Abstract
Trichoderma harzianum secretes α-1,3-glucanases when it is grown on polysaccharides, fungal cell walls, or autoclaved mycelium as a carbon source (simulated antagonistic conditions). We have purified and characterized one of these enzymes, named AGN13.1. The enzyme was monomeric and slightly basic. AGN13.1 was an exo-type α-1,3-glucanase and showed lytic and antifungal activity against fungal plant pathogens. Northern and Western analyses indicated that AGN13.1 is induced by conditions that simulated antagonism. We propose that AGN13.1 contributes to the antagonistic response of T. harzianum.
Biological control by antagonistic organisms is a potential nonchemical tool for crop protection against phytopathogenic fungi (33). Several strains from the genus Trichoderma have been described as antagonistic fungi able to control a wide range of phytopathogenic fungi. The antifungal activity of Trichoderma involves production of antibiotics, including compounds affecting the integrity of fungal membranes, competition for key nutrients, and production of fungal cell wall-degrading enzymes (CWDEs) (14, 23). Although none of these mechanisms have been convincingly proven, the degradation and further assimilation of fungal structures and contents have been proposed as the major mechanism accounting for the antagonistic process against fungal plant pathogens (7).
A number of Trichoderma isolates produce a wide variety of CWDEs, among them, chitinases, β-1,3- and β-1,6-glucanases, and proteases, when grown on polysaccharides, fungal cell walls, or autoclaved mycelium as a carbon source (4, 10, 29). These conditions have been described as “simulated antagonism” (13, 14, 30, 45). These observations, together with the fact that chitin and β-1,3-glucan are the main skeletal polysaccharides of fungal cell walls (except those from Oomycetes, which contain β-glucans and cellulose) (3, 34), suggest that chitinases and β-1,3-glucanases act as the key enzymes in the lysis of phytopathogenic fungal cell walls during the antagonistic action of Trichoderma. However, other CWDEs, including those hydrolyzing minor polymers (i.e., β-1,6-glucans or proteins), may also be involved in the antagonistic response of Trichoderma (10, 11, 18).
Polysaccharides, which consist of α-glycosidic linkages, also appear to be important in the cell wall composition and architecture. The so-called alkali-soluble glucans (S-glucans) range from polymers containing nearly pure α-1,3-linkages to polymers regularly alternating α-1,3- and α-1,4-linkages (2, 3). S-glucans represent the major matrix polysaccharides for most fungi (3, 34); in some instances, like in Aspergillus nidulans, S-glucans account for approximately 25% of the dry weight of the cell wall (47). Despite the importance of S-glucans, there have been few reports describing the presence of crude enzyme activities capable of hydrolyzing this polysaccharide, and there is almost no biochemical and molecular information on α-1,3-glucanases (17, 19, 22, 41, 43, 49).
Different applications have been envisaged for antifungal CWDEs and their genes from Trichoderma strains, ranging from the improvement of biocontrol agents (4, 16, 28) to their use as heterologous genes for plant resistance against phytopathogenic fungi (31). In the search for new CWDEs, no attention has been devoted to α-1,3-glucanases to date.
In this article, we report on the purification and molecular characterization of an exo-α-1,3-glucanase (glucan 1,3-α-glucosidase [EC 3.2.1.84]), namely AGN13.1, from the antagonistic fungus T. harzianum. During the progress of this work, AGN13.1 from T. harzianum was identified independently as an enzyme able to degrade the mutan, an extracellular α-1,3-glucan produced by tooth-colonizing streptococci (17). Herein, we show that the expression of the gene and the enzyme secretion occur when T. harzianum grows under conditions that simulate antagonism. The enzyme is able to degrade cell walls of some phytopathogenic fungi. Antifungal assays reinforce the possible role of AGN13.1 during the antagonistic action of T. harzianum.
Microorganisms and culture conditions.
T. harzianum CECT 2413 was obtained from the Spanish Type Culture Collection (Burjasot, Valencia, Spain) and maintained on potato-dextrose-agar medium (Difco, Detroit Mich.). For enzyme production, this strain was grown in two-step liquid cultures as exactly described in reference 13.
To obtain cell walls, other fungi were also purchased from CECT: Aspergillus niger CECT 2574, Botrytys cinerea CECT 2100, Colletotrichum acutatum 74, Fusarium oxysporum, Penicillium aurantiogriseum IMI 374515, Phytophthora syringae CECT 2351, Rhizoctonia solani CECT 2815, Schizosaccharomyces pombe and Saccharomyces cerevisiae (La Cinta Roja, Spain). S-glucan from A. niger was prepared as described previously (22, 47). The composition of this S-glucan was determined by infrared (2) and 13C nuclear magnetic resonance (13C-NMR) spectra (5, 44), resulting in a polymer with nearly pure α-1,3 linkages. Mutan from Streptococcus mutans (an α-1,3-glucan with some α-1,6-glucan side chains) was prepared exactly as described previously (17). Fungal cell walls and β-glucan from S. cerevisiae were prepared as previously described (15, 37).
α-1,3-Glucanase assay and protein determination.
α-1,3-Glucanase activity was routinely assayed by incubating 0.2 ml of 5 mg of S-glucan per ml from A. niger (α-1,3-glucan) in 50 mM potassium acetate buffer (pH 5.5) (buffer A) with 50 μl of enzyme solution appropriately diluted in the same buffer. Reaction mixtures were incubated at 37°C for 30 min and were stopped by boiling for 5 min. Samples were centrifuged (5,000 × g, 5 min), 0.15 ml of supernatant per reaction was taken, and reducing sugars were determined (32, 40), with glucose as a standard. Enzyme and substrate blanks were included. One unit of α-1,3-glucanase activity was defined as the amount of enzyme that releases 1 μmol of reducing sugar equivalents (expressed as glucose) per min under the standard assay conditions. Protein concentration was determined by the method of Bradford (6), with ovalbumin as a standard.
