Abstract
In recent years, breast cancer attracts more and more attention because of its high incidence. To explore the molecular functions and mechanisms, we performed RNA sequencing on the tumor tissues and their paired normal tissues from three breast cancer patients. By differential expression analysis, we found 3764 differentially expressed (DE) mRNAs, 5416 DE lncRNAs, and 148 DE circRNAs. Enrichment analysis suggested that the DE lncRNAs and DE circRNAs were enriched in mitochondria and nucleus, which indicated that they may participate in the vital metabolism directly or indirectly, such as fatty acid metabolism. Subsequently, the protein–protein interaction (PPI) network was constructed and we got 8 key proteins, of which the matrix metalloproteinase-9 (MMP9; degree 5) draws our attention. Based on the 38 up-regulated circRNAs and 14 down-regulated circRNAs, we constructed competing endogenous RNA (ceRNA) networks, from which the has-miR-6794-5p has been identified to enriched in the up-regulated network and correlated with the circNFIX directly. At this point, we presented that the circNFIX and MMP9 may play a significant role by regulating fatty acid metabolism in breast cancer.
Keywords: breast cancer, ceRNA, fatty acid metabolism, molecular mechanism, RNA sequencing
Introduction
Breast cancer, the most threatening malignancy to female worldwide, caused about 2.3 million new cases according to the statistics in 2020 [1]. For the early-stage breast cancer, surgery remains preferred for local–regional therapy. As for systemic therapy, adjuvant endocrine therapy and adjuvant chemotherapy are advised for nearly all patients with invasive breast cancer [2]. Although these adjuvant treatments bring patients survival benefit, recurrence, metastases and treatment resistance are still unavoidable in some patients. Focusing on these problems, increased understanding of the molecular mechanisms in breast cancer is required urgently.
High-throughput next-generation sequencing (NGS) technologies, which allow sequencing millions of DNA or RNA sequence simultaneously, have a wide range of applications in diagnosis, treatments, and prognosis prediction for oncology [3–5]. Competing endogenous RNAs (ceRNAs) are RNAs that can compete for microRNAs binding and form regulatory networks across the transcriptome, thereby regulating varieties of pathological processes via distinct molecular mechanisms [6–8]. Recently, Wang et al. [9] reported that circRNA Hsa_circ_0005273 served as a sponging for miR-200-3p to facilitate breast cancer progression by regulating YAP1-hippo signaling pathway. Furthermore, Yu et al. [10] reviewed that circRNAs act as ceRNAs participate in lipid metabolism. The reported study not only indicated that the ceRNA cross-talk will affect the molecular functions and induce cancer progression but also represented a potential regulating mechanism in cancer. Therefore, RNA sequencing, which accurately reveals the gene expression profile, can be applied to investigate the molecular functions and mechanisms in breast cancer.
In our study, RNA sequencing was performed based on three tumor tissues (C) and their paired normal tissues (CP) from three breast cancer patients with different molecular subtypes. Subsequently, differentially expressed (DE) mRNAs, lncRNAs, and circRNAs were examined on the C and CP groups, followed by functional enrichment analysis, which were investigated through Gene Ontology (GO) categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. The present study aims to explore the potential molecules for breast cancer diagnosis and treatment.
Materials and methods
Patients and tissue collection
The tissue samples were collected from three female breast cancer patients who underwent mastectomy at the First Affiliated Hospital of Jinan University, Guangzhou, between 2019 and 2020 and have no preoperative chemotherapy or radiation. All experimental procedures were approved by the Ethics Committee of the First Affiliated Hospital of Jinan University. The patients included in the present study signed inform consents. The three paired normal tissues used as control were obtained 5 cm away from the primary tumor sites. All patients were free of any other form of cancer and all their specimens were examined by pathologists before preservation in liquid nitrogen. Detailed patients’ information is shown in Supplementary Table 1.
RNA extraction and quality control
Total RNAs were extracted from frozen tumor tissues and their paired normal tissues by TRIzol following the manufacturer's protocol. The preliminary RNA concentration and purity were evaluated with the NanoDrop 2000 Spectrophotometer (Thermo Scientific) and the precise quantification were performed by Qubit (ABI). Purified RNA was stored at −80°C.
Library construction and sequencing
The RNA-seq library was generated using VAHTS Universal V8 RNA-seq Library Prep Kit for Illumina according to the manufacturer’s protocol. The quality of each library was controlled using an Agilent 2100 TapeStation (Agilent). The libraries were pooled and sequenced on an Illumina NovaSeq 6000 platform following to the manufacturer’s protocol and finally 150bp paired-end reads were generated.
Quality control and alignment of RNA-sequencing reads
The raw RNA-sequencing reads were filtered to remove r-RNA using fastp to get the clean reads. Q20, Q30, and GC contents of the clean reads were calculated to evaluate the quality of sequencing. The clean reads were mapped to the reference genome using Hisat2. To defined the novel RNAs with or without coding potential, the transcript of each sample was assembled from mapped reads by StringTie. To estimate the expression levels of mRNAs and lncRNAs, the FPKM (Fragments Per Kilobases per Million reads) and TPM (Transcripts Per Million reads) were calculated through RSEM (http://deweylab.github.io/RSEM/) (Supplementary Table 2).
