Abstract
Nuclear copies of mitochondrial genes (numts) are commonplace in vertebrate genomes and have been characterized in many species. However, relatively little attention has been paid to understanding their evolutionary origins and to disentangling alternative sources of insertions. Numts containing genes with intact mitochondrial reading frames represent good candidates for this purpose. The sequences of the genes they contain can be compared with their mitochondrial homologs to characterize synonymous to nonsynonymous substitution rates, which can shed light on the selection pressures these genes have been subjected to. Here, we characterize 25 numts in the Antarctic fur seal (Arctocephalus gazella) genome. Among those containing genes with intact mitochondrial reading frames, three carry multiple substitutions in comparison to their mitochondrial homologs. Our analyses reveal that one represents a historic insertion subjected to strong purifying selection since it colonized the Otarioidea in a genomic region enriched in retrotransposons. By contrast, the other two numts appear to be more recent and their large number of substitutions can be attributed to noncanonical insertions, either the integration of heteroplasmic mtDNA or hybridization. Our study sheds new light on the evolutionary history of pinniped numts and uncovers the presence of hidden sources of mitonuclear variation.
Keywords: nuclear DNA sequences of mitochondrial origin (numts), genome evolution, mitochondrial DNA, pinniped, Antarctic fur seal, Arctocephalus gazella
Significance.
Eukaryotic organelles, such as mitochondria or plastids, possess their own genomes. Intriguingly, fragments of organelle DNA can be found in the nuclear genomes of many species. These are known as numts and represent mitochondrial DNA that has been copied into a nuclear genome. These insertions are assumed to be nonfunctional and little is known about their evolutionary fates. We conducted a genome-wide scan of numts in the Antarctic fur seal genome and found one numt that has been subjected to strong purifying selection. We further identified two numts that appear to have originated from noncanonical sources. Our study shows that numts can have complex evolutionary histories and that they need not necessarily be nonfunctional.
Introduction
Mitochondria are organelles that are believed to share a common ancestor with α-protobacteria (Sagan 1967). During their evolution as endosymbionts of eukaryotic cells, their genomes dramatically reduced in size as the vast majority of their functional genes transferred into the nucleus over the last 1.5 billion years (Bock and Timmis 2008). Mitochondrial colonization of the nuclear genome is a continuous process that is still ongoing (Ricchetti et al. 2004). This is reflected by the presence of numerous nuclear sequences revealing homology to contemporary mitochondrial DNA (mtDNA), usually referred to as “nuclear mtDNA sequences” or numts, across a wide range of eukaryotic species (Hazkani-Covo et al. 2010; Calabrese et al. 2017).
With the increasing number of available reference genomes, considerable effort has been invested over the past decade into characterizing numts, especially in mammalian species (Calabrese et al. 2017; Schiavo et al. 2017; Grau et al. 2020). These studies have shown that the number of numts present in a nuclear genome can vary substantially across species, from as few as 13 numts in the polar bear (Ursus maritius) (Lammers et al. 2017) to over 1,000 in the opossum (Monodelphis domestica) (Hazkani-Covo et al. 2010) and platypus (Ornithorhynchus anatinus) (Calabrese et al. 2017). However, limited attention has so far been paid to understanding whether numts could be functional or to distinguishing among possible alternative sources of mitonuclear integrations.
It has been shown that certain numts, depending on their insertion position, can have deleterious effects and be associated with diseases (Borensztajn et al. 2002; Turner et al. 2003). Yet, numts do not appear to be functional per se, as most of them become pseudogenes upon their integration into the nuclear genome due to the presence of premature stop codons resulting from the difference between the nuclear and mitochondrial genetic codes (Bensasson et al. 2001). After their insertion into the nuclear genome, numts are then expected to accumulate mutations over time, causing their mitochondrial reading frame to quickly degrade. Numts deviating from this assumption, namely ancient mitonuclear insertions (i.e. insertions predating the split between major taxonomic groups such as orders) presenting intact mitochondrial reading frames despite the presence of numerous synonymous mutations, could then be seen as potential evidence for a numt sequence being under functional constraint. Although it is unlikely that most numts are expressed (Bensasson et al. 2001) or play a role in RNA–RNA interactions (Cloonan 2015), they could nevertheless exercise a functional role as regulatory elements or through the creation of novel exons (Noutsos et al. 2007).
As described above, numts that carry numerous mutations in comparison to their mitochondrial homologs may intuitively be regarded as ancient insertions, because they will have had more time to diverge. However, appreciable numbers of substitutions might also be observed for more recent insertions (i.e. insertions that occurred after the split between major taxonomic groups and that are observed only in a single species or genus) when they derive from noncanonical sources, such as heteroplasmic mitochondrial variants (Wei et al. 2020; Formenti et al. 2021; Parakatselaki and Ladoukakis 2021) or mtDNA that has introgressed via hybridization (Baldo et al. 2011). Genes residing in these recent insertions might be expected to contain intact mitochondrial reading frames because they were functional in the mitochondrion before their recent insertion into the nuclear genome. By contrast, assuming that numts are selectively neutral, ancient insertions should have a greater likelihood of containing genes with premature stop codons and/or out of frame mutations, as defined by the mitochondrial genetic code.
To identify numts that are possibly functional and/or which may have originated from alternative sources, one could search for genes residing in mitonuclear insertions that show intact mitochondrial reading frames and compare them to their mitochondrial homologs. Intact mitochondrial reading frames in numts containing mutations would then be indicative of either ancient insertions that violate the assumption of numts being nonfunctional or recent insertions from alternative sources. The amounts of synonymous substitutions (dS), together with phylogenetic analysis, could then be used to distinguish recent mitonuclear insertions from ancient ones. Finally, the ratio of synonymous and nonsynonymous substitutions (dN/dS) could be analyzed to reveal the evolutionary pressures that these potentially coding sequences have been subjected to Kryazhimskiy and Plotkin (2008) and to formally test for the presence of nonneutral numts.
The Antarctic fur seal (Arctocephalus gazella) represents an ideal candidate to search for numts that might contain functional genes and/or genes that have originated from noncanonical sources. First, A. gazella is known to have recently hybridized with other two Arctocephalus species (Lancaster et al. 2006). This may have introduced novel mitochondrial variation that, if integrated into the nuclear genome, would appear divergent from homologous A. gazella mitochondrial sequences. Second, both a high quality, chromosome-level, nuclear reference genome (Humble et al. 2018, 2016; Peart et al. 2021) and a mitochondrial (Nagel et al. 2019) genome are available for this species.
Here, we performed a scan of the nuclear reference genome of the Antarctic fur seal for mitonuclear insertions, identifying a total of 25 numts. Four of these contain genes with intact mitochondrial reading frames. Among those containing mutations, one appears to be an ancient insertion exhibiting strong signatures of purifying selection, whereas two numts appear to represent recent integrations of highly divergent mtDNA sequences. Taken together, our results provide new insights into the evolutionary forces shaping numts in pinnipeds and show how the study of mitonuclear genes with intact mitochondrial reading frames can uncover hidden sources of mitonuclear variation.
