After initiating synthesis of RNA at a promoter, RNA polymerase (RNAP) normally continues to elongate the transcript until it reaches a termination site. Important elements of termination sites are transcribed before polymerase translocation stops, and the resulting RNA is an active element of the termination pathway. Nascent transcripts of intrinsic sites can halt transcription without the assistance of additional factors, and those of Rho-dependent sites recruit the Rho termination protein to the elongation complex. In both cases, RNAP, the transcript, and the template dissociate (reviewed in references 76 and 80).
Termination is rarely, if ever, completely efficient, and the expression of downstream genes can be controlled by altering the efficiency of terminator readthrough. Two distinct mechanisms of elongation control have been reported for bacterial RNA polymerases. In one, exemplified by attenuation of the his and trp operons of Salmonella typhimurium and Escherichia coli, respectively, a single terminator is inactivated by interaction with an upstream sequence in the transcript, with a terminator-specific protein, or with a translating ribosome that follows closely behind RNAP (reviewed in references 35 and 104). In a second, whose prototype is antitermination of phage λ early transcription, polymerase is stably modified to a terminator-resistant form after it leaves the promoter. In this case, the modified enzyme not only transcribes through sequential downstream terminators, but also it is less sensitive to the pause sites that normally delay transcript elongation. Both pathways are widespread in nature, but in this minireview we consider only the second, which is known as processive antitermination (for previous reviews, see references 22, 23, 27, and 32). The recent explosive growth in our understanding of transcription elongation (reviewed in references 57, 96, and 99) make this an especially appropriate time to survey regulatory elements that target the transcription elongation complex.
Antitermination in λ is induced by two quite distinct mechanisms. The first is the result of interaction between λ N protein and its targets in the early phage transcripts, and the second is the result of an interaction between the λ Q protein and its target in the late phage promoter. We describe the N mechanism first. Lambda N, a small basic protein of the arginine-rich motif (ARM) (Fig. 1) family of RNA binding proteins, binds to a 15-nucleotide (nt) stem-loop called BOXB (17) (Fig. 2). (We will capitalize the names of sites in RNA and italicize the names of the corresponding DNA sequences; e.g., BOXB and boxB.) boxB is found twice in the λ chromosome, once in each of the two early operons (82, 83). It is close to the start point of the PL operon transcript and just downstream of the first translated gene of the PR operon. Neither the distance between the transcription start site and boxB, nor the nature of the promoter (at least in the case of sigma-70-dependent promoters), nor the nature of the terminator is relevant to N action. Although the boxB sequence is not well conserved in other bacteriophages of the λ family, most of these phages encode proteins that are analogous to λ N and have sequences capable of forming BOXB-like structures in their PL and PR operons. In some cases, it has been shown that these structures are recognized by the cognate N analogs. It is believed that this accounts for the phage specificity of N-mediated antitermination.
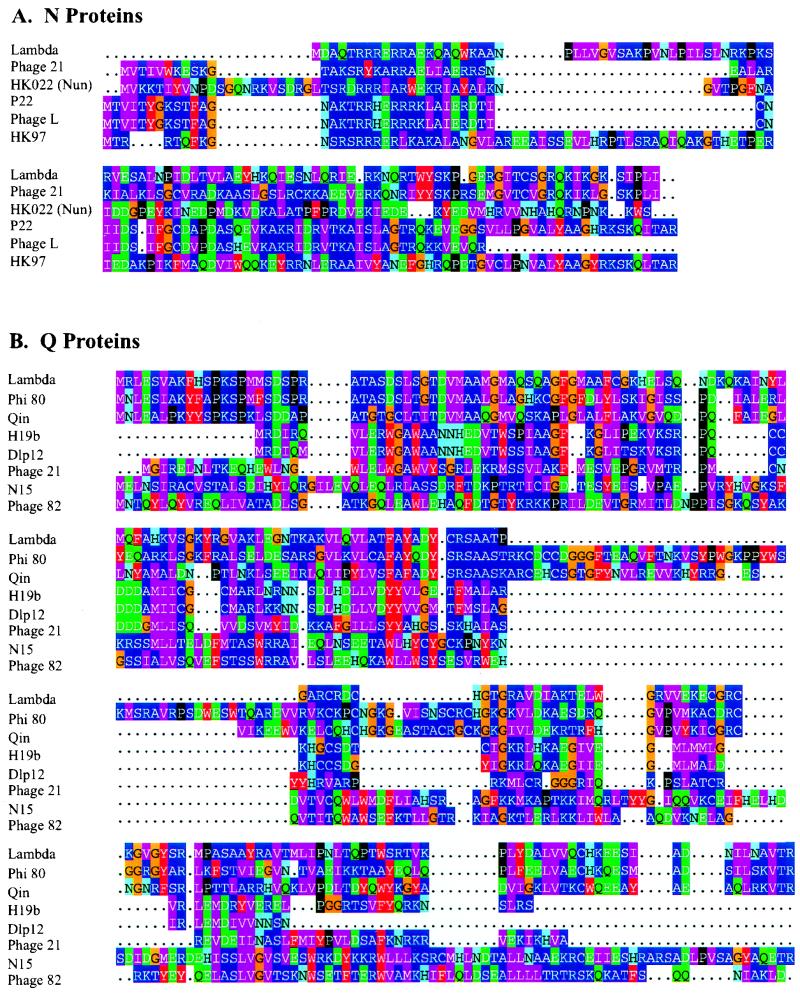
FIG. 1.
(A) Alignment of phage N proteins and the HK022 Nun protein. The color groupings reflect the frequency of amino acid substitutions in evolutionarily related protein domains: an amino acid is more likely to be replaced by one in the same color group than by one in a different color group in related proteins (34). The amino-proximal ARM regions were aligned by eye and according to the structures of the P22 and λ ARMs complexed to their cognate nut sites (see text and Fig. 2), and the remainder of the proteins was aligned by ClustalW (38). The dots indicate gaps introduced to improve the alignment. Aside from the ARM regions, the proteins fall into three very distantly related (or unrelated) families: (i) λ and phage 21; (ii) P22, phage L, and HK97; and (iii) HK022 Nun. The divergence of Nun from the N proteins is unsurprising because of their different functions. The sequence database was searched for additional N homologs with the PSI-BLAST program (3), using each of the listed sequences as a query, but none were found. Two N proteins were omitted from the alignment: that of phage H19b, because it differs by only three conservative substitutions from N of HK97 (E60D, K80E, and R100K) (3), and that of lambdoid phage φ80 (Phi 80), because it shows no resemblance to any of the other N proteins, lacking even an ARM (42, 69). (B) Alignment of phage Q proteins. The alignments were generated by ClustalW, and the database was searched for Q homologs as described above. These proteins fall into three very distantly related (or unrelated) families: (i) λ and Qin; (ii) H19b, Dlp12, and phage 21; and (iii) N15 and phage 82. Qin and Dlp12 are defective lambdoid prophages of E. coli, but it is likely that their Q proteins are active (see reference 16). The Q proteins of phages HK022 and P22 were omitted from the alignment because of their close similarity to that of λ. A putative and possibly defective Q, encoded by a sequence located upstream of Shiga-like toxin I genes in an E. coli isolate (72) and found by a BLAST search of the translated nucleotide sequence database, was omitted from the alignment because of its close similarity to the Q of phage H19B (61).