Enzyme production and purification.
T. harzianum was grown on rich medium and transferred to fresh minimal medium supplemented with different carbon sources (induced conditions). Enzyme production was determined in filtrates from T. harzianum transferred to the induced conditions. As reported for other CWDEs (13, 46), we found that the α-1,3-glucanase activity is repressed at a high glucose concentration (2 to 10% glucose, 50 mU/mg of protein) and produced under carbon starvation conditions (0.1% glucose, 100 mU/mg of protein) and under induced conditions with 0.5% (wt/vol) fungal cell walls or their polymers (100 to 400 mU/mg of protein). We conclude that production of α-1,3-glucanase by T. harzianum is dependent on the carbon source available.
Three consecutive steps were used for the purification of AGN13.1. (i) T. harzianum cultures were grown as described above and transferred for 48 h to minimal medium with 0.5% A. niger cell walls, filtrated through Whatman no. 1 paper, and centrifuged (8,000 × g, 10 min). The supernatant (about 600 ml) was precipitated with solid ammonium sulfate (80% saturation), and the pellet was recovered by centrifugation (12,000 × g, 20 min), resuspended in distilled water, and dialyzed against 50 mM buffer A. The dialyzed fraction was centrifuged (12,000 × g, 20 min), and the supernatant (about 18 ml) was recovered. (ii) Aliquots of 2 ml of this sample were adsorbed to S-glucan (5 mg/ml) for 2 h with magnetic stirring, and the pellet was collected by centrifugation at 12,000 × g for 10 min. Adsorbed fractions were washed three times with 50 mM buffer A containing 1 M NaCl, resuspended in 50 mM buffer A with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 mM sodium azide, and incubated overnight at 37°C for S-glucan digestion. (iii) Aliquots of 0.5 ml of S-glucan digestion were repeatedly applied to a PAK125 gel filtration column (7.8 mm by 30 cm; Millipore) equilibrated with 100 mM buffer A containing 0.5 M NaCl, and eluted with the same buffer at a flow rate of 0.2 ml/min. Fractions of 0.2 ml were collected and monitored for protein (A280) and for α-1,3-glucanase activity. Most active fractions were pooled, washed in 100 mM buffer A, and concentrated to approximately 1 ml on Centricon 10 concentrators (Amicon, Beverley, Mass.). The purified protein was stored in 100 mM buffer A at −20°C.
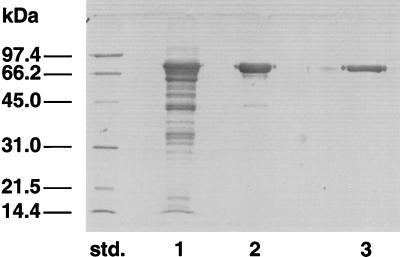
Following this procedure, AGN13.1 was purified sevenfold with a minimal estimated recovery of approximately 10%. As shown in Fig. 1, the final purified preparation migrated as a single band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE [12% polyacrylamide]) with an apparent molecular mass of 72 kDa after Coomassie staining (27). When the molecular mass was calculated by gel filtration, the value obtained was 67 kDa (data not shown). We conclude that this procedure achieved purification of AGN13.1 to apparent electrophoretic homogeneity and that the enzyme is a monomeric protein.
FIG. 1.
Purification of AGN13.1 from T. harzianum. Samples containing α-1,3-glucanase from each purification step were analyzed by SDS-PAGE. Lanes: std., molecular mass standards; 1, ammonium sulfate precipitation from a culture filtrate from T. harzianum grown on 0.5% A. niger cell walls (10 μg of protein); 2, adsorption and digestion of S-glucan (5 μg of protein); 3, gel filtration pool (5 μg of protein).
Biochemical properties.
The pIs of the pure enzyme, determined by isoelectrofocusing (36) and basic chromatofocusing (11), were estimated to be 6.7 and 7.5, respectively (data not shown). The Michaelis-Menten constant resulted in Kms of 1.76 mg/ml (S-glucan) and 1.69 mg/ml (mutan). The optimal temperature for the enzyme was 55°C at pH 5.5. After 30 min of preincubation time at different temperatures, the inactivation temperature was calculated as 50°C. Therefore, S-glucan seems to stabilize the enzyme.
Substrate specificity and action pattern.
The activity of AGN13.1 was tested on different polymers with α- or β-glycosidic bonds with the standard assay described above. Chitinase and protease activities were determined as previously described (9, 39). As shown in Table 1, the maximal activity was detected against mutan and S-glucan, which are linear α-1,3-glucans (17) (and data not shown). No activity was detected with nigeran as substrate, which is a linear polyglucan containing alternating α-1,3 and α-1,4 bonds (34) or against other α- or β-glucans or chitin. Furthermore, the enzyme preparation lacks protease activity (data not shown). We conclude that the AGN13.1 enzyme is a specific α-1,3-glucanase.
TABLE 1.
