Abstract
The Asian citrus psyllid, Diaphorina citri, is the insect vector of the causal agent of huanglongbing (HLB), a devastating bacterial disease of commercial citrus. Presently, few genomic resources exist for D. citri. In this study, we utilized PacBio HiFi and chromatin confirmation contact (Hi-C) sequencing to sequence, assemble, and compare three high-quality, chromosome-scale genome assemblies of D. citri collected from California, Taiwan, and Uruguay. Our assemblies had final sizes of 282.67 Mb (California), 282.89 Mb (Taiwan), and 266.67 Mb (Uruguay) assembled into 13 pseudomolecules—a reduction in assembly size of 41–45% compared with previous assemblies which we validated using flow cytometry. We identified the X chromosome in D. citri and annotated each assembly for repetitive elements, protein-coding genes, transfer RNAs, ribosomal RNAs, piwi-interacting RNA clusters, and endogenous viral elements. Between 19,083 and 20,357 protein-coding genes were predicted. Repetitive DNA accounts for 36.87–38.26% of each assembly. Comparative analyses and mitochondrial haplotype networks suggest that Taiwan and Uruguay D. citri are more closely related, while California D. citri are closely related to Florida D. citri. These high-quality, chromosome-scale assemblies provide new genomic resources to researchers to further D. citri and HLB research.
Keywords: citrus greening disease, huanglongbing, California, Taiwan, Uruguay
1. Introduction
The Asian citrus psyllid, Diaphorina citri, Kuwayama, 1908 (Hemiptera: Liviidae), is a phloem-feeding insect and serious pest of Citrus spp. and other related plants of the Rutaceae. It is a current threat to worldwide commercial citrus production due to its role in vectoring the causal agent of huanglongbing (HLB, also known as citrus greening disease), Candidatus Liberibacter asiaticus (CLas).1Diaphorina citri likely originated from the Indian subcontinent and radiated into southwestern and southeastern Asia. Diaphorina citri was first described in 1908 from specimens collected on the island of Taiwan2 and later appeared in mainland China in 1934.3 By 1942, D. citri had established in Brazil.4 The first report of D. citri in the USA occurred in 1998 in Florida,5 although HLB was not detected there until 2004.6 Since appearing in Florida, D. citri has now been detected across much of the southern USA and Mexico; in 2008, the insect was discovered in southern California.7CLas-infected D. citri and infected citrus were later discovered in residential southern California in 2012.8 So far, effective quarantine and control of HLB in southern California has kept D. citri and HLB out of the citrus producing Central Valley.9
Diaphorina citri and HLB are now nearly ubiquitous in most citrus producing regions worldwide,1 causing massive economic losses to citrus growers.9 Control of HLB in endemic areas usually includes removal of infected trees, control of D. citri, and proper management of citrus nurseries10,11—an approach applied in Brazil with some limited success.12 Growers in Florida, however, largely chose to focus efforts on control of D. citri, rather than removal of HLB-affected citrus. While control of D. citri has been shown to be effective at improving citrus yields,13 the high cost of insecticidal treatments, risk posed to beneficial insects, and the development of insecticide-resistant D. citri have made chemical control difficult in Florida.13–19 The near-100% incidence of HLB and D. citri in south Florida groves poses a threat to northern Florida and southern Georgia groves which have thus far maintained low incidence of the disease.9 The HLB epidemic has been slow growing in Texas relative to Florida, yet without intervention, the disease is predicted to reach 100% incidence by the mid-2030s.20 California’s Central Valley, the leading producer of fresh citrus in the USA, remains threatened by HLB and D. citri present in non-commercial citrus in the nearby Los Angeles basin. Together, these highlight the need to develop better control methods to keep D. citri and HLB from spreading further.
Genomic resources for D. citri have been limited. In 2014, a short-read D. citri genome assembly, complemented by PacBio RS long-reads (Diaci v1.1, NCBI BioProject PRJNA29447), was published.21 This assembly was generated from D. citri collected in Florida and has a total assembly size of 485,705,082 bp in 161,988 scaffolds and has a low contig N50 of 34,407 bp. Additionally, BUSCO analysis of this assembly revealed that over one-third of conserved single-copy genes were absent or fragmented.22 Since then, sequencing technology has rapidly advanced, providing longer, more accurate sequencing reads, improved methods of scaffolding contigs, and lower DNA input requirements.23–25 In 2018, the Diaci v2.0 reference assembly was released, utilizing PacBio long-reads and Dovetail sequencing (Scott’s Valley, California, USA) to increase the contig N50 to 759 Kbp (www.citrusgreening.org, July 2021, date last accessed). While a chromosome-length reference assembly of Floridian D. citri was produced by Hosmani et al.22 (Diaci v3.0), chromosome-scale genome assemblies and comparisons of D. citri genomes from different populations are still lacking.
In this work, we sequenced and assembled the first high-quality chromosome-scale genome assemblies for D. citri that were collected from three geographically distinct populations and maintained in inbred laboratory colonies: California (CRF-CA), Taiwan (CRF-TW), and Uruguay (CRF-UY). PacBio HiFi sequencing of a single adult male specimen from each population coupled with chromatin confirmation capture (Hi-C) scaffolding revealed a haploid genome size of 282.67 Mb for CRF-CA, 282.89 Mb for CRF-TW, and 266.76 Mb for CRF-UY. Genome size estimates from flow cytometry further supported this haploid assembly size. Nearly all scaffolds of each D. citri assembly were organized into 12 autosomes and one X chromosome typical of other psyllids,26 massively improving upon the contig N50, scaffold N50, and BUSCO completeness scores of previous assemblies. We annotated protein-coding genes, ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), piwi-interacting RNA (piRNA) clusters, and identified putative endogenous viral elements (EVEs) in each D. citri genome assembly. Additionally, we assembled the mitochondrial genomes (mitogenomes) from our psyllids and constructed haplotype networks to better understand the genetic relationships between these assemblies and worldwide D. citri. These highly accurate and complete chromosome-scale D. citri assemblies will provide a solid foundation for further studies and more potential effective applications for HLB control strategies.
2. Materials and methods
2.1. Maintenance of Diaphorina citri insects
Diaphorina citri insects were maintained on Citrus macrophylla plants inside mesh cages (BugDorm, Taichung, Taiwan) at 25 ± 2°C using a 14-h/10-h (light/dark) photoperiod and 60–70% relative humidity at the Contained Research Facility of the University of California, Davis (CRF; https://crf.ucdavis.edu, July 2021, date last accessed).27 Live D. citri insects from California were collected from populations in urban Los Angeles in 2011. Live D. citri insects from Taiwan and Uruguay were imported under USDA APHIS-PPQ permit P526P-17-02906 in 2015 and 2019, respectively.
2.2. Pacbio library construction and sequencing
A single adult male D. citri was collected from each colony and high-molecular weight DNA was extracted using the MagAttract HMW DNA Kit (Qiagen, Germany) following the manufacturer’s protocol. Pacific Biosciences (Menlo Park, California, USA) Single Molecule Real-Time (SMRT) sequencing libraries were prepared using the HiFi Ultra-Low Input DNA protocol (Pacific Biosciences, USA). Libraries were sequenced on the PacBio Sequel II platform using either two 8M SMRT cells (CRF-CA) or one 8M SMRT cell (CRF-TW and CRF-UY) at the UC Davis Genome Center. Generation of circular consensus sequence (CCS) reads and adapter trimming were performed using PacBio SMRTLink 10. Duplicate CCS reads were removed from each library using pbmarkdup (v1.0.2) (https://github.com/PacificBiosciences/pbmarkdup/, 1 July 2021, date last accessed) prior to assembly.