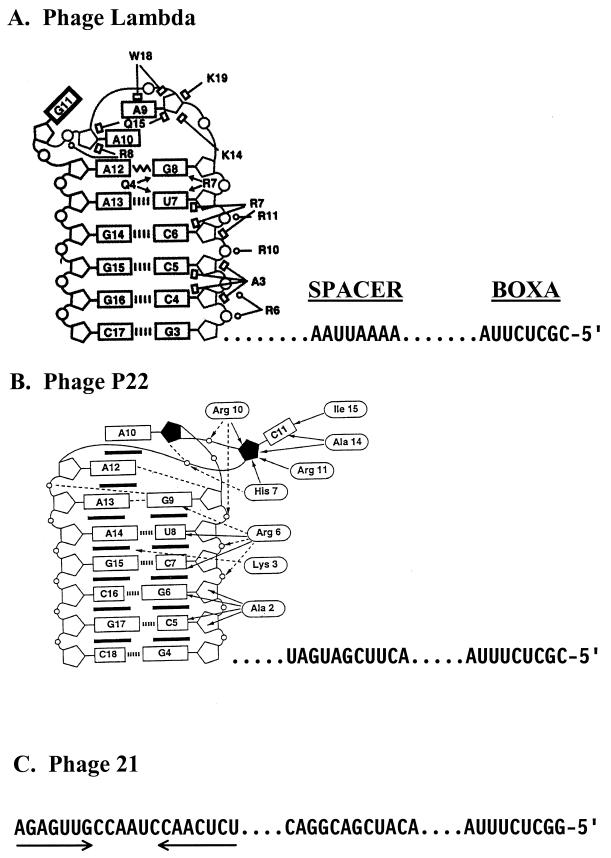
FIG. 2.
BOXA and BOXB RNAs and their interaction with the ARM of their cognate N proteins. The amino acid-nucleotide interactions are shown to the left except for BOXB of phage 21, for which the structure of the complex is unknown. The sequences of BOXA and BOXA-BOXB spacer are shown to the right. The dots to the left and right of the spacer sequences are for alignment. (A) λ N-ARM-BOXB complex (adapted from reference 48 with permission of the publisher). Open circles, pentagons, and rectangles represent phosphates, riboses, and bases, respectively. Watson-Crick base pairs (||||) are indicated. The zigzag line denotes a sheared G · A base pair. Open circles, open rectangles, and arrowheads depict ionic, hydrophobic, and hydrogen-bonding interactions, respectively. Guanine-11, indicated by a bold rectangle, is extruded from the BOXB loop (see text). (B) P22 N-ARM-BOXB complex (adapted from reference 15 with permission of the publisher). Open circles, pentagons, rectangles, and ovals represent phosphates, riboses, bases, and amino acids, respectively. The solid pentagons indicate riboses with a C2′-endo pucker. Base stacking ( ), intermolecular hydrogen bonding or electrostatic interactions (<-----), intermolecular hydrophobic or van der Waals interactions (←), intramolecular hydrogen bonds (––––) and Watson-Crick base pairs (|||||) are indicated. Cytosine-11 is extruded from the loop (see text). Note that the amino-terminal amino acid residue in the complex corresponds to Asn-14 in the complete protein (Fig. 1), and the displayed amino acids are numbered accordingly. (C) NUTL site of phage 21. The arrows indicate the inverted sequence repeats of BOXB.
), intermolecular hydrogen bonding or electrostatic interactions (<-----), intermolecular hydrophobic or van der Waals interactions (←), intramolecular hydrogen bonds (––––) and Watson-Crick base pairs (|||||) are indicated. Cytosine-11 is extruded from the loop (see text). Note that the amino-terminal amino acid residue in the complex corresponds to Asn-14 in the complete protein (Fig. 1), and the displayed amino acids are numbered accordingly. (C) NUTL site of phage 21. The arrows indicate the inverted sequence repeats of BOXB.
The structures of complexes between the ARMs of λ N (residues 1 to 22) and P22 N (residues 14 to 32) and their cognate BOXBs have recently been determined by nuclear magnetic resonance (15, 48) (Fig. 2). The two complexes, although similar, show differences that account for the specificity of N-BOXB recognition (92, 101). Upon binding, the λ and P22 ARMs adopt a bent α-helix conformation that packs against the BOXB hairpin through hydrophobic and ionic interactions. Residues in the amino-proximal segments make multiple base, ribose, and phosphate contacts in the 5′-ascending stem of BOXB without disrupting its regular A-form. The two stem-proximal residues of both RNA loops form a sheared G · A base pair which is contacted by R7 of λ N and R19 of P22 N (note that R19 corresponds to Arg 6 of Fig. 2; see Fig. 2 legend). In the λ complex, the fourth residue (G) of the GAAGA loop is extruded and the remaining residues form a GNRA fold similar to the base-stacked GAAA tetraloop reported in a number of important RNA structures (Fig. 2). P22 N also creates a GNRA fold, but this is accomplished by extrusion of the third residue (C) of the GACAA loop. In contrast to the extruded G residue in BOXB-λ, which is not close to λ N, the extruded C residue in BOXB-P22 makes contacts with residues in the carboxyl-proximal segment of P22 N. The structure of the λ N complex is stabilized by an important stacking interaction between W18 and the second residue (A) in the BOXB-λ loop. This interaction is not found in the P22 complex. Formation of the GNRA fold is essential for λ N binding, and mutations in loop residues 1, 3, and 5 that prevent tetraloop formation block N binding (17). The structure of the complex between the BOXB of phage 21 and its cognate N protein must be substantially different than the complexes described above because the BOXB-21 loop (TCTAACCG) cannot be folded into a GNRA tetraloop. However, the HK022 Nun protein, which recognizes λ BOXB (see below), probably does so in a way that resembles that of λ (18). If so, A3, R7, and W18 of λ N, all of which make base contacts in BOXB, probably correspond to S27, R31, and Y42 of Nun (Fig. 1).
Promoter proximal to each of the boxB sequences is boxA, which is also important for antitermination by λ N (70) (Fig. 2). boxA-like sequences are also found in corresponding positions in several other lambdoid phages. Together, the two boxes constitute a nut (for N utilization) site, which contains all of the cis-acting elements required for N-dependent antitermination. In the presence of N, polymerase becomes termination resistant after transcription of nut (7a). Antitermination can still be detected after polymerase has read through thousands of base pairs and many sequential terminators. This implies that the N-dependent modification to polymerase that occurs after transcription of nut is stable. BOXA is the loading site for the E. coli S10 (or NusE) and NusB proteins (62). In association with two additional E. coli factors, NusA and NusG, an antitermination complex containing N and λ NUT is formed with RNAP. It is clear that N is the active factor in the complex, since at sufficiently high concentrations, N suppresses transcription termination in vitro in the absence of nut or the Nus factors (24, 75). However, the additional components increase the stability of the antitermination complex and reduce the level of N that is needed (24, 54). The RNAP α subunit might also have a specific role in N-mediated antitermination. Mutations that alter the carboxyl-terminal domain of the RNAP α subunit have been reported to enhance or inhibit N antitermination in vivo, according to the nature and location of the mutation (68, 85). However, deletion of the carboxyl-terminal domain of α does not affect N-dependent antitermination in vitro, perhaps indicating that some regulatory component is missing from the reaction (53).
