Abstract
Triple-negative breast cancer (TNBC) is the most aggressive breast cancer subtype with the worst prognosis and few effective therapies. Here we identified MS023, an inhibitor of type I protein arginine methyltransferases (PRMTs), which has antitumor growth activity in TNBC. Pathway analysis of TNBC cell lines indicates that the activation of interferon responses before and after MS023 treatment is a functional biomarker and determinant of response, and these observations extend to a panel of human-derived organoids. Inhibition of type I PRMT triggers an interferon response through the antiviral defense pathway with the induction of double-stranded RNA, which is derived, at least in part, from inverted repeat Alu elements. Together, our results represent a shift in understanding the antitumor mechanism of type I PRMT inhibitors and provide a rationale and biomarker approach for the clinical development of type I PRMT inhibitors.

Subject terms: Cancer therapy, Chemical genetics, RNA splicing, Screening
Type I PRMT inhibition elicits potent antitumor activity associated with increased interferon response and intron-retained dsRNA accumulation, suggesting its potential combination with immune checkpoint inhibitors for cancer treatment.
Main
Breast cancer is the world’s most commonly diagnosed cancer1. Triple-negative breast cancer (TNBC) is a heterogeneous breast cancer subtype lacking the expression of estrogen receptor, progesterone receptor and human epidermal growth factor receptor 2 (ref. 2). Although TNBC accounts for 15–20% of breast cancer cases, it leads to 25% of deaths2. Individuals with TNBC experience worse prognosis and overall survival rates than other subtypes due to both higher rates of recurrence (over 30%) and shorter survival following metastatic recurrence3. Due to the lack of targetable receptors that define other subtypes, options for TNBC treatment are limited, with chemotherapy representing the mainstay of current treatment. Despite some recent progress, new and effective targeted therapies for TNBC are urgently needed.
Epigenetic mechanism changes, such as DNA hypermethylation and chromatin modulator alteration, can disrupt regular development and lead to cancer4. Such evidence, coupled with recent progress in the development of drug-like small molecules capable of modulating epigenetic regulation, has fueled interest in exploiting the therapeutic potential of epigenetic targets5. Targeting epigenetic regulators, such as histone deacetylase inhibitors, BET bromodomain inhibitors and protein arginine methyltransferase 5 (PRMT5) inhibitors, has shown antitumor effects in TNBC and other cancers6–9. Despite these advances, the systematic understanding of epigenetic vulnerabilities in TNBC remains unclear.
To better understand how epigenetic regulators contribute to TNBC development and survival, we screened a collection of TNBC cell lines with a library of validated chemical probes that selectively target specific epigenetic regulators. Our screen revealed several previously reported epigenetic targets for TNBC, including enhancer of zeste homolog 2 (EZH2), BET bromodomains and PRMT5 (refs. 6,7,10). In addition to these, our results also revealed that type I PRMTs, the enzymes that catalyze asymmetric ω-NG,NG-dimethylarginine (ADMA) of histones and non-histone proteins, are important for TNBC growth. Among type I PRMTs, PRMT1 catalyzes around 85% of asymmetric dimethylation of proteins11. PRMT1-mediated arginine methylation is required for multiple cellular processes, including transcription, RNA splicing, cell communication and DNA repair12. mRNA splicing proteins have been identified as PRMT1 substrates, thus suggesting a role of PRMT1-dependent methylation in the regulation of RNA splicing13. Increasing evidence also demonstrates the functional roles of PRMT1 in DNA damage response by directly methylating DNA repair proteins, such as BRCA1, MRE11 and 53BP1 (refs. 14,15). Although cancer-associated mutations in PRMT1 are rare, its aberrant expression often correlates with poor outcome in various cancer types, including breast cancer16. Hence, the oncogenic potential of PRMT1 has been recognized, and it has recently received considerable attention as a potential therapeutic target in cancer17. Notably, GSK3368715, a type I PRMT inhibitor, has entered human clinical trials (NCT03666988), potentially opening up new avenues for the treatment of solid and hematological malignancies17. However, little is known about the therapeutic potential of type I PRMT inhibition in TNBC, and reliable biomarkers to identify tumors susceptible to type I PRMT inhibition are not known.
In this study, we developed a chemical screening approach to systematically investigate the epigenetic dependencies of TNBC and identified a specific vulnerability to the inhibition of type I PRMTs. We then examined the molecular factors underlying cell line-specific dependencies and validated our findings in human-derived models. Our data show that type I PRMT inhibition alters mRNA splicing, which leads to the expression of Alu sequences that can form cytosolic double-stranded RNA (dsRNA). This in turn triggers an antiviral interferon (IFN)-mediated response that pushes TNBC cells that are already stressed due to the elevated preexisting IFN response signature over a threshold to induce cell death.
Results
Type I PRMTs are key mediators of TNBC growth
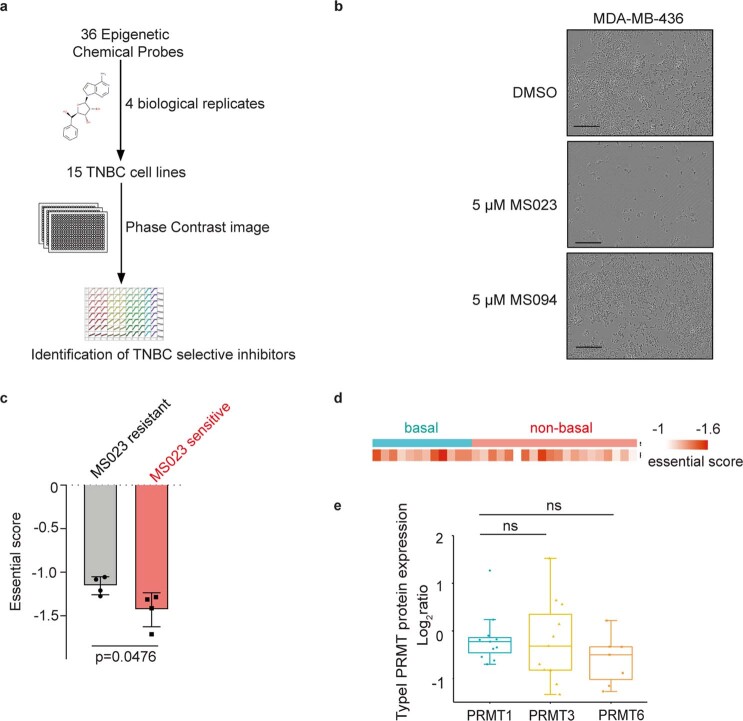
We first performed a cell-based chemical screen using a collection of 36 epigenetic chemical probes from the Structural Genomics Consortium (SGC) collection (https://www.thesgc.org/chemical-probes) to identify epigenetic regulators of TNBC cell growth9,18 (Supplementary Table 1). This chemical probe library includes tool compounds that selectively target key epigenetic regulatory proteins, including several epigenetic targets for which there are drugs currently in preclinical or clinical development. Each compound was evaluated in 15 TNBC cell lines, and cell confluency was assessed over 6 d using a live-cell imaging platform (Extended Data Fig. 1a). The primary screen identified six inhibitors that substantially reduced cell proliferation by more than 50% in at least one-third of the TNBC cell lines screened (Fig. 1a). BET bromodomain inhibitors (JQ1 and PFI-1) showed sensitivity across almost all the TNBC cell lines, as previously reported6. H3K27me3 methyltransferase and demethylase inhibitors (UNC1999 and GSKJ4) as well as a PRMT5 inhibitor (GSK591) also had antiproliferative effects on a wide variety of cancer cells, including TNBC7,19,20. Interestingly, a selective and cell-active inhibitor of type I PRMTs, MS023, showed growth-inhibitory properties across many TNBC cell lines, whereas its chemically similar but inactive control compound MS094 showed no effect (Fig. 1a and Extended Data Fig. 1b)21. Because type I PRMT inhibitors have potential broad applicability in diverse human cancers13 with at least one drug currently in clinical trials (NCT03666988), we sought to further characterize the action of type I PRMT inhibition in TNBC.
Extended Data Fig. 1. Dependency and expression of PRMTs across TNBC cell lines.
a. Flow chart of the epigenetic-focused chemical screen: 36 epigenetic probes at 5 μM were tested in the indicated cell lines and images were recorded by an Incucyte Zoom System in order to calculate cell proliferation rate. b. A representative experiment showing effects of in vitro treatment of MDA-MB-436 cells with indicated compound. Images were captured by an Incucyte Zoom. Scale bar represents 300 µM (representative n = 4 independent experiments). c. Essential score of PRMT1 in MS023 sensitive and resistant cell lines (mean ± s.d., n = 4 independent TNBC cell line per group, Student’s two-tailed t-test). d. Essential score of PRMT1 in breast cancer cell lines (n = 32 breast cancer cell lines were tested). e. Protein expression of Type I PRMTs including PRMT1, PRMT3 and PRMT8 in Cancer Dependency Map dataset (https://depmap.org/portal/) datasets. Protein expression is reported as log2 ratio. In the boxplots, the centre lines mark the median, the box limits indicate the 25th and 75th percentiles, and the whiskers extend to 1.5× the interquartile range from the 25th and 75th percentiles (n = 11 TNBC cell line per group, unpaired two-tailed Student’s t-test).
Fig. 1. Chemical screen of 36 epigenetic probes identifies type I PRMTs as therapeutic targets in TNBC.
a, Heat map showing the average cell proliferation values of the indicated epigenetic chemical probes at 6 d in 15 TNBC cell lines (data are shown as mean ± s.d. of n = 4); KDM, lysine demethylase. b, Viable cell counts of three TNBC cell lines treated with the indicated chemical probes for 7 d (data are shown as mean ± s.d. of n = 4); data were analyzed by one-way analysis of variance (ANOVA) with Dunnett’s test for multiple comparisons. c, Essential score of type I PRMTs across TNBC cell lines from the Cancer Dependency Map dataset (https://depmap.org/portal/). d, Type I PRMT mRNA expression in TNBC cell lines (n = 28 TNBC cell lines per group, each dot as an individual line); data were analyzed by one-way ANOVA with Dunnett’s test for multiple comparisons; TPM, transcripts per million.