Results
Discovery and Characterization of numts
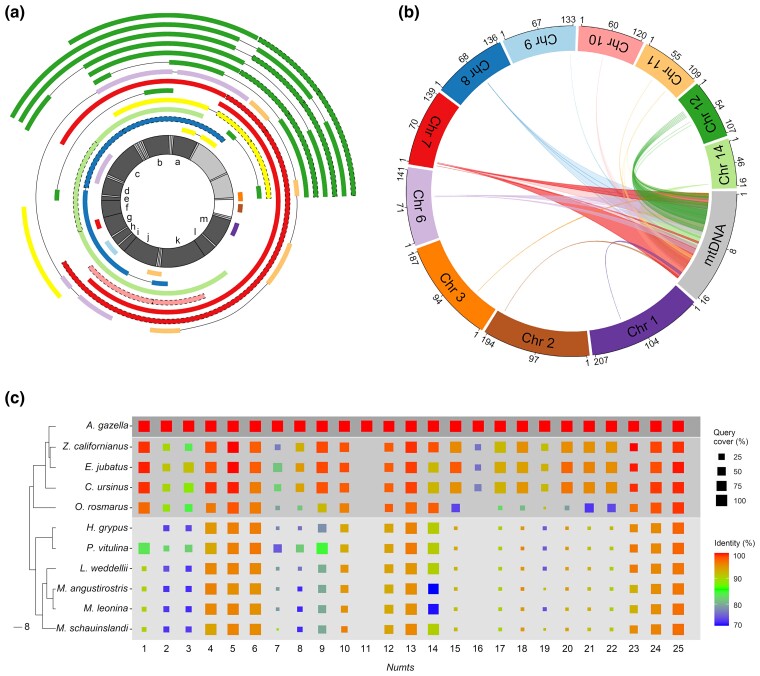
Sequence homology searches between the mitochondrial (Nagel et al. 2019) and nuclear genome (Peart et al. 2021) of the Antarctic fur seal reveal a total of 25 putative numts varying in length between 294 and 14,199 bp and consisting of between one and four separate fragments (see Materials and Methods, fig. 1 and table 1). These are located on 11 different chromosomes and three unplaced genomic scaffolds, each of which carries between one and eight numts (fig. 1b and table 1). The majority of these numts contain multiple full-length mitochondrial genes, and collectively they represent the entire mitochondrial genome (fig. 1a). The first 5,000 bp of the mitochondrial genome shows the highest average depth of coverage, reflecting the presence of multiple closely related numts on chromosome 12 (fig. 1a).
Fig. 1.
Summary of 25 numts characterized in A. gazella including their mitochondrial origins, locations in the nuclear genome, and patterns of phylogenetic conservation. (a) The mitochondrial origin of each numt (outer colored arcs). The innermost circle represents the A. gazella mitochondrial genome, where light grey sectors refer to tRNA and rRNA genes, dark grey sectors represent other mitochondrial genes (a: ND1, b: ND2, c: COX1, d: COX2, e: ATP8, f: ATP6, g: COX3, h: ND3, i: ND4L, j: ND4, k: ND5, l: ND6, m: CYTB) and the white sector represents the D-loop region. Numts are color coded according to the nuclear chromosome in which they are located as shown in (b). Yellow numts, absent in (b), were found in unplaced genomic scaffolds. Fragments belonging to the same numt are connected by solid black lines, whereas fragments belonging to the same numt and originating from overlapping mitochondrial regions are indicated with dashed lines. (b) The location of the numts in the A. gazella nuclear genome. Each colored arc connects the relevant region of the mitochondrial genome (grey) to the corresponding chromosome in the nuclear genome. Numbers indicate base-pair positions along the mitochondrial genome (kb) and along the nuclear chromosomes (Mb). (c) The results of blast searches against the nuclear genomes of A. gazella and other pinniped species (Z. californianus, E. jubatus, C. ursinus, O. rosmarus, H. grypus, P. vitulina, L. weddellii, M. angustirostris, M. leonina, and M. schauinslandi), whose phylogenetic relationships are indicated by the tree on the left (redrawn from Higdon et al., 2007). Each column represents a single numt. The colored squares indicate both the query cover (which is proportional to the size of square, see caption) and the percentage of sequence identity (indicated by the color gradient, see caption) when a given numt was blasted against the nuclear genome of another species. The circular plots were drawn using the R package circlize (Gu et al. 2014).
Table 1.
Summary Information for All 25 numts Characterized in the Antarctic Fur Seal Nuclear Genome
| Numt ID | Nuclear Scaffold | Start | End | Chromosome | Number of Fragments | Numt Length | mtDNA Coordinates | Expected PCR Product |
|---|---|---|---|---|---|---|---|---|
| 1 | ScWAj4l_1;HRSCAF = 3 | 127934607 | 127935176 | 9 | 1 | 569 | 9,566–10,139 | Yes |
| 2 | ScWAj4l_2;HRSCAF = 13 | 74612525 | 74621691 | 8 | 4 | 9166 | 2,107–5,497; 7,736–10,840; 5,454–7,653; 11,628–12,127 | Yes |
| 3 | ScWAj4l_18;HRSCAF = 239 | 84876333 | 84889827 | 14 | 2 | 13494 | 3,018–13,905; 6,301–8,931 | No |
| 4 | ScWAj4l_37;HRSCAF = 538 | 184440549 | 184440843 | 2 | 1 | 294 | 15,767–16,061 | Yes |
| 5 | ScWAj4l_43;HRSCAF = 587 | 54190362 | 54194849 | 10 | 2 | 4487 | 9,871–11,748; 11,733–13,078 | Yes |
| 6 | ScWAj4l_44;HRSCAF = 590 | 2316056 | 2316387 | 7 | 1 | 331 | 8,723–9,062 | No |
| 7 | ScWAj4l_44;HRSCAF = 590 | 7718809 | 7728008 | 7 | 2 | 9199 | 1–2,801; 10,088–16,156 | No |
| 8 | ScWAj4l_44;HRSCAF = 590 | 7977588 | 7991731 | 7 | 4 | 14,143 | 9,436–16,156; 1–1,616; 2,738–6,806; 1,597–2,721 | Yes |
| 9 | ScWAj4l_46;HRSCAF = 602 | 45289204 | 45289714 | 11 | 1 | 510 | 11,396–11,907 | Yes |
| 10 | ScWAj4l_46;HRSCAF = 602 | 85808165 | 85817409 | 11 | 4 | 9,244 | 104–428; 1,757–2,257; 11,770–12,369; 14,387–15,304 | Yes |
| 11 | ScWAj4l_49;HRSCAF = 609 | 50930992 | 50931325 | 3 | 1 | 333 | 1–326 | Yes |
| 12 | ScWAj4l_82;HRSCAF = 715 | 87894123 | 87902946 | 6 | 4 | 8,823 | 3,942–5,516; 2,319–3,843; 10,185–10,984; 9,671–9,955 | Yes |
| 13 | ScWAj4l_82;HRSCAF = 715 | 88838208 | 88839332 | 6 | 1 | 1,124 | 6,301–7,449 | Yes |
| 14 | ScWAj4l_84;HRSCAF = 721 | 138510899 | 138511378 | 1 | 1 | 479 | 14,905–15,381 | Yes |
| 15 | ScWAj4l_133;HRSCAF = 810 | 20791 | 26530 | Unplaced scaffold | 3 | 5,739 | 2,968–5,471; 1,597–2,721; 74–1,616 | Yes |
| 16 | ScWAj4l_2350;HRSCAF = 3197 | 2 | 2125 | Unplaced scaffold | 2 | 2,123 | 2,177–2,819; 3,037–3,535 | No |
| 17 | ScWAj4l_2441;HRSCAF = 3292 | 24002125 | 24007610 | 12 | 3 | 5,485 | 74–1,616; 2,968–4,007; 5,120–5,471 | Yes |
| 18 | ScWAj4l_2441;HRSCAF = 3292 | 24086242 | 24091805 | 12 | 3 | 5,563 | 74–1,616; 2,738–5,471; 1,597–2,721 | Yes |
| 19 | ScWAj4l_2441;HRSCAF = 3292 | 24285509 | 24299708 | 12 | 3 | 14,199 | 74–1,198; 2,738–5,471; 6,202–6,875 | Yes |
| 20 | ScWAj4l_2441;HRSCAF = 3292 | 24425761 | 24432730 | 12 | 3 | 6,969 | 74–1,616; 2,968–6,806; 1,597–2,671 | Yes |
| 21 | ScWAj4l_2441;HRSCAF = 3292 | 24605035 | 24611940 | 12 | 3 | 6,905 | 2,968–6,806; 74–1,198; 1,597–2,671 | Yes |
| 22 | ScWAj4l_2441;HRSCAF = 3292 | 24665417 | 24670984 | 12 | 3 | 5,567 | 79–1,616; 2,740–5,471; 1,597–2,721 | Yes |
| 23 | ScWAj4l_2441;HRSCAF = 3292 | 53287344 | 53289309 | 12 | 2 | 1,965 | 1,917–2,215; 78–432 | No |
| 24 | ScWAj4l_2441;HRSCAF = 3292 | 60968082 | 60971080 | 12 | 2 | 2,998 | 3,905–4,582; 7,807–8,060 | Yes |
| 25 | ScWAj4l_4416;HRSCAF = 5370 | 222 | 1965 | Unplaced scaffold | 1 | 1,743 | 8,175–9,955 | Yes |
Note.— Locations in the A. gazella nuclear genome are reported together with information on the number of numt fragments, total length, mitochondrial location of origin, and whether PCR amplification using nuclear genome-specific primers yielded a product of the expected size.