Substrate specificity of purified AGN13.1 from T. harzianum
| Substratea | Main linkage type (monomer)b | α-1,3-Glucanase activity (%)c |
|---|---|---|
| Non-cell wall | ||
| S-glucan (A. niger) | α-1,3 (Glc) | 100 |
| IO4− S-glucan | α-1,3 (Glc) | <1 |
| Mutan | α-1,3 (Glc) | 142 |
| Nigeran | α-1,3:α-1,4 (Glc) | 0 |
| Soluble starch | α-1,4:α-1,6 (Glc) | 0 |
| Dextran | α-1,6 (Glc) | 0 |
| Pachyman | β-1,3 (Glc) | 0 |
| Glucan (S. cerevisiae) | β-1,3:β-1,6 (Glc) | 0 |
| Laminaran | β-1,3:β-1,6 (Glc) | <1 |
| Carboxymethyl cellulose | β-1,4 (Glc) | 0 |
| Pustulan | β-1,6 (Glc) | 0 |
| Colloidal chitin | β-1,4 (GlcNAc) | 0 |
| Cell walls | ||
| A. niger | β-Glucan:chitin | 83 |
| P. aurantogriseum | β-Glucan:chitin | 71 |
| B. cinerea | β-Glucan:chitin | 59 |
| C. acutatum | β-Glucan:chitin | 13 |
| F. oxysporum | β-Glucan:chitin | 10 |
| R. solani | β-Glucan:chitin | 0 |
| P. syringae | β-Glucan:cellulose | 0 |
| S. cerevisiae | β-Glucan | 0 |
| S. pombe | β-Glucan | 0 |
All substrates were used at a final concentration of 4 mg/ml.
α-Glucans have been reported as matrix components of at least the cell walls of A. niger and Penicillium sp. (3, 34).
An activity level of 100% corresponds to 3.5 U/mg of protein. Each value is relative to the mean of four independent experiments, with a relative standard deviation of less than 1%.
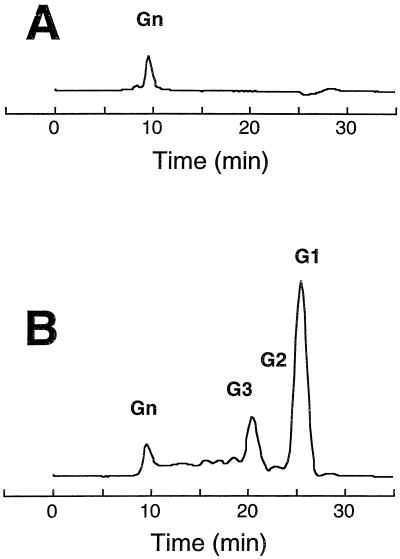
The action pattern of AGN13.1 toward S-glucan was examined by comparing the rate of glucose production to the rate of reducing sugar production at different times, comparing the activities of the purified enzyme against S-glucan and periodate-oxidized S-glucan (unable to be hydrolyzed by exo-α-1,3-glucanases), and finally analyzing the corresponding hydrolysis products by high-performance liquid chromatography (HPLC). The reaction products were separated on an Aminex HPX 42-A column (Bio-Rad) maintained at 40°C. Water was used as an eluant at a flow rate of 0.4 ml/min. Products were detected on the basis of their refraction index and identified by comparison with glucose and cellulose oligosaccharide. Incubations of S-glucan with the enzyme resulted in a 1:1 correlation between the production of reducing sugars and glucose (data not shown). Therefore, glucose accounts for almost all of the reducing sugar and is the major hydrolysis product of the enzyme. AGN13.1 was unable to hydrolyze periodate-oxidized S-glucan (Table 1). By HPLC analysis, glucose was detected at the initial stage of the enzymatic reaction (data not shown). At longer time points, the peak of glucose increased, since glucose was the final product of hydrolysis (Fig. 2). Identical results were obtained when mutan was used as the substrate (data not shown). Altogether, these results indicate an exo-type mode of action for the purified AGN13.1 enzyme.
FIG. 2.
HPLC profiles of the reaction products of AGN13.1 acting on S-glucan. S-glucan (4 mg/ml) was incubated for 14 h at 37°C in the absence of enzyme (A) or with 2 μg of the purified enzyme (B). In the abbreviation “Gn,” n represents the degree of polymerization.
Antifungal properties.
AGN13.1 was assayed for fungal cell wall binding and degrading activity as previously described (13). The enzyme was able to bind to cell walls of various phytopathogenic fungi, such as A. niger, B. cinerea, C. acutatum, F. oxysporum, P. aurantiogriseum, or R. solani, but not those of P. syringae, S. cerevisiae, or S. pombe (data not shown). In addition, the enzyme alone was quite active against cell walls from A. niger, P. aurantiogriseum, B. cinerea, C. acutatum, and F. oxysporum (in descending order of efficacy), but showed no activity against cell walls from R. solani, P. syringae, S. cerevisiae or S. pombe (Table 1). Moreover, when the mode of action of AGN13.1 toward cell walls from A. niger or P. aurantiogriseum was studied, an exo-type mechanism was detected, with glucose as the major final product of hydrolysis (data not shown). We conclude that AGN13.1 acts as a lytic enzyme against phytopathogenic fungal cell walls, which may therefore contain α-1,3-glucans.