PRMT1 is a major target among type I PRMTs in TNBC
Given the potency of MS023 against all type I PRMT members, including PRMT1, PRMT3, PRMT4, PRMT6 and PRMT8, we sought to understand whether a specific PRMT is the dominant target in TNBC. As illustrated in the screening heat map of cell confluence (Fig. 1a), inhibitors specifically targeting PRMT3 (SGC707) or PRMT4 (TP-064) and PRMT6 (MS049) showed no significant antiproliferative effect across the TNBC cell lines (Fig. 1a and Supplementary Table 2). Likewise, assessment of cell viability by CellTiter-Glo luminescent assay confirmed the profound inhibitory effect of MS023 on the representative TNBC cell lines, which was not observed with the other three inhibitors (Fig. 1b). These data rule out a major role for PRMT3, PRMT4 and PRMT6 in sustaining TNBC cell proliferation, individually.
Analysis of the public human Cancer Dependency Map dataset (https://depmap.org/portal/)22,23 revealed that among type I PRMTs, PRMT1 knockout gives the highest essential gene score across a collection of TNBC cell lines, suggesting a critical role for PRMT1 in TNBC (Fig. 1c). Across these TNBC cells, the essentiality score for PRMT1 was highest among those sensitive as opposed to resistant to MS023 treatment (Extended Data Fig. 1c). Nevertheless, some non-basal breast cancer cell lines are also dependent on PRMT1, indicating the potential of PRMT1 as a therapeutic target in these breast cancer types (Extended Data Fig. 1d). We then compared the expression level of type I PRMTs in our 15 TNBC cell lines and found that PRMT1 mRNA was expressed at a higher level than other type I PRMTs (Fig. 1d and Extended Data Fig. 1e). Notably, PRMT8 is barely expressed in TNBC, which is consistent with evidence of PRMT8 having brain-specific expression patterns (Fig. 1d)24. Taken together, we hypothesize that the observed efficacy of MS023 in TNBC is primarily due to inhibition of PRMT1 catalytic activity.
PRMT1 inhibition suppresses tumor growth
Through reviewing clinical data from two large The Cancer Genome Atlas (TCGA) and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohorts, we further evaluated TNBC’s dependency on PRMT1 and compared the expression levels of PRMT1 in TNBC to other subtypes25,26. The analysis demonstrated that PRMT1 mRNA expression is significantly higher in the basal-like subtype13 than in other breast tumor subtypes (Fig. 2a,b). This finding is further confirmed in a small cohort of independent breast cancer human-derived xenograft (PDX) and cell line27 cohorts (Fig. 2c,d).
Fig. 2. Type I PRMT inhibition suppresses tumor growth in a subset of TNBC.
a–d, PRMT1 gene expression in TCGA breast cancer datasets (a), METABRIC breast cancer datasets (b), Princess Margaret Hospital PDX datasets (PM-PDXs; c) and Princess Margaret Hospital cell line datasets (PM-cell lines; d). According to PAM50 classification, the cohorts were designated as basal and non-basal subtypes. Gene expression is reported as log2 (TPM + 0.001). In the box plots, the center lines mark the median, the box limits indicate the 25th and 75th percentiles, and the whiskers extend to 1.5× the interquartile range from the 25th and 75th percentiles. The numbers of individuals (n) per group are indicated, and the fold change (FC) values are as labeled. Data were analyzed by unpaired two-tailed Student’s t-test. e, Heat map of responsiveness to MS023 in the indicated cell lines. AAC was calculated from dose–response assays across 17 TNBC cell lines. Data are normalized to DMSO. A higher AAC indicates greater sensitivity. Colored cell lines are studied in more detail in this paper. Data are shown as mean ± s.d.; n = 4. f, Growth curves of Hs578-T and MDA-MB-468 cells treated with MS023 for 5 d. Data are shown as mean ± s.d.; n = 4. g, Immunoblots of MDA-MD-468 and Hs578-T cells following 5 d of treatment with the indicated doses of MS023 and the negative control MS094. Data are representative of n = 3 independent experiments. h, Immunoblots showing the doxycycline-inducible shRNA knockdown of PRMT1 or luciferase control in MDA-MB-468 and Hs578-T cells. Data are representative of n = 3 independent experiments; SDMA, symmetric dimethylarginine; Luc, luciferase. i, Individual tumor growth of the MDA-MB-468 xenograft model with once-daily administration of 60 mg kg–1 MS023 when tumors reach 2 mm in diameter. Data are shown as mean ± s.d.; n = 3. Data were analyzed by two-way ANOVA with Dunnett’s test for multiple comparisons. j, Tumor weight was measured as a surrogate for tumor burden from the control (CTL) and MS023-treated mice. Data are shown as mean ± s.d. (n = 3) and were analyzed by one-way ANOVA with Dunnett’s test. k, Immunoblot of tumor tissue from mice treated with control or MS023 at the experimental endpoint. Data are representative of n = 3 independent technical experiments.
Next, we performed dose–response assays with nine concentrations (ranging from 40 nM to 10 µM) of MS023 across 17 TNBC cell lines and assayed the response using a live-cell imaging platform to monitor cell confluence over time (Extended Data Fig. 2a). We calculated the area above the curve (AAC) to capture the efficacy and potency of the inhibitor, with a higher AAC value indicating greater sensitivity to MS023 treatment (Fig. 2e). These highly reproducible responses identified both sensitive (for example, MDA-MB-468) and resistant (for example, Hs578-T) cell lines, suggesting that MS023 is not universally cytostatic and that specific determinants of sensitivity exist within TNBC models. A representative experiment shows the effects of MS023 on TNBC cell growth in both sensitive and resistant cell lines over 5 d (Fig. 2f). A similar differential inhibitory pattern was observed with the clinical inhibitor GSK3368715 in MS023-sensitive and MS023-resistant cells (Extended Data Fig. 2b).
Extended Data Fig. 2. PRMT1 inhibition suppresses cell growth in a subset of TNBC cell lines.
a. Growth curves of 17 TNBC cell lines with MS023 at indicated concentrations for 5 days treatment (means±s.d., n = 4). b. Cell confluency of TNBC cells with 5 μM GSK3368715 treatment for 5 days (means±s.d., n = 3). c. Immunoblots showing two more shRNA knockdown of PRMT1 in both MDA-MB-468 and Hs578-T cells (representative of n = 3 independent experiments). d. Normalized cell confluence of PRMT1 knockdown for the indicated time post doxycycline induction (means±s.d., n = 4, two-way ANOVA with Dunnett’s test for multiple comparisons, ns: not significant). e. Growth curves of 4 TNBC cell lines with MS023 at indicated concentrations for 5 days treatment (means±s.d., n = 4). f. Representative immunoblots showing shRNA knockdown of PRMT1 in TNBC cell lines (red: MS023 sensitive; black: MS023 resistant) (representative of n = 3 independent experiments). g. Normalized cell confluence of PRMT1 knockdown for the indicated time post doxycycline induction (means±s.d., n = 4, two-way ANOVA with Dunnett’s test for multiple comparisons, ns: not significant). h. Body weights measurement of individual mouse over the course of treatment with control or MS023 (means±s.d., n = 3).
Given that PRMT1 is the most abundant type 1 PRMT enzyme, we investigated whether the antiproliferative effect of MS023 coincides with a reduction of the ADMA mark. To address this, we measured ADMA levels in a sensitive cell line (MDA-MB-468) following MS023 treatment for 5 d (Fig. 2g). We observed decreased ADMA levels in a dose-dependent manner over the dose range from 0.1 µM to 3 µM, which had a significant suppressive effect on proliferation of the MDA-MB-468 cell line. The observed PRMT1-dependent effect of MS023 inhibition was not due to changes in PRMT1 levels, as its expression was not altered following treatment (Fig. 2g). No significant differences in symmetric dimethylarginine levels were observed between sensitive and resistant cells in response to type I PRMT inhibition, indicating that the resistance to MS023 is not due to symmetric dimethylarginine compensation (Fig. 2g). MS023 treatment also reduced the ADMA mark in the resistant cell line Hs578-T, ruling out the possibility that differential sensitivity was attributable to drug efflux pumps or other mechanisms that might simply prevent target modulation by MS023.
We then performed PRMT1 genetic knockdown assays to confirm the on-target effect of type I PRMT inhibition. Knockdown of PRMT1 using three different inducible short hairpin RNAs (shRNAs) reduced PRMT1 expression and ADMA marks in both cell lines (Fig. 2h and Extended Data Fig. 2c). As observed with MS023 treatment, PRMT1 knockdown substantially suppressed cell growth in the sensitive cell lines (MDA-MB-468, HCC1143 and SUM149-PT) but not in the resistant cell lines (Hs578-T, CAL-120 and HCC1806; Extended Data Fig. 2d–g). Taken together, these data support that the antiproliferative effect of MS023 in a subset of TNBC cell lines is due to inhibition of PRMT1 activity.
To determine whether the in vitro effects observed above can translate to antitumor activity in vivo, we next evaluated the efficacy and tolerability of MS023 in vivo. Mice bearing MDA-MB-468 xenografts were treated with control (water) or MS023, respectively. MS023 was well tolerated, with no difference in body weight of animals treated with MS023 versus control-treated animals over 35 d (Extended Data Fig. 2h). Once-daily dosing of 60 mg kg–1 MS023 initiated in mice with palpable tumors significantly reduced tumor growth and final tumor weight (Fig. 2i,j)28. Notably, MS023 treatment reduced the ADMA mark of the xenograft tissue, suggesting that the reduction of tumor size coincides with disruption of the enzymatic function of PRMT1 (Fig. 2k). While this exploratory pilot assay requires further confirmation with more in vivo studies, our data collectively suggest that type I PRMT inhibition, primarily through PRMT1 inhibition, may be a promising therapeutic strategy for the treatment of a subset of TNBCs.