The formation of the complete antitermination complex can occur in discrete steps (56). Initially, NusA binds to an N-BOXB complex. This binding requires extrusion of the loop residue of BOXB. Thus, a BOXB tetraloop mutant (GAAGA → GAAA) binds N but does not form an N-BOXB-NusA complex, as demonstrated by supershift experiments (17, 55). A core complex of BOXB, N, NusA, and RNAP can read through terminators located close to the NUT site in vitro. In the absence of N and NUT, NusA binds to the elongation complex near the 3′-OH terminus of the nascent RNA chain and enhances pausing and termination (53).
Processive antitermination requires the complete antitermination complex. The assembly of NusB, S10, and NusG onto the core complex involves nt 2 to 7 of λ BOXA (CGCUCUUACACA), as well as the carboxyl-terminal region of N, which interacts with RNAP (56). The role of NusG in the N antitermination reaction is not clear. NusG binds to termination factor Rho and to RNAP (49, 54). It stimulates the rate of transcription elongation and is required for the activity of certain Rho-dependent terminators (12, 93). NusG is a component of the complete antitermination complex and enhances N antitermination in vitro. However, alteration of λ BOXA to a variant called BOXA consensus (CGCUCUUUAACA) allows NusB and S10 to assemble in the absence of NusG (56). Furthermore, depletion of NusG has no effect on λ N antitermination in vivo, and unlike nusA, nusB, and nusE, no point mutations in nusG that block N activity have been isolated. A NusG homolog, RfaH, enhances elongation of several transcripts in E. coli and S. typhimurium (see below). The possibility that RfaH and NusG are redundant for N antitermination has not yet been tested, although for several other functions, the two proteins are not interchangeable.
The function of BOXA in λ N-mediated antitermination is likewise not entirely clear. Point mutations in boxA that decrease or increase antitermination efficiency have been isolated (70, 73, 84). On the other hand, deletion of the boxA region does not inhibit antitermination in vivo. Instead, antitermination no longer requires NusB (73). To account for this, it has been proposed that BOXA is not directly required for antitermination but instead is the site of interplay between inhibitory and anti-inhibitory factors. According to this model, boxA point mutations that reduce antitermination eliminate binding of the anti-inhibitor but not the inhibitor. boxA deletions eliminate binding of the inhibitor, and therefore, the anti-inhibitor, presumably NusB, is no longer needed. This notion is supported by an experiment in which high-level transcription of an antitermination-defective boxA point mutant activated growth in trans of a λ phage carrying the same mutation in a nusB mutant host, presumably by titrating the inhibitor (73). In a similar experiment, high-level transcription of a consensus BOXA inhibited growth of a phage carrying a wild-type boxA, probably by titrating NusB (28). However, the role of NusB is likely to go beyond that of an anti-inhibitor, and that of BOXA is likely to go beyond that of a site for the interplay of inhibitory and anti-inhibitory factors. In vitro studies with purified proteins show that point mutations in boxA impede the assembly of the antitermination complex even in the absence of a known inhibitor (56), and NusB stimulates processive antitermination in such a system (24, 54). In addition, the role of BOXA in antitermination of Rho-dependent terminators in bacterial rRNA operons appears to be more central than it is in λ, raising the possibility that λ BOXA contributes to antitermination in a way that is at least partially independent of BOXB (32) (see below).
Surprisingly, the λ nut sites are also components in a transcription termination pathway. In this pathway, N is replaced by Nun, a protein encoded by a relative of λ, phage HK022 (see below) (67, 78). Nun converts antitermination into termination. Other components of the two pathways, notably NusA, NusB, S10, NusG, BOXA, and BOXB, are shared. The sequence similarity of Nun to proteins of the N family, although weak, includes the amino-proximal ARM region (Fig. 1). This is unsurprising because Nun, like λ N, binds specifically to BOXB and requires the same BOXB nucleotides for biological function (8, 18). In vivo, Nun terminates transcription just distal to the nut sites (78, 88). In vitro, Nun arrests RNAP translocation at several positions downstream of nut (39). The arrested elongation complexes contain the 3′ ends of the nascent transcripts in the polymerase active center, and this site remains enzymatically active: the 3′ nucleotide can be removed by pyrophosphorolysis and restored by addition of the appropriate nucleoside triphosphate (40). However, forward and backward translocation of RNAP is blocked. The Nus factors increase the efficiency of transcription arrest but are not essential if the concentration of Nun is elevated. Nun-dependent release of arrested RNAP from the template and transcript has not been observed in a purified transcription system, presumably because a factor(s) is missing. The differences between N and Nun that lead to their opposed biological activities are unknown. However, the amino-proximal regions, which contain the ARMs, can be interchanged between the two proteins without altering their functions (36). Therefore, the functional differences are in the carboxyl-terminal 50 to 75% of the proteins. Of particular note is the presence of three C-terminal His residues, specific to Nun. These residues form part of a Zn2+ binding motif that is required for Nun activity (100). The carboxyl-terminal regions of N and Nun may bind different RNAP subunits; certain rpoC (β′) mutations block Nun but not N activity (81).
A second phage-encoded factor, λ Q protein, induces antitermination in the λ late operon (25, 37). Lambda Q, like λ N, has functional analogs in other phages (Fig. 1). These late antiterminators probably act by a similar mechanism, although some are only distantly related or unrelated to λ Q (30, 33, 102). Initially, Q binds to a region within the λ PR′ promoter (105). Interaction with RNAP can be detected when the transcription complex pauses at +16, downstream of a site similar to the extended −10 sequence of some sigma-70 promoters (31, 77). The presence of the sigma-70 subunit of RNAP holoenzyme is essential for pausing and for Q-mediated antitermination: RNAP core enzyme that has been artificially paused at +16 by omission of the appropriate nucleoside triphosphate cannot be modified by Q. In addition, sigma-70 mutants that are unable to support Q-mediated antitermination have been isolated (43b). However, once Q has interacted with RNAP holoenzyme, sigma-70 is no longer needed for stable association of Q with the elongation complex. Although antitermination by Q is enhanced by NusA in vitro, it is not clear if the Q reaction has additional requirements in vivo. How Q modifies RNAP function is likewise unknown.
Processive antitermination can be mediated by RNA as well as proteins. Coliphage HK022, alone among the known lambdoid phages, does not encode an analog to λ N (66). Instead, it promotes antitermination of early phage transcription through the direct action of transcribed sequences called put (for polymerase utilization) sites (Fig. 3) (20, 43). There are two closely related put sites, one located in the PL operon and the other located in the PR operon, roughly corresponding to the positions of the nut sequences in λ and in other λ relatives. put sites act in cis to promote readthrough of downstream terminators in the absence of all HK022 proteins. The put transcripts are predicted to form two stem-loops separated by a single unpaired nucleotide. This prediction is supported by mutational studies and the pattern of sensitivity of the two RNAs to cleavage with single- and double-strand-specific endoribonucleases (7). RNA structure is critical to antitermination because mutations that prevent the formation of base pairs in the stems reduce function, and these mutations can be suppressed by additional mutations that restore base pairing (43). Like λ N and Q, the PUT sequences suppress polymerase pausing and promote processive antitermination in a purified in vitro transcription system. In contrast to λ N, no phage or auxiliary bacterial factors are required. The only mutations known to block PUT-mediated antitermination change highly conserved amino acids located in a cysteine-rich amino-proximal domain of the RNAP β′ subunit (20). Strains carrying these mutations are unable to support lytic growth of HK022 but are normal in all other respects tested, including lytic growth of λ and other λ relatives. The phage-restricted phenotypes conferred by these mutations suggests that they alter a domain of RNAP-β′ that interacts specifically with nascent PUT RNA in the transcription elongation complex, but this idea has not been directly tested. The stability of the putative PUT-RNAP interaction and the nature of the PUT-induced modification to the elongation complex are unknown.
