1. INTRODUCTION
Sarcoidosis is a systemic inflammatory disorder characterized by non-caseating granulomas with variable involvement of the lungs, central nervous system (CNS), skin, liver, bone, and eye (Drent et al., 2021). CNS disease, also known as neurosarcoid, is similarly heterogenous with diverse anatomical manifestations (Arun et al., 2020, Bathla et al., 2020, Kidd, 2018) and response to therapy (Bradshaw et al., 2021, Voortman et al., 2019). This diversity creates diagnostic challenges, especially since histopathologic confirmation is often difficult to obtain due to the invasiveness of a CNS biopsy. Furthermore, this heterogeneity has complicated prior attempts to identify shared aspects of pathophysiology across patients and to prescribe targeted therapy.
Previous work suggested that CD4 T cells play a role in sarcoidosis (Drent, Crouser, 2021). For example, lungs in sarcoidosis contain an expansion of T cell clones, consistent with an antigen-driven inflammatory response (Fu et al., 2017, Silver et al., 1996, Trentin et al., 1997). Furthermore, Lofgren’s syndrome, a subtype of sarcoidosis characterized by fever, erythema nodosa, arthritis, and hilar lymphadenopathy, is genetically linked to the MHC class II allele HLA-DRB1*03:01 (Grunewald and Eklund, 2009) and is associated with shared T cell receptor (TCR) sequences in bronchoalveolar lavage fluids across individuals (Greaves et al., 2021, Grunewald et al., 1994, Mitchell et al., 2017), suggesting a shared pathogenic antigen that is recognized by CD4 T cells. In support of this, recent work found these shared TCRs recognize a common peptide presented by HLA-DRB1*03:01 with strong homology to NAD-dependent histone deacetylase hst4 of Aspergillus nidulans (Greaves, Ravindran, 2021). Thus, pulmonary sarcoidosis appears driven by antigen-specific CD4 T cell responses, at least in a subset of patients.
It is unclear, however, if these findings in pulmonary sarcoidosis can be extrapolated to other organs, such as the CNS. In a prior study, we employed single-cell RNA-sequencing (scRNAseq) to evaluate the ocular cells from granulomatous uveitis – the clinical phenotype of sarcoid uveitis (Hassman et al., 2021). Pairing scRNAseq with TCR and B cell receptor (BCR) sequencing allowed us to identify T and B cell clonal expansion and infer the type of antigen driving each individual’s uveitis. In contrast to the findings of Lofgren’s syndrome, we found clonotypic expansion of CD4 T cells in some patients, but B and CD8 T cells in others (Hassman, Paley, 2021), raising the possibility that antigenic drivers of inflammation may vary across patients or tissues. We therefore sought to examine whether individuals with neurosarcoid contained a shared clonotypic expansion of T or B cells in the cerebrospinal fluid (CSF).
Here, we performed scRNAseq on paired CSF and blood samples from neurosarcoid and control participants. We complemented our scRNAseq data with TCR and B cell receptor (BCR) sequencing to identify tissue-specific, antigen-driven T or B cell responses as measured by clonal expansion. In contrast to prior studies in pulmonary disease (Greaves, Ravindran, 2021, Grunewald, Olerup, 1994, Mitchell, Kaiser, 2017), neurosarcoid participants had consistent clonal expansion of CD8 T cells that were enriched in the CSF. In addition, neurosarcoid blood samples showed a specific decline of CD8 T cell subsets that were enriched in the CSF, suggesting tissue-specific recruitment out of the blood to inflamed organs. Lastly, interferon response genes were the dominant cytokine gene signature, which paralleled higher IFNγ levels in the CSF of neurosarcoid participants.
2. METHODS
2.1. Participants Details
Paired samples of CSF and peripheral blood were obtained from study participants with a diagnosis of Probable Neurosarcoidosis, based on the 2018 Consensus Diagnostic Criteria for Neurosarcoidosis by the Neurosarcoidosis Consortium Consensus Group (Stern et al., 2018), or from participants who had a diagnostic work-up that identified an alternative etiology (non-neurosarcoid controls, i.e., participants NS010 and UV181). Active neurosarcoid or multiple sclerosis was defined by onset of new neurological signs or symptoms with corresponding findings on imaging that necessitated initiation or escalation of immunomodulatory therapy. Inactive neurosarcoid was defined as an absence of new signs or symptoms and no change in medical therapy. For flow cytometry experiments, peripheral blood samples were obtained from additional healthy participants who were recruited separately through the Volunteers for Health registry and were free of any autoimmune disease. For cytokine analysis, CSF samples from multiple sclerosis and normal pressure hydrocephalus were collected through the John L. Trotter MS Center, while CSF samples from idiopathic intracranial hypertension were collected through the Yokoyama Laboratory.
2.2. Sample Processing and Preservation:
CSF samples were centrifuged at 300g for 10 min. The CSF supernatant was removed and frozen at −80°C. Blood samples were obtained by venipuncture, collected into EDTA tubes, and centrifuged at 200g for 10 min. Plasma was removed, centrifuged at 1,000g for 10 min, and frozen at −80°C. Peripheral blood mononuclear cells (PBMCs) were purified via Ficoll-Hypaque density gradient centrifugation. The CSF cell pellet and PBMCs were cryopreserved in FBS with 10% DMSO and stored at −140°C.
2.3. Library preparation, single-cell 5′, and TCR/BCR sequencing:
Frozen CSF cells were thawed and washed once with FBS and once with PBS with 0.1% BSA. Due to low cell numbers (5 of 8 samples had <60,000 cells), minimal processing was performed to reduce cell loss. PBMCs were thawed and washed twice with 10% RPMI and then once with PBS with 0.1% BSA. Viability was >95% by Trypan Blue exclusion. Single-Cell 5’ Gene Expression cDNA libraries were generated per the 10x Genomics Chromium Single-Cell 5′ Library and Gel Bead Kit v1 and the 10x Chromium Controller (10x Genomics, Pleasanton, CA) platform for microdroplet-based, single-cell barcoding, by the Genome Technology Access Center at the McDonnell Genome Institute (GTAC@MGI, Washington University in St. Louis). T cell and B cell enrichment libraries were generated per the Chromium Single-Cell V(D)J Enrichment Kits for Human T cell and Human B cell (10x Genomics) using the same input samples. All libraries were sequenced at the GTAC@MGI on the NovaSeq Sequencing System (Illumina, San Diego, CA).
2.4. Single-cell RNA expression analysis:
Sequencing reads were aligned to the human genome using Cell Ranger v3.1.0 (10x Genomics). The publicly available Seurat R software package version 3.2.3 (Stuart et al., 2019) was used for downstream analysis. Cells that had more than 11% of mitochondrial gene content or less than 200 detectable genes were excluded from analysis. Doublets were identified using DoubletFinder v2.0.3 (McGinnis et al., 2019) and excluded from analysis. Samples were normalized using SCTransform (Hafemeister and Satija, 2019). The SCT-transformed data was integrated with 3000 anchor features and PrepSCTIntegration. Dimensionality reduction was done with runPCA using default settings and runUMAP with the top 30 calculated dimensions. Clustering was performed with a resolution of 2.5. Cells were initially assigned to a leukocyte lineage (CD4 T cell, CD8 T cell, B cell, Myeloid cell, or NK cell) based on canonical gene expression of the entire cluster (Fig S1A), as previously described (Hassman, Paley, 2021). Clusters with genes from red blood cells (e.g., HBA1, HBA2, HBB), platelets (e.g., PPBP, TUBB1, PF4), or multiple cell types (e.g., CD3D & CD79A) were labelled as “Other.” This clustering resolution of 2.5 divided CD8 T cells into 10 separate clusters (labelled CD8T_0 through CD8T_9), which were used for downstream analysis. Differential gene expression utilized the Wilcoxon Rank Sum test on normalized and scaled RNA count data. For differential gene expression across clusters, subclusters, or tissue-enriched clonotypes, FindAllMarkers function in the Seurat package was employed with a log-fold change threshold >0.25, minimum group percentage = 10%, and the pseudocount = 1. Gene set enrichment analysis was performed using the escape R package (v1.3.1) (Borcherding et al., 2021). Gene sets were derived from the Hallmark library of the Molecular Signature Database (v7.0).
2.4.1. Pseudotime Analysis:
The Monocle2 package within publicly available Seurat Wrappers packages was used for the calculation of pseudotime of CD8 T cells (Qiu et al., 2017, Trapnell et al., 2014). Raw RNA counts, gene names, and metadata were extracted from the Seurat object containing CD8 T cell clusters and used to generate a CellDataSet for Monocle2. Monocle2 was used to estimate size factors, estimate dispersions, perform tSNE dimensionality reduction, and calculate a dispersion table for pseudotime trajectories.
2.4.2. VDJ analysis:
The Gini coefficient was calculated with the publicly available reldist R software package (Handcock and Morris, 1999). T and B cell clonotypes were defined by Cell Ranger. Assignment of CD4 or CD8 T cell lineage to T cell clonotypes was based on whether the clonotype had a greater number of cells in the CD4 or CD8 T cell clusters, respectively. Evenly split clonotypes were labelled as “double positive” and not used for downstream calculations. To compare CD4 and CD8 T cell clonotype frequency across tissues, clonotypes were defined by having the same TRAV and TRBV gene segment usage with the same amino acid sequence for CDR3a and CDR3b. Clonotypes were assigned on the integrated Seurat object. To compare CD8 T cell clonotypes with TCR sequences of known specificity, we downloaded human TRA and TRB paired sequences from a curated TCR database, (VDJdb, vdjdb.cdr3.net) (Bagaev et al., 2020). TCRs were matched if they had the same TRAV and TRBV gene segment usage with the same amino acid sequence for CDR3a and CDR3b.
