Abstract
Basal cell carcinoma (BCC) is the most common skin cancer, with over 2 million cases diagnosed annually in the United States. Conventionally, BCC is diagnosed by naked eye examination and dermoscopy. Suspicious lesions are either removed or biopsied for histopathological confirmation, thus lowering the specificity of non-invasive BCC diagnosis. Recently, reflectance confocal microscopy (RCM), a non-invasive diagnostic technique that can image skin lesions at cellular level resolution, has shown to improve specificity in BCC diagnosis and reduced the number needed to biopsy by 2-to-3 times. In this study, we developed and evaluated a deep learning-based artificial intelligence model to automatically detect BCC in RCM images. The proposed model achieved an area under the curve (AUC) for the receiver operator characteristic (ROC) curve of 89.7% (stack-level) and 88.3% (lesion level), a performance on par with that of RCM experts. Furthermore, the model achieved an AUC of 86.1% on a held-out test set from international collaborators, demonstrating the reproducibility and generalizability of the proposed automated diagnostic approach. These results provide a clear indication that the clinical deployment of decision support systems for the detection of BCC in RCM images has the potential for optimizing the evaluation and diagnosis of skin cancer patients.
Keywords: Reflectance Confocal Microscopy, Artificial Intelligence, Basal Cell Carcinoma
Graphical Abstract
INTRODUCTION
Basal cell carcinoma (BCC) is the most common skin cancer with over 2 million cases diagnosed annually in the United States [Rogers et al. 2015]. Conventionally, BCC is diagnosed by naked eye examination, along with the aid of dermoscopy [Navarrete-Dechent et al. 2016]. While the sensitivity of BCC diagnosis via dermoscopy is very high, the specificity can be in some cases as low as 53.8% [Reiter et al. 2019]. A low specificity entails a high number of invasive diagnostic biopsies; these rates are concerning since BCCs are commonly located on cosmetically and functionally concerning sites (e.g. face), and since patients may have multiple BCCs.
Recently, reflectance confocal microscopy (RCM), a non-invasive diagnostic technique that can image skin lesions at cellular level resolution, has been shown to improve specificity in BCC diagnosis [Kadouch et al. 2015] and reduce the number of lesions to be biopsied or treated by 2-to-3 times [Rajadhyaksha et al. 2017]. Unfortunately, image interpretation, due to the horizontal (en-face) view of the skin on RCM images and gray-scale contrast compared with the vertical view and purple and pink colored H&E stained images of histopathology, remains challenging for novices and requires extensive training [Jain et al. 2018].
Lately, artificial intelligence (AI) has been widely applied to the analysis of medical images for cancer diagnosis. Esteva et al. (2017) have shown that dermatologist-level accuracy can be achieved for the automated analysis of dermoscopy images. Campanella et al. (2019) described clinical-grade decision support systems for the pathology analysis of biopsied lesions, including for BCC. Recently, Tschandl et al. (2019) showed that human-computer collaboration can be beneficial in clinical scenarios. The research in applicability of AI for the RCM diagnosis of BCC is still limited [Wodzinski et al. 2019]. Yet, decision support tools that can guide the interpretation of RCM images could be immensely beneficial to increase the specificity of BCC diagnosis.
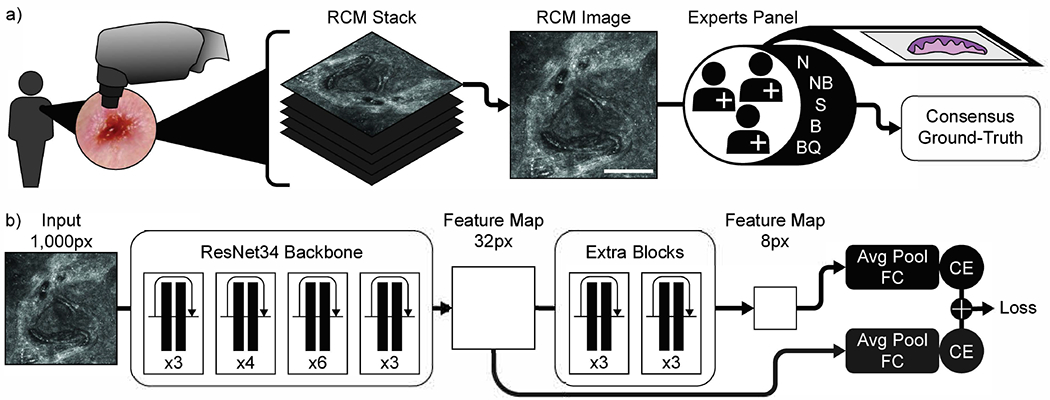
In this study, we developed and evaluated a deep learning-based AI model to automatically detect BCC in RCM images. For this, 312 stacks of RCM images were collected retrospectively at Memorial Sloan Kettering Cancer Center (MSKCC) from 66 lesions in 52 consecutive patients that were clinically equivocal for BCC and underwent RCM imaging after clinical and dermoscopy evaluations as part of their routine clinical evaluation. Stacks are small single field-of-view (0.5-1mm2) images acquired at consecutive depths (3.0 μm apart) starting from stratum corneum up to superficial dermis (i.e. 200 μm depth). All images within a stack were annotated by five expert RCM readers and confirmed by biopsy (Figure 1a). Each RCM image (within a stack) was labeled with one of the 5 labels: BCC (B), not-BCC (NB), suspicious for BCC (S), normal (N), and bad quality (BQ). Stacks containing malignant lesions other than BCC were removed, as well as low quality stacks. A total of 267 stacks were used for training and testing a predictive model via a 5-fold cross validation. For 131 of the stacks, the annotators were initially blinded to the histopathology diagnosis to compare the performance of human experts to that of the proposed method. See Methods for further details.
