Abstract
Background
Tumor-associated macrophages (TAM) are known to facilitate colorectal cancer (CRC) growth. High macrophage infiltration in thymidine phosphorylase (TYMP) expressing CRC may correspond to poor prognosis. The prognostic impact of the expression CD163, a receptor associated with TAM, and TYMP in stroma, respectively, tumor tissue is not yet established. The aim of this study was to identify the potential associations between TYMP and CD163 expression levels and relapse-free survival (RFS) of patients with stage II CRC, and if microdissection is of importance.
Methods
Stage II CRC patients, radically resected with relapse (n = 104), were matched to patients with a 5-year relapse-free follow-up (n = 206). Gene expression of TYMP and CD163 was analyzed in snap-frozen tumor tissues and in microdissected formalin-fixed tumor tissues separated into tumor epithelium and stroma.
Results
TYMP expression was high in poorly differentiated tumors, right-sided CRC, and tumors with high microsatellite instability CD163-expressing macrophages near tumor epithelial cells had high expression in poorly differentiated and T4 tumors. High TYMP expression in tumor epithelial cells was in the multivariate analyses associated with shorter relapse-free survival (hazard ratio 1.66; 95% confidence interval: 1.09–2.56; p < 0.05).
Conclusions
TYMP expression in tumor epithelial cells was associated with RFS and emphasizes the need for tissue microdissection. Additional studies are needed to establish whether TYMP and CD163 could add clinically relevant information to identify high-risk stage II patients that could benefit from adjuvant chemotherapy.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12094-022-02839-2.
Keywords: Macrophages, Thymidine phosphorylase, Colorectal neoplasm, Biomarkers, Microdissection, Microsatellite instability
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide [1]. Its curative treatment is surgery, which sometimes is complemented with chemotherapy based on 5-fluorouracil (5-FU) [2]. Mutations in tumor-suppressor genes, oncogenes, and genes related to DNA repair mechanisms play an important part in the development of CRC, as does the tumor microenvironment with its heterogenous composition of tumor and non-tumor cells [3].
Adjuvant chemotherapy is usually not recommended to patients with stage II CRC unless high-risk features have been identified, including < 12 analyzed lymph nodes, perineural invasion, tumor perforation, poorly differentiated tumors, and T4 stage tumors [2]. Studies have shown that some patients in the high-risk group and those with stage III CRC may benefit from adjuvant chemotherapy [4]. To better individualize the treatment strategy, it is important to identify relevant and informative biomarkers.
Whether or not the tumor has a defect in the DNA repair mechanisms, such as mismatch repair genes, may be of prognostic value. Tumors can be sub-grouped according to microsatellite instability (MSI) status: i.e., as microsatellite—stable (MSS), microsatellite instability—low (MSI-L), or microsatellite instability—high (MSI-H). MSI-H is associated with a better prognosis in the early stages of CRC, as well as a worse response to 5-FU-based chemotherapy [5].
The enzyme thymidine phosphorylase is encoded by the gene TYMP [6]. TYMP has a proangiogenic character and has been shown to cause resistance to apoptosis [7, 8]. In CRC, TYMP is often elevated compared with non-neoplastic tissues, and its high level may correspond to a poor prognosis [6, 9]. Epithelial cells, as well as stromal cells, thrombocytes, endothelial cells, and tumor-infiltrating macrophages express TYMP. In TYMP-positive CRC, high macrophage infiltration correlates with worse prognosis [10].
Depending on stimuli, monocytes may develop into traditional M1-polarized macrophages, which are bactericidal, or into M2-polarized macrophages with proangiogenic and anti-inflammatory properties. In colorectal tumors, a higher M2/M1 macrophage ratio correlates with a worse prognosis [11, 12]. The cluster of differentiation 163 (CD163) protein is a macrophage-specific hemoglobin scavenger receptor used for identification of M2-polarized macrophages among tumor-associated macrophages (TAMs) [13, 14]. Increased levels of TAM have been observed in stages III–IV compared with stages I–II, and correspond to a more aggressive CRC [15]. Controversially, the strong infiltration of TAM in CRC has also been considered as a predictor of low tumor grade and less lymph node metastasis [16]. Furthermore, the location of TAM relative to the tumor epithelium and stroma is of interest because high levels of TAM in the tumor epithelium indicate a worse prognosis [17].
The tumor microenvironment plays a crucial role in the pathogenesis of CRC. A better understanding of the relationship between gene expression levels in different cell types in the microenvironment and tumorigenesis is needed [18, 19]. Stromal cells stimulate the tumor’s invasive and metastatic abilities. Therefore, the heterogenous composition of stromal cells and tumor epithelial cells may by itself be of prognostic value. Gene expression analysis is usually conducted with microdissected tumor tissues consisting of epithelial cells, thus excluding the influence of stromal cells. In macrodissected tumor tissues, both epithelial cells and stromal cells are included. However, it has been shown that the RNA yield from stromal cells is lower than that from epithelial cells, which means that the contribution of the stroma may have a minor effect on the gene expression profile [20]. Thus, the effect of TYMP and CD163 gene expression on CRC carcinogenesis may vary depending on whether these genes are mostly expressed in tumor epithelial or stromal cells [10]. For example, in patients with primary, operable CRC, a high stromal TYMP gene expression has been shown to be associated with a favorable prognosis [7]. Whether macrodissected tissue is the best choice for analysis of gene expression, or if more accurate results will be obtained from microdissected tissues separated into epithelial and stromal cells, is presently unknown.