Identification of DE transcripts
DE analysis was performed on C and CP using DESeq2. A P-value<0.05 and |log2 foldchange (FC)| >1 were considered as the thresholds for identifying significantly DE mRNAs, DE lncRNAs, and DE circRNAs. The volcano plots and heatmaps were drawn on DE mRNAs, DE lncRNAs, and DE circRNAs using the R package.
Analysis of target mRNAs regulated by DE lncRNAs
DE lncRNAs-targeted mRNAs were predicted using the StarBase (http://starbase.sysu.edu.cn/) database. The lncRNAs–mRNAs interaction networks were constructed between DE lncRNAs and their targeted mRNAs by igraph package in R.
Analysis of functional enrichment pathways
Analysis of the GO and KEGG enrichment pathways of DE mRNAs, DE lncRNAs, and DE circRNAs were performed by clusterProfiler package in R. The GO terms and KEGG pathways with the enriched gene count ≥ 2 and the significance threshold P≤0.05 were considered significant.
Protein–protein interaction network construction
The STRING database (version 11.5) was used to predict the potential interactions among proteins translated by top 300 DE mRNAs. The threshold was selected as score ≥ 0.7. The protein–protein interaction (PPI) network was built by the STRING (https://string-db.org/).
CircRNAs–miRNAs–mRNAs network construction
circRNA-–miRNA–mRNA interactions were predicted by miRanda (version 3.3a). The up-regulated and down-regulated circRNAs/mRNAs were selected to construct a ceRNA network by igraph package in R.
Results
Differential expression analysis
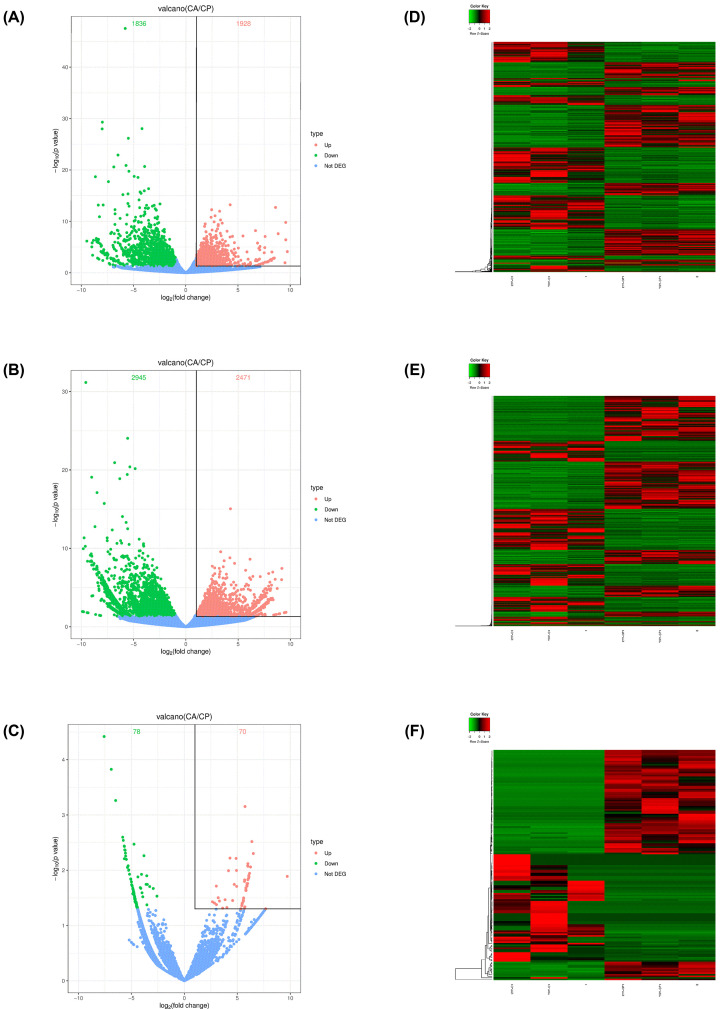
The volcano plots showed the DE genes were delimited clearly in the C and CP groups (Figure 1A–C). According to the screening criteria, a total of 3764 DE mRNAs were determined, of which 1928 were up-regulated and 1836 were down-regulated. DE lncRNAs analysis indicated 5416 deregulated lncRNAs, among those, significantly up-regulated and down-regulated RNAs were 2471 and 2945, respectively. For circRNAs, out of 148 dysregulated, 70 up-regulated and 78 down-regulated were observed. The hierarchical clustering analysis of DE mRNAs, DE lncRNAs, and DE circRNAs, which showed the expression profiles between the C and CP groups (Figure 1D–F), further explained the reliability of differential expression analysis.
Figure 1. Analysis of DE mRNAs, lncRNAs, and circRNAs.
(A–C) Volcano plots of DE mRNAs (A), DE lncRNAs (B), and DE circRNAs (C). (D–F) Heatmaps of DE mRNAs (D), DE lncRNAs (E), and DE circRNAs (F). Red indicates up-regulation and green indicates down-regulation.