2.5.1. Flow cytometry:
Reagents used for flow cytometry are listed in Table S1. Frozen PBMCs were thawed and washed twice in RPMI with 10% FBS to remove residual DMSO. Cells were washed with PBS and stained with Live/Dead (ThermoFisher, Waltham, MA) for 20 minutes at room temperature. Cells were then washed with PBS containing 2% FBS and stained with surface antibodies. Cells were washed with PBS containing 2% FBS and stained with streptavidin-PE-Cy5 conjugate. Data were collected on a BD AriaFusion (BD Biosciences) and analyzed with FlowJo v10 (Tree Star, Ashland, OR, USA). To exclude the influence of invariant T cells on the analysis, mucosal-associated invariant T cells and invariant natural killer T cells were excluded by selecting on Vα7.2 and Vα24 negative cells.
2.5.2. Cytokine Analysis:
Cytokine measurements were performed by the CHiiPs core facility (Washington University in St. Louis) using the Human Luminex Discovery Assay (R&D Systems, Minneapolis, MN) per manufacturer’s instructions.
2.5.3. HLA Typing:
HLA genotyping was performed by Histogenetics LLC (Osining, NY).
2.6. Statistical Analysis:
Statistical testing for flow cytometry (2-way ANOVA followed by Sidak’s multiple comparisons test) and cytokine concentrations (Ratio paired t test; Brown-Forsythe and Welch ANOVA followed by Dunnett’s T3 multiple comparisons test) were performed using Prism 8 (GraphPad, San Diego, CA).
2.7. Data and Code Availability:
Aligned sequencing data is available at figshare.com (https://doi.org/10.6084/m9.figshare.c.5761613). Code will be available at https://github.com/RobersonLab/.
2.8. Study approval:
All human participants were enrolled in accordance with Declaration of Helsinki principles and the institutional review board of Washington University in St. Louis (IRB 201704141, 201912043, and 201104379). Written informed consent was received from participants prior to inclusion in the study.
3. RESULTS
3.1. The majority of cerebrospinal fluid cells in neurosarcoid are CD4 and CD8 T cells
To define the cellular composition and clonality of cerebrospinal fluid (CSF) cells in neurosarcoid, we isolated paired CSF and peripheral blood mononuclear cells (PBMCs) from five participants with active neurosarcoid. Three participants (NS001, NS008, & NS016) had a new diagnosis of neurosarcoid, while two (NS006 & NS015) had a diagnosis of at least 2 years prior to sampling. Four participants were not on immunosuppressive therapy while the fifth (NS015) was on a combination of low dose corticosteroids and TNF inhibitor therapy. Longitudinal sampling was performed on one participant (NS001) during active disease (NS001–1) and three months later, after control of disease with weekly intravenous corticosteroids (NS001–2). Paired CSF and PBMCs from two participants were also obtained and served as controls after a neurologic evaluation led to a diagnosis of Parkinson’s dementia (NS010) and birdshot chorioretinopathy (UV181) (Table 1).
Table 1.
Clinical features of neurosarcoid and control participants.
| Group | Neurosarcoid | Control | ||||||
|---|---|---|---|---|---|---|---|---|
| Participant | NS001–1 | NS001–2 | NS006 | NS008 | NS015 | NS016 | NS010 | UV181 |
| Diagnosis | Probable | Probable | Probable | Probable | Probable | PD | BSCR | |
| Time of Sampling after Initial Diagnosis | new diagnosis | 3 months | 16 years | new diagnosis | 2 years | new diagnosis | new diagnosis | new diagnosis |
| Symptom Duration | 1 year | N/A | 1 month | 9 months | 2 weeks | 9 years | 3 months | 4 months |
| Disease Activity | Active | Inactive | Active | Active | Active | Active | N/A | N/A |
| Age (y) | 60 | 48 | 54 | 35 | 37 | 69 | 39 | |
| Sex | F | M | M | F | M | F | F | |
| Immunosuppressive Medications | None | MP 1g IV weekly | None | None | IFX 5mg/kg Q6weeks PDN 10mg daily | None | None | None |
| Duration of Current Immunosuppression | N/A | MP: 2 months | N/A | N/A | IFX: 6 months PDN: 1 week | N/A | N/A | N/A |
| CSF Protein (mg/dL) | 304 | 65 | 81 | 60 | 36 | 56 | 21 | 36 |
| CSF Glucose (mg/dL) | 40 | 58 | 48 | 38 | 48 | 63 | 58 | 62 |
| CSF Nucleated Cells (#/uL) | 194 | 7 | 9 | 14 | 34 | 9 | 44 | 1 |
| CSF Lymphocytes (%) | 92 | n.d. | 93 | 97 | 95 | 94 | 95 | n.d. |
| CSF Monocytes (%) | 7 | n.d. | 7 | 3 | 4 | 6 | 5 | n.d. |
| CSF PMN (%) | 1 | n.d. | 0 | 0 | 1 | 0 | 0 | n.d. |
| CSF RBC (#/uL) | 0 | 113 | 4 | 0 | 53 | 9 | 0 | 0 |
PD, Parkinson’s Disease; BSCR, Birdshot chorioretinitis (i.e., isolated uveitis); MP, methyprednisolone; IFX, infliximab; PDN, prednisone; PMN, polymorphonuclear cells; RBC, red blood cells; n.d., not done
We performed single-cell RNA sequencing (scRNAseq) on the above eight paired CSF-PBMC samples (Fig 1 A & B). Using expression of lineage-defining genes, we identified the five major leukocyte lineages [CD4 T cells, CD8 T cells, myeloid cells, natural killer (NK) cells, and B cells] in the blood and CSF in a combined analysis of neurosarcoid cases and controls (Fig 1B, S1A). For both neurosarcoid cases and controls, the predominant cell type in the CSF was CD4 T cells (45–60%), followed by CD8 T cells, myeloid cells, NK cells, and B cells (Fig 1C). In this cohort, the CD4:CD8 T cell ratio in the CSF ranged from 1.9 to 2.6 (Fig S1B). We also observed a high variance in the concentration of CSF cells ranging from 1 to 194 cells per ul of CSF (Table 1), which led to parallel variability in the concentration of the five major immune cells (Fig 1D). Thus, neurosarcoid CSF appears to contain consistent proportions of leukocyte lineages across patients.
Figure 1. Leukocyte lineage proportions are consistent and T cell predominant across neurosarcoid and control participants.
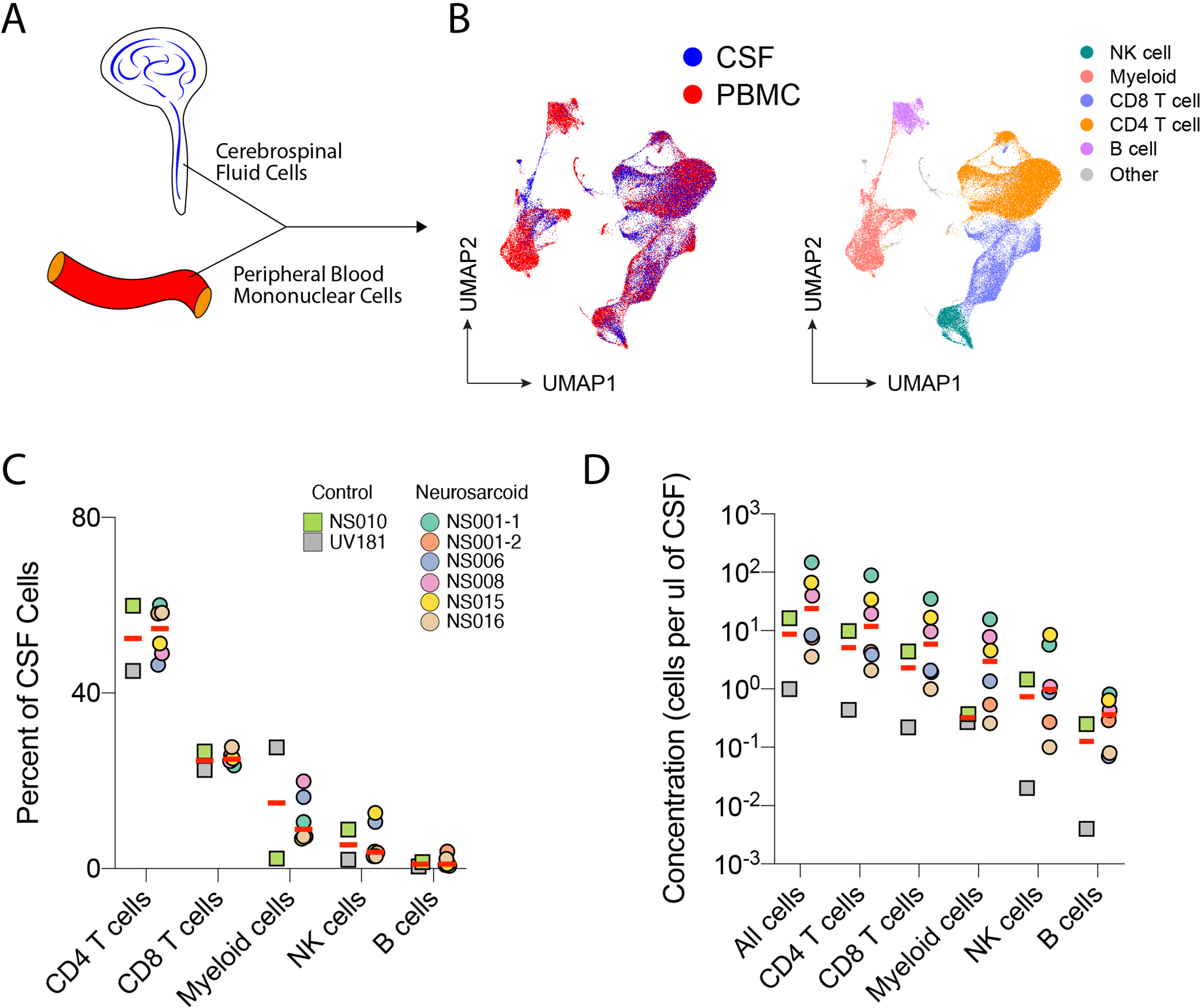
(A) Cells were collected from the CSF and Blood for scRNAseq. (B) Uniform Manifold Approximation and Projection (UMAP) plot of all samples colored by tissue source (left) and major cell lineage (right). (C) Frequency and (D) concentration of major immune cell lineages in the CSF for neurosarcoid and control participants. Clinical diagnosis for controls were Parkinson’s dementia for NS010 and uveitis for UV181.