The aim of study was to identify the potential association between TYMP and CD163 gene expression and relapse-free survival (RFS), along with the clinicopathological factors of patients with stage II CRC, as well as the variation in expression in macro- and microdissected tumor tissues, the latter being divided into tumor epithelial cells and stroma cells.
Materials and methods
Study population
From 2002 until 2015, 1105 patients underwent surgery for CRC stage II at the Sahlgrenska University Hospital/Östra, Gothenburg, Sweden. Stage II CRC was defined as tumor growth through the muscularis propria into the subserosa or through all layers of the colon possibly invading nearby organs [21], no presence of tumor cells in regional lymph nodes or near the colon, and no distant metastases. All radically resected stage II patients who relapsed within 5 years of follow-up, and did not receive neoadjuvant treatment, were identified and included in the study if tissue samples could be retrieved (n = 104). These patients were matched according to tumor stage, tumor differentiation, and age to 208 patients who were relapse free after a 5-year follow-up. Two patients from this control group were excluded due to unmeasurable gene expression.
Macro- and microdissection of tumor tissues
Macroscopically dissected tumor tissues were snap frozen in liquid nitrogen and stored at –80 °C until further analysis. Formalin-fixed tumor tissues were microdissected and separated into tumor epithelial cells and stromal cells. If the tumor cell area to be analyzed was homogenous and large enough (≥ 80% tumor cells), a scalpel blade was used to manually collect the cells. Otherwise, tumor and/or stroma cells were microdissected using the PALM MicroBeam microscope (Carl Zeiss) at the CCI, Core Facilities, University of Gothenburg, Sweden. In 26 of the tumors, the stroma was lacking completely or was present in a very small area and, thus, could not be excised.
Total RNA extraction, cDNA preparation, and real-time PCR
Total RNA was isolated from snap-frozen tumor biopsies (10–30 mg) using the RNeasy kit (Cat # 74104, Qiagen, Sollentuna, Sweden) according to the manufacturer´s instructions. Total RNA was also isolated from formalin-fixed paraffin-embedded (FFPE) 10 µm sections using the RNeasy FFPE kit (Cat # 73504, Qiagen, Sollentuna, Sweden) according to the manufacturer’s instructions. The TissueLyser (Qiagen, Hilden, Germany) was used to disrupt and homogenize the tissue. Conditions for cDNA synthesis are presented in Supplemetary file 1. Real-time qPCR was performed using the 7500 Fast Real-time PCR system (Applied Biosystems, Foster City, USA). Assay details and PCR conditions are described in Supplementary file 2.
Microsatellite status
DNA was isolated from snap-frozen tumor tissues using an All-prep DNA/RNA mini kit (Cat # 80204 Qiagen, Sollentuna, Sweden), or from FFPE tumor tissue using an All-prep DNA/RNA FFPE kit (Cat # 80234; Qiagen, Sollentuna, Sweden). The MSI status was analyzed using the MSI Analysis System, version 1.2 (Promega, Madison, USA), which examined five microsatellites. The PCR was run on the Perkin-Elmer Gene Amp PCR system 9600 Thermal Cycler (Perkin Elmer, USA) according to the manufacturer’s instructions using 2 ng of DNA. The MSI markers were detected on an ABI prism 3730 instrument at KI Gene using the PowerPlex 4C matrix Standard (Cat # DG4800; Applied Biosystems, USA). MSI was defined as peak alterations in the marker electropherogram when tumor tissue was compared with matching mucosa. When more than one marker showed instability, the tumor was defined as MSI-H. If only one marker showed instability, it was defined as MSI-L. If no instability was detected, the tumor was designated as MSS.
Filter-dense multicolor microscopy
Filter-dense multicolor microscopy (FDMM) was used to visualize the distribution of TYMP and CD163. FDMM is an enhanced multifluorescence setup, which enables the visualization of several proteins simultaneously in one tissue sample [22]. The FFPE colorectal tumor tissues were cut into 4-µm-thick sections, deparaffinized with xylene, and rehydrated with an ethanol series (100%, 70%, 50%). Antigen retrieval was carried out in a pH 9 buffer (Agilent DAKO, Santa Clara, USA) in a pressure cooker. Tissue sections were stained with an anti-EpCAM antibody (VU1D9 LSBio, Seattle USA) for 1 h at RT. Tyramide amplification with Opal570 was applied according to the manufacturer’s instructions (Perkin Elmer, Waltham, USA). Antibodies were stripped away by incubating the slides in pH 9 buffer at a high temperature (just before boiling) in the microwave for 5 min. After cooling to room temperature, the tissue sections were stained with an anti-CD163 antibody (10D6, Novus Bio, Centennial, USA) for 1 h at RT, followed by tyramide amplification with Opal520. Subsequently, the tissue sections were stained with an anti-thymidine phosphorylase antibody (abcam180783, Cambridge, UK) at RT for 1 h, followed by anti-rabbit-AF647 Fab2 (Jackson Immuno West Grove, USA) at RT for 40 min. The tissues were mounted on the DAPI ProLong Diamond Antifade and were scanned with the Metasystems Scanner (Axio Imager.Z2 Microscope (Zeiss), 20 ×, Oberkochen, Germany) [22].
