Abstract
We investigated the influence of plant species, soil type, and plant development time on the shaping of microbial communities in soil and in association with roots. The sample group consisted of a total of 32 microcosms in three habitats: soil, rhizosphere, and rhizoplane. Communities were represented by the patterns of a sequence-specific separation of rRNA target sequences. Effects of experimental parameters were classified by a cluster analysis of pattern similarities. The type of plant species (clover, bean, or alfalfa) had the greatest effect in plant-associated habitats and also affected soil patterns. Plant development had a minor habitat-dependent effect that was partly obscured by replicate variation. The results stress the applicability of biased community representations in an analysis of induced variation.
A great variety of abiotic and biotic factors shape soil- and plant-associated habitats and modify the compositions and activities of their microbial communities, which in turn bear upon the quality of their environment, the growth of plants, and the production of root exudates (2). Bacterial communities in root-associated habitats respond with respect to density, composition, and activity to the abundance and great diversity of organic root exudates, eventually yielding plant species-specific microfloras which may also vary during plant development stages (3, 4, 17, 18). Due in part to the scarcity of convenient methods for exploration, our understanding of the different degrees and dynamics of microbial community variation as induced by soil type, plant type, or plant development is limited so far (5, 18).
As one step towards a better understanding of the relative variations of complex microbial communities in response to common conditions of agricultural practice, we studied the effects of plant type, soil type, and temporal development on the compositions and activities of microbial communities in soil and in association with leguminous plants. Therefore, we compared banding patterns of amplified 16S rRNA target sequences from soil, rhizosphere, and rhizoplane fractions by temperature gradient gel electrophoresis (TGGE). This technique offers a culture-independent method for tracking dominant bacterial populations in space and time (21). Unlike with the use of 16S ribosomal DNA target sequences for such investigations, the use of rRNA modifies the observation in favor of metabolically active microorganisms, because the abundance of ribosomes in a community can be viewed as a species-dependent function of cell numbers and their growth rates (9, 24, 28, 31). To the best of our knowledge, this study represents the first approach using culture-independent methods to systematically monitor the degree of variation of active, dominant bacterial populations in response to soil type, plant type, and the state of development of the plant.
In detail, soil samples from two different agricultural field sites in Braunschweig, Lower Saxony, Germany, were used: BBA soil (loamy sand) and FAL soil (para brown earth, silty sand). After removal of stones and root material, both soils were homogeneously mixed, and 36 kg of soil was added to each microcosm container (size, 40 by 30 by 20 cm).
To examine the variability of the bacterial community due to plant development, four replicates from microcosm containers with FAL soil seeded with 2 g of alfalfa seeds (Medicago sativa cv. Europa) were sampled at intervals of two weeks from week 2 to week 10. Four containers received BBA soil and were seeded with alfalfa and sampled after 10 weeks to analyze the influence of the soil type. To examine the influence of the crop species, four microcosm containers with FAL soil were seeded with 2.5 g of clover seeds (Trifolium pratense), and four more were seeded with 24 seeds of beans (Phaseolus vulgaris cv. Tilla). A total of 32 microcosm containers were placed in a randomized arrangement in the same greenhouse compartment and watered in the same way with tap water. The arrival of the shoot stage of the plants marked the end of the experiments and sampling, occurring after 8 weeks for beans, 10 weeks for alfalfa, and 11 weeks for clover.
From each microcosm, fractions of soil, rhizosphere, and rhizoplane were analyzed. Plants were removed from the containers and shaken carefully to remove nonadhering soil. After the complete removal of the plants, the remaining soil was homogeneously mixed and then sieved (0.8-mm mesh width). Samples of 2 g (wet weight) were used for the analysis of the soil fraction.
Plant roots were cut in smaller pieces of 1- to 2-cm lengths, and 4 g (fresh weight) of roots from each microcosm was washed twice for 2 min in 40 ml of saline (0.85% NaCl solution). The combined washing solutions were centrifuged (8,000 × g, 10 min, 4°C), and the resulting pellets were defined as the rhizosphere fraction.