Experimental Validation
PCR amplification using nuclear genome-specific primers yields PCR products of the approximate expected size in the majority of cases (n = 20, 80%, supplementary fig. S1, Supplementary Material online). Three of the PCR products are larger than expected, whereas a further two primer pairs yield unclear products with the suggestion of multiple bands (supplementary fig. S1, Supplementary Material online). The fact that most of the numts can be validated in vitro suggests that most if not all of them are genuine mitonuclear integrations rather than genome assembly artifacts.
Phylogenetic Patterns
Blast searches of the 25 numts against available pinniped reference genomes produce highly variable profiles in terms of query cover and percentage of identity (fig. 1c). We interpret blast results characterized by high query cover as hits to homologous genomic regions and blast results showing low query cover as hits to nonhomologous regions. In fact, even in the case of highly diverged numts characterized by low percentage of identity, we would expect query cover to be relatively high also in the presence of gaps, as these can be identified during a blast search. The patterns summarized in figure 1c are then suggestive of an ongoing process of mitonuclear integration. Specifically, we find that some of the numts appear to have been integrated into the nuclear genome of the Antarctic fur seal rather recently, as these are unique to this species (e.g., numts 11 and 16, fig. 1c). These represent insertions that took place not earlier than the split between Arctocephalus species and Zalophus californianus and Eumetopias jubatus (fig. 1c), which occurred around 5.2 Ma. By contrast, other numts, which are shared among all of the pinniped species (e.g. numts 5 and 25, fig. 1c), are likely to represent ancient insertions that occurred before the split between the three pinniped families and are thus at least 25 Myr old (Deméré et al. 2003; Higdon et al. 2007). Finally, numts present in a subset of the pinniped species (e.g. numts 1 and 9, fig. 1c) were probably integrated at intermediate time points. Repeating this analysis separately for each numt fragment yields comparable results (supplementary fig. S2, Supplementary Material online). Moreover, this analysis also reveals a highly consistent pattern of query cover and percentage of identity among fragments clustered within the same numt (supplementary fig. S2, Supplementary Material online), suggesting that they can be considered part of the same mitonuclear insertion.
Numt Coding Genes
To understand the evolutionary origins and fates of the identified mitonuclear insertions, we focus on numt fragments showing sequence homology to mitochondrial protein-coding DNA. Because we are specifically interested in genes showing intact mitochondrial reading frames, we focus on the separate numt fragments, as opposed to whole numts, in order to exclude the nuclear sequences located between numt fragments. We identify six numt fragments containing a total of 21 genes that contain intact mitochondrial reading frames (table 2), although all of them contain premature stop codons as defined by the nuclear genetic code. The majority of these genes are identical or nearly identical to their mitochondrial homologs and are therefore likely to represent recent integrations from the A. gazella mitochondrial genome. However, we also identify fragments in numts 1, 2, and 3 that contain genes differing by more than five nucleotide substitutions from their mitochondrial homologs. Pairwise dN/dS analysis of genes located in these numt fragments reveal strong signatures of purifying selection, indicated by dN/dS < 1 (table 2). Synonymous substitution rates (dS), a proxy for neutral divergence, for the fragments in numts 2 and 3 are modest, indicating that the substitutions observed at these genes are likely to have resulted from relatively recent insertions of highly divergent mtDNA sequences, as compared with the Antarctic fur seal reference mitochondrial genome. By contrast, dS for the gene located in numt 1, ND3, exceeds one (table 2), indicating that this numt shows substantial neutral divergence at synonymous sites relative to its mitochondrial homolog.
Table 2.
The Number of Synonymous and Nonsynonymous Substitutions, Together with Relative dN and dS Measures, Between Protein-Coding Genes Residing in the numt Fragments and Their mtDNA Homologs
| Numt | Fragment | Gene | Accession Number | Synonymous Substitutions | Nonsynonymous Substitutions | dN | dS | dN/dS | P-value |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | ND3 | DAC80253.1 | 48 | 14 | 0.0564 | 1.1062 | 0.051 | <0.01 |
| 2 | 1 | ATP8 | DAC80250.1 | 0 | 0 | — | — | — | — |
| 2 | 1 | ATP6 | DAC80251.1 | 0 | 0 | — | — | — | — |
| 2 | 1 | COX3 | DAC80252.1 | 0 | 0 | — | — | — | — |
| 2 | 1 | ND3 | DAC80253.1 | 0 | 0 | — | — | — | — |
| 2 | 1 | ND4L | DAC80254.1 | 1 | 0 | — | 0.0148 | 0 | — |
| 2 | 3 | ND2 | DAC80247.1 | 46 | 16 | 0.0218 | 0.1881 | 0.1160 | <0.01 |
| 3 | 1 | COX1 | DAC80248.1 | 0 | 1 | 0.0009 | — | — | — |
| 3 | 1 | ATP8 | DAC80250.1 | 0 | 0 | — | — | — | — |
| 3 | 1 | COX3 | DAC80252.1 | 13 | 4 | 0.0066 | 0.0835 | 0.0794 | <0.01 |
| 3 | 1 | ND3 | DAC80253.1 | 5 | 2 | 0.0075 | 0.0719 | 0.1042 | <0.01 |
| 3 | 1 | ND4L | DAC80254.1 | 4 | 0 | — | 0.0683 | 0 | — |
| 3 | 1 | ND4 | DAC80255.1 | 24 | 5 | 0.0052 | 0.0615 | 0.085 | <0.01 |
| 3 | 1 | ND5 | DAC80256.1 | 30 | 5 | 0.0038 | 0.0667 | 0.0562 | <0.01 |
| 3 | 2 | COX2 | DAC80249.1 | 7 | 1 | 0.0019 | 0.0472 | 0.0404 | <0.01 |
| 3 | 2 | ATP8 | DAC80250.1 | 0 | 1 | 0.0068 | — | — | — |
| 3 | 2 | ATP6 | DAC80251.1 | 4 | 5 | 0.0103 | 0.022 | 0.4668 | 0.2145 |
| 8 | 3 | ND4L | DAC80254.1 | 4 | 0 | — | 0.0683 | 0 | — |
| 8 | 3 | ND4 | DAC80255.1 | 0 | 0 | — | — | — | — |
| 8 | 3 | ND5 | DAC80256.1 | 0 | 0 | — | — | — | — |
| 8 | 3 | CYTB | DAC80258.1 | 0 | 0 | — | — | — | — |
Note.—Identities and NCBI accession numbers are provided for each gene.