GO and KEGG enrichment analysis of DE mRNAs
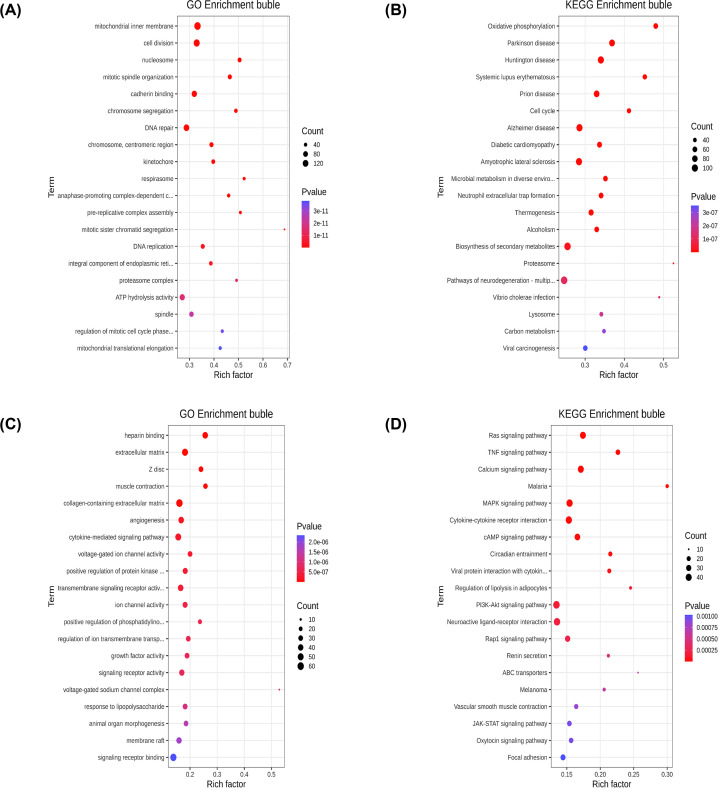
To predict the potential biological function of DE mRNAs, we performed GO and KEGG enrichment analysis to them. Figure 2 displayed top 20 enriched terms by up-regulated or down-regulated DE mRNAs. From the results, we found that most up-regulated DE mRNAs were related to cell division in biological process and the significantly enriched signaling pathway was biosynthesis of secondary metabolites (Figure 2A,B). For down-regulated DE mRNAs, several genes involved in cytokine-mediated signaling pathway (Figure 2C). Additionally, pathway analysis indicated that down-regulated DE mRNAs were mainly associated with MAPK signaling pathway and cytokine–cytokine receptor interactions (Figure 2D).
Figure 2. The GO and KEGG enrichment analysis of DE mRNAs.
(A) Top 20 GO terms enriched by up-regulated DE mRNAs. (B) Top 20 pathways enriched by up-regulated DE mRNAs. (C) Top 20 GO terms enriched by down-regulated DE mRNAs. (D) Top 20 pathways enriched by down-regulated DE mRNAs.
PPI network
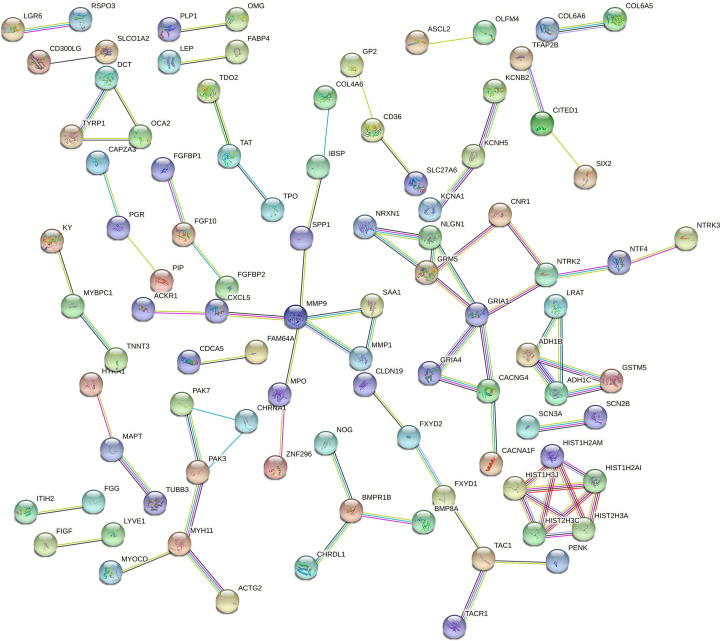
To better investigate the effect of protein interactions in breast cancer, a PPI network, which consisted of 93 nodes and 82 interaction pairs, was constructed using the STRING based on the top 300 DE mRNAs (Figure 3). The nodes with high degree can be regarded as key proteins, including GRIA1 (degree 5), matrix metalloproteinase-9 (MMP9; degree 5), GRM5 (degree 4), HIST1H2AI (degree 4), HIST1H2AM (degree 4), HIST1H3J (degree 4), HIST2H3A (degree 4), and HIST2H3C (degree 4). Since the proteins were involved in several essential signaling pathways, the dysregulation of them may cause the progression of breast cancer (Table 1).
Figure 3. The PPI network in breast cancer using top 300 DE mRNAs.
The nodes in the network represent proteins and the color of edges indicates known or predicted interactions.