Statistical analysis
All statistical analyses were conducted using the commercial software JMP Pro 13.1.0 (SAS Institute Inc. Cary, NC, USA). Descriptive statistics and t test/ANOVA were used to evaluate the data. Values were presented as means ± standard deviations (SD), or as medians and ranges. The ΔΔCt method was applied to calculate the gene expression values, which were then transformed logarithmically, as the data were not normally distributed. Contingency tables with the nonparametric Chi-square/Kruskal–Wallis test was used to assess the differences between the groups. The Pearson’s correlation coefficient (r) was used to compare the sets of continuous parameters measured in the same tissue. RFS was defined as the time period from primary surgery to any recurrence of CRC, thus censoring death of any cause. To assess the putative relation of classical risk factors and gene expression on outcome, in terms of hazard ratios with 95% confidence intervals, univariate Cox proportional hazards regression analysis was applied. A multivariate proportional hazards regression analysis was used to adjust for possible confounding factors. The Wald test was used to evaluate significance in multivariate analyses. P value < 0.05 was considered significant. No correction for multiple testing was done.
Results
Patient and tumor characteristics
Patient and tumor characteristics are presented in Table 1. As shown, there was an even gender and age distribution between the relapse and relapse-free groups. Two hundred and sixty-seven patients had colon cancer; 41 had rectal cancer; and 2 had tumors in both the rectum and the colon. Of all the patients with colon cancer, 140 had tumors on the right side, whereas 127 had tumors on the left side. There was no significant difference in tumor location between the two groups. Twenty patients received adjuvant chemotherapy: 8 from the relapse group and 12 from the relapse-free group. Patients receiving adjuvant chemotherapy had as expected a higher degree of high-risk features: 75% had a T4 tumor; 30% had low tumor differentiation; and 20% had emergency surgery. Adjuvant chemotherapy was given with 5-FU/leucovorin (FLV) as single treatment (n = 12) or as combination treatment (n = 8).
Table 1.
Characteristics of the study population
| Relapse (n = 104) | No relapse (n = 206) | All patients (n = 310) | |
|---|---|---|---|
| Age, median (IQR) | 70 (60–79) | 70 (61–78) | 70 (61–78) |
| Gender, n (%) | |||
| Female | 50 (48.1) | 103 (50.0) | 153 (49.4) |
| Male | 54 (51.9) | 103 (50.0) | 157 (50.6) |
| Differentiation, n (%) | |||
| Well/moderate (G1–G2) | 89 (85.6) | 171 (83.0) | 260 (83.9) |
| Poor (G3–G4) | 11 (10.6) | 22 (10.7) | 33 (10.6) |
| Mucinous | 4 (3.8) | 13 (6.3) | 17 (5.5) |
| Tumor location, n (%) | |||
| Colon | 84 (80.8) | 183 (88.8) | 267 (86.1) |
| Rectum | 20 (19.2) | 21 (10.2) | 41 (13.2) |
| Colon and rectum | 0 | 2 (1.0) | 2 (0.64) |
| No. of examined lymph nodes, median (IQR) | 19 (14–23) | 23 (18–27) | 21 (16–26) |
| T-stage, n (%) | |||
| T3 | 81 (77.9) | 190 (92.2) | 271 (87.4) |
| T4 | 23 (22.1) | 16 (7.8) | 39 (12.6) |
| MSI status (%)a | |||
| MSI-H | 15 (15) | 45 (22) | 60 (19.8) |
| MSS/MSI-L | 84 (85) | 159 (78) | 243 (80.2) |
IQR interquartile range, MSI microsatellite instability, MSI-H microsatellite instability—high, MSS microsatellite—stable, MSI-L microsatellite instability—low, n number of patients
aMSI status could not be obtained for seven patients
Microsatellite status
The results of the MSI analysis are presented in Table 1. Sixty patients (19.8%) had MSI-H; 4 (1.3%) MSI-L; and 239 (78.9%) MSS tumors. Out of the 267 colon cancer patients, 23% had MSI-H tumors. All MSI-H tumors were located in the colon, of which 50 were on the right side and 10 on the left. There was no association between MSI and age or T-stage, but more female than male patients had MSI-H tumors (p < 0.01). There was a difference in MSI status according to tumor differentiation: 53% of the mucinous tumors, 50% of the well/moderately differentiated, but only 14% of the poorly differentiated tumors were MSI-H (p < 0.01).
TYMP gene expression
The mean TYMP gene expression in macrodissected tumor tissues (macTYMP), microdissected tumor epithelial cells (tecTYMP), and microdissected tumor stroma (stromaTYMP) was 0.52 ± 0.56, 0.24 ± 0.23, and 0.32 ± 0.36, respectively. There was a positive correlation between macTYMP and tecTYMP (r = 0.35; p < 0.01) and between stromaTYMP and tecTYMP (r = 0.51; p < 0.01); however, there was no correlation between macTYMP and stromaTYMP (r = 0.13; NS). TYMP gene expression did not correlate with age, gender, or T-stage, nor with relapse variables (Table 2).
Table 2.