Phylogenetic Analysis of Fragments in numt 1, 2, and 3
We implement phylogenetic analysis of the fragments in numts 1, 2, and 3 that contain genes with intact reading frames in order to better understand when these insertions might have colonized the Antarctic fur seal nuclear genome. Our phylogenetic reconstruction is based on the Antarctic fur seal numt fragments and on homologous sequences from the mtDNA of A. gazella, 26 other pinniped species (see Materials and Methods) and the dog (Canis lupus). We additionally include sequences of homologous numts from other pinnipeds when present. Consistent with our previous results, numt 1 appears to be a relatively old insertion (supplementary fig. S3a, Supplementary Material online) that predates the split between the walrus and the otariids, which occurred around 18 Ma (Higdon et al. 2007), whereas numts 2 and 3 appear to be more recent (supplementary fig. S3b and c, Supplementary material online). Notably, the numt 2 sequences from E. jubatus, Z. californicus, Callorhinus ursinus, and Odobenus rosmarus cluster separately and more distantly from the numt 2 sequence of A. gazella, suggesting that these sequences may not be homologous (supplementary fig. S3b, Supplementary material online). For numt 3, we only analyzed the nuclear sequence of A. gazella because we could not identify homologous sequences in other pinnipeds (blast hits were obtained to multiple locations with low query cover and percentage of identity).
Selection Analysis of numt 1
Phylogenetic analysis of numt 1 suggests that this integration likely occurred around 18 Ma (fig. 2). We therefore wanted to know if the mitochondrial reading frames of the gene in numt 1 are also conserved in other pinnipeds, which is indeed the case. We investigated the possibility of a potential functional conservation of numt 1 on the protein-coding level since it was integrated into the nuclear genome by conducting substitution rate analyses to identify the evolutionary forces that have shaped the sequence composition of the gene ND3 located in numt 1 (indicated by the red branch in fig. 2). By estimating dN/dS ratios for the predefined branches (red vs. black, fig. 2), we find that dN/dS ratios are significantly different between the numt 1 gene sequences and their mitochondrial counterparts (P < 0.001, likelihood ratio test [LRT], one-ratio vs. two-ratio model, table 3). This pattern cannot be solely attributed to differences in the intensity of purifying selection (P = 0.99, LRT, two-ratio vs. two-ratio fixed model, table 3). Instead, neutral evolution and/or positive selection may have played a pervasive role in the evolution of numt 1.
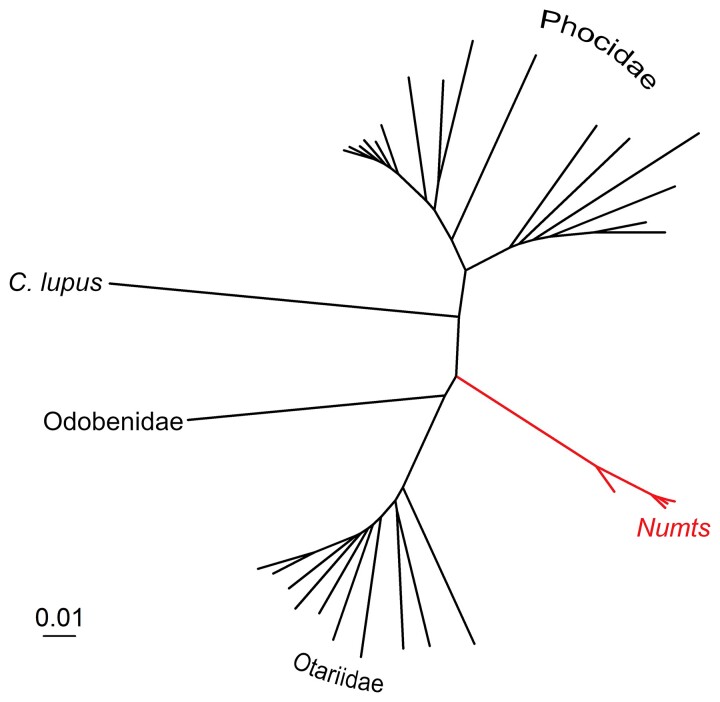
Fig. 2.
Phylogenetic reconstruction of numt 1. The phylogenetic tree was built using the nuclear sequences of numt 1 found in A. gazella, Z. californianus, E. jubatus, C. ursinus, and O. rosmarus together with homologous mtDNA sequences from all pinnipeds with an available mitochondrial reference genome (see Materials and Methods). The branch containing the nuclear sequences is indicated in red.
Table 3.
Summary Statistics of the Substitution Rate Ratio Models Implemented to Test for Functional Conservation of the ND3 Gene Located in numt 1
| Model | Site Class | Proportion | dN/dS (mitochondria) | dN/dS (NUMTs) | LogL |
|---|---|---|---|---|---|
| One-ratio | / | / | 0.01613 | −1828.804375 | |
| Two-ratio | / | / | 0.01105 | 0.99424 | −1811.746294 |
| Two-ratio (fixed) | / | / | 0.01105 | 1 | −1811.746326 |
| Clade C | 1 | 0.57666 | 0 | 0 | −1802.187057 |
| 2 | 0 | 1 | 1 | ||
| 3 | 0.42334 | 0.02947 | 2.7993 | ||
| Clade C (fixed) | 1 | 0.55786 | 0 | 0 | −1803.236605 |
| 2 | 0 | 1 | 1 | ||
| 3 | 0.44214 | 0.02856 | 1 | ||
| M1A | 1 | 0.97937 | 0.02063 | −1827.229958 | |
| 2 | 0.02063 | 1 | |||
| M22 | 1 | 0.90499 | 0.00806 | −1815.988374 | |
| 2 | 0 | 1 | |||
| 3 | 0.09501 | 0.13246 | |||
Note.—For each model, we report dN/dS ratios for mitochondrial and nuclear sequences separately, as well as the associated log likelihood value. Models “Clade C,” “Clade C (fixed),” “M1A,” and “M22” take into account heterogeneity of the dN/dS ratio across the protein sequence and the sites are therefore partitioned into three classes. Site class 1 corresponds to sites under purifying selection (dN/dS < 1), site class 2 corresponds to neutral sites (dN/dS = 1), and site class 3 corresponds to unrestricted sites.
To test this, we refined our model by accounting for heterogeneity in the dN/dS ratio across the protein sequence, which is a realistic assumption for mammalian proteins (Afanasyeva et al. 2018). Specifically, we assume that differences in the dN/dS ratio between the numt 1 genes and their mitochondrial counterparts can be attributed to differences in selection pressures on particular codons. A model that incorporates such differences (model Clade C) fits the data significantly better than branch models (P < 0.001, LRT, Clade C vs. two-ratio, table 3) as well as site models (P < 0.001, LRT, Clade C vs. M1a and LRT, Clade C vs. M22, P < 0.001, table 3), illustrating that part of the numt 1 coding sequences are evolving under strong purifying selection (site class 1, dN/dS = 0). Interestingly, the Clade C model estimates that positive selection also plays a role (dN/dS = 2.8 for class 2, table 3), although this is marginally nonsignificant when compared with a Clade C model that assumes site class 2 to be evolving under drift (P = 0.15, LRT, Clade C vs. Clade with fixed site class 2: dN/dS = 1, table 3).