3.2. Clonal expansion and CSF-enrichment are seen in CD8 T cell clonotypes in neurosarcoid
We previously reported that the proportion of distinct immune lineages in granulomatous disease may not reflect the degree of antigen-specific responses, as measured by clonal expansion (Hassman, Paley, 2021). We therefore sought to define the clonality of T and B cells in the CSF for neurosarcoid and controls. We performed TCR and BCR sequencing of the paired CSF and PBMC samples in order to define T and B cell clonotypes, i.e., sets of clonal progeny from a common ancestor. We first used the Gini coefficient to measure the diversity and clonality of T and B cells, with 0 representing no clonal expansion and 1 representing a monoclonal population. While two neurosarcoid participants had some CD4 T cell clonality with a Gini coefficient of 0.13–0.23, the remaining samples had a Gini coefficient of ~0.05, indicating minimal clonality of CD4 T cells in neurosarcoid (Fig 2A). By contrast, all neurosarcoid samples contained CD8 T cell clonal expansion (Gini coefficient 0.2–0.5). We did not detect any significant clonal expansion of B cells in neurosarcoid samples. In the control samples, the participant with isolated uveitis, UV181, had minimal CD4 or CD8 T cell clonality (Fig 2A). While the subject with Parkinson’s dementia, NS010, had evidence of CD8 T cell clonal expansion (Gini 0.19), this is consistent with previous reports of Parkinson’s disease that identified clonal CSF T cells, some of which may recognize α-synuclein (Cebrian et al., 2014, Gate et al., 2020, Sulzer et al., 2017, Wang et al., 2021). Thus, compared to CD4 T cells and B cells, CD8 T cells may have the greatest degree of clonal expansion in neurosarcoid CSF.
Figure 2. Clonal expansion and CSF-enrichment are seen in CD8 T cells in neurosarcoid.
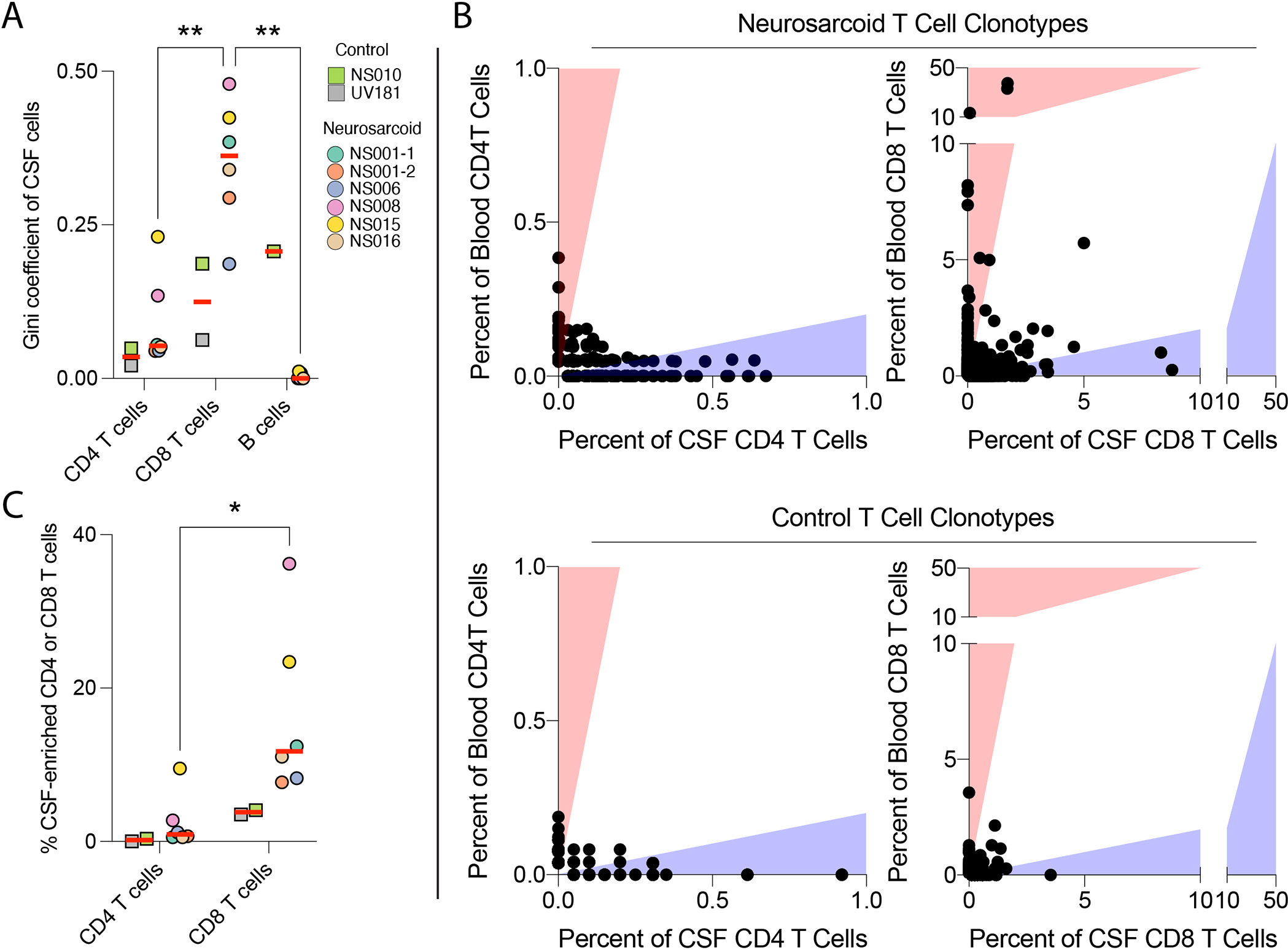
(A) Diversity and clonality of indicated lymphocyte lineages within the CSF as measured by the Gini coefficient, where on a scale from 0 to 1, 0 indicates all clonotypes have the same frequency and 1 indicates a monoclonal population. (B) Frequency of CD4 (left) and CD8 (right) T cell clonotypes in the blood and CSF for neurosarcoid (top) and control (bottom) participants. Each dot represents a single clonotype defined by V-gene segments and CDR3 amino-acid sequences for both alpha and beta chains. Red and blue shading indicates clonotypes with a 5-fold increase in frequency in one tissue over the other, either blood (red) or CSF (blue). (C) Percent of CSF-enriched CD4 or CD8 T cells for neurosarcoid and control participants. A CSF-enriched cell has a clonotype with at least 5 cells in the CSF and a 5-fold increased frequency in the CSF over the blood. * p < 0.05; ** p < 0.01, 2-way ANOVA followed by Sidak’s multiple comparisons test.
Similar to the CSF, CD8 T cells in the blood exhibited increased clonal expansion compared to CD4 T cells and B cells in neurosarcoid patients (Fig S2A). To determine whether the T cell clonal expansion was specific to the CSF or overlapped with blood clonal expansion, we compared the frequency of specific CD4 and CD8 T cell clonotypes in the blood and CSF for each individual. There were multiple expanded CD8 T cell clonotypes in neurosarcoid where the frequency in the CSF was 5x greater than the frequency in the blood, with some clonotypes accounting for 8–9% of all CSF CD8 T cell clonotypes in those individuals, suggesting CSF-specific clonal expansion (Fig 2B). However, we found minimal overlap of CD8 TCR sequences between participants (both neurosarcoid and controls), although we did observe a 15–30% overlap in the TCR repertoire from longitudinal CSF samples from NS001 (Fig S2B). In contrast, while a few CD4 T cell clonotypes were also specifically expanded in neurosarcoid CSF, they were not expanded to the same degree as CD8 T cells (Fig 2B). Furthermore, in longitudinal analysis of participant NS001, we found only a 3–8% overlap of the CSF CD4 TCR repertoire in these samples (Fig S2B). Similar to CD8 T cells, we did not observe shared TCRs across individuals. Thus, the clonal expansion of CD8 T cells in the CSF appears distinct from expanded blood clonotypes, suggesting CSF clonotypes may be driven by tissue-specific antigens with subsequent retention in the CNS despite treatment with immunosuppression and clinical remission.
We next evaluated the frequency of all CSF-enriched CD4 and CD8 T cells for neurosarcoid and control samples. We defined “CSF-enrichment” based on two criteria: (1) a 5-fold increase in frequency of the CSF over the blood and (2) an expansion of 5 cells or more in the CSF, as previously described (Gate, Saligrama, 2020). By that metric, we found 8–36% of CD8 T cells were CSF-enriched in neurosarcoid (Fig 2C), whereas a lower proportion (0.5–10%) of CD4 T cells were CSF-enriched. In control samples, only 3–4% of CD8 T cells and 0–0.5% of CD4 T cells were CSF-enriched (Fig 2C). Thus, the CSF-enrichment of CD8 T cells suggests an antigen-specific role in patrolling the CNS during neurosarcoid.