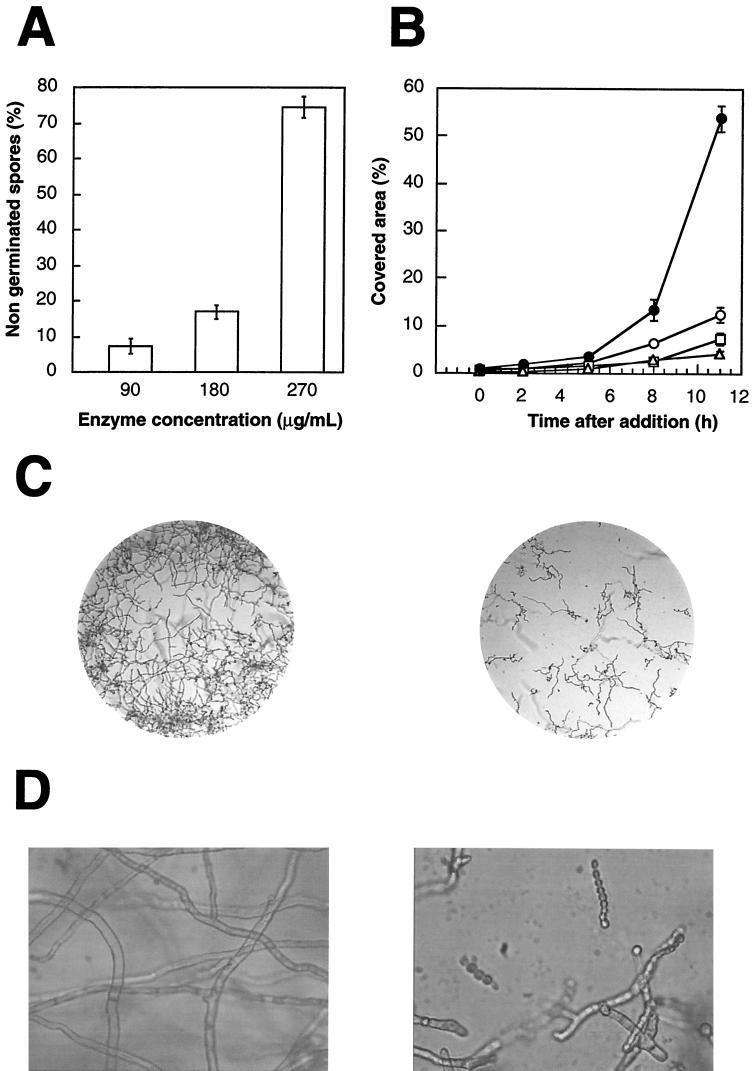
Antifungal activity for conidia or hyphae from A. niger, B. cinerea, and P. aurantiogriseum was also detected. Antifungal assays were performed in microtiter plates (Nunclon Microwell Minitray; 60 by 80 mm) at 24°C. Each microwell contained 1 μl of 5× potato dextrose broth, 2 μl of a spore suspension of different phytopathogenic fungi (60 total spores), and 13 μl of distilled water (control) or purified enzyme in distilled water. To determine the effects of the enzyme on fungal germination, the enzyme solution was added simultaneously to the spore suspension. At different times, the number of germinated spores was counted and compared to that of the control. To determine the effects of the enzyme on hyphal growth, the enzyme solution was added after most spores were germinated. At different times, microscopic observations of the microwells were done and compared to those of the control. In this case, hyphal growth was calculated automatically as a percentage of the covered area of the microwells (Microimage program 3.0; Windows). The results represent the mean of three replicates. Our results indicate that although germination was slightly affected when A. niger and B. cinerea were used, around 70% inhibition of P. aurantiogriseum spore germination was determined in the presence of the enzyme at concentrations around 270 μg/ml (Fig. 3A). However, growth was efficiently inhibited after the addition of AGN13.1 to germinated spores from the three fungi, at concentrations around and greater than 90 μg/ml (data not shown) (Fig. 3B and C). Moreover, aberrant morphology of hyphal tips was observed under the light microscope, as shown for A. niger in Fig. 3D.
FIG. 3.
Antifungal properties of AGN13.1. (A) Inhibition of spore germination of P. aurantiogriseum by AGN13.1 at different concentrations. (B) Inhibition of hyphal growth of A. niger by AGN13.1 at different concentrations: solid circles, 0 μg/ml; open circles, 90 μg/ml; open squares, 180 μg/ml; open triangles, 270 μg/ml. In panels A and B, error bars indicate standard deviations. The experiment was performed three times with similar results. (C) Photographs of A. niger under the light microscope growing in the absence (left panel) or presence (right panel) of 270 μg of AGN13.1 per ml. Magnification, 100-fold. (D) Detail of the photographs shown in panel C at 400-fold magnification. As indicated above, the left panel corresponds to the control without AGN13.1, and the right panel corresponds to 270 μg of AGN13.1 per ml.
Expression pattern of agn13.1 mRNA and AGN13.1 protein.
We have determined that the N-terminal amino acid sequence of the purified AGN13.1 is ASSADRLVFSHFMIGIVGD. Based on this sequence, we have cloned a complete agn13.1 cDNA (M. Rey, unpublished results). The nucleotide sequence of the full agn13.1 cDNA was identical to that of a previously described cDNA coding for a mutanase (GenBank/EBI accession no. AF214480) (17). Northern and Western blot analyses were not covered by Fuglsang et al. (17), but they are pertinent to establish whether the induction of agn13.1 preferentially occurs under conditions similar to those involving the antagonistic action by T. harzianum.