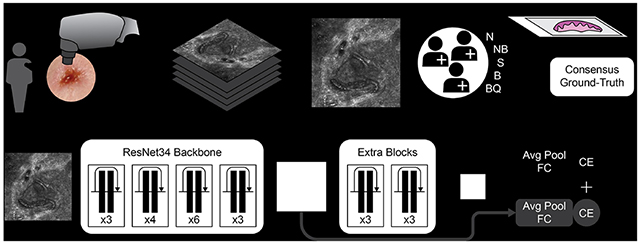
Figure 1. Experimental workflow.
a) Image stacks generation by RCM devices and biopsy validated consensus ground-truth generation by a panel of expert confocalists. b) CNN model used in this study. A ResNet34 pretrained backbone was used along with two extra layers of residual blocks to increase the receptive field of the last feature map. The numbers indicate the number of residual blocks in each of the layers. In addition to the final classifier, during training, a classification loss was backpropagated also from intermediate activation maps. Abbreviations: Avg Pool: average pooling; FC: fully connected layer; CE: Cross-entropy loss. Ground-truth abbreviations: N: normal skin; NB: not BCC; S: suspicious; B: BCC; BQ: bad quality image. Scale bar: a) 250 μm.
A convolutional neural network (CNN) (Figure 1b) was trained to classify each image within a stack to match the ground- truth by RCM experts for BCC detection. The image predictions were aggregated into stack predictions via max-pooling. For the lesion-based analysis, stack predictions were aggregated via average-pooling. Training was performed using a five-fold cross-validation strategy. The final cross-validation performance is obtained by concatenating the results from the validation split of each training fold. To test the generalization performance, an ensemble of models was trained using the full cohort. The trained ensemble was then applied to an external test dataset collected from international collaborators in Italy and Australia consisting of 53 stacks from 34 lesions.
RESULTS
Patients and Tumors Characteristics
Mean age was 60.8 years (SD 16.6; range 25 – 87 years); 47.1% (24/51) were males. In all, 25/66 (37.9%) of lesions were located on the head and neck. A total of 41/66 (62.1%) lesions had a diagnosis of BCC on final histopathology. Demographics of the patients and lesion diagnosis and anatomic location are available in Table 1.
Table 1.
Demographics and clinical characteristics of the sample
| Variable | Total (66 lesions) |
|---|---|
| Age, mean (SD), y * | 60.1 (16.6) |
| Sex, n(%) * | |
| Male | 24 (47.1%) |
| Female | 27 (52.9%) |
|
| |
| Diagnosis, n(%) | |
| BCC | 41 (62.1%) |
| Nodular | 21 (51.2%) |
| Superficial | 13 (31.7%) |
| Infiltrative | 3 (7.3%) |
| Micronodular | 2 (4.9%) |
| NOS | 2 (4.9%) |
| LPLK | 6 (9.1%) |
| Actinic keratosis | 4 (6.1%) |
| Fibrous papule | 3 (4.5%) |
| IDN | 3 (4.5%) |
| Desmoplastic trichoepithelioma | 1 (1.5%) |
| Clear cell acanthoma | 1 (1.5%) |
| Folliculitis | 1 (1.5%) |
| Foreign body granuloma | 1 (1.5%) |
| Melanoma | 1 (1.5%) |
| Sebaceous carcinoma | 1 (1.5%) |
| Seborrheic keratosis | 1 (1.5%) |
| Dysplastic nevus | 1 (1.5%) |
| Epidermolytic acanthoma | 1 (1.5%) |
|
| |
| Location, n(%) | |
| Head and neck | 25 (37.9%) |
| Nose | 12 (18.2%) |
| Cheek | 5 (7.6%) |
| Jaw | 2 (3.0%) |
| Other | 6 (9.1%) |
| Upper extremities | 18 (27.3%) |
| Back | 9 (13.6%) |
| Arm | 5 (7.6%) |
| Forearm | 4 (6.1%) |
| Trunk | 15 (22.7%) |
| Chest | 8 (12.1%) |
| Shoulder | 4 (6.1%) |
| Abdomen | 3 (4.5%) |
| Lower extremities | 3 (4.5%) |
| Leg | 3 (4.5%) |
| Other trunk and extremities | 5 (7.6%) |
Abbreviations: SD = standard deviation; BCC = basal cell carcinoma; LPLK = lichen planus-like keratosis; IDN = intradermal nevus; NOS = not otherwise specified.