We investigated whether the CSF-enriched TCRs in neurosarcoid matched previously described TCRs with known cognate antigens. To accomplish this, we utilized VDJdb, which is a curated database of TCR sequences with known antigen specificities (Bagaev, Vroomans, 2020). Out of the 70 CSF-enriched TCRs, we found a single, clonally expanded TCR sequence from participant NS006 that mapped to the database (Table S2). This TCR recognizes an Epstein Barr Virus (EBV) epitope (FLRGRAYGL from EBNA3A) in the context of HLA-B*08:01 as well as two self-peptides (EEYLKAWTF from MLANA and EEYLQAFTY from ABCD3) in the context of HLA-B*44:05 (Table S3). Of note, NS006 has HLA-B*08:01, but not HLA-B*44:05 (Table S4). Thus, at least some CSF-enriched CD8 T cells are specific for viral epitopes.
3.3. CSF-enriched CD8 T cell clonotypes have a shared transcriptional signature
To determine the nature of the CSF-enriched CD8 T cell clonotypes, we performed a transcriptional analysis of all samples and divided the CD8 T cells into 10 clusters (CD8T_0 though CD8T_9) (Fig 3A). Compared to controls, neurosarcoid CSF samples demonstrated numerically higher frequencies of clusters CD8T_3 and CD8T_4, although this did not reach statistical significance (Fig S3A). CSF-enriched CD8 T cell clonotypes mapped to two regions, which we labelled Set A and Set B. Set A contained CSF-enriched CD8 T cell clonotypes from all samples and could be found within neighboring clusters CD8T_3 and CD8T_4, suggesting a shared transcriptional program (Fig 3B, S3B). Set B, however, mainly came from participant NS015 and mapped to cluster CD8T_9 (Fig 3B, S3B). Notably, NS015 was the only one with active disease while on immunosuppressive therapy (infliximab and prednisone). Thus, while CSF-enriched CD8 T cells may express a shared transcriptional program in neurosarcoid, immunosuppressive medications or patient-specific features may contribute to different proportions of Set A and B clonotypes.
Figure 3. CSF-enriched CD8 T cell clonotypes have a shared transcriptional signature.
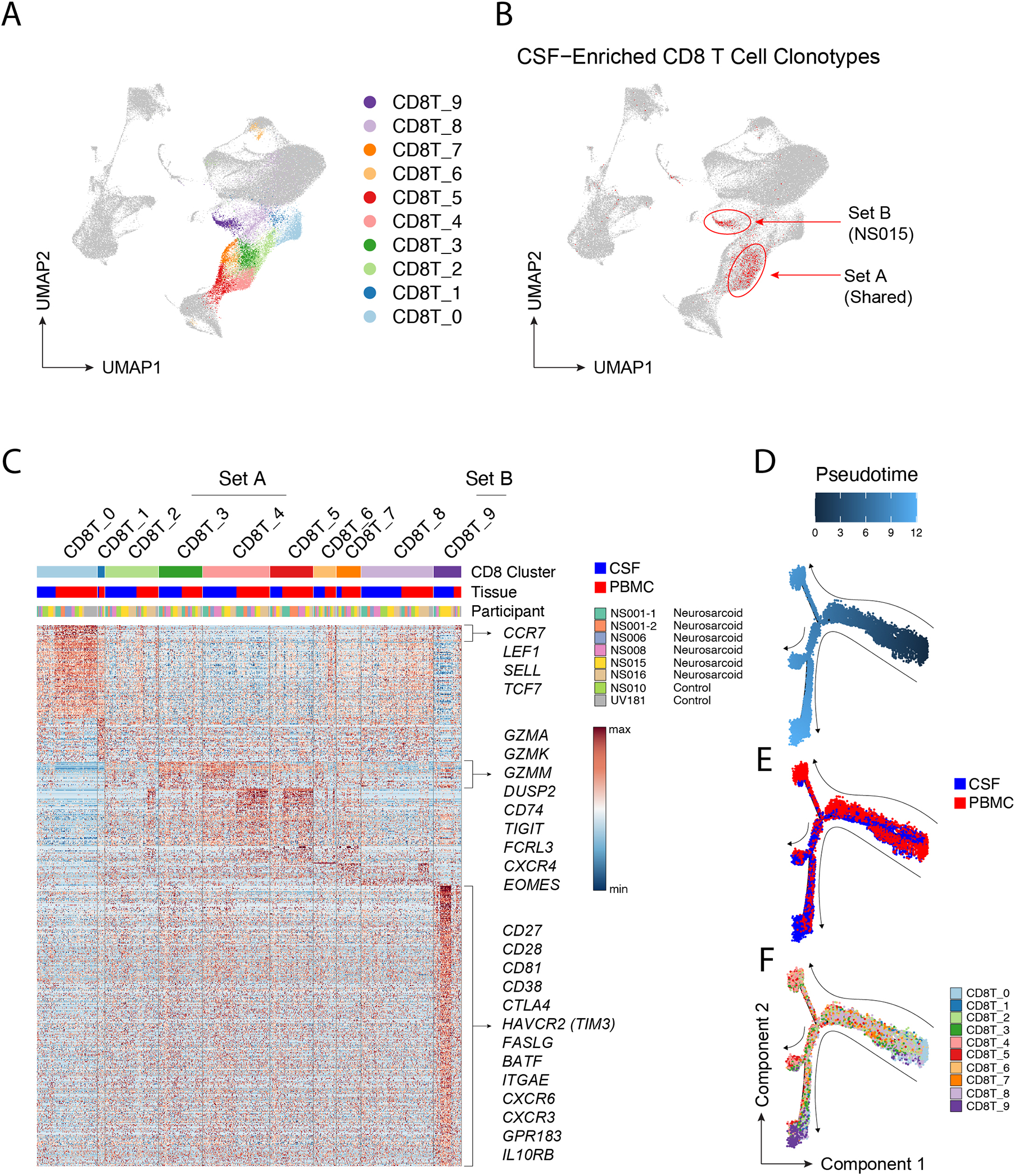
(A) Ten CD8 T cell clusters are highlighted on UMAP visualization. (B) Mapping of CSF-enriched CD8 T cell clonotypes (highlighted in red) to clusters 3, 4 and 9 on UMAP. Whether the CSF-enriched CD8 T cell clonotypes are from multiple participants (Set A) or predominantly a single participant (NS015; Set B) is annotated. (C) Heatmap of genes upregulated in each CD8 T cell cluster. Each column is cell annotated by cluster, tissue, and participant. Genes upregulated in CD8 T cells from the CSF in clusters 3, 4, and 9 are annotated. (D-F) Lineage trajectory of CD8 T cells colored by (D) pseudotime, (E) tissue, or (F) CD8 cluster.
To determine the transcriptional program of CSF-enriched CD8 T cells, we next defined the differentially expressed genes (DEGs) amongst the CD8 T cell clusters. CD8T_0 contained largely naïve T cells as evidenced by the high expression of CCR7, LEF1, SELL, & TCF7 (Fig 3C). CSF CD8 T cells in Set A clusters (CD8T_3 and CD8T_4) expressed higher levels of granzymes (GZMA, GZMK, & GZMM), chemokine receptors (CD74 and CXCR4), the inhibitory receptor TIGIT, and the transcription factor EOMES. Unlike their counterparts in the CSF, blood cells in Set A clusters, which were expanded in neurosarcoid samples (Fig S3A), had high expression of NKG7, GZMB, PRF1, and GNLY. This is consistent with an expansion of cytotoxic CD8 T effector-memory CD45RA+ cells in pulmonary sarcoidosis, as previously reported (Parasa et al., 2018).
In contrast to CSF cells in Set A clusters, CD8 T cells in the Set B cluster CD8T_9, which was mainly composed of cells from the CSF (Fig 3C), expressed higher levels of activation co-receptors (CD27 and CD28), chemokine receptors (CXCR3 and CXCR6), the oxysterol receptor GPR183 (EBI2), inhibitory receptors (CTLA4 and HAVCR2), and the transcription factor BATF. The co-expression of CD103 (ITGAE) and CXCR6 in the CD8T_9 cluster raises the possibility of tissue-resident CD8 T cells in the CSF in neurosarcoid, similar to other tissues in health and disease (Sasson et al., 2020).
Previous reports have suggested a linear CD8 T cell differentiation process from Granzyme K+ (GZMK+) intermediate CD8 T cells to Granzyme B+ (GZMB+) terminally differentiated CD8 T cells in response to viral infections or other antigenic stimuli (Mogilenko et al., 2021). As GZMK expression was upregulated in CSF CD8 T cells in neurosarcoid, we investigated whether our dataset would also suggest a lineage relationship between blood and CSF CD8 T cells. Similar to prior reports (Mogilenko, Shpynov, 2021), we generated a pseudotime analysis that takes cross-sectional scRNAseq data and calculates a putative differentiation path. We found that instead of a linear differentiation path, our pseudotime analysis generated a branching process from one shared path into two major branches (Fig 3D). These major branches were predominantly, albeit not completely, composed of either CSF or blood cells (Fig 3E). Without pre-specifying the root of differentiation, naïve cells in CD8T_0 were calculated to have a pseudotime of 0 (Fig 3F). One major branch was composed of blood cells from clusters CD8T_2 through CD8T_5, while the second major branch contained mostly CSF cells from clusters CD8T_2 through CD8T_9, with most cells from CD8T_9 accumulating at the terminal end of this projected differentiation (Fig 3F). These results suggest that, in the context of neurosarcoid, CSF CD8 T cells progress down a pathophysiologic process that is distinct from traditional antiviral immunity found in the blood.
