolf JM, Gehlen M, Soares TDS, Halon ML. Improvement of Mycobacterium tuberculosis detection in sputum using DNA extracted by sonication. Braz J Infect Dis. 2020;24(5):398–404. doi: 10.1016/j.bjid.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JU, Cha CH, An HK. Direct identification of mycobacteria from clinical specimens by multiplex real-time PCR. J Appl Microbiol. 2015;118(6):1498–1506. doi: 10.1111/jam.12780. [DOI] [PubMed] [Google Scholar]
- 16.Kim JU, Ryu DS, Cha CH, Park SH. Paradigm for diagnosing mycobacterial disease direct detection and differentiation of Mycobacterium tuberculosis complex and non-tuberculous mycobacteria in clinical specimens using multiplex real-time PCR. J Clin Pathol. 2018;71(9):774–780. doi: 10.1136/jclinpath-2017-204945. [DOI] [PubMed] [Google Scholar]
- 17.Michelon CT, Rosso F, Schmid KB, Sperhacke RD, Oliveira MM, Kritski AL. Colorimetric microwell plate reverse-hybridization assay for Mycobacterium tuberculosis detection. Mem Inst Oswaldo Cruz. 2011;106(2):194–199. doi: 10.1590/s0074-02762011000200013. [DOI] [PubMed] [Google Scholar]
- 18.Jagielski T, Minias A, van Ingen J, Rastogi N, Brzostek A, Zaczek A. Methodological and clinical aspects of the molecular epidemiology of Mycobacterium tuberculosis and other mycobacteria. Clin Microbiol Rev. 2016;29(2):239–290. doi: 10.1128/CMR.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Telenti A, Marchesi F, Balz M, Bally F, Böttger EC, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31(2):175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenthal A, Coutelle O, Craxton M. Large-scale production of DNA sequencing templates by microtitre format PCR. Nucleic Acids Res. 1993;21(1):173–174. doi: 10.1093/nar/21.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barros MCS. Produção, caracterização e purificação de colagenases. Recife: Universidade Federal de Pernambuco; 2011. [Google Scholar]
- 22.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–174. [PubMed] [Google Scholar]
- 23.MA - Ministério da Agricultura . Manual de verificação de desempenho de métodos para diagnóstico molecular de doenças infecciosas na rede nacional de laboratórios agropecuários. Brasília: Ministério da Agricultura; 2015. [Google Scholar]
- 24.Soini H, Musser JM. Molecular diagnosis of mycobacteria. Clin Chem. 2001;47(5):809–814. [PubMed] [Google Scholar]
- 25.Peixoto ADS, Montenegro LML, Lima AS, Melo FL, Barbosa WL, Jr, Neves MMC. Identification of nontuberculous mycobacteria species by multiplex real-time PCR with high-resolution melting. Rev Soc Bras Med Trop. 2020;53:e20200211. doi: 10.1590/0037-8682-0211-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casaril AE, Oliveira LP, Alonso DP, Oliveira EF, Barrios SPG, Infran JOM. Standardization of DNA extraction from sand flies application to genotyping by next generation sequencing. Exp Parasitol. 2017;177:66–72. doi: 10.1016/j.exppara.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 27.Gómez AV, González JM, García BAL. Xpert(r) MTB/RIF utilidade nel diagnóstico de la tuberculosis y de la resistencia a la rifampicina. Med Clin (Barc) 2017;1(1):1–7. [Google Scholar]
- 28.Costa RR, Silva MR, Gonçalves IC. Laboratory diagnosis of tuberculosis literature review. Rev Med Minas Gerais. 2018;28(5):197–206. [Google Scholar]
- 29.Calsolari RAO. Diagnóstico laboratorial de Mycobacterium spp. em Botucatu e região, utilizando a técnica Reação em Cadeia da Polimerase (PCR) em material biológico e avaliação de condições associadas [PhD Thesis] Faculdade de Medicina de Botucatu. 2016 [Google Scholar]
- 30.Andrade LK, Belotti NCU, Cury MRCO, Souza AR, Pedro HSP, Chimara E. Mycobacterium tuberculosis e micobactérias não tuberculose em paciente com síndrome da imunodeficiência adquirida e câncer relato de caso. Arq Cienc. 2018;22(1):49–53. [Google Scholar]
- 31.Hoza AS, Mfinanga SG, Rodloff AC, Moser I, König B. Increased isolation of nontuberculous mycobacteria among TB suspects in Northeastern, Tanzania public health and diagnostic implications for control programmes. BMC Res Notes. 2016;9:109–109. doi: 10.1186/s13104-016-1928-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Lima CAM, Gomes HM, Oelemann MAC, Ramos JP, Caldas PC, Campos CED. Nontuberculous mycobacteria in respiratory samples from patients with pulmonary tuberculosis in the state of Rondônia, Brazil. Mem Inst Oswaldo Cruz. 2013;108(4):457–462. doi: 10.1590/0074-0276108042013010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim K, Lee H, Lee MK, Lee SA, Shim TS, Lim SY. Development and application of multiprobe real-time PCR method targeting the hsp65 gene for differentiation of Mycobacterium species from isolates and sputum specimens. J Clin Microbiol. 2010;48(9):3073–3080. doi: 10.1128/JCM.00939-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poroca DR, Lima AS, Lima JF, Cruz HL, Montenegro RA, Melo FL. Diferenciação de micobactérias por PCR multiplex [Differentiation of micobacteria by multiplex PCR] Rev Soc Bras Med. Trop. 2009;42(6):716–722. doi: 10.1590/s0037-86822009000600020. [DOI] [PubMed] [Google Scholar]
- 35.Cerca PAR. Identificação de micobactérias não tuberculosas através de métodos moleculares não comerciais. Lisboa: Universidade Nova de Lisboa; 2010. [Google Scholar]
- 36.Xu Y, Liang B, Du C, Tian X, Cai X, Hou Y. Rapid identification of clinically relevant Mycobacterium species by multicolor melting curve analysis. J Clin Microbiol. 2019;57(1):e01096–e01018. doi: 10.1128/JCM.01096-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazumder SA, Hicks A, Norwood J. Mycobacterium gordonae pulmonary infection in an immunocompetent adult. N Am J Med Sci. 2010;2(4):205–207. [PMC free article] [PubMed] [Google Scholar]
- 38.ANVISA Nota Técnica Conjunta Nº 01/2009. Infecções por micobactérias de crescimento rápido: fluxo de notificações, diagnósticos clínico, microbiógicos e tratamento. Agência Nacional de Vigilância Sanitária. 2009 [Google Scholar]
- 39.Shin S, Yoo IY, Shim HJ, Kang OK, Jhun BW, Koh WJ. Diagnostic performance of the GENEDIA MTB/NTM detection Kit for detecting Mycobacterium tuberculosis and nontuberculous mycobacteria with sputum specimens. Ann Lab Med. 2020;40(2):169–173. doi: 10.3343/alm.2020.40.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ueki SYM, Martins MC, Telles MAS, Virgilio MC, Giampaglia CMS, Chimara E. Micobactérias não-tuberculosas diversidade das espécies no estado de São Paulo. J Bras Patol Med Lab. 2005;41(1):1–8. [Google Scholar]