2.3. Hi-C library construction and sequencing
Approximately 0.5 g of adult D. citri were collected from each colony and ground into a fine powder in liquid nitrogen using a mortar and pestle. The resulting powder was then used as input for Hi-C library construction using the Phase Genomics Proximo Hi-C Kit (Animal) version 2.0 (Seattle, Washington, USA). Hi-C libraries were pooled and sequenced on an Illumina NovaSeq at the UC Berkeley Vincent J. Coates Genomics Sequencing Laboratory. Sequencing adapters and low-quality reads were trimmed using Trimmomatic (v0.39).28
2.4. BGIseq whole-genome shotgun library construction and sequencing
Twenty adult D. citri were collected at random from each colony and ground into a fine powder in liquid nitrogen using a mortar and pestle before suspending in 300 µl of DNA extraction buffer containing 100 mM Tris HCl pH 8.0, 50 mM EDTA pH 8.0, 500 mM NaCl, and 1% N-Lauryl-Sarcosine. Three microliters of Rnase A (10 mg/ml) were added to each homogenate before incubating in a 55°C water bath for 1 h. DNA was then extracted following standard phenol: chloroform: isoamyl alcohol extraction protocol. Purified genomic DNA was then shipped overnight to BGI Group (BGI Group, Shenzhen, China) for library construction and sequencing on a BGIseq 500.
2.5. Transcriptome library construction and sequencing
Two samples of total RNA were extracted from 20 pooled CRF-CA D. citri, per sample, using TRIzol (Invitrogen, USA) following the manufacturer’s protocol. Purified RNAs were used to construct poly-A transcriptome RNA sequencing libraries. The transcriptome libraries were prepared and sequenced on an Illumina Hi-Seq 2500 Illumina by LC Sciences (Houston, Texas, USA). Transcriptome reads were trimmed of adapter sequences and low-quality reads using Trimmomatic (v 0.39).28
2.6. De novo genome assembly and evaluation
We first filtered endosymbiont reads from the deduplicated CCS reads by using minimap2 (v 2.19)29 to map reads to published D. citri endosymbiont and CLas genomes (NCBI accession numbers: NZ_CP012591.1, NZ_CP012592.1, NZ_CP019943.1, KB223528 to KB223540, and NZ_JMIL00000000.2) and retained unmapped reads using samtools (v1.9).30 Filtered CCS reads were then assembled using hifiasm (v 0.15.2 r334)31 with default haplotig purging parameters and ‘–primary’. Trimmed Hi-C reads were mapped to their respective primary assemblies using BWA (v 0.7.17)32 and chromosomal contact frequency maps were generated with Juicer (v 1.5.6).33 Primary assembly contigs were then scaffolded into superscaffolds (pseudomolecules/chromosomes) using 3D-DNA (v 180922)34 with default parameters and ‘–editor-repeat-coverage 25’ followed by manual review in Juicebox Assembly Tools (v 1.11.08).35 The remaining small scaffolds not assigned to a superscaffold were manually screened for contaminating sequence by web BLASTn and removed if they had significant non-arthropod hits.
QUAST (v 5.0.2)36 was used to evaluate general assembly statistics. Genome completeness was determined using BUSCO (v 5.1.2),37 with the following ortholog datasets: Hemiptera_odb10, Insecta_odb10, and Arthropoda_odb10 (all last accessed 14 June 2021). Genome assemblies were visualized using shinyCircos.38
2.7. Flow cytometry genome size estimation
Flow cytometric genome size estimates were made following the methods used by Johnston et al.39 In brief, the head of a single male or female CRF-CA D. citri was placed into 1 ml of cold Galbraith buffer in a 2-ml Dounce grinding tube along with ½ head of a female yw Drosophila melanogaster (1C = 175 Mb). Nuclei were released by 15 strokes of the loose (A) pestle at a rate of 3 strokes every 2 s. The resulting mixture was filtered through 0.45 µm nylon filter and stained with propidium iodide to a final concentration of 1 µl/ml. Following 2+ hours of staining in the cold and dark, the mean red fluorescence for a minimum of 1,000 standard and 1,000 sample nuclei was determined for the 2C (diploid) peaks of the standard and sample using a Beckman/Coulter Cytoflex flow cytometer and associated software. The 1C amount of DNA in the psyllid is estimated as the ratio, mean red fluorescence (expressed as a mean channel number) of the 2C peak of the psyllid divided by mean red fluorescence of the 2C peak of the standard, times the 1C amount of DNA in the standard.
2.8. Repeat element identification, genome annotation, and structural variant analysis
We generated de novo repeat libraries for each assembly using RepeatModeler2 (v 2.0.1)40 with default settings and ‘-LTRStruct’ to identity long terminal repeats (LTRs) as well. The resulting repeat libraries were used to predict transposable elements (TEs) and mask complex repeats with RepeatMasker (v 4.1.2).41 The transcriptome reads were mapped to each assembly using Hisat2 (v 2.2.1)42 and used as evidence along with the OrthoDB43 arthropod protein dataset (arthropoda_odb10, last accessed 14 June 2021) to train ProtHint, GeneMark-ETP+, and AUGUSTUS, as implemented in the BRAKER2 (v 2.1.6) gene prediction and annotation pipeline.44–54 Predicted protein-coding genes have been deposited in GenBank under BioProject PRJNA800468. rRNA and tRNAs were identified and annotated using RNAmmer (v 1.2)55 and tRNAscan-SE (v 1.3.1),56 respectively.
Completeness of protein predictions were evaluated using BUSCO (v 5.1.2) with the Hemipteran_odb10, Insecta_odb10, and Arthropoda_odb10 datasets (all last accessed 14 June 2021). Predicted proteins were annotated and assigned gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)57 terms using EnTAP (v 0.10.8)58 in ‘–runP’ mode using the RefSeq Invertebrate and UniProt SwissProt databases (last accessed 11 February 2022). Assigned KEGG terms were mapped to pathways using KEGG Mapper (v 5).59,60 Predicted proteins were grouped into orthologous clusters and then compared across all three assemblies using the OrthoVenn2 webserver.61
The CRF-CA genome assembly was selected as the reference genome for structural variant calling. CRF-TW and CRF-UY genomes were mapped to CRF-CA using minimap2 (v 2.19)29 and then sorted into bam files using samtools (v 1.9).30 Structural variants were then called using SVIM-asm (v 1.0.2) using default settings.62
2.9. Sex chromosome identification
Flow cytometric analyses suggests that D. citri have a XX/X0 sex determination system, whereby males possess only a single X chromosome copy. To confirm this, we mapped each mixed-sex short-read library and each male-only HiFi library to their respective assemblies using either BWA-MEM (v 0.7.17)32 or minimap2 (v 2.19).29 Mapped reads were then sorted and indexed using samtools (v 1.9).30 Each genome was divided into 10 kb sliding windows using bedtools (v 2.30.0)63 and sequencing depth was calculated using mosdepth (v 0.3.2).64
2.10. piRNA cluster and EVE identification
Small RNA (sRNA) libraries from Nigg et al.65 were downloaded from the NCBI sequence read archive (SRA accessions in Supplementary Table S1) and trimmed of adapter sequences and low-quality reads using Trimmomatic (v 0.39),28 retaining reads as short as 18 nucleotides. Trimmed sRNA libraries for each D. citri population were concatenated and used with proTRAC (v 2.4.2)66 under default settings to predict and define piRNA clusters in each assembly.