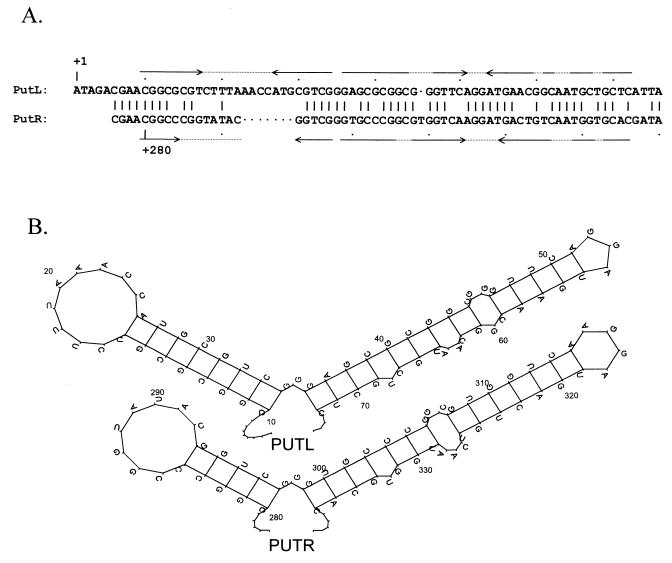
FIG. 3.
HK022 put sites and folded PUT RNAs. (A) Alignment of putL and putR (43). The numbers give distances from the start sites of the PL and PR promoters, respectively, and the pairs of arrows indicate inverted sequence repeats. (B) Folded PUTL and PUTR RNAs. The structures, which were generated by energy minimization as described (43), have been partially confirmed by genetic and biochemical studies (7, 43).
So far, factor-independent antitermination is unique to phage HK022. Both factor-independent and -dependent modes of antitermination are efficient, processive, and well-suited to their tasks. Indeed, the isolation and characterization of a hybrid phage that contains the early promoters and put sites of phage HK022 fused to the PL and PR operons of λ show that the HK022 antitermination pathway suppresses the λ terminators to the extent necessary for normal lytic growth and lysogen formation by the hybrid (66). In addition, late gene expression in phage HK022 is activated by a Q-dependent antitermination pathway that is very closely related to that of λ (4). The relative advantages of the PUT and N-NUT antitermination mechanisms and the evolutionary reasons for their adoption by different closely related phages are obscure. Recent work suggests that the activity of λ N-dependent antitermination is autoregulated so as to couple the lysogeny-lysis decision of infected cells to their growth phase (21a). It is not clear if the put-dependent antitermination activity of HK022 is regulated. However, HK022 has devoted the resources saved by the dispensability of N to the production of Nun, a protein that prevents the growth of a potential competitor by coopting a component of its antitermination system (see above).
Processive antitermination was first discovered in a bacteriophage, but examples have since been found in bacterial operons. The E. coli rrn operons are regulated by an antitermination mechanism that is dependent on sites that are closely related to λ boxA and located promoter proximal to the 16S and 23S structural genes in each operon (1, 50, 58; reviewed in reference 21). The sequences of the rrn BOXA sites are more similar to the bacteriophage consensus than is that of λ, and they bind NusB-S10 more efficiently (62). Although stem-loop structures analogous to BOXB are found promoter proximal to the BOXA sites, they are not essential for antitermination. An rrn BOXA sequence confers full antitermination activity against Rho-dependent but not against intrinsic terminators (2, 9). BOXA also increases the rate of transcription elongation by RNAP (97). Point mutations in BOXA induce premature transcription termination. rrn antitermination requires NusB in vivo, as shown by a NusB depletion experiment (89). NusA stimulates the elongation rate of rrn RNA chains carrying BOXA (98). A role for NusA is further suggested by the observation that the nusA10(Cs) mutation inhibits both antitermination and the rate of transcription elongation in an rrn operon (98). The role of other Nus factors in rrn regulation in vivo is not clear. In vitro, an antitermination complex that includes NusA, NusB, S10, and NusG forms at the BOXA sequence of rrnG, but these components are not sufficient for antitermination by themselves (89). An additional factor or factors that can be supplied by a cellular extract are required, but their identities are unknown.
A second bacterial elongation control pathway depends on the RfaH protein, a NusG homolog (5, 6, 91). RfaH and a cis-acting promoter-proximal sequence element, ops (for operon polarity suppressor, also called JUMPstart), increase the expression of several bacterial operons. The products of these operons affect the production and transport of components located on the outside of the inner membrane, such as lipopolysaccharide core, exopolysaccharide, F pili, and hemolysin. These operons are relatively long, with several genes and intergenic terminators. RfaH and ops appear to function together, since mutational inactivation of both elements does not have an additive effect on gene expression. The two elements are thought to act by suppressing termination. First, transcription is increased distal to the promoter but not proximal to a terminator. Second, stimulation of gene expression is not promoter specific. Third, an rfaH mutation can be suppressed by a mutation that reduces the activity of transcription termination factor Rho (26). Purified RfaH stimulates transcription promoter distal to an intrinsic terminator when added to a crude bacterial extract programmed with an ops-containing template. However, it has not been shown that the increase results from elongation of transcripts that would otherwise have been terminated. Nevertheless, the requirement for a cis-acting site, the ability of the site to act at a distance from terminators, and the homology of RfaH and NusG suggest common elements with the mechanisms of N-dependent antitermination.
Phage P4 has an entirely different mechanism of controlling elongation. It encodes a protein, Psu (for polarity suppressor), that reduces termination by E. coli Rho factor (51, 94). Unlike λ N and Q, Psu does not require cis-acting sites to antiterminate and is specific for Rho-dependent terminators. Extracts of cells that contain Psu are deficient in termination at Rho-dependent terminators, and termination can be restored by adding Rho to the extracts (52). Psu does not act by reducing the level of Rho protein, but it interferes, directly or indirectly, with Rho action. The importance of Rho inactivation in the life cycle of P4 is unclear. Psu stimulates lytic growth of P4, but this is likely to be the result of incorporation of Psu into the P4 capsid rather than (or in addition to) activation of transcription of essential genes that lie downstream of Rho-dependent terminators (41). The only known protein that is similar to Psu is encoded by a P4 relative, retronphage φR73 (41).
How do terminators and antiterminators act? Do the antitermination pathways described here have common steps? We cannot yet answer these questions, but a brief discussion of what we think we know about the structure and stability of the elongation complex should limit the possibilities and provide a basis for speculation. The active bacterial elongation complex consists of core RNAP, template, and RNA product. The 3′ end of the RNA is engaged in the active site of the enzyme, the following ∼8 nt are hybridized to the template strand of the DNA, and the next ∼9 nt remain closely associated with RNAP (64). About 17 nt of the nontemplate DNA strand are separated from the template strand in the transcription bubble. Elongation complexes can also contain NusA and/or NusG. These proteins, which increase the stability of the N-mediated antitermination complex (see above), have different effects on elongation. NusA decreases and NusG increases the elongation rate, and both proteins alter termination efficiency in a terminator-specific manner (13, 14, 86; see reference 76).