Table 1. Top eight key proteins in PPI network.
| DE mRNAs | Express | Degree | Pathways |
|---|---|---|---|
| GRIA1 | Down | 5 | cAMP signaling pathway |
| MMP9 | Up | 5 | Lipid and atherosclerosis Transcriptional misregulation in cancer |
| GRM5 | Up | 4 | Phospholipase D signaling pathway Calcium signaling pathway |
| HIST1H2AI | Up | 4 | Necroptosis Neutrophil extracellular trap formation |
| HIST1H2AM | Up | 4 | Necroptosis Neutrophil extracellular trap formation |
| HIST1H3J | Up | 4 | Transcriptional misregulation in cancer Neutrophil extracellular trap formation |
| HIST2H3A | Up | 4 | Transcriptional misregulation in cancer Neutrophil extracellular trap formation |
| HIST2H3C | Up | 4 | Transcriptional misregulation in cancer Neutrophil extracellular trap formation |
Functional enrichment analysis of lncRNAs and circRNAs
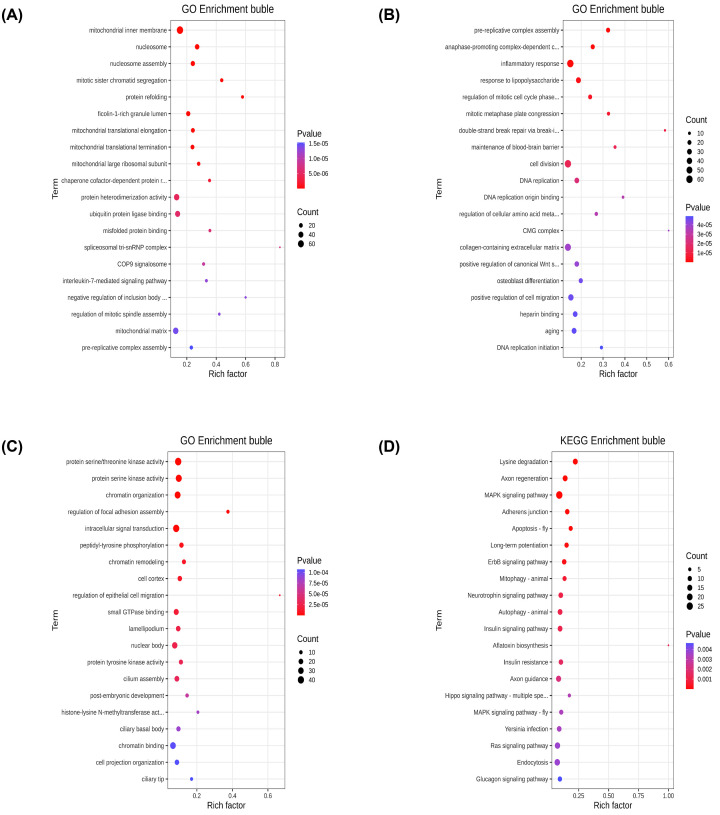
The diverse function of noncoding RNAs achieved by acting on the target genes in cis or trans. Depending on the location of the sites of transcription, cis-acting lncRNAs can activate or repress the expression of neighboring target genes in the linear genome [11,12]. The genes within a 100 kb of upstream or downstream in the DE lncRNAs were regarded as cis-acting lncRNAs, and the bubble diagram showed that the target genes were largely enriched in mitochondrial inner membrane (Figure 4A). Based on the absolute Pearson correlation coefficient over 0.99 and P-values less than 0.01, the possible trans-regulatory relationship was identified. The results suggested that the vast of majority trans-target mRNAs were involved in cell division (Figure 4B).
Figure 4. Bubble maps of enrichment analysis of lncRNAs and circRNAs.
(A) GO analysis of cis-target for DE lncRNAs. (B) GO analysis of trans-target for DE lncRNAs. (C) GO analysis of DE circRNAs. (D) Pathways analysis of DE circRNAs.
In order to explore the function of circRNA, GO and KEGG enrichment analysis were performed on DE circRNAs. As showed in Figure 4C, GO annotations revealed that many biological processes of DE circRNAs were related to intracellular signal transduction and chromatin organizations. KEGG pathway analysis showed that most relevant pathways of DE circRNAs were largely involved in MAPK signaling pathway (Figure 4D).
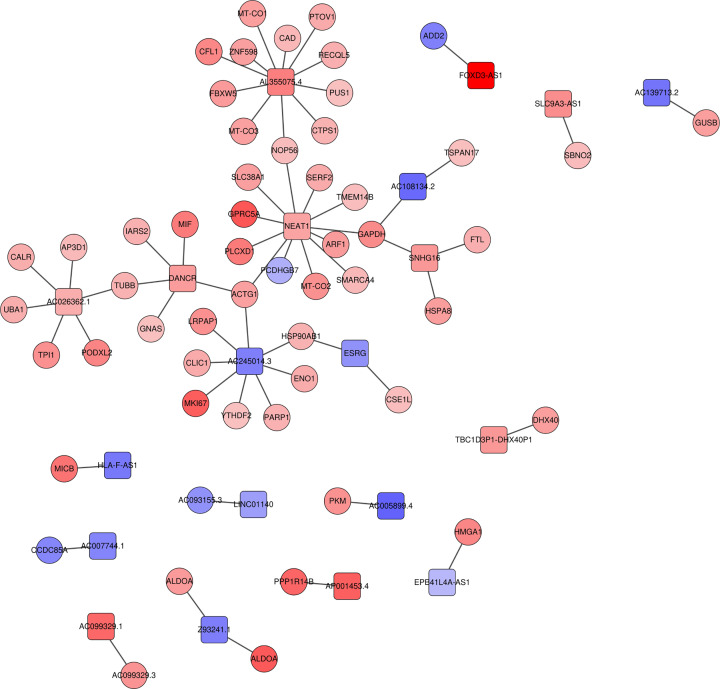
LncRNAs–mRNAs interaction networks
In our study, 63 targeted mRNAs were predicted by DE lncRNAs using StarBase. To analyze the relationships between them, the lncRNAs–mRNAs interaction networks were constructed (Figure 5). Among the relationships, the number of up-lncRNAs were accounted for more than half and the proportion of up-mRNAs exceeded 90%, which suggested that the relationships in the interaction networks were mostly in the state of co-activation.