Data calculated based on total number of patients not lesions.
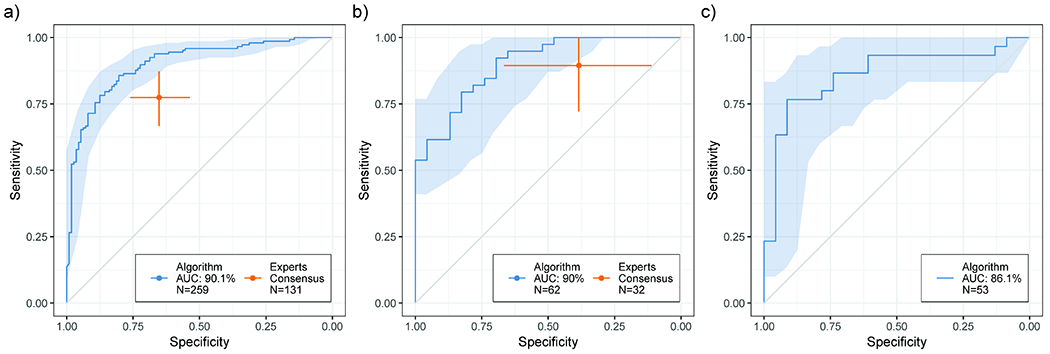
The aggregated results over the five-fold of cross-validation were used to generate a ROC curve of the model performance. The model achieved an area under the curve of 90.1% on the entire dataset of 259 stacks. To compare the performance with the panel of experts, we restricted the result to 131 stacks for which a blinded reading was performed. The AUC achieved in this subset of the data was 89.7%. The blinded consensus of experts achieved a sensitivity of 77.4% (95% CI: 67.3% - 87.3%) and specificity of 65.2% (95% CI: 53.7% - 76.6%). The confidence interval analysis for the model ROC and the experts performance shows some overlap between the two, suggesting that the proposed model is at least comparable to the performance of the human experts (Figure 2a). Analyzing the model’s performance at the lesion level, the AUC for the full validation cohort of 62 lesions was 90.0%. In comparison, the experts performed with 89.5% sensitivity (95% CI: 73.6% - 100%) and 38.5% specificity (95% CI: 12.5% - 64.3%) on 32 of the lesions. On that same subset, the algorithm achieved and AUC of 88.3% (Figure 2b).
Figure 2. Performance of proposed method and comparison with a consensus panel of human experts.
95% CI calculated via bootstrapping. a) Stack-level performance on the MSKCC dataset. The algorithm (N=259 stacks), reported as a ROC curve, achieved an AUC of 90.1. The experts (N=131 stacks) achieved a sensitivity of 77.4%, (95% CI: 67.3% - 87.3%) and a specificity of 65.2% (95% CI: 53.7% - 76.6%). b) Lesion-level performance on the MSKCC dataset. The algorithm (N=62 lesions) achieved an AUC of 90.0%. The experts (N=32 lesions) obtained a sensitivity of 89.5% (95% CI: 73.6% - 100%) and a specificity of 38.5% (95% CI: 12.5% - 64.3%). c) Generalization performance on an external dataset. N=53 stacks. The proposed algorithm achieved an AUC of 86.1%.
