Summary
Background
To investigate a vaccine technology with potential to protect against coronavirus disease 2019 (COVID-19) and reduce transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with a single vaccine dose, we developed a SARS-CoV-2 candidate vaccine using the live vesicular stomatitis virus (VSV) chimeric virus approach previously used to develop a licensed Ebola virus vaccine.
Methods
We generated a replication-competent chimeric VSV-SARS-CoV-2 vaccine candidate by replacing the VSV glycoprotein (G) gene with coding sequence for the SARS-CoV-2 Spike glycoprotein (S). Immunogenicity of the lead vaccine candidate (VSV∆G-SARS-CoV-2) was evaluated in cotton rats and golden Syrian hamsters, and protection from SARS-CoV-2 infection also was assessed in hamsters.
Findings
VSV∆G-SARS-CoV-2 delivered with a single intramuscular (IM) injection was immunogenic in cotton rats and hamsters and protected hamsters from weight loss following SARS-CoV-2 challenge. When mucosal vaccination was evaluated, cotton rats did not respond to the vaccine, whereas mucosal administration of VSV∆G-SARS-CoV-2 was found to be more immunogenic than IM injection in hamsters and induced immunity that significantly reduced SARS-CoV-2 challenge virus loads in both lung and nasal tissues.
Interpretation
VSV∆G-SARS-CoV-2 delivered by IM injection or mucosal administration was immunogenic in golden Syrian hamsters, and both vaccination methods effectively protected the lung from SARS-CoV-2 infection. Hamsters vaccinated by mucosal application of VSV∆G-SARS-CoV-2 also developed immunity that controlled SARS-CoV-2 replication in nasal tissue.
Funding
The study was funded by Merck Sharp & Dohme, Corp., a subsidiary of Merck & Co., Inc., Rahway, NJ, USA, and The International AIDS Vaccine Initiative, Inc. (IAVI), New York, USA. Parts of this research was supported by the Biomedical Advanced Research and Development Authority (BARDA) and the Defense Threat Reduction Agency (DTRA) of the US Department of Defense.
Keywords: Vesicular stomatitis virus, VSV, SARS-CoV-2 vaccine, Mucosal vaccination, Cotton rat, Syrian hamster
Research in context.
Evidence before this study
The target for protective humoral immunity against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the trimeric surface glycoprotein Spike, which directs essential functions including binding to the angiotensin converting enzyme-2 (ACE2) cellular receptor and virus entry. An ebolavirus Zaire (ZEBOV) vaccine (rVSV∆G-ZEBOV-GP marketed as ERVEBO®) that immunises against the ZEBOV surface glycoprotein (GP) had been developed before using the VSV∆G chimeric virus vaccine approach, and it was shown to be safe and highly effective for vaccinating people in an infectious disease outbreak environment. Intramuscular (IM) injection with live rVSV∆G-ZEBOV-GP was shown to rapidly induce antibodies against the trimeric GP complex that are associated with protection. Thus, we followed this approach to develop a vaccine candidate delivering the SARS-CoV-2 Spike (VSV∆G-SARS-CoV-2).
Added value of this study
Preclinical evaluation of VSV∆G-SARS-CoV-2 showed that vaccination by IM injection elicited serum antibodies in hamsters, including antibodies capable of neutralising SARS-CoV-2. Vaccine-elicited systemic humoral responses also were associated with reduced SARS-CoV-2 replication in the hamster lung and prevention of weight loss in challenged animals. This preclinical data supported development of a vaccine candidate for intramuscular injection that was advanced for a phase 1 clinical trial. Importantly, data from additional preclinical studies also showed that hamsters developed substantial systemic immune responses when VSV∆G-SARS-CoV-2 was used for mucosal vaccination and that the vaccinated animals were able to control SARS-CoV-2 replication in both the lung and nasal tissue. Additional investigation of intranasal (IN) vaccination revealed that lower doses of VSV∆G-SARS-CoV-2 elicited higher neutralising antibody titres in hamsters when compared with IM injection.
Implications of all the available evidence
The data demonstrated that hamsters developed neutralising serum antibodies and were protected from SARS-CoV-2 infection in the lung when vaccinated by IM injection or by applying VSV∆G-SARS-CoV-2 to upper respiratory tract mucosal surfaces. The data from the hamster studies also indicated that mucosal administration of the vaccine induced protective immunity that controlled SARS-CoV-2 replication in the nasal cavity and nasopharynx, which are the key portals of entry for initiation of host infection and an important point of virus egress for transmission to others. Furthermore, compared with IM injection, IN vaccination was found to be strongly immunogenic even when lower vaccine doses were used, which may enable dose-sparing regimens.
Alt-text: Unlabelled box
Introduction
The coronavirus infectious disease pandemic that began in 2019 (coronavirus disease 2019 [COVID-19]) and continues today illustrates the threat caused by emerging RNA viruses.1,2 COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is a member of the Coronaviradae family of enveloped RNA viruses that contain single-stranded, nonsegmented, positive-sense RNA genomes2 and is genetically related to coronaviruses that circulate in bats.3
The multimeric Spike (S) glycoprotein (GP) complex on the SARS-CoV-2 surface is the major target of the protective humoral immune response.4,5 Spike directs cell attachment by binding the angiotensin-converting enzyme 2 (ACE2) cellular receptor, after which cellular protease cofactors such as transmembrane serine protease 2 (TMPRSS2), or endosomal proteases such as Cathepsin B or L, cleave the Spike to activate fusion between the viral and cellular membranes that allows the virus core to enter the cell cytoplasm.4,6 Vaccine developers have focused primarily on candidates that will induce humoral responses against Spike, and remarkable progress has been made adapting and advancing various technologies for Spike immunogen delivery.7, 8, 9 Spike-specific binding antibodies and neutralising antibodies (nAbs) induced by vaccination have been correlated with a reduced risk of infection and disease.10, 11, 12, 13
An ideal vaccine for use in a pandemic is one that is safe and effective following a single dose, and therefore, we focused on developing a SARS-CoV-2 vaccine using the vesicular stomatitis virus (VSV) chimeric virus approach used for the ebolavirus Zaire vaccine (VSV∆G-ZEBOV-GP marketed as ERVEBO®), which was shown to be effective against ebolavirus disease following a single immunisation.14,15 In this vaccine design, VSV is modified by substituting the VSV G surface glycoprotein gene with a gene encoding a functional glycoprotein from a heterologous enveloped virus that can provide essential functions needed for virus replication, including cell attachment and entry.16,17 Thus, the VSV∆G-based vaccine is designed to deliver a native functional viral glycoprotein target in the context of a nonpathogenic viral infection. Motivated by the efficacy observed with VSV∆G-ZEBOV-GP, we applied similar technology to SARS-CoV-2 and developed a VSV∆G chimera encoding the SARS-CoV-2 Spike (VSV∆G-SARS-CoV-2).
We have shown that VSV∆G-SARS-CoV-2 was immunogenic in cotton rats and Syrian hamsters when administered with a single intramuscular (IM) injection, and that vaccination protected hamsters from disease following SARS-CoV-2 challenge. The vaccine for IM injection was then advanced to a phase 1 clinical trial (ClinicalTrials.gov Identifier: NCT04569786).18 We also evaluated mucosal vaccination and found that hamsters developed anti-Spike serum antibody titres that exceeded those in animals vaccinated by IM injection, but that cotton rats did not respond, suggesting that immunogenicity following mucosal VSV∆G-SARS-CoV-2 application was associated with ability of the Spike-dependent chimeric virus to infect and replicate in the respiratory tract mucosa of hamsters. Immunity induced by mucosal vaccination also protected vaccinated hamsters from SARS-CoV-2 replication in the lung and, notably, the nose as well. Moreover, strong serum antibody responses could be elicited in hamsters by intranasal (IN) vaccination using much lower doses of VSV∆G-SARS-CoV-2 than were necessary to elicit responses following IM vaccination.
Methods
Cell culture
Vero cells used to rescue and propagate VSV∆G-SARS-CoV-2 chimeras were derived from a working cell bank generated from a master cell bank prepared for IAVI (unpublished). The master cell bank was generated from cells that originated from The World Health Organisation (WHO) working cell bank (WHO 10-87) that was deposited at the European Collection of Authenticated Cell Cultures (Vero [WHO], ECACC 88020401). The IAVI cell bank was used earlier to manufacture clinical trial material.19
Vero cell monolayers were propagated in Dulbecco's modified Eagle medium (DMEM; Thermo Fisher Scientific, Waltham, MA) supplemented with 10% gamma irradiated fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO) with additional additives, including 2 mM L-glutamine, 1 mM sodium pyruvate, 0.1 mM MEM nonessential amino acids and 50 µg/mL gentamicin (all from Thermo Fisher Scientific, Waltham, MA). Cells were grown in incubators maintained at 37 °C, 5% carbon dioxide (CO2), and 85% humidity. Monolayers were dissociated for subculturing by treatment with TrypLE™ Select (Thermo Fisher Scientific, Waltham, MA) and cells were counted using a Countess™ 3 Automated Cell Counter (Thermo Fisher Scientific, Waltham, MA).
Vero cells used to prepare VSV∆G-SARS-CoV-2 vaccine material were derived from the WHO 10-87 working cell bank (Merck & Co., Inc., Rahway, NJ, USA, unpublished). Cells used for infection were grown in buffered serum-free medium (VP-SFM, Thermo Fisher Scientific, Waltham, MA) in sealed roller bottles. Medium from infected cultures was harvested 48 h after infection, after which, virus was purified by ultrafiltration and stored in buffer formulated with 10 mM Tris (pH 7.4), 10% sucrose, and 2.5 mg/L recombinant human serum albumin (Cellastim™, InVitria, Aurora, CO).
Molecular cloning and recombinant VSV
A gene encoding the full-length SARS-CoV-2 Spike (2019-nCoV/USA-WA1/2020, GenBank MN985325.1)20 was designed using a codon frequency consistent with VSV,21 after which it was synthesised by GenScript, Inc. The Spike gene was inserted into the VSV genomic clone, which was kindly provided by the Public Health Agency of Canada,17 between the M and L genes (Figure 1a).
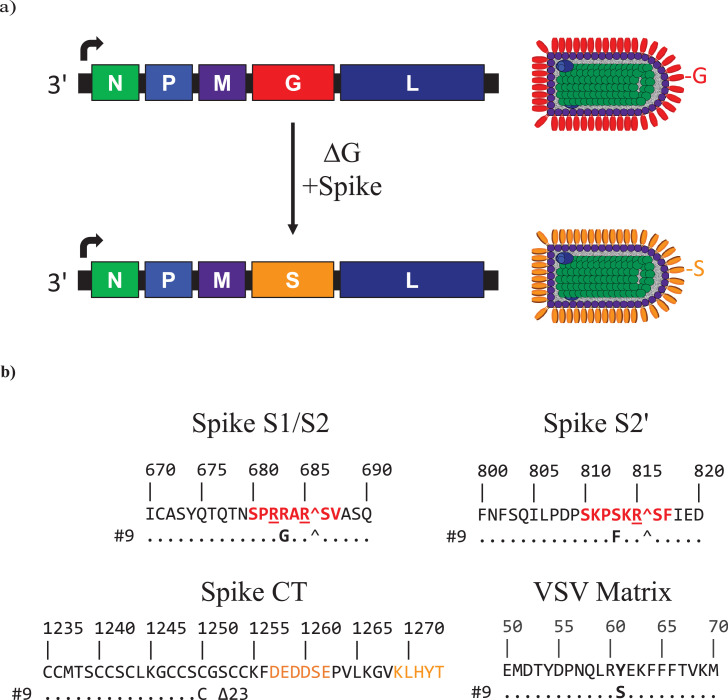
Figure 1.
VSV∆G-SARS-CoV-2 vaccines. (a) Genome maps for vesicular stomatitis virus (VSV) and the VSV∆G-SARS-CoV-2 chimera illustrate that the chimeric virus was generated by replacing the VSV G gene (red) with a gene encoding the SARS-CoV-2 (WA1/2020) Spike (orange). Adjacent to the genome maps, the proteins in the virion particle illustrations are coloured to correspond with the genomes. VSV encodes five structural proteins: N, nucleocapsid protein; P, phosphoprotein polymerase subunit and nucleocapsid assembly chaperone; M, matrix; G, glycoprotein; L, large protein, which is the catalytic domain of the RNA-dependent RNA polymerase.102 The VSV genome is a single-stranded, negative-sense, nonsegmented RNA. VSV mRNAs are transcribed from single promoter at the 3′ terminus of the genome indicated by an arrow. Transcription termination and reinitiation signals in each intergenic region allow synthesis of individual mRNAs.102 (b) Amino acid sequences where adaptive mutations developed in rVSV∆G-SARS-CoV-2 #9. The Spike amino acid coordinates correspond to the Wuhan reference strain (Genbank NC_045512). The VSV matrix sequence positions refer to laboratory-adapted recombinant VSV Indiana (Genbank AR123015).103 Spike S1/S2: cleavage site between the S1 and S2 subunits recognised by furin. The core furin cleavage site53 is indicated in red. Spike S2′: protease cleavage site adjacent to the fusion peptide with a conserved potential protease recognition site shown in red.104 Spike CT; S cytoplasmic tail (CT) sequence with the acidic and KxHxx motifs51, 52 shown in orange text. (c and d) Western blot analyses were conducted using (c) infected cell lysates or (d) purified VSV∆G-SARS-CoV-2 #9. Spike was detected using rabbit polyclonal antisera prepared against S2 or the soluble ectodomain of Spike. VSV N was detected with a rabbit polyclonal antiserum. The blot was cropped to remove lanes containing irrelevant samples (original blots are provided in Supplementary Figures S2 and S3) (e) Cryo-electron microscopy image of purified VSV∆G-SARS-CoV-2 #9 produced during manufacturing process development.
Recovery of VSV∆G-SARS-CoV-2 from plasmid DNA was executed using Vero cells as described previously21,22 with modification. In brief, 2.5 × 107 Vero cells were electroporated with the VSV∆G-SARS-CoV-2 genomic clone (10 µg) and six expression plasmids based on a modified pCI-neo vector (Promega, Madison, WI) lacking a T7 promoter. The six expression plasmids provided the five VSV structural proteins (Figure 1a; 8 µg N, 4 µg P, 1 µg M, 1 µg G, and 1 µg L) and T7 RNA polymerase (50 µg). Conditions for electroporation using the BTX830 apparatus (Harvard Bioscience, BTX, Holliston, MA) were described earlier.21,22 After electroporation, the cells were cultured in DMEM supplemented with 10% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 0.1 mM MEM nonessential amino acids and 220 µM 2-Mercaptoethanol (Thermo Fisher Scientific, Waltham, MA) for 3 h at 37 °C, after which the cultures were subjected to heat shock at 42 °C (5% CO2 and 85% humidity) for 2 h before incubation was continued for 3 days at 37 °C (5% CO2 and 85% humidity).
Following virus rescue, the first round of VSV∆G-SARS-CoV-2 amplification was conducted with VSV G complementation. Monolayers expressing G were prepared by electroporating Vero cells with the G expression plasmid (BTX830 electroporator, 50 µg of G expression plasmid, 2.5 × 107 cells) followed by incubation for 3 h at 37 °C in DMEM supplemented with 10% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 0.1 mM MEM nonessential amino acids and 220 µM 2-Mercaptoethanol. The cultures were then heat shocked at 42 °C for 2 h and returned to 37 °C for another 2 h before infection was initiated with virus supernatant harvested from the initial virus rescue. Passage-1 VSV∆G-SARS-CoV-2 was harvested 48 h after pseudotyping with VSV G and was then used to initiate a subsequent virus passage (passage-2) using Vero cell monolayers without providing VSV G complementation. Because the virus amplified without G complementation during passage-2 achieved low titres, six additional serial passages without G complementation were conducted to allow the development of adaptive mutations that improved titres. Virus from the 8th passage was used for one round of plaque isolation performed using monolayers overlayed with serum-free medium (VP-SFM; Thermo Fisher Scientific, Waltham, MA) containing 0.5% agarose (SeaPlaque™ agarose, Lonza, Bend, OR). Plaque isolates were amplified in Vero cells grown in VP-SFM, and virus harvested in medium from infected cultures was stored at less than –60 °C. Plaque-isolate VSV∆G-SARS-CoV-2 #9 was selected for further vaccine development.
Western blot analysis
Vero cells were infected with VSV∆G-SARS-CoV-2 at a multiplicity of infection of 0.01 plaque-forming units (PFUs) per cell. At about 72 h after infection, cells were harvested and collected by low-speed centrifugation at 400 × g then resuspended in Dulbecco's phosphate buffered saline (DPBS, Thermo Fisher Scientific, Waltham, MA) before repeating the centrifugation step. Cell lysates were prepared using CelLytic M buffer (Sigma-Aldrich, St. Louis, MO) supplemented with 1% protease inhibitors (Sigma-Aldrich, St. Louis, MO) and then were clarified by centrifugation at 15, 700 × g for 10 min at 5 °C.