Preexisting IFN signaling correlates with MS023 sensitivity
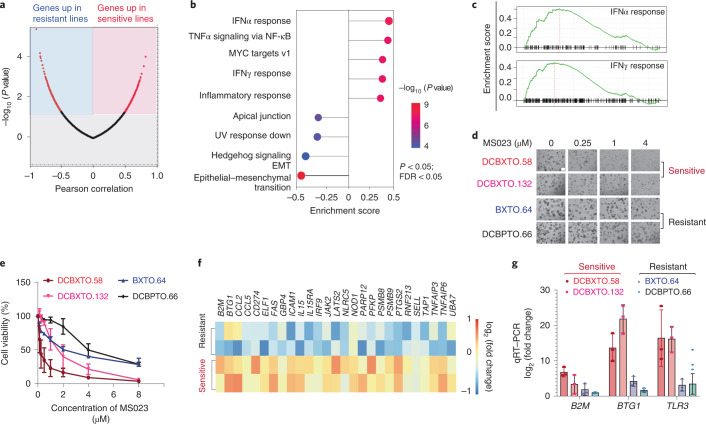
Although PRMT1 has been implicated in multiple biological processes, such as transcriptional modulation, pre-mRNA splicing and receptor signaling13, the extent to which any of these pathways contributes to the PRMT1 dependency of TNBC tumor cells is not clear. Previous studies have identified the association between sensitivity of PRMT1 inhibition and methylthioadenosine phosphorylase (MTAP) level, although this correlation in breast cancer is not as strong as in lymphoma17. Our data show no correlation between the response to PRMT1 inhibition and MTAP deficiency (Extended Data Fig. 3a). A recent report showed that mutation of serine- and arginine-rich splicing factor 2 (SRSF2) sensitizes cells to type I PRMT inhibition in leukemia28. However, we found that SRSF status or other splicing factor mutations appear unrelated to PRMT1 sensitivity in TNBC (Extended Data Fig. 3b). Given the synergistic effect between PRMT1 and PRMT5 (ref. 29), we investigated PRMT5 expression and MS023 sensitivity. Neither the mRNA nor protein level of PRMT5 was predictive of MS023 response (Extended Data Fig. 3c,d). Having ruled out these candidates, we performed systematic global profiling of basal gene expression for our panel of 17 TNBC cell lines27 and calculated the Pearson correlation coefficients of MS023 activity (as measured by the AAC value of the MS023 dose–response curve in Fig. 2e) with individual gene expression levels (Fig. 3a). We then subjected these rank-ordered gene lists to gene set enrichment analysis (GSEA). The IFNα response pathway was the most enriched pathway, with several additional immune-related pathways, including IFNγ response and tumor necrosis factor-α signaling via NF-κB pathways, also among the most enriched in MS023-sensitive lines (Fig. 3b,c). Given the link between replication stress and IFN response, we calculated the doubling time of each TNBC cell line and found no correlation between the proliferation rate and MS023 sensitivity (Extended Data Fig. 3e). Altogether, our results suggest that a preexisting enhanced expression of genes involved in the IFN signaling pathway might predict the responsiveness to MS023.
Extended Data Fig. 3. No significant identified molecular signature correlates with MS023 sensitivity in TNBC.
a. Correlation of MTAP expression and MS023 sensitivity. b. Oncoplot showing the mutation status of genes involved in RNA splicing across TNBC cell lines. Color indicates different mutation type. Arrow indicates the sensitivity of cell lines to MS023 treatment. c. PRMT5 gene expression in the Princess Margaret Hospital cell line datasets (PM-Cell lines) datasets. Gene expression is reported as log2(TPM + 0.001). d. PRMT5 protein expression in TNBC cell lines. Arrow indicates the sensitivity of cell lines to MS023 treatment. e. Correlation of cell doubling time and MS023 sensitivity.
Fig. 3. Increased IFN responses underlie the responsiveness to type I PRMT inhibition.
a, Pearson correlation analysis of individual gene expression and MS023 activity. A Volcano plot of log2 fold change values for all genes significantly upregulated in sensitive lines (red, left) or in resistant lines (blue, right) is shown. Data were analyzed by unpaired two-tailed Student’s t-test for multiple comparisons. b, Top nine pathways significantly correlated with MS023 sensitivity. Data were analyzed by one-tailed Fisher’s exact test for multiple comparisons; FDR, false discovery rate; EMT, epithelial–mesenchymal transition. c, GSEA for gene sets associated with IFN responses enriched in sensitive TNBC lines. Data were analyzed by one-tailed Fisher’s exact test. d, Images of different organoid models with either DMSO or MS023 treatment; scale bar, 100 μm. The image shown is representative of n = 4 independent experiments. e, Differential response of organoids to MS023 treatment. Data are shown as mean ± s.d.; n = 2. f, Heat map showing the differential expression of IFN-responsive genes between sensitive and resistant models. g, qRT–PCR validation of the expression of the indicated genes in both MS023-sensitive and MS023-resistant organoid models. Data are shown as mean ± s.d.; n = 3.
To better address this hypothesis in more clinically relevant TNBC models, we tested whether the correlation between IFN signaling gene signatures and MS023 sensitivity was also evident across human-derived TNBC samples. Three-dimensional cancer organoid models are thought to better recapitulate clinical disease and epithelial heterogeneity in TNBC and are therefore regarded as a superior tool for the evaluation of drug responses30. Four different organoid models were treated with either DMSO or MS023 at dose ranges from 0 µM to 10 µM and assessed by microscopic evaluation and PrestoBlue staining. Representative images of these organoid models (Fig. 3d) and the quantified percent cell viability of each model and treatment conditions are shown (Fig. 3e). Using the transcriptomic data of tumors matching these organoid models, the differential expression of IFN response genes was studied between the MS023-sensitive and MS023-resistant organoid models (Fig. 3f). Consistently, organoids with higher basal IFN gene expression were more sensitive to MS023 treatment, while organoids with lower IFN gene expression were more resistant. Quantitative real-time PCR (qRT–PCR) of RNA extracted from organoids further confirmed the upregulation of type 1 helper T cell chemokines (for example, CCL2 and CCL5) and antigen presentation genes (for example, B2M and BTG1) in sensitive models (Fig. 3g). Taken together, our findings across a range of human-derived models suggest that the preexisting levels of IFN response signatures are correlated with the degree of sensitivity to type I PRMT inhibition.
Type I PRMT inhibition triggers IFN responses
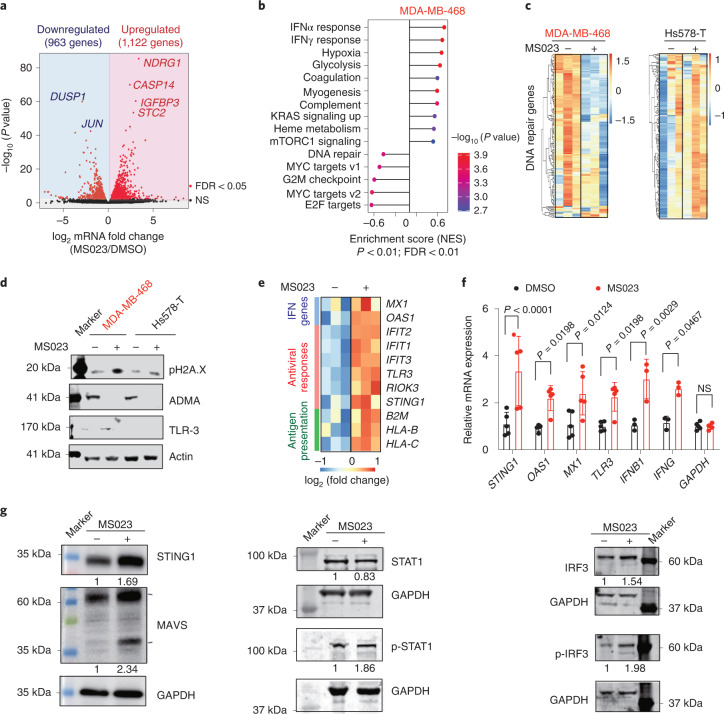
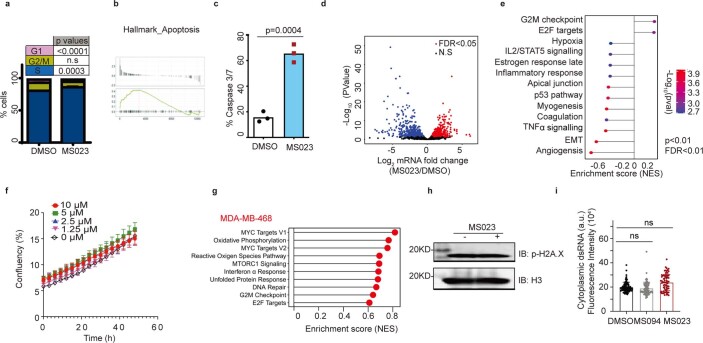
To better understand the potential mechanisms underlying the effects of type I PRMT inhibition in TNBC, we performed RNA sequencing (RNA-seq) on MDA-MB-468 cells following 5 d of treatment with MS023 and identified 2,085 genes as significantly differentially expressed (P < 0.05, false discovery rate < 0.05; Fig. 4a). Hallmark enrichment analysis revealed that E2F targets and G2M checkpoint pathways were downregulated after MS023 treatment, suggesting that type I PRMT inhibition affects the expression of genes that function in cell cycle regulation (Fig. 4b). In agreement, MS023 treatment leads to a modest but significant decrease in S phase and a concurrent increase in G1 phase (Extended Data Fig. 4a)17. Also consistent with the antitumor effect of MS023 observed in sensitive cell lines, GSEA revealed induction of the apoptosis pathway following MS023 treatment (Extended Data Fig. 4b). This was confirmed by the increased number of apoptotic cells after MS023 treatment (Extended Data Fig. 4c). In addition to the above noted effects, which likely reflect the downstream consequences of type I PRMT inhibition, a comparison of the differential gene expression patterns in both MDA-MB-468 (sensitive) and Hs578-T (resistant) lines showed that 5 d of treatment with MS023 decreased the expression of DNA repair genes in the sensitive line but not in the resistant line (Fig. 4c and Extended Data Fig. 4d,e). Western blotting of MDA-MB-468 cells revealed that the phosphorylation of histone H2AX at Ser 139 (pH2AX), a marker of DNA damage, was enhanced after MS023 treatment (Fig. 4d). To rule out potential non-specific effects induced by compound toxicity, we again treated MDA-MB-468 cells with MS023 but only for 2 d. No significant cell growth suppression (Extended Data Fig. 4f), E2F/G2M pathway downregulation (Extended Data Fig. 4g), DNA damage or dsRNA accumulation (Extended Data Fig. 4h,i) was observed at this earlier time point, consistent with a growth-suppressive mechanism other than acute toxicity.