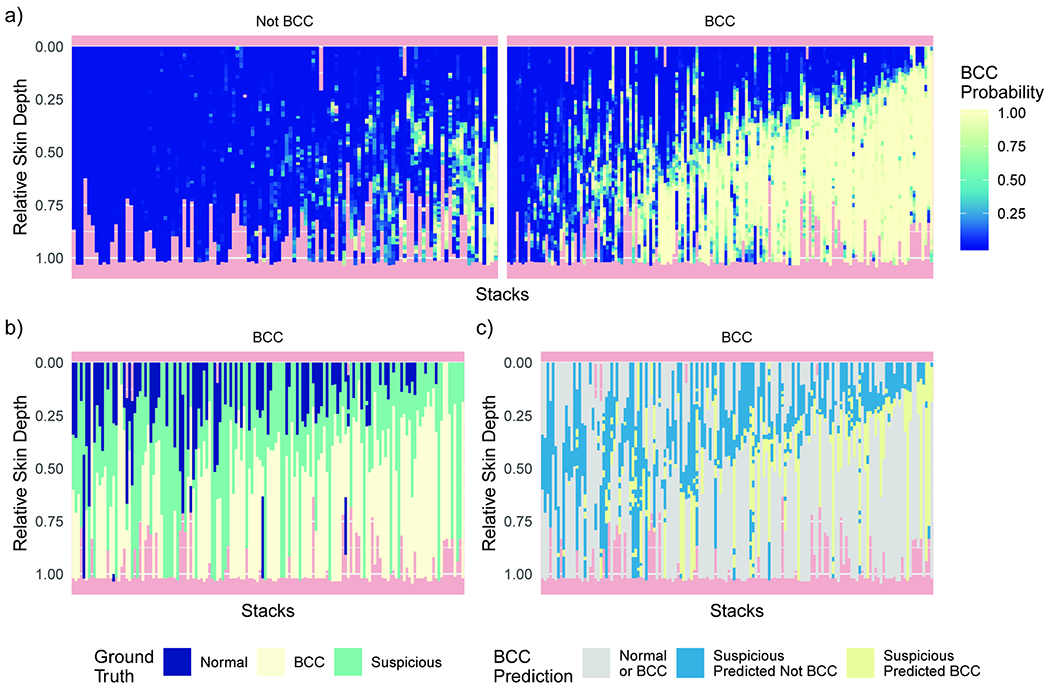
We analyzed the predictions also at the level of singular RCM images (Figure 3). Without explicitly training the model with depth information, the general relationship between depth and BCC occurrence was learned. We can observe how the positive predictions of BCC tend to be present together in deeper sections of the skin (Figure 3a). This is in good agreement with the true image status as annotated by a consensus of RCM experts (Figure 3b). A large number of images were found to be suspicious by the experts, especially at the interface between surely not BCC and surely BCC images. Focusing the analysis on the suspicious images, we found that our model usually assigned high probability of BCC in deeper levels of the stack (Figure 3c).
Figure 3. RCM image level validation predictions for all stacks and their relation to skin depth.
a) Predicted probability of each image given the proposed model for biopsy confirmed “Not BCC” and “BCC” stacks. Strong support for BCC tends to be found in deeper skin levels. b, c) Analysis performed on “BCC” stacks. The order is preserved from panel a. b) Ground-truth annotations from the consensus of RCM experts. Of note is the number of suspicious images usually at the interface between normal and BCC images. c) Analysis of the proposed model prediction on the suspicious images. Normal and BCC images are in gray. The model favors BCC predictions on images at deeper skin lesions.
We further validated the proposed method on an external test dataset from three institutions in Italy and Australia. This external set of stacks provides a better estimate of real-world performance and generalization in clinical settings given the inclusion of various types of technical variability. The proposed method achieved and AUC of 86.1%, only a modest drop in performance compared to the cross-validation result on a single center dataset.
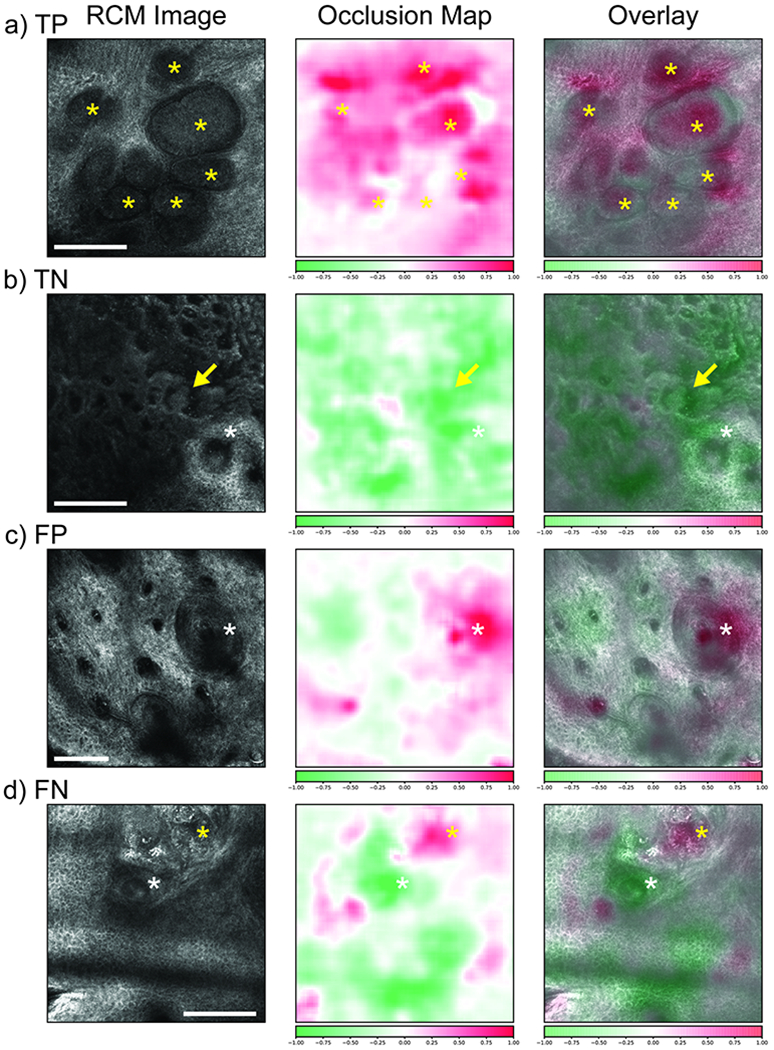
Finally, we developed occlusion maps [Zeiler & Fergus 2014] to gain insight into what features of an image are important for classification and to aid interpretation of the CNN results. In this technique, regions of an image are occluded and the model’s change in prediction is measured. If the occlusion of a region yields a large drop in probability of a certain label (e.g. BCC) then that region is of high importance for that label. In Figure 4, we show representative RCM images with true positive (TP, Panel A), true negative (TN, Panel B), false positive (FP, Panel C), and false negative (FN, Panel D) model prediction for BCC along with their corresponding occlusion maps. RCM images with TP model prediction showed an excellent correspondence of the high positive (BCC) attribution areas on occlusion map to the tumor nodules. Similarly, RCM images with TN model prediction showed an excellent correspondence to the high negative (not-BCC) attribution areas on occlusion map to the benign structures such as elongated cords and bulbous projection of a lichen planus-like keratosis (LPLK). Interestingly, in RCM image from an actinic keratosis lesion, occlusion map showed high positive attribution areas corresponding to hair follicles (Panel C). Hair follicles often mimics BCC tumor nodule to a novice RCM reader, causing false positive results. On the other hand, we noticed that in RCM image from a nodular BCC that had a FN model prediction (Panel D), occlusion map showed high positive attribution areas in a small foci of BCC tumor nodules, despite a FN model prediction.