Purified virus particles for Western blot analysis were prepared by collecting serum-free medium supernatant from infected Vero cell cultures and purifying virions by centrifugation through a 20% sucrose cushion (58,000 × g; SW-28 rotor). The viral pellet was resuspended with Hank's Balanced Salt Solution (HBSS, Thermo Fisher Scientific, Waltham, MA) supplemented with 15% trehalose before virus was stored at less than –60 °C.
Samples were denatured in lithium dodecyl sulfate (LDS) loading buffer (Thermo Fisher Scientific, Waltham, MA) and Reducing Buffer (Thermo Fisher Scientific, Waltham, MA) by heating at 80 °C for 10 min. Denatured samples were electrophoresed in denaturing sodium dodecyl sulfate polyacrylamide gels (NuPage™ 4-12% Bis-Tris Gel, Thermo Fisher Scientific, Waltham, MA), and then the separated proteins were electroblotted to nitrocellulose membranes (iBlot™ Gel Transfer Stacks Nitrocellulose, Thermo Fisher Scientific, Waltham, MA). Membranes were incubated overnight at 4 °C in blocking buffer (StartingBlock™ T20 Blocking Buffer prepared in phosphate-buffered saline [PBS], Thermo Fisher Scientific, Waltham, MA) before antibody detection. Primary antibodies used for detection included mouse monoclonal antibody 1A9 specific for the Spike S2 subunit (GeneTex, Irvine, CA) or rabbit polyclonal antisera specific for the Spike S1 receptor binding domain or the S2 subunit (Sino Biological, Wayne, PA). Rabbit polyclonal antisera specific for VSV N also was used.21 Membranes were washed with deionised water five times and with PBS (Thermo Fisher Scientific, Waltham, MA) containing 0.1% Tween 20 (Bio-Rad, Hercules, CA) three times for 5 min each. Washed membranes then were incubated with anti-mouse or anti-rabbit IgG conjugated to horse radish peroxidase (HRP; Santa Cruz Biotechnology, Dallas, TX) followed by washing steps. The blot was developed using an ECL kit (Thermo Fisher Scientific, Waltham, MA) and signals were detected with a Bio-Rad ChemiDoc™ Touch Imaging System (Bio-Rad, Hercules, CA).
Spike protein used in vaccination studies
An expression plasmid was generated containing a mammalian codon-optimised gene for expression of the Spike ectodomain. The soluble ectodomain contained two proline substitutions, a mutated furin cleavage site, and a C-terminal foldon trimerisation domain as previously described,23 followed by a thrombin digestion site and a His-Tag sequence. The plasmid was used to transfect Expi293F™ cells (Invitrogen, Thermo Fisher Scientific, Waltham, MA) to express soluble Spike, which was then harvested in cell culture supernatant that was clarified by centrifugation. The Spike protein was then purified using Ni-Sepharose chromatography (Cytiva, Marlborough, MA) and then the His-tag was cleaved by digestion with thrombin overnight. The spike protein was further purified by a second Ni-Sepharose chromatography step to remove immobilised metal ion affinity contaminants and uncleaved Spike. Final purification was carried out by gel filtration chromatography (Superdex 200, Cytiva, Marlborough, MA). Adju-Phos® (InvivoGen, San Diego) was added to the Spike protein solution and inverted 10 times prior to IM injection of cotton rats or hamsters.
Animal studies
Studies were conducted at three facilities in accordance with the US Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals. Studies conducted at Merck & Co., Inc., West Point, PA, USA, were approved by the Institutional Animal Care and Use Committee (IACUC) of Merck & Co., Inc., West Point, PA, USA. The immunogenicity and efficacy studies conducted in hamsters were performed at Bioqual, Inc. (Rockville, MD, USA) and approved by the Bioqual IACUC. The IN vaccination dose range study performed in hamsters was conducted at IAVI facilities within the State University of New York Downstate Health Sciences University in Brooklyn and were approved by the University Institutional Biosafety Committee and IACUC. Additionally, all studies adhered to the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines of the National Centre for the Replacement, Refinement & Reduction of Animals in Research (https://www.nc3rs.org.uk/arrive-guidelines).
Cotton rat vaccination
Female cotton rats (Sigmodon hispidus) that were 3 to 7 weeks old were purchased from Envigo (Indianapolis, IN) and assigned randomly to study groups (Figure 2) by individuals with no direct knowledge of the test materials. Upon arrival, cotton rats underwent an acclimation period for 9 days before procedures were performed. Five animals were placed in each study group as we had conducted pilot studies with an earlier VSVΔG-SARS-CoV-2 clonal isolate, which indicated that a group size of five was adequate to assess differences in immunogenicity when the vaccine dose was varied. The animals were immunised with a single dose of VSV∆G-SARS-CoV-2 by one of three different methods: IM injection (100 μL split equally over right and left quadriceps); 10 μL delivered to the oral mucosa (OM), as described by Munoz-Wolf and colleagues24; or 10 μL IN drops in sedated animals (ketamine/xylazine 50–100 mg/kg/2–5 mg/kg).25 Immunisations were performed by group identification rather than by test article to reduce general awareness of each treatment group by the operator. Animals were pair-housed and were dosed or sampled in the order they were caught and removed from the cage, with several cages containing animals in different groups. Blood samples were collected and clarified by centrifugation at approximately 2000 g for 10 min in serum separator tubes, aliquoted, frozen, and stored at –70 °C. Study outcomes were limited to serology (ELISA and neutralisation assays). Assayists received samples labelled by animal number and were unaware of the test article associated with the samples.
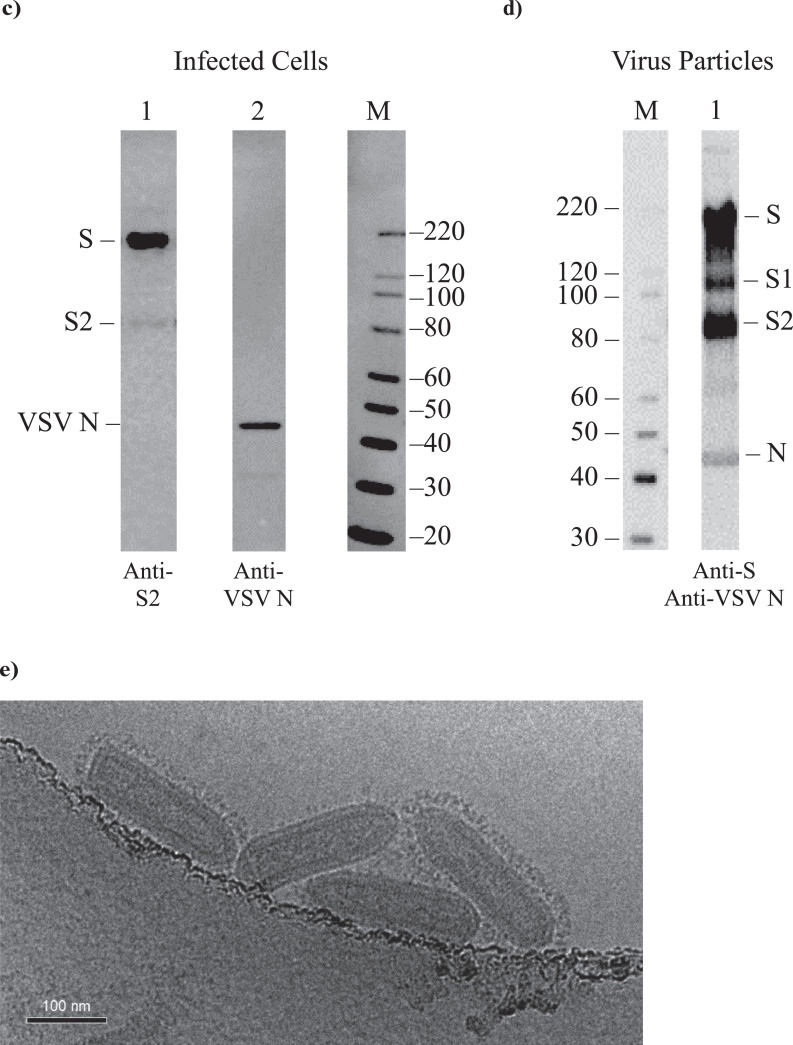
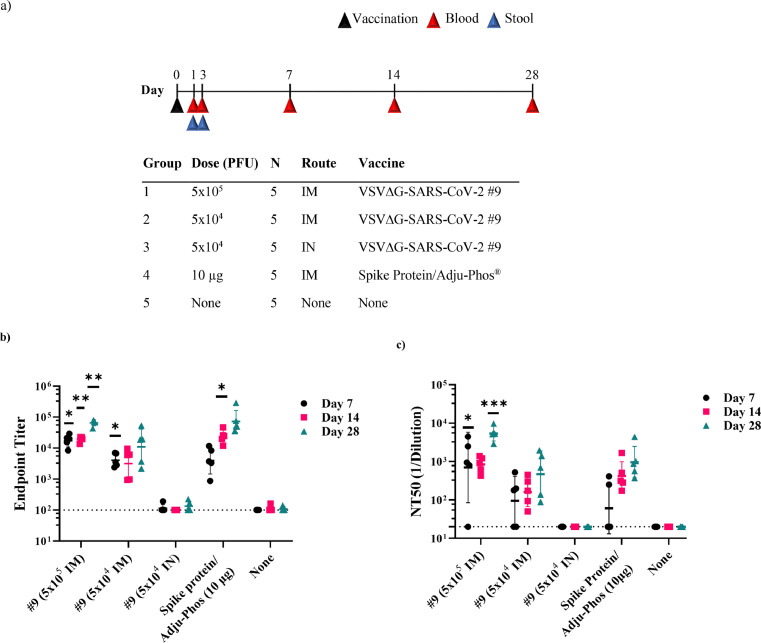
Figure 2.
Immunogenicity of VSV∆G-SARS-CoV-2 in cotton rat. (a) Design of the cotton rat study. Groups of five animals were immunised with a single dose of VSVΔG-SARS-CoV-2 #9. Control groups included naïve animals or cotton rats vaccinated with a soluble Spike trimer formulated with Adju-Phos. Doses and vaccination routes are indicated in the table. Stool and blood samples were collected on days 1 and 3 following immunisation for viral RNA quantification, and blood was collected on days 7, 14, and 28 for analysis of the serological response. (b) Sera collected on days 7 (black), 14 (red), and 28 (teal) were analysed for binding IgG using ELISA plates coated with soluble Spike and the endpoint titres are plotted. (c) Serum neutralisation titres were quantified using VSV∆G-SARS-CoV-2 #9 plaque reduction. The neutralising titres that reduced plaques by 50% (NT50) were graphed plotted as in b. In b and c, the plots show the geometric mean with geometric standard deviation. Dashed lines indicate lower limit of assay detection for Endpoint Titre (LOD 100) and NT50 (LOD 20) analysis. Statistical analysis was performed comparing groups of vaccinated animals to control unvaccinated animals (none) using a 2-way ANOVA multiple comparison with Dunnett correction (n = 5, *p ≤ 0·05, **p ≤ 0·005, ***p ≤ 0·0001). Symbols indicating comparisons to controls that were not significant were omitted for clarity. Grp, group; IM, intramuscular; IN, intranasal; LOD, limit of detection; PFU, plaque-forming unit.
Hamster vaccination and SARS-CoV-2 challenge
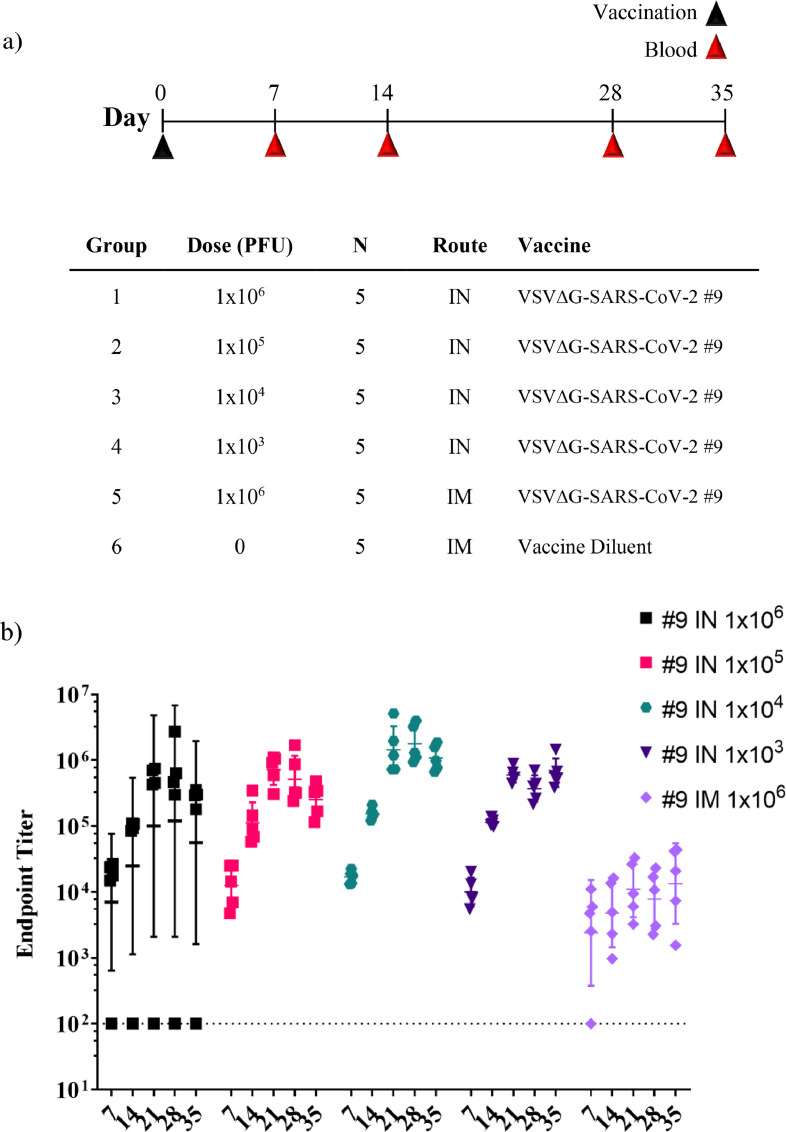
Male and female golden Syrian hamsters (6–10 weeks old) were purchased from Envigo (Indianapolis, IN). Animals underwent a 7-day acclimation period prior to any procedures being performed. The hamsters were randomly assigned to each group by individuals with no direct knowledge of the test materials, maintaining a male-to-female ratio that was as close as possible to 1:1. The hamsters were group-housed. Formal power analyses were not performed when the rodent studies were designed. Our objective was to use a minimum number of animals and still detect sizable quantitative differences between study groups in which substantial vaccination variables were introduced such as alternative vaccination routes or 10-fold changes in vaccine dose. Group size was study-specific and selected using data from our pilot cotton rat study (Figure 2; n=5 per group) and guidance provided by preprint reports available early during the pandemic.26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38 The designs of these early studies allowed assessment of vaccine candidate immunogenicity and efficacy in a variety of models with group sizes ranging from 4-15 depending on the species being used, and informed designs of our experiments, presented important comparators for our study data, and provided sufficient power for statistical inference.
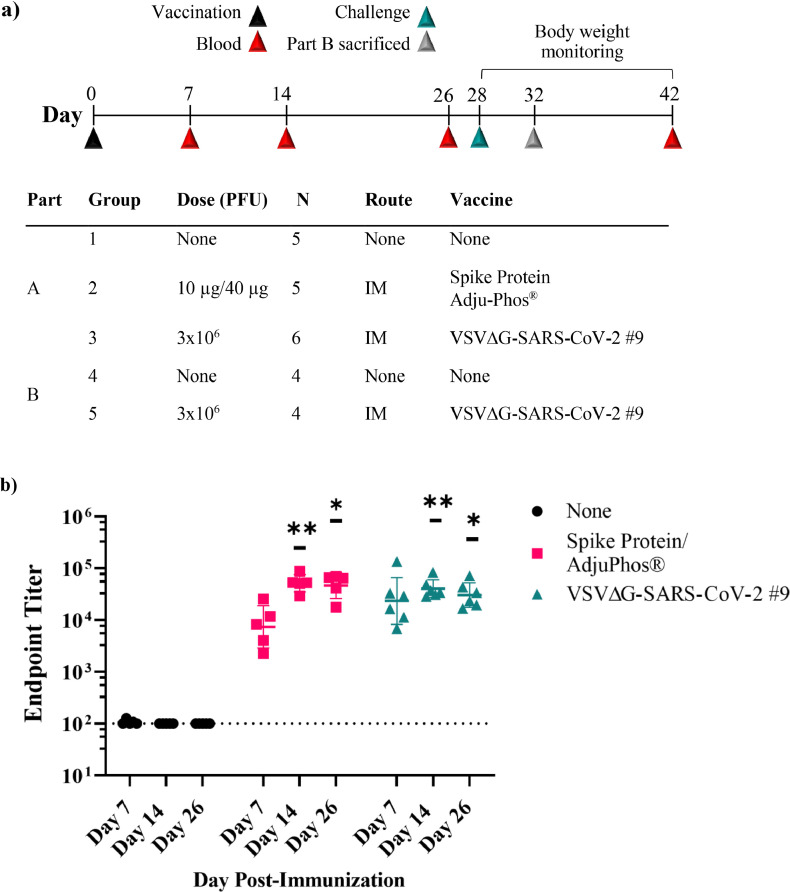
The first study conducted in hamsters (Figure 3a) was composed of two parts (Part A and Part B). In Part A of the study (Figure 3a), hamsters (5–6 animals per group) were monitored for weight loss following SARS-CoV-2 challenge presented in Figure 4a, and in Part B (Figure 3a) hamsters (4 animals per group) were sacrificed 4 days after challenge to assess infectious SARS-CoV-2 in tissues (Figure 4b and c). The total number of animals used, group size, and number of groups included in this study was considered to be the minimum required to characterise the effects of the test article.