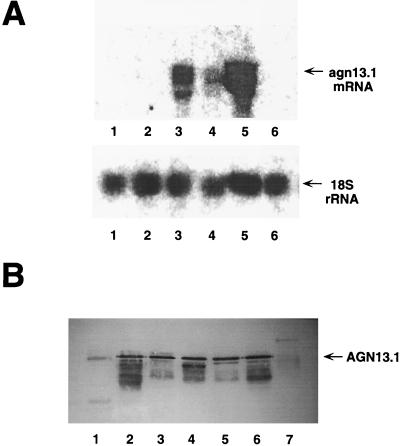
T. harzianum was grown on rich medium and transferred to fresh minimal medium supplemented with different carbon sources. Samples of mycelia were then collected at different times, and RNA was extracted and subjected to Northern blot analyses according to standard procedures (8, 38). The radioactivity of the bands was quantified by using a Cyclone device with OptiQuant software (Packard Instrument Co.). Expression levels were normalized against the signal obtained by hybridizing the blots with 18S radish ribosomal DNA (rDNA). As shown in Fig. 4A, the agn13.1 mRNA is detectable as early as 9 h after transfer to chitin or fungal cell walls (Fig. 4A, lanes 3 to 5). Other lower and less intense bands were also detected, but they might correspond to partial degradation of the full-length agn13.1 mRNA. Densitometric analyses of the Northern blots indicated that the agn13.1 mRNA accumulated at its highest levels when A. niger cell walls were used as an inducer (relative intensity, 0.5), instead of chitin (relative intensity, 0.25) or P. syringae cell walls (relative intensity, 0.15). Other fungal cell walls induced agn13.1 expression to intermediate levels between those from A. niger and P. syringae cell walls (data not shown). No expression was found at this time on glucose-supplemented media and with carbon or nitrogen starvation (Fig. 4A, lanes 1, 2, and 6). However, when the expression was investigated at longer time points, agn13.1 mRNA began to accumulate in mycelia transferred to carbon starvation conditions (relative intensity, 0.10) (data not shown).
FIG. 4.
Expression pattern of agn13.1. (A) Northern blot analysis. Total RNA was extracted from cultures grown for 9 h on 2% glucose (lane 1), 0.1% glucose (lane 2), 1.5% chitin (lane 3), 0.5% P. syringae cell walls (lane 4), 0.5% A. niger cell walls (lane 5), and with nitrogen starvation (lane 6). Total RNA (50 μg) was then subjected to electrophoresis on an agarose gel under denaturing conditions, transferred to a nylon membrane, and hybridized against a 1.9-kb probe of the coding sequence of agn13.1 (upper panel) or 18S rDNA (bottom panel). (B) Western blot analysis. Filtrates were obtained from cultures grown for 48 h on 2% glucose (lane 1), 0.1% glucose (lane 2), 1.5% chitin (lane 3), 0.5% P. syringae cell walls (lane 4), 0.5% P. aurantiogriseum cell walls (lane 5), 0.5% A. niger cell walls (lane 6), and with nitrogen starvation (lane 7). Samples (25 μg) were subjected to SDS-PAGE on a 12% polyacrylamide gel, and protein was transferred to a nitrocellulose membrane and probed with specific rabbit polyclonal AGN13.1 antibodies (dilution 1:1,000). AGN13.1 was visualized with peroxidase-conjugated goat anti-rabbit secondary antibodies (dilution of 1:1,000).
The steady-state levels of extracellular AGN13.1 protein were measured by Western blot analyses in culture filtrates from cultures growing in the presence of different carbon sources. Western blots were done according to standard procedures (1) with rabbit polyclonal antibodies raised against the purified AGN13.1 protein. Figure 4B shows that AGN13.1 is present at almost similar levels in filtrates from T. harzianum grown for 48 h on chitin and several fungal cell walls. Some protein was detected in filtrates from T. harzianum transferred to carbon starvation conditions. In all of these cases, increased α-1,3-glucanase activity in filtrates correlates with the presence of the AGN13.1 protein. However, no signal at an AGN13.1 position, determined with purified preparations, was detected in samples of T. harzianum grown on 2% glucose or under nitrogen starvation conditions. Therefore, AGN13.1 did not account for the basal extracellular activity found under these conditions (data not shown) (Fig. 4B, lanes 1 and 7). Other minor bands were also detected, but they might correspond to partial degradation of the full-length AGN13.1 protein and/or unspecific binding of the primary antibody. We conclude that agn13.1 expression is mainly triggered at the transcriptional level by products derived from fungal cell walls or isolated polymers such as chitin.
Conclusions.
T. harzianum antagonizes a large variety of plant pathogenic fungi responsible for major crop diseases (7, 33). Among different mechanisms for antagonism, T. harzianum is able to produce lytic enzymes for the degradation of the fungal cell wall to further assimilate the intracellular contents of its hosts (4, 7). Physically, the fungal cell wall is a fabric of interwoven microfibrils embedded in or cemented by amorphous matrix substances (3). In most fungi, chitin and non-cellulosic β-glucans are the most abundant skeletal or microfibrilar components, while proteins and α-glucans are the main cementing components (34). A considerable amount of recent research has been devoted to study of the lytic systems produced by T. harzianum, including chitinases (9, 21, 30), β-glucanases (4, 11, 12), and proteases (18), and the relative importance of any of these systems in the antagonistic process. However, the α-glucanolytic system has neither been biochemically well characterized nor tested for its antifungal potential.