TYMP expression according to pathological characteristics, MSI status, and relapse in stage II colorectal cancer
| n | macTYMPd | p value | n | tecTYMPd | p value | n | stromaTYMPd | p value | |
|---|---|---|---|---|---|---|---|---|---|
| T-stage | |||||||||
| T3 | 235 | 0.50 ± 0.53 | 268 | 0.24 ± 0.23 | 247 | 0.32 ± 0.37 | |||
| T4 | 27 | 0.69 ± 0.77 | NS | 39 | 0.26 ± 0.21 | NS | 35 | 0.29 ± 0.27 | NS |
| Tumor localizationa | |||||||||
| Colon | 221 | 0.55 ± 0.59 | 264 | 0.25 ± 0.24 | 242 | 0.34 ± 0.38 | |||
| Rectum | 39 | 0.33 ± 0.26 | < 0.01 | 41 | 0.17 ± 0.11 | NS | 38 | 0.20 ± 0.17 | < 0.01 |
| Tumor localization in colon | |||||||||
| Right-sided colon | 108 | 0.62 ± 0.68 | 140 | 0.28 ± 0.25 | 131 | 0.35 ± 0.42 | |||
| Left-sided colon | 113 | 0.48 ± 0.48 | < 0.05 | 124 | 0.22 ± 0.23 | < 0.01 | 111 | 0.33 ± 0.33 | NS |
| Differentiationb | |||||||||
| Well/moderate (G1–G2) | 223 | 0.44 ± 0.35 | 257 | 0.21 ± 0.20 | 236 | 0.29 ± 0.34 | |||
| Poor (G3–G4) | 26 | 1.07 ± 1.20 | < 0.01 | 33 | 0.48 ± 0.32 | < 0.01 | 32 | 0.48 ± 0.39 | < 0.01 |
| Mucinousb | 13 | 0.80 ± 0.85 | < 0.01 | 17 | 0.28 ± 0.17 | < 0.01 | 14 | 0.48 ± 0.55 | < 0.01 |
| MSI statusc | |||||||||
| MSI-H | 49 | 0.71 ± 0.63 | 60 | 0.30 ± 0.26 | 58 | 0.39 ± 0.50 | |||
| MSS/MSI-L | 212 | 0.47 ± 0.53 | < 0.01 | 240 | 0.23 ± 0.22 | < 0.05 | 217 | 0.30 ± 0.32 | NS |
| Relapse | |||||||||
| Yes | 75 | 0.48 ± 0.46 | 103 | 0.25 ± 0.23 | 93 | 0.29 ± 0.29 | |||
| No | 187 | 0.54 ± 0.59 | NS | 204 | 0.24 ± 0.23 | NS | 189 | 0.33 ± 0.39 | NS |
SD standard deviation, MSI microsatellite instability, MSI-H microsatellite instability—high, MSS microsatellite—stable, MSI-L: microsatellite instability—low, NS not significant, TYMP thymidine phosphorylase, macTYMP TYMP expression in tumor sample, tecTYMP TYMP expression in tumor epithelial cells, stromaTYMP TYMP expression in stroma
aTwo tumors with localization in both the colon and rectum were excluded
bMucinous tumors were compared with highly/moderately and poorly differentiated tumors
cUpon exclusion of the 20 patients that received adjuvant chemotherapy, there was no significant difference in terms of MSI status
dValues are expressed as the mean ± SD
The gene expression of TYMP was higher in colon tumors compared to rectal tumors, and both macTYMP and tecTYMP, but not stromaTYMP expression was significantly higher in right-sided compared to left-sided colon tumors (Table 2). Poorly differentiated tumors had a higher mean TYMP gene expression compared with well/moderately differentiated tumors, regardless of sampling method. The TYMP gene expression in mucinous tumors was higher than well/moderately but lower than poorly differentiated tumors, with the exception of stromaTYMP (Table 2). The mean expression of both macTYMP and tecTYMP was higher in MSI-H compared to MSI-L/MSS tumors; however, there was no difference in stromaTYMP expression according to MSI (Table 2).
CD163 gene expression
The mean CD163 gene expression in macrodissected tumor tissues (macCD163), microdissected tumor epithelial cells (tecCD163), and microdissected tumor stroma (stromaCD163) was 0.13 ± 0.25, 0.06 ± 0.09, and 0.11 ± 0.22, respectively. There was no correlation between macCD163 and tecCD163 (r = 0.12; NS), but there was a weak correlation between stromaCD163 and tecCD163 (r = 0.14; p < 0.05), and between macCD163 and stromaCD163 (r = 0.22; p < 0.01). CD163 gene expression did not correlate with age, gender, tumor location, or relapse variables (Table 3). There was no difference in stromaCD163 or macCD163 expression with regard to the T-stage.
Table 3.
CD163 expression according to pathological characteristics, MSI status, and relapse in stage II colorectal cancer
| n | macCD163d | p value | n | tecCD163d | p value | n | stromaCD163d | p value | |
|---|---|---|---|---|---|---|---|---|---|
| T-stage | |||||||||
| T3 | 233 | 0.11 ± 0.21 | 265 | 0.056 ± 0.072 | 244 | 0.12 ± 0.23 | |||