Figure 3.
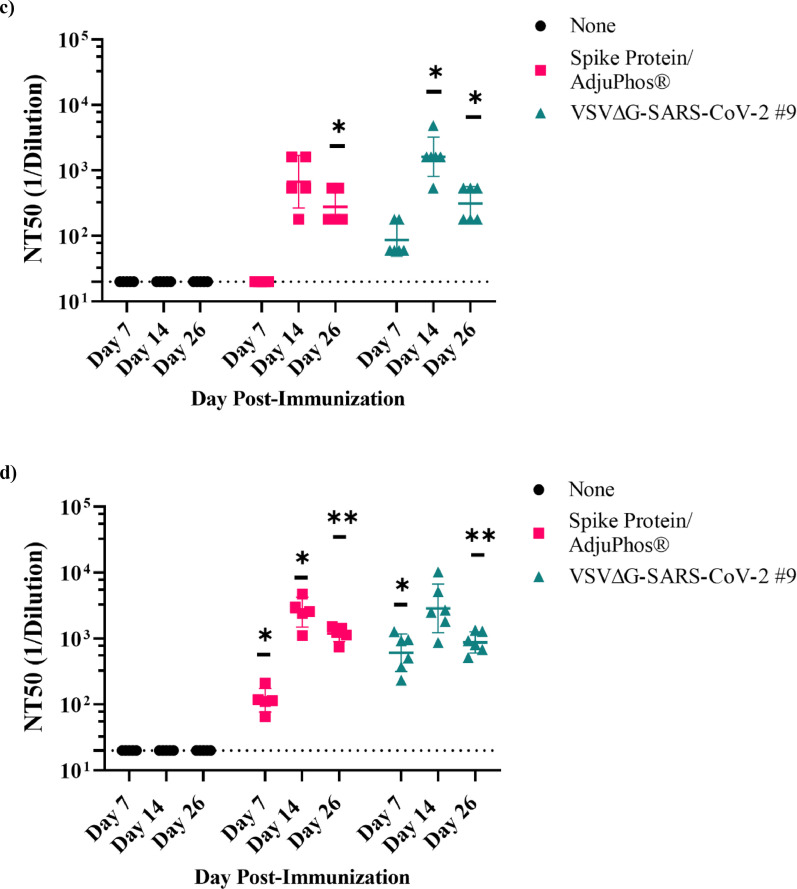
Immunogenicity of VSV∆G-SARS-CoV-2 in golden Syrian hamster following intramuscular (IM) immunisation. (a) Design of the immunogenicity and efficacy study conducted in golden Syrian hamsters. In Part A of the study, three groups of five or six hamsters were immunised with a single IM injection of VSV∆G-SARS-CoV-2 #9, or soluble Spike protein formulated with Adju-Phos® at the indicated dose. Control hamsters were unimmunised. Blood samples were collected on days 7, 14, and 26 for analysis of the serological response. Two days after the third blood draw (day 28), the animals were challenged with SARS-CoV-2 (USA-WA1/2020) by intranasal inoculation, and body weights were measured daily through day 42 (see Figure 4). In Part B of the study, two groups of four animals were either unimmunised or given a single IM injection with VSV∆G-SARS-CoV-2 #9. These animals were challenged on day 28 as described for Part A above but were euthanised 4 days after challenge for quantification of live SARS-CoV-2 in lung and nasal tissue (Figure 4). (b) Sera samples collected on days 7, 14, and 26 from animals in Part A were analysed for binding to the Spike ectodomain by ELISA. The endpoint titres for each animal are graphed and the geometric mean titres are plotted. Serum neutralising antibody titres against (c) authentic SARS-CoV-2 (USA-WA1/2020) or (d) VSV∆G-SARS-CoV-2 #9 were quantified for each animal. Neutralising titres (NT50) are defined as the dilution at which there is a 50% reduction in plaques compared with controls. For b-d, the geometric mean with geometric standard deviation were plotted. Dashed lines indicate the lower limit of assay detection for Endpoint Titre (LOD 100) and NT50 (LOD 20) analysis. Statistical analysis was performed comparing vaccinated group responses to control unvaccinated animals (none) using a two-way ANOVA multiple comparison with Dunnett correction (n = 4–6, *p ≤ 0·05, **p ≤ 0·005, ***p ≤ 0·0001). Symbols indicating comparisons to controls that were not significant were omitted for clarity. LOD, limit of detection.
Figure 4.
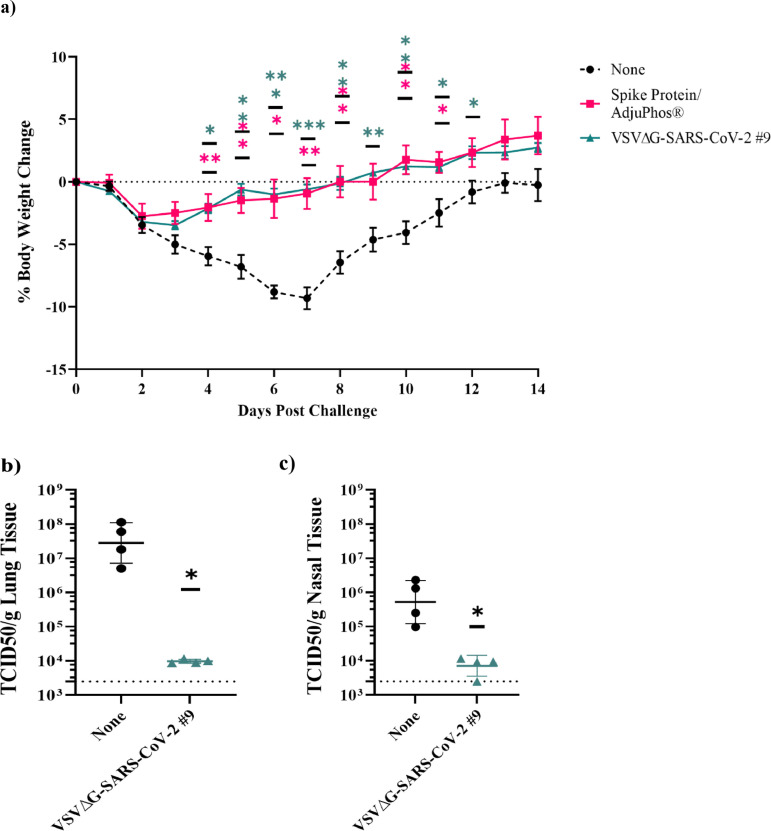
Protection of hamsters from challenge with SARS-CoV-2 by intramuscular immunisation with VSV∆G-SARS-CoV-2. (a) Body weight was monitored for animals in Part A of the study described in Figure 3. Day of challenge is day 0. (b and c) In Part B of the study in Figure 3, unimmunised controls or animals immunised with VSV∆G-SARS-CoV-2 #9 (3 × 106 PFUs) were euthanised on day 4 following challenge. Lung (b) and nasal (c) tissues were isolated, and live SARS-CoV-2 was quantified by TCID50. Virus titres per gram of tissues were plotted. The plots in a-c include the geometric mean with geometric standard deviation. Dashed lines in b and c indicate lower limit of assay detection for lung and nasal tissue TCID50 (LOD 2500). Statistical analysis was performed comparing vaccine cohort responses to control unvaccinated animals (none) using a two-way ANOVA multiple comparison with Dunnett correction for weight change in (a; n = 5–6, *p ≤ 0.05,**p ≤ 0.005, ***p ≤ 0.0005) and an unpaired nonparametric Mann-Whitney test (n = 4, *p ≤ 0.05) for TCID50 in b and c. Symbols indicating comparisons to controls that were not significant have been omitted for clarity. LOD, limit of detection; PFU, plaque-forming units; TCID50, tissue culture infectious dose 50.
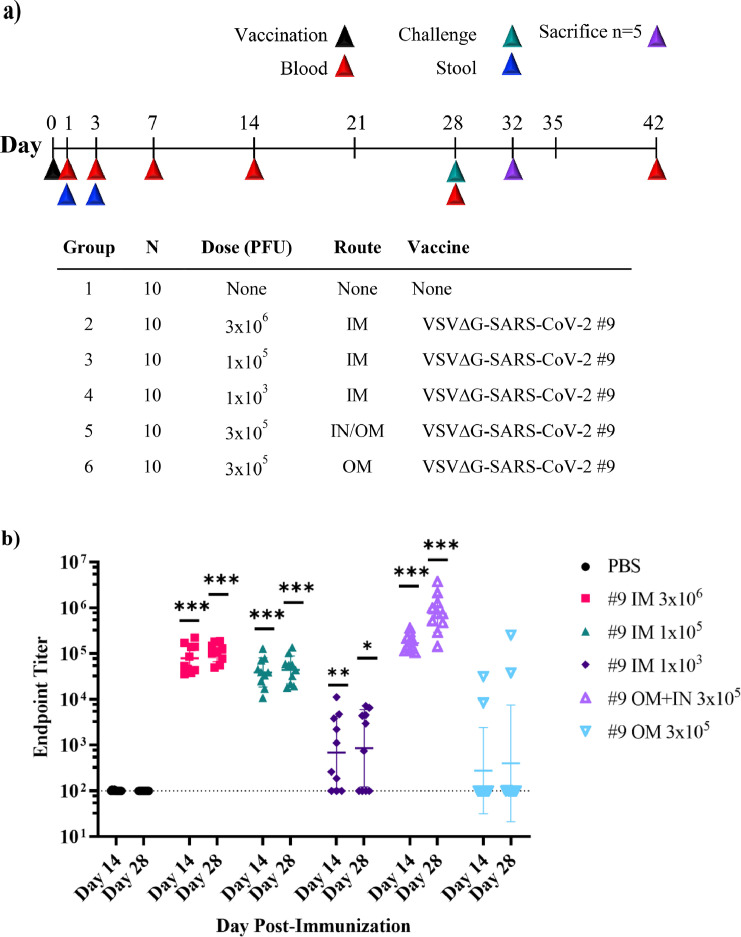
In the second immunogenicity and efficacy study conducted in hamsters (Figures 5 and 6), vaccination variables included dose as well as route. Ten animals were included per group and were monitored for immunogenicity. Following SARS-CoV-2 challenge, the groups were divided with five animals from each group sacrificed to assess infectious SARS-CoV-2 in tissues while the remaining animals were monitored for weight loss. The animals were vaccinated by IM injection in two hind legs (50 μL per leg) or by OM application (10 μL), as described above for cotton rats. Some animals were vaccinated by a combination of OM plus IN application (10 μL oral cavity and 50 μL per nostril). Immunisations were done by group identification, so the operator was not aware of which groups received which test article for those test articles delivered intramuscularly. Animals were housed with multiple animals per cage and were dosed or sampled in the order they were caught and removed from the cage. Blood was collected via the retro-orbital or saphenous veins from anesthetised animals following immunisation.
Figure 5.
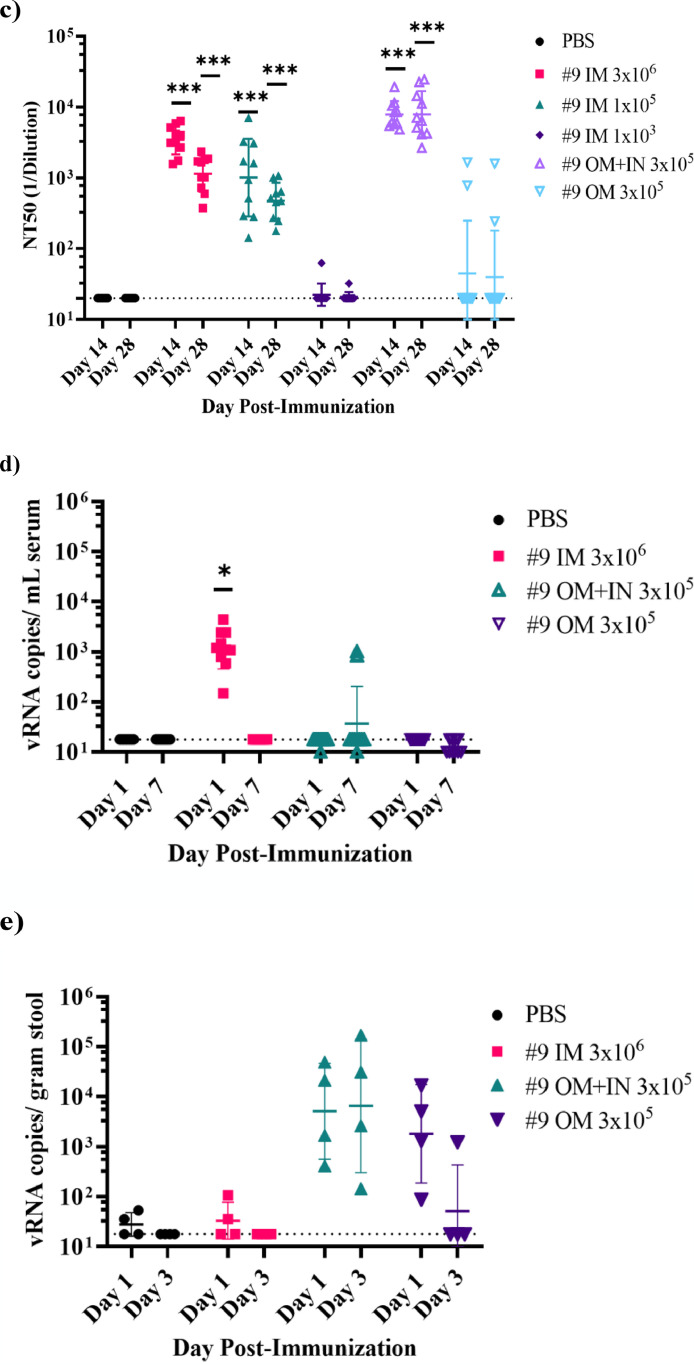
Immunogenicity of VSV∆G-SARS-CoV-2 #9 in hamsters following intramuscular, intranasal/oral mucosa, or oral mucosa immunisation. The design of the second efficacy study is shown in (a) where six groups of ten hamsters were vaccinated once with VSV∆G-SARS-CoV-2 #9 using the indicated dose and vaccination route. Control animals were unvaccinated. Stool samples were collected from cages on days 1 and 3 following vaccination and blood samples were drawn on days 1, 3, 7, 14, 28, and 42 from all animals. Hamsters were challenged with SARS-CoV-2 on day 28 after which five animals from each group of 10 were euthanised on day 32 (4 days after challenge) to assess SARS-CoV-2 titres in tissues (Figure 6). In (b), sera samples collected on days 14 and 28 were analysed for IgG titres using ELISA plates coated with soluble Spike. The endpoint titres for each animal are graphed and the geometric mean titre and geometric standard deviation (SD) are indicated for each group. (c) Serum neutralising antibodies were quantified by plaque reduction assay using VSV∆G-SARS-CoV-2 #9 and the NT50 titres are graphed for days 14 and 28. The geometric mean titre and geometric standard deviation are indicated for both time points within each group. Neutralising titres (NT50) are defined as the dilution at which there is a 50% reduction in plaques compared with controls. (d) VSV∆G-SARS-CoV-2 RNA copies per millilitre of blood collected on days 1 and 7 after vaccination were quantified by RT-qPCR using primers specific for the VSV matrix gene. Similarly, in (e) VSV∆G-SARS-CoV-2 RNA copies in stool collected from the cages on days 1 and 3 after immunisation were quantified by RT-qPCR. Two hamsters were cohoused per cage thus the stool samples could not be associated with an individual animal. The geometric mean and geometric standard deviation are indicated for both time points within each group. Dashed lines indicate lower limit of assay detection (LOD Endpoint Titre 100, NT50 20, viral RNA stool and serum 17.8 RNA copies). Statistical analysis was performed comparing vaccine cohort responses to control unvaccinated animals (PBS) using an unpaired nonparametric Mann-Whitney test for Endpoint Titre and NT50 (n = 10, *p ≤ 0.05, **p ≤0.005, ***p ≤0.0005) and a two-way ANOVA multiple comparison with Dunnett correction for vRNA copies (n = 4, *p ≤ 0.05). Symbols indicating comparisons to controls that were not significant have been omitted for clarity. Grp, group; IM, intramuscular; IN, intranasal; LOD, limit of detection; OM, oral mucosa; PBS, phosphate-buffered saline; PFU, plaque-forming unit; RT-qPCR, reverse transcriptase quantitative polymerase chain reaction; vRNA, viral ribonucleic acid; VSV, vesicular stomatitis virus.