EVEs were identified in each assembly using modified scripts derived from ter Horst et al.67 In brief, we created a BLAST database of all non-retroviral ssRNA, dsRNA, and ssDNA virus protein sequences deposited in GenBank (last accessed 9 August 2021), excluding prokaryotic and chordate-infecting viruses. Viral sequences were then clustered at 100% amino acid identity using USEARCH (v 8.1.1861)68 to create the final non-redundant viral sequence database. Each assembly was then searched for matches to viral proteins using BLASTx with default parameters and ‘-evalue 0.001 -outfmt 5’. The resulting XML files were then parsed and filtered to remove duplicate and overlapping viral hits, retaining hits with higher bitscores, using the custom Python script ‘Parse_EVE_XML.py’. Nucleotide sequences for each filtered putative EVE were extracted from each assembly and reverse BLASTx searched against the highly complete D. melanogaster proteome (Uniprot proteome accession number UP000000803) using default parameters and ‘-max_target_seqs 1 -max_hsps 1 -evalue 0.001 -outfmt 10’ with the custom Python script ‘Run_Drosophila_BLASTx.py’. EVEs with hits to the D. melanogaster proteome were removed. Viral proteins corresponding to remaining putative EVEs were manually compared with the nonredundant protein database using web-based BLASTp with default settings. Any EVE whose corresponding viral protein sequence hit a nonviral or conserved domain not exclusive to viruses was removed from the list.
2.11. Mitogenome assembly and haplotype network construction
Mitochondrial genomes for each inbred D. citri population were assembled from adapter-cleaned WGS short-reads using MEGAHIT (v 1.1.3)69 implemented in MitoFinder (v 1.4),70 with the complete D. citri mitogenome sequence from Wu et al.71 used as a reference (NCBI Reference Sequence: NC_030214.1). Mitochondrial tRNAs were annotated with MitFi (v 0.1).72 Mitochondrial genomes were deposited in GenBank (accession numbers OM181945-OM181947).
We downloaded all available complete D. citri mitogenomes published on NCBI (accession numbers in Supplementary Table S2) as well as partial or complete D. citri cytochrome c oxidase I (COI) sequences (accession numbers in Supplementary Table S3). Complete coding sequences from this study’s mitogenomes and those published on NCBI were extracted and then aligned using ClustalW73 implemented in MEGAX (v 10.0.4).74 DnaSP6 (v 6.12.01)75 was then used to call haplotypes and calculate nucleotide diversity,76 haplotype diversity, Strobeck’s S Statistic,77 and Fu and Li’s D* and F* statistics78 from the alignments. PopART (v 1.7)79 was used to generate parsimony informative TCS haplotype networks.80 The same process was repeated for all partial COI sequences.
3. Results
3.1. Diaphorina citri genome assembly and size estimation
3.1.1. De novo sequencing and assembly
We generated 33.7 Gb (∼119× theoretical coverage), 27.9 Gb (∼98×), and 26.8 Gb (∼100×) of clean PacBio HiFi reads for CRF-CA, CRF-TW, and CRF-UY, respectively. For scaffolding assemblies, we generated 38.4, 69.4, and 39.4 Gb of adapter-trimmed in vivo chromatin conformation capture (Hi-C) reads. Additionally, 60.8, 61.0, and 61.0 Gb of clean WGS short-reads were generated for CRF-CA, CRF-TW, and CRF-UY, respectively.
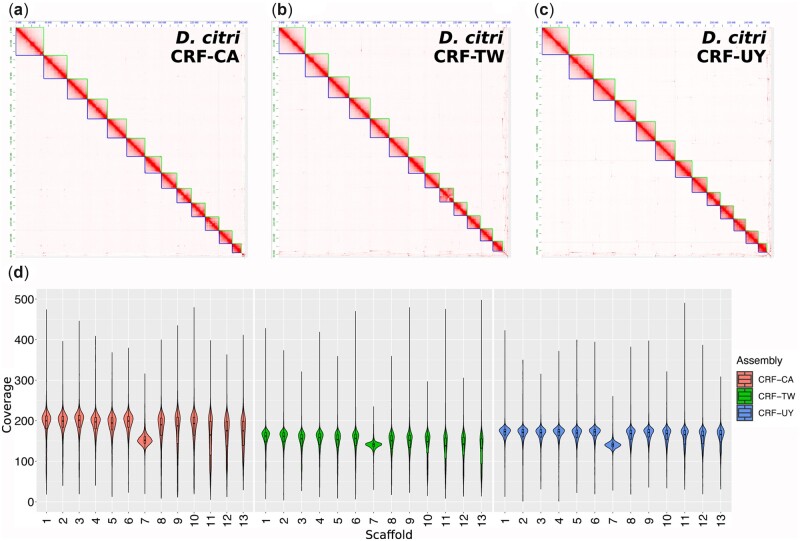
Following assembly of PacBio HiFi reads and subsequent Hi-C scaffolding, we obtained final primary assembly sizes of 282.67 Mb for CRF-CA, 282.89 Mb for CRF-TW, and 266.67 Mb for CRF-UY (Table 1). Each assembly was scaffolded into 13 chromosome-length pseudomolecules (Fig. 1a–c) containing 97.61–98.33% of all assembled contigs (Table 1). Our assemblies are highly contiguous, with contig N50s of 1.34 Mb for CRF-CA, 0.6 Mb for CRF-TW, and 0.4 Mb for CRF-UY. BUSCO analysis using the Hemipteran_odb10 dataset indicate that our assemblies have high levels of completeness, ranging from 93.7% to 94.4% complete (Tables 1 and 2), and low BUSCO duplication rates, from 2% to 3.4% (Table 2). Additional high completeness and low duplication rates were observed for the Insecta_odb10 and Arthropoda_odb10 datasets (Table 2).
Table 1.
Assembly metrics for each D. citri population
| Assembly | CRF-CA | CRF-TW | CRF-UY |
|---|---|---|---|
| Assembly size (Mb) | 282.67 | 282.89 | 266.67 |
| % Ns | 0.10 | 0.18 | 0.22 |
| # of contigsa | 899 | 1526 | 1615 |
| Contig N50 (Mb) | 1.34 | 0.60 | 0.40 |
| # of scaffolds | 345 | 452 | 382 |
| Scaffold N50 (Mb) | 22.62 | 23.56 | 22.57 |
| # of chromosomes | 12A + 1X | 12A + 1X | 12A + 1X |
| % of assembly in chromosome-length scaffolds | 98.36 | 97.57 | 98.07 |
| Protein coding genes | 20184 | 20357 | 19083 |
| BUSCO % completenessb | 94.3 | 94.2 | 93.7 |
Note: A, Autosome; X, X chromosome.
Scaffolds split on runs of 10 or more Ns.
Complete and duplicated conserved Hemipteran benchmarking universal single-copy orthologs (BUSCO).
Figure 1.
Assembly chromatin contact frequency maps and sequencing coverage. (a–c) Hi-C chromatin confirmation contact frequency heatmaps of D. citri collected from geographically distinct populations maintained in laboratory colonies: California (CRF-CA) (a), Taiwan (CRF-TW) (b), and Uruguay (CRF-UY) (c). (d) Short-read, mixed-sex whole-genome shotgun sequencing average coverage per superscaffold (chromosome) across 10 kb sliding windows of CRF-CA, CRF-TW, and CRF-UY.
Table 2.