An elongation complex, unless located at a terminator, is extraordinarily stable, even when translocation is prevented by removal of substrates. Recent observations suggest that this stability depends mainly on interactions between RNAP and the RNA-DNA hybrid as well as between polymerase and the downstream duplex DNA template (63, 87). Nascent RNA emerging from the hybrid region and upstream duplex DNA do not appear to be required. The strength of the RNA-DNA hybrid is believed to assure the lateral stability of the complex. Reducing the strength of the RNA-DNA bonds, for example by incorporation of nucleotide analogs, favors backsliding of RNAP on the template, with consequent disengagement of the 3′ RNA end from the active site, and concerted retreat of the RNA-DNA hybrid region from the 3′ end (65). Such a disengaged complex retains its resistance to dissociation and is capable of resuming elongation if the original or a newly created 3′ end reengages with the active site (10, 44, 45, 65, 71, 95).
Intrinsic terminators consist of a guanine- and cytosine-rich RNA hairpin stem immediately followed by a short uracil-rich segment within which termination can occur. If termination does not occur at this point, polymerase continues to elongate the transcript with normal processivity until it reaches the next terminator. Neither the stem nor the uracil-rich segment is sufficient for termination, although either can transiently slow elongation. The weakness of base pairing between rU and dA destabilizes the RNA-DNA hybrid in the uracil-rich segment, and this probably contributes to termination. Formation of the hairpin stem as nascent terminator RNA emerges from polymerase destabilizes the RNA-DNA hybrid and interrupts contacts between the emerging nascent RNA and RNAP (62a). It might also interfere with the stabilizing interactions between RNAP and the hybrid or those between RNAP and the downstream region of the template. Cross-linking of nucleic acid to RNAP suggests that both the downstream DNA and the nascent RNA that emerges from the hybrid region, and within which the terminator hairpin might form, are located close to the same regions of the enzyme (64). Conversely, modifications that render RNAP termination resistant could prevent the terminator stem from destabilizing one or more of these targets, at least while the 3′ end of the RNA is within the uracil-rich segment of the terminator.
The λ N and Q proteins and HK022 PUT RNA also suppress Rho-dependent terminators (43a, 79, 103) which, in contrast to intrinsic terminators, lack a precisely determined termination point. Rho is an RNA-dependent ATPase that binds to cytosine-rich, unstructured regions in nascent RNA and acts preferentially to terminate elongation complexes that are paused at nearby downstream sites (19, 29, 46, 47, 59, 60). Rho possesses RNA-DNA helicase activity, and this activity is directional, unwinding DNA paired to the 3′ end of the RNA molecule (11, 90). This corresponds to the location of the hybrid and of RNAP in an active ternary elongation complex. The ability of antiterminators to suppress Rho-dependent and -independent terminators suggests that they prevent a step that is common to both classes. Given the helicase activity of Rho, a likely candidate for this step is disruption of the RNA-DNA hybrid. However, other candidates, such as destabilization of RNAP-template or RNAP-hybrid interactions, are also plausible. Alternatively, the ability of N, Q, and PUT to suppress RNAP pausing (31, 43, 54, 74) suggests that they prevent Rho-dependent termination by accelerating polymerase away from Rho bound at upstream RNA sites. This explanation raises the problem of why NusG, which also accelerates polymerase, enhances rather than suppresses Rho-dependent termination (see above). Clearly, the molecular details of processive antitermination remain poorly understood despite the 30 years that have elapsed since its discovery.
ACKNOWLEDGMENTS
Thanks to Donald Court, Asis Das, David Friedman, Rodney King, Orna Resnekov, and Randy Watnick for their comments on the manuscript.
REFERENCES
- 1.Aksoy S, Squires C L, Squires C. Evidence for antitermination in Escherichia coli rRNA transcription. J Bacteriol. 1984;159:260–264. doi: 10.1128/jb.159.1.260-264.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrechtsen B, Squires C L, Li S, Squires C. Antitermination of characterized transcriptional terminators by the Escherichia coli rrnG leader region. J Mol Biol. 1990;213:123–134. doi: 10.1016/S0022-2836(05)80125-1. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkinson B L, Gottesman M E. The Escherichia coli rpoB60 mutation blocks antitermination by coliphage HK022 Q-function. J Mol Biol. 1992;227:29–37. doi: 10.1016/0022-2836(92)90679-e. [DOI] [PubMed] [Google Scholar]
- 5.Bailey M J, Hughes C, Koronakis V. Increased distal gene transcription by the elongation factor RfaH, a specialized homologue of NusG. Mol Microbiol. 1996;22:729–737. doi: 10.1046/j.1365-2958.1996.d01-1726.x. [DOI] [PubMed] [Google Scholar]
- 6.Bailey M J, Hughes C, Koronakis V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol Microbiol. 1997;26:845–851. doi: 10.1046/j.1365-2958.1997.6432014.x. [DOI] [PubMed] [Google Scholar]
- 7.Banik-Maiti S, King R A, Weisberg R A. The antiterminator RNA of phage HK022. J Mol Biol. 1997;272:677–687. doi: 10.1006/jmbi.1997.1251. [DOI] [PubMed] [Google Scholar]
- 7a.Barik S, Ghosh B, Whalen W A, Lazinski D, Das A. An antitermination protein engages the elongating transcription apparatus at a promoter-proximal recognition site. Cell. 1987;50:885–899. doi: 10.1016/0092-8674(87)90515-0. [DOI] [PubMed] [Google Scholar]
- 8.Baron J, Weisberg R A. Mutations of the phage lambda nutL region that prevent the action of Nun, a site-specific transcription termination factor. J Bacteriol. 1992;174:1983–1989. doi: 10.1128/jb.174.6.1983-1989.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg K L, Squires C, Squires C L. Ribosomal RNA operon anti-termination. Function of leader and spacer region box B-box A sequences and their conservation in diverse micro-organisms. J Mol Biol. 1989;209:345–358. doi: 10.1016/0022-2836(89)90002-8. [DOI] [PubMed] [Google Scholar]
- 9a.Beutin L, Manning P A, Achtman M, Willetts N. sfrA and sfrB products of Escherichia coli K-12 are transcriptional control factors. J Bacteriol. 1981;145:840–844. doi: 10.1128/jb.145.2.840-844.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 11.Brennan C A, Dombroski A J, Platt T. Transcription termination factor rho is an RNA-DNA helicase. Cell. 1987;48:945–952. doi: 10.1016/0092-8674(87)90703-3. [DOI] [PubMed] [Google Scholar]
- 12.Burns C M, Richardson J P. NusG is required to overcome a kinetic limitation to Rho function at an intragenic terminator. Proc Natl Acad Sci USA. 1995;92:4738–4742. doi: 10.1073/pnas.92.11.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burns C M, Richardson L V, Richardson J P. Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J Mol Biol. 1998;278:307–316. doi: 10.1006/jmbi.1998.1691. [DOI] [PubMed] [Google Scholar]
- 14.Burova E, Hung S C, Sagitov V, Stitt B L, Gottesman M E. Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J Bacteriol. 1995;177:1388–1392. doi: 10.1128/jb.177.5.1388-1392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Z, Gorin A, Frederick R, Ye X, Hu W, Majumdar A, Kettani A, Patel D J. Solution structure of P22 transcriptional antitermination N peptide-boxB RNA complex. Nat Struct Biol. 1998;5:203–212. doi: 10.1038/nsb0398-203. [DOI] [PubMed] [Google Scholar]
- 16.Campbell A M. Cryptic prophages. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 2041–2046. [Google Scholar]
- 17.Chattopadhyay S, Garcia-Mena J, DeVito J, Wolska K, Das A. Bipartite function of a small RNA hairpin in transcription antitermination in bacteriophage lambda. Proc Natl Acad Sci USA. 1995;92:4061–4065. doi: 10.1073/pnas.92.9.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chattopadhyay S, Hung S C, Stuart A C, Palmer III A G, Garcia-Mena J, Das A, Gottesman M E. Interaction between the phage HK022 Nun protein and the nut RNA of phage lambda. Proc Natl Acad Sci USA. 1995;92:12131–12135. doi: 10.1073/pnas.92.26.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C Y, Galluppi G R, Richardson J P. Transcription termination at lambda tR1 is mediated by interaction of rho with specific single-stranded domains near the 3′ end of cro mRNA. Cell. 1986;46:1023–1028. doi: 10.1016/0092-8674(86)90701-4. [DOI] [PubMed] [Google Scholar]
- 20.Clerget M, Jin D J, Weisberg R A. A zinc binding region in the β′ subunit of RNA polymerase is involved in antitermination of early transcription of phage HK022. J Mol Biol. 1995;248:768–780. doi: 10.1006/jmbi.1995.0259. [DOI] [PubMed] [Google Scholar]
- 21.Condon C, Squires C, Squires C L. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Court, D. Personal communication.