3.4. Circulating CSF-enriched CD8 T cell clonotypes express CD27, EBI2, and GZMK
To identify the gene expression that marked CSF-enriched CD8 T cell clonotypes in the blood, we first defined CSF- and PBMCs-enriched clonotypes as a 5-fold increase in frequency of one tissue over the other and a clonal expansion of at least 5 cells. We next identified the DEGs for CSF- and PBMC-enriched clonotypes on cells in the blood (Fig 4A). We found that CD27 and GZMK were upregulated in CSF-enriched clonotypes regardless of the tissue from which they were collected (Fig 4B). GPR183 (EBI2) was also upregulated in CSF-enriched clonotypes, although this was not seen in all cells, either due to inefficient detection of expression or limited expression to a subset of cells. In contrast, PBMC-enriched clonotypes had upregulated expression of KLRD1 (CD94), HOPX, GPR56, GZMB, and GNLY regardless of the tissue from which the cells were collected (Fig 4B). CX3CR1 was also upregulated in PBMC-enriched clonotypes, but only in the blood. We next evaluated whether the expression of DEGs from tissue-enriched clonotypes would reflect the putative branching of CD8 T cell differentiation from our pseudotime analysis. We found that CD27, EBI2, and GZMK expression was most concentrated in the “southward” branch of CSF CD8 T cells, with KLRD1, HOPX, GPR56, CX3CR1, GZMB, and GNLY concentrated in the “northward” branch of blood CD8 T cells (Fig S4). Thus, the DEGs from tissue-specific clonotypes likely reflect differences in the differentiation of CD8 T cells, as indicated by the pseudotime analysis.
Figure 4. CSF-enriched CD8 T cell clonotypes in the blood express CD27, EBI2, and Granzyme K (GZMK).
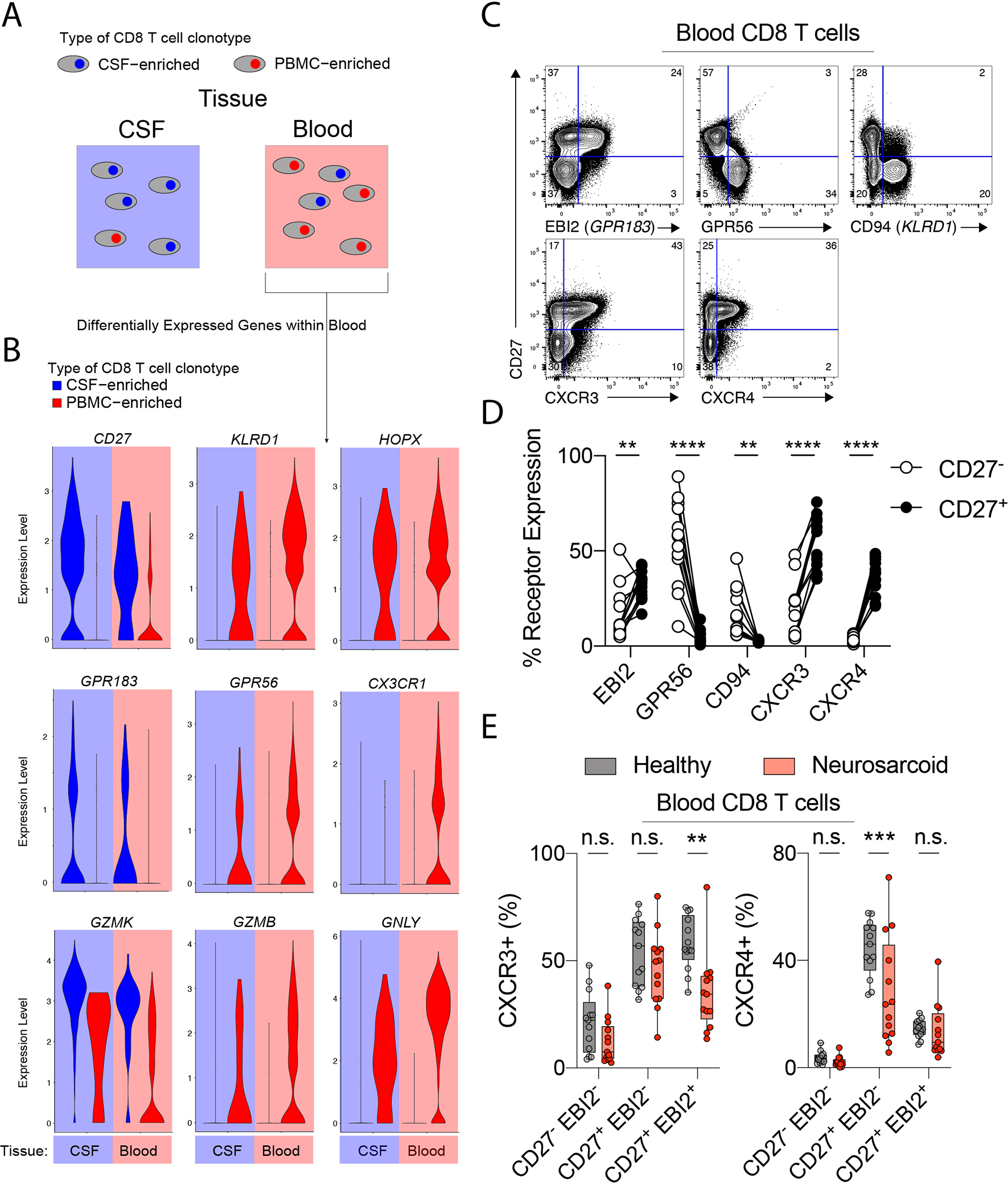
(A) CSF- and PBMC-enriched CD8 T cell clonotypes within the CSF and blood were identified as in Figure 2. Differential gene expression was then calculated for CSF- and PBMC-enriched clonotypes within the blood. (B) Violin plots of indicated differentially expressed genes for CSF-enriched (blue fill) and PBMC-enriched (red fill) CD8 T cell clonotypes with the CSF (blue background) and blood (red background). (C) Flow cytometry of CD8 T cells from a healthy donor. CD8 T cells are TCRab+CD8a+CD8b+CD4−CD19−Va7.2−Va24−. (D) Quantification of expression of indicated receptors on CD27+ (solid) and CD27− (clear) CD8 T cells from healthy donors (n=13). (E) Expression of CXCR3 and CXCR4 on indicated CD27/EBI2 CD8 T cell populations for neurosarcoid and healthy participants. Boxes show 25th to 75th percentiles. ** p < 0.01; *** p < 0.001; **** p < 0.0001, 2-way ANOVA followed by Sidak’s multiple comparisons test.
We next determined whether the DEGs from tissue-enriched clonotypes were differentially expressed at the protein level by flow cytometry. Analysis of PBMCs from healthy donors demonstrated high expression of EBI2 in CD27+ but not CD27− CD8 T cells, while GPR56 and CD94 were highly expressed in CD27− CD8 T cells (Fig 4C, 4D). Additionally, the chemokine receptors upregulated in CSF CD8 T cells, CXCR3 and CXCR4, both showed high expression in CD27+ CD8 T cells (Fig 4C, 4D). As the expression of GPR183 (EBI2) was associated with Set B clonotypes (CD8T_9) (Fig 3C), we asked whether the expression of CXCR4 and CXCR3, which were associated with Set A and Set B CSF cells, respectively (Fig 3C), were altered on EBI2+ and EBI2− CD8 T cells in neurosarcoid. We found that, in neurosarcoid participants, CXCR3 had lower expression on CD27+EBI2+ CD8 T cells, but not other subpopulations (Fig 4E). By contrast, CXCR4 expression was lower in CD27+EBI2− CD8 T cells, but not CD27+EBI2+ CD8 T cells, in neurosarcoid participants (Fig 4E). Thus, distinct subpopulations of CD8 T cells may be recruited to sites of inflammation in neurosarcoid via parallel mechanisms.
3.5. Interferon-stimulated genes are a core signature in CSF CD8 T cells during active neurosarcoid.
We next sought to determine a core transcriptional signature of CSF CD8 T cells in neurosarcoid. We performed single-cell gene set enrichment analysis of several cytokine signatures on the CD8 T cells from the blood and CSF in both control and neurosarcoid samples. We found that IFNα and IFNγ response signatures were enriched in CSF neurosarcoid CD8 T cells compared to CSF control CD8 T cells (Fig 5A). As our dataset contained the capacity to make multiple parallel comparisons, neurosarcoid vs control, CSF vs blood, and active vs inactive disease, we determined a core set of DEGs to define CSF CD8 T cells in active neurosarcoid. We identified 12 genes that were upregulated in (1) neurosarcoid vs control CSF CD8 T cells, (2) CSF vs blood neurosarcoid non-naïve CD8 T cells, and (3) active vs inactive neurosarcoid CSF CD8 T cells within the longitudinal collection of a single participant, NS001 (Fig 5B). Of these 12 genes, eight were interferon-stimulated genes (ISGs). Thus, interferon signaling appears to elicit a core transcriptional signature in CSF CD8 T cells in neurosarcoid.
Figure 5. Interferon stimulated genes are a core signature in CSF CD8 T cells during active neurosarcoid.