Benchmarking universal single-copy ortholog (BUSCO) statistics for each D. citri assembly and protein set
| CRF-CA genome |
CRF-CA protein |
CRF-TW genome |
CRF-TW protein |
CRF-UY genome |
CRF-UY protein |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | # | % | # | % | # | % | # | % | # | % | # | ||
| Arthropoda (n: 1,013) | Completeness | 92 | 932 | 94.3 | 955 | 91.7 | 929 | 94.7 | 959 | 90.9 | 921 | 94.9 | 962 |
| Complete and single copy | 89.8 | 910 | 79.1 | 801 | 87.8 | 890 | 77.6 | 786 | 88.9 | 901 | 82.1 | 832 | |
| Complete and duplicated | 2.2 | 22 | 15.2 | 154 | 3.8 | 39 | 17.1 | 173 | 1.9 | 20 | 12.8 | 130 | |
| Fragmented | 4.2 | 43 | 2.3 | 23 | 4.6 | 47 | 1.7 | 17 | 5.2 | 53 | 2.1 | 21 | |
| Missing | 3.8 | 38 | 3.4 | 35 | 3.6 | 37 | 3.6 | 37 | 3.8 | 39 | 3 | 30 | |
| Insecta (n: 1,367) | Completeness | 91.8 | 1254 | 94.4 | 1291 | 91.8 | 1255 | 95.1 | 1300 | 91.3 | 1249 | 95.5 | 1305 |
| Complete and single copy | 89.8 | 1227 | 80.3 | 1098 | 87.5 | 1197 | 78.1 | 1067 | 89.3 | 1222 | 81.9 | 1119 | |
| Complete and duplicated | 2 | 27 | 14.1 | 193 | 4.2 | 58 | 17 | 233 | 1.9 | 27 | 13.6 | 186 | |
| Fragmented | 4.5 | 61 | 2 | 27 | 4.4 | 61 | 1.7 | 23 | 4.9 | 67 | 1.8 | 24 | |
| Missing | 3.7 | 52 | 3.6 | 49 | 3.7 | 51 | 3.2 | 44 | 3.7 | 51 | 2.7 | 38 | |
| Hemiptera (n: 2,510) | Completeness | 94.3 | 2368 | 95.3 | 2392 | 94.2 | 2366 | 95.4 | 2395 | 93.7 | 2352 | 96.1 | 2411 |
| Complete and single copy | 91.8 | 2304 | 77.7 | 1951 | 90.8 | 2280 | 77.4 | 1942 | 91.6 | 2301 | 80.7 | 2025 | |
| Complete and duplicated | 2.5 | 64 | 17.6 | 441 | 3.4 | 86 | 18 | 453 | 2 | 51 | 15.4 | 386 | |
| Fragmented | 3.3 | 82 | 1.6 | 40 | 3.3 | 85 | 1.4 | 34 | 3.5 | 90 | 0.8 | 21 | |
| Missing | 2.4 | 60 | 3.1 | 78 | 2.3 | 59 | 3.2 | 81 | 2.7 | 68 | 3.1 | 78 | |
3.1.2. Flow cytometry genome size estimation
Diaphorina citri genome size estimation was further confirmed by flow cytometric analyses using adult CRF-CA D. citri. Heads from CRF-CA D. citri yielded a mean 1C genome size estimate of 274.4 Mb for males (n = 6, s.e. = 1.4 Mb) and 287.0 Mb for females (n = 6, s.e. = 1.7 Mb), closely consistent with our assembly sizes. The difference between male and female 2C values was 25.4 Mb ± 3.6 Mb.
3.2. Genome annotation, gene prediction, and structural variant analyses
In total, repeat masking of CRF-CA, CRF-TW, and CRF-UY assemblies resulted in 38.26% (108,248,856 bp), 37.91% (107,246,725 bp), and 36.87% (98,311,742 bp) being identified as repeat regions, respectively (Table 3). Among major classes of repeat elements, LTRs and DNA transposon elements were the most abundant, at 4.05% and 4.03% of CRF-CA, 3.4% and 3.97% of CRF-TW, and 4.2% and 2.89% of CRF-UY, respectively. Approximately 44–47% of all repeat regions identified in each assembly remain unclassified (Table 3).
Table 3.
Repetitive element content for each D. citri population
| Assembly |
CRF-CA |
CRF-TW |
CRF-UY |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Class | Number | Length (bp) | % of assembly | Number | Length (bp) | % of assembly | Number | Length (bp) | % of assembly |
| SINE | 35,857 | 5,465,588 | 1.93 | 48,090 | 5,839,290 | 2.06 | 27,534 | 3,696,644 | 1.39 |
| LINE | 27,413 | 8,029,488 | 2.84 | 25,870 | 8,086,480 | 2.86 | 21,579 | 6,817,727 | 2.56 |
| LTR | 28,907 | 11,446,977 | 4.05 | 19,453 | 9,607,570 | 3.4 | 24,155 | 11,199,711 | 4.2 |
| DNA | 61,270 | 11,407,505 | 4.03 | 59,803 | 11,233,280 | 3.97 | 39,645 | 7,715,657 | 2.89 |
| RC | 39,608 | 6,659,795 | 2.35 | 45,897 | 7,218,552 | 2.55 | 31,826 | 5,408,708 | 2.03 |
| Other | 245,324 | 18,068,785 | 6.39 | 242,703 | 17,972,967 | 6.35 | 235,091 | 17,102,413 | 6.47 |
| Unclassified | 350,011 | 47,170,718 | 16.67 | 335,817 | 47,288,586 | 16.72 | 330,443 | 46,190,882 | 17.32 |
| Total | 788,390 | 108,248,856 | 38.26 | 777,633 | 107,246,725 | 37.91 | 710,273 | 98,311,742 | 36.87 |
SINE, short interspersed nuclear elements; LINE, long interspersed nuclear elements; LTR, long terminal repeats; DNA, DNA elements; RC, rolling circle elements; Other, small RNA, satellite DNA, simple repeats, and low complexity repeats; bp, base pair.
BRAKER2 predicted 20,184 protein-coding gene models in the CRF-CA assembly, 20,357 gene models in the CRF-TW assembly, and 19,083 gene models in the CRF-UY assembly (Table 1). BUSCO analysis of predicted proteins using the Hemipteran_odb10 dataset showed high completeness with scores of 95.3% (CRF-CA), 95.4% (CRF-TW), and 96.1% (CRF-UY) complete (Table 2). Similar BUSCO scores were observed using the Insecta_odb10 and Arthropoda_odb10 datasets (Table 2).
From our predicted gene models, we used EnTAP to successfully annotate 16,047 gene models in CRF-CA, 15,985 gene models in CRF-TW, and 15,198 gene models in CRF-UY. 895,968 GO terms were assigned to 12,462 CRF-CA genes, 870,081 GO terms were assigned to 12,462 CRF-TW genes, and 844,334 GO terms were assigned to 11,909 CRF-UY genes. A total of 4,508, 4,417, and 4,223 genes from CRF-CA, CRF-TW, and CRF-UY, respectively, were successfully assigned KEGG orthology (KO) terms and accordingly mapped to 309, 312, and 309 biological pathways. Predicted proteins clustered into a total of 15,598 orthologous groups, with 14,554, 14,501, and 14332 orthologous groups belonging to CRF-CA, CRF-TW, and CRF-UY, respectively (Supplementary Fig. S1). A total of 12,612 orthologous groups were shared between all three D. citri protein sets and 417 groups were unique to at least one population set.