Figure 5. The lncRNAs–mRNAs interaction networks.
The interaction networks between DE lncRNAs and their targeted mRNAs. Quadrilateral represents the DE lncRNAs, circle represents the mRNAs.
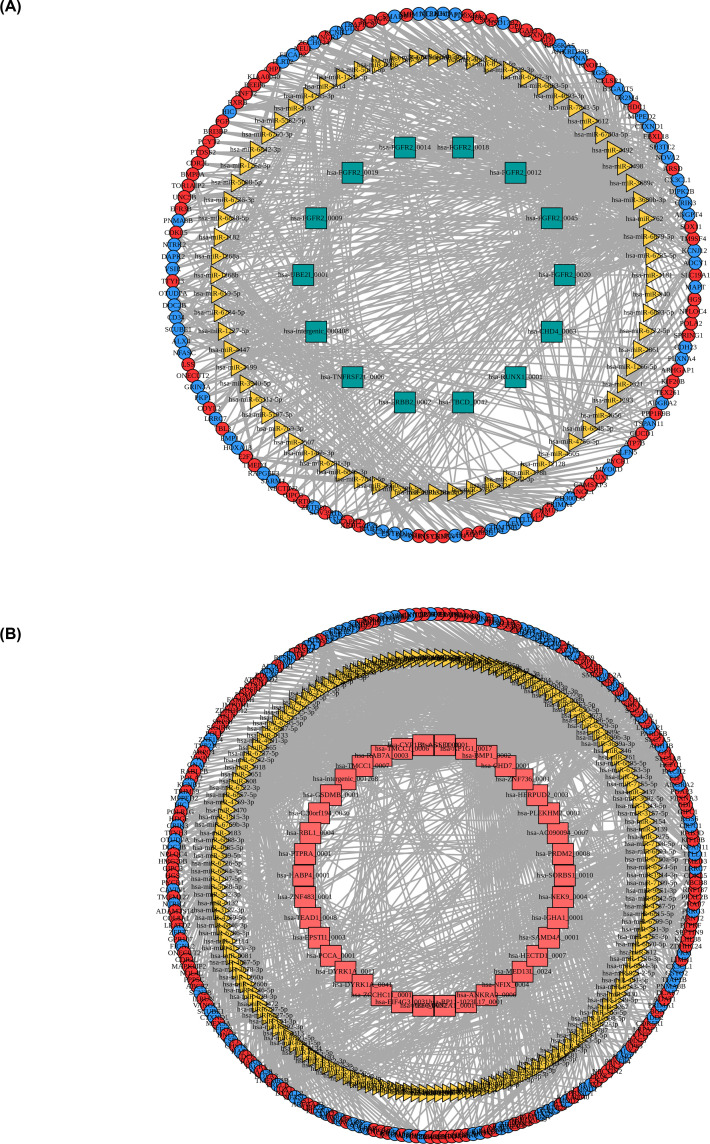
ceRNA networks construction
circRNAs can act as endogenous RNAs of miRNAs to regulate the expression of mRNAs indirectly, further promoting or inhibiting the biological processes, such as proliferation, migration, or chemoresistance of breast cancer [13–16]. Based on the DE circRNAs and DE mRNAs, we predicted the miRNAs targets of DE circRNAs via miRanda and TargetScan, therefore constructing the circRNAs–miRNAs–mRNAs regulatory networks and found that has-miR-6794-5p was significantly enriched in the ceRNA network. A suppressed network is shown in Figure 6A, including 14 down-circRNAs, 72 miRNA, 150 down-mRNAs, and 125 up-mRNAs. Activated interaction pairs were obtained in Figure 6B, of which 38 up-circRNAs, 175 miRNA, 197 down-mRNAs, and 242 up-mRNAs were included.
Figure 6. The circRNAs–miRNAs–mRNAs networks.
The ceRNA networks of down- (A) or up-regulated circRNAs (B). Green quadrilateral represents the down-regulated circRNA, yellow triangle represents the miRNA, red circle represents up-regulated mRNA, blue circle represents down-regulated mRNA, and red quadrilateral indicates the up-regulated circRNA.