The rhizoplane fraction was extracted according to a protocol developed by B. Mogge et al. (unpublished data). Briefly, the washed roots of the rhizosphere preparation were suspended in 20 ml of 0.1% sodium-cholate in a sterile plastic bag and twice treated mechanically in a Stomacher blender (model 400; Seward Medical, London, England) for 120 s each time at the highest speed. After transfer of the suspension into Erlenmeyer flasks, 0.5 g of polyethylene glycol 600 (Sigma, Deisenhofen, Germany) and 0.4 g of Chelex 100 (Sigma) were added and the solution was stirred (50 rpm) for 1 h at 4°C. The supernatant was stored on ice, while the roots were treated again by Stomacher blending with sodium-cholate solution. This Stomacher-stirring procedure to separate bacteria from roots was repeated two times, but only 0.5 g of polyethylene glycol 600 was added to the Erlenmeyer flasks after the first cycle. The supernatants were pooled, and the remaining roots or soil particles were removed by filtration through gauze (70-μm mesh width). The suspensions were centrifuged (8,000 × g, 10 min, 4°C), yielding the pellets that constituted the rhizoplane fraction.
Ribosomes were extracted from 2 g of soil, and the rhizosphere and rhizoplane fractions were extracted from 4 g of roots, as described by Felske et al. (8) with the modifications noted in reference 19. Reverse transcription-PCR (RT-PCR) of extracted 16S rRNA was performed with Moloney murine leukemia virus reverse transcriptase and RNase H− (Promega, Madison, Wis.). A 7.5-μl sample of the RT reaction mixture, containing 1 μl of template rRNA and 20 pmol of the reverse primer R1346, was denaturated at 70°C for 5 min, cooled on ice, and mixed with 12.5 μl of a solution containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 250 μM each deoxynucleoside triphosphate, and 100 U of Moloney murine leukemia virus reverse transcriptase. After incubation for 1 h at 42°C, 2 μl of this reaction mixture was used as a template for amplification of the resulting cDNA (as described by Felske et al. [8]) with the primer pair F968-GC and R1346, which is specific for highly conserved 16S rRNA of procaryotes amplifying the V6-V8 region of 16S rRNA. Amplifications omitting the RT step served as a negative control for the exclusive presence of the RNA template and negligible contamination with DNA.
Amplified fragments were separated by TGGE and stained with silver as described previously (19). Image analysis and clustering of the different habitat fractions were performed with Diversity Database software (MWG-Biotech, Ebersberg, Germany). Through the use of standard samples (PCR products from a DNA mixture of several bacterial species) on each gel as well as through reduction of the background signal of the gel in each lane, it became possible to compare patterns from different gels with each other.
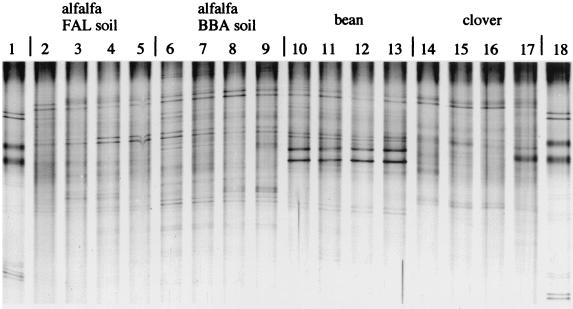
PCR-amplifiable rRNA was recovered from almost all soil, rhizosphere, and rhizoplane samples. Replicates of amplification of the same RNA extract or TGGE runs of the same RNA product produced highly similar banding profiles, demonstrating a good reproducibility for these procedures. Examples of the similarity of experimental microcosm replicates are shown in Fig. 1 for alfalfa, bean, and clover at the shoot stage.
FIG. 1.
TGGE separation patterns of rhizosphere samples from microcosm replicates. Samples were taken after 10 weeks from containers with alfalfa in FAL and BBA soil (lanes 2 to 5 and 6 to 9, respectively), after 8 weeks from containers with bean (lanes 10 to 13), and after 11 weeks from containers with clover (lanes 14 to 17). Amplification products for analysis were obtained after RT of rRNA target sequences of the samples through the use of the primers F984-GC and R1346. Lanes 1 and 18 show separation of the product mix used as a marker.