Sequence Context of numt 1 in other Pinnipeds
To better understand the functional context of the genomic regions surrounding numt 1, we extract 150 kb of flanking sequences from each side of this numt from all five Otaroidea species where the numt is present (with the exception of the walrus, where only ∼110 kb of sequence is available on the 5′ side of the numt). Blastx searches against the available mammalian genome sequences in the NCBI nonredundant database reveal that in all five species, the genomic regions flanking numt 1 are enriched with retrotransposable elements (supplementary fig. S4, Supplementary Material online). Specifically, we retrieve several partial hits to the ORF2p gene of C. lupus (accession number: QOV08757.1) in four species. Top hits in the walrus are to the LORF gene of Crocuta crocuta (accession number: KAF0872869.1) and to the endonuclease/reverse transcriptase of Sus scrofa (accession number: ABR01162.1). Furthermore, the genomic regions surrounding numt 1 appear to have a very similar organization in at least four of the pinnipeds (supplementary fig. S4, Supplementary Material online), suggesting that these regions are homologous. This is harder to evaluate for the walrus due to the smaller number of retrieved annotations and the fact that these produce hits to different species.
Discussion
To shed light on the origin and evolution of nuclear copies of mitochondrial genes, we searched for the presence of mitochondrial sequences in the nuclear genome of the Antarctic fur seal, identifying a total of 25 numts. By focusing on mitonuclear insertions containing genes whose mitochondrial reading frames are intact, we identified one potentially functional numt and two mitonuclear fragments that represent good candidates for noncanonical sources of mitonuclear integrations. The first clearly represents an ancient insertion that we can characterize in homologous genomic regions across five divergent pinniped species and that appears to have been subject to strong purifying selection since the time of insertion. By contrast, the other two mitonuclear insertions represent recent integrations that likely originate from the integration of heteroplasmic mtDNA or as a result of hybridization.
Because their integration into eukaryotic cells, mitochondrial genomes have substantially reduced in size, as their functional genes were transferred into the nuclear genome (Bock and Timmis 2008). Our results are consistent with the notion that this is a continuous and still ongoing process (Ricchetti et al. 2004), given that we found evidence for numts having originated at different time points in the evolutionary history of pinnipeds. On the one hand, certain numts were present in all of the investigated species and may therefore be more than 25 Myr old, which is when pinnipeds diverged from the other arctoid carnivores (Deméré et al. 2003; Higdon et al. 2007). On the other hand, some of the numts were only found in the Antarctic fur seal, meaning that they were probably integrated into this species’ nuclear genome sometime during the past 5.2 Myr.
Nuclear copies of mitochondrial genes have been characterized in a variety of mammals (Ning et al. 2017; Schiavo et al. 2017). With its 25 numts comprising a total of 57 separate fragments, the Antarctic fur seal is a species characterized by a relatively low numt content, as the number of identified numts exceeds 100 in most of the species examined so far (Calabrese et al. 2017). Nevertheless, the numts that we identify collectively represent the entire A. gazella mitochondrial genome. In line with previous studies (Tsuji et al. 2012), we also found that different mtDNA regions are integrated to varying extents into the nuclear genome, with the D-loop region being the least represented.
We identified multiple numts spanning mitochondrial regions containing genes. However, based on the mitochondrial genetic code, most of them harbor premature stop codons and out of frame mutations, suggesting that they are ancient insertions. By contrast, fragments in numts 1, 2, 3, and 8 contain genes with intact mitochondrial reading frames. In numt 2 (only fragment 1) and 8, these are identical or nearly identical to their mitochondrial homologs and may represent very recent insertions that have not yet accumulated mutations. Given that the PCR amplification of these numts yielded a product of the expected size, we believe they are genuine recent insertions and not genome assembly artifacts. By contrast, the remaining genes in numt 1, 2 (only fragment 3), and 3 harbor substantially more substitutions and we therefore investigated them in greater detail.
Numt 1 is characterized by a surprisingly high synonymous substitution rate. Our phylogenetic analysis shows that this numt is an ancient insertion, which explains the high level of observed sequence divergence. Specifically, this numt appears to be present in homologous genomic regions across five pinnipeds belonging to the family Otaroidea. The ND3 gene in numt 1 contains an intact mitochondrial reading frame in all of these species and our substitution rate analyses reveals that it has undergone strong purifying selection since the time of insertion. Previous studies have shown that numts are unlikely to be transcribed (Bensasson et al. 2001). In line with this, we found that the ND3 gene in numt 1 contains premature stop codons as defined by the nuclear genetic code. It also appears doubtful that numts play a role in the generation of small mitochondrial RNAs (Pozzi and Dowling 2019), small noncoding RNAs of mitochondrial origin that have been suggested to exert a functional role, possibly via RNA interference (Cloonan 2015). Nonetheless, the fact that the ND3 gene in numt 1 deviates from expectations under selective neutrality suggests that it is likely to be somehow functional. Noutsos et al. (2007) showed that nuclear insertions of organellar DNA can create novel exons, but this was observed uniquely in plants. Moreover, such novel exons appeared to derive from markedly reshaped protein domains, which is not the case for numt 1. Thus, the most likely explanation for our findings is that numt 1 might have a function as regulatory element. An alternative but arguably less likely possibility could be that numt 1 inserted into a region of an ancestral Otaroidea genome characterized by a very low mutation rate, which would have prevented disruption of the mitochondrial reading frame. In this scenario, the large number of observed substitutions would have been caused by mutations in the mtDNA rather than in the numt itself. Such a numt would then represent a “genomic fossil,” which provides a picture of the sequence of the Otaroidea ND3 mitochondrial gene prior to the radiation of the Odobenidae and Otariidae. Regardless of the exact explanation for our findings, our study paves the way for future research aimed at understanding the functional role of numts.
Functional annotation of the flanking sequences of numt 1 across five Otaroid species shows that this numt is located in a genomic region rich in retrotransposable elements. Specifically, most of the hits we retrieved were to the ORF2p gene, an endonuclease of the LINE-1 retrotrasposon (Kines et al. 2016). This is not surprising as retrotransposable elements are frequently found in the flanking sequences of numts (Tsuzuki et al. 1983; Zullo et al. 1991; Tsuji et al. 2012). LINE-1 retrotransposons in particular are often overrepresented (Zullo et al. 1991; Tsuji et al. 2012) to the extent that Tsuji et al. (2012) speculated that the LINE-1 endonuclease might play a role in both retrotransposon and numt expansion.
Numts 2 and 3 appear to have resulted from more recent insertion events, as their sequences cluster closely to the A. gazella mitochondrial homologs in our phylogenetic reconstructions. Nevertheless, the genes in these numts show high dissimilarity to the homologous A. gazella mitochondrial sequence, which poses the question of which mtDNA lineage(s) they originated from. One possibility is that they could have originated from a mitochondrial sequence belonging to a closely related pinniped species that introgressed into A. gazella as a result of hybridization. In support of this hypothesis, it is known that Antarctic fur seals hybridize with A. tropicalis and, to a lesser extent, with males of A. forsteri (Lancaster et al. 2006). Under this scenario, it is possible that a recent mitonuclear insertion that was already present in A. tropicalis or A. forsteri introgressed into the A. gazella nuclear genome as a consequence of hybridization. Alternatively, introgressed mtDNA haplotypes might have been inserted into the A. gazella nuclear genome after hybridization. However, both hypotheses cannot yet be formally tested as mitochondrial reference genomes are currently lacking for A. tropicalis and A. forsteri. Alternatively, these numts may represent mitonuclear insertions of highly divergent A. gazella heteroplasmic mtDNA variants (Formenti et al. 2021; Parakatselaki and Ladoukakis 2021). These could potentially derive from the inheritance of paternal mtDNA that, despite occurring rarely, has been documented in humans (Wei et al. 2020) and other vertebrates (Kvist et al. 2003; Zhao et al. 2004). In support of both of these hypotheses, the genes in numts 2 and 3 appear to be under purifying selection, which probably reflects the fact that until recently they were expressed as part of the mitochondrial genome. Finally, we cannot exclude the possibility that numt 3 (but not numt 2) could be a genome assembly artifact given that the PCR product for this numt was not of the expected size.