| T4 | 27 | 0.25 ± 0.50 | NS | 39 | 0.11 ± 0.15 | < 0.05 | 35 | 0.075 ± 0.099 | NS |
| Tumor localizationa | |||||||||
| Colon | 220 | 0.14 ± 0.27 | 262 | 0.062 ± 0.087 | 239 | 0.11 ± 0.23 | |||
| Rectum | 38 | 0.067 ± 0.082 | NS | 40 | 0.061 ± 0.086 | NS | 38 | 0.097 ± 0.15 | NS |
| Tumor localization in colon | |||||||||
| Right-sided colon | 108 | 0.15 ± 0.30 | 138 | 0.062 ± 0.081 | 128 | 0.11 ± 0.18 | |||
| Left-sided colon | 112 | 0.13 ± 0.25 | NS | 124 | 0.063 ± 0.094 | NS | 111 | 0.11 ± 0.28 | NS |
| Differentiationb | |||||||||
| Well/moderate (G1–G2) | 221 | 0.11 ± 0.23 | 255 | 0.058 ± 0.088 | 234 | 0.10 ± 0.21 | |||
| Poor (G3–G4) | 26 | 0.21 ± 0.37 | NS | 32 | 0.10 ± 0.082 | < 0.01 | 31 | 0.16 ± 0.28 | < 0.05 |
| Mucinousb | 13 | 0.27 ± 0.32 | NS | 17 | 0.050 ± 0.042 | < 0.01 | 14 | 0.14 ± 0.24 | NS |
| MSI status | |||||||||
| MSI-H | 49 | 0.20 ± 0.37 | 59 | 0.058 ± 0.056 | 57 | 0.13 ± 0.20 | |||
| MSS/MSI-L | 210 | 0.11 ± 0.22 | 0.05 | 238 | 0.063 ± 0.093 | NS | 215 | 0.10 ± 0.23 | < 0.05 |
| Relapsec | |||||||||
| Yes | 75 | 0.13 ± 0.26 | 100 | 0.071 ± 0.092 | 89 | 0.085 ± 0.13 | |||
| No | 185 | 0.13 ± 0.25 | NS | 204 | 0.057 ± 0.084 | NS | 190 | 0.12 ± 0.25 | NS |
SD standard deviation, MSI microsatellite instability, MSI-H microsatellite instability—high, MSS microsatellite—stable, MSI-L microsatellite instability—low, NS not significant, CD163 cluster of differentiation 163, macCD163 CD163 expression in tumor sample, tecCD163 CD163 expression in tumor epithelial cells, stromaCD163 CD163 expression in stroma
aTwo tumors with localization in both the colon and rectum were excluded
bMucinous tumors were compared with highly/moderately and poorly differentiated tumors
cUpon exclusion of the 20 patients that received adjuvant chemotherapy, tecCD163 expression was significantly higher in the relapse group (p < 0.05)
dValues are expressed as the mean ± SD
However, T4-tumors had a higher tecCD163 gene expression compared with T3-tumors (Table 3). There was also a significant difference between tecCD163 and stromaCD163 in terms of tumor differentiation. The mucinous tumors expressed lower tecCD163 compared to well/moderately and poorly differentiated tumors. MacCD163 and stromaCD163 gene expression was higher in MSI-H compared to MSI-L/MSS tumors; however, there was no difference in tecCD163 according toMSI status (Table 3).
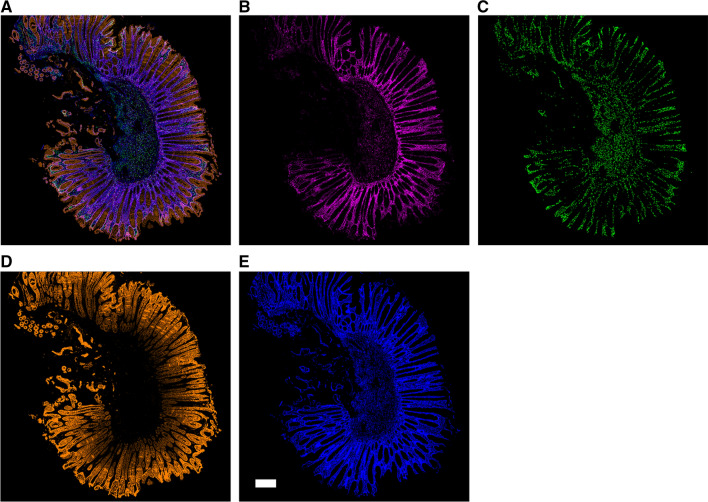
There was a positive correlation between TYMP and CD163 (p < 0.01) expression, comparing macTYMP with macCD163 (r = 0.37), tecTYMP with tecCD163 (r = 0.26) and stromaTYMP with stromaCD163 (r = 0.45). To visualize the distribution of TYMP and CD163-expressing macrophages in the tumor, the enhanced multifluorescence setup FDMM was performed (n = 12). FDMM revealed that TYMP and CD163 protein expression was heterogenous within and between samples. FDMM also showed that CD163 protein expression was not detected within, but near the tumor cells (Fig. 1).
Fig. 1.
Filter-dense multicolor microscopy of stage II colorectal cancer. 500 μm scale bar. a Merged image, b thymidine phosphorylase (magenta), c CD163 macrophages (green), d tumor epithelium immunolabeled with antibodies against EpCAM (orange), e nuclei counterstained with DAPI (blue)
TYMP and CD163 gene expression in relation to other risk factors
Cox regression univariate analysis of known risk factors showed that the tumor location, number of examined lymph nodes, T-stage, and whether the surgery was planned or acute were risk factors associated with RFS (Table 4). However, in the multivariate analysis, only T-stage and planned/acute surgery were found to be independent of other covariates included in the model. More advanced T-stage and acute surgery were associated with worse RFS. TecTYMP expression was als an independent variable associated with RFS in the multivariate analysis. The risk of relapse increased with increased expression of TYMP in the epithelium.
Table 4.