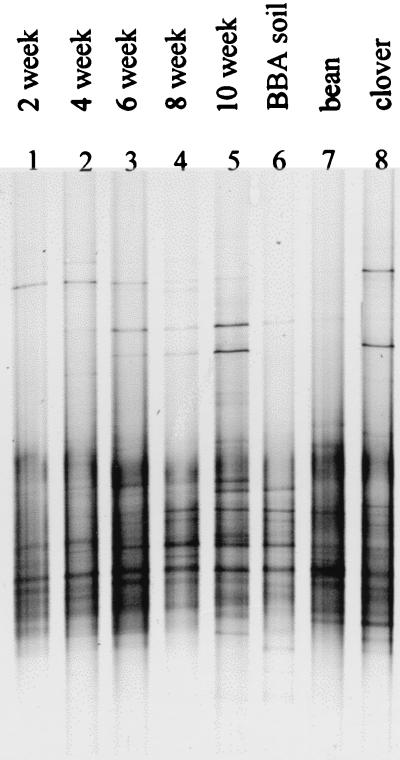
Variations among TGGE patterns corresponding to plant development time were minor in all habitats. Systematic community shifts could not be identified in samples from bulk soil, whereas some pattern variation could be correlated to development time in the rhizosphere and rhizoplane habitats. Figure 2 shows TGGE patterns from the rhizoplane in the time course experiment, with one replicate being taken from each sampling time. The variety of band intensities, predominantly in the upper part of the patterns, suggests time-dependent community shifts (Fig. 2, lanes 1 to 5).
FIG. 2.
TGGE separation patterns of rhizoplane samples. Individual examples of samples taken after 2, 4, 6, 8, and 10 weeks from microcosms with alfalfa in FAL soil (lanes 1 to 5), alfalfa in BBA soil after 10 weeks (lane 6), bean after 8 weeks (lane 7), and clover after 11 weeks (lane 8). After sampling and rRNA isolation, the target sequence amplification was done according to the general protocol (see Fig. 1 legend and the text).
More-apparent variations of the complete patterns were observed in response to soil and plant type. A comparison of how BBA soil and FAL soil affect the patterns of alfalfa plants at the shoot stage is shown in Fig. 1 (lanes 2 to 9) and Fig. 2 (lanes 5 and 6). The patterns of clover and bean microcosm samples in these figures demonstrate the degree of pattern variation depending on the plant type in the root-associated habitats.
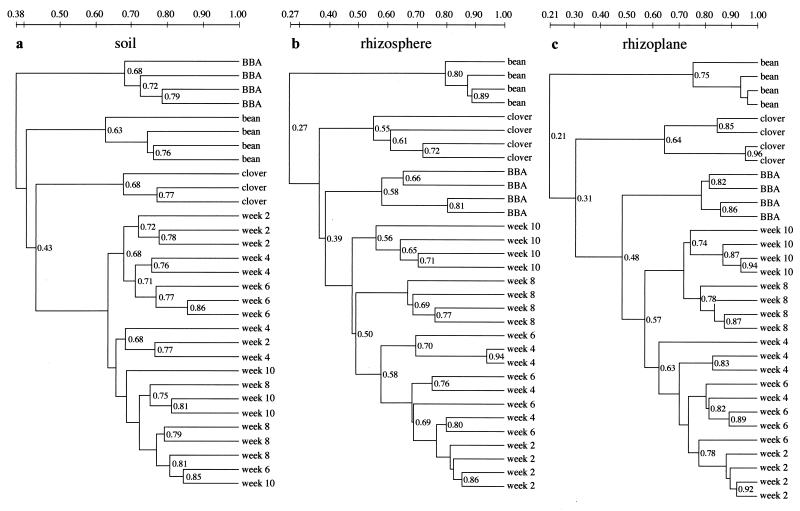
Similarity dendrograms serve to represent a comparison of all TGGE pattern scans from one habitat by cluster analysis (Fig. 3). After a series of independent test runs and comparisons, we used the Dice coefficient with weighting of band intensities as the distance measure and unweighted pair grouping with mathematical averages for clustering, as provided by Diversity Database software. These methods are conventionally regarded as suitable for such analyses (26).