Fig. 4. MS023 treatment triggers IFN responses.
a, Volcano plot of log2 fold change for genes significantly upregulated (red, right) or downregulated (blue, left) following 5 d of MS023 treatment in the MDA-MB-468 cell line. Data were analyzed by unpaired two-tailed Student’s t-test; NS, not significant. b, GSEA of all ranked differentially expressed genes. Data were analyzed by one-tailed Fisher’s exact test; NES, normalized enrichment score. c, Heat map showing genes associated with DNA repair after MS023 treatment for 5 d in both MDA-MB-468 and Hs578-T cell lines. d, Immunoblots showing the expression level of pH2AX, a DNA damage marker, with either DMSO or MS023 treatment in both MDA-MB-468 and Hs578-T cell lines. Data are representative of n = 3 independent experiments. e, Heat map showing RNA-seq data for indicated genes in MDA-MB-468 cells with either DMSO or MS023 treatment for 5 d. f, Expression of indicated IFN-responsive genes in MDA-MB-468 cells with either DMSO or MS023 treatment for 5 d analyzed by qRT–PCR. Data are shown as mean ± s.d.; n = 5 for STING1, OAS1, MX1, TLR3 and GAPDH; n = 3 for IFNB1 and IFNG. Data were analyzed by two-way ANOVA with Dunnett’s test for multiple comparisons. g, Immunoblots of the indicated proteins in MDA-MB-468 cells after 5 d of MS023 treatment. Normalized band intensity is labeled. Data are representative of n = 3 independent experiments; p-STAT1, phospho-STAT1; p-IRF3, phospho-IRF3.
Extended Data Fig. 4. Type I PRMT inhibition stimulates interferon responses in a subset of TNBC cell lines.
a. Flow cytometry cell cycle analysis for MDA-MB-468 cells cultured with or without MS023 for 5 days (means±s.d., n = 3, two-way ANOVA with Dunnett’s test for multiple comparisons, ns: not significant). b. GSEA of the apoptosis pathway in MDA-MB-468 cells after MS023 treatment for 5 days. (one-tailed Fisher’s exact test for multiple comparisons). c. Apoptotic cell counts of MS023 treated for 5 days by caspase 3/7 staining (means±s.d., n = 3, two-sided Student’s t test). d. Volcano plot of log2 fold change for genes significantly upregulated (red in the right panel) or downregulated (blue in left panel) upon MS023 treatment (n = 3) in Hs578-T cell line (unpaired two-tailed Student’s t-test). e. Gene set enrichment analysis (GSEA) of all ranked differential expressed genes in Hs578-T after 5 days MS023 treatment (one-tailed Fisher’s exact test for multiple comparisons). f. Growth curves of MDA-MB-468 cell with indicated concentration of MS023 for 2 days culture (means±s.d., n = 4). g. Top 10 gene set enrichment analysis (GSEA) of all ranked differential expressed genes in MDA-MB-468 after 2 days MS023 treatment. h. Representative immunoblots showing DNA damage after MS023 treatment for 2 days (one of three independent experiments are shown). i. Quantification of cytoplasmic dsRNA signal intensity in MDA-MB-468 cells with indicated compound treatment for 2 days (means±s.d., n = 3 independent biologically experiments of at least 86 cells per group, one-way ANOVA with Dunnett’s test for multiple comparisons).
GSEA further suggested an innate immune response mechanism, which could contribute to the antiproliferative phenotype. Upregulation of the IFNα and IFNγ innate immune response pathways were prominent GSEA signatures triggered by type I PRMT inhibition in MDA-MB-468 cells (Fig. 4b,c). Among the top upregulated genes after MS023 treatment, we observed increased expression of genes such as IFN-responsive genes of IFNβ and IFNγ and dsRNA-sensing pathway genes of STING1 and TLR-3. These markers of an antiviral stress response were observed both in RNA-seq and subsequent qRT–PCR validation (Fig. 4e,f). Similarly, western blotting identified activation of the dsRNA-sensing pathway, as reflected by increased STAT1 and IRF3 phosphorylation as well as the expression of STING1 and MAVS after MS023 treatment in the sensitive cell line MDA-MB-468 (Fig. 4g). Together, these results identified a stress response to elevated dsRNA as a potential result of type I PRMT inhibition, which could underlie its antiproliferative and cytotoxic effects.
Type I PRMT inhibition induces dsRNA accumulation
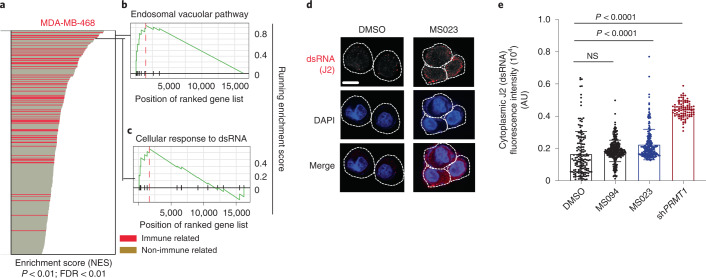
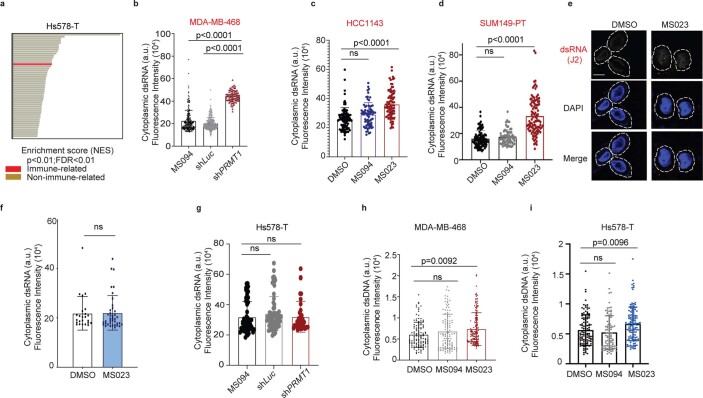
To investigate whether and how MS023 treatment may trigger a viral mimicry response in TNBCs, gene ontology (GO) analysis was performed in both MDA-MB-468 (sensitive) and Hs578-T (resistant) cell lines. MS023 induced immune-related responses in sensitive lines to a much greater extent than the resistant line (Fig. 5a and Extended Data Fig. 5a). Gene sets including cellular response to dsRNA and dsRNA-sensing signaling (endosomal vacuolar pathway) were two of the top significant positively enriched pathways (Fig. 5b,c). To further validate dsRNA formation, immunofluorescence staining with a J2 antibody (a gold standard for dsRNA detection31) identified that either genetic or pharmacological inhibition of PRMT1 induced a robust increase of cytoplasmic dsRNA in MS023-sensitive cell lines (Fig. 5d,e and Extended Data Fig. 5b–d) but not in the MS023-resistant lines (Extended Data Fig. 5e–g). We also compared dsDNA formation in both MS023-sensitive (MDA-MB-468) and MS023-resistant (Hs578-T) cell lines and found that dsDNA is accumulated to a similar extent in both cell lines after MS023 treatment, ruling out a major role for dsDNA in differential IFN activation in MDA-MB-468 cells (Extended Data Fig. 5h,i).
Fig. 5. Type I PRMT inhibition induces cytoplasmic dsRNA formation due to increased intron retention.
a–c, Type I PRMT inhibition leads to activation of immune signatures in MS023-sensitive cells. a, Bar plot of upregulated gene sets enriched in the MDA-MB-468 cell line after MS023 treatment. Gene sets associated with immune response are red. Data were analyzed by one-tailed Fisher’s exact test. b, GSEA for gene sets associated with the dsRNA-sensing endosomal vacuolar pathway. Data were analyzed by one-tailed Fisher’s exact test. c, GSEA for gene sets associated with cellular response to dsRNA. Data were analyzed by one-tailed Fisher’s exact test. d, Cellular dsRNA was evaluated by anti-dsRNA (J2) immunofluorescence; scale bar, 10 μm. The images shown are representative of n = 3 independent experiments. e, Quantification of cytoplasmic dsRNA signal intensity; AU, arbitrary units. Data are shown as mean ± s.d.; n = 3 independent biological experiments of at least 85 cells per group analyzed. Data were analyzed by one-way ANOVA with Dunnett’s test for multiple comparisons.
Extended Data Fig. 5. MS023 triggers interferon response through dsRNA accumulation.
a. Upregulated gene sets enriched in Hs578-T cell after MS023 treatment for 5 days. Gene sets associated with immune response are red (one-tailed Fisher’s exact test). b. Quantification of cytoplasmic dsRNA signal intensity in MDA-MB-468 cells with indicated compound or shPRMT1 for 5 days (means±s.d., n = 3 independent biologically experiments of at least 91 cells per group analyzed). c. Quantification of cytoplasmic dsRNA signal intensity in HCC1143 cells with indicated compound treatment for 5 days (means±s.d., n = 3 independent biologically experiments of at least 70 cells per group analyzed). d. Quantification of cytoplasmic dsRNA signal intensity in SUM149-PT cells with indicated compound treatment for 5 days (means±s.d., n = 3 independent biologically experiments of at least 70 cells per group analyzed). e. Images of cellular dsRNA staining in Hs578-T cell after MS023 treatment for 5 days. Scale bar represents 10 μm (Representative images of 3 independent experiments are shown). f. Quantification of cytoplasmic dsRNA signal intensity in Hs578-T cell after MS023 treatment for 5 days (means±s.d., n = 3 of at least 40 cells per group analyzed, one-way ANOVA with Dunnett’s test). g. Quantification of cytoplasmic dsRNA signal intensity in Hs578-T cells with indicated compound or shPRMT1 for 5 days (means±s.d., n = 3 independent biologically experiments of at least 45 cells per group analyzed). h. Quantification of cytoplasmic dsDNA signal intensity in MDA-MB-468 cells with indicated compound treatment for 5 days (means±s.d., n = 3 independent biologically experiments of at least 120 cells per group analyzed). i. Quantification of cytoplasmic dsDNA signal intensity in Hs578-T cells with indicated compound treatment for 5 days (means±s.d., n = 3 independent biologically experiments of at least 43 cells per group analyzed). For panels b-d, and g-i, P values were calculated using one-way ANOVA with Dunnett’s test for multiple comparisons.