In conclusion, by focusing on functionally intact genes in numts and by characterizing their synonymous and nonsynonymous substitution rates, we uncovered the presence of a potentially functional numt in the Antarctic fur seal genome. Future research is needed to ground truth the functionality of the nuclear version of the ND3 gene and to better understand how this functionality may be exerted. We also identified two mitonuclear insertions that do not appear to originate from the mitochondrial genome DNA of the Antarctic fur seal and which probably represent insertions from either heteroplasmic mtDNA variants or mtDNA introgressed via hybridization. Taken together, our findings reveal not only the promise of our approach for better understanding the origin and evolution of numts, but also the potential of mitonuclear integrations as genomic fossils to pinpoint historic mitochondrial variation.
Materials and Methods
Numt Discovery
We searched for numts using the methodology of Lammers et al. (2017). Briefly, the complete mitochondrial genome of the Antarctic fur seal (Nagel et al. 2019) (NCBI accession number: BK010918) was searched against the nuclear reference genome of the same species (Peart et al. 2021) (NCBI accession number: GCA_900642305.1) using BLAST (Camacho et al. 2009) with a word_size of 20 bp. Matching sequences were then filtered for length and only those longer than 200 bp were classified as putative numts and retained for further analysis. Multiple mitochondrial sequence fragments that mapped to within 10 kb of one another in the nuclear genome are clustered into a single numt. This was done to avoid artificially inflating the number of detected numts, as many of these fragments are likely to be part of the same mitonuclear insertions.
Experimental Validation
We attempted to experimentally validate the presence of all 25 numts in the Antarctic fur seal nuclear genome by designing PCR primers specific to the nuclear genome. Because many of these numts are very large, we did not attempt to PCR amplify full-length sequences. Instead, we designed one primer inside of the numt and the other primer either outside of the numt or overlapping the boundary between the numt and the adjacent nuclear genome. We used in silico PCR to check that none of the primer pairs would amplify a region of the mitochondrial genome. PCR amplification was consequently expected to yield a product only if both of the primers annealed to the nuclear genome. Primer design was implemented using default parameters in PRIMER3 (Untergasser et al. 2007).
Genomic DNA was extracted from two Antarctic fur seal individuals using a standard chloroform/isoamylalcohol extraction protocol (Sambrookand et al. 1989). PCR amplification with each primer pair was undertaken using a Type-it microsatellite PCR kit (QIAGEN) with a temperature profile of initial melting for 5 min at 95 °C, followed by 40 amplification cycles of 30 s at 95 °C, 30 s at 56 °C, and 50 s at 72 °C, and concluding with a elongation step of 30 min at 60 °C. An established primer pair targeting the MC1R gene was included as positive control (Peters et al. 2016). PCR products were visualized on a 2% agarose gel.
Phylogenetic Characterization of numts
In order to shed light on the timings of the identified mitonuclear integrations, we carried out blast searches of the numt sequences characterized in the Antarctic fur seal against the nuclear genomes of other pinniped species. Specifically, we focused on all ten species for which a nuclear reference genome is currently available (Z. californianus, GCF_009762345.2; E. jubatus, GCA_004028035.1; C. ursinus, GCA_003265705.1; O. rosmarus, GCA_000321225.1; Halichoerus grypus, GCA_012393455.1; Phoca vitulina, GCA_004348235.1; Leptonychotes weddellii, GCA_000349705.1; Mirounga angustirostris, https://www.dnazoo.org/assemblies/Mirounga_angustirostris; M. leonina, GCA_011800145.1; and Monachus schauinslandi, GCA_002201575.1). Blast searches were carried out separately for each species using blastn default parameters and an E-value cutoff of 0.01. For each numt, we recorded the percentage of query cover and the percentage of identity of the top hit to characterize its similarity to the corresponding A. gazella numt. This analysis was additionally repeated considering all numt fragments separately. This was done with the aim of verifying whether different mitonuclear fragments clustered within the same numt can be considered part of the same mitonuclear insertion.
Identification of Potential Protein-Coding Numts
Pairwise alignments between the sequences of 13 genes present in the A. gazella mitochondrial genome (Nagel et al. 2019) and each separate numt fragment were performed to identify numt fragments containing genes with intact mitochondrial reading frames. Gap-free alignments where the respective numt fragment sequence did not contain a premature stop codon were extracted. Pairwise nonsynonymous (dN) and synonymous (dS) substitution rates were then calculated using KaKs Calculator v2.0 (Wang et al. 2010) to assess the magnitude of divergence between the nuclear and mitochondrial homologs.
Comparative Substitution Rate Analysis
To better understand the evolutionary forces that shaped the observed sequence divergence, we carried out pairwise dN/dS analyses (Kryazhimskiy and Plotkin 2008) between the mitochondrial genes and their respective homologous in the relevant numt fragments. Specifically, dN/dS ratios close to one indicate the random fixation of mutations. By contrast, ratios below one are indicative of purifying selection, whereas ratios above one are suggestive of the presence of beneficial mutations.
Phylogenetic Analysis of numts Containing Potential Coding Genes
High dS values (>1) along with low dN values (<<1) could result from historic numt integrations. We therefore implemented phylogenetic analyses of numts showing this signature to better understand their possible origin(s). To do so, we first retrieved homologous numt sequences from other pinniped nuclear genomes using BLAST. Second, we identified homologous mitochondrial sequences from other pinniped species by BLASTing the A. gazella numt sequences against the complete mitochondrial genomes of 26 pinniped species (A. australis, MG023139.1; A. forsteri, NC_004023.1; A. townsendi, NC_008420.1; A. pusillus, NC_008417.1; C. ursinus, NC_008415.3; Cystophora cristata, NC_008427.1; Erignathus barbatus, NC_008426.1; E. jubatus, NC_004030.2; H. grypus, NC_001602.1; P. fasciata, NC_008428.1; P. groenlandica, KP942529.1; P. larga, NC_008430.1; P. vitulina, NC_001325.1; Hydrurga leptonyx, NC_008425.1; L. weddellii, NC_008424.1; Lobodon carcinophaga, NC_008423.1; M. leonina, NC_008422.1; M. monachus, NC_044972.1; M. schauinslandi, NC_008421.1; Neophoca cinerea, NC_008419.1; Phocarctos hookeri, NC_008418.1; Pusa caspica, NC_008431.1; P. hispida, NC_008433.1; P. sibirica, NC_008432.1; O. rosmarus, NC_004029.2; Z. californianus, NC_008416.1) as well as the dog (C. lupus familiaris, NC_002008.4) using a word_size of 20 and an E-value cutoff of 0.01. Next, we aligned the pinniped numts and the corresponding mitochondrial sequences of the Antarctic fur seal, the other 26 pinnipeds species and the dog. This was implemented using the MUSCLE algorithm in MEGA (Kumar et al. 2018). Finally, phylogenetic trees were constructed within MEGA as neighbour-joining trees using the Tajima-Nei model method, which allows for the mutation rate to be heterogeneous among different lineages (Nei and Kumar 2000).