- 22.Das A. How the phage lambda N gene product suppresses transcription termination: communication of RNA polymerase with regulatory proteins mediated by signals in nascent RNA. J Bacteriol. 1992;174:6711–6716. doi: 10.1128/jb.174.21.6711-6716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das A. Control of transcription termination by RNA-binding proteins. Annu Rev Biochem. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- 24.DeVito J, Das A. Control of transcription processivity in phage lambda: Nus factors strengthen the termination-resistant state of RNA polymerase induced by N antiterminator. Proc Natl Acad Sci USA. 1994;91:8660–8664. doi: 10.1073/pnas.91.18.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dove W F. Action of the lambda chromosome. I. Control of functions late in bacteriophage development. J Mol Biol. 1966;19:187–201. doi: 10.1016/s0022-2836(66)80060-8. [DOI] [PubMed] [Google Scholar]
- 26.Farewell A, Brazas R, Davie E, Mason J, Rothfield L I. Suppression of the abnormal phenotype of Salmonella typhimurium rfaH mutants by mutations in the gene for transcription termination factor Rho. J Bacteriol. 1991;173:5188–5193. doi: 10.1128/jb.173.16.5188-5193.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman D I, Court D L. Transcription antitermination: the lambda paradigm updated. Mol Microbiol. 1995;18:191–200. doi: 10.1111/j.1365-2958.1995.mmi_18020191.x. [DOI] [PubMed] [Google Scholar]
- 28.Friedman D I, Olson E R, Johnson L L, Alessi D, Craven M G. Transcription-dependent competition for a host factor: the function and optimal sequence of the phage lambda boxA transcription antitermination signal. Genes Dev. 1990;4:2210–2222. doi: 10.1101/gad.4.12a.2210. [DOI] [PubMed] [Google Scholar]
- 29.Galloway J L, Platt T. Signals sufficient for rho-dependent transcription termination at trp t′ span a region centered 60 base pairs upstream of the earliest 3′ end point. J Biol Chem. 1988;263:1761–1767. [PubMed] [Google Scholar]
- 30.Goliger J A, Roberts J W. Bacteriophage 82 gene Q and Q protein. Sequence, overproduction, and activity as a transcription antiterminator in vitro. J Biol Chem. 1987;262:11721–11725. [PubMed] [Google Scholar]
- 31.Grayhack E J, Yang X J, Lau L F, Roberts J W. Phage lambda gene Q antiterminator recognizes RNA polymerase near the promoter and accelerates it through a pause site. Cell. 1985;42:259–269. doi: 10.1016/s0092-8674(85)80121-5. [DOI] [PubMed] [Google Scholar]
- 32.Greenblatt J, Nodwell J R, Mason S W. Transcriptional antitermination. Nature. 1993;364:401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- 33.Guo H C, Kainz M, Roberts J W. Characterization of the late-gene regulatory region of phage 21. J Bacteriol. 1991;173:1554–1560. doi: 10.1128/jb.173.4.1554-1560.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henikoff S, Henikoff J G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henkin T M. Control of transcription termination in prokaryotes. Annu Rev Genet. 1996;30:35–57. doi: 10.1146/annurev.genet.30.1.35. [DOI] [PubMed] [Google Scholar]
- 36.Henthorn K S, Friedman D I. Identification of functional regions of the Nun transcription termination protein of phage HK022 and the N antitermination protein of phage gamma using hybrid nun-N genes. J Mol Biol. 1996;257:9–20. doi: 10.1006/jmbi.1996.0142. [DOI] [PubMed] [Google Scholar]
- 37.Herskowitz I, Signer E R. A site essential for expression of all late genes in bacteriophage lambda. J Mol Biol. 1970;47:545–556. doi: 10.1016/0022-2836(70)90321-9. [DOI] [PubMed] [Google Scholar]
- 38.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 39.Hung S C, Gottesman M E. Phage HK022 Nun protein arrests transcription on phage lambda DNA in vitro and competes with the phage lambda N antitermination protein. J Mol Biol. 1995;247:428–442. doi: 10.1006/jmbi.1994.0151. [DOI] [PubMed] [Google Scholar]
- 40.Hung S C, Gottesman M E. The Nun protein of bacteriophage HK022 inhibits translocation of Escherichia coli RNA polymerase without abolishing its catalytic activities. Genes Dev. 1997;11:2670–2678. doi: 10.1101/gad.11.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isaksen M L, Rishovd S T, Calendar R, Lindqvist B H. The polarity suppression factor of bacteriophage P4 is also a decoration protein of the P4 capsid. Virology. 1992;188:831–839. doi: 10.1016/0042-6822(92)90538-z. [DOI] [PubMed] [Google Scholar]
- 42.Kanemoto K, Tanaka S, Miyashita T, Matsushiro A. Identification and purification of the N gene product of bacteriophage phi 80. Mol Gen Genet. 1986;205:523–529. doi: 10.1007/BF00338092. [DOI] [PubMed] [Google Scholar]
- 43.King R A, Banik-Maiti S, Jin D J, Weisberg R A. Transcripts that increase the processivity and elongation rate of RNA polymerase. Cell. 1996;87:893–903. doi: 10.1016/s0092-8674(00)81996-0. [DOI] [PubMed] [Google Scholar]
- 43a.King, R. A., and R. A. Weisberg. Unpublished results.