Discussion
RNA sequencing, which can be applied to identify the tumor driver genes, not only brought incredible changes and advantages in the field of tumor [17–19], but expands understanding and provides potential treatment strategies for some special types of tumors, such as inflammatory breast cancer and juvenile papillomatosis with co-existing breast carcinoma [20,21]. In our study, we found 3764 DE mRNAs, 5416 DE lncRNAs, and 148 DE circRNAs by RNA sequencing in three breast cancer tissues compared with their paired normal tissues. Above all, enrichment analysis indicated that DE lncRNAs and DE circRNAs were mainly enriched in mitochondria and nucleus. Apparently, nucleus is the control center of various cell metabolisms and mitochondria produce ATP for them. Recently, fatty acid oxidation (FAO) has been proved to be a crucial energy pathway in metastatic breast cancer, and the key enzyme of FAO exist in mitochondria, such as CPT1 (carnitine palmitoyl transferase 1), limit the rate of reaction thus restricting the proliferation of malignant cells [22–24]. Moreover, ATP citrate lyase (ACLY), a key enzyme responsible for de novo fatty acid synthesis, was considered to enhance triple-negative breast cancer (TNBC) stemness and metastasis via miR-206/ITGA2/ACLY-CCND1 signaling axis [25]. It follows that the exploration of noncoding RNAs, which can regulate the key enzyme of fatty acid metabolism, may reveal the mechanisms of the breast cancer progression in the future.
In our study, we found that up-circRNA NFIX, which directly correlated with has-miR-6794-5p, was a significant role in the circRNAs–miRNAs–mRNAs network. Previous evidence demonstrated that circNFIX can promote progression in several types of cancer, such as glioma, lung cancer, and pituitary adenoma [26–28]. Recent study has reported that circNFIX can function as a ceRNA of has-miR-3064-5p to facilitate the cell growth, migration, and invasion of hepatocellular carcinoma (HCC), further enhance glutaminolysis [29]. To our knowledge, the role of circNFIX in breast cancer is still unknown. Since fatty acid metabolism can induce breast cancer progression as well as therapy resistance [30–34], whether circNFIX regulates fatty acid metabolism in breast cancer is worthy further investigation in the future.
Additionally, our PPI network identified several hub proteins from vital DE mRNAs, which were speculated to be potential factors in regulating breast cancer. For example, MMP9, an important enzyme to the degradation of extracellular matrix (ECM), was considered to be an onset of invasion or metastasis in breast cancer [35–37]. Furthermore, MMP9 was suggested can be regulated by free fatty acids receptor 1 (FFA1), which indicated that fatty acid metabolism may be a potential mechanism of MMP9 activity [38]. In addition, Coilly et al. [39] identified MMP9 as a predictive factor for poor prognosis of nonalcoholic fatty liver. Kwapisz et al. [40] reported that fatty acid can activate MMP9 and further induce epithelial–mesenchymal transition (EMT), which can even lead to the occurrence of HCC. Thus, we predicted that MMP9 is related to fatty acid in our study. Based on the previous study, the mechanism research, which about MMP9 promotes breast cancer progression by regulating fatty acid metabolism, possibly serving as a therapeutic strategy in breast cancer.
Elevated expression of MMP9 was correlates with more aggressive subtypes of breast cancer, such as TNBC and HER2-positive breast cancer [41]. In our study, two pairs of HER2-positive breast cancer and one pair of TNBC samples were used for RNA-sequencing and further analyses revealed that MMP9 was one of the most up-regulated expressed mRNA and may promote breast cancer progression by regulating fatty acid metabolism. As the most aggressive molecular subtypes in breast cancer, TNBC and HER2-positive breast cancer remained obstacles in the therapy of recurrence, metastasis, or chemotherapy resistance. The metabolic regulatory roles of MMP9 and the upstream regulatory mechanism of the elevated expression of MMP9 in TNBC and HER2-positive breast cancer require further study. In the present study, we found that circNFIX may act as a ceRNA through regulating MMP9 expression and fatty acid metabolism in breast cancer, which may be a novel mechanism of elevated MMP9 expression.
In conclusion, our study provides insights into the molecular mechanisms of breast cancer by analyzing the expression profiles of mRNAs, lncRNAs, and circRNAs. circNFIX and MMP9 may play a unique role by regulating fatty acid metabolism in breast cancer, which suggest a potential therapy in the future. However, our study has lack of analysis of miRNAs to provide a whole-transcriptome sequencing for breast cancer. In addition, based on our present study, further experimental validations are required to perform.
Supplementary Material
Acknowledgements
Breast cancer samples and normal tissues were stored at Biobank of the first affiliated hospital of Jinan University. All patients had signed informed consent for donating their samples to Biobank of the first affiliated hospital of Jinan University.
Abbreviations
- ACLY
ATP citrate lyase
- ceRNA
competing endogenous RNA
- DE
differentially expressed
- FAO
fatty acid oxidation
- FC
foldchange
- FPKM
fragments per kilobases per million
- GC
The proportion of the bases G and C account for the total bases A, T, C, G.