FIG. 3.
Dendrogram structures of TGGE pattern comparisons from the habitats soil, rhizosphere, and rhizoplane. Through the use of Diversity Database software, the similarity of patterns was calculated using the Dice coefficient with weighting of band intensities. Unweighted pair grouping with mathematical averages was used for clustering. Experimental replicates are given the same designation, and their positions within the dendrogram do not indicate their identities. Bean and clover were sampled after 8 and 11 weeks, respectively. Week 2 through week 10, alfalfa microcosms sampled after the times indicated; BBA, alfalfa in BBA soil. All microcosms except that for alfalfa in BBA soil contained FAL soil (sampling after 10 weeks).
In the bulk soil samples, a relatively minor variation due to sampling time was confounded by replicate variation, and systematic community shifts were barely indentifiable (Fig. 3a). Apparently, the experimental variation of some replicates exceeded systematic shifts of community composition here. The replicate pattern variability was within the range of the total variability over time, which in turn was similar to the interreplicate variation of the other samples from the soil habitat (BBA soil, bean, and clover) (Fig. 3a). Time-dependent shifts, however, appear to be represented in the clustering of the rhizosphere and rhizoplane TGGE patterns. Patterns of 2-week-old roots were separated from those of 4- and 6-week-old roots here, and an even more pronounced separation of the clustered 8- and 10-week samples from the patterns at earlier times was visible in these habitats (Fig. 3b and c).
Our results are in accordance with the observations of Mahaffe and Klöpper (18), who used a fatty acid methyl ester analysis of isolates to demonstrate community shifts in the rhizosphere of growing cucumber, shifts that contrasted to what was observed in bulk soil. In our study, the time-dependent shifts of microorganism communities were clearly minor compared to the effects of soil type or plant species in all examined habitats. Apparently, major shifts in the microbial community did not occur during the plant development time investigated, and our method of using universal primers for rRNA target amplification produced results close to its limits of detection. Duineveld et al. (5, 6) also noted a high similarity among denaturing gradient gel electrophoresis patterns of rhizosphere samples from growing chrysanthemum; the degree of similarity demonstrated some correlation to sampling time, depending on the analysis of DNA or rRNA target sequences. Only an altered window of observation generated by the use of specific primers might reveal a stronger time-dependent stimulation of certain bacterial groups than has been observed (25, 27). Furthermore, it must be kept in mind that more-pronounced shifts might occur during the first days after the emergence of the root from the seed, when the root surface appears to be a rapidly changing substrate (11).
Differences in soil type and plant species affected the TGGE patterns more strongly, again depending on the habitat. The effect of the soil type exceeded that of the plant type in the soil habitat only, as it was expressed in a clustering of alfalfa plants in BBA soil that was clearly distinct from the clustering of the same plants in FAL soil. Griffiths et al. (14) also described differences in microbial communities depending on the soil type. However, an effect of the plant species on the microbial communities is also clearly visible in our soil samples. The extent of variation introduced by the growth of bean or clover instead of alfalfa almost matches the extent of the soil type effect (Fig. 3a). Thus, in spite of the experimental procedure separating soil from root pieces, the samples drawn from the rather large volume of the microcosm containers demonstrate the influence of plant density and intense rooting. As the rhizosphere is generally defined as the zone of soil that is directly influenced by the plant roots, the sampled bulk soil might, by definition, be regarded as belonging to that habitat (16).