Figure 4. Occlusion maps.
Left: RCM image; center: occlusion map; right: overlay. A) True positive example of a nodular BCC showing tumor nodules (yellow *). High positive attribution over the nodules. B) True negative example of an LPLK with elongated cords and bulbous projections (arrows), and hair follicle (white *). High negative attribution over these benign structures. C) False positive example of an AK showing hair follicles (white *). High positive attribution over a hair follicle. D) False negative example of a nodular BCC with a small tumor foci (yellow *) and a hair follicle (white *). High positive attribution over the BCC foci and high negative attribution over the hair follicle. Color scalebar: Red color, high attribution for BCC; Green color, low attribution for BCC. Scale bars: a, b, d) 250 μm; c) 125 μm.
DISCUSSION
In this study, we evaluated the efficacy of a deep learning model in the evaluation of cutaneous lesions imaged by RCM stacks for the diagnosis of BCC in clinically equivocal lesions. The proposed method achieved almost comparable AUCs of 89.7% at the stack level and 88.3% at the lesion level in the internal dataset. A similar AUC of 86.1% was achieved in the external datasets, demonstrating the generalizability of the system’s performance. The performance was also compared to that of expert RCM readers and was at least on par with the human experts.
In our study, contrary to the previous studies where mosaics (large field-of-view images ~10mm2 acquired by stitching together individual small FOV images at a given depth) were used to develop AI algorithms [Wodzinski et al. 2019, Bozkurt et al. 2018, Kose et al. 2021a, Kose et al 2021b, Kose et al. 2020], only RCM stacks were included. The use of mosaics could be a major limitation in some cases since these images can be acquired with an arm-mounted device only. For example, on the facial sites where the majority of BCC and their mimickers occur [Castro et al. 2015], attaching the arm-mounted device to the lesion is often difficult due to its size and the curvature of the imaging surface. Conversely, hand-held RCM (HH-RCM) devices are more practical for evaluating facial lesions and have shown high sensitivity and specificity for diagnosing BCC which further strengthens their utility [Castro et al. 2015, Que et al. 2016]. Furthermore, an AI algorithm developed on RCM stacks will have a wider applicability as it can be applied not only to the HH-RCM device but also to stacks obtained using arm-mounted devices. We believe that a combination of prediction maps such as occlusion map presented in this study along with the model prediction could aid a dermatologist in real-time bedside diagnosis of BCC and help to explain CNN prediction. Indeed, these maps could increase the interpretability of CNN considered as “black box” regarding RCM images. They could also be used to train novices RCM skills and can increase the use of CNN in their clinical setting.
The major limitation of this study was the relatively small number of lesions included. In the next steps we will validate our preliminary results on a larger cohort of lesions and compare with human reading at various expertise level. In addition, sampling errors within the lesion due to the limited field of view provided by stacks could have led to the acquisition of sub-optimal RCM images. In the future, we envision combining stack acquisition guided by real-time dermoscopy using an integrated camera within the hand-held device to overcome this limitation [Dickensheets et al. 2016].
These results provide a clear indication that deep learning-powered decision support systems can be trained to detect BCC on RCM images with accuracy at least on par with a consensus of experts. These decision support systems could eventually be deployed clinically as guidance tools that will optimize the evaluation and diagnosis of various types of skin cancer.
MATERIAL AND METHODS
This was an IRB (#17-078) approved, retrospective study performed at a tertiary cancer center.