- 43b.Ko D C, Marr M T, Guo J, Roberts J W. A surface of Escherichia coli sigma-70 required for promoter function and antitermination by phage lambda Q protein. Genes Dev. 1998;12:3276–3285. doi: 10.1101/gad.12.20.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 46.Lau L F, Roberts J W, Wu R. Transcription terminates at lambda tR1 in three clusters. Proc Natl Acad Sci USA. 1982;79:6171–6175. doi: 10.1073/pnas.79.20.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lau L F, Roberts J W, Wu R. RNA polymerase pausing and transcript release at the lambda tR1 terminator in vitro. J Biol Chem. 1983;258:9391–9397. [PubMed] [Google Scholar]
- 48.Legault P, Li J, Mogridge J, Kay L E, Greenblatt J. NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- 49.Li J, Mason S W, Greenblatt J. Elongation factor NusG interacts with termination factor rho to regulate termination and antitermination of transcription. Genes Dev. 1993;7:161–172. doi: 10.1101/gad.7.1.161. [DOI] [PubMed] [Google Scholar]
- 50.Li S C, Squires C L, Squires C. Antitermination of E. coli rRNA transcription is caused by a control region segment containing lambda nut-like sequences. Cell. 1984;38:851–860. doi: 10.1016/0092-8674(84)90280-0. [DOI] [PubMed] [Google Scholar]
- 51.Linderoth N A, Calendar R L. The Psu protein of bacteriophage P4 is an antitermination factor for rho-dependent transcription termination. J Bacteriol. 1991;173:6722–6731. doi: 10.1128/jb.173.21.6722-6731.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linderoth N A, Tang G, Calendar R. In vivo and in vitro evidence for an anti-Rho activity induced by the phage P4 polarity suppressor protein Psu. Virology. 1997;227:131–141. doi: 10.1006/viro.1996.8325. [DOI] [PubMed] [Google Scholar]
- 53.Liu K, Zhang Y, Severinov K, Das A, Hanna M M. Role of Escherichia coli RNA polymerase alpha subunit in modulation of pausing, termination and anti-termination by the transcription elongation factor NusA. EMBO J. 1996;15:150–161. [PMC free article] [PubMed] [Google Scholar]
- 54.Mason S W, Li J, Greenblatt J. Host factor requirements for processive antitermination of transcription and suppression of pausing by the N protein of bacteriophage lambda. J Biol Chem. 1992;267:19418–19426. [PubMed] [Google Scholar]
- 55.Mogridge J, Mah T F, Greenblatt J. A protein-RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the lambda N protein. Genes Dev. 1995;9:2831–2845. doi: 10.1101/gad.9.22.2831. [DOI] [PubMed] [Google Scholar]
- 56.Mogridge J, Mah T F, Greenblatt J. Involvement of boxA nucleotides in the formation of a stable ribonucleoprotein complex containing the bacteriophage lambda N protein. J Biol Chem. 1998;273:4143–4148. doi: 10.1074/jbc.273.7.4143. [DOI] [PubMed] [Google Scholar]
- 57.Mooney R A, Artsimovitch I, Landick R. Information processing by RNA polymerase: recognition of regulatory signals during RNA chain elongation. J Bacteriol. 1998;180:3265–3275. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morgan E A. Antitermination mechanisms in rRNA operons of Escherichia coli. J Bacteriol. 1986;168:1–5. doi: 10.1128/jb.168.1.1-5.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morgan W D, Bear D G, von Hippel P H. Rho-dependent termination of transcription. II. Kinetics of mRNA elongation during transcription from the bacteriophage lambda PR promoter. J Biol Chem. 1983;258:9565–9574. [PubMed] [Google Scholar]
- 60.Morgan W D, Bear D G, von Hippel P H. Rho-dependent termination of transcription. I. Identification and characterization of termination sites for transcription from the bacteriophage lambda PR promoter. J Biol Chem. 1983;258:9553–9564. [PubMed] [Google Scholar]
- 61.Neely M N, Friedman D I. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of Shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol Microbiol. 1998;28:1255–1267. doi: 10.1046/j.1365-2958.1998.00890.x. [DOI] [PubMed] [Google Scholar]
- 62.Nodwell J R, Greenblatt J. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell. 1993;72:261–268. doi: 10.1016/0092-8674(93)90665-d. [DOI] [PubMed] [Google Scholar]
- 62a.Nudler, E. Personal communication.
- 63.Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Transcription processivity: protein-DNA interactions holding together the elongation complex. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 64.Nudler E, Gusarov I, Avetissova E, Kozlov M, Goldfarb A. Spatial organization of transcription elongation complex in Escherichia coli. Science. 1998;281:424–428. doi: 10.1126/science.281.5375.424. [DOI] [PubMed] [Google Scholar]
- 65.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 66.Oberto J, Clerget M, Ditto M, Cam K, Weisberg R A. Antitermination of early transcription in phage HK022. Absence of a phage-encoded antitermination factor. J Mol Biol. 1993;229:368–381. doi: 10.1006/jmbi.1993.1040. [DOI] [PubMed] [Google Scholar]
- 67.Oberto J, Weisberg R A, Gottesman M E. Structure and function of the nun gene and the immunity region of the lambdoid phage HK022. J Mol Biol. 1989;207:675–693. doi: 10.1016/0022-2836(89)90237-4. [DOI] [PubMed] [Google Scholar]
- 68.Obuchowski M, Wegrzyn A, Szalewska-Palasz A, Thomas M S, Wegrzyn G. An RNA polymerase alpha subunit mutant impairs N-dependent transcriptional antitermination in Escherichia coli. Mol Microbiol. 1997;23:211–222. doi: 10.1046/j.1365-2958.1997.2101576.x. [DOI] [PubMed] [Google Scholar]
- 69.Ogawa T, Masukata H, Tomizawa J. Transcriptional regulation of early functions of bacteriophage phi 80. J Mol Biol. 1988;202:551–563. doi: 10.1016/0022-2836(88)90285-9. [DOI] [PubMed] [Google Scholar]
- 70.Olson E R, Flamm E L, Friedman D I. Analysis of nutR: a region of phage lambda required for antitermination of transcription. Cell. 1982;31:61–70. doi: 10.1016/0092-8674(82)90405-6. [DOI] [PubMed] [Google Scholar]
- 71.Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci USA. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paton A W, Paton J C, Goldwater P N, Heuzenroeder M W, Manning P A. Sequence of a variant Shiga-like toxin type-I operon of Escherichia coli O111:H−. Gene. 1993;129:87–92. doi: 10.1016/0378-1119(93)90700-d. [DOI] [PubMed] [Google Scholar]
- 73.Patterson T A, Zhang Z, Baker T, Johnson L L, Friedman D I, Court D L. Bacteriophage lambda N-dependent transcription antitermination. Competition for an RNA site may regulate antitermination. J Mol Biol. 1994;236:217–228. doi: 10.1006/jmbi.1994.1131. [DOI] [PubMed] [Google Scholar]
- 74.Rees W A, Weitzel S E, Das A, von Hippel P H. Regulation of the elongation-termination decision at intrinsic terminators by antitermination protein N of phage lambda. J Mol Biol. 1997;273:797–813. doi: 10.1006/jmbi.1997.1327. [DOI] [PubMed] [Google Scholar]
- 75.Rees W A, Weitzel S E, Yager T D, Das A, von Hippel P H. Bacteriophage lambda N protein alone can induce transcription antitermination in vitro. Proc Natl Acad Sci USA. 1996;93:342–346. doi: 10.1073/pnas.93.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Richardson J P, Greenblatt J. Control of RNA chain elongation and termination. In: Neidhardt F C, Curtiss III R C, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 822–848. [Google Scholar]