- GO
gene ontology
- HCC
hepatocellular carcinoma
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MMP9
matrix metalloproteinase-9
- NGS
next-generation sequencing
- PPI
protein–protein interaction
- TNBC
triple-negative breast cancer
- TPM
transcripts per million reads
Data Availability
The data generated or analyzed to support the current study are included within the article.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The present work was supported by Youth Science Foundation of National Natural Science Foundation of China [grant number 81702598]; The Science Foundation of Guangdong Province [grant number 2017A030313803]; The Science and Technology Program of Guangzhou [grant number 201804010011]; and Flagship specialty construction project-General surgery [grant number 711003].
CRediT Author Contribution
Yuan Li: Writing—original draft. Chiseng Lei: Writing—original draft. Yude Xie: Writing—original draft. Jie Zhang: Methodology. Ningxia Wang: Resources. Weili He: Resources. Shaohua Qu: Supervision, Funding acquisition, Writing—review & editing.
References
- 1.Sung H., Ferlay J., Siegel R.L.et al. (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Burstein H.J., Curigliano G., Thurlimann B.et al. (2021) Customizing local and systemic therapies for women with early breast cancer: the St. Gallen International Consensus Guidelines for treatment of early breast cancer 2021. Ann. Oncol. 32, 1216–1235 10.1016/j.annonc.2021.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong Y., Xu F., Wu J.et al. (2021) Application of next generation sequencing in laboratory medicine. Ann. Laboratory Med. 41, 25–43 10.3343/alm.2021.41.1.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamps R., Brandao R.D., Bosch B.J.et al. (2017) Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int. J. Mol. Sci. 18, 308–364 10.3390/ijms18020308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morganti S., Tarantino P., Ferraro E.et al. (2019) Complexity of genome sequencing and reporting: next generation sequencing (NGS) technologies and implementation of precision medicine in real life. Crit. Rev. Oncol. Hematol. 133, 171–182 10.1016/j.critrevonc.2018.11.008 [DOI] [PubMed] [Google Scholar]
- 6.Yuan H., Xiaowei W., Yiran Z.et al. (2021) Construction of an mRNA-miRNA-lncRNA network prognostic for triple-negative breast cancer. Aging 13, 1153–1175 10.18632/aging.202254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H. and Lu B. (2020) The roles of ceRNAs-mediated autophagy in cancer chemoresistance and metastasis. Cancers (Basel) 12, 2926–2948 10.3390/cancers12102926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L., Cho K.B., Li Y.et al. (2019) Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int. J. Mol. Sci. 20, 5758–5783 10.3390/ijms20225758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X., Ji C., Hu J.et al. (2021) Hsa_circ_0005273 facilitates breast cancer tumorigenesis by regulating YAP1-hippo signaling pathway. J. Exp. Clin. Cancer Res. 40, 29–45 10.1186/s13046-021-01830-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu G., Yang Z., Peng T.et al. (2021) Circular RNAs: rising stars in lipid metabolism and lipid disorders. J. Cell. Physiol. 236, 4797–4806 10.1002/jcp.30200 [DOI] [PubMed] [Google Scholar]
- 11.Gil N. and Ulitsky I. (2020) Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 21, 102–117 10.1038/s41576-019-0184-5 [DOI] [PubMed] [Google Scholar]
- 12.Hansen T.B., Kjems J. and Damgaard C.K. (2013) Circular RNA and miR-7 in cancer. Cancer Res. 73, 5609–5612 10.1158/0008-5472.CAN-13-1568 [DOI] [PubMed] [Google Scholar]
- 13.Zhang M., Bai X., Zeng X.et al. (2021) circRNA-miRNA-mRNA in breast cancer. Clin. Chim. Acta 523, 120–130 10.1016/j.cca.2021.09.013 [DOI] [PubMed] [Google Scholar]
- 14.Sang Y., Chen B., Song X.et al. (2019) circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol. Ther. 27, 1638–1652 10.1016/j.ymthe.2019.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L., Yi J., Lu L.Y.et al. (2021) Estrogen-induced circRNA, circPGR, functions as a ceRNA to promote estrogen receptor-positive breast cancer cell growth by regulating cell cycle-related genes. Theranostics 11, 1732–1752 10.7150/thno.45302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L., Zhou Y., Jiang L.et al. (2021) CircWAC induces chemotherapeutic resistance in triple-negative breast cancer by targeting miR-142, upregulating WWP1 and activating the PI3K/AKT pathway. Mol. Cancer 20, 43–57 10.1186/s12943-021-01332-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H., Wang C., Song H.et al. (2019) RNA-Seq profiling of circular RNAs in human colorectal cancer liver metastasis and the potential biomarkers. Mol. Cancer 18, 8–13 10.1186/s12943-018-0932-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong M., Tao S., Zhang L.et al. (2020) RNA sequencing: new technologies and applications in cancer research. J. Hematol. Oncol. 13, 166–181 10.1186/s13045-020-01005-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vo J.N., Cieslik M., Zhang Y.et al. (2019) The landscape of circular RNA in cancer. Cell 176, 869–881 10.1016/j.cell.