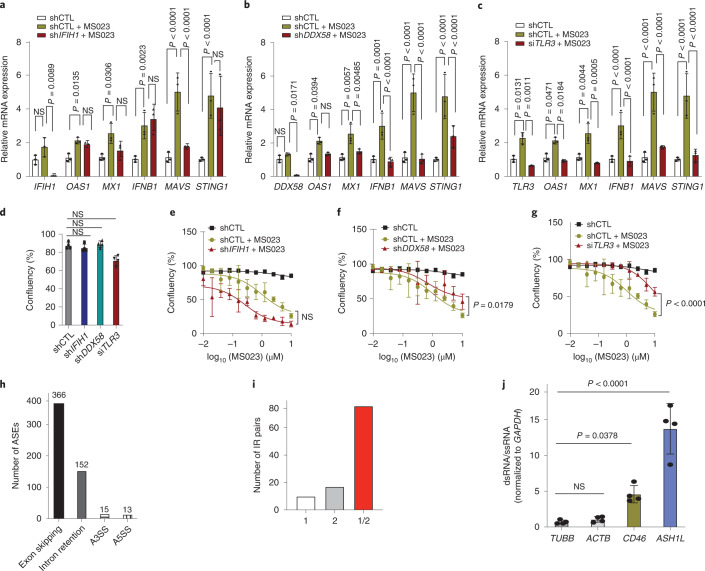
MDA5 (IFIH1), RIG-I (DDX58) and TLR-3 are receptors that bind to intracellular dsRNA32. To understand the individual contributions of these dsRNA sensors in IFN activation, we reduced their expression in MDA-MB-468 cells with validated shRNAs or short interfering RNA. Knockdown of DDX58 or TLR-3 decreased MS023-mediated upregulation of IFN genes IFNB1, MX1 and OAS1 and rescued the MS023 growth inhibitory effect (Fig. 6a–g). However, IFIH1 knockdown had no significant effect on MS023-induced IFN gene expression and cell growth (Fig. 6a,e). These results suggest that RIG-I (DDX58) and TLR-3, which preferentially recognize distinct pools of dsRNA32, are responsible for IFN activation in response to MS023 treatment.
Fig. 6. MS023 activates dsRNA sensors to induce IFN signaling.
a, The expression of IFIH1 and indicated IFN-responsive genes in scramble-treated (shCTL), MS023-treated and IFIH1-knockdown/MS023-treated MDA-MB-468 cells analyzed by qRT–PCR. b, The expression of DDX58 and indicated IFN-responsive genes in scramble-treated, MS023-treated and DDX58-knockdown/MS023-treated MDA-MB-468 cells analyzed by qRT–PCR. c, The expression of TLR3 and indicated IFN-responsive genes in scramble-treated, MS023-treated and TLR3-knockdown/MS023-treated MDA-MB-468 cells analyzed by qRT–PCR. Data in a–c are shown as mean ± s.d. (n = 3) and were analyzed by one-way ANOVA with Dunnett’s test for multiple comparisons. d, Cell confluence of scramble-treated, IFIH1-knockdown, DDX58-knockdown and TLR3-knockdown MDA-MB-468 cells. Data are shown as mean ± s.d. (n = 4) and were analyzed by one-way ANOVA with Dunnett’s test for multiple comparisons. e, Cell confluence of scramble-treated, MS023-treated and IFIH1-knockdown/MS023-treated MDA-MB-468 cells. f, Cell confluence of scramble-treated, MS023-treated and DDX58-knockdown/MS023-treated MDA-MB-468 cells. g, Cell confluence of scramble-treated, MS023-treated and TLR3-knockdown/MS023-treated MDA-MB-468 cells. Data in e–g are shown as mean ± s.d. (n = 4) and were analyzed by two-way ANOVA with Dunnett’s test for multiple comparisons. h, Bar plot showing the number of ASEs belonging to each of the main alternative splicing categories. i, Bar plot showing the count of IR pairs in which the intron intersects with only the first Alu in the pair (1; white), with only the second Alu in the pair (2; gray) or with the two Alus in the pair (1/2; red). j, qRT–PCR analysis of the indicated genes after J2 immunoprecipitation. Data are shown as mean ± s.d. (n = 4) and were analyzed by one-way ANOVA with Dunnett’s test for multiple comparisons; ssRNA, single-stranded RNA.
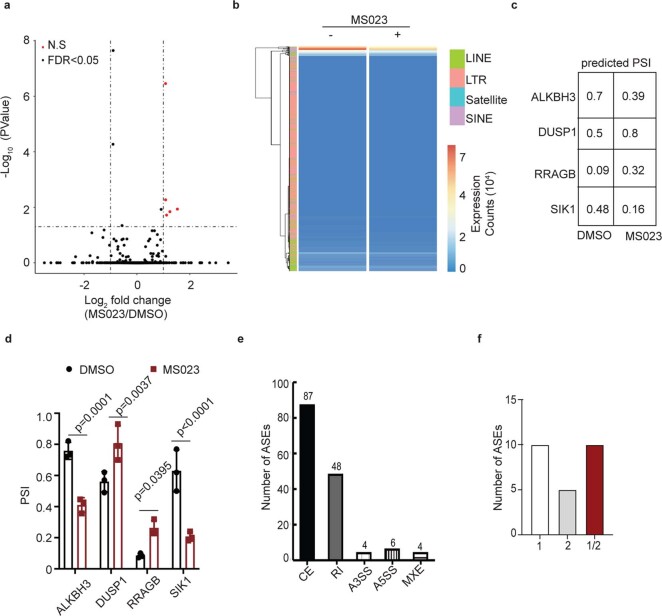
Having observed cytoplasmic dsRNA accumulation in response to type I PRMT inhibition, we then tried to identify the source of dsRNA and its relationship to inhibition of type I PRMTs. Unlike other epigenetic inhibitors, such as DNA methyltransferase inhibitors and lysine-specific demethylase inhibitors33–35 that induce endogenous retrovirus (ERV) expression, MS023 treatment did not significantly increase ERV expression (Extended Data Fig. 6a,b). It was recently reported that disruption of mRNA splicing through inhibition of the splicing regulatory pathway could trigger an antiviral response through the induction of misspliced RNAs. In particular, misspliced mRNAs with retained introns formed double-stranded structures that accumulated in the cytoplasm36. Given the broad effects of type I PRMTs on mRNA splicing12,28, we hypothesized that induction of dsRNA formation in response to MS023 treatment was due to direct deregulation of RNA splicing. To test this hypothesis, RNA-seq data were analyzed to identify disruptive alternative splicing events (ASEs) associated with MS023 treatment. A subset of predicted ASEs from splicing analysis was validated by qRT–PCR (Extended Data Fig. 6c,d). In total, 546 statistically significant differentially spliced events, including exon skipping, alternative splicing at the 3′ or 5′ site (A3SS/A5SS) and retained introns, distributed across 422 genes were identified following MS023 treatment (Fig. 6h). Importantly, retained introns are one of the major ASEs, suggesting that the dominant effect of type I PRMT inhibition on splicing is to induce intron retention.
Extended Data Fig. 6. Retained introns contribute to dsRNA accumulation in MS023 sensitive cells.
a. Volcano plot depicting expression changes of transposable elements in MDA-MB-468 cells after MS023 treatment for 5 days (unpaired two-tailed Student’s t-test). b. Heatmap depicting expression changes of transposable elements in MDA-MB-468 cells after MS023 treatment for 5 days (unpaired two-tailed Student’s t-test). c. Predicted PSI (percent spliced in) of indicated genes in MDA-MB-468 cells with indicated treatment for 5 days. d. PSI values derived from RT-PCR analysis for indicated genes from MDA-MB-468 cells after MS023 treatment for 5 days (means±s.d., n = 3, one-way ANOVA with Dunnett’s test for multiple comparisons). e. Barplot showing the number of alternative splicing events belonging to each of the main ASEs in Hs578-T cells with MS023 treatment for 5 days. f. Barplot showing the count of inverted-repeat (IR)-pairs in which the intron intersects with only the first Alu in the pair (1; white), or the only the second Alu in the pair (2; grey), or the two Alus in the pair (1/2; red).
Recent studies have shown that intronic SINE elements, specifically inverted repeat (IR) Alu elements (IR-Alus), induced after treatment with DNA-hypomethylating agents, lead to immunogenic dsRNA35–38. We asked whether IR-Alus within retained introns could be the source of dsRNA induced by MS023 treatment. A search for sequences capable of forming IR-Alus within the MS023-associated retained introns revealed that 62% of the total 152 retained introns had sequences that intersected with IR-Alus, and about 50% of the retained introns had IR-Alu sequence pairs that were bidirectionally transcribed in both sense and antisense directions (Fig. 6i). However, only 10 IR-Alu sequences residing in 48 retained introns were identified in resistant Hs578-T cells, which is much less than in MDA-MB-468 cells (66 IR-Alus of 152 retained introns; Extended Data Fig. 6e,f). This low number of IR-Alu pairs might be insufficient to trigger dsRNA accumulation and subsequent antiviral-mediated IFN response. Immunopurification of dsRNA followed by qRT–PCR in MDA-MB-468 cells revealed enrichment for selected retained intron mRNAs with IR-Alus (CD46 and ASH1L) but not two representative host mRNAs (TUBB and ACTB; Fig. 6j), confirming their contributions to the pool of dsRNA in response to type I PRMT inhibition in MS023-sensitive cell lines. Together, these results suggest that type I PRMT inhibition caused retained introns with IR-Alus, leading to dsRNA accumulation. Because we did not detect any preexisting difference of dsRNA accumulation or retained intron enrichment between MS023-sensitive and MS023-resistant cell lines, further investigations are required to identify the factors contributing to the basal IFN response gene expression signature and how these factors may prime TNBC cells to MS023 sensitivity (Extended Data Fig. 7a,b).
Extended Data Fig. 7. No correlation between IR-Alus induced dsRNA and basal IFN signature.
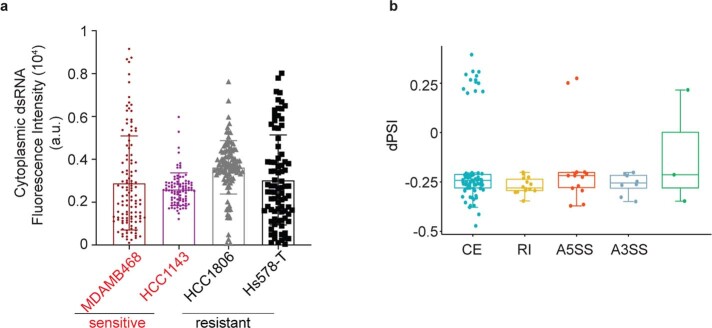
a. Quantification of cytoplasmic dsRNA signal intensity across four TNBC cell lines (means±s.d., n = 3 independent biologically experiments of at least 85 cells per group analyzed). b. Distribution of dPSI for classes of alternative splicing events that correlates with MS023 sensitivity. Each dot represents a potential splicing event from n = 3 biological independent experiments. In the boxplots, the centre lines mark the median, the box limits indicate the 25th and 75th percentiles, and the whiskers extend to 1.5× the interquartile range from the 25th and 75th percentiles (unpaired two-tailed Student’s t-test).