Association of covariates with relapse-free survival in stage II colorectal cancer
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p | HR | 95% CI | p | |
| Age | 1.01 | 0.98–1.02 | NS | 1.01 | 0.99–1.04 | NS |
| Gender | ||||||
| Female | 1 | 1 | ||||
| Male | 1.11 | 0.75–1.64 | NS | 1.43 | 0.81–2.60 | NS |
| Differentiation | ||||||
| Well/moderate | 1 | 1 | 0.39–9.74 | |||
| Poor | 0.77 | 0.38–1.41 | 0.84 | 0.28–2.22 | ||
| Mucinous | 0.66 | 0.20–1.59 | NS | 0.68 | 0.10–2.58 | NS |
| Tumor location* | ||||||
| Colon | 1 | 1 | ||||
| Rectum | 1.94 | 1.16–3.1 | < 0.05 | 1.88 | 0.93–3.70 | NS |
| No. of examined lymph nodes | 0.94 | 0.92–0.97 | < 0.05 | 1 | 0.96–1.03 | NS |
| T-stage | ||||||
| T3 | 1 | 1 | ||||
| T4 | 2.27 | 1.39–3.58 | < 0.05 | 2.60 | 1.05–5.84 | < 0.05 |
| Planned surgery | ||||||
| Yes | 1 | 1 | ||||
| No | 3.5 | 2.05–5.61 | < 0.05 | 5.44 | 1.29–19.22 | < 0.05 |
| Adjuvant chemotherapy | ||||||
| No | 1 | 1 | ||||
| Yes | 1.10 | 0.46–2.20 | NS | 1.26 | 0.33–6.4 | NS |
| MSI status | ||||||
| MSI-H | 1 | 1 | ||||
| MSI-L/MSS | 1.55 | 0.92–2.79 | NS | 1.55 | 0.33–6.40 | NS |
| TYMPa | ||||||
| Log macTYMP | 0.82 | 0.62–1.08 | NS | 0.69 | 0.46–1.05 | NS |
| Log tecTYMP | 1.06 | 0.85–1.33 | NS | 1.66 | 1.09–2.56 | < 0.05 |
| Log stromaTYMP | 0.86 | 0.69–1.07 | NS | 0.92 | 0.65–2.56 | NS |
| CD163a | ||||||
| Log macCD163 | 1.04 | 0.89–1.21 | NS | 1.03 | 0.83–1.28 | NS |
| Log tecCD163 | 1.15 | 0.98–1.34 | NS | 1.10 | 0.84–1.45 | NS |
| Log stromaCD163 | 0.93 | 0.79–1.09 | NS | 0.95 | 0.75–1.23 | NS |
Patients were matched by age, tumor stage, and differentiation, and the number of patients included in the univariate analysis were the same as in Tables 1, 2, 3, whereas 225 patients were included in the multivariate analysis
CI confidence interval, CD163 cluster of differentiation 163, Log macCD163 logarithmized CD163 expression in tumor sample, Log tecCD163 logarithmized CD163 expression in tumor epithelial cells, Log stromaCD163 logarithmized CD163 expression in stroma, HR hazard ratio, NS not significant, MSI microsatellite instability, MSI-H microsatellite instability—high, MSS microsatellite—stable, MSI-L microsatellite instability—low, TYMP thymidine phosphorylase, Log macTYMP logarithmized TYMP expression in tumor sample, Log tecTYMP logarithmized TYMP expression in tumor epithelial cells, Log stromaTYMP logarithmized TYMP expression in stroma
*When excluding adjuvantly treated patients (n = 20), multivariate analysis showed that patients with rectal tumors had an increased risk of relapse (p < 0.05)
aGene expression values were not normally distributed and therefore, logarithmized in the statistical analysis
Discussion
In this study, we analyzed TYMP and CD163 expression in micro- and macrodissected tumor tissue from 312 patients with stage II CRC. The effect of an active gene within a tumor may depend on whether it is expressed in stromal or epithelial cells and to what extent. Comparing micro- and macrodissected tumor tissues, the highest TYMP gene expression was found in macrodissected tissues, including both stromal and epithelial cells. There was a correlation between macTYMP and tecTYMP, suggesting that macTYMP could be used as a surrogate for gene expression in tumor epithelial cells. These results were in concordance with those of a previous study comparing mRNA levels in micro- and macrodissected tissues. The authors concluded that mRNA levels of stromal cells were low, and reliable tumor-specific gene expression profiles could be obtained from macrodissected tissues [20]. However, in this study, multivariate analysis showed that only tecTYMP was significantly associated with RFS. This association would not have been detected if only macTYMP had been analyzed. The tecTYMP expression was independent of the other two covariates associated with RFS namely, acute surgery and T4-tumors, which are known risk factors for CRC [2]. However, tecTYMP was not significant in the univariate analysis, which suggests that there were interactions between TYMP gene expression in epithelial cells and other risk factors. There was also a positive correlation between tecTYMP and stromaTYMP, possibly reflecting the overall TYMP gene expression in poorly differentiated tumors.
Although some studies evaluating TYMP as a predictive marker for chemotherapy have shown comparable results between microdissected tumor epithelial cells and macrodissected tissues, other studies show contradicting results [7, 20]. For example, when the expression levels of several 5-FU-related genes in micro- and macrodissected tumor tissues of patients with locally advanced rectal cancer who subsequently received radiotherapy were compared, a significant difference in TYMP gene expression was found [19]. The authors concluded that microdissection of tumor tissues after irradiation was important because of the changed tumor/stroma ratio induced by irradiation. However, in the present study no patient received radiotherapy, and there was a difference between macTYMP and tecTYMP which might indicate the importance of microdissection also in non-radiated CRC.
TYMP is expressed not only in the tumor epithelial cells but also in macrophages [6]. TYMP positive tumors and macrophage infiltration have previously been associated with a worse prognosis in CRC [10]. Furthermore, macrophages may polarize into an M2 macrophage subtype, considered as TAM, thus evolving pro-tumoral properties. In the study, it was possible to identify TAM both in micro- and macrodissected tissue by including the macrophage-specific marker CD163, and further, to stratify the influence of TAM in the tumor microenvironment and its correlation to TYMP expression. The results showed a positive correlation between TYMP and CD163 gene expression, both in macro- and microdissected tumor tissue. However, as visualized by FDMM, high TYMP protein expression could not be explained by high infiltration of CD163-positive macrophages expressing TYMP.