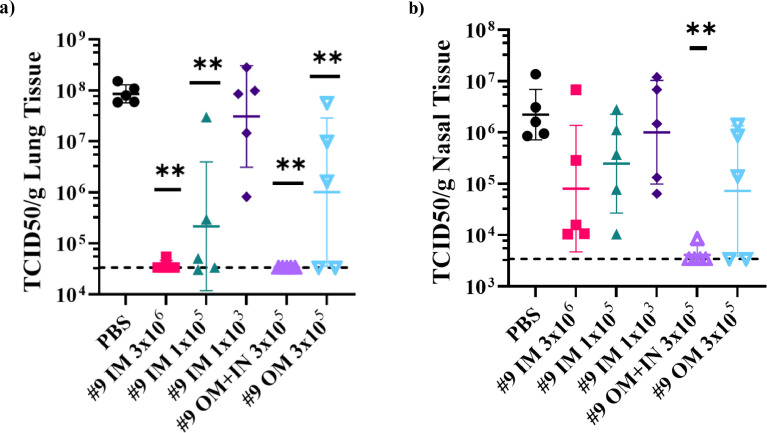
Figure 6.
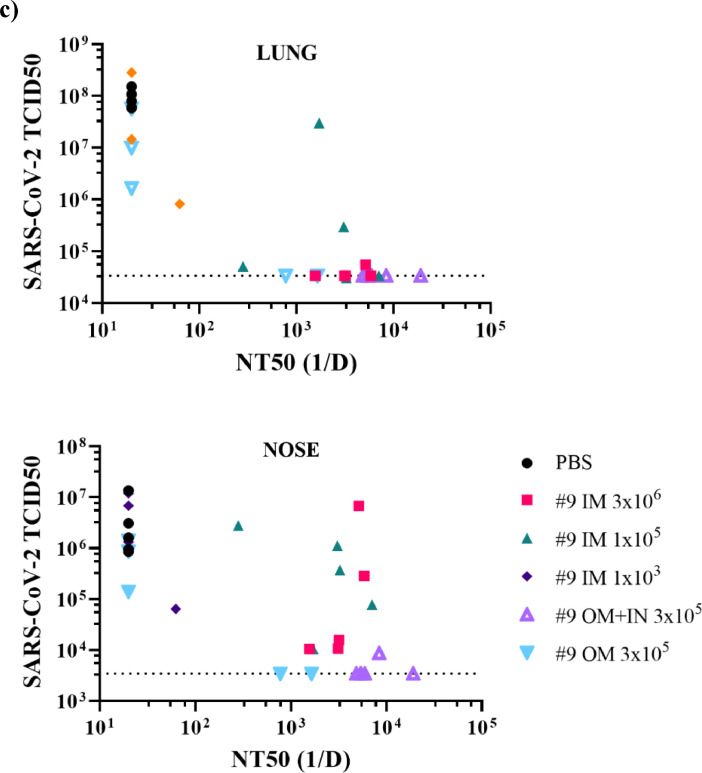
Protection of hamsters from challenge with SARS-CoV-2 by intramuscular, oral mucosa, or intranasal/oral mucosa immunisation with VSVΔGΔ-SARS-CoV-2. Five of 10 hamsters per group from the second immunogenicity and efficacy study described in Figure 5a were euthanised on day 4 after SARS-CoV-2 challenge. Lung (a) and nasal tissue (b) were processed to quantify infectious SARS-CoV-2 by TCID50 assay and the data are plotted as geometric mean with geometric standard deviation. Statistical analysis was performed comparing vaccine group responses to control unvaccinated animals (PBS) using an unpaired nonparametric Mann-Whitney test (n = 5, **p ≤ 0.005). Symbols indicating comparisons to controls that were not significant have been omitted for clarity. Dashed lines indicate lower limit of assay detection. In (c), SARS-CoV-2 titres in lung and nasal tissues for each animal are graphed (y-axis) against the nAb titre for that animal (x-axis) to illustrate the relationship between nAb titre and effect on SARS-CoV-2 replication. 1/D, 1/dilution; IM, intramuscular; IN, intranasal; OM, oral mucosa; PBS, phosphate-buffered saline; TCID50, tissue culture infectious dose 50; nAb, neutralizing antibody.
For the efficacy study described in Figure 3, pathogen challenge was conducted using working-virus stocks that were amplified once in Vero E6 cells from a SARS-CoV-2 USA-WA1/2020 seed stock (BEI Resources, Manassas, VA; virus stock NR-52281) and assigned lot no. 061620-1000. Next-generation sequencing analysis (NeoGenomics, Fort Meyers, FL) on working-virus stocks showed that the challenge virus used in the study included in Figure 3 was largely wild-type in the S1/S2 cleavage site, although variants were detectable in the population that had developed amino acid substitution in the furin cleavage site like those reported by others.39, 40, 41, 42, 43, 44, 45 For the experiment described in Figure 5, virus from the original BEI NR-52281 stock was amplified and passaged four times in Vero E6 cells, and next-generation sequencing analysis retrospectively showed that less than 5% of the challenge virus used in the study had a wild-type furin cleavage site (NCBI BioProject accession: PRJNA666696). Because most of the challenge virus population had a defect in the furin cleavage site, the virus stock was less virulent, as shown by weight loss in control animals that was less than 5% (Supplementary Figure S1a and S1b), although the virus did replicate to high titres in hamsters (see control animals Figure 6).
SARS-CoV-2 challenge virus was administered (2 × 104 PFU) to anesthetised animals dropwise into each nostril (100 μL per animal). Necropsy was performed on selected animals at 4 days after challenge to collect lung and nasal turbinate tissues, which were homogenised using a handheld tissue homogeniser for 20 s (Omni International, Kennesaw, GA, USA) in solution containing either 500 μL medium (DMEM/10% FBS/gentamicin) for virus quantification by tissue culture infectious dose 50 (TCID50) or RNA-Stat-60 (AMSBIO, Cambridge, MA) for RNA isolation. Homogenates were then clarified by centrifugation and processed for viral quantification by TCID50 (described below) or for isolation of RNA according to the manufacturer's instructions. Remaining animals were weighed and observed twice daily for 2 weeks after challenge.
The final study conducted in hamsters (Figure 7) was an immunogenicity study that was performed with some modifications of the procedures. Animals were anesthetised with a cocktail of ketamine (150 mg/kg) and xylazine (5 mg/kg) delivered by the intraperitoneal (IP) route. Vaccination by IM injection were performed in a single hind leg. IN vaccination was dropwise using a P20 pipettor delivering 10 µL per nostril. Five animals were included per group to allow comparison of effects of vaccination route and vaccine dose.
Figure 7.
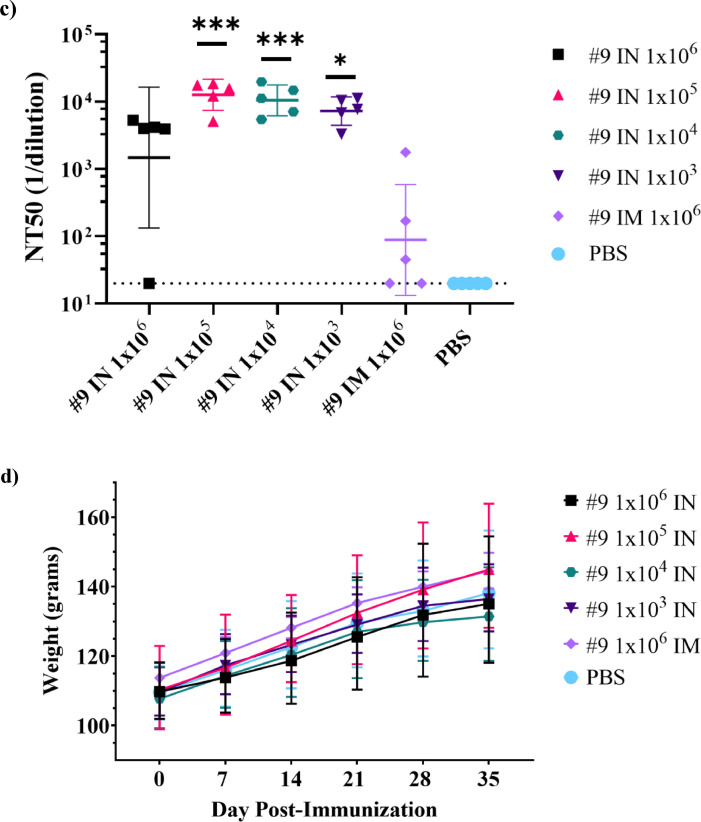
Immunogenicity of intranasal VSVΔG-SARS-CoV-2 vaccination in hamsters. (a) Design of the hamster IN immunogenicity dose-range study. Four groups of five animals were immunised with a single IN inoculation of VSV∆G-SARS-CoV-2 #9 using the indicated dose. As a comparator, a group of five animals were immunised with a single IM injection in one hind leg using the highest dose (1 × 106 PFUs) used in the earlier studies (Figures 3 and 5). Blood samples were collected on days 0, 7, 14, 28, and 35 and serum IgG titres were analysed using ELISA plates coated with soluble Spike (b) and neutralising antibody titres (c) were quantified by plaque reduction assay using VSV∆G-SARS-CoV-2 #9 and sera from day 28. (d) Hamster weights also were monitored weekly. The data is plotted as geometric mean with geometric SD. Dashed lines indicate lower limit of assay detection for the ELISA Endpoint Titre (LOD 100) and neutralisation assay (LOD 20). Statistical analysis was performed for the ELISA (b) and neutralisation assay (c) by comparing vaccine cohort values to negative control assays conducted with PBS using a two-way ANOVA multiple comparison with Dunnett correction. For the ELISA values (b) there was no significant difference between the intranasal groups so symbols indicating significance are omitted for clarity. Similarly, hamster weights were not significantly different between groups and symbols indicating significance are omitted. IM, intramuscular; IN, intranasal; LOD, limit of detection; NT50, NT50, neutralising titres that reduced plaques by 50%; OM, oral mucosa; PBS, phosphate-buffered saline; PFU, plaque-forming units. (n = 5, *p ≤ 0·05, **p ≤ 0·005, ***p ≤ 0·0001).
In the various studies conducted in hamsters, analyses included serology (ELISA and virus neutralisation assays), quantification of infectious SARS-CoV-2 in lung and nasal tissue (TCID50), VSV∆G-SARS-CoV-2 #9 viral RNA (vRNA) detection in serum and stool, and body weight monitoring. For the serological, viral load, and vRNA detection, assayists received samples labelled by animal number and were unaware of the test article associated with the samples. For the assessment of body weight loss, animal handlers were unaware of the test article associated with the animals being weighed.
RT-qPCR detection of VSV∆G-SARS-CoV-2 RNA in blood and stool
RNA from hamster blood and stool was isolated with the High Pure Viral RNA Kit (Roche LifeScience, Indianapolis, IN) and RNeasy PowerMicrobiome kit (Qiagen, Germantown, MD), respectively. In brief, 200 µL hamster serum was mixed with 400 µL binding buffer supplemented with poly(A) from the kit; the mixture was added into the binding column, washed once with 500 µL inhibitor removal buffer and twice with 450 µL washing buffer. RNA was eluted with 40 µL RNase-free water. For hamster stool samples, approximately 250 mg of stool was soaked with 650 µL solution PM1 with 1:100 diluted β-Mercaptoethanol. Stool was then homogenised with a TissueLyser (Qiagen, Germantown, MD) using 5 mm stainless steel beads for 2 min. Supernatant was collected after centrifugation at 20,000 g for 5 min for RNA isolation according to the kit's instruction. DNase digestion was performed on the column and RNA was eluted with 50 µL RNase-free water. Reverse transcription (RT) was conducted with 30 µL of purified RNA from either serum or stool using the Ambion Cells-to-Ct kit (Thermo Fisher Scientific, Waltham, MA) following the manufacturer's instructions. RT products were concentrated by precipitation with 3 volumes of 100% ethanol with 0.1 volume of 3M sodium acetate, incubation at –80 °C overnight, and centrifuging at 20,000 g at 4 °C for 30 min. RNA pellets were washed twice with 500 μL ice-cold 75% ethanol and vacuum dried, and then the concentrated RT product was resuspended with 4 µL nuclease-free water for polymerase chain reaction (PCR). The VSV matrix gene was used as the quantitative polymerase chain reaction (qPCR) target. The primers and probe used in qPCR were: Forward primer: 5′-GCTGCAGTGGACATGTTCTTC-3′, Reverse primer: 5′-GCAGCACAATCTTTGAATCTGGAAA-3’, Probe: 5′- FAM-CTGAACGAGGCACATTC-TAMRA-3′ (Thermo Fisher Scientific, Waltham, MA). PCR was performed in QuantStudio 12K flex real-time PCR system (Thermo-Fisher Scientific, Waltham, MA) with the default PCR thermocycling parameters (denature at 95 °C for 20 s, then 95 °C for 1 second and 60 °C for 20 s for 40 cycles). vRNA copies were determined using a standard curve generated from the purified VSV genomic RNA.
Serologic analysis
ELISA protocol
Binding antibodies specific for SARS-CoV-2 Spike were quantified by indirect ELISA. Corning Costar® high-binding 96-well plates (Corning, Tewksbury, MA) were coated with 50 μL of soluble trimeric SARS-CoV-2 spike protein (Lake Pharma) diluted to 1 μg/mL in 1x ELISA Coating Buffer (BioLegend, San Diego, CA) overnight at 2 to 8 °C. Unbound coating antigen was removed by washing three times with 150 μL of PBS containing 0.05% Tween-20 (PBST), after which the wells were blocked with 100 μL of PBS containing 2% Blotting-Grade Blocker (Bio-Rad, Hercules, CA). Test and positive control samples were diluted in PBS containing 1% Tween 20 and 2% Blotting-Grade Blocker starting at 1:100 followed by three-fold serial dilutions across the plate. Plates were then incubated for 1 h at 37 °C before samples were removed and the plates were washed three times with 150 μL PBST. Anti-hamster secondary antibody (goat anti-Syrian hamster IgG-heavy chain and IgG-light chain antibody conjugated to HRP, Rockland Immunochemicals, Pottstown, PA) diluted 1:6,000 in PBST/2% Blotting-Grade Blocker was added (50 µL) to each well and incubated for 60 min at 37 °C. Secondary antibody was then removed, and the plates were washed three times with 150 μL PBST. Signal was developed by adding 50 μL of 1-Step™ Ultra TMB-ELISA (Thermo Fisher Scientific) and incubating for 10 min at room temperature in the dark before the reaction was stopped by addition of 50 μL of 2N sulphuric acid (Thermo Fisher Scientific, Waltham, MA). Plates were read within 30 min at 450 nm with a Molecular Devices’ (San Jose, CA) VersaMax Microplate Reader using SoftMax Pro GxP Data Acquisition Software.
For assessing SARS-CoV-2 specific binding antibodies in cotton rat serum the above method was used with changes in the coating protein and secondary antibody used. SARS-CoV-2 S protein (ACRO Biosystems, Newark, DE) was used to coat plates at 1 µg/mL. A chicken anti-cotton rat IgG HRP secondary antibody (Immunology Consultants Laboratory, Portland, OR) was used at a dilution of 1:1000.