In this paper, we have described the induction of α-1,3-glucanase activity in filtrates of T. harzianum growing on simulated antagonistic conditions. Moreover, we have purified and biochemically characterized an α-1,3-glucanase protein, AGN13.1. The purification method was based on the strong affinity of AGN13.1 to insoluble S-glucan from A. niger cell walls. After gel filtration, the enzyme was recovered to apparent electrophoretic homogeneity. The yield of the purification was only 10%; this apparent low yield can be explained by the overestimation of the α-1,3-glucanase activity in crude preparations, perhaps due to synergistic effects of AGN13.1 with other α-1,3-glucanolytic enzymes present in the filtrates. The enzyme was purified only seven times with respect to the crude filtrates. This result indicates that AGN13.1 is an abundant protein in the filtrates of T. harzianum growing on A. niger cell walls, as demonstrated by visualizing the profiles of protein secreted by T. harzianum under these conditions (Fig. 1, lane 1). The purified protein has a molecular mass of 72 kDa, as calculated by SDS-PAGE. This value was 67 kDa according to gel filtration, strongly suggesting a native monomeric form, as previously reported for a large variety of extracellular CWDEs from T. harzianum (4, 9, 25, 42). These data are not too disparate from the predicted molecular mass for the mature protein (ca. 63.8 kDa) and data published by Fuglsang and coworkers (17). The optimal and inactivation temperatures (both around 50°C) were similar to those found for other CWDEs from T. harzianum and other fungi. The Km values for S-glucan and mutan (1.76 and 1.69 mg/ml, respectively) were high, but these values are common among enzymes attacking insoluble substrates. The Km values are in the range calculated for other bacterial and fungal α-1,3-glucanases (17, 19, 22, 41, 43, 49).
The purified AGN13.1 enzyme was found to be highly specific for α-1,3-linkages in polysaccharides, hydrolyzing S-glucan and mutan very efficiently. Our experiments show unequivocally that AGN13.1 exhibits an exo type of action, with glucose as the main early and final hydrolytic product. Oligosaccharides containing two or three residues of glucose were also detected as final reaction products by HPLC. This result strongly suggests that AGN13.1 cannot recognize and/or split α-1,3-glucans with a degree of polymerization less than 3. Both, the high specificity for α-1,3-linkages and the exo-type mechanism explain the lack of activity against α-1,3-α-1,4-glucans as nigeran.
Finally, our results strongly support that AGN13.1 plays an important role in the antagonism of T. harzianum. (i) First, expression of agn13.1 mRNA and protein is repressed by glucose and induced in cultures with chitin or fungal cell walls as a nutrient source, which may represent a good simulation of antagonism. This expression pattern is almost identical to that found for other CWDEs from T. harzianum (4, 13, 18, 21), and it may reflect a coordinate induction of CWDEs for optimal establishment of antagonism, as discussed in references 12 and 13. In addition, AGN13.1 is derepressed late by low glucose levels, suggesting some mobilization of α-glucans from the cell wall of T. harzianum under carbon starvation conditions, as previously indicated for A. nidulans (48). (ii) AGN13.1 is able to bind and release reducing sugar from the cell walls of a variety of fungal phytopathogens. This activity depends on the cell wall source and may reflect different proportions of accessible α-1,3-glucan in those cell walls. Intriguingly, AGN13.1 bound efficiently, but failed to attack the cell walls from R. solani. Rather than the lack of an α-1,3-glucan substrate in these cell walls, this result is likely due to the presence of substances, such as melanin-like compounds, able to inhibit CWDEs (35). (iii) Most importantly, we observed different rates of spore germination and growth inhibition by AGN13.1 of phytopathogenic fungi A. niger, B. cinerea, and P. aurantiogriseum. Differences in sensitivity to the enzyme among the fungi might be due to the different molecular architectures of the spore and hyphal cell walls and/or the presence of specific inhibitors of CWDEs.
Altogether, the expression pattern of agn13.1 gene and protein, the lytic properties of AGN13.1 against fungal cell walls, and AGN13.1's antifungal activity strongly suggest that AGN13.1 contributes to the antagonism of T. harzianum. In addition to its possible medical applications against dental caries (17, 20, 24), important agricultural applications can be envisaged for this enzyme. Overexpression in recombinant T. harzianum could result in the generation of more effective biocontrol strains. Plant cell walls lack α-1,3-glucans (26), and to our knowledge, no α-1,3-glucanase activity has been described among the defense response systems in plants. Expression of the agn13.1 gene in transgenic plants might therefore improve resistance to fungal pathogens.
Acknowledgments
We gratefully acknowledge F. Domínguez for helpful assistance during the first period of the experimental work. We thank R. Sánchez for help with HPLC experiments and J. M. García and M. López-Reyes for 13C-NMR. J. de la Cruz thanks A. Vioque for encouragement.
A. Soler is a recipient of a fellowship from the MEC. J. de la Cruz thanks the MEC (Spain) for financial support. This work was supported by grants FAIR6-CT98–4140 from EU and FEDER IFD97–0843-C05–01 from EU and Plan Nacional I+D.