RCM Stacks Acquisition
Stacks of RCM images with 0.5-1μm resolution covering a field-of-view of 0.5-1 mm2 were collected at consecutive depths (3.0 μm apart) starting from stratum corneum up to superficial dermis (i.e. 200 μm depth). The imaging was done using either the arm-mounted RCM (Vivascope 1500) or handheld RCM (HH-RCM; Vivascope 3000, Caliber I.D., Rochester, NY), depending on the accessibility of the lesion. RCM imaging was conducted within the lesion area. The lesion area was delineated using a paper ring during acquisition, a standard operating procedure when using a HH-RCM device. For each lesion, we included all stacks that were available. The number of stacks per lesion was determined by the RCM experts at the time of imaging. We collected an average of 6.1 (SD 2.7; range 1–12) RCM stacks per lesion. Although RCM stacks were obtained from the lesional area, we had few stacks which showed background normal skin and were excluded after reviewing them for consensus (i.e. images probably obtained outside the lesional area).
Data Annotation
A total of 66 unique lesions from 52 patients comprising a final dataset of 312 stacks were included in this study. Each RCM image was labeled with one of the 5 labels: BCC (B), not-BCC (NB), suspicious for BCC (S), normal (N), and bad quality (BQ). A panel of five expert RCM readers (C.N-D., K.L., S.A., J.M., and M.J.) analyzed all the RCM images within each RCM stack and voted for these images as B, NB, S, and BQ. A final label was rendered when at least 3/5 experts agreed on a given label. In case of lack agreement, a sixth reviewer (A.S.), with more than 15 years of experience, was consulted and his verdict was used as the final label.
The annotation was conducted in two phases. The first phase was the “standardization phase” where a subset of stacks (n=153 stacks; 7,578 individual images) was analyzed by the readers to homogenize the labeling criteria. During this phase the ground-truth histopathology was made available to the readers during labeling (not blinded). In the second phase (n= 159 stacks; 8,395 images), the readers labeled individual images (using the standardized criteria from the first phase) in a blinded manner (without knowing the histopathology diagnosis). For this second phase, the same voting decision rules, as detailed above, were used to determine the final clinician label. This phase was primarily done to assess the performance of human reading and was later used to compare against the AI performance (Figure 2). To generate the ground-truth for the AI analysis, the stacks used in the second phase were cross-checked with the final diagnosis on histopathology for that lesions and then re-labeled (B, NB, S, N, BQ) according to the ground-truth. In addition, stacks containing other malignant skin diseases (e.g. melanoma) were removed. The final dataset analyzed consisted of 267 stacks, 131 of which were used during the blinded consensus read from human experts.
External Dataset
An external dataset was obtained from equivocal lesions for BCC to assess the generalization of the proposed methods. A total of 53 stacks from 34 lesions were collected from three international institutions in Italy and Australia: Università degli Studi di Modena e Reggio Emilia, Modena, Italy (13 lesions and 25 stacks); University of Modena and Reggio Emilia, Reggio Emilia, Italy (16 lesions and 22 stacks); Sydney Melanoma Diagnostic Centre, Australia (5 lesions and 6 stacks). The external dataset was labeled at the stack level with histopathology confirmation. The diversity of sources ensures a high degree of technical variability in terms of acquisition protocols.
Neural Network Architecture
The CNN model used is based on a ResNet34 [He et al. 2015] and was adapted to analyze gray-scale images of size 1000px. This model provides a 32-by-32px wide feature representation after its final convolutional layer. To increase the receptive field, two additional residual blocks were added, obtaining a feature representation of size 8-by-8px. The 32px and the 8px feature representations were connected to independent average pooling and fully connected classification layers that classifies each RCM image into BCC (positive) vs not-BCC (negative) labels. The final loss is the sum of cross-entropy losses from each classification layer (Figure 1b). At test time only the last classification layer (8px) was used for prediction.
Training and Testing
Training was performed following a five-fold cross-validation strategy. The internal dataset was split into five folds at the stack level so that all images from a particular stack only appear in one of the folds. Bad quality (BQ) images were removed. Suspicious (S) images were removed from training but were kept at inference time. NB and normal skin images were considered negative while BCC images were considered positive. In 8 stacks, the consensus ground-truth did not match the biopsy ground-truth. These stacks were used for training but were removed for validation (259 total stacks). For each fold, a predictive model was trained to correctly classify each image in a stack. At validation, the image level predictions were aggregated into a stack level prediction by max-pooling. Each network was trained for 40 epochs with mini-batch stochastic gradient descent with an initial learning rate of 0.001 which was annealed every 20 epochs by a factor of 10. During training, an augmentation pipeline consisting of random 90-degree rotations, random horizontal flips, intensity jittering, and gaussian blur, was performed on the fly.
Given the above training parameters chosen via cross-validation, five models were trained on the full training cohort. Stack predictions on the external test dataset were obtained via max-pooling as already described. Stack outputs were averaged to obtain the final lesion level test prediction.