Several studies have shown that high CD163 expression correlates with worse survival, and higher CD163 expression has been reported in stages III–IV compared to earlier stages [13, 23, 24]. It has been suggested that in advanced cancer, the tumor epithelial cells can fuse with macrophages thereby adopting some of their abilities (such as migration) thus making them more prone to metastasize [13, 25]. If increased expression of CD163 is a late event during CRC development, this may explain the lack of association between CD163 expression and recurrence or RFS in the present study on stage II CRC.
It is known that some tumor characteristics vary depending on the tumor location. For example, right-sided colon cancer most often has been associated with a worse prognosis, higher rate of MSI, BRAF mutations, and CpG island methylation [26, 27]. In contrast, left-sided CRC has been associated with a higher grade of p53 and KRAS mutations [26]. In the present study, almost 20% of the patients had MSI-H tumors, and as expected, these were preferentially located in the right side of the colon and more common among female than male patients. However, in support of previous studies in terms of survival, the MSI status was not of prognostic value for RFS [14, 28].
A limitation of the study was that non-microdissected sections of FFPE tissue were not analyzed as a complement to the macrodissected snap-frozen tissue. Another limitation was that we did not include any pan-macrophage marker for comparison of infiltration of M1 and M2 macrophages, which might be of importance, since a high M2/M1 ratio is associated with worse survival [24, 29]. It might be of interest to address these issues in future studies.
In conclusion, our findings revealed that CD163-expressing macrophages near tumor epithelial cells had high expression in poorly differentiated and T4 tumors. High TYMP gene expression was seen in poorly differentiated tumors, right-sided CRC, and MSI-H tumors. In tumor epithelial cells, high TYMP gene expression was associated with shorter RFS, independent of known risk factors. This emphasizes the need of further studies using microdissection to establish whether TYMP and CD163 could add clinically relevant information to identify high risk stage II patients that could benefit from adjuvant chemotherapy.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Jaqueline Flach and Marianne Åkerström for their technical assistance, Hillevi Björkqvist and Ann-Louise Helminen for the collection of surgical samples, and Lena Munro for the work with the clinical database. We acknowledge the Centre for Cellular Imaging at the University of Gothenburg and the National Microscopy Infrastructure, NMI (VR-RFI 2016-00968), for providing assistance in microscopy.
Author contributions
DK, LS, UY, YW and EBL contributed equally to this work. EBL, and YW conceptualized the study; EBL supervised the work as well as provided resources, conducted the research and investigation process; EBL, DK, LS, and YW wrote the original draft of the manuscript; EBL. DK, and YW analyzed the study data; EBL, and DK, provided funding acquisition; UY and LS designed the FDMM methodology and verified its reproducibility; all authors have read and approved the manuscript.
Funding
Open access funding provided by University of Gothenburg. This work was funded by the Swedish state under the LUA/ALF agreement (ALFGBG-784211); the Swedish Cancer Society (CAN 2015/4999); the Lions Cancer Research Foundation (LCV 2017;38); the Assar Gabrielsson Foundation (FB15-51); the Swedish Society of Medicine (SLS-689001); the Göteborg Medical Society (GLS-499861); and the Anna-Lisa and Bror Björnsson Foundation. The funding sources had no role in the design or execution of the study.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
This article has been read and approved in the present form for submission by all authors.
Ethics approval
The study was conducted in line with the Declaration of Helsinki, and the regional ethical review board in Gothenburg approved the study (ethical board number 590-15).
Informed consent
Written informed consent to participate was obtained from all participants.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ferlay J, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2014 doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Labianca R, Nordlinger B, Beretta GD, et al. Early colon cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24(Suppl 6):vi64–vi72. doi: 10.1093/annonc/mdt354. [DOI] [PubMed] [Google Scholar]
- 3.Marmol I, Sanchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18:197. doi: 10.3390/ijms18010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van de Velde CJ, Boelens PG, Borras JM, et al. EURECCA colorectal: multidisciplinary management: European consensus conference colon and rectum. Eur J Cancer. 2014;50:1.e1–1.e34. doi: 10.1016/j.ejca.2013.06.048. [DOI] [PubMed] [Google Scholar]
- 5.Grady WM, Pritchard CC. Molecular alterations and biomarkers in colorectal cancer. Toxicol Pathol. 2014;42:124–139. doi: 10.1177/0192623313505155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bronckaers A, Gago F, Balzarini J, Liekens S. The dual role of thymidine phosphorylase in cancer development and chemotherapy. Med Res Rev. 2009;29:903–953. doi: 10.1002/med.20159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yasuno M, Mori T, Koike M, et al. Importance of thymidine phosphorylase expression in tumor stroma as a prognostic factor in patients with advanced colorectal carcinoma. Oncol Rep. 2005;13:405–412. [PubMed] [Google Scholar]