The sampling of root-associated habitats, then, clearly reverses the order of similarity clustering by shifting the dominant role for pattern formation to the plant type (Fig. 3b and c). Patterns of bean habitats are well separated from those of all others in the rhizosphere and rhizoplane, whereas a distinct clustering of clover samples apart from the rest is more apparent in the latter habitat. Obviously, plants select for divergent communities on their roots, even if they belong to the same plant family. The soil remained a factor with similar levels of importance for community composition and activity for both of the alfalfa root-associated habitats. The soil effect well exceeds the time-dependent effect, as is apparent in the clustering of alfalfa in BBA soil that is distinct from that observed at all sampling times in the FAL soil microcosms.
The results of this TGGE pattern comparison with respect to plant and soil effects correspond with those of a previous greenhouse experiment which included other approaches for inference of community composition and activity associated with rye and alfalfa plants (19). Similar results were found by a fatty acid methyl ester analysis of isolates from microbial communities in the rhizospheres of canola and wheat in different soils by Germida et al. (13). Other investigators, however, found soil factors to be more important for determining the compositions of microbial rhizosphere communities (1). Latour et al. (15) identified the soil type as the most important factor affecting fluorescent Pseudomonas populations in the rhizospheres of flax and tomato. Such heterogeneous results may be explained either by case-specific differences (i.e., differences in plant and soil type) or by different sampling and analysis methods. In this study, the influence of the soil type diminished in a gradient from soil to rhizoplane, while the divergent influence of the plant species on the TGGE profile of microbial communities became increasingly dominant, in spite of the close relationship of the plants to one another.
Our results could be regarded as a classification of variations in microbial diversity, because TGGE patterns represent the frequency distribution of dominant sequence species from the habitat in a first approximation (10, 21). However, the patterns are a poor representation of the species diversity of complex microbial communities due to a variety of biases. First, an incomplete and selective extraction of nucleic acids from environmental samples is followed by an amplification step introducing additional biases (30). Second, sequence identity does not necessarily reflect an origin from the same species (12), and heterogeneous bands of a pattern may originate from one bacterial strain due to its heterogeneity in rRNA genes (22). These biases are shared by other approaches in molecular microbial ecology. Third, a one-dimensional separation of heterogeneous sequences must result in a limited potential to represent the informational content present in the mixture, with respect to detection sensitivity and separation potential. It was determined that the band intensities of the silver-stained gels generally fell into the range of proportionality of DNA content and band intensity (data not shown). Sensitivity was consistent with a detection limit of sequences comprising about 1% of the most abundant sequences as a general limitation of the method (20). One band of a TGGE pattern may also be composed of several nonidentical sequences, due to characteristics of the separation principle which cannot be compensated for by calculation (23, 29). An analysis of the informational content of TGGE profiles is further compromised by the fact that a fraction of the sequences will escape analysis due to the formation of a “cloudy” distribution during denaturation and subsequent migration, thus contributing to background noise but not to their identification as a signal for pattern comparison (32, 33).
Though TGGE patterns represent biased sequence frequency distributions with an unknown variable relationship to the species diversity of complex communities, the patterns are well suited for monitoring the variation of complex communities in response to a variety of experimental conditions with large sets of samples, as demonstrated in this study. The comparative approach compensates for some of the quantitative biases of the representation of sequence abundances in TGGE patterns; the variation of signal intensity between two samples represents a real change if the types of bias in their generation are similar in both. Some degree of similarity (in composition and thus in their TGGE profiles) among the communities under investigation must be mentioned as one prerequisite for meaningful comparisons. The sensitivity of the analysis then depends on pattern complexity and the reproducibility of experimental replicates. In this study, the experimental procedure, together with a minor variation among microcosm replicates, was suitable for observing an effect from the parameter under investigation in most cases. From previous experiments (unpublished), we tentatively infer that the use of activity-related rRNA templates for amplification and analysis increased the sensitivity of our approach.