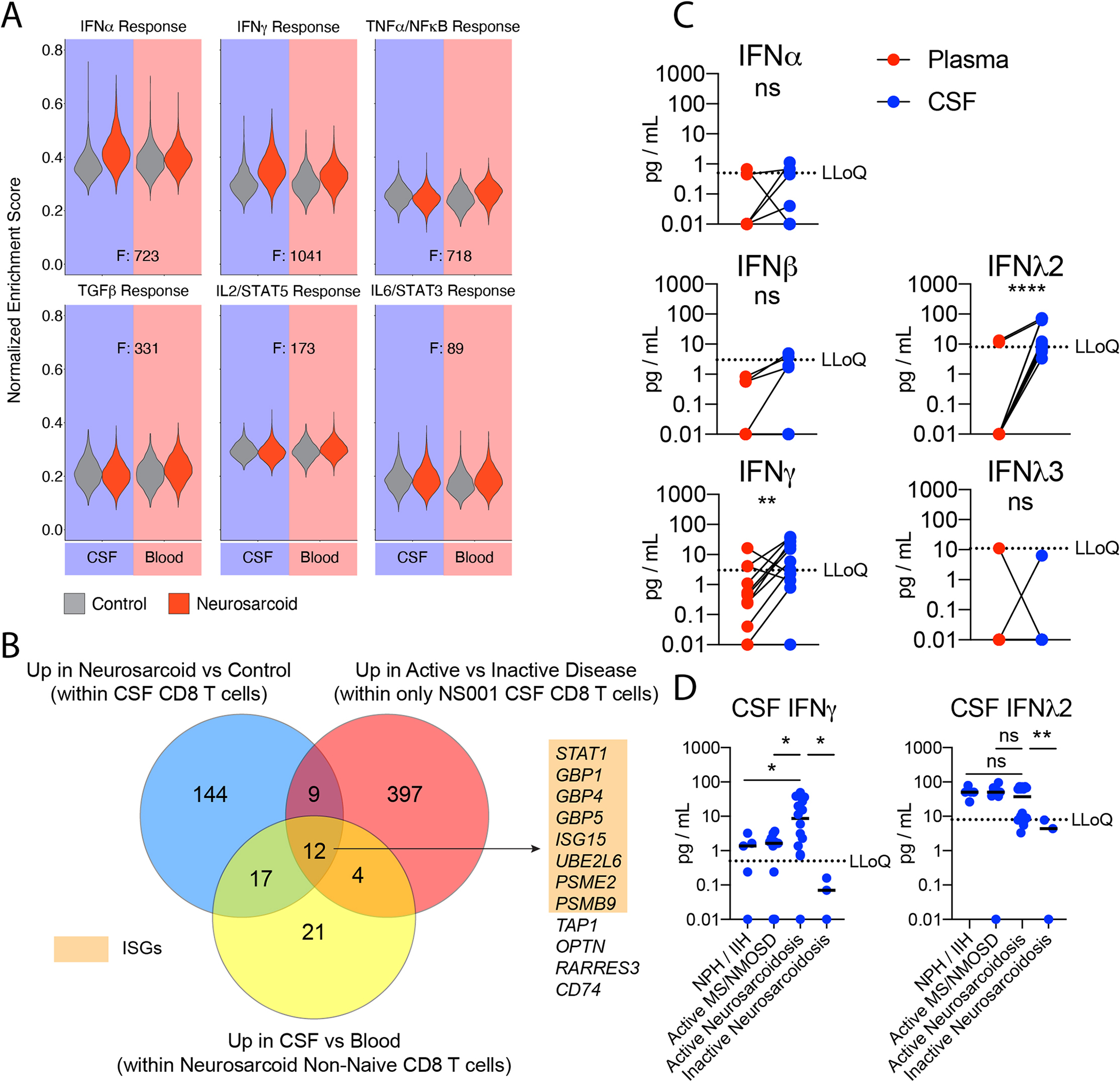
(A) Violin plots of Normalized Enrichment Scores in CD8 T cells for indicated Hallmark gene sets from MSigDB. Each plot compares control (gray) and neurosarcoid (red) for CSF (left) and blood (right). F values as a measure of variance between tissue-groups is provided. (B) Venn diagram depicting the overlap of three sets of differentially expressed genes: (blue) upregulated genes within CSF CD8 T cells in neurosarcoid vs control participants, (yellow) upregulated genes within neurosarcoid non-naive CD8 T cells in CSF vs blood, and (red) upregulated genes within CSF CD8 T cells from a single participant (NS001) during active vs inactive disease. Numbers represent the quantity of genes within each region. Genes within the central overlapping region are listed. Interferon-stimulated genes (ISGs) are highlighted in orange. (C) Quantification of indicated interferons (IFN) in the Blood (red) and CSF (blue) of active Neurosarcoidosis (n=13). (Ratio paired t test). (D) Quantification of indicated IFN in the CSF of normal pressure hydrocephalus (NPH), idiopathic intracranial hypertension (IIH), active multiple sclerosis (MS) or neuromyelitis optica spectrum disorder (NMOSD), or active or inactive neurosarcoidosis. (Brown-Forsythe and Welch ANOVA followed by Dunnett’s T3 multiple comparisons test). LLoQ, lower limit of quantification. * p < 0.05; ** p < 0.01; **** p < 0.0001; ns p > 0.05
As multiple types of interferons can activate a shared set of ISGs, we sought to examine whether specific interferons were upregulated in the CSF of neurosarcoid. We examined the concentration of Type 1, 2, and 3 interferons in the CSF and blood on neurosarcoid participants. We found higher levels of both IFNγ and IFNλ2 in the CSF (Fig 5C). To examine whether the higher concentration of IFNγ and IFNλ2 was specific to neurosarcoid, we compared the CSF levels in (1) noninflammatory disease (normal pressure hydrocephalus and idiopathic intracranial hypertension), (2) active demyelinating disease (multiple sclerosis and neuromyelitis spectrum disorder), (3) active neurosarcoid and (4) inactive neurosarcoid. We found higher concentrations of IFNγ in active neurosarcoid CSF compared to uninflamed diseases, demyelinating disease, and inactive neurosarcoid (Fig 5D). In contrast, IFNλ2 was not elevated in active neurosarcoid compared to uninflamed and demyelinating diseases (Fig 5D). Furthermore, IFNγ but not IFNλ2 levels correlated with soluble indicators of neurosarcoid disease, e.g., TNFα, soluble CD25, and IL-6 (Fig S5) (Otto et al., 2020). Thus, IFNγ is likely the major cytokine driving the IFN-signature in neurosarcoid.
4. DISCUSSION
This is the first report to determine the clonality of T and B cells enriched in the CSF in neurosarcoid patients. Unlike investigations into pulmonary sarcoidosis, we observed a consistent clonal expansion of CD8 T cells, but not CD4 T cells, in the CSF. We therefore defined the transcriptional profile of two subtypes of clonally expanded CD8 T cells in the CNS, tracked CSF-enriched CD8 T cell clonotypes in the blood, and tested their relevance in neurosarcoid participants compared to healthy controls using flow cytometry. Finally, we provide evidence that IFNγ is likely a major bioactive cytokine in neurosarcoid compared to other neurologic disorders.
Prior studies of human CD8 T cells have identified clonal expansion in the CSF in both healthy and Alzheimer’s disease participants (Gate, Saligrama, 2020, Pappalardo et al., 2020). The transcriptional signature of those CD8 T cells, however, were more similar to PBMC-enriched than CSF-enriched clonotypes in neurosarcoid, expressing higher levels of HOPX as well as NKG7, CCL4, CCL5, which have been previously associated with antiviral CD8 T effector memory CD45RA+ cells (Gate, Saligrama, 2020, Willinger et al., 2005). By contrast, clonally expanded CD8 T cells from multiple sclerosis are more similar to CSF-enriched clonotypes in neurosarcoid, with high expression of GZMA, GZMK, CD74, and EOMES (Pappalardo, Zhang, 2020). Unlike neurosarcoid CSF, which was devoid of B cells, CSF from multiple sclerosis contains clonally expanded B cells undergoing somatic hypermutation, suggesting a strong component of an antigen-driven B cell response (Ramesh et al., 2020). Accordingly, the presence of oligoclonal bands in the CSF can help discriminate between neurosarcoid and multiple sclerosis (Arun, Pattison, 2020, Heming et al., 2020). Thus, coupling an analysis of T and B cell clonal expansion to gene expression may help distinguish pathophysiologic processes of CNS disorders.
The pseudotime analysis with Monocle suggested that GZMK+ CSF-enriched CD8 T cell clonotypes undergo a distinct differentiation process compared to GZMB+ PBMC-enriched CD8 T cell clonotypes. Granzyme K+ CD8 T cells have also been seen in rheumatoid arthritis (Stephenson et al., 2018, Zhang et al., 2019), non-infectious uveitis (Hassman, Paley, 2021), and inflammation associated with aging (Mogilenko, Shpynov, 2021). The expression of granzyme K in CD8 T cells inversely correlates with cytotoxicity against viral antigens (Harari et al., 2009), but granzyme K can potentiate the inflammatory functions of non-immune cells (Mogilenko, Shpynov, 2021). Additionally, prior studies have reported that granzyme K enables T cell entry into tissues such as the CNS via transendothelial diapedesis (Herich et al., 2019). Thus, granzyme K+ CD8 T cells may have a pathogenic role in multiple inflammatory disorders, via mechanisms other than perforin-mediated cytotoxicity.