Analysis of Functional Conservation of the ND3 Gene Located on numt 1
Genes located in an ancient mitonuclear insertion showing signatures of purifying selection might reveal evidence of functional conservation at the protein-coding level since their integration into the nuclear genome. We therefore investigated whether the gene located on numt 1, ND3, shows signals of such functional conservation across the Otaroidea. To do so, we divided the branches of the phylogeny constructed as described above into one set of branches forming the clade consisting of all numt sequences since the integration into the nuclear genome, and one set consisting of the rest of the tree (fig. 2, red and black branches, respectively). We then performed a series of substitution rate ratio model comparisons using codeml (Yang 2007), where different substitution rates on coding DNA and estimates of dS were used for nuclear and mitochondrial branches to account for the difference in mutation rates between the mitochondrial and the nuclear genome. We implemented a series of branch models (Yang 1998; Yang and Nielsen 1998). First, we assumed a model with a single dN/dS (“one-ratio model”) over the entire phylogeny. Second, we assumed free dN/dS for the two branch classes (“two-ratio model”) and third, we implemented a model where we fixed the dN/dS for the numt clade to 1, but kept the other class free (e.g. neutral evolution in the numt clade, “two-ratio model fixed”). Finally, we conducted likelihood ratio tests to check whether the models with varying dN/dS between the two branch classes (e.g. two-ratio models) provide a better fit to the data than the model with a single dN/dS or a model of neutral evolution. Next, to account for heterogeneity of selective pressures within the protein sequence, we implemented branch-site models, using the same branch classification as above (numt branches vs. the rest of the tree), but allowing three site classes (class 1: dN/dS < 1, class 2: dN/dS = 1, and site class 3: unrestricted). We implemented the Clade model C (Bielawski and Yang 2004), where only site class 2 can be different between the branch classes. The Clade C model was compared with site models M1A and M22 using LRTs, as suggested by the PAML user manual.
Annotation of numt 1 Flanking Sequences
The flanking sequences of numt 1, which is believed to represent an ancient insertion that underwent strong purifying selection, were extracted from all of the Otaroidea reference genomes and annotated. This had the double aim of checking whether this numt was present in homologous genomic regions and of functionally characterizing the DNA sequences surrounding it. Specifically, we extracted 150 kb of flanking sequences from each side of the numt and blasted these against the nonredundant NCBI database, whereas limiting our searches to mammals. The top hit of each search was then used for annotation.
Supplementary Material
Supplementary materials are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
This research was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) in the framework of a Sonderforschungsbereich (project numbers 316099922 and 396774617–TRR 212) and the priority program “Antarctic Research with Comparative Investigations in Arctic Ice Areas” (project number 424119118). It was also supported by core funding from the Natural Environment Research Council to the British Antarctic Survey’s Ecosystems Program. This work contributes to the Ecosystems project of the British Antarctic Survey, Natural Environmental Research Council, and is part of the Polar Science for Planet Earth Programme. Finally, we acknowledge support for the publication costs by the Open Access Publication Fund of Bielefeld University and the Deutsche Forschungsgemeinschaft (DFG).
Contributor Information
David L J Vendrami, Department of Animal Behaviour, Bielefeld University, 33501 Bielefeld, Germany.
Toni I Gossmann, Department of Animal Behaviour, Bielefeld University, 33501 Bielefeld, Germany.
Nayden Chakarov, Department of Animal Behaviour, Bielefeld University, 33501 Bielefeld, Germany.
Anneke J Paijmans, Department of Animal Behaviour, Bielefeld University, 33501 Bielefeld, Germany.
Vivienne Litzke, Department of Animal Behaviour, Bielefeld University, 33501 Bielefeld, Germany.
Adam Eyre-Walker, School of Life Science, University of Sussex, Brighton BN1 9QG, UK.
Jaume Forcada, British Antarctic Survey, High Cross, Madingley Road, Cambridge CB3 OET, UK.
Joseph I Hoffman, Department of Animal Behaviour, Bielefeld University, 33501 Bielefeld, Germany; British Antarctic Survey, High Cross, Madingley Road, Cambridge CB3 OET, UK.
Author Contributions
Conceived the study: J.I.H. and T.I.G. Sample collection and logistics: J.F. Laboratory work: N.C., A.J.P., and V.L. Analyzed data: D.L.J.V., T.G., and A.E.-W. Wrote the paper: D.L.J.V., T.G., and J.I.H. All of the authors commented upon and approved the final manuscript.
Data Availability
All of the data analyzed in this study are already publicly available. The characterized numt sequences have been deposited in figshare (https://figshare.com/articles/dataset/Antarctic_fur_seal_NUMTs_sequences_SPP_1158_/16399938, keyword: SPP 1158). The code relevant for this study is available at: https://github.com/DavidVendrami/Antarctic_fur_seal_numts.
Animal Ethics
Samples were collected as part of the Polar Science for Planet Earth program of the British Antarctic Survey under the authorization of the Senior Executive and the Environment Officers of the Government of South Georgia and the South Sandwich Islands. Samples were collected and retained under Scientific Research Permits for the British Antarctic Survey field activities on South Georgia, and in accordance with the Convention on International Trade in Endangered Species of Wild Fauna and Flora. All field procedures were approved by the British Antarctic Survey Animal Welfare and Ethics Review Body (reference no. PEA6).