Statistical analysis
All statistical analyses were performed in R [R Core Team 2019] version 3.5.1. The positive class probability inferred by the model was used to generate ROC curves using the package “pROC”[Robin et al. 2011] (version 1.15.0). 95% confidence intervals for the ROC curves and the experts’ sensitivity and specificity measures were generated using the package “boot”[Canty et al. 2020] (version 1.3.20). ROC plots were generated with package “ggplot2”[Wickham et al. 2009] (version 3.1.0).
Occlusion maps
Occlusion is a perturbation-based technique for visualizing importance across an image [Zeiler & Fergus 2014]. The attribution is the normalized change in probability for a specific class when occluding a square patch. Occlusion maps were generated using python package “captum” [Kokhlikyan et al. 2020].
ACKNOWLEDGEMENTS
The authors are grateful for the generous computational support given by the Warren Alpert foundation, and the project management support of Christina M. Virgo. We are also grateful to Dr. Angela Lobato Williams and Ms. Marica Mirra who helped with data collection. We would like to thank Ms. Ucalene Harris and Dr. Cristian Fischer for aiding in data management and data transfer. This research was funded in part through the NIH/NCI Cancer Center Support Grant [grant number P30 CA008748] and NM grants R01CA199673, R01CA240771.
FUNDING SOURCE:
National Institutes of Health grant, R01CA199673, R01CA240771 from NCI and MSKCC’s Cancer Center core support National Institutes of Health grant P30CA008748 from NCI.
Abbreviations:
- BCC
Basal Cell carcinoma
- RCM
Reflectance Confocal Microscopy
- AI
Artificial Intelligence
- ROC
Receiver Operator Characteristic
- AUC
Area Under The Curve
- CI
Confidence Interval
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFILCT OF INTERESTS
Dr. Thomas J. Fuchs is a founder, equity owner, and Chief Scientific Officer of Paige.
DATA AVAILABILITY
“Datasets related to this article can be found at, http://dx.doi.org/10.17632/csvwf6mvr9.1, http://dx.doi.org/10.17632/n9y9x42bk2.1, and https://github.com/MSKCC-Computational-Pathology/MSKCC_RCM_BCC and hosted at public repositories of Mendeley and Github. Citations: Campanella, Gabriele et al (2021), “MSKCC RCM BCC Part 2”, Mendeley Data, V1, doi: 10.17632/n9y9x42bk2.1; Campanella, Gabriele et al. (2021), “MSKCC RCM BCC Part 1”, Mendeley Data, V1, doi: 10.17632/csvwf6mvr9.1
REFERENCES
- Bozkurt A, Kose K, Alessi-Fox C, Gill M, Brooks DH, Dy J et al. A Multiresolution Convolutional Neural Network with Partial Label Training for Annotating Reflectance Confocal Microscopy Images of Skin. In Frangi AF, Schnabel JA, Davatzikos C, Alberola-López C & G. Fichtinger (eds.) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018, vol. 11071, 292–299, DOI: 10.1007/978-3-030-00934-2_33 (Springer International Publishing, Cham, 2018). Series Title: Lecture Notes in Computer Science. [DOI] [Google Scholar]
- Campanella G, Hanna MG, Miraflor A, Silva VWK, Busam KJ, Brogi E et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Medicine 25, 1301–1309, DOI: 10.1038/s41591-019-0508-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canty A & Ripley BD. boot: Bootstrap R (S-Plns) Functions (2020).
- Castro RP, Stephens A, Fraga-Braghiroli NA, Oliviero MC, Rezze GG, Rabinovitz H et al. Accuracy of in vivo confocal microscopy for diagnosis of basal cell carcinoma: a comparative study between handheld and wide-probe confocal imaging. J. Eur. Acad. Dermatol. Venereol 29, 1164–1169, DOI: 10.1111/jdv.12780 (2015). [DOI] [PubMed] [Google Scholar]
- Dickensheets DL, Kreitinger S, Peterson G, Heger M & Rajadhyaksha M. Dermoscopy-guided reflectance confocal microscopy of skin using high-NA objective lens with integrated wide-field color camera. 96890U, DOI: 10.1117/12.2213332 (2016). [DOI] [PMC free article] [PubMed]
- Esteva A, Kuprel B, Novoa RA, KO J, Swetter SM, Blau HM et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118, DOI: 10.1038/nature21056 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S & Sun J. Deep Residual Learning for Image Recognition. arXiv: 1512.03385 [cs] (2015). ArXiv: 1512.03385. J. Am. Acad. Dermatol 80, 1380–1388, DOI: 10.1016/j.jaad.2018.12.026 (2019). [DOI] [Google Scholar]
- Jain M, Pulijal SV, Rajadhyaksha M, Halpern AC & Gonzalez S. Evaluation of Bedside Diagnostic Accuracy, Learning Curve, and Challenges for a Novice Reflectance Confocal Microscopy Reader for Skin Cancer Detection In Vivo. JAMA Dermatol. 154, 962, DOI: 10.1001/jamadermatol.2018.1668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadouch D, Schram ME, Leeflang MM, Limpens J, Spuls PI, de Rie MA et al. In vivo confocal microscopy of basal cell carcinoma: a systematic review of diagnostic accuracy. J. Eur. Acad. Dermatol. Venereol 29, 1890–1897, DOI: 10.1111/jdv.13224 (2015). [DOI] [PubMed] [Google Scholar]
- Kokhlikyan N, Miglani V, Martin M, Wang E, Alsallakh B, Reynolds J, Melnikov A, Kliushkina N, Araya C, Yan S, & Reblitz-Richardson O. (2020). Captum: A unified and generic model interpretability library for PyTorch.