- 77.Ring B Z, Yarnell W S, Roberts J W. Function of E. coli RNA polymerase sigma factor sigma 70 in promoter-proximal pausing. Cell. 1996;86:485–493. doi: 10.1016/s0092-8674(00)80121-x. [DOI] [PubMed] [Google Scholar]
- 78.Robert J, Sloan S B, Weisberg R A, Gottesman M E, Robledo R, Harbrecht D. The remarkable specificity of a new transcription termination factor suggests that the mechanisms of termination and antitermination are similar. Cell. 1987;51:483–492. doi: 10.1016/0092-8674(87)90644-1. [DOI] [PubMed] [Google Scholar]
- 79.Roberts J W. Termination factor for RNA synthesis. Nature. 1969;224:1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- 80.Roberts J W. Transcription termination and its control. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. New York, N.Y: Chapman Hall; 1996. pp. 27–45. [Google Scholar]
- 81.Robledo R, Atkinson B L, Gottesman M E. Escherichia coli mutations that block transcription termination by phage HK022 Nun protein. J Mol Biol. 1991;220:613–619. doi: 10.1016/0022-2836(91)90104-e. [DOI] [PubMed] [Google Scholar]
- 82.Rosenberg M, Court D L, Shimatake H, Brady C, Wulff D L. The relationship between function and DNA sequence in an intercistronic regulatory region in phage lambda. Nature. 1978;272:414–423. doi: 10.1038/272414a0. [DOI] [PubMed] [Google Scholar]
- 83.Salstrom J S, Szybalski W. Coliphage lambda nutL−: a unique class of mutants defective in the site of gene N product utilization for antitermination of leftward transcription. J Mol Biol. 1978;124:195–221. doi: 10.1016/0022-2836(78)90156-0. [DOI] [PubMed] [Google Scholar]
- 84.Schauer A T, Carver D L, Bigelow B, Baron L S, Friedman D I. Lambda N antitermination system: functional analysis of phage interactions with the host NusA protein. J Mol Biol. 1987;194:679–690. doi: 10.1016/0022-2836(87)90245-2. [DOI] [PubMed] [Google Scholar]
- 85.Schauer A T, Cheng S W, Zheng C, St. Pierre L, Alessi D, Hidayetoglu D L, Costantino N, Court D L, Friedman D I. The alpha subunit of RNA polymerase and transcription antitermination. Mol Microbiol. 1996;21:839–851. doi: 10.1046/j.1365-2958.1996.451409.x. [DOI] [PubMed] [Google Scholar]
- 86.Schmidt M C, Chamberlin M J. Amplification and isolation of Escherichia coli nusA protein and studies of its effects on in vitro RNA chain elongation. Biochemistry. 1984;23:197–203. doi: 10.1021/bi00297a004. [DOI] [PubMed] [Google Scholar]
- 86a.Sharrock R A, Gourse R L, Nomura M. Defective antitermination of rRNA transcription and derepression of rRNA and tRNA synthesis in the nusB5 mutant of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5275–5279. doi: 10.1073/pnas.82.16.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sidorenkov I, Komissarova N, Kashlev M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 88.Sloan S B, Weisberg R A. Use of a gene encoding a suppressor tRNA as a reporter of transcription: analyzing the action of the Nun protein of bacteriophage HK022. Proc Natl Acad Sci USA. 1993;90:9842–9846. doi: 10.1073/pnas.90.21.9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Squires C L, Greenblatt J, Li J, Condon C. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc Natl Acad Sci USA. 1993;90:970–974. doi: 10.1073/pnas.90.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Steinmetz E J, Brennan C A, Platt T. A short intervening structure can block rho factor helicase action at a distance. J Biol Chem. 1990;265:18408–18413. [PubMed] [Google Scholar]
- 91.Stevens M P, Clarke B R, Roberts I S. Regulation of the Escherichia coli K5 capsule gene cluster by transcription antitermination. Mol Microbiol. 1997;24:1001–1012. doi: 10.1046/j.1365-2958.1997.4241780.x. [DOI] [PubMed] [Google Scholar]
- 92.Su L, Radek J T, Hallenga K, Hermanto P, Chan G, Labeots L A, Weiss M A. RNA recognition by a bent alpha-helix regulates transcriptional antitermination in phage lambda. Biochemistry. 1997;36:12722–12732. doi: 10.1021/bi971408k. [DOI] [PubMed] [Google Scholar]
- 93.Sullivan S L, Gottesman M E. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell. 1992;68:989–994. doi: 10.1016/0092-8674(92)90041-a. [DOI] [PubMed] [Google Scholar]
- 94.Sunshine M, Six E. Relief of P2 bacteriophage amber mutant polarity by the satellite bacteriophage P4. J Mol Biol. 1976;106:673–682. doi: 10.1016/0022-2836(76)90258-8. [DOI] [PubMed] [Google Scholar]
- 95.Surratt C K, Milan S C, Chamberlin M J. Spontaneous cleavage of RNA in ternary complexes of Escherichia coli RNA polymerase and its significance for the mechanism of transcription. Proc Natl Acad Sci USA. 1991;88:7983–7987. doi: 10.1073/pnas.88.18.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Uptain S M, Kane C M, Chamberlin M J. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 97.Vogel U, Jensen K F. Effects of the antiterminator BoxA on transcription elongation kinetics and ppGpp inhibition of transcription elongation in Escherichia coli. J Biol Chem. 1995;270:18335–18340. doi: 10.1074/jbc.270.31.18335. [DOI] [PubMed] [Google Scholar]
- 98.Vogel U, Jensen K F. NusA is required for ribosomal antitermination and for modulation of the transcription elongation rate of both antiterminated RNA and mRNA. J Biol Chem. 1997;272:12265–12271. doi: 10.1074/jbc.272.19.12265. [DOI] [PubMed] [Google Scholar]
- 99.von Hippel P H. An integrated model of the transcription complex in elongation, termination, and editing. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 100.Watnick R S, Gottesman M E. Escherichia coli NusA is required for efficient RNA binding by phage HK022 nun protein. Proc Natl Acad Sci USA. 1998;95:1546–1551. doi: 10.1073/pnas.95.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weiss M A. RNA-mediated signaling in transcription. Nat Struct Biol. 1998;5:329–333. doi: 10.1038/nsb0598-329. [DOI] [PubMed] [Google Scholar]
- 102.Yang X J, Goliger J A, Roberts J W. Specificity and mechanism of antitermination by Q proteins of bacteriophages lambda and 82. J Mol Biol. 1989;210:453–460. doi: 10.1016/0022-2836(89)90122-8. [DOI] [PubMed] [Google Scholar]
- 103.Yang X J, Roberts J W. Gene Q antiterminator proteins of Escherichia coli phages 82 and lambda suppress pausing by RNA polymerase at a rho-dependent terminator and at other sites. Proc Natl Acad Sci USA. 1989;86:5301–5305. doi: 10.1073/pnas.86.14.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yanofsky C, Konan K V, Sarsero J P. Some novel transcription attenuation mechanisms used by bacteria. Biochimie. 1996;78:1017–1024. doi: 10.1016/s0300-9084(97)86725-9. [DOI] [PubMed] [Google Scholar]
- 105.Yarnell W S, Roberts J W. The phage lambda gene Q transcription antiterminator binds DNA in the late gene promoter as it modifies RNA polymerase. Cell. 1992;69:1181–1189. doi: 10.1016/0092-8674(92)90639-t. [DOI] [PubMed] [Google Scholar]