Discussion
In this study, we took an unbiased epigenetic chemical screening approach and identified the type I PRMT inhibitor MS023 as capable of suppressing the proliferation of a subset of TNBC cells. Type I PRMTs have recently come into focus as promising targets due to their overexpression in many cancer types and the finding that their inhibition is tumor suppressive in many of these settings12,17. Specifically, in TNBC, our studies show that genetic and pharmacological inhibition of PRMT1, the main target among type I PRMTs, induces cell cycle arrest and apoptosis, leading to tumor suppression in a subset of TNBC. Moreover, preclinical models showed that type I PRMT inhibition suppressed tumor growth in human-derived models in vitro and in vivo.
Molecular-based prediction of drug response is a major goal of precision oncology. Given the complexity and heterogeneity of TNBC, the development of robust molecular predictors of drug response represents both an opportunity and a challenge. Although several factors, such as genomic deletion or epigenetic silencing of MTAP or SRSF mutation, have been linked to the responsiveness to type I PRMT inhibition, none of these molecular markers correlated with sensitivity to MS023 in the context of TNBC17,28. However, our gene expression data revealed that TNBC cells that are most sensitive to MS023 treatment have gene expression signatures enriched for IFN response and antiviral signaling. This correlation, which was confirmed in human-derived organoids, identifies an opportunity for the development of markers of innate immune response to stratify individuals most likely to benefit from type I PRMT inhibition. Our growth-inhibitory results align with reports of MS023 sensitivity in a subset of colorectal cancer organoids and human-derived glioblastoma stem cell lines39,40, which provide additional models to assess the dependency toward the innate immune response for sensitivity to PRMT1 inhibitors, such as MS023, but the underlying mechanisms were not investigated. Further study is needed to assess whether our observations in TNBC are generalizable to other human tumors.
Emerging evidence supports that some epigenetic-targeting molecules, such as inhibitors targeting DNA methyltransferase34,35, EZH2 (ref. 41), LSD1 (ref. 33), SETDB1 (ref. 42) and CARM1 (ref. 43), are capable of inducing robust antitumor immune responses. These effects are the result of induced ‘viral mimicry effects’, increased tumor antigen expression/presentation, T cell activation, a remodeled tumor microenvironment or combinations thereof. Here, we discovered a new link between type I PRMT inhibition and innate antitumor immunity. Pharmacological inhibition of type I PRMT amplifies preexisting IFN responses by inducing cytosolic dsRNA accumulation in sensitive TNBC lines. Unlike many of the other epigenetic inhibitors that induce ERV expression, MS023 treatment triggers dsRNA accumulation from intron-retained RNAs. While PRMT enzymes are often classified as epigenetic targets based on their histone substrates, PRMTs have a much broader set of targets, especially RNA-binding proteins involved in mRNA splicing. Given these broad roles of type I PRMT on RNA splicing, our discovery is consistent with the recent report showing that spliceosome-targeted therapies induce intron-retained transcripts and dsRNA formation36.
In summary, our results show that type I PRMT inhibition results in potent antitumor activity associated with increased IFN response and dsRNA accumulation derived from misspliced RNAs. Notably, the innate immune effects of type I PRMT inhibition are restricted to TNBC cells with preexisting elevated IFN response gene expression signatures, suggesting that MS023 treatment may push TNBC cells that are already stressed over a threshold to induce cell death. The identification of biomarkers of type I inhibition sensitivity provides important insights into the targeted development of this class of therapy in TNBC and other cancers. Furthermore, given the general role of type I PRMT inhibitors in stimulating dsRNA and IFN response signals, targeting type I PRMTs combined with immune checkpoint inhibition may offer potential new opportunities for cancer immunotherapy.
Methods
Cell culture
TNBC cell lines were obtained from ATCC and cultured according to ATCC’s recommendations with Dulbecco’s modified Eagle’s medium (DMEM; Gibco) or RPMI 1640 (Life Technologies, 11965) with 10% FBS (Merck, F12103) and 1% penicillin/streptomycin (Gibco, 15140122). Cells were maintained mycoplasma free by using a MycoAlert mycoplasma detection kit (Lonza) and were passaged no more than 25 times.
Epigenetic chemical screen
All compounds were purchased from Cayman Chemical, Millipore-Sigma or MedChemExpress. The detailed resources for each compound are listed in the Supplementary Information. Chemical purity was validated at the SGC at more than 99%. TNBC cell lines were plated at 500 cells per well on 384-well plates. After cells adhered to the plates, compounds were dissolved in DMSO and added to achieve a final concentration of 5 μM. Each plate contained four replicates for each compound and DMSO as a control. Cells were then placed in an IncuCyte ZOOM live-cell analysis system (Essen492 Biosciences) for 6 d. Confluency was imaged with a ×10 objective in phase-contrast mode and analyzed using Incucyte 2016 integrated software, according to manufacturer’s instructions. Data were normalized to the DMSO control wells, and the log2 average confluency values are presented.
Western blotting
Cells and tumor tissues were collected and lysed as described before44. Briefly, 20–100 μg of protein was boiled at 95 °C for 5 min, loaded onto NuPAGE 4–12% Bis-Tris protein gels (Invitrogen) and transferred to nitrocellulose membranes (Bio-Rad) using the Mini-PROTEAN Tetra electrophoresis system (Bio-Rad). Primary antibodies used for membrane staining were anti-PRMT1 (Millipore, 7404; 1:1,000), anti-ADMA (Cell Signaling, 13522S; 1:1,000), anti-GAPDH (Santa Cruz Biotechnology, sc-32233; 1:5,000), anti-pH2A.X (Cell Signaling, 9718S; 1:1,000), anti-β-actin (Santa Cruz Biotechnology, sc-47778; 1:3,000) and anti-dsRNA (J2) (Scicons, 10010200; 1:5,000). Images were collected on an Odyssey scanner (LiCor) or Amershan ImageQuant 80 and analyzed with ImageQuant TL (v8.2.0) and ImageJ (v1.53a).
Cell cycle assay
Cells were plated in six-well plates and treated with 1 μM MS023 or DMSO as a control. An allophycocyanin BrdU flow kit (BD Pharmingen) was used for the following analysis. Cells were incubated for 6 h with 10 μM BrdU and fixed and permeabilized with BD Cytofix/Cytoperm buffer. Immunofluorescent cell staining with fluorochrome-conjugated anti-BrdU and 7-aminoactinomycin D was performed following the manufacturer’s instructions and analyzed using a BD FACScan flow cytometer. The number of live cells in each stage was determined (FlowJo software, version 9.3.1).
Cell apoptosis assay
Cells were plated at 3,000 cells per well in 96-well plates with DMSO or MS023 for 5 d of treatment. Caspase-3/caspase-7 green apoptosis assay reagent (Sartorius, 4440) was added to the cells and imaged and analyzed with an Incucyte ZOOM 2FLR (v.1.00) system according to manufacturer’s instruction.
Cell viability assay
Cells were seeded and treated with prescribed conditions in 384-well plates accordingly. At the endpoint, 25 µl of Promega CellTiter-Glo luminescent cell viability assay reagent (Promega, G7572) was added to each well. Plates were mixed gently for 2 min on an orbital shaker and incubated at room temperature for 20 min protected from light. The luminescent signal intensity was subsequently read on a CLARIOstar plate reader (BMG LABTECH).
Gene set enrichment analysis
Compound activity and cell viability data were collected and processed using an IncuCyte ZOOM live-cell analysis system (Essen492 Biosciences). Data were normalized to the DMSO control and were analyzed using R (v3.5.1) with Bioconductor (v.3.14). Cell half-maximal inhibitory concentration (IC50) and AAC45 values were analyzed using PharmacoGx R package (v. 2.6.0). We obtained the transcriptome data of the 17 cell lines through RNA-seq and processed the data with the Kallisto pipeline3. For all 17 cell lines, genes were ranked based on Pearson correlation coefficients between the measured compound activity (IC50) and individual gene expression level. Hallmarks gene sets were downloaded from MsigDB (https://www.gsea-msigdb.org/gsea/msigdb/)46. The piano R package was used to generate the GSEA results, and the fgsea R package was used to create pathway enrichment plots47,39.
Proteomics analysis
Princess Margaret Cancer Centre protein expression data were downloaded from ref. 27, and MD Anderson protein expression data were downloaded from ref. 48. The relative correlations between individual protein expression level and compound effectiveness (IC50) were analyzed by Pearson correlation coefficients using GraphPad Prism 8.0.
Splicing data analysis
ASEs were identified for the human genome (hg38) using the vast-tools pipeline49,50 with |dPSI | ≥ 0.2 and MV | dPSI_at_95 | ≥ 0.05 for significance, and additional non-default parameters for vast-tools diff module include -S 3, -e 10, -m 0.01. ASEs were further categorized into functional classifications according to their predicted impact on the open reading frame (ORF) as ‘neutral’ for events that generate known functional isoforms or that do not alter the protein sequence (for example, an alternative exon), ‘protective’ for events that reduce the occurrence of deleterious nucleotide sequences and therefore generate a functional protein (for example, removal of an intron/exon containing a premature stop codon) and ‘deleterious’, which denotes events that increase the frequency of disruptive sequences in the ORF (for example, inclusion of introns/exons containing premature stop codons or removal of essential exons for protein function)51. All postprocessing data analysis and figure generation steps were conducted using custom Python 3.7 scripts, which are available upon request.
Lentiviral mRNA targets
PRMT1-targeting shRNA vector was a kind gift from the laboratory of C. Strahdee, and the sequences are listed in Supplementary Table 4. Target and packaging plasmid were cotransfected into HEK293 cells to produce lentivirus. Lentivirus was transfected into cell lines along with polybrene (8 μg ml–1). Fresh medium was replaced after 16 h, and puromycin selection (0.2–0.6 μg ml–1) began after 48 h. After 72 h, the selection was complete, and stable transduced cells were generated.
dsRNA immunoprecipitation
MDA-MB-468 cells were seeded at 1.0 × 106 in 10-cm dishes. After cells adhered for 24 h, 1 μM MS023 or DMSO was added to treat cells for 5 d. Cells were then trypsinized and washed with ice-old PBS. Cells were centrifuged (4 °C,180g, 5 min), and the supernatant was discarded. Cell pellets were lysed in 1 ml of RIP buffer (25 mM HEPES pH 7.2, 150 mM NaCl, 5 mM MgCl2, 0.1% Igepal CA-630 and 1 U µl–1 Rnasin Plus) for 5 min on ice. After cell lysis, tubes were centrifuged, and the supernatant was transferred into a new Eppendorf tube; 10% of the supernatant was used for total RNA extraction with Trizol, and the rest was used for dsRNA immunoprecipitation.