- 8.Matsuura T, Kuratate I, Teramachi K, Osaki M, Fukuda Y, Ito H. Thymidine phosphorylase expression is associated with both increase of intratumoral microvessels and decrease of apoptosis in human colorectal carcinomas. Can Res. 1999;59:5037–5040. [PubMed] [Google Scholar]
- 9.Lindskog EB, Wettergren Y, Odin E, Gustavsson B, Derwinger K. Thymidine phosphorylase gene expression in stage III colorectal cancer. Clin Med Insights Oncol. 2012;6:347–353. doi: 10.4137/CMO.S10226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumura M, Chiba Y, Lu C, et al. Platelet-derived endothelial cell growth factor/thymidine phosphorylase expression correlated with tumor angiogenesis and macrophage infiltration in colorectal cancer. Cancer Lett. 1998;128:55–63. doi: 10.1016/s0304-3835(98)00051-2. [DOI] [PubMed] [Google Scholar]
- 11.Cui YL, Li HK, Zhou HY, Zhang T, Li Q. Correlations of tumor-associated macrophage subtypes with liver metastases of colorectal cancer. Asian Pac J Cancer Prev. 2013;14:1003–1007. doi: 10.7314/apjcp.2013.14.2.1003. [DOI] [PubMed] [Google Scholar]
- 12.Zhong X, Chen B, Yang Z. The role of tumor-associated macrophages in colorectal carcinoma progression. Cell Physiol Biochem. 2018;45:356–365. doi: 10.1159/000486816. [DOI] [PubMed] [Google Scholar]
- 13.Shabo I, Olsson H, Elkarim R, Sun XF, Svanvik J. Macrophage infiltration in tumor stroma is related to tumor cell expression of CD163 in colorectal cancer. Cancer Microenviron. 2014;7:61–69. doi: 10.1007/s12307-014-0145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y, Wen X, Bae JM, Kim JH, Cho NY, Kang GH. The distribution of intratumoral macrophages correlates with molecular phenotypes and impacts prognosis in colorectal carcinoma. Histopathology. 2018;73:663–671. doi: 10.1111/his.13674. [DOI] [PubMed] [Google Scholar]
- 15.Kang JC, Chen JS, Lee CH, Chang JJ, Shieh YS. Intratumoral macrophage counts correlate with tumor progression in colorectal cancer. J Surg Oncol. 2010;102:242–248. doi: 10.1002/jso.21617. [DOI] [PubMed] [Google Scholar]
- 16.Koelzer VH, Canonica K, Dawson H, et al. Phenotyping of tumor-associated macrophages in colorectal cancer: impact on single cell invasion (tumor budding) and clinicopathological outcome. Oncoimmunology. 2016;5:e1106677. doi: 10.1080/2162402X.2015.1106677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrera M, Herrera A, Dominguez G, et al. Cancer-associated fibroblast and M2 macrophage markers together predict outcome in colorectal cancer patients. Cancer Sci. 2013;104:437–444. doi: 10.1111/cas.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park JH, McMillan DC, Powell AG, et al. Evaluation of a tumor microenvironment-based prognostic score in primary operable colorectal cancer. Clin Cancer Res. 2015;21:882–888. doi: 10.1158/1078-0432.CCR-14-1686. [DOI] [PubMed] [Google Scholar]
- 19.Inoue Y, Tanaka K, Yokoe T, et al. Microdissection is essential for gene expression analysis of irradiated rectal cancer tissues. Oncol Rep. 2009;22:901–906. doi: 10.3892/or_00000515. [DOI] [PubMed] [Google Scholar]
- 20.de Bruin EC, van de Pas S, Lips EH, et al. Macrodissection versus microdissection of rectal carcinoma: minor influence of stroma cells to tumor cell gene expression profiles. BMC Genom. 2005;6:142. doi: 10.1186/1471-2164-6-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertero L, Massa F, Metovic J, et al. Eighth edition of the UICC classification of malignant tumours: an overview of the changes in the pathological TNM classification criteria—what has changed and why? Virchows Arch. 2018;472:519–531. doi: 10.1007/s00428-017-2276-y. [DOI] [PubMed] [Google Scholar]
- 22.Kijani S, Yrlid U, Heyden M, Levin M, Boren J, Fogelstrand P. Filter-dense multicolor microscopy. PLoS ONE. 2015;10:e0119499. doi: 10.1371/journal.pone.0119499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding D, Yao Y, Yang C, Zhang S. Identification of mannose receptor and CD163 as novel biomarkers for colorectal cancer. Cancer Biomark Sect A Dis Mark. 2018;21:689–700. doi: 10.3233/CBM-170796. [DOI] [PubMed] [Google Scholar]
- 24.Yang C, Wei C, Wang S, et al. Elevated CD163(+)/CD68(+) ratio at tumor invasive front is closely associated with aggressive phenotype and poor prognosis in colorectal cancer. Int J Biol Sci. 2019;15:984–998. doi: 10.7150/ijbs.29836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shabo I, Midtbo K, Andersson H, et al. Macrophage traits in cancer cells are induced by macrophage-cancer cell fusion and cannot be explained by cellular interaction. BMC Cancer. 2015;15:922. doi: 10.1186/s12885-015-1935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrelli F, Tomasello G, Borgonovo K, et al. Prognostic survival associated with left-sided vs right-sided colon cancer: a systematic review and meta-analysis. JAMA Oncol. 2017;3:211–219. doi: 10.1001/jamaoncol.2016.4227. [DOI] [PubMed] [Google Scholar]
- 27.Natsume S, Yamaguchi T, Takao M, et al. Clinicopathological and molecular differences between right-sided and left-sided colorectal cancer in Japanese patients. Jpn J Clin Oncol. 2018;48:609–618. doi: 10.1093/jjco/hyy069. [DOI] [PubMed] [Google Scholar]
- 28.Gkekas I, Novotny J, Pecen L, Strigard K, Palmqvist R, Gunnarsson U. Microsatellite instability as a prognostic factor in stage II colon cancer patients, a meta-analysis of published literature. Anticancer Res. 2017;37:6563–6574. doi: 10.21873/anticanres.12113. [DOI] [PubMed] [Google Scholar]
- 29.Pinto ML, Rios E, Duraes C, et al. The two faces of tumor-associated macrophages and their clinical significance in colorectal cancer. Front Immunol. 2019;10:1875. doi: 10.3389/fimmu.2019.01875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.