EBI2 (GPR183) is an oxysterol G-protein-coupled receptor that is critical for T and B cell migration and localization (Baptista et al., 2019, Li et al., 2016, Liu et al., 2011, Pereira et al., 2009). In our analysis, EBI2 was upregulated in CSF-enriched CD8 T cell clonotypes irrespective of their location in the blood or CSF, which suggested that EBI2 may be an important receptor in the pathogenic potential of CD8 T cells in neurosarcoid. In line with this hypothesis, in multiple sclerosis, EBI2 is found on CD4 T cells within inflammatory CNS lesions, blood CD4 T cells excluded from the CNS after natalizumab treatment, and clonally expanded B cells in the CSF (Clottu et al., 2017, Ramesh, Schubert, 2020, Wanke et al., 2017). Additionally, in experimental autoimmune encephalitis, EBI2 is expressed by pathogenic CD4 T cells in the CNS and is required for T-cell-driven disease (Tischner et al., 2017, Wanke, Moos, 2017). Thus, EBI2 may be a shared molecule to identify pathogenic T and B cells in inflammatory CNS disorders.
In addition to EBI2, we identified two other G protein-coupled receptors expressed on CSF CD8 T cells, CXCR3 and CXCR4. Moreover, we found lower frequencies of CD8 T cells expressing either CXCR3 or CXCR4 in the blood in neurosarcoid participants, consistent with recruitment of these CD8 T cell populations out of the blood and into the CNS. Prior studies of pulmonary sarcoid have reported an increase in the CXCR3 ligands CXCL9 and CXCL11 (and possibly CXCL10) within the bronchoalveolar lavage fluid (BALF) and a concurrent upregulation of CXCR3 on multiple immune cells, including T cells, in both the BALF and granulomas (Busuttil et al., 2009, Dyskova et al., 2015, Miyara et al., 2006). Expression of CXCR4 and its ligand CXCL12, by contrast, were not enriched in BALF cells from sarcoid patients compared to controls (Dyskova, Fillerova, 2015). Thus, CNS immune cell recruitment may share mechanisms with other organs affected by sarcoidosis but may also utilize tissue-specific chemokines.
In our dataset, the strongest gene signature for active neurosarcoid came from interferon-stimulated genes, and we provide evidence that this is likely driven by IFNγ. Moreover, IFNγ induces expression of the chemokines, CXCL9, CXCL10, and CXCL11, which – as noted above – are ligands for CXCR3, which is expressed by CSF-enriched CD8 T cells. Elevated IFNγ transcripts and protein have also been found in both BALF and blood in sarcoidosis (Dyskova, Fillerova, 2015, Gharib et al., 2016, Prior and Haslam, 1991). In addition, IFNγ levels are predictive of a response to corticosteroid therapy (Gharib, Malur, 2016). These findings suggests that IFNγ-driven JAK/STAT signaling may play a central pathogenic role across multiple tissues in sarcoidosis. In support of this, multiple case reports or small case series have described the successful treatment of cutaneous, pulmonary, or multiorgan sarcoidosis with the JAK inhibitor tofacitinib or baricitinib (Alam et al., 2021, Damsky et al., 2018, Damsky et al., 2020, Friedman et al., 2021, Kerkemeyer et al., 2021, Scheinberg et al., 2020, Singh et al., 2021, Talty et al., 2021). Thus, JAK inhibitors may offer an additional treatment option to the current set of therapeutics by targeting IFN-signaling.
One limitation of this study is the small sample size used for scRNAseq. As a result, we could not deconvolute the impact of different classes of immunosuppression, such as corticosteroid and TNF inhibition. Larger cohorts will need to be examined in future studies to determine generalizability of the findings presented here. In addition, our analysis focused on the CSF and not the granulomas within the CNS tissue. Due to the invasive nature of CNS tissue sampling, we anticipate that long-term recruitment will be required to accumulate sufficient neurosarcoid biospecimens to test the clonality of T and B cells within the CNS parenchyma. Despite this limitation, we note that shared CD4 T cell clonality was readily detected in the BALF in pulmonary sarcoid without direct evaluation of granulomas (Greaves, Ravindran, 2021). Finally, although our control samples for scRNAseq were not obtained from healthy participants as in other reports (Pappalardo, Zhang, 2020), we were able to test our scRNAseq findings via flow cytometry of healthy and neurosarcoid blood samples and via CSF cytokine measurements of several CNS disorders.
5. CONCLUSIONS
In summary, this initial evaluation of T and B cell antigen receptor sequencing in neurosarcoid revealed that, unlike pulmonary sarcoid, CNS disease may be associated with a tissue-specific CD8 T cell clonal expansion. Furthermore, IFNγ appears to be bioactive in the CSF and may drive an interferon signature in CD8 T cells. This work provides the foundation for larger and longitudinal studies to define the antigenic triggers of pathogenic T cells, establish predictive biomarkers for response to therapy, and identify novel treatment targets.
Supplementary Material
6. ACKNOWLEDGEMENTS
The authors thank the study participants for their generous and enthusiastic participation. We thank Gregory F. Wu, Anne H. Cross, Laura Piccio, and Dana C. Perantie for support with sample collection and constructive comments. We thank Naresha Saligrama and Tarin Bigley for manuscript review and critical feedback. MAP was supported by the Rheumatology Research Foundation and the Arthritis National Research Foundation. SRD was supported by the NeuroNEXT Network. CC was supported by the National Multiple Sclerosis Foundation. LMH was supported by an unrestricted grant from Research to Prevent Blindness. Funding for this project was provided by the Barnes-Jewish Hospital Foundation (WMY), a grant from the McDonnell Center for Cellular and Molecular Neurobiology (WMY), the Arthritis National Research Foundation (MAP), and the Washington University Rheumatic Diseases Research Resource-based Center (EDOR; P30-AR073752). The funding organizations had no role in the design or conduct of this research.
Footnotes
Declaration of Interest:
MAP receives research support from Lilly paid to the institution and received consultant fees from AbbVie, JK Market Research, and Priovant Therapeutics. The authors have no other declarations of interest.
10. REFERENCES
- Alam M, Fang V, Rosenbach M. Treatment of cutaneous sarcoidosis with tofacitinib 2% ointment and extra virgin olive oil. JAAD Case Rep. 2021;9:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun T, Pattison L, Palace J. Distinguishing neurosarcoidosis from multiple sclerosis based on CSF analysis: A retrospective study. Neurology. 2020;94:e2545–e54. [DOI] [PubMed] [Google Scholar]
- Bagaev DV, Vroomans RMA, Samir J, Stervbo U, Rius C, Dolton G, et al. VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res. 2020;48:D1057–D62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista AP, Gola A, Huang Y, Milanez-Almeida P, Torabi-Parizi P, Urban JF, Jr., et al. The Chemoattractant Receptor Ebi2 Drives Intranodal Naive CD4(+) T Cell Peripheralization to Promote Effective Adaptive Immunity. Immunity. 2019;50:1188–201 e6. [DOI] [PubMed] [Google Scholar]
- Bathla G, Freeman CW, Moritani T, Song JW, Srivastava S, Soni N, et al. Retrospective, dual-centre review of imaging findings in neurosarcoidosis at presentation: prevalence and imaging sub-types. Clin Radiol. 2020;75:796 e1–e9. [DOI] [PubMed] [Google Scholar]
- Borcherding N, Vishwakarma A, Voigt AP, Bellizzi A, Kaplan J, Nepple K, et al. Mapping the immune environment in clear cell renal carcinoma by single-cell genomics. Commun Biol. 2021;4:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw MJ, Pawate S, Koth LL, Cho TA, Gelfand JM. Neurosarcoidosis: Pathophysiology, Diagnosis, and Treatment. Neurol Neuroimmunol Neuroinflamm. 2021;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busuttil A, Weigt SS, Keane MP, Xue YY, Palchevskiy V, Burdick MD, et al. CXCR3 ligands are augmented during the pathogenesis of pulmonary sarcoidosis. Eur Respir J. 2009;34:676–86. [DOI] [PubMed] [Google Scholar]
- Casiraghi C, Dorovini-Zis K, Horwitz MS. Epstein-Barr virus infection of human brain microvessel endothelial cells: a novel role in multiple sclerosis. J Neuroimmunol. 2011;230:173–7. [DOI] [PubMed] [Google Scholar]
- Cebrian C, Zucca FA, Mauri P, Steinbeck JA, Studer L, Scherzer CR, et al. MHC-I expression renders catecholaminergic neurons susceptible to T-cell-mediated degeneration. Nat Commun. 2014;5:3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clottu AS, Mathias A, Sailer AW, Schluep M, Seebach JD, Du Pasquier R, et al. EBI2 Expression and Function: Robust in Memory Lymphocytes and Increased by Natalizumab in Multiple Sclerosis. Cell Rep. 2017;18:213–24. [DOI] [PubMed] [Google Scholar]
- Damsky W, Thakral D, Emeagwali N, Galan A, King B. Tofacitinib Treatment and Molecular Analysis of Cutaneous Sarcoidosis. N Engl J Med. 2018;379:2540–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damsky W, Young BD, Sloan B, Miller EJ, Obando JA, King B. Treatment of Multiorgan Sarcoidosis With Tofacitinib. ACR Open Rheumatol. 2020;2:106–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drent M, Crouser ED, Grunewald J. Challenges of Sarcoidosis and Its Management. N Engl J Med. 2021;385:1018–32. [DOI] [PubMed] [Google Scholar]