VSV∆G-SARS-CoV-2 plaque reduction microneutralisation assay
Vero cells (ATCC CCL-81) were seeded into 96-well plates at 6.25 × 104 cells per well and incubated overnight at 37 °C, 5% CO2. Heat-inactivated hamster or cotton rat serum samples were four-fold serially diluted in infection medium composed of DMEM supplemented with 2% FBS (Cytiva HyClone™, Marlborough, MA), penicillin/streptomycin and non-essential amino acids (Gibco™, Thermo Fisher Scientific, Waltham, MA). Diluted serum samples (25 µL) were then combined with an equal volume of VSV∆G-SARS-CoV-2 containing a quantity of virus previously shown to produce about 50 plaques per well, followed by incubation for 1 h at 37 °C. Subsequently, the virus–serum sample mixtures were transferred to wells containing Vero cell monolayers and the plates were incubated for 1 h at 37 °C. After incubation, 100 µL of a 0.75% methylcellulose overlay prepared in DMEM supplemented with 2% heat-inactivated FBS (Cytiva Hyclone™, Marlborough, MA) and 1x penicillin/streptomycin (Gibco) was added for a total volume of 150 µL per well, and the infection was allowed to proceed for 2 days at 37 °C. The methylcellulose overlay (Sigma, St. Louis, MO) then was removed and cells were fixed for 10 to 30 min with 3.7% formaldehyde (Polysciences, Inc., Warrington, PA) prepared in PBS after which the fixative was removed, and the cells were permeabilised with PBS containing 0.1% Triton-X (Sigma-Aldrich, St. Louis, MO). After permeabilising the cells, 200 µL per well of blocking solution (PBS, 1% BSA, 0.05% Tween 20) was added and the plates were incubated overnight at 4 °C. After removing the blocking solution, primary anti-S polyclonal antibody (Sino Biologicals, Wayne, PA) diluted 1:1000 in blocking solution was added and incubated for 1 h at room temperature. Plates were then washed with PBST and subsequently incubated with labelled secondary antibody (goat anti-rabbit IgG Alexa 488; Life Technologies, Thermo Fisher Scientific, Waltham, MA) diluted 1:200 in blocking buffer for 1 h. Secondary antibody was removed, and the plates were washed with PBST before adding 200 µL of PBS per well and quantifying fluorescent plaques using the Virus Plaque Analysis for Kaleido 2.0 programme on the EnSight™ Multimode Plate Reader (PerkinElmer, Waltham, MA). Plaque count data were exported to Microsoft Excel for analysis. Neutralising titres (NT50) are defined as the dilution at which there is a 50% reduction in plaques compared with controls. The NT50 values are determined by four-parameter curve fit with GraphPad Prism software v.8.1.1 (San Diego, CA) by plotting the log transformed sample dilution (x-axis) by the per cent neutralisation (y-axis). Per cent neutralisation is calculated with the following equation: % Neutralisation = [1 – (sample plaque count – average cell control plaque count) / (average virus control plaque count – average cell control plaque count)] (100).
SARS-CoV-2 neutralisation assay
Serum samples were heat inactivated at 56 °C for 30 min, then diluted in PBS containing 5% BSA (PBS/BSA). Diluted serum samples (200 µL) were subsequently mixed with an equal volume of SARS-CoV-2 (USA-WA1/2020) that was prepared in PBS/BSA to contain 40 PFU per 200 μL. Serum-virus mixtures were incubated for 1 h at 37 °C before the 400 µL mixture was distributed between duplicate wells of a six-well plate containing monolayers of Vero E6 cells (ATCC, CRL-1586). Infection was allowed to proceed for 1 h at 37 °C before the plates were overlayed with 3 mL of medium containing 0.9% agarose (Lonza, Rockland, ME). Plaques were allowed to develop at 37 °C in a 5% CO2 incubator for 2 days. A second overlay medium containing neutral red and 1% agarose was then added, and the plates were incubated overnight to visualise plaques for counting. The nAb titre was identified as the highest serum dilution that reduced the number of virus plaques in the test by 90% or greater.
Hamster tissue virus load quantification
TCID50 was used to quantify SARS-CoV-2 in homogenised tissue samples. Vero E6 cells were seeded at 25,000 cells per well in a 96-well plate in DMEM supplemented with 10% FBS and gentamicin. Cells were cultured overnight at 37 °C, 5.0% CO2, and were 80% to 100% confluent the following day, at which time the media was aspirated and replaced with 180 μL of DMEM containing 2% FBS and gentamicin. Lung or nose tissue homogenate (20 µL) was added to the top row in quadruplicate and mixed using a P200 pipettor 5 times before transferring 20 μL to the adjacent well in the row and repeating the process across the plate. Positive and negative (medium only) control wells were included in each assay. Cytopathic effect was allowed to develop at 37 °C for 4 days, after which virus-positive wells were counted. A TCID50 value was calculated using the Reed-Muench method.46
Statistical analysis section
All statistical analysis was performed using GraphPad Prism (9.0.0) software (GraphPad, San Diego, CA). Endpoint ELISA Titer and NT50 neutralisation data was analyzed by comparing vaccine cohort responses to control unvaccinated animals (none or PBS) using either a two-way ANOVA multiple comparison with Dunnett correction or an unpaired nonparametric Mann-Whitney test. Viral load data was analysed using an unpaired nonparametric Mann-Whitney test. Statistical differences in weight changes associated with viral challenge or vaccination determined using a two-way ANOVA multiple comparison with Dunnett correction. No a priori power analyses were conducted in the design of these studies. Post hoc power analyses were conducted based on the statistical tests used for each analysis. Using an 80% power of detection for differences between groups yielded an approximate minimum detectable difference of 103-4 for immunologic assessments for endpoint titers and virus neutralization, approximately 3.6% difference in weight change, approximately 104 for viral RNA copies, and approximately 108 for differences is tissus virus loads. Immunogenicity studies in cotton rats included n = 5 animals per group. Golden Syrian hamster immunogenicity and efficacy studies included n = 5–10 animals, with n = 4–5 animals for measurement of viral load and weight. Repeated measure (RM) two-way ANOVA with Geissier-Greenhouse and Dunnett correction were performed when appropriate and balanced data were available. A review of the ANOVA residual QQ-plots showed linearity and subsequently that the data followed a normal distribution and the assumptions held for these analyses.
Role of the funding source
The funders did not have a role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
Results
Development of VSV∆G-SARS-CoV-2 chimeras
To generate a VSV∆G-SARS-CoV-2 vaccine candidate, a modified VSV (serotype Indiana) genomic clone 17,47 was prepared by inserting a gene encoding the full-length SARS-CoV-2 spike in place of the VSV G gene (Figure 1a). Recombinant virus recovered from electroporated Vero cells (Methods) initially grew slowly, caused extensive cell-to-cell fusion, and released low quantities of infectious virus into the medium. Growth was improved substantially by serially passaging the recombinant virus eight times in Vero cells cultured in DMEM containing FBS during which a virus population developed that caused more rapid and extensive cytopathology, lessened cell-to-cell fusion, and released increased titres of virus into the culture medium. Plaque isolates were selected from infected Vero monolayers overlaid with serum-free medium (VP-SFM) containing 0.5% agarose, and they subsequently were studied to assess growth properties, Spike expression, and genomic nucleotide sequence. Multiple plaque isolates with improved replicative fitness and unique amino acid substitutions were isolated (Parks et al, unpublished), and consistent with the experience of others,38,48, 49, 50 we found that the Spike must be modified, particularly by truncation of the cytoplasmic tail, to lessen cell-to-cell fusion and allow improved growth of VSV∆G-SARS-CoV-2 chimeras. A plaque-isolate of VSV∆G-SARS-CoV-2 (#9) was selected for continued development that had the following characteristics: 1) it consistently produced more than 1 × 107 PFUs per millilitre of culture medium harvested from infected Vero cells grown in VP-SFM; 2) virions incorporated abundant Spike as seen in virus purified by ultrafiltration (Figure 1e); 3) the virus was genetically stable upon serial passage; and 4) it contained relatively few adaptive mutations (Figure 1b).
VSV∆G-SARS-CoV-2 #9 contained three amino acid substitutions in Spike as well as one in the VSV matrix protein (Figure 1b). Like the Spike cytoplasmic tail deletions described by others working with similar chimeric viruses,38,48, 49, 50 VSV∆G-SARS-CoV-2 #9 developed a stop codon that resulted in deletion of 23 C-terminal amino acid residues (Figure 1b). To achieve improved growth, truncation of the Spike C-terminus appears to be obligatory to remove an acidic element (DEDDSE) and a KxHxx motif (Figure 1b), which control Spike membrane trafficking during coronavirus infection. These trafficking signals may be incompatible with the VSV life cycle since they direct Spike localisation largely to internal cellular membranes, and these signals must be removed to allow greater accumulation of Spike in the cell surface membrane where VSV assembles and buds.51,52 Two additional Spike mutations developed in the VSV∆G-SARS-CoV-2 #9 protease cleavage sites. Notably, there was an S1/S2 cleavage site substitution (R683G) that replaced an amino acid residue in the conserved core of the furin cleavage recognition site.53 The R683G substitution was expected to decrease cleavage by furin, and Western blot analyses conducted using infected cell lysates (Figure 1c) or virions purified by centrifugation (Figure 1d) both confirmed that uncleaved Spike precursor was abundant, although the analysis conducted with purified virus showed that bands consistent with the S1 and S2 cleavage products also were present (Figure 1d), indicating that proteolytic processing was not entirely blocked by R683G. Mutations that reduce or prevent Spike cleavage by furin have been reported before for VSV∆G-Spike chimeras38,49 as well as SARS-CoV-2 passaged in Vero cell lines,39,41, 42, 43, 44, 45,54, 55, 56 indicating that S1/S2 cleavage is not essential for infection in Vero cells and that virus entry likely proceeds predominantly through an endocytic pathway.4,6 Mutations that result in reduced or no cleavage at the S1/S2 site might also be favoured in vitro because they stabilise the multimeric Spike complex on the virion.57
VSV∆G-SARS-CoV-2 #9 additionally had a unique substitution in the S2′ cleavage recognition site in which a polar serine was substituted by a bulky aromatic phenylalanine residue (Figure 1b, S813F). Interestingly, this Spike mutation is not presently found in the GISAID58 SARS-CoV-2 genome database, implying that it might be favoured specifically by the VSV∆G-SARS-CoV-2 chimera grown in vitro. Although this was a nonconservative substitution, the modified cleavage site must still allow cleavage at S2′ because proteolytic processing at this site is essential to activate membrane fusion potential.4,6 Possibly, the large hydrophobic residue substitution enhances cleavage by endosomal Cathepsin B or L59 and facilitates entry by the endocytic pathway preferred in Vero cells.4,6 Finally, VSV∆G-SARS-CoV-2 #9 contained a stable mutation in the VSV matrix gene encoding a substitution at amino acid position 61 (Y61S). The M functions that this substitution might affect are unknown, although Y61S has been observed before when VSV has been adapted to different cell lines,60 suggesting that it may reflect some adjustment in the matrix protein interaction with host cell proteins. The VSV∆G-SARS-CoV-2 #9 constellation of mutations was stable during the manufacturing of vaccine material to support the phase 1 trial.18
Immunogenicity of VSV∆G-SARS-CoV-2 in cotton rats
Cotton rats can be infected with VSV,61 and they are a commonly used model for studying respiratory viruses;62,63 therefore, we evaluated their use for investigating the immunogenicity of VSV∆G-SARS-CoV-2 administered by IM injection or IN drops. Two groups of cotton rats were immunised once by IM injection with either 5 × 105 or 5 × 104 PFUs of VSV∆G-SARS-CoV-2 #9 split across both hind legs (Figure 2a). A third group was vaccinated by applying 5 × 104 PFUs of VSV∆G-SARS-CoV-2 #9 dropwise to IN mucosal surfaces. In this experiment, IN administration was conducted with a small total volume (10 µL) split across both nostrils to limit distribution of the virus in the inoculum primarily to the upper respiratory tract.25 Group 4 was included as a comparator in which animals were vaccinated by IM injection with 10 µg of a soluble trimeric form of SARS-CoV-2 Spike formulated with 40 µg of Alum (Adju-Phos). This soluble immunogen (Methods) contained the 2P mutations and a furin cleavage site knockout to stabilise the trimeric Spike in a native-like prefusion state.23,64 The final study group included unvaccinated animals.
Animals were vaccinated once on day 0 and blood was collected on days 7, 14, and 28 for immunologic analysis (Figure 2a). ELISA conducted with the Spike ectodomain showed that a single IM vaccination with either dose of VSV∆G-SARS-CoV-2 #9 induced substantial serum IgG titres detectable on day 7 (Figure 2b) that increased modestly by day 28. The IgG titres induced by the higher IM dose (5 × 105 PFUs) of VSV∆G-SARS-CoV-2 #9 were statistically significant on days 7, 14, and 28 when compared with the sera from control animals. At day 28, the titres also were similar to those in animals vaccinated with trimeric Spike protein formulated with Adju-Phos, although the humoral response induced by the subunit vaccine may have developed more gradually. At the lower dose of VSV∆G-SARS-CoV-2 #9 (5 × 104 PFUs; Figure 2b), it was clear that the sera samples were positive for Spike antibodies on all 3 days that samples were collected, but the group data reached significance compared with control animals only on day 7 probably because of the small group size and increased scatter in the data on days 14 and 28. Notably, cotton rats vaccinated by the IN route with VSV∆G-SARS-CoV-2 #9 did not develop anti-Spike serum antibodies.
Serum nAb titres were quantified using a plaque-reduction microneutralisation assay based on neutralisation of VSV∆G-SARS-CoV-2 #9 (Figure 2c). Cotton rats vaccinated by IM injection with the higher dose (Group 1; 5 × 105 PFUs) developed considerable neutralising titres with geometric mean NT50 values of 1000 or more by day 28. Animals vaccinated with 5 × 104 PFU also developed nAbs, although the geometric mean titres were reduced, and the group data did not reach significance due to the lower titres and increased variability within the group. The Spike protein/Adju-Phos vaccine also induced serum nAbs, and by day 28 the titres were similar in magnitude to those in cotton rats vaccinated with the lower dose of VSV∆G-SARS-CoV-2 #9 (5 × 104 PFUs). Mucosal vaccination failed to induce serum nAbs (Figure 2c), as expected based on the ELISA data (Figure 2b).
Additional sampling at days 1 and 3 (Figure 2a) was performed to assess if VSV∆G-SARS-CoV-2 #9 genomic RNA (vRNA) was detectable in blood or stool early after infection. VSV∆G-SARS-CoV-2 RNA was not detectable in blood by RT-qPCR on either day (Supplementary Table S1). Stool samples collected from some cages of cohoused animals did produce positive RT-qPCR signals on days 1 and 3, but the genome copies were minimal and near detection limits (Supplementary Table S2). Thus, if VSV∆G-SARS-CoV-2 #9 replicated substantially after IM or IN vaccination in cotton rats, it did not produce a systemic infection accompanied by viremia or significant shedding of genomes in faeces.
Immunogenicity and efficacy of VSV∆G-SARS-CoV-2 in golden Syrian hamsters
Golden Syrian hamsters were evaluated as a second model for VSV∆G-SARS-CoV-2 immunogenicity and efficacy following publication of data demonstrating the animal's susceptibility to respiratory tract infection with SARS-CoV-2.65, 66, 67 The study was divided into two parts. In Part A (Figure 3a), the animals were vaccinated and later challenged with SARS-CoV-2 (USA-WA1/2020), after which their weight and clinical signs were monitored for 14 days. In Part B (Figure 3a), vaccinated hamsters were challenged with SARS-CoV-2 and then sacrificed 4 days later to evaluate tissue virus loads.
Part A (Figure 3a) included two groups of vaccinated animals (n = 5 per group) and an unvaccinated control group (n = 6). One group was vaccinated with VSV∆G-SARS-CoV-2 #9 (3 × 106 PFU total, IM) split between both hind legs and a second group was injected with soluble Spike protein (10 µg) formulated with Adju-Phos (40 µg), as used in the cotton rat study described above (Figure 2a). Blood was collected on days 7, 14, and 26 after vaccination (Figure 3a) for analysis of serum antibodies, and on day 28 the animals were challenged by IN inoculation with 2 × 104 PFUs of SARS-CoV-2 (USA-WA1/2020), after which they were monitored for 14 days for clinical signs and weight loss.
Serum antibodies induced in animals in Part A were quantified and characterised by anti-Spike ELISA (Figure 3b), neutralisation of authentic SARS-CoV-2 (USA-WA1/2020; Figure 3c), or neutralisation of VSV∆G-SARS-CoV-2 #9 (Figure 3d). As shown in cotton rats (Figure 2b), the Spike protein/Adju-Phos vaccine (Figure 3b, red) elicited a strong response that exceeded titres of 1 × 104 by days 14 and 26. Hamsters immunised with VSV∆G-SARS-CoV-2 #9 (Figure 3b, teal) also rapidly developed substantial serum IgG titres by day 7 that increased modestly at the later time points. The binding titres in both vaccine groups were statistically significant on days 14 and 28 when compared with controls.
When virus neutralisation was evaluated using authentic SARS-CoV-2 (Figure 3c) or VSV∆G-SARS-CoV-2 #9 (Figure 3d), it was evident that both vaccines induced serum nAbs detectable with either assay, and that the highest mean titres were observed on day 14. The neutralising titres were higher in the assay based on VSV∆G-SARS-CoV-2 #9 and were statistically significant compared with controls, except on day 14 in the group vaccinated with VSV∆G-SARS-CoV-2 #9 due to increased scatter in the data points. Both neutralisation assays (Figure 3c and d) also detected some decrease in nAb titres at day 26 while the ELISA titres were relatively stable (Figure 3a), indicating that the neutralisation assay may be a more sensitive indicator of titre fluctuations by the subset of antibodies that primarily target the Spike receptor binding domain. This observation also indicated that it will be informative to extend future studies to assess whether the nAb titres stabilise, and whether a boost affects the magnitude and persistence of nAbs.