Protein A Dynabeads were prepared by using NT-2 buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1 mM MgCl2 and 0.1% Igepal CA-630) for washing and resuspending. Five micrograms of anti-dsRNA (J2) (monoclonal) was added to 100 µl of beads and incubated at 4 °C overnight. The next day, cell lysate was co-incubated with 100 µl of J2-bound Protein A Dynabeads at 4 °C for 3 h. Beads were washed with NT-2 buffer three times, followed by washing with high-salt wash buffer (50 mM Tris-Cl, pH 7.4, 300 mM NaCl, 1 mM MgCl2, 0.5% Igepal CA-630 and 0.1% SDS) three times. Trizol was used to collect J2-bound dsRNA from beads. Chloroform was added at a 1:5 ratio, and RNA was cleaned using an RNA Clean and Concentrator kit (Zymo).
Quantitative real-time PCR
An iScript gDNA Clear cDNA Synthesis kit (Bio-Rad) was used for synthesizing cDNA. According to the manufacturer’s protocol, 1 µg of total RNA or dsRNA was used to generate a 20-µl reaction mixture. A 10× dilution was then made by adding 180 µl of double-distilled water, and 2 µl of synthesized cDNA was used per reaction. During qRT–PCR, cDNA template and PowerUp SYBR Green master mix (Applied Biosystems) were added into white PCR reaction tubes and placed in a CFX Maestro (Bio-Rad). All experiments were performed in at least three biological replicates. Primers were designed to measure fully spliced transcripts and intron-containing transcripts, and all primer sequences are provided in Supplementary Table 3. For data analysis, threshold cycle (Ct) values obtained during qRT–PCR were used to calculate the ratio of intron-containing transcripts to fully spliced transcripts. Data were collected on a Bio-Rad CFX96 touch and analyzed by Bio-Rad CFX Manager (v3.1.1517.0823).
Transposable elements analysis
FASTQ files were aligned to the human genome (Grch38) with Bowtie2 (v2.2.5). After converting the .sam files to .bam files using samtools (v1.10), we used the Repenrich2_subset function to generate discrete files for uniquely and multimapped reads. These reads were further analyzed to estimate the expression level of repetitive elements by the Repenrich2 function. DESeq2 (v1.34) was used to analyze the differential expression of estimated repeats counts.
dsRNA and dsDNA immunofluorescence staining
Cells were grown on poly-l-ornithine-pretreated coverslips placed in 12-well plates. Knockout cells were induced by 1 μg ml–1 doxycycline for 3 d, and compound-treated cells were treated with 1 μM MS023 or DMSO for 5 d. After treatment, cells were washed in PBS and fixed in 4% formaldehyde for 15 min at room temperature. Cells were then washed three times with PBS, permeabilized in 0.5% (vol/vol) Triton X-100 for 10 min and blocked in 5% bovine serum albumin in PBS for 1 h at room temperature. Primary antibody was added (anti-dsRNA J2 (Scicons, 10010200) diluted 1:500 and anti-dsDNA (Abcam, ab27156) diluted 1:1,000) and incubated at 4 °C overnight. Secondary anti-mouse IgG Alexa 647 (Cell Signaling Technology, 4410S) was diluted 1:1,000 and incubated for 1 h in a black container at room temperature. Coverslips were washed with PBS, and DAPI-containing mountant (Invitrogen ProLong Gold Antifade Mountant, P36930) was used. Images were collected on a A1 HD25 Single-Photon confocal microscope and analyzed by NIS-Elements (v5.21.03).
Organoid assay
BXTO.64, DCBXTO.58 and DCBXTO.132 organoids were derived from PDX tumors. DCBPTO.66 organoids were derived from human tumors. Human tumors were collected with informed participant consent according to University Health Network-approved Research Ethics Board protocols (14-8358). Tumor tissue was minced and digested in 5–10 ml of Advanced DMEM containing 1× GlutaMAX, 10 mM HEPES, 1× antibiotic–antimycotic (AdDF+++) and 250–500 μg ml–1 Liberase TH for 45 min at 37 °C with gentle rocking. Tissue was filtered over a 100-μm cell strainer and pelleted by centrifugation at 400g for 10 min at 4 °C. Pellets were washed once with AdDF+++, repelleted and treated with Red Cell Lysis Buffer Hybri-Max for 5 min on ice before cell counting. Organoids were cultured in medium as previously described30. PDX-derived organoids were regularly evaluated for human and mouse cell content by flow cytometry to ensure purity. Short tandem repeat analysis was used to confirm that the organoids matched their tumor of origin, and mycoplasma testing was performed as a quality control step.
For drug assays, organoids were dissociated into single cells, and 2,000 cells were plated per well in duplicate in 25 µl of basement membrane extracts (BME) in 48-well plates. Once the BME had solidified, the organoid/BME domes were overlaid with 475 µl of medium with or without drug. Fresh medium and drug were applied every 5 d. A well containing BME only (no cells) was included as a medium-only control. The cells were cultured for 12–21 d (depending on the growth rate of the model) until the untreated controls had formed organoids of >50 mm in diameter. Medium was removed, and the organoids were incubated with 1× PrestoBlue HS Reagent (Thermo Fisher) in Breast Organoid Media30 overnight at 37 °C. The following day, aliquots of the medium supernatant were transferred to a 384-well plate, and fluorescence readings were taken at 560/590-nm excitation/emission wavelength using a CLARIOstar Plus microplate reader. The medium-only control was used for background correction, and cell viability for each organoid model was normalized to its respective no-drug control. Three independent assays were done per organoid model.
Mouse studies
All animal experiments were reviewed and approved by the Animal Care Committee at the University Health Network in Toronto. For in vivo dosing experiments, 7 × 106 MDA-MB-468 cells were injected into the mammary fat pad of severe combined immunodeficient mice following standard procedures. MS023 was prepared at 5% N-Methyl-2-pyrrolidone (NMP), 20% captisol (wt/vol), 20% PEG-400 and 55% normal saline and administered to the mice by intraperitoneal injection. The MS023-sensitive cell line (MDA-MB-468) used in this experiment has a tendency to ulcerate through the skin when it reaches a modest size, which precludes continuation of experiments to larger tumor volumes due to humane endpoints. Thus, we allowed tumors to grow to a detectable size, as assessed by palpation (~2 mm in diameter), and started to treat with 60 mg kg–1 MS023 daily for a total of 5 weeks. Animals were randomized to treatment via random number generation/assignment. Body weights and tumor growth were measured once a week over the course of treatment until the endpoint was reached.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41589-022-01024-4.
Supplementary information
Supplementary Tables 1–5 and Fig. 1.
Acknowledgements
We thank the following organizations for supporting our study: the Terry Fox Research Institute (New Frontiers Research Program PPG-1064 to D.W.C., B.H.-K., M.L. and C.H.A.), the Canadian Institute for Health Research (CEEHRC team grant 158225 to M.L. and C.H.A., grant 363288 to B.H.-K., grant FDN154328 to C.H.A., grant 136963 to M.L. and fellowship award 430943 to Q.W.). This work was also supported by the Princess Margaret Cancer Foundation (M.L.), the Gattuso-Slaight Personalized Cancer Medicine Fund (B.H.-K.), National Natural Science Foundation of China (grant 82103287 to Q.W.), Zhejiang Provincial Natural Science Foundation (grant LR22B050001 to Q.W.), FAPESP (grants 2012/0195-3, 2014/50897-0 and 20/02006-0 to K.B.M. and fellowships 2016/25521-1 and 2015/25134-5 to F.E.C.), Ontario Institute for Cancer Research (Investigator Award to M.L.), Canadian Cancer Society (the Bernard and Francine Dorval Award for Excellence to M.L.) and the SGC. The SGC is a charity (register number 1097737) that received funding from AbbVie, Bayer AG, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Genome Canada through Ontario Genomics Institute [OGI-196], Janssen, Merck KGaA (aka EMD in Canada and USA), Pfizer, Takeda and the Innovative Medicines Initiative 2 Joint Undertaking (IMI2 JU; EUbOPEN grant 875510) from the European Union and European Federation of Pharmaceutical Industries and Associations (EFPIA). We thank the Princess Margaret Living Biobank for organoid development and the Princess Margaret Bioinformatics group for assisting with the bioinformatic analysis. We acknowledge the use of instruments at the Shared Instrumentation Core Facility at the Institute of Basic Medicine and Cancer (IBMC), Chinese Academy of Sciences.
Extended data
Source data
Statistical source data.
Statistical source data.
Unprocessed western blots.
Statistical source data.
Statistical source data.
Unprocessed western blots.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Unprocessed western blots.
Statistical source data.
Statistical source data.
Unprocessed western blots.
Statistical source data.
Statistical source data.
Statistical source data.
Author contributions
Q.W., C.H.A. and M.L. coordinated and managed the overall project. B.H.-K., G.D., P.P., J.J., K.B.M., D.B.-L., D.D.D.C., X.J.W., D.W.C., M.L. and C.H.A. supervised the research. Q.W. planned the experiments, designed and prepared figures and performed the bulk of the assays with D.Y.N. W.B. and Y.S.J. performed the RNA-seq analysis. F.E.C. performed the splicing analysis. J. Chen performed dsRNA staining assays. qRT–PCR, cell growth, western blotting and genetic knockdown studies were performed by S.D., Z.Z.W., M.M.S. and S.A.M. P.M. conducted Alu analysis. Y.D.S. and J.L. provided the MS023 compound. Organoid assays were performed by J. Cruickshank. J.H. performed the mouse studies. N.D.-A., W.J.C. and L.X.Z. performed cell culturing, cell knockdown and immunoblotting assays. Q.W., M.L. and C.H.A. wrote the manuscript. All authors assisted in writing the manuscript, reviewed the final version and approved the content and submission.
Peer review
Peer review information
Nature Chemical Biology thanks Steven Corsello, Ernesto Guccione and Stephane Richard for their contribution to the peer review of this work.
Data availability
Gene dependency data are available at depmap bioportal (https://depmap.org/portal/). RNA/protein expression data for TNBC cell lines are available in the datasets GSE73526 and GSE74702. RNA expression data for human breast cancer samples are available at TCGA (https://portal.gdc.cancer.gov/) and METABRIC (https://www.cbioportal.org/) and for the cell lines at GSE73526 and GSE74702. RNA expression data for PDX models are available upon request to protect participant privacy. The gene sets used for GSEA analysis are available at the molecular signatures database (https://www.gsea-msigdb.org/gsea/msigdb/). The RNA-seq data generated in this article are available from http://neellab.github.io/bfg/. Other data are available from the corresponding authors upon reasonable request. Source data are provided with this paper.