Relationships between changes in the environment and the response of TGGE patterns of microbial communities are providing some comprehensive insight into their natural dynamics and variability. Determining the TGGE patterns may be a useful step towards a better understanding of the dynamics of complex microbial communities in demonstrating a sensitive monitoring of the responses of the microbes to common natural conditions in agricultural practice. For more focused analyses of particular factors affecting variation, the observation can be modified by choosing specific primers for target sequence selection and by transforming pattern information through the characterization of bands. In such cases, however, univariate comparisons usually cannot lead to the unequivocal identification of responsive microbial or sequence species representative of community reactions, as indicated by replicate variation-dependent clustering topology. For subtle community shifts, this objective will generally unravel as a problem of characterizing complex factors (principal components) in a multivariate analysis as used for community-level physiological profiling with BIOLOG.
Whereas a biased view of their composition suffices to sensitively determine a relative degree of variation among complex microbial communities, the parameter of variation does not appear to be sufficient for evaluating the effect of stress factors on diversity and long-term adaptability in a straightforward manner. Thus, the use of microbial community variation as an alternative endpoint in ecotoxicology for evaluating the hazard of human interference deserves another critical inspection (7).
Acknowledgments
This work was supported by grants of the German Ministry for Education and Research, BMBF.
REFERENCES
- 1.Bachman G, Kinzel H. Physiological and ecological aspects of the interactions between plant roots and rhizosphere soil. Soil Biol Biochem. 1992;24:5543–5552. [Google Scholar]
- 2.Bever J D, Westover K M, Antonovics J. Incorporating the soil community into plant population dynamics: the utility of the feedback approach. J Ecol. 1997;85:561–573. [Google Scholar]
- 3.Bowen G D, Rovira A D. The rhizosphere, the hidden half. In: Waisel Y, Eshel A, Kafkafi U, editors. Plant roots—the hidden half. New York, N.Y: Marcel Dekker; 1991. pp. 641–649. [Google Scholar]
- 4.Curl E A, Truelove B. The rhizosphere. Berlin, Germany: Springer-Verlag; 1986. [Google Scholar]
- 5.Duineveld B M, Rosado A S, van Elsas J D, van Veen J A. Analysis of the dynamics of bacterial communities in the rhizosphere of the chrysanthemum via denaturing gradient gel electrophoresis and substrate utilization patterns. Appl Environ Microbiol. 1998;64:4950–4957. doi: 10.1128/aem.64.12.4950-4957.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duineveld B M, Kowalchuk G A, Keijzer A, van Elsas J D, van Veen J A. Analysis of bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA as well as DNA fragments coding for 16S rRNA. Appl Environ Microbiol. 2001;67:172–178. doi: 10.1128/AEM.67.1.172-178.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelen B, Meinken K, von Wintzingerode F, Heuer H, Malkomes H-P, Backhaus H. Monitoring impact of a pesticide treatment on bacterial soil communities by metabolic and genetic fingerprinting in addition to conventional testing procedures. Appl Environ Microbiol. 1998;64:2814–2821. doi: 10.1128/aem.64.8.2814-2821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felske A, Engelen B, Nübel U, Backhaus H. Direct ribosome isolation from soil to extract bacterial rRNA for community analysis. Appl Environ Microbiol. 1996;62:4162–4167. doi: 10.1128/aem.62.11.4162-4167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felske A, Rheims H, Wolterink A, Stackebrandt E, Akkermans A D L. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology. 1997;143:2983–2989. doi: 10.1099/00221287-143-9-2983. [DOI] [PubMed] [Google Scholar]
- 10.Ferris M J, Ward D M. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fokkema N J, Schippers B. Phyllosphere versus rhizosphere as environments for saprophytic colonisation. In: Fokkema N J, van den Heuvel J, editors. Microbiology of the phyllosphere. Cambridge, England: Cambridge University Press; 1986. pp. 137–159. [Google Scholar]