When the hamsters described above from Part A of the study (Figure 3a) were challenged with SARS-CoV-2 by IN inoculation, animals vaccinated with either VSV∆G-SARS-CoV-2 #9 or with Spike protein/Adju-Phos were protected from weight loss after SARS-CoV-2 challenge (Figure 4a). All vaccinated animals exhibited transient weight loss out to day 2 after challenge that may have been influenced by anaesthesia and handling, but vaccinated hamsters soon resumed gaining weight normally. In contrast, unvaccinated animals lost on average 9% of their body weight over a period of 7 days after which they recovered and steadily gained weight consistent with earlier data on SARS-CoV-2 infection in hamsters.67 Overall, hamsters vaccinated with VSV∆G-SARS-CoV-2 #9 or the subunit vaccine lost significantly less weight than unvaccinated animals on days 4 to 13, but by the end of the study, body weights were not significantly different for all three groups.
Part B of the study (Figure 3a) included four animals vaccinated by IM injection with VSV∆G-SARS-CoV-2 #9 (3 × 106 PFU) and four unvaccinated controls. Blood was not collected for serological analysis in Part B, however, animals were challenged on day 28 using the same SARS-CoV-2 challenge stock and IN challenge methods used in Part A. Four days after challenge, the hamsters were euthanised and tissues were collected to assess SARS-CoV-2 virus load. Infectious SARS-CoV-2 in lung tissue (Figure 4b) and nasal tissue (Figure 4c) homogenates were quantified by TCID50 using Vero E6 cells. In unvaccinated animals, high virus loads with a geometric mean of 4.9 × 107 TCID50 per gram of lung tissue were detected (Figure 4b) as expected based on the prolonged weight differential in the unvaccinated animals in Part A (Figure 4a). In contrast, animals vaccinated with VSV∆G-SARS-CoV-2 #9 had about 3000 times less infectious SARS-CoV-2 in the lung, with a geometric mean TCID50 titre of 9.8 × 103. In unvaccinated animals, infectious titres of SARS-CoV-2 in nasal tissue (Figure 4c) also were substantial with a geometric mean value of 9.8 × 105 TCID50 per gram of tissue. In animals vaccinated with VSV∆G-SARS-CoV-2 #9, 20 times less infectious virus was observed demonstrating that systemic immunity induced by IM vaccination did reduce SARS-CoV-2 replication in nasal tissue although not as strongly as it did in the lower respiratory tract.
Immunogenicity and efficacy of VSV∆G-SARS-CoV-2 in golden Syrian hamsters vaccinated by alternative routes
Although cotton rats did not mount an immune response to VSV∆G-SARS-CoV-2 following IN immunisation (Figure 2b and c), we reinvestigated mucosal vaccination with VSV∆G-SARS-CoV-2 in hamsters for multiple reasons. First, hamsters were shown to be susceptible to Spike-dependent infection when SARS-CoV-2 was administered intranasally.67 Second, various examples of VSV vectors have previously demonstrated immunogenicity when delivered by mucosal routes.21,68, 69, 70, 71 Third, effective mucosal vaccination has the potential to stimulate systemic immune responses as well as establish protective mucosal tissue-resident memory immunity that can respond rapidly to SARS-CoV-2 exposure in the upper airways and provide greater control over local SARS-CoV-2 replication.72 Finally, cells expressing the cofactors needed for productive Spike-dependent infection, including ACE2, TMPRSS2, and furin, have been shown to be relatively abundant in multiple mucosal sites, including the nasal cavity, oral cavity, and gastrointestinal tract.73, 74, 75, 76, 77, 78, 79, 80 Therefore, we compared mucosal and IM vaccination with VSV∆G-SARS-CoV-2 #9.
In the hamster study outlined in Figure 5a, 10 hamsters per group were vaccinated either by application of VSV∆G-SARS-CoV-2 #9 (3 × 105 PFUs) to mucosal surfaces or by IM injection using three different dose levels (3 × 106, 1 × 105, 1 × 103 PFUs). IM injection in this study was performed with the vaccine dose split across both hind legs as in the earlier hamster study (Figures 3 and 4). Mucosal vaccination was conducted by applying 10 µL virus dropwise to OM along the buccal surfaces or by a combined IN/OM method dividing mucosal vaccination across the OM (10 µL of virus) and IN surfaces (50 µL of virus per nare). The relatively large volume of vaccine used for IN/OM administration was selected to ensure the virus formulation made maximal contact with mucosal surfaces in the hamster upper respiratory tract. However, it should be noted that this approach likely allowed some of the inoculum to reach the lower respiratory tract.
Serum IgG titres (Figure 5b) indicated that IM injection with 3 × 106 or 1 × 105 PFUs was strongly immunogenic, consistent with the prior hamster study (Figure 3b). The lowest IM dose of 1 × 103 PFUs induced lower IgG titres in some animals, whereas other animals failed to mount a detectable antibody response, which indicated that consistent seroconversion after a single IM vaccination required a dose greater than 1 × 103 PFU. Hamsters vaccinated by the IN/OM method using 3 × 105 PFUs also developed a strong systemic antibody response, as shown by ELISA titres (Figure 5b). The binding titres induced by IN/OM vaccination increased between days 14 and 28, whereas titres induced by IM injection levelled off after day 14, which resulted in IN/OM vaccination producing seven-fold higher serum IgG titres at day 28 compared with those produced by IM injection with a 10-fold higher dose (Figure 5b). Vaccination using only the OM method was less effective and not all animals developed detectable serum IgG, although responding animals did achieve substantial binding titres at day 28.
Serum nAbs where quantified by VSV∆G-SARS-CoV-2 #9 plaque-reduction assay (Figure 5c). The results showed that IM injection or mucosal vaccination elicited substantial quantities of serum nAbs and that the titres generally aligned with the magnitude of IgG ELISA titres (Figure 5b), although there were several differences worth mentioning. For example, the magnitude of the ELISA titres at days 14 and 28 did not change substantially in animals vaccinated by IM injection with either 3 × 106 or 1 × 105 PFUs (Figure 5b), whereas the nAbs titres declined modestly (Figure 5c) as also seen in Figure 3b and c. Notably, a similar decline in the nAb titres was not evident in the IN/OM group (Figure 5c). It was also interesting that low-dose IM injection (1 × 103 PFU) did not result in development of nAbs much above the limit of detection even in animals with detectable ELISA titres. In contrast, animals with similar modest ELISA titres after OM vaccination (Figure 5b) did develop considerable serum nAb titres (Figure 5c). Taken together, these results highlight that both IM and mucosal vaccination can induce potent serum nAbs, but that the routes of administration may result in qualitative differences in the humoral responses.
To generate some initial data on how the different vaccination methods may affect the distribution of VSV∆G-SARS-CoV-2 #9 RNA in hamsters, we isolated RNA from blood samples at days 1 and 7 after vaccination and from stool samples on days 1 and 3 after vaccination (Figure 5d and e). Note that for this part of the study, only the high-dose IM vaccination group (3 × 106 PFU; Figure 4a) was analysed for VSV∆G-SARS-CoV-2 #9 vRNA, and stool for RNA extraction was collected from cages of animals that were housed together within the same vaccine group.
Analysis of RNA extracted from blood (Figure 5d) showed that animals vaccinated by IM injection had low transient RT-qPCR signals (∼100–1000s copies per millilitre of serum of VSV matrix gene sequence) 1 day after vaccination that were absent by day 7. In the OM or IN/OM groups, only two animals in the OM group had a delectable signal produced from blood (∼1000 copies per millilitres) at day 7. When stool was analysed, RNA was detectable in samples from animals vaccinated by the IN/OM or OM route that were above background signals observed in samples from unvaccinated controls on day 1, and the positive signals were observed again on day 3 in the IN/OM group and to a lesser extent in the OM group. Although blood and stool samples were identified with positive RT-PCR signals, the genome copies were generally low, suggesting it would be challenging to demonstrate that live virus was present in these samples. Further studies will be needed to determine if the RNA signals were indicative of low levels of viremia or infectious virus shedding in stool. The presence of live VSV∆G-SARS-CoV-2 in stool seems unlikely because detection of infectious SARS-CoV-281 or VSV in feces82 is not common.
Hamsters from the study described above (Figure 5) were challenged (Figure 6) at 28 days post-vaccination with 2.3 × 104 PFUs of SARS-CoV-2 (USA-WA1/2020) delivered by IN drops. Four days post challenge, five animals from each group were euthanised to analyse tissue virus loads and infectious SARS-CoV-2 in homogenised lung or nasal tissue were quantified using a TCID50 method, as in the previous study (Figure 4).
It is important to mention that the SARS-CoV-2 challenge stock used in this study was subjected to additional passage in Vero cells compared with the stock used in the hamster study described Figures 3 and 4. The stock used for the study in Figure 6 was passaged four times in Vero E6 cells, which resulted in Spike substitutions in R682 (Supplementary Figure S1b; 681-PRRAR-685), which is a highly conserved R residue in the furin cleavage site consensus.53 Based on next-generating sequencing, we estimate that about 95% of the challenge virus had a substitution at R682 (L, W, or Q; Supplementary Figure S1b) and this likely reduced challenge stock virulence, resulting in modest weight loss (<5%) even in control hamsters (Supplementary Figure S1a). Interestingly, even though the expected weight loss was not observed, live SARS-CoV-2 titres in lung tissue (Figure 6a; PBS) or nasal tissue (Figure 6b; PBS) confirmed that the challenge virus stock was capable of substantial virus replication in the upper and lower respiratory tract, consistent with prior observations showing that SARS-CoV-2 strains with furin cleavage site mutations maintain the ability to replicate in hamsters and ferrets.40,41,83,84
In animals vaccinated with the high-dose (3 × 106 PFUs) by the IM route, significantly reduced SARS-CoV-2 challenge virus replication was observed in the lungs (Figure 6a). In four of five hamsters vaccinated by IM injection, infectious SARS-CoV-2 was undetectable in lung tissue samples and one animal had virus quantities just above the lower limit of detection. IM vaccination with lower doses (1 × 105 or 1 × 103 PFUs) was less effective with breakthrough SARS-CoV-2 replication in 2 of 5 animals in the 1 × 105 PFU group and considerable breakthrough in all animals vaccinated with 1 × 103 PFUs.
Like high-dose IM vaccination (Figure 6a), mucosal vaccination by the IN/OM method with 10-fold less vaccine (3 × 105 PFUs) was effective and protected the lung of all animals in this group. OM-only vaccination presented mixed results in line with the ELISA (Figure 5b) and nAb (Figure 5c) titres, with seropositive hamsters protected from SARS-CoV-2 replication in the lung.
When SARS-CoV-2 quantities in nasal tissue samples were evaluated (Figure 6b), it was evident that the vaccination routes had a noticeable effect on immunity in the nasal mucosa. Despite all IM-vaccinated hamsters in the 3 × 106 PFU group being seropositive (Figure 5b and c) there was variable prevention of challenge virus replication in the nasal cavity, with all five hamsters having TCID50 titres of 1 × 104 or greater in nasal tissues (Figure 6b). Also, as the IM dose was decreased, there was increasing SARS-CoV-2 breakthrough replication in the nasal tissue. In contrast, four of five IN/OM-vaccinated hamsters had undetectable TCID50 titres of SARS-CoV-2, and the fifth animal had titres of less than 1 × 104 per gram of nasal tissue. Consistent with the IN/OM findings, the two OM-vaccinated seropositive animals also strongly controlled SARS-CoV-2 replication in the nasal cavity. Collectively, these results suggest that mucosal vaccination, even when provided at a 10x lower dose than used for the highest dose of IM injection, has increased potential to protect the upper airways.
Plotting SARS-CoV-2 tissue virus loads against serum nAbs (Figure 6c) further highlighted the point that mucosal vaccination provided an additional protective element to the immune response not offered by parenteral vaccination. Serum nAb titres appeared to correlate well with protection in the lung by all routes of vaccine administration. All but two vaccinated animals that developed serum nAbs (x-axis) titres of greater than 100 were protected from SARS-CoV-2 replication in the lung, including the two responding animals from the OM vaccinated group (Figure 5b and c, Figure 6c). In contrast, protection from SARS-CoV-2 replication in the nasal cavity was less correlated with serum nAb levels, as all animals vaccinated by IM injection had higher quantities of SARS-CoV-2 in nasal tissue samples (Figure 6c). Animals vaccinated by the IN/OM route or the seropositive animals vaccinated by the OM-only route were able to control SARS-CoV-2 replication in the nose, perhaps suggesting local immunity might be contributing to protection in the upper respiratory tract (Figure 6c).
Immunogenicity of intranasal VSV∆G-SARS-CoV-2 vaccination in hamsters
The immunogenicity and efficacy of IN/OM and OM vaccination in the previous study (Figures 5 and 6) indicated that mucosal vaccination-induced immunity could protect both lung and nasal tissues from SARS-CoV-2. Because the OM-only route of vaccination was inconsistently immunogenic and protective (Figures 5 and 6) and to better define the route of vaccination driving the strong immunogenicity observed in IN/OM-vaccinated animals, we further evaluated the immunogenicity of IN vaccination with VSV∆G-SARS-CoV-2 #9 in the hamsters.
Four groups of 5 hamsters were vaccinated by IN administration using doses from 1 × 106 down to 1 × 103 PFUs (Figure 7a). IN vaccination was conducted dropwise on anesthetised animals using 10 µL per nostril to help limit spread of the initial inoculum.25 Controls injected with buffer and a group vaccinated by IM injection (1 × 106 PFU) were included as comparators. Blood was collected weekly for 5 weeks for evaluation by ELISA and VSV∆G-SARS-CoV-2 plaque-reduction assay.
IN vaccination across all doses was strongly immunogenic (Figure 7a). ELISA titres peaked around days 21 to 28 in all animals except one in the 106 PFU group that failed to respond. Noticeably, the titres were relatively unaffected by dose, with all groups exceeding endpoint titres of 3 × 105 on day 28. The serum IgG titres induced in all IN vaccination groups also were clearly higher than in the IM vaccination (1 × 106 PFUs) comparator group. However, it should be noted that the titres produced by IM injection in this experiment were somewhat lower than those seen earlier (Figures 3 and 5), possibly because IM injection was performed in a single hind leg, whereas in the studies described earlier the dose was split between both hind legs. Even when compared with the lowest IN vaccine dose of 1 × 103 PFU that produced a geometric mean titre of 370779 at day 28, the IgG titres induced by IM vaccination (mean 7821) were 47x lower. This result might suggest IN vaccination provides a cell environment that allows for increased VSV∆G-SARS-CoV-2 infection and replication, which drives a stronger immune response than is possible when the virus is injected into muscle tissue. Even though IN vaccination likely supported increased VSV∆G-SARS-CoV-2 replication, it appeared to have little effect on the hamsters, as they gained weight steadily during the study (Figure 7d). Future studies will specifically investigate VSV∆G-SARS-CoV-2 replication in the animals vaccinated by the IN route as well as assess if the live vaccine elicits nAbs that are active against genetically diverse stains of SARS-CoV-2.
Discussion
Our original objective was to rapidly develop a vaccine that would deliver an authentic SARS-CoV-2 transmembrane glycoprotein Spike immunogen using the VSV∆G chimeric virus approach that was used for development of the successful ebolavirus Zaire vaccine.14,15 Our initial plan was to compare IM and mucosal vaccination, since we expected mucosal delivery would be advantageous due to the abundance of ACE2 receptors at respiratory tract mucosal surfaces75 and the potential for this route of vaccination to elicit immunity resident at vulnerable mucosal sites.72 However, due to the urgent need to develop an effective COVID-19 vaccine when this programme was initiated, we expedited the development of VSV∆G-SARS-CoV-2 for IM injection by making use of the substantial nonclinical and clinical data associated with the ebolavirus Zaire vaccine ERVEBO®, which is delivered by the IM route.14,15,85 Moreover, advancing a vaccine for administration by standard IM injection was well supported by immunogenicity and efficacy data produced here using the hamster model, as well as a full nonclinical safety assessment that was completed before advancing to human trials (unpublished data); thus, VSV∆G-SARS-CoV-2 #9 (called V590) was advanced and evaluated in a phase 1 clinical trial,18 which showed that a single IM injection with V590 was safe but not sufficiently immunogenic to continue with the product development path based on a vaccine for IM injection. Some of the preclinical data presented here provide useful context for considering the basis for the low immunogenicity of the single IM dose of VSV∆G-SARS-CoV-2 #9 observed in the phase 1 clinical trial.18 Yet, more importantly, the research described here, as well as by others50,86 also provides encouraging data to support the development and further evaluation of an intranasal VSV∆G-SARS-CoV-2 vaccine that could help prevent upper respiratory tract infection by SARS-CoV-2 and reduce transmission from asymptomatic infected individuals.