Code availability
Code to reproduce the RNA-seq bioinformatics analyses and the related data are available at https://github.com/bhklab/TNBC_PRMT1i, and code to reproduce the RNA splicing analyses is available at https://gist.github.com/Fciamponi/ef8aae813575a4947c1f9cb87a1c7330. Code to reproduce the ERV analysis is available at https://github.com/WuEpiLab/TNBC_MS023_ERV.
Competing interests
J.J. is a cofounder, consultant, shareholder and scientific advisory board member in Cullgen, Inc. J.J. is also a consultant at Accent Therapeutics, Inc., and EpiCypher, Inc. The J.J. lab received research funds from Cullinan Oncology, Inc., Celgene Corporation, Cullgen, Inc., and Levo Therapeutics, Inc. D.D.D.C. is a cofounder, employee and shareholder at DNAMx, Inc. D.D.D.C. received funding unrelated to this project from Pfizer and Nektar Therapeutics. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Qin Wu, David Y. Nie.
Contributor Information
Qin Wu, Email: wuqin@ibmc.ac.cn.
Mathieu Lupien, Email: mathieu.lupien@uhnresearch.ca.
Cheryl H. Arrowsmith, Email: cheryl.arrowsmith@uhnresearch.ca
Extended data
is available for this paper at 10.1038/s41589-022-01024-4.
Supplementary information
The online version contains supplementary material available at 10.1038/s41589-022-01024-4.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Bianchini G, De Angelis C, Licata L, Gianni L. Treatment landscape of triple-negative breast cancer—expanded options, evolving needs. Nat. Rev. Clin. Oncol. 2022;19:91–113. doi: 10.1038/s41571-021-00565-2. [DOI] [PubMed] [Google Scholar]
- 3.Kim C, et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell. 2018;173:879–893. doi: 10.1016/j.cell.2018.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 5.Burridge S. Drugging the epigenome. Nat. Rev. Drug Discov. 2013;12:92–93. doi: 10.1038/nrd3943. [DOI] [PubMed] [Google Scholar]
- 6.Shu S, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529:413–417. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vinet M, et al. Protein arginine methyltransferase 5: a novel therapeutic target for triple-negative breast cancers. Cancer Med. 2019;8:2414–2428. doi: 10.1002/cam4.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fedele P, Orlando L, Cinieri S. Targeting triple negative breast cancer with histone deacetylase inhibitors. Expert Opin. Investig. Drugs. 2017;26:1199–1206. doi: 10.1080/13543784.2017.1386172. [DOI] [PubMed] [Google Scholar]
- 9.Wu Q, et al. A chemical toolbox for the study of bromodomains and epigenetic signaling. Nat. Commun. 2019;10:1915. doi: 10.1038/s41467-019-09672-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deblois G, et al. Epigenetic switch-induced viral mimicry evasion in chemotherapy-resistant breast cancer. Cancer Discov. 2020;10:1312–1329. doi: 10.1158/2159-8290.CD-19-1493. [DOI] [PubMed] [Google Scholar]
- 11.Tang J, et al. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 2000;275:7723–7730. doi: 10.1074/jbc.275.11.7723. [DOI] [PubMed] [Google Scholar]
- 12.Guccione E, Richard S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 2019;20:642–657. doi: 10.1038/s41580-019-0155-x. [DOI] [PubMed] [Google Scholar]
- 13.Wu, Q., Schapira, M., Arrowsmith, C. H. & Barsyte-Lovejoy, D. Protein arginine methylation: from enigmatic functions to therapeutic targeting. Nat. Rev. Drug Discov. 20, 509–530 (2021). [DOI] [PubMed]
- 14.Boisvert F-M, Déry U, Masson J-Y, Richard S. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 2005;19:671–676. doi: 10.1101/gad.1279805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boisvert F-M, Rhie A, Richard S, Doherty AJ. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle. 2005;4:1834–1841. doi: 10.4161/cc.4.12.2250. [DOI] [PubMed] [Google Scholar]
- 16.Yoshimatsu M, et al. Dysregulation of PRMT1 and PRMT6, type I arginine methyltransferases, is involved in various types of human cancers. Int. J. Cancer. 2011;128:562–573. doi: 10.1002/ijc.25366. [DOI] [PubMed] [Google Scholar]
- 17.Fedoriw A, et al. Anti-tumor activity of the type I PRMT inhibitor, GSK3368715, synergizes with PRMT5 inhibition through MTAP loss. Cancer Cell. 2019;36:100–114. doi: 10.1016/j.ccell.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Scheer S, et al. A chemical biology toolbox to study protein methyltransferases and epigenetic signaling. Nat. Commun. 2019;10:19. doi: 10.1038/s41467-018-07905-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Y, et al. Epigenetic co-deregulation of EZH2/TET1 is a senescence-countering, actionable vulnerability in triple-negative breast cancer. Theranostics. 2019;9:761–777. doi: 10.7150/thno.29520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan N, et al. GSKJ4, an H3K27me3 demethylase inhibitor, effectively suppresses the breast cancer stem cells. Exp. Cell. Res. 2017;359:405–414. doi: 10.1016/j.yexcr.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 21.Eram MS, et al. A potent, selective, and cell-active inhibitor of human type I protein arginine methyltransferases. ACS Chem. Biol. 2016;11:772–781. doi: 10.1021/acschembio.5b00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsherniak A, et al. Defining a cancer dependency map. Cell. 2017;170:564–576. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behan FM, et al. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature. 2019;568:511–516. doi: 10.1038/s41586-019-1103-9. [DOI] [PubMed] [Google Scholar]
- 24.Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J. Biol. Chem. 2005;280:32890–32896. doi: 10.1074/jbc.M506944200. [DOI] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcotte R, et al. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell. 2016;164:293–309. doi: 10.1016/j.cell.2015.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fong JY, et al. Therapeutic targeting of RNA splicing catalysis through inhibition of protein arginine methylation. Cancer Cell. 2019;36:194–209. doi: 10.1016/j.ccell.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao G, et al. PRMT1 loss sensitizes cells to PRMT5 inhibition. Nucleic Acids Res. 2019;47:5038–5048. doi: 10.1093/nar/gkz200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sachs N, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–386. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 31.Schönborn J, et al. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 1991;19:2993–3000. doi: 10.1093/nar/19.11.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 33.Sheng W, et al. LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell. 2018;174:549–563. doi: 10.1016/j.cell.2018.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiappinelli KB, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roulois D, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowling EA, et al. Spliceosome-targeted therapies trigger an antiviral immune response in triple-negative breast cancer. Cell. 2021;184:384–403. doi: 10.1016/j.cell.2020.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehdipour P, et al. Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature. 2020;588:169–173. doi: 10.1038/s41586-020-2844-1. [DOI] [PubMed] [Google Scholar]
- 38.Šulc, P. et al. Repeats mimic immunostimulatory viral features across a vast evolutionary landscape. Preprint at bioRxiv10.1101/2021.11.04.467016 (2021).
- 39.Lima-Fernandes E, et al. Targeting bivalency de-represses Indian Hedgehog and inhibits self-renewal of colorectal cancer-initiating cells. Nat. Commun. 2019;10:1436. doi: 10.1038/s41467-019-09309-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sachamitr P, et al. PRMT5 inhibition disrupts splicing and stemness in glioblastoma. Nat. Commun. 2021;12:979. doi: 10.1038/s41467-021-21204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morel KL, et al. EZH2 inhibition activates a dsRNA–STING–interferon stress axis that potentiates response to PD-1 checkpoint blockade in prostate cancer. Nat. Cancer. 2021;2:444–456. doi: 10.1038/s43018-021-00185-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffin, G. K. et al. Epigenetic silencing by SETDB1 suppresses tumour intrinsic immunogenicity. Nature595, 309–314 (2021). [DOI] [PMC free article] [PubMed]
- 43.Kumar, S. et al. CARM1 inhibition enables immunotherapy of resistant tumors by dual action on tumor cells and T cells. Cancer Discov. 11, 2050–2071 (2021). [DOI] [PMC free article] [PubMed]
- 44.Wu Q, et al. GLUT1 inhibition blocks growth of RB1-positive triple negative breast cancer. Nat. Commun. 2020;11:4205. doi: 10.1038/s41467-020-18020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smirnov P, et al. PharmacoGx: an R package for analysis of large pharmacogenomic datasets. Bioinformatics. 2016;32:1244–1246. doi: 10.1093/bioinformatics/btv723. [DOI] [PubMed] [Google Scholar]
- 46.Liberzon A, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korotkevich, G. et al. Fast gene set enrichment analysis. Preprint at bioRxiv10.1101/060012 (2016).
- 48.Li J, et al. Characterization of human cancer cell lines by reverse-phase protein arrays. Cancer Cell. 2017;31:225–239. doi: 10.1016/j.ccell.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tapial J, et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 2017;27:1759–1768. doi: 10.1101/gr.220962.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han H, et al. Multilayered control of alternative splicing regulatory networks by transcription factors. Mol. Cell. 2017;65:539–553. doi: 10.1016/j.molcel.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 51.Irimia M, et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159:1511–1523. doi: 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1–5 and Fig. 1.
Data Availability Statement
Gene dependency data are available at depmap bioportal (https://depmap.org/portal/). RNA/protein expression data for TNBC cell lines are available in the datasets GSE73526 and GSE74702. RNA expression data for human breast cancer samples are available at TCGA (https://portal.gdc.cancer.gov/) and METABRIC (https://www.cbioportal.org/) and for the cell lines at GSE73526 and GSE74702. RNA expression data for PDX models are available upon request to protect participant privacy. The gene sets used for GSEA analysis are available at the molecular signatures database (https://www.gsea-msigdb.org/gsea/msigdb/). The RNA-seq data generated in this article are available from http://neellab.github.io/bfg/. Other data are available from the corresponding authors upon reasonable request. Source data are provided with this paper.
Code to reproduce the RNA-seq bioinformatics analyses and the related data are available at https://github.com/bhklab/TNBC_PRMT1i, and code to reproduce the RNA splicing analyses is available at https://gist.github.com/Fciamponi/ef8aae813575a4947c1f9cb87a1c7330. Code to reproduce the ERV analysis is available at https://github.com/WuEpiLab/TNBC_MS023_ERV.