- Kose Kivanc, Bozkurt Alican, Christi Alessi-Fox Melissa Gill, Longo Caterina, Pellacani Giovanni, Dy Jennifer G., Brooks Dana H., Rajadhyaksha Milind, Segmentation of cellular patterns in confocal images of melanocytic lesions in vivo via a multiscale encoder-decoder network (MED-Net), Medical Image Analysis, Volume 67, pp. 101841, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alonzo M, Bozkurt A, Alessi-Fox C et al. Semantic segmentation of reflectance confocal microscopy mosaics of pigmented lesions using weak labels. Sci Rep 11, 3679 (2021). 10.1038/s41598-021-82969-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kose K, Bozkurt A, Alessi-Fox C, Brooks DH, Dy J, Rajadhyaksha M. et al. Utilizing Machine Learning for Image Quality Assessment for Reflectance Confocal Microscopy. J. Investig. Dermatol 140, 1214–1222, DOI: 10.1016/j.jid.2019.10.018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete-Dechent C, Bajaj S, Marchetti MA, Rabinovitz H, Dusza SW, Marghoob A. Association of Shiny White Blotches and Strands With Nonpigmented Basal Cell Carcinoma: Evaluation of an Additional Dermoscopic Diagnostic Criterion. JAMA Dermatol. 152, 546, DOI: 10.1001/jamadermatol.2015.5731 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que SKT, Grant-Kels JM, Rabinovitz HS, Oliviero M & Scope A. Application of Handheld Confocal Microscopy for Skin Cancer Diagnosis. Dermatol. Clin 34, 469–475, DOI: 10.1016/j.det.2016.05.009 (2016). [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2019). [Google Scholar]
- Rajadhyaksha M, Marghoob A, Rossi A, Halpern AC & Nehal KS. Reflectance confocal microscopy of skin in vivo: From bench to bedside: REFLECTANCE CONFOCAL MICROSCOPY OF SKIN IN VIVO. Lasers Surg. Medicine 49, 7–19, DOI: 10.1002/lsm.22600 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter O, Mimouni I, Gdalevich M, Marghoob A, Levi A, Hodak E et al. The diagnostic accuracy of dermoscopy for basal cell carcinoma: A systematic review and meta-analysis. J Am Acad Dermatol. 2019. May;80(5):1380–1388. doi: 10.1016/j.jaad.2018.12.026. Epub 2018 Dec 21. [DOI] [PubMed] [Google Scholar]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinforma. 12, DOI: 10.1186/1471-2105-12-77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HW, Weinstock MA, Feldman SR & Coldiron BM. Incidence Estimate of Nonmelanoma Skin Cancer (Keratinocyte Carcinomas) in the US Population, 2012. JAMA Dermatol. 151, 1081, DOI: 10.1001/jamadermatol.2015.1187 (2015). [DOI] [PubMed] [Google Scholar]
- Tschandl P, Codella N, Akay BN, Argenziano G, Braun RP, Cabo H et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: an open, web-based, international, diagnostic study. The Lancet Oncol. 20, 938–947, DOI: 10.1016/S1470-2045(19)30333-X (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H ggplot2 (Springer New York, New York, NY, 2009). [Google Scholar]
- Wodzinski M, Skalski A, Witkowski A, Pellacani G & Ludzik J. Convolutional Neural Network Approach to Classify Skin Lesions Using Reflectance Confocal Microscopy. 4754–4757, DOI: 10.1109/EMBC.2019.8856731 (IEEE, 2019). [DOI] [PubMed] [Google Scholar]
- Zeiler Matthew D., and Fergus Rob. “Visualizing and understanding convolutional networks”. European conference on computer vision. Springer, Cham, 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
“Datasets related to this article can be found at, http://dx.doi.org/10.17632/csvwf6mvr9.1, http://dx.doi.org/10.17632/n9y9x42bk2.1, and https://github.com/MSKCC-Computational-Pathology/MSKCC_RCM_BCC and hosted at public repositories of Mendeley and Github. Citations: Campanella, Gabriele et al (2021), “MSKCC RCM BCC Part 2”, Mendeley Data, V1, doi: 10.17632/n9y9x42bk2.1; Campanella, Gabriele et al. (2021), “MSKCC RCM BCC Part 1”, Mendeley Data, V1, doi: 10.17632/csvwf6mvr9.1