The cotton rat immunogenicity study (Figure 2) demonstrated that VSV∆G-SARS-CoV-2 #9 elicited binding antibodies and nAbs after a single IM injection, supporting the feasibility of the IM route for vaccination. Interestingly, when cotton rats were vaccinated by the IN route (Figure 2a), humoral responses were undetectable (Figure 2b and c). One interpretation of this data was that IN vaccination was ineffective because infection by VSV∆G-SARS-CoV-2 #9 at the mucosal barrier was inefficient, whereas IM injection allowed sufficient infection and replication to induce an antibody response. However, mucosal vaccination studies conducted later in hamsters (Figures 3, 5 and 7) showed that IN vaccination induced robust antibody responses, indicating that there was no innate barrier to infection at the nasal mucosal in naïve hamsters. Instead, it seemed more likely that cotton rats failed to develop immune responses after IN vaccination because VSV∆G-SARS-CoV-2 was unable to efficiently infect and replicate in the nasal mucosa perhaps because the interaction with cotton rat ACE2 was not optimal. Although we do not have direct evidence to show that the interaction between cotton rat ACE2 and Spike is suboptimal, computational studies indicate that multiple rodent ACE2 receptors are likely to interact weakly with Spike.87 The explanation for the lack of immunogenicity after IN vaccination due to a weak interaction with cotton rat ACE2 further implies that antibody responses elicited by IM injection (Figure 2) may have been primarily driven by exposure to Spike arrayed on VSV∆G-SARS-CoV-2 particles (Figure 1e) in the vaccine inoculum rather than infection and replication, which has been described previously in studies with inactivated VSV.88,89 Interestingly, this interpretation may also agree with earlier results from cotton rat studies in which exposure to the original SARS-CoV led to the development of Spike antibodies without any signs of infection or virus replication.90
Our initial study (Figure 3) conducted in hamsters was part of our evaluation of IM vaccination and efficacy. Consistent with the data in cotton rats, IM injection with VSV∆G-SARS-CoV-2 #9 (3 × 106 PFUs) elicited a strong binding antibody response (Figure 3b), and the animals developed potent serum antibodies that could neutralise VSV∆G-SARS-CoV-2 #9 (Figure 3d) as well as authentic SARS-CoV-2 (Figure 3c). Moreover, when the animals were challenged with SARS-CoV-2, they were protected from weight loss (Figure 4a) and virus replication was prevented in the lower respiratory tract (Figure 4b), demonstrating that IM injection was efficacious in hamsters. Similar encouraging results in hamsters vaccinated with a single IM injection of VSV∆G-based SARS-CoV-2 vaccines also have been observed by others,38,50,86 further supporting use of this route of vaccination. Accordingly, administration of a single IM dose of VSV∆G-SARS-CoV-2 #9 (V590) was evaluated in a phase 1 clinical trial.18
Although we had encouraging preclinical data from studies in cotton rats and hamsters supporting use of a single IM injection of VSV∆G-SARS-CoV-2, these positive results failed to translate to vaccine responses observed in humans.18 The reason for the conflicting results between rodents and humans is speculative and is yet to be fully defined; however, as mentioned above, our cotton rat data (Figure 2) suggested that it was possible to induce humoral immune responses in rodents by IM injection in the absence of significant infection and replication primarily through immune responses induced by the regular arrays of Spikes on the input VSV∆G-SARS-CoV-2 virions (Figure 1e) 91 and perhaps this is augmented by virion components interacting with viral sensors such as Spike binding to toll-like receptor 4.92 The possibility that IM injection was immunogenic in rodents without substantial virus infection also may be extended to our hamster data (Figures 3 and 5), because skeletal muscle tissue is not a rich source of cells expressing ACE2 receptors74,93 that could support infection. The hypothesis that skeletal muscle is not an effective site for Spike-dependent VSV∆G-SARS-CoV-2 replication also has been discussed by others based on their research with similar chimeric viruses.50,86,94 Thus, although the VSV∆G-SARS-CoV-2 particles displaying Spike arrays might be quite immunogenic after IM injection in rodents in the absence of significant infection and replication, it is conceivable that a similar quantity of virion particles injected a single time into a much larger macaque94 or human muscle18 would be less immunostimulatory in the absence of a potent adjuvant, administration of a booster dose,95 or immune system priming, for example, by previous infection with SARS-CoV-2.18
Our investigation of differing routes of vaccine administration (Figures 3 and 5) suggests that mucosal vaccination results in a greater immune response to VSV∆G-SARS-CoV-2 in hamsters compared with IM injection and that IN vaccination may be optimal (Figure 7). The likely reason for this result is that cells expressing ACE2 as well as the other cofactors needed for productive Spike-dependent infection, are abundant in the upper respiratory tract mucosa and not the muscle.73, 74, 75, 76, 77, 78, 79, 80 Consistent with this explanation, it also is clear that VSV vectors encoding Spike immunogens are immunogenic by other vaccination routes if they are designed to use infection pathways that are independent of ACE2.27,50,94,96, 97, 98, 99 Our initial hamster study using alternative routes assessed two mucosal vaccination methods. The high volume IN/OM combination was used initially to enable infection across multiple mucosal surfaces in the upper respiratory tract (Figure 5a), whereas another group was vaccinated by applying virus only to OM surfaces because data were emerging on ACE2 abundance and the susceptibility of cells to SAR-CoV-2 infection in the oral cavity.76,78,79 Notably, IN/OM vaccination was highly immunogenic and induced serum nAbs in all animals in this study (Figure 5b and c), whereas a single dose delivered by just the OM route was immunogenic but not consistently (two of five OM-vaccinated animals responded), which may be similar to the lack of immunogenicity seen in macaques after a single oral dose.94 Together these results indicated that the application of VSV∆G-SARS-CoV-2 to the mucosal epithelium in the nasal cavity, nasopharynx, and possibly the lower respiratory tract was the main contributor to the strong immunogenicity seen with the IN/OM vaccination method. A follow-up hamster study (Figure 7) was then conducted using a small volume of VSV∆G-SARS-CoV-2 #9 applied only to the nasal cavity to restrict most of the vaccine inoculum to the nasal epithelium. This method of IN vaccination caused no observable adverse reactions in the hamsters (Figure 7d) and produced a robust systemic immune response with doses as low as 1000 PFU (Figure 7b and c), supporting the interpretation that IN vaccination is an effective method for VSV∆G-SARS-CoV-2 delivery in hamsters, as also concluded by others.50,86
The potential benefit of using mucosal vaccination also was evident in greater control of upper respiratory tract SARS-CoV-2 infection. All IN/OM-vaccinated and seropositive OM-vaccinated hamsters strongly suppressed SARS-CoV-2 replication in the nasal cavity (Figure 6b). In contrast, parenteral vaccine delivery, even at a 10-fold higher dose, was unable to abort upper respiratory tract infection (Figure 6b), suggesting that mucosal vaccination may afford protection against severe SARS-CoV-2 disease in the lung (Figure 6a) and decrease viral transmission. It was also notable that protection from SARS-CoV-2 replication in the lung had a strong association with serum nAb titres (Figure 6c), while in nasal tissue this correlation was less convincing (Figure 6c), indicating that local immune responses elicited following mucosal immunisation may contribute to preventing replicating in the nose. Although these data clearly point to mucosal delivery of VSV∆G-SARS-CoV-2 providing important advantages, recent interesting immunogenicity and efficacy data following mucosal vaccination with VSV-based vectors suggests that the immune response profile may be substantially influenced by the glycoprotein that is determining cell tropism, replicative capacity of the VSV vector delivering the Spike, and the model animal. Notably, results in hamsters indicate that VSV∆G chimeras designed to coexpress functional ebolavirus GP and a SARS-CoV-2 Spike immunogen or chimeras expressing Spike only50,86 (Figures 5 and 6) are efficacious after mucosal vaccination, whereas the VSV∆G chimera expressing both GP and Spike was not effective in macaques after IN vaccination.100 Additional research is needed to understand this observation; however, one interpretation is that glycoprotein antigen immune dominance driven by the ebolavirus GP following IN vaccination in macaques might shift the balance of neutralising and non-neutralising Spike antibodies more towards binding antibodies that cannot contribute to protection from SARS-CoV-2.
Some limitations to the interpretation of our studies should be noted. One confounding variable for IN vaccination studies involving small animals is the volume of vaccine applied, with larger volumes delivered to anesthetised rodents allowing vaccine to descend to the lung where greater vascularisation and infiltration by antigen-presenting cells can result in a more robust immune response than strict IN immunisation.101 In studies of cotton rats, we previously demonstrated that applying a light anaesthetic and using a volume of 5 µL per nare ensures that the vaccine does not descend to the lung.25 Although we do not have imaging data to confirm the localisation of our mucosally-applied VSV∆G-SARS-CoV-2 in hamsters, it is likely that the OM/IN vaccination with 50 µL vaccine formulation applied per nare (Figure 5) resulted in deposition of the vaccine in both the nose and lung. In the follow-up immunogenicity study (Figure 7), IN administration to hamsters was conducted with 10 µL of vaccine applied per nare, which improved the likelihood of retaining vaccine in the nose, but the possibility of some lung deposition cannot be excluded.
In conclusion, we have demonstrated that VSV∆G-SARS-CoV-2 is immunogenic and protective against SARS-CoV-2 in golden Syrian hamsters following parenteral (IM) or mucosal (IN/OM or IN) administration, and that mucosal routes of administration induce stronger immunogenicity even with lower vaccine doses. Mucosal vaccination also conferred greater upper respiratory protection compared with parenteral administration. The data suggest that if VSV∆G-SARS-CoV-2 can replicate in mucosal tissues of the human nasal cavity and nasopharynx, mucosal administration of VSV∆G-SARS-CoV-2 may result in better clinical outcomes than have recently been observed following a phase 1 study with IM vaccination.18
Contributors
A.J.B., A.S.E., C.C., C.L.P., D.D.R., E.Sa., E.Str., G.M., J.D., L.W., M.C., M.B.F., M.Y., S.S., T.K.M. contributed to the conception, design or planning of the study. A.W., B.L., F.H., F.L., G.H., G.M., G.W., K.K., L.R., M.H., M.R., M.Y., S.L., S.S., T.K.M., Y.C. contributed to the acquisition of the data. A.S.E., A.W., C.C., C.L.P., F.H., G.M., G.W., K.K., L.R., L.Z., M.C., M.B.F., M.H., M.R., M.Y., S.L., S.S., T.K.M., Y.C. contributed to the analysis of the data. A.J.B., A.S.E., A.W., B.L., C.C., C.L.P., D.D.R., F.H., G.M., G.W., J.D., L.R., L.W., L.Z., M.C., M.B.F., M.H., M.Y., S.G., S.L., TKM contributed to the interpretation of the results. A.S.E., B.L., C.C., C.L.P., G.M., G.W., M.H., M.Y. drafted the manuscript. A.J.B., A.S.E., A.W., B.L., C.C., D.D.R., E.Sa., E.Str., F.H., F.L., G.H., G.M., K.K., J.D., L.R., L.W., L.Z., M.C., M.B.F., M.R., S.G., S.L., S.S., T.K.M., Y.C. critically reviewed or revised the manuscript for important intellectual content.
All authors had full access to all the data in the study and accept responsibility to submit for publication. The underlying data was verified by A.S.E., C.C., and C.L.P.
Data sharing statement
Data will be made available upon reasonable request through the corresponding author.
Declaration of interests
C.L.P., M.Y., M.B.F., A.J.B., A.S.E. are listed as inventors on a patent application submitted jointly by IAVI, NY, USA and Merck & Co., Inc., Rahway, NJ, USA (Vaccine Compositions for Preventing Coronavirus Disease). MBF holds stock in Merck Sharpe & Dohme, Corp. and is a consultant for Sanofi Pasteur. M.B.F. is a member of the board of directors at IAVI. No other potential conflicts of interest are declared. D.D.R., S.S., L.Z., A.J.B., M.C., A.S.E., G.H., T.K.M. and M.H. are employees of Merck Sharpe & Dohme, Corp., and division of Merck & Co., Inc., Rahway, NJ, USA and hold stock options in the Company. K.K., E.S., G.W., B.L. and F.L. are employees of Merck Sharpe & Dohme, Corp., and division of Merck & Co., Inc., Rahway, NJ, USA.
Acknowledgements
The vesicular stomatitis virus vector was licensed from the Public Health Agency of Canada (PHAC) for use in development of a COVID-19 vaccine. The authors wish to thank and acknowledge Paul Duncan, Irene Chang, Michael McNevin, Jennifer Galli, Eberhard Durr, Bonnie Howell, Stephen Pacchione, Walter Glaab, Christopher Ton, Michael Winters, Christopher Tubbs, Melissa Whiteman, Lisa Plitnick, Nickolas Murgolo, Arthur Fridman, and Daria Hazuda of Merck & Co., Inc., Rahway, NJ, USA, for their contributions to this study. In addition, the authors thank and acknowledge Jennifer Martinez, Megan Downey, Yutdelia Santana, Brooke Sanchez, Shima Nikkhah, Denise Wagner, Alexei Carpov, Olivia Wallace-Selman, Kristie Valentin, John Coleman, Finora Franck, Gretchen Meller, Kevin Wright, Devin Hunt, and Anne Ercolini of IAVI, New York, USA, and Swagata Kar and Hanne Andersen from BioQual, Maryland, USA, for their contributions to this study. Professional editorial assistance was provided by Christina Balle, PhD, of ApotheCom (London, UK) and was funded by Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Rahway, NJ, USA. The study was funded by Merck Sharp & Dohme, Corp., a subsidiary of Merck & Co., Inc., Rahway, NJ, USA. Part of this research was supported by the Biomedical Advanced Research and Development Authority (BARDA) of the US Department of Health and Human Services and the Defense Threat Reduction Agency (DTRA) of the US Department of Defense.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104203.