- Dyskova T, Fillerova R, Novosad T, Kudelka M, Zurkova M, Gajdos P, et al. Correlation Network Analysis Reveals Relationships between MicroRNAs, Transcription Factor T-bet, and Deregulated Cytokine/Chemokine-Receptor Network in Pulmonary Sarcoidosis. Mediators Inflamm. 2015;2015:121378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman MA, Le B, Stevens J, Desmarais J, Seifer D, Ogle K, et al. Tofacitinib as a Steroid-Sparing Therapy in Pulmonary Sarcoidosis, an Open-Label Prospective Proof-of-Concept Study. Lung. 2021;199:147–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Li Y, Xu L, Liu S, Wang M, Xiao L, et al. Immunology repertoire study of pulmonary sarcoidosis T cells in CD4+, CD8+ PBMC and tissue. Oncotarget. 2017;8:89515–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gate D, Saligrama N, Leventhal O, Yang AC, Unger MS, Middeldorp J, et al. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature. 2020;577:399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharib SA, Malur A, Huizar I, Barna BP, Kavuru MS, Schnapp LM, et al. Sarcoidosis activates diverse transcriptional programs in bronchoalveolar lavage cells. Respir Res. 2016;17:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves SA, Ravindran A, Santos RG, Chen L, Falta MT, Wang Y, et al. CD4+ T cells in the lungs of acute sarcoidosis patients recognize an Aspergillus nidulans epitope. J Exp Med. 2021;218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald J, Eklund A. Lofgren’s syndrome: human leukocyte antigen strongly influences the disease course. Am J Respir Crit Care Med. 2009;179:307–12. [DOI] [PubMed] [Google Scholar]
- Grunewald J, Olerup O, Persson U, Ohrn MB, Wigzell H, Eklund A. T-cell receptor variable region gene usage by CD4+ and CD8+ T cells in bronchoalveolar lavage fluid and peripheral blood of sarcoidosis patients. Proc Natl Acad Sci U S A. 1994;91:4965–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handcock MS, Morris M. Relative distribution methods in the social sciences. New York: Springer; 1999. [Google Scholar]
- Harari A, Bellutti Enders F, Cellerai C, Bart PA, Pantaleo G. Distinct profiles of cytotoxic granules in memory CD8 T cells correlate with function, differentiation stage, and antigen exposure. J Virol. 2009;83:2862–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani A, Corboy JR, Al-Salam S, Khan G. Epstein-Barr virus is present in the brain of most cases of multiple sclerosis and may engage more than just B cells. PLoS One. 2018;13:e0192109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassman LM, Paley MA, Esaulova E, Paley GL, Ruzycki PA, Laurent J, et al. Clinico-molecular identification of conserved and individualized features of granulomatous uveitis. Ophthalmol Sci. 2021;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heming M, Lohmann L, Schulte-Mecklenbeck A, Brix T, Gross CC, Wiendl H, et al. Leukocyte profiles in blood and CSF distinguish neurosarcoidosis from multiple sclerosis. J Neuroimmunol. 2020;341:577171. [DOI] [PubMed] [Google Scholar]
- Herich S, Schneider-Hohendorf T, Rohlmann A, Khaleghi Ghadiri M, Schulte-Mecklenbeck A, Zondler L, et al. Human CCR5high effector memory cells perform CNS parenchymal immune surveillance via GZMK-mediated transendothelial diapedesis. Brain. 2019;142:3411–27. [DOI] [PubMed] [Google Scholar]
- Kerkemeyer KL, Meah N, Sinclair RD. Tofacitinib for cutaneous and pulmonary sarcoidosis: A case series. J Am Acad Dermatol. 2021;84:581–3. [DOI] [PubMed] [Google Scholar]
- Kidd DP. Sarcoidosis of the central nervous system: clinical features, imaging, and CSF results. J Neurol. 2018;265:1906–15. [DOI] [PubMed] [Google Scholar]
- Li J, Lu E, Yi T, Cyster JG. EBI2 augments Tfh cell fate by promoting interaction with IL-2-quenching dendritic cells. Nature. 2016;533:110–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Yang XV, Wu J, Kuei C, Mani NS, Zhang L, et al. Oxysterols direct B-cell migration through EBI2. Nature. 2011;475:519–23. [DOI] [PubMed] [Google Scholar]
- McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019;8:329–37 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AM, Kaiser Y, Falta MT, Munson DJ, Landry LG, Eklund A, et al. Shared alphabeta TCR Usage in Lungs of Sarcoidosis Patients with Lofgren’s Syndrome. J Immunol. 2017;199:2279–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyara M, Amoura Z, Parizot C, Badoual C, Dorgham K, Trad S, et al. The immune paradox of sarcoidosis and regulatory T cells. J Exp Med. 2006;203:359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilenko DA, Shpynov O, Andhey PS, Arthur L, Swain A, Esaulova E, et al. Comprehensive Profiling of an Aging Immune System Reveals Clonal GZMK(+) CD8(+) T Cells as Conserved Hallmark of Inflammaging. Immunity. 2021;54:99–115 e12. [DOI] [PubMed] [Google Scholar]
- Otto C, Wengert O, Unterwalder N, Meisel C, Ruprecht K. Analysis of soluble interleukin-2 receptor as CSF biomarker for neurosarcoidosis. Neurol Neuroimmunol Neuroinflamm. 2020;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappalardo JL, Zhang L, Pecsok MK, Perlman K, Zografou C, Raddassi K, et al. Transcriptomic and clonal characterization of T cells in the human central nervous system. Sci Immunol. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasa VR, Forsslund H, Enger T, Lorenz D, Kullberg S, Eklund A, et al. Enhanced CD8(+) cytolytic T cell responses in the peripheral circulation of patients with sarcoidosis and non-Lofgren’s disease. Respir Med. 2018;138S:S38–S44. [DOI] [PubMed] [Google Scholar]
- Pereira JP, Kelly LM, Xu Y, Cyster JG. EBI2 mediates B cell segregation between the outer and centre follicle. Nature. 2009;460:1122–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior C, Haslam PL. Increased levels of serum interferon-gamma in pulmonary sarcoidosis and relationship with response to corticosteroid therapy. Am Rev Respir Dis. 1991;143:53–60. [DOI] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14:309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh A, Schubert RD, Greenfield AL, Dandekar R, Loudermilk R, Sabatino JJ, Jr., et al. A pathogenic and clonally expanded B cell transcriptome in active multiple sclerosis. Proc Natl Acad Sci U S A. 2020;117:22932–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasson SC, Gordon CL, Christo SN, Klenerman P, Mackay LK. Local heroes or villains: tissue-resident memory T cells in human health and disease. Cell Mol Immunol. 2020;17:113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinberg M, Maluf F, Wagner J. Steroid-resistant sarcoidosis treated with baricitinib. Ann Rheum Dis. 2020;79:1259–60. [DOI] [PubMed] [Google Scholar]
- Serafini B, Rosicarelli B, Veroni C, Mazzola GA, Aloisi F. Epstein-Barr Virus-Specific CD8 T Cells Selectively Infiltrate the Brain in Multiple Sclerosis and Interact Locally with Virus-Infected Cells: Clue for a Virus-Driven Immunopathological Mechanism. J Virol. 2019;93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver RF, Crystal RG, Moller DR. Limited heterogeneity of biased T-cell receptor V beta gene usage in lung but not blood T cells in active pulmonary sarcoidosis. Immunology. 1996;88:516–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Wang A, Heald P, McNiff JM, Suozzi K, King B, et al. Treatment of angiolupoid sarcoidosis with tofacitinib ointment 2% and pulsed dye laser therapy. JAAD Case Rep. 2021;7:122–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson W, Donlin LT, Butler A, Rozo C, Bracken B, Rashidfarrokhi A, et al. Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat Commun. 2018;9:791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern BJ, Royal W 3rd, Gelfand JM, Clifford DB, Tavee J, Pawate S, et al. Definition and Consensus Diagnostic Criteria for Neurosarcoidosis: From the Neurosarcoidosis Consortium Consensus Group. JAMA Neurol. 2018;75:1546–53. [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–902 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulzer D, Alcalay RN, Garretti F, Cote L, Kanter E, Agin-Liebes J, et al. Erratum: T cells from patients with Parkinson’s disease recognize alpha-synuclein peptides. Nature. 2017;549:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talty R, Damsky W, King B. Treatment of cutaneous sarcoidosis with tofacitinib: A case report and review of evidence for Janus kinase inhibition in sarcoidosis. JAAD Case Rep. 2021;16:62–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischner D, Grimm M, Kaur H, Staudenraus D, Carvalho J, Looso M, et al. Single-cell profiling reveals GPCR heterogeneity and functional patterning during neuroinflammation. JCI Insight. 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentin L, Zambello R, Facco M, Tassinari C, Sancetta R, Siviero M, et al. Selection of T lymphocytes bearing limited TCR-Vbeta regions in the lung of hypersensitivity pneumonitis and sarcoidosis. Am J Respir Crit Care Med. 1997;155:587–96. [DOI] [PubMed] [Google Scholar]
- Voortman M, Drent M, Baughman RP. Management of neurosarcoidosis: a clinical challenge. Curr Opin Neurol. 2019;32:475–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Yao L, Luo M, Zhou W, Jin X, Xu Z, et al. Single-cell transcriptome and TCR profiling reveal activated and expanded T cell populations in Parkinson’s disease. Cell Discov. 2021;7:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanke F, Moos S, Croxford AL, Heinen AP, Graf S, Kalt B, et al. EBI2 Is Highly Expressed in Multiple Sclerosis Lesions and Promotes Early CNS Migration of Encephalitogenic CD4 T Cells. Cell Rep. 2017;18:1270–84. [DOI] [PubMed] [Google Scholar]
- Willinger T, Freeman T, Hasegawa H, McMichael AJ, Callan MF. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J Immunol. 2005;175:5895–903. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wei K, Slowikowski K, Fonseka CY, Rao DA, Kelly S, et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol. 2019;20:928–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Aligned sequencing data is available at figshare.com (https://doi.org/10.6084/m9.figshare.c.5761613). Code will be available at https://github.com/RobersonLab/.