Literature Cited
- Afanasyeva A, Bockwoldt M, Cooney CR, Heiland I, Gossmann TI. 2018. Human long intrinsically disordered protein regions are frequent targets of positive selection. Genome Res. 28:975–982. doi: 10.1101/gr.232645.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, De Queiroz A, Hedin M, Hayashi CY, Gatesy J. 2011. Nuclear-mitochondrial sequences as witnesses of past interbreeding and population diversity in the jumping bristletail mesomachilis. Mol Biol Evol. 28:195–210. doi: 10.1093/molbev/msq193 [DOI] [PubMed] [Google Scholar]
- Bensasson D, Zhang DX, Hartl DL, Hewitt GM. 2001. Mitochondrial pseudogenes: evolution’s misplaced witnesses. Trends Ecol Evol. 16:314–321. doi: 10.1016/S0169-5347(01)02151-6 [DOI] [PubMed] [Google Scholar]
- Bielawski JP, Yang Z. 2004. A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J Mol Evol. 59:121–132. doi: 10.1007/s00239-004-2597-8 [DOI] [PubMed] [Google Scholar]
- Bock R, Timmis JN. 2008. Reconstructing evolution: gene transfer from plastids to the nucleus. BioEssays 30:556–566. doi: 10.1002/bies.20761 [DOI] [PubMed] [Google Scholar]
- Borensztajn K, et al. 2002. Characterization of two novel splice site mutations in human factor VII gene causing severe plasma factor VII deficiency and bleeding diathesis. Br J Haematol. 117:168–171. doi: 10.1046/j.1365-2141.2002.03397.x [DOI] [PubMed] [Google Scholar]
- Calabrese FM, et al. 2017. NumtS colonization in mammalian genomes. Sci Rep. 7:1–10. doi: 10.1038/s41598-017-16750-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinform. 10:1–9. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N. 2015. Re-thinking miRNA–mRNA interactions: intertwining issues confound target discovery. BioEssays 37:379–388. doi: 10.1002/bies.201400191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deméré T, Berta A, Adam P. 2003. Chapter 3 pinnipedimorph evolutionary biogeography. Nat Hist. 279:32–76. http://sdnhm.org/research/paleontology/demere_2003.pdf. [Google Scholar]
- Formenti G, et al. 2021. Complete vertebrate mitogenomes reveal widespread repeats and gene duplications. Genome Biol. 22:1–22. doi: 10.1186/s13059-021-02336-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau ET, et al. 2020. Survey of mitochondrial sequences integrated into the bovine nuclear genome. Sci Rep. 10:1–11. doi: 10.1038/s41598-020-59155-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, Brors B. 2014. Circlize implements and enhances circular visualization in R. Bioinformatics 30:2811–2812. doi: 10.1093/bioinformatics/btu393 [DOI] [PubMed] [Google Scholar]
- Hazkani-Covo E, Zeller RM, Martin W. 2010. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet. 6:e1000834. doi: 10.1371/journal.pgen.1000834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higdon JW, Bininda-Emonds ORP, Beck RMD, Ferguson SH. 2007. Phylogeny and divergence of the pinnipeds (Carnivora: Mammalia) assessed using a multigene dataset. BMC Evol Biol. 7:1–19. doi: 10.1186/1471-2148-7-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humble E, et al. 2016. A draft fur seal genome provides insights into factors affecting SNP validation and how to mitigate them. Mol Ecol Resour. 16:909–921. doi: 10.1111/1755-0998.12502 [DOI] [PubMed] [Google Scholar]
- Humble E, et al. 2018. RAD sequencing and a hybrid Antarctic fur seal genome assembly reveal rapidly decaying linkage disequilibrium, global population structure and evidence for inbreeding. Genes, Genomes, Genetics 8:2709–2722. doi: 10.1534/g3.118.200171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kines KJ, et al. 2016. The endonuclease domain of the LINE-1 ORF2 protein can tolerate multiple mutations. Mob DNA 7:1–16. doi: 10.1186/s13100-016-0064-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S, Plotkin JB. 2008. The population genetics of dN/dS. PLoS Genet. 4:e1000304. doi: 10.1371/journal.pgen.1000304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist L, Martens J, Nazarenko AA, Orell M. 2003. Paternal leakage of mitochondrial DNA in the great tit (Parus major). Mol Biol Evol. 20:243–247. doi: 10.1093/molbev/msg025 [DOI] [PubMed] [Google Scholar]
- Lammers F, Janke A, Rücklé C, Zizka V, Nilsson MA. 2017. Screening for the ancient polar bear mitochondrial genome reveals low integration of mitochondrial pseudogenes (numts) in bears. Mitochondrial DNA Part B Resour. 2:251–254. doi: 10.1080/23802359.2017.1318673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster ML, Gemmell NJ, Negro S, Goldsworthy S, Sunnucks P. 2006. Ménage à trois on Macquarie Island: hybridization among three species of fur seal (Arctocephalus spp.) following historical population extinction. Mol Ecol. 15:3681–3692. doi: 10.1111/j.1365-294X.2006.03041.x [DOI] [PubMed] [Google Scholar]
- Nagel R, Forcada J, Hoffman JI. 2019. Complete mitochondrial genome of the Antarctic fur seal (Arctocephalus gazella). Mitochondrial DNA Part B Resour. 4:2936–2937. doi: 10.1080/23802359.2019.1662751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press. [Google Scholar]
- Ning FY, Fu J, Du ZH. 2017. Mitochondrial DNA insertions in the nuclear Capra hircus genome. Genet Mol Res. 16:1–8. doi: 10.4238/gmr16018266 [DOI] [PubMed] [Google Scholar]
- Noutsos C, Kleine T, Armbruster U, DalCorso G, Leister D. 2007. Nuclear insertions of organellar DNA can create novel patches of functional exon sequences. Trends Genet. 23:597–601. doi: 10.1016/j.tig.2007.08.016 [DOI] [PubMed] [Google Scholar]
- Parakatselaki ME, Ladoukakis ED. 2021. mtDNA heteroplasmy: origin, detection, significance, and evolutionary consequences. Life 11:633. doi: 10.3390/life11070633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart CR, et al. 2021. Hi-C scaffolded short- and long-read genome assemblies of the California sea lion are broadly consistent for syntenic inference across 45 million years of evolution. Mol Ecol Resour. 1–16. doi: 10.1111/1755-0998.13443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters L, et al. 2016. Born blonde: a recessive loss-of-function mutation in the melanocortin 1 receptor is associated with cream coat coloration in Antarctic fur seals. Ecol Evol. 6:5705–5717. doi: 10.1002/ece3.2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi A, Dowling DK. 2019. The genomic origins of small mitochondrial RNAs: are they transcribed by the mitochondrial DNA or by mitochondrial pseudogenes within the nucleus (NUMTs)?. Genome Biol. Evol 11:1883–1896. doi: 10.1093/gbe/evz132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricchetti M, Tekaia F, Dujon B. 2004. Continued colonization of the human genome by mitochondrial DNA. PLoS Biol. 2:e273. doi: 10.1371/journal.pbio.0020273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan L. 1967. On the origin of mitosing cells. J Theor Biol. 14:225–274. doi: 10.1016/0022-5193(67)90079-3 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. 1989, editors. Molecular cloning: a laboratory manual (No. Ed. 2): Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schiavo G, et al. 2017. A genomic landscape of mitochondrial DNA insertions in the pig nuclear genome provides evolutionary signatures of interspecies admixture. DNA Res. 24:487–498. doi: 10.1093/dnares/dsx019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji J, Frith MC, Tomii K, Horton P. 2012. Mammalian NUMT insertion is non-random. Nucleic Acids Res. 40:9073–9088. doi: 10.1093/nar/gks424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T, Nomiyama H, Setoyama C, Maeda S, Shimada K. 1983. Presence of mitochondrial-DNA-like sequences in the human nuclear DNA. Gene 25:223–229. doi: 10.1016/0378-1119(83)90226-3 [DOI] [PubMed] [Google Scholar]
- Turner C, et al. 2003. Human genetic disease caused by de novo mitochondrial-nuclear DNA transfer. Hum Genet. 112:303–309. doi: 10.1007/s00439-002-0892-2 [DOI] [PubMed] [Google Scholar]
- Untergasser A, et al. 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35:71–74. doi: 10.1093/nar/gkm306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. 2010. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom Proteom Bioinform. 8:77–80. doi: 10.1016/S1672-0229(10)60008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, et al. 2020. Nuclear-mitochondrial DNA segments resemble paternally inherited mitochondrial DNA in humans. Nat Commun. 11:1–11. doi: 10.1038/s41467-020-15336-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957 [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. doi: 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. 1998. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J Mol Evol. 46:409–418. doi: 10.1007/PL00006320 [DOI] [PubMed] [Google Scholar]
- Zhao X, et al. 2004. Further evidence for paternal inheritance of mitochondrial DNA in the sheep (Ovis aries). Heredity (Edinb) 93:399–403. doi: 10.1038/sj.hdy.6800516 [DOI] [PubMed] [Google Scholar]
- Zullo S, Sieu LC, Slightom JL, Hadler HI, Eisenstadt JM. 1991. Mitochondrial D-loop sequences are integrated in the rat nuclear genome. J Mol Biol. 221:1223–1235. doi: 10.1016/0022-2836(91)90930-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data analyzed in this study are already publicly available. The characterized numt sequences have been deposited in figshare (https://figshare.com/articles/dataset/Antarctic_fur_seal_NUMTs_sequences_SPP_1158_/16399938, keyword: SPP 1158). The code relevant for this study is available at: https://github.com/DavidVendrami/Antarctic_fur_seal_numts.