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo R., Chong W., Wei Q.et al. (2021) Whole-exome sequencing identifies somatic mutations and intratumor heterogeneity in inflammatory breast cancer. NPJ Breast Cancer 7, 72–79 10.1038/s41523-021-00278-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Alfonso T.M., Pareja F., Da Cruz Paula A.et al. (2021) Whole-exome sequencing analysis of juvenile papillomatosis and coexisting breast carcinoma. J. Pathol. Clin. Res. 7, 113–120 10.1002/cjp2.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo X., Cheng C., Tan Z.et al. (2017) Emerging roles of lipid metabolism in cancer metastasis. Mol. Cancer 16, 76–85 10.1186/s12943-017-0646-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Y., Temkin S.M., Hawkridge A.M.et al. (2018) Fatty acid oxidation: an emerging facet of metabolic transformation in cancer. Cancer Lett. 435, 92–100 10.1016/j.canlet.2018.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y.N., Zeng Z.L., Lu J.et al. (2018) CPT1A-mediated fatty acid oxidation promotes colorectal cancer cell metastasis by inhibiting anoikis. Oncogene 37, 6025–6040 10.1038/s41388-018-0384-z [DOI] [PubMed] [Google Scholar]
- 25.Adorno-Cruz V., Hoffmann A.D., Liu X.et al. (2021) ITGA2 promotes expression of ACLY and CCND1 in enhancing breast cancer stemness and metastasis. Genes Dis. 8, 493–508 10.1016/j.gendis.2020.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding C., Wu Z., You H.et al. (2019) CircNFIX promotes progression of glioma through regulating miR-378e/RPN2 axis. J. Exp. Clin. Cancer Res. 38, 506–517 10.1186/s13046-019-1483-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu J., Zhu Y., Qin Y.et al. (2020) CircNFIX acts as a miR-212-3p sponge to enhance the malignant progression of non-small cell lung cancer by up-regulating ADAM10. Cancer Manag. Res. 12, 9577–9587 10.2147/CMAR.S272309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng J., Nie D., Li B.et al. (2021) CircNFIX promotes progression of pituitary adenoma via CCNB1 by sponging miR-34a -5p. Mol. Cell. Endocrinol. 525, 111140–111148 10.1016/j.mce.2020.111140 [DOI] [PubMed] [Google Scholar]
- 29.Erhui X., Dongmei Z., Weili Z.et al. (2021) circNFIX facilitates hepatocellular carcinoma progression by targeting miR-3064-5p/HMGA2 to enhance glutaminolysis. Am. J. Transl. Res. 13, 8697–8710 [PMC free article] [PubMed] [Google Scholar]
- 30.Hoy A.J., Nagarajan S.R. and Butler L.M. (2021) Tumour fatty acid metabolism in the context of therapy resistance and obesity. Nat. Rev. Cancer 21, 753–766 10.1038/s41568-021-00388-4 [DOI] [PubMed] [Google Scholar]
- 31.Currie E., Schulze A., Zechner R.et al. (2013) Cellular fatty acid metabolism and cancer. Cell Metab. 18, 153–161 10.1016/j.cmet.2013.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z. and Zhang H. (2016) Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell. Mol. Life Sci. 73, 377–392 10.1007/s00018-015-2070-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang T., Fahrmann J.F., Lee H.et al. (2018) JAK/STAT3-regulated fatty acid β-oxidation is critical for breast cancer stem cell self-renewal and chemoresistance. Cell Metab. 27, 136–150 10.1016/j.cmet.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camarda R., Zhou A.Y., Kohnz R.A.et al. (2016) Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat. Med. 22, 427–432 10.1038/nm.4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bi X., Lou P., Song Y.et al. (2021) Msi1 promotes breast cancer metastasis by regulating invadopodia-mediated extracellular matrix degradation via the Timp3-Mmp9 pathway. Oncogene 40, 4832–4845 10.1038/s41388-021-01873-8 [DOI] [PubMed] [Google Scholar]
- 36.Dong H., Diao H., Zhao Y.et al. (2019) Overexpression of matrix metalloproteinase-9 in breast cancer cell lines remarkably increases the cell malignancy largely via activation of transforming growth factor beta/SMAD signalling. Cell Prolif. 52, e12633 10.1111/cpr.12633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owyong M., Chou J., van den Bijgaart R.J.et al. (2019) MMP9 modulates the metastatic cascade and immune landscape for breast cancer anti-metastatic therapy. Life Sci. Alliance 2, e201800226 10.26508/lsa.201800226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manosalva C., Alarcon P., Gonzalez K.et al. (2020) Free fatty acid receptor 1 signaling contributes to migration, MMP-9 activity, and expression of IL-8 induced by linoleic acid in HaCaT cells. Front. Pharmacol. 11, 595–608 10.3389/fphar.2020.00595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coilly A., Desterke C., Guettier C.et al. (2019) FABP4 and MMP9 levels identified as predictive factors for poor prognosis in patients with nonalcoholic fatty liver using data mining approaches and gene expression analysis. Sci. Rep. 9, 19785–19800 10.1038/s41598-019-56235-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwapisz O., Gorka J., Korlatowicz A.et al. (2021) Fatty acids and a high-fat diet induce epithelial-mesenchymal transition by activating TGFbeta and beta-catenin in liver cells. Int. J. Mol. Sci. 22, 1272–1287 10.3390/ijms22031272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yousef E.M., Tahir M.R., St-Pierre Y.et al. (2014) MMP-9 expression varies according to molecular subtypes of breast cancer. BMC Cancer 14, 609–620 10.1186/1471-2407-14-609 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated or analyzed to support the current study are included within the article.