- 12.Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 13.Germida J J, Siciliano S D, Renato de Freitas J, Seib A M. Diversity of root-associated bacteria associated with field-grown canola (Brassica napus L.) and wheat (Triticum aestivum L.) FEMS Microbiol Ecol. 1998;26:43–50. [Google Scholar]
- 14.Griffiths B S, Ritz K, Glover L A. Broad-scale approaches to the determination of soil microbial community structure: application of the community DNA hybridization technique. Microb Ecol. 1996;31:269–280. doi: 10.1007/BF00171571. [DOI] [PubMed] [Google Scholar]
- 15.Latour X, Corberand T, Laguerre G, Allard F, Lemanceau P. The composition of fluorescent pseudomonad populations associated with roots is influenced by plant and soil type. Appl Environ Microbiol. 1996;62:2449–2456. doi: 10.1128/aem.62.7.2449-2456.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch J M. The rhizosphere. Chichester, England: John Wiley & Sons; 1990. [Google Scholar]
- 17.Lynch J M, Whipps J M. Substrate flow in the rhizosphere. Plant Soil. 1990;129:1–10. [Google Scholar]
- 18.Mahaffe W F, Klöpper J W. Temporal changes in the bacterial communities of soil, rhizosphere, and endorhiza associated with field-grown cucumber (Cucumis sativus L.) Microb Ecol. 1997;34:210–223. doi: 10.1007/s002489900050. [DOI] [PubMed] [Google Scholar]
- 19.Miethling R, Wieland G, Backhaus H, Tebbe C C. Variation of microbial communities in response to crop species, soil origin and inoculation with Sinorhizobium meliloti L 33. Microb Ecol. 2000;40:43–56. doi: 10.1007/s002480000021. [DOI] [PubMed] [Google Scholar]
- 20.Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- 21.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R I, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nübel U, Garcia-Pichel F, Kühl M, Muyzer G. Quantifying microbial diversity: morphotypes, 16S rRNA genes, and carotenoids of oxygenic phototrophs in microbial mats. Appl Environ Microbiol. 1999;65:422–430. doi: 10.1128/aem.65.2.422-430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rooney-Varga J N, Devereux R, Evans R S, Hines M E. Seasonal changes in the relative abundance of uncultivated sulfate-reducing bacteria in a salt marsh sediment and in the rhizosphere of Spartina alterniflora. Appl Environ Microbiol. 1997;63:3895–3901. doi: 10.1128/aem.63.10.3895-3901.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sackin M J, Jones D. Computer-assisted classification. In: Goodfellow M, O'Donnel A G, editors. Handbook of new bacterial systematics. London, England: Academic Press Ltd.; 1993. pp. 281–313. [Google Scholar]
- 27.Sato K, Jiang H-Y. Gram-positive bacterial flora on the root surface of wheat (Triticum aestivum L.) grown under different soil conditions. Biol Fertil Soils. 1996;23:121–125. [Google Scholar]
- 28.Teske A, Wawer C, Muyzer G, Ramsing N B. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vallaeys T, Topp E, Muyzer G, Macheret V, Laguerre G, Rigaud A, Soulas G. Evaluation of denaturing gradient gel electrophoresis in the detection of 16S rDNA sequence variation in rhizobia and methanotrophs. FEMS Microbiol Ecol. 1997;24:279–285. [Google Scholar]
- 30.Von Wintzingerode F, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 31.Ward D M, Bateson M M, Weller R, Ruff-Roberts A L. Ribosomal RNA analysis of microorganisms as they occur in nature. In: Marshall K C, editor. Advances in microbial ecology. Vol. 12. New York, N.Y: Plenum Press; 1992. pp. 219–286. [Google Scholar]
- 32.Wieland G. Analysis of microbial communities and their natural and induced shifts by separation of ribosomal nucleic acids. Ph.D. dissertation. 2000. http://www.biblio.tu-bs.de/ediss/data/20000808a/20000808a.html [Online.] University of Braunschweig, Braunschweig, Germany. http://www.biblio.tu-bs.de/ediss/data/20000808a/20000808a.html. . [Google Scholar]
- 33.Wu Y, Hayes V M, Osinga J, Mulder I M, Looman M W G, Buys C H C, Hofstra R M W. Improvement of fragment and primer selection for mutation detection by denaturing gradient gel electrophoresis. Nucleic Acids Res. 1998;26:5432–5440. doi: 10.1093/nar/26.23.5432. [DOI] [PMC free article] [PubMed] [Google Scholar]