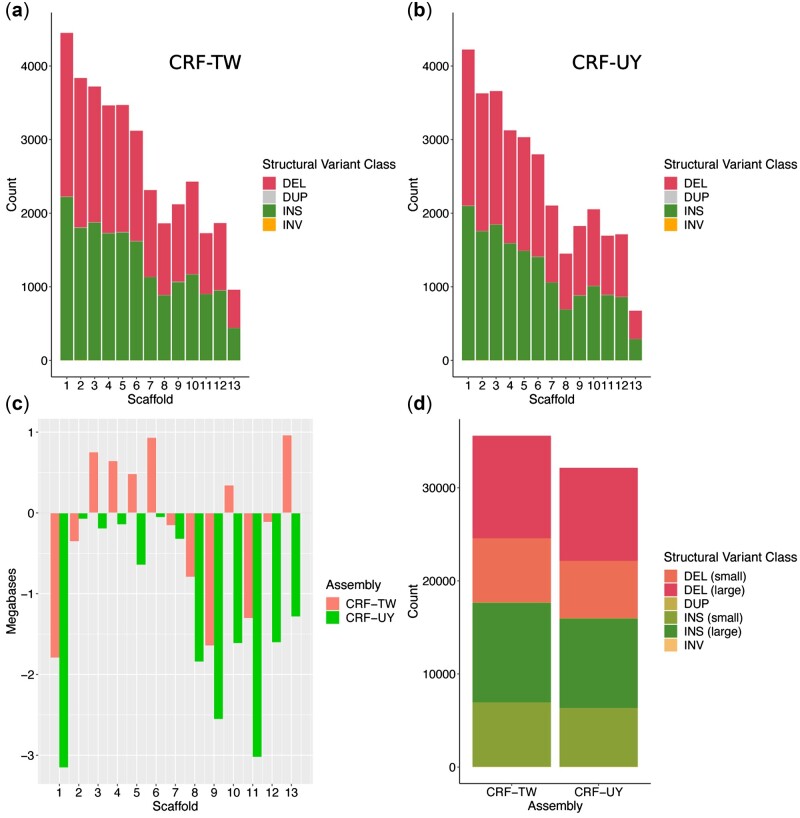
SVIM-asm identified 17,642 insertions, 17,890 deletions, 41 duplications, and 9 inversions in CRF-TW relative to CRF-CA, and 15,950 insertions, 16,150 deletions, 26 duplications, and 10 inversions in CRF-UY relative to CRF-CA (Fig. 2a, b, d). The largest insertion identified in CRF-TW was 91,041 bp on Scaffold 13, while the largest insertion in CRF-UY was 93,516 bp on Scaffold 5. A 99,815 bp deletion on Scaffold 12 was identified in CRF-TW, and a 93,591-bp deletion was identified in CRF-UY on Scaffold 9. CRF-UY’s Scaffold 1 showed the largest size discrepancy relative to CRF-CA, with a size difference of ∼−3.15 Mb, followed by Scaffold 11 with a difference of ∼−3.02 Mb (Fig. 2c).
Figure 2.
Structural variants and superscaffold size discrepancies of CRF-TW and CRF-UY relative to CRF-CA. (a) CRF-TW structural variant counts per superscaffold. (b) CRF-UY structural variant counts per superscaffold. (c) Superscaffold size differences in megabases per assembly relative to CRF-CA. (d) Overall structural variant counts per assembly, relative to CRF-CA. DEL (small), deletions ≤100 bp; DEL (large), deletions >100 bp; DUP: duplications; INS (small): Insertions ≤100 bp; INS (large), insertions > 100 bp; INV, inversions. DEL, deletions; DUP, duplications; INS, insertions; INV, inversions.
3.3. Sex chromosome identification
Flow cytometry results supports a XX/X0 sex determination system in D. citri, with females carrying two X chromosomes and males carrying a single copy. Here the difference in mean 2C values between males and females suggested a size of 25.4 Mb ± 3.6 Mb for the X chromosome. Based on this information, we expected to find one chromosome-scale scaffold with approximately 50% coverage in our male-only PacBio HiFi libraries and approximately 75% coverage in our mixed-sex short-read libraries relative to other chromosome-scale scaffolds.
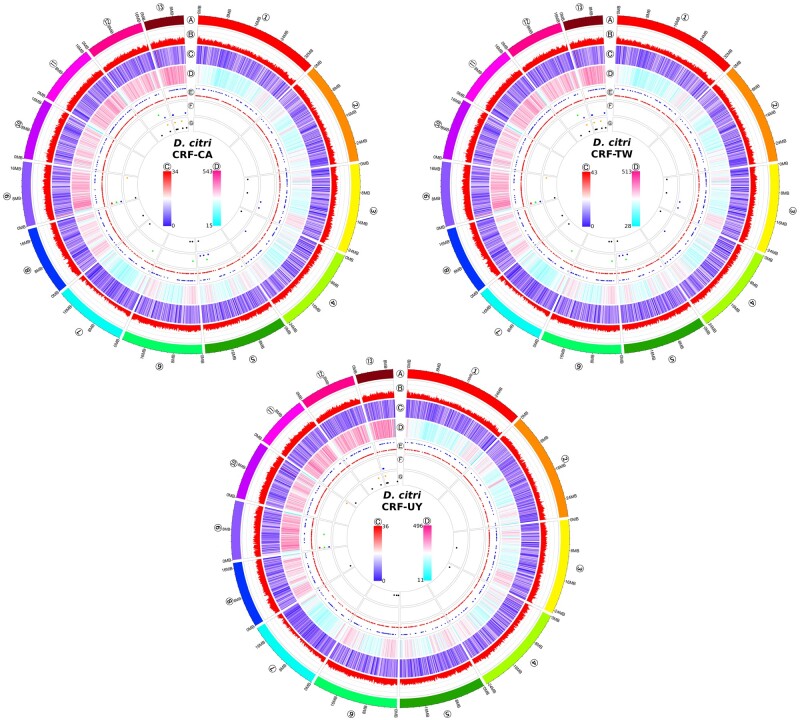
We found scaffold 7 (∼20 Mb) to have approximately 50% coverage relative to scaffolds that have similar repeat density (Fig. 3, scaffolds 1 through 6) when mapping male PacBio HiFi reads (Supplementary Fig. S2); however, because of high repeat density in scaffolds 8 through 13 (Fig. 3, track D) and the subsequent difficulties in polymerase processivity during PCR-based library construction, these scaffolds also have lower coverage relative to scaffolds 1 through 6. For this reason, we mapped mixed-sex short-read libraries to each assembly and calculated coverage. Again, scaffold 7 was found to have lower coverage than all others, at approximately 75% coverage (Fig. 1d). Finally, syntenic analysis between CRF-CA D. citri and another recently published psyllid genome, Pachypsylla venusta (NCBI accession GCA_012654025.1; Li et al.81) showed high synteny between D. citri scaffold 7 and P. venusta scaffold 2870, identified by Li et al.81 as the putative X chromosome (Supplementary Fig. S3).
Figure 3.
Circos plots of chromosomal features of each Diaphorina citri geographic population, excludingunplaced scaffolds. Tracks are, from outermost to innermost: (A) ideogram of the 13 D. citri chromosome-scale scaffolds. (B) GC percentage in 100 kb sliding windows. (C) Gene density in 100 kb sliding windows; heatmap scale in circle centres showing local min to local max. (D) Repeat DNA density in 100 kb sliding windows; heatmap scale in circle centres from local min to local max. (E) rRNA (outer row dots) and tRNA (inner row dots). (F), piRNA clusters and their direction (antisense: outer row dots; ambisense: middle row dots; sense: inner row dots). (G) Endogenous viral element (EVE) locations (D. citri-specific virus-derived EVEs: outer row dots; all other EVEs: inner row dots). The circos plots represent D. citri collected from geographically distinct populations maintained in laboratory colonies: California (CRF-CA), Taiwan (CRF-TW), and Uruguay (CRF-UY).