Appendix. Supplementary materials
References
- 1.Plante JA, Mitchell BM, Plante KS, Debbink K, Weaver SC, Menachery VD. The variant gambit: COVID-19′s next move. Cell Host Microbe. 2021;29(4):508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peacock TP, Penrice-Randal R, Hiscox JA, Barclay WS. SARS-CoV-2 one year on: evidence for ongoing viral adaptation. J Gen Virol. 2021;102(4) doi: 10.1099/jgv.0.001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boni MF, Lemey P, Jiang X, et al. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol. 2020;5(11):1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- 4.Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir Res. 2020;178 doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castro Dopico X, Ols S, Lore K, Karlsson Hedestam GB. Immunity to SARS-CoV-2 induced by infection or vaccination. J Intern Med. 2022;291(1):32–50. doi: 10.1111/joim.13372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murgolo N, Therien AG, Howell B, et al. SARS-CoV-2 tropism, entry, replication, and propagation: considerations for drug discovery and development. PLoS Pathog. 2021;17(2) doi: 10.1371/journal.ppat.1009225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraborty S, Mallajosyula V, Tato CM, Tan GS, Wang TT. SARS-CoV-2 vaccines in advanced clinical trials: where do we stand? Adv Drug Deliv Rev. 2021;172:314–338. doi: 10.1016/j.addr.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connors M, Graham BS, Lane HC, Fauci AS. SARS-CoV-2 vaccines: much accomplished, much to learn. Ann Intern Med. 2021;174(5):687–690. doi: 10.7326/M21-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Creech CB, Walker SC, Samuels RJ. SARS-CoV-2 Vaccines. JAMA. 2021;325(13):1318–1320. doi: 10.1001/jama.2021.3199. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert PB, Montefiori DC, McDermott AB, et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375(6576):43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng S, Phillips DJ, White T, et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat Med. 2021;27(11):2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Earle KA, Ambrosino DM, Fiore-Gartland A, et al. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine. 2021;39(32):4423–4428. doi: 10.1016/j.vaccine.2021.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldblatt D, Fiore-Gartland A, Johnson M, et al. Towards a population-based threshold of protection for COVID-19 vaccines. Vaccine. 2022;40(2):306–315. doi: 10.1016/j.vaccine.2021.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolfe DN, Taylor MJ, Zarrabian AG. Lessons learned from Zaire ebolavirus to help address urgent needs for vaccines against Sudan ebolavirus and Marburg virus. Hum Vacc Immunotherap. 2020;16(11):2855–2860. doi: 10.1080/21645515.2020.1741313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monath TP, Fast PE, Modjarrad K, et al. rVSVDeltaG-ZEBOV-GP (also designated V920) recombinant vesicular stomatitis virus pseudotyped with Ebola Zaire Glycoprotein: Standardized template with key considerations for a risk/benefit assessment. Vaccine X. 2019;1 doi: 10.1016/j.jvacx.2019.100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boritz E, Gerlach J, Johnson JE, Rose JK. Replication-competent rhabdoviruses with human immunodeficiency virus type 1 coats and green fluorescent protein: entry by a pH-independent pathway. J Virol. 1999;73(8):6937–6945. doi: 10.1128/jvi.73.8.6937-6945.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garbutt M, Liebscher R, Wahl-Jensen V, et al. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J Virol. 2004;78(10):5458–5465. doi: 10.1128/JVI.78.10.5458-5465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robbins JA, Tait D, Huang Q, et al. Safety and immunogenicity of intramuscular, single-dose V590 (rVSV-SARS-CoV-2 Vaccine) in healthy adults: Results from a phase 1 randomised, double-blind, placebo-controlled, dose-ranging trial. EBioMedicine. 2022;82 doi: 10.1016/j.ebiom.2022.104138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nyombayire J, Anzala O, Gazzard B, et al. First-in-human evaluation of the safety and immunogenicity of an intranasally administered replication-competent sendai virus-vectored HIV Type 1 Gag vaccine: induction of potent T-Cell or antibody responses in prime-boost regimens. J Infect Dis. 2017;215(1):95–104. doi: 10.1093/infdis/jiw500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harcourt J, Tamin A, Lu X, et al. Severe acute respiratory syndrome coronavirus 2 from patient with Coronavirus disease, United States. Emerg Infect Dis. 2020;26(6):1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabinovich S, Powell RL, Lindsay RW, et al. A novel, live-attenuated vesicular stomatitis virus vector displaying conformationally intact, functional HIV-1 envelope trimers that elicits potent cellular and humoral responses in mice. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0106597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witko SE, Kotash CS, Nowak RM, et al. An efficient helper-virus-free method for rescue of recombinant paramyxoviruses and rhadoviruses from a cell line suitable for vaccine development. J Virol Methods. 2006;135(1):91–101. doi: 10.1016/j.jviromet.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Hsieh CL, Goldsmith JA, Schaub JM, et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369(6510):1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munoz-Wolf N, Rial A, Saavedra JM, Chabalgoity JA. Sublingual immunotherapy as an alternative to induce protection against acute respiratory infections. J Vis Exp. 2014;(90) doi: 10.3791/52036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Citron MP, Patel M, Purcell M, et al. A novel method for strict intranasal delivery of non-replicating RSV vaccines in cotton rats and non-human primates. Vaccine. 2018;36(20):2876–2885. doi: 10.1016/j.vaccine.2018.02.110. [DOI] [PubMed] [Google Scholar]
- 26.Corbett KS, Flynn B, Foulds KE, et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. New Engl J Med. 2020;383(16):1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Case JB, Rothlauf PW, Chen RE, et al. Replication-competent vesicular stomatitis virus vaccine vector protects against SARS-CoV-2-mediated pathogenesis in mice. Cell Host Microbe. 2020;28(3):465–474.e4. doi: 10.1016/j.chom.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erasmus JH, Khandhar AP, O'Connor MA, et al. An Alphavirus-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci Transl Med. 2020;12(555) doi: 10.1126/scitranslmed.abc9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassan AO, Kafai NM, Dmitriev IP, et al. A single-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell. 2020;183(1):169–184.e13. doi: 10.1016/j.cell.2020.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horner C, Schurmann C, Auste A, et al. A highly immunogenic and effective measles virus-based Th1-biased COVID-19 vaccine. Proc Natl Acad Sci USA. 2020;117(51):32657–32666. doi: 10.1073/pnas.2014468117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercado NB, Zahn R, Wegmann F, et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586(7830):583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J, Tostanoski LH, Peter L, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369(6505):806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith TRF, Patel A, Ramos S, et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun. 2020;11(1):2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solforosi L, Kuipers H, Jongeneelen M, et al. Immunogenicity and efficacy of one and two doses of Ad26.COV2.S COVID vaccine in adult and aged NHP. J Exp Med. 2021;218(7) doi: 10.1084/jem.20202756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun W, Leist SR, McCroskery S, et al. Newcastle disease virus (NDV) expressing the spike protein of SARS-CoV-2 as a live virus vaccine candidate. EBioMedicine. 2020;62 doi: 10.1016/j.ebiom.2020.103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Doremalen N, Lambe T, Spencer A, et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586(7830):578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu S, Zhong G, Zhang J, et al. A single dose of an adenovirus-vectored vaccine provides protection against SARS-CoV-2 challenge. Nat Commun. 2020;11(1):4081. doi: 10.1038/s41467-020-17972-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yahalom-Ronen Y, Tamir H, Melamed S, et al. A single dose of recombinant VSV-G-spike vaccine provides protection against SARS-CoV-2 challenge. Nat Commun. 2020;11(1):6402. doi: 10.1038/s41467-020-20228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogando NS, Dalebout TJ, Zevenhoven-Dobbe JC, et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol. 2020;101(9):925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson BA, Xie X, Bailey AL, et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021;591(7849):293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lau SY, Wang P, Mok BW, et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg Microbes Infect. 2020;9(1):837–842. doi: 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z, Zheng H, Lin H, et al. Identification of common deletions in the spike protein of severe acute respiratory syndrome Coronavirus 2. J Virol. 2020;94(17):e00790–20. doi: 10.1128/JVI.00790-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davidson AD, Williamson MK, Lewis S, et al. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12(1):68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klimstra WB, Tilston-Lunel NL, Nambulli S, et al. SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J Gen Virol. 2020;101(11):1156–1169. doi: 10.1099/jgv.0.001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sasaki M, Uemura K, Sato A, et al. SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog. 2021;17(1) doi: 10.1371/journal.ppat.1009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reed E, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 47.Schnell MJ, Buonocore L, Whitt MA, Rose JK. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J Virol. 1996;70(4):2318–2323. doi: 10.1128/jvi.70.4.2318-2323.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Case JB, Rothlauf PW, Chen RE, et al. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe. 2020;28(3):475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dieterle ME, Haslwanter D, Bortz RH, 3rd, et al. A replication-competent vesicular stomatitis virus for studies of SARS-CoV-2 spike-mediated cell entry and its inhibition. Cell Host Microbe. 2020;28(3):486–496.e6. doi: 10.1016/j.chom.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Donnell KL, Clancy CS, Griffin AJ, et al. Optimization of single-dose VSV-based COVID-19 vaccination in hamsters. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.788235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cattin-Ortola J, Welch LG, Maslen SL, Papa G, James LC, Munro S. Sequences in the cytoplasmic tail of SARS-CoV-2 Spike facilitate expression at the cell surface and syncytia formation. Nat Commun. 2021;12(1):5333. doi: 10.1038/s41467-021-25589-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ke Y, Yu D, Zhang F, et al. Recombinant vesicular stomatitis virus expressing the spike protein of genotype 2b porcine epidemic diarrhea virus: A platform for vaccine development against emerging epidemic isolates. Virology. 2019;533:77–85. doi: 10.1016/j.virol.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Remacle AG, Shiryaev SA, Oh ES, et al. Substrate cleavage analysis of furin and related proprotein convertases. A comparative study. J Biol Chem. 2008;283(30):20897–20906. doi: 10.1074/jbc.M803762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lamers MM, Mykytyn AZ, Breugem TI, et al. Human airway cells prevent SARS-CoV-2 multibasic cleavage site cell culture adaptation. Elife. 2021;10 doi: 10.7554/eLife.66815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou W, Xiong M, Hao S, et al. The SARS-CoV-2 transcriptome and the dynamics of the S gene furin cleavage site in primary human airway epithelia. mBio. 2021;12(3) doi: 10.1128/mBio.01006-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Funnell SGP, Afrough B, Baczenas JJ, et al. A cautionary perspective regarding the isolation and serial propagation of SARS-CoV-2 in Vero cells. NPJ Vaccines. 2021;6(1):83. doi: 10.1038/s41541-021-00346-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen HT, Zhang S, Wang Q, et al. Spike glycoprotein and host cell determinants of SARS-CoV-2 entry and cytopathic effects. J Virol. 2020;95(5) doi: 10.1128/JVI.02304-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22(13) doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biniossek ML, Nagler DK, Becker-Pauly C, Schilling O. Proteomic identification of protease cleavage sites characterizes prime and non-prime specificity of cysteine cathepsins B, L, and S. J Proteome Res. 2011;10(12):5363–5373. doi: 10.1021/pr200621z. [DOI] [PubMed] [Google Scholar]
- 60.Remold SK, Rambaut A, Turner PE. Evolutionary genomics of host adaptation in vesicular stomatitis virus. Mol Biol Evol. 2008;25(6):1138–1147. doi: 10.1093/molbev/msn059. [DOI] [PubMed] [Google Scholar]
- 61.Sun CS, Wyde PR, Wilson SZ, Knight V. Efficacy of aerosolized recombinant interferons against vesicular stomatitis virus-induced lung infection in cotton rats. J Interf Res. 1984;4(4):449–459. doi: 10.1089/jir.1984.4.449. [DOI] [PubMed] [Google Scholar]
- 62.Boukhvalova MS, Prince GA, Blanco JC. The cotton rat model of respiratory viral infections. Biologicals. 2009;37(3):152–159. doi: 10.1016/j.biologicals.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kolappaswamy K. Susceptibility of Sigmodon hispidus. Lab Anim. 2015;44(6):199. doi: 10.1038/laban.767. [DOI] [PubMed] [Google Scholar]
- 64.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosenke K, Meade-White K, Letko M, et al. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. Emerg Microbes Infect. 2020;9(1):2673–2684. doi: 10.1080/22221751.2020.1858177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sia SF, Yan LM, Chin AWH, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583(7818):834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chan JF, Zhang AJ, Yuan S, et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71(9):2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qiu X, Fernando L, Alimonti JB, et al. Mucosal immunization of cynomolgus macaques with the VSVDeltaG/ZEBOVGP vaccine stimulates strong ebola GP-specific immune responses. PLoS One. 2009;4(5):e5547. doi: 10.1371/journal.pone.0005547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rose NF, Marx PA, Luckay A, et al. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell. 2001;106(5):539–549. doi: 10.1016/s0092-8674(01)00482-2. [DOI] [PubMed] [Google Scholar]
- 70.Kahn JS, Roberts A, Weibel C, Buonocore L, Rose JK. Replication-competent or attenuated, nonpropagating vesicular stomatitis viruses expressing respiratory syncytial virus (RSV) antigens protect mice against RSV challenge. J Virol. 2001;75(22):11079–11087. doi: 10.1128/JVI.75.22.11079-11087.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ryder AB, Buonocore L, Vogel L, Nachbagauer R, Krammer F, Rose JK. A viable recombinant rhabdovirus lacking its glycoprotein gene and expressing influenza virus hemagglutinin and neuraminidase is a potent influenza vaccine. J Virol. 2015;89(5):2820–2830. doi: 10.1128/JVI.03246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gallo O, Locatello LG, Mazzoni A, Novelli L, Annunziato F. The central role of the nasal microenvironment in the transmission, modulation, and clinical progression of SARS-CoV-2 infection. Mucosal Immunol. 2021;14(2):305–316. doi: 10.1038/s41385-020-00359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suresh V, Parida D, Minz AP, Sethi M, Sahoo BS, Senapati S. Tissue distribution of ACE2 protein in syrian golden hamster (Mesocricetus auratus) and its possible implications in SARS-CoV-2 related studies. Front Pharmacol. 2020;11 doi: 10.3389/fphar.2020.579330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hou YJ, Okuda K, Edwards CE, et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182(2):429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang N, Perez P, Kato T, et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat Med. 2021;27(5):892–903. doi: 10.1038/s41591-021-01296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu L, Wei Q, Alvarez X, et al. Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques. J Virol. 2011;85(8):4025–4030. doi: 10.1128/JVI.02292-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu H, Zhong L, Deng J, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12(1):8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee AC, Zhang AJ, Chan JF, et al. Oral SARS-CoV-2 inoculation establishes subclinical respiratory infection with virus shedding in golden syrian hamsters. Cell Rep Med. 2020;1(7) doi: 10.1016/j.xcrm.2020.100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sungnak W, Huang N, Becavin C, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jones DL, Baluja MQ, Graham DW, et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci Total Environ. 2020;749 doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Howerth EW, Mead DG, Mueller PO, Duncan L, Murphy MD, Stallknecht DE. Experimental vesicular stomatitis virus infection in horses: effect of route of inoculation and virus serotype. Vet Pathol. 2006;43(6):943–955. doi: 10.1354/vp.43-6-943. [DOI] [PubMed] [Google Scholar]
- 83.Peacock TP, Goldhill DH, Zhou J, et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol. 2021;6(7):899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- 84.Wong YC, Lau SY, Wang To KK, et al. Natural transmission of bat-like severe acute respiratory syndrome Coronavirus 2 without proline-arginine-arginine-alanine variants in coronavirus disease 2019 patients. Clin Infect Dis. 2021;73(2):e437–e444. doi: 10.1093/cid/ciaa953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tell JG, Coller BG, Dubey SA, et al. Environmental Risk Assessment for rVSVDeltaG-ZEBOV-GP, a genetically modified live vaccine for ebola virus disease. Vaccines. 2020;8(4) doi: 10.3390/vaccines8040779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O’Donnell KL, Gourdine T, Fletcher P, et al. VSV-Based vaccines reduce virus shedding and viral load in hamsters infected with SARS-CoV-2 variants of concern. Vaccines. 2022;10(3) doi: 10.3390/vaccines10030435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Damas J, Hughes GM, Keough KC, et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci USA. 2020;117(36):22311–22322. doi: 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hangartner L, Zinkernagel RM, Hengartner H. Antiviral antibody responses: the two extremes of a wide spectrum. Nat Rev Immunol. 2006;6(3):231–243. doi: 10.1038/nri1783. [DOI] [PubMed] [Google Scholar]
- 89.Bachmann MF, Kundig TM, Kalberer CP, Hengartner H, Zinkernagel RM. Formalin inactivation of vesicular stomatitis virus impairs T-cell- but not T-help-independent B-cell responses. J Virol. 1993;67(7):3917–3922. doi: 10.1128/jvi.67.7.3917-3922.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Watts DM, Peters CJ, Newman P, et al. Evaluation of cotton rats as a model for severe acute respiratory syndrome. Vector Borne Zoonotic Dis. 2008;8(3):339–344. doi: 10.1089/vbz.2007.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bachmann MF, Jennings GT. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat Rev Immunol. 2010;10(11):787–796. doi: 10.1038/nri2868. [DOI] [PubMed] [Google Scholar]
- 92.Zhao Y, Kuang M, Li J, et al. SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res. 2021;31(7):818–820. doi: 10.1038/s41422-021-00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hikmet F, Mear L, Edvinsson A, Micke P, Uhlen M, Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol. 2020;16(7):e9610. doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peng KW, Carey T, Lech P, et al. Boosting of SARS-CoV-2 immunity in nonhuman primates using an oral rhabdoviral vaccine. Vaccine. 2022;40(15):2342–2351. doi: 10.1016/j.vaccine.2021.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yahalom-Ronen Y, Erez N, Fisher M, et al. Neutralization of SARS-CoV-2 variants by rVSV-DeltaG-Spike-elicited human sera. Vaccines. 2022;10(2) doi: 10.3390/vaccines10020291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim GN, Choi JA, Wu K, et al. A vesicular stomatitis virus-based prime-boost vaccination strategy induces potent and protective neutralizing antibodies against SARS-CoV-2. PLoS Pathog. 2021;17(12) doi: 10.1371/journal.ppat.1010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Malherbe DC, Kurup D, Wirblich C, et al. A single dose of replication-competent VSV-vectored vaccine expressing SARS-CoV-2 S1 protects against virus replication in a hamster model of severe COVID-19. NPJ Vaccines. 2021;6(1):91. doi: 10.1038/s41541-021-00352-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lu M, Zhang Y, Dravid P, et al. A methyltransferase-defective vesicular stomatitis virus-based SARS-CoV-2 vaccine candidate provides complete protection against SARS-CoV-2 infection in hamsters. J Virol. 2021;95(20) doi: 10.1128/JVI.00592-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kapadia SU, Simon ID, Rose JK. SARS vaccine based on a replication-defective recombinant vesicular stomatitis virus is more potent than one based on a replication-competent vector. Virology. 2008;376(1):165–172. doi: 10.1016/j.virol.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Furuyama W, Shifflett K, Pinski AN, et al. Rapid protection from COVID-19 in nonhuman primates vaccinated intramuscularly but not intranasally with a single dose of a vesicular stomatitis virus-based vaccine. mBio. 2022 doi: 10.1128/mbio.03379-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lund FE, Randall TD. Scent of a vaccine. Science. 2021;373(6553):397–399. doi: 10.1126/science.abg9857. [DOI] [PubMed] [Google Scholar]
- 102.Lyles DS, Kuzmin I, Rupprecht CE. In: Fields Virology. 6th ed. Knipe DM, Howley PM, editors. Lippincott Williams and Wilkins; Philadelphia: 2013. Rhabdoviridae; pp. 885–922. [Google Scholar]
- 103.Lawson ND, Stillman EA, Whitt MA, Rose JK. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci USA. 1995;92(10):4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bestle D, Heindl MR, Limburg H, et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance. 2020;3(9) doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.