H. Ait-Lahsen and A. Soler contributed equally to this work.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Analysis of proteins. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 2.Bacon J S D, Jones D, Farmer V C, Webley D M. The occurrence of α-(1,3) glucan in Cryptococcus, Schizosaccharomyces and Polyporus species, and its hydrolysis by a Streptomyces culture filtrate lysing cell walls of Cryptococcus. Biochim Biophys Acta. 1968;158:313–315. doi: 10.1016/0304-4165(68)90153-0. [DOI] [PubMed] [Google Scholar]
- 3.Bartnicki-García S. Cell wall chemistry, morphogenesis and taxonomy. Annu Rev Microbiol. 1968;22:87–109. doi: 10.1146/annurev.mi.22.100168.000511. [DOI] [PubMed] [Google Scholar]
- 4.Benítez T, Limón C, Delgado-Jarana J, Rey M. Glucanolytic and other enzymes and their genes. In: Harman G E, Kubicek C, editors. Trichoderma and Gliocladium: enzymes, biological control and commercial applications. Vol. 2. London, United Kingdom: Taylor and Francis, Ltd.; 1998. pp. 101–127. [Google Scholar]
- 5.Bock K, Pedersen C, Pedersen H. Carbon-13 nuclear magnetic resonance data for oligosaccharides. Adv Carbohydr Chem Biochem. 1984;42:193–225. [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Chet I, Benhamou N, Haran S. Mycoparasitism and lytic enzymes. In: Harman G E, Kubicek C, editors. Trichoderma and Gliocladium: enzymes, biological control and commercial applications. Vol. 2. London, United Kingdom: Taylor and Francis, Ltd.; 1998. pp. 153–172. [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.de la Cruz J, Hidalgo-Gallego A, Lora J M, Benítez T, Pintor-Toro J A, Llobell A. Isolation and characterization of three chitinases from Trichoderma harzianum. Eur J Biochem. 1992;206:856–867. doi: 10.1111/j.1432-1033.1992.tb16994.x. [DOI] [PubMed] [Google Scholar]
- 10.de la Cruz J, Llobell A. Purification and properties of a basic endo-β-1,6-glucanase (BGN16.1) from the antagonistic fungus Trichoderma harzianum. Eur J Biochem. 1999;265:145–151. doi: 10.1046/j.1432-1327.1999.00698.x. [DOI] [PubMed] [Google Scholar]
- 11.de la Cruz J, Pintor-Toro J A, Benítez T, Llobell A. Purification and characterization of an endo-β-1,6-glucanase from Trichoderma harzianum related to its mycoparasitism. J Bacteriol. 1995;177:1864–1871. doi: 10.1128/jb.177.7.1864-1871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Cruz J, Pintor-Toro J A, Benítez T, Llobell A, Romero L C. A novel endo-β-1,3-glucanase, BGN13.1, involved in the mycoparasitism of Trichoderma harzianum. J Bacteriol. 1995;177:6937–6945. doi: 10.1128/jb.177.23.6937-6945.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Cruz J, Rey M, Lora J M, Hidalgo-Gallego A, Domínguez F, Pintor-Toro J A, Llobell A, Benítez T. Carbon source control on β-glucanase, chitobiase and chitinase from Trichoderma harzianum. Arch Microbiol. 1993;159:316–322. [Google Scholar]
- 14.Elad Y, Chet I, Henis Y. Degradation of plant pathogenic fungi by Trichoderma harzianum. Can J Microbiol. 1982;28:719–725. [Google Scholar]
- 15.Fleet G H, Phaff H J. Glucanases in Schizosaccharomyces. Isolation and properties of the cell wall-associated β-1,3-glucanases. J Biol Chem. 1974;249:1717–1728. [PubMed] [Google Scholar]
- 16.Flores A, Chet I, Herrera-Estrella A. Improved biocontrol activity of Trichoderma harzianum by over-expression of the proteinase-encoding gene pbr1. Curr Genet. 1997;31:30–37. doi: 10.1007/s002940050173. [DOI] [PubMed] [Google Scholar]
- 17.Fuglsang C C, Berka R M, Wahleithner J A, Kauppinen S, Shuster J R, Rasmussen G, Halkier T, Dalbøge H, Henrissat B. Biochemical analysis of recombinant fungal mutanases. A new family of α-1,3-glucanases with novel carbohydrate-binding domains. J Biol Chem. 2000;275:2009–2018. doi: 10.1074/jbc.275.3.2009. [DOI] [PubMed] [Google Scholar]
- 18.Geremía R, Goldman G H, Jacobs D, Ardiles W, Vila S B, van Montagu M, Herrera-Estrella A. Molecular characterization of the proteinase-encoding gene, prb1, related to mycoparasitism by Trichoderma harzianum. Mol Microbiol. 1993;8:603–613. doi: 10.1111/j.1365-2958.1993.tb01604.x. [DOI] [PubMed] [Google Scholar]
- 19.Guggenheim B, Haller R. Purification and properties of an α-(1→3) glucanohydrolase from Trichoderma harzianum. J Dent Res. 1972;51:394–402. doi: 10.1177/00220345720510022701. [DOI] [PubMed] [Google Scholar]
- 20.Guggenheim B, Regolati B, Schmid R, Mühlemann H R. Effects of the topical applications of mutanase on rat caries. Caries Res. 1980;14:128–135. doi: 10.1159/000260447. [DOI] [PubMed] [Google Scholar]
- 21.Haran S, Schickler H, Oppenheim A, Chet I. Differential expression of Trichoderma harzianum chitinases during mycoparasitism. Phytopathology. 1996;86:980–985. [Google Scholar]
- 22.Hasewaga S, Nordin J H. Enzymes that hydrolyze fungal cell wall polysaccharides. Purification and properties of an endo-α-d-(1→3)-glucanase from Trichoderma viride. J Biol Chem. 1969;244:5460–5470. [PubMed] [Google Scholar]
- 23.Hjeljord L, Tronsmo A. Trichoderma and Gliocladium in biological control: an overview. In: Harman G E, Kubicek C, editors. Trichoderma and Gliocladium: enzymes, biological control and commercial applications. Vol. 2. London, United Kingdom: Taylor and Francis, Ltd.; 1998. pp. 131–151. [Google Scholar]