3.4. piRNA cluster prediction and EVE identification
We predicted 29 piRNA clusters in CRF-CA (305,270 bp), 41 piRNA clusters in CRF-TW (569,585 bp), and 11 piRNA clusters in CRF-UY (121,000 bp) (Fig. 3, tracks F). In total, 30 EVEs were identified in CRF-CA, 40 EVEs in CRF-TW, and 32 in CRF-UY by BLASTx searches against our viral database followed by filtering steps (Supplementary Table S4). Putative EVEs with closest BLASTx hits to densoviruses account for 28–36% of all EVEs. Nine EVEs identified in CRF-CA were located inside piRNA clusters, 10 EVEs in CRF-TW fell within a piRNA cluster, and 3 EVEs identified in CRF-UY were within a predicted piRNA cluster. Most putative EVEs are located among the repeat-dense chromosomes 12 and 13 (Fig. 3, tracks G). In contrast to ter Horst et al.,67 we did not identify any plant-infecting, virus-derived EVEs within any of our assemblies. We identified EVEs with closest BLASTx hits to two D. citri-specific viruses: Diaphorina citri densovirus (DcDV) and Diaphorina citri flavi-like virus (DcFLV). The DcFLV-derived EVE was the largest integration identified by our pipeline, spanning 2,696 nucleotides and sharing 77.07% identity at the deduced amino acid level. The DcDV-derived EVE reported by Nigg et al.65 could not be identified in any of our assemblies, their alternate haplotigs, nor their raw PacBio subreads; however, we were able to PCR amplify this EVE from CRF-CA and CRF-TW D. citri DNA.
3.5. Mitogenome assembly and worldwide D. citri haplotype networks
We assembled complete mitogenomes for each inbred D. citri population. CRF-CA, CRF-TW, and CRF-UY mitogenomes each possesses the typical 13 protein coding genes, 2 ribosomal RNAs, 22 tRNAs, and variable length control region and are 15,038, 15,145, and 14,965 bp in length, respectively.
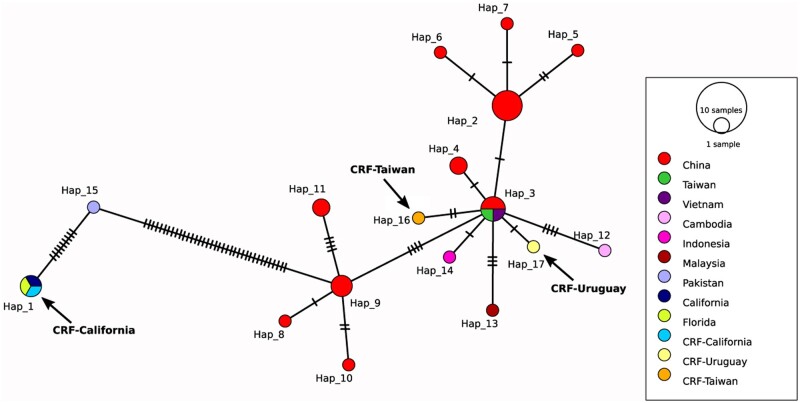
Analysis of full mtDNA coding sequence alignments (14,060 bp) from 31 D. citri mitogenomes revealed 72 variable sites resulting in 17 unique haplotypes. Haplotype 2 was most common, shared by six D. citri from China (Fig. 4). Each haplotype is separated from its neighbouring haplotype by nucleotide substitutions ranging from 1 to 37. CRF-TW (haplotype 16) and CRF-UY (haplotype 17) are separated from haplotype 3 by two and one nucleotide substitution, respectively. CRF-CA shares haplotype 1 with a Floridian mitogenome and another Californian mitogenome. Haplotype 1 neighbours haplotype 15, containing a single Pakistani mitogenome, and is separated by 9 nucleotide substitutions. In turn, haplotype 15 neighbours haplotype 9, separated by 37 nucleotide substitutions.
Figure 4.
Full mitochondrial DNA coding sequence TCS haplotype network coded by geographic region. Circle size is scaled to the number of haplotypes. Hatch marks represent single-nucleotide substitutions. CRF-CA is in haplotype 1, CRF-TW is in haplotype 16, and CRF-UY is in haplotype 17. Arrows added for emphasis. Haplotype 2 is most common (n = 6). Regions labelled ‘Taiwan’ and ‘California’ represent mitogenomes downloaded from NCBI. Accession numbers available in Supplementary Table S2.
Nucleotide diversity (π) for full mtDNA coding sequences was low, at 0.00099 ± 0.00028, while haplotype diversity (h) was high, at 0.938 ± 0.025, indicating a high number of closely related haplotypes. Strobeck’s S statistic (probability that Nhap ≤ 17) was 0.625. Fu and Li’s D* and F* statistics were 0.08828 and −0.28099, respectively, though neither was significant (P > 0.10).
In addition to analyzing the full mitochondrial coding sequences from our assemblies, we performed the same analyses using alignments of our mitogenomes and partial D. citri COI sequences published on NCBI (accession numbers in Supplementary Table S3). Alignment of 240 partial D. citri COI sequences (563 bp) from 21 countries revealed six variable sites and seven haplotypes (Supplementary Fig. S4). CRF-CA belonged to Haplotype 1, the most common haplotype, shared by 123 samples, including all Floridian samples. CRF-TW and CRF-UY belonged to Haplotype 2, the second most abundant haplotype with 90 samples. The remaining haplotypes (3–7) were common to 1–14 samples. Each COI haplotype is separated from its neighbouring haplotype by one nucleotide substitution.
Diaphorina citri partial COI nucleotide diversity (π) was low (0.00126 ± 0.00007) and haplotype diversity (h) was moderate (0.594 ± 0.019). Strobeck’s S statistic (probability that Nhap ≤ 7) was 0.926. Fu and Li’s D* and F* test statistics were, respectively, −1.06103 and −1.05941, though neither was significant (P > 0.10).
4. Discussion
We present in this work, three high-quality, chromosome-scale genome assemblies of D. citri laboratory colonies originally collected from California, Taiwan, and Uruguay—all citrus producing regions either affected or threatened by HLB.1,9,82 These assemblies are the first chromosome-scale genome assemblies for these D. citri populations and are the second chromosome-scale assemblies for a psyllid species.81 Our assemblies are approximately 55–58% the length of the Diaci v1.1 assembly21 and 56–59% the length of the Diaci v3.0 assembly.22 Due to the large discrepancy between our assembly sizes and the previously published assemblies, we utilized flow cytometry to estimate the genome size of male and female D. citri39—the first flow cytometric estimates for this species. These estimates were within ±2–4% of our assembly sizes (Table 1) and in combination with the observed high completeness and low duplicate BUSCO scores (Table 2), support our assembly sizes as close to the true genome size of D. citri. The inflated Diaci v1.1 and v3.0 assemblies could be due to under-collapsed heterozygosity—an issue for sequencing small-bodied organisms that often requires the pooling of many individuals for sufficient sequencing input. It is unclear how the Diaci v1.1 libraries were initially constructed,21 nor how many adult D. citri were pooled to construct long-read libraries by Hosmani et al.22 Assemblies utilizing long read technology from heterozygous organisms can suffer from assembly software failing to recognize and remove haplotigs from the assemblies, thus leading to artifactual duplications in the final genome.83 While Hosmani et al.22 applied redundans84 to remove redundancy in unplaced scaffolds in Diaci v3.0, no purging of haplotigs in their putative chromosome-scale scaffolds was described.
While CRF-CA and CRF-TW’s final assembly sizes are in close agreement, our CRF-UY assembly is ∼16.14 Mb shorter overall, with ∼11.9 Mb of this difference arising from repeat-dense Scaffolds 8–13 (Fig. 2c). CRF-UY had the fewest annotated genes (Table 1), the lowest repeat DNA content (Table 3), and the lowest contiguity (Table 1). Despite this, CRF-UY showed roughly equivalent BUSCO scores compared with CRF-CA and CRF-TW (Table 2). The shorter assembly could be attributed to PCR bias during library preparation for that sample, or to over-purging of haplotigs during assembly. The CRF-UY assembly size could also be a case of genome size plasticity, as large, intraspecific genome size variation has been observed from extensive sampling within flea populations.85 Additional sampling and resequencing of individuals within and among D. citri populations, particularly using non-PCR amplified libraries, could help resolve whether these differences in genome size are real or artifacts.
The difference between the mean 2C values for female and male D. citri indicated that males are the heterogametic sex, possessing a single X chromosome ∼25.4 Mb ± 3.6 Mb in size. Mapping of PacBio reads from our male-only libraries pointed towards Scaffold 7 (∼20 Mb) as the putative X chromosome with ∼50% coverage compared with Scaffolds 1–6, however, Scaffolds 8–13 also showed lower coverage (Supplementary Fig. S2) likely due to their higher repeat content affecting the PCR-based library construction (Fig. 3, tracks D). Mapping our mixed-sex short-read libraries identified Scaffold 7 as having approximately 75% coverage relative to all others (Fig. 1d), indicative of the sex chromosome in an XX/X0 sex-determination system, common to other psyllids.26 Scaffold 7 also shares strong synteny with the X chromosome recently identified in the hackberry petiole gall psyllid81 (Supplementary Fig. S3).
Nouri et al.86 identified six novel insect-specific viruses infecting D. citri populations through metagenomic sequencing and another novel D. citri virus, DcFLV87 was discovered later that year. After the identifications of the D. citri viruses, ter Horst et al.67 identified EVEs within the D. citri genome, utilizing the Diaci v1.1 assembly. Interestingly, EVEs with relatively high deduced amino acid identity to two D. citri-specific viruses were identified: DcFLV and DcDV. Nigg et al.65 explored the possible role of the DcDV-derived EVE in generating piRNAs to target infection from DcDV. They found this DcDV-derived EVE to be unevenly distributed amongst D. citri from different geographic backgrounds. Our updated EVE pipeline identified several EVEs with closest BLASTx hits to DcDV. The DcDV-derived EVE reported by Nigg et al.65 was not identified in our assemblies, though we were able to PCR amplify this EVE from CRF-CA and CRF-TW D. citri. This could be due to deficiencies in PCR-based library preparation, as most EVEs are located in the higher GC percentage, repeat-rich Scaffolds 12 and 13 (Fig. 3, tracks B and G), or could represent a polymorphism among D. citri used in this study.
Far fewer EVEs were identified in our new assemblies, compared with those discovered by ter Horst et al.,67 possibly due to utilizing an updated viral protein sequence database and the highly fragmented and inflated size of the genome used in that work. Further investigation into the role assembly contiguity may play in EVE identification is warranted.
In this work, we identified a variable number and size of piRNA clusters among populations. piRNA clusters are often located in heterochromatic regions of genomes,88 and here the majority of predicted piRNA clusters fall in the repeat-rich Scaffolds 9, 12, and 13 (Fig. 3, tracks F). Investigations using Drosophila have revealed widespread variation in TE landscapes and rapid evolution of piRNA clusters even among the same fly cell strains.89–91 Natural intraspecific variation could account for the differences in piRNA cluster counts among our assemblies, however, only a single sRNA library was available for CRF-UY (Supplementary Table S1) compared with three sRNA libraries each for CRF-CA and CRF-TW, which may have affected piRNA cluster prediction. Additionally, CRF-UY’s assembly may be missing sequence from Scaffolds 8–13 (Fig. 2c) where most piRNA clusters reside in the other assemblies.
Studies attempting to resolve D. citri invasion history and genetic structure utilizing the mitochondrial cytochrome c oxidase I (COI) gene have revealed 44 haplotypes amongst D. citri worldwide.92–95 These haplotypes largely cluster into two distinct lineages: a southeast Asian lineage (lineage A) and a southwest Asian lineage (lineage B). These works showed that D. citri in the USA are related to lineage B and that both lineages A and B are present in South America, though the predominant lineage there is A.
Metagenomic sequencing from Nouri et al.86 revealed uneven distribution of D. citri viruses between geographic populations. Nigg and Falk96 found CRF-CA D. citri to be resistant to DcDV infection while D. citri from CRF-TW and CRF-UY maintain persistent infections, speculating that different genetic backgrounds may play a role in viral susceptibility. A recent surveillance report on D. citri in Florida identified five of the D. citri-specific viruses from Nouri et al.86 present in grove-collected psyllids,97 which may suggest both haplogroups are now present there.
To better understand our D. citri genetic backgrounds, we constructed TCS haplotype networks using full mitochondrial coding sequence alignments and partial COI alignments from our study and publicly available sequences deposited in GenBank (Fig. 4 and Supplementary Fig. S4, respectively). Full mitochondrial coding sequence alignments revealed three closely related southeast Asian haplotypes (haplotypes 2, 3, and 9) and a more distant southwest Asian haplotype from a Pakistani D. citri (haplotype 15) (Fig. 4). CRF-TW and CRF-UY are closely related to haplotype 3. CRF-CA shares the same haplotype with D. citri from Florida, suggesting that D. citri in southern California were introduced from Floridian populations.71 A haplotype network of 240 partial COI sequences from 21 localities again shows D. citri cluster into two lineages, with D. citri in North America clustering into haplotype 1 (lineage B) and most D. citri from South America clustering with southeast Asian D. citri (Supplementary Fig. S4). Observed moderate-to-high haplotype diversity (0.594–0.938) coupled with low nucleotide diversity (0.00099–0.00126) suggests a high number of closely related haplotypes, indicative of a recent population expansion98 congruent with the invasion history of D. citri.
5. Conclusion
We produced high-quality, chromosome-level genome assemblies for three D. citri geographic populations from California (CRF-CA), Taiwan (CRF-TW), and Uruguay (CRF-UY) from single male specimens. These D. citri assemblies are the most complete and contiguous to date and represent D. citri present on three continents. Importantly, our assemblies have a reduction from previous D. citri assemblies of approximately 41–45% yet have higher contiguity and BUSCO completeness. This reduction was validated using flow cytometry which highlights the importance of accurate genome size estimations in de novo arthropod assemblies. These new D. citri genome assemblies will provide a better foundation for genomic research of this important agricultural pest and will allow for improved gene annotations and comparative genomic studies with other arthropod pests. Our improved D. citri genomes will serve as references for local HLB control strategies and can be used to build a D. citri pangenome.
Supplementary Material
Acknowledgements
We thank Dr Kris Godfrey for assistance with and access to CRF-CA D. citri. We thank Dr José Buenahora and Dr Hsin-Hung Yeh for providing D. citri samples. We thank Dr Joanna Chiu, Kyle Lewald, and Dr Jared Nigg for their critical reading and input on the manuscript.