- 24.Kelstrup J, Holm-Pedersen P, Poulsen S. Reduction of the formation of dental plaque and gingivitis in humans by crude mutanase. Scand J Dent Res. 1978;86:93–102. doi: 10.1111/j.1600-0722.1978.tb00613.x. [DOI] [PubMed] [Google Scholar]
- 25.Knowles J, Lehtovaara P, Teeri T. Cellulases families and their genes. Trends Biotechnol. 1987;5:255–261. [Google Scholar]
- 26.Krishnamurthy K V. Methods in cell wall cytochemistry. London, United Kingdom: CRC Press; 1999. pp. 1–24. [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Limón M C, Pintor-Toro J A, Benítez T. Increased antifungal activity of Trichoderma harzianum that overexpress a 33-kDa chitinase. Phytopathology. 1999;89:254–261. doi: 10.1094/PHYTO.1999.89.3.254. [DOI] [PubMed] [Google Scholar]
- 29.Lorito M. Chitinolytic enzymes and their genes. In: Harman G E, Kubicek C P, editors. Trichoderma and Gliocladium: enzymes, biological control and commercial applications. Vol. 2. London, United Kingdom: Taylor and Francis, Ltd.; 1998. pp. 153–172. [Google Scholar]
- 30.Lorito M, Harman G E, Hayes C K, Brodway R M, Tronsmo A, Wo S L, di Pietro A. Chitinolytic enzymes produced by Trichoderma harzianum: antifungal activity of purified endochitinase and chitobiosidase. Phytopathology. 1993;83:302–307. [Google Scholar]
- 31.Lorito M, Woo S L, García-Fernández I, Colucci G, Harman G E, Pintor-Toro J A, Filippone E, Mucciflora S, Lawrence C, Zoina A, Tuzun S, Scala F. Genes from mycoparasitic fungi as a source for improving plant resistance to fungal pathogens. Proc Natl Acad Sci USA. 1998;95:7860–7865. doi: 10.1073/pnas.95.14.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson N J. Colorimetric analysis of sugars. Methods Enzymol. 1955;3:85–86. [Google Scholar]
- 33.Papavizas G C. Trichoderma and Gliocladium: biology, ecology and potential for biocontrol. Annu Rev Phytopathol. 1985;23:23–54. [Google Scholar]
- 34.Peberdy J F. Fungal cell walls—a review. In: Kuhn P J, Trinci A P J, Jung M J, Goosey M W, Copping L G, editors. Biochemistry of cell walls and membranes in fungi. Heidelberg, Germany: Springer-Verlag; 1990. pp. 5–24. [Google Scholar]
- 35.Potgieter H J, Alexander M. Susceptibility and resistance of several fungi to microbial lysis. J Bacteriol. 1966;91:1526–1532. doi: 10.1128/jb.91.4.1526-1532.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson E F, Dannelly H K, Malloy P J, Reeve H C. Rapid isoelectrofocusing in a vertical polyacrylamide minigel system. Anal Biochem. 1987;167:290–294. doi: 10.1016/0003-2697(87)90166-7. [DOI] [PubMed] [Google Scholar]
- 37.Rombouts F M, Phaff H J. Lysis of yeast cell walls. Lytic β-1,6-glucanase from Bacillus circulans WL-12. Eur J Biochem. 1976;63:109–120. doi: 10.1111/j.1432-1033.1976.tb10213.x. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Schwartz W, Barrett A J. Human cathepsin H. Biochem J. 1980;191:487–497. doi: 10.1042/bj1910487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somogyi M. Notes in sugar determination. J Biol Chem. 1952;195:19–23. [PubMed] [Google Scholar]
- 41.Takehara T, Inoue M, Morioka T, Yokogawa K. Purification and properties of endo-α-1,3-glucanase from a Streptomyces chartreusis strain. J Bacteriol. 1981;145:729–735. doi: 10.1128/jb.145.2.729-735.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torronen A, Mach R, Messner M, Gonzalez R, Kalkkinen N, Harkki A, Kubicek C. The two major xylanases from Trichoderma reesei: characterization of both enzymes and genes. Bio/Technology. 1992;10:1461–1465. doi: 10.1038/nbt1192-1461. [DOI] [PubMed] [Google Scholar]
- 43.Tsunoda A, Nagaki T, Sakano Y, Kobayashi T. Purification and properties of an exo-α-1,3-glucanase from Trichoderma viride. Agric Biol Chem. 1977;41:939–943. [Google Scholar]
- 44.Tvaroska I, Tarabvel F. Carbon-carbon coupling constants in the conformational analysis of sugar molecules. Adv Carbohydr Chem Biochem. 1995;51:15–61. doi: 10.1016/s0065-2318(08)60191-2. [DOI] [PubMed] [Google Scholar]
- 45.Ulhoa C J, Peberdy J F. Regulation of chitinase synthesis in Trichoderma harzianum. J Gen Microbiol. 1991;137:2163–2169. doi: 10.1099/00221287-137-9-2163. [DOI] [PubMed] [Google Scholar]
- 46.Vazquez-Garcidueñas S, Leal-Morales C A, Herrera-Estrella A. Analysis of the β-1,3-glucanolytic system of the biocontrol agent Trichoderma harzianum. Appl Environ Microbiol. 1998;64:1442–1446. doi: 10.1128/aem.64.4.1442-1446.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zonneveld B J M. Biochemical analysis of the cell wall of Aspergillus nidulans. Biochim Biophys Acta. 1971;249:506–514. doi: 10.1016/0005-2736(71)90126-x. [DOI] [PubMed] [Google Scholar]
- 48.Zonneveld B J M. Morphogenesis in Aspergillus nidulans. The significance of α-1,3-glucan of the cell wall and α-1,3-glucanase for cleistothecium development. Biochim Biophys Acta. 1972;273:174–187. doi: 10.1016/0304-4165(72)90205-x. [DOI] [PubMed] [Google Scholar]
- 49.Zonneveld B J M. A new type of enzyme, and exo-splitting α-1,3 glucanase from non-induced cultures of Aspergillus nidulans. Biochim Biophys Acta. 1972;258:541–547. doi: 10.1016/0005-2744(72)90245-8. [DOI] [PubMed] [Google Scholar]