Funding
This work was funded by USDA APHIS Huanglongbing Multiagency Coordination Group (AP19PPQS&T00C234 to B.W.F.) and USDA National Institute of Food and Agriculture (2020-70029-33200 to B.W.F. and Y.-W.K.).
Conflict of interest
None declared.
Contributor Information
Curtis R Carlson, Department of Plant Pathology, University of California Davis, Davis, CA 95616, USA.
Anneliek M ter Horst, Department of Plant Pathology, University of California Davis, Davis, CA 95616, USA.
J Spencer Johnston, Department of Entomology, Texas A&M University, College Station, TX 77843, USA.
Elizabeth Henry, Department of Plant Pathology, University of California Davis, Davis, CA 95616, USA.
Bryce W Falk, Department of Plant Pathology, University of California Davis, Davis, CA 95616, USA.
Yen-Wen Kuo, Department of Plant Pathology, University of California Davis, Davis, CA 95616, USA.
Data accessibility
All raw sequencing datasets have been deposited in the NCBI Sequence Read Archive (BioProject PRJNA800468). Genome assemblies and protein-coding genes have been deposited in DDBJ/ENA/GenBank under BioProject PRJNA800468, with accession numbers JAKMOW000000000, JAKMOV000000000, and JAKMOU000000000. The versions described in this article are versions JAKMOW010000000, JAKMOV010000000, and JAKMOU010000000. Mitogenomes have been deposited in GenBank, accession numbers OM181945-OM181947. All genome annotation files and the custom viral protein database have been deposited in Dryad (https://datadryad.org/, doi:10.25338/B8MW7P). The custom EVE Python scripts produced for this study are available on GitHub (https://github.com/Mrbland/Dcitri_genome_EVE_scripts_2021).
Author contributions
C.R.C. designed and performed research, analyzed data, and wrote the manuscript. A.M.t.H. wrote the EVE scripts and analyzed data. J.S.J. performed flow cytometry. E.H. performed research and assisted in the design of this work. B.W.F. assisted in the design of this work. Y.-W.K. assisted in the design of this work, analyzed data, and wrote the manuscript. All authors reviewed and approved the manuscript.
Supplementary data
Supplementary data are available at DNARES online.
References
- 1. Bové J.M. 2006, Huanglongbing: a destructive, newly-emerging, century-old disease of citrus, J. Plant Pathol., 88, 7–37. [Google Scholar]
- 2. Kuwayama S. 1908, Die psylliden Japans, Trans. Sopporo Nat. Hist. Soc., 2, 149–89. [Google Scholar]
- 3. Hoffmann W.E. 1936, Diaphorina citri Kuw. (Homoptera: Chernidae), a citrus pest in Kwangtung, Lingnan Sci. J., 15, 127–32. [Google Scholar]
- 4. Lima A.C. 1942, Insetos do Brasil, Homopteros, Ser. Didat. 4 Esc. Nac. Agron., 3, 327. [Google Scholar]
- 5. Halbert S.E., Niblett C.L., Manjunath K.L., Lee R.F., Brown L.G.. 2002, Establishment of two new vectors of citrus pathogens in Florida, Proc. Intl. Soc. Citric., IX Congress, 1016–7. [Google Scholar]
- 6. Halbert S.E. 2005, The discovery of huanglongbing in Florida. In: Proceedings of the international citruscanker and huanglongbing research workshop, Orlando FL, H-3.
- 7. California Department of Food and Agriculture. Asian Citrus Psyllid Pest Profile. https://www.cdfa.ca.gov/citrus/pests_diseases/acp/PestProfile.html (7 December 2021, date lastaccessed).
- 8. Kumagai L.B., LeVesque C.S., Blomquist C.L., et al. 2013, First report of Candidatus Liberibacter asiaticus associated with citrus huanglongbing in California, Plant Dis., 97, 283. [DOI] [PubMed] [Google Scholar]
- 9. Graham J., Gottwald T., Setamou M.. 2020, Status of huanglongbing (HLB) outbreaks in Florida, California and Texas, Trop. Plant Pathol., 45, 265–78. [Google Scholar]
- 10. Tsai C.-H., Hung T.-H., Su H.-J.. 2013, An integrated management of citrus huanglongbing in Taiwan. In: Proceedings of the 2013 international symposium on insect vectors and insect-borne diseases, TaiwanAgricultural Research Institute, pp. 193–210.
- 11. Zheng Z., Chen J., Deng X.. 2018, Historical perspectives, management, and current research of citrus HLB in Guangdong Province of China, where the disease has been endemic for over a hundred years, Phytopathology, 108, 1224–36. [DOI] [PubMed] [Google Scholar]
- 12. Bassanezi R.B., Lopes S.A., de Miranda M.P., Wulff N.A., Volpe H.X.L., Ayres A.J.. 2020, Overview of citrus huanglongbing spread and management strategies in Brazil, Trop. Plant Pathol., 45, 251–64. [Google Scholar]
- 13. Stansly P.A., Arevalo H.A., Qureshi J.A., et al. 2014, Vector control and foliar nutrition to maintain economic sustainability of bearing citrus in Florida groves affected by huanglongbing, Pest Manag. Sci., 70, 415–26. [DOI] [PubMed] [Google Scholar]
- 14. Chen X.D., Gill T.A., Pelz-Stelinski K.S., Stelinski L.L.. 2017, Risk assessment of various insecticides used for management of Asian citrus psyllid, Diaphorina citri in Florida citrus, against honey bee, Apis mellifera, Ecotoxicology, 26, 351–9. [DOI] [PubMed] [Google Scholar]
- 15. Chen X.D., Gill T.A., Ashfaq M., Pelz‐Stelinski K.S., Stelinski L.L.. 2018, Resistance to commonly used insecticides in Asian citrus psyllid: stability and relationship to gene expression, J. Appl. Entomol., 142, 967–77. [Google Scholar]
- 16. Monzo C., Qureshi J.A., Stansly P.A.. 2014, Insecticide sprays, natural enemy assemblages and predation on Asian citrus psyllid, Diaphorina citri (Hemiptera: Psyllidae), Bull. Entomol. Res., 104, 576–85. [DOI] [PubMed] [Google Scholar]
- 17. Monzo C., Stansly P.A.. 2017, Economic injury levels for Asian citrus psyllid control in process oranges from mature trees with high incidence of huanglongbing, PLoS One, 12, e0175333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tansey J.A., Vanaclocha P., Monzo C., Jones M., Stansly P.A.. 2017, Costs and benefits of insecticide and foliar nutrient applications to huanglongbing-infected citrus trees: insecticide and foliar nutrient applications to HLB-infected citrus trees, Pest Manage. Sci., 73, 904–16. [DOI] [PubMed] [Google Scholar]
- 19. Tiwari S., Mann R.S., Rogers M.E., Stelinski L.L.. 2011, Insecticide resistance in field populations of Asian citrus psyllid in Florida, Pest Manage. Sci., 67, 1258–68. [DOI] [PubMed] [Google Scholar]
- 20. Sétamou M., Alabi O.J., Kunta M., Dale J., da Graça J.V.. 2020, Distribution of Candidatus Liberibacter asiaticus in citrus and the Asian citrus psyllid in Texas over a decade, Plant Dis., 104, 1118–26. [DOI] [PubMed] [Google Scholar]
- 21. Hunter W.B., Reese J.;. The International Psyllid Genome Consortium. 2014, The Asian citrus psyllid genome (Diaphorina citri, Hemiptera), J. Citrus Pathol., 1, 143. [Google Scholar]
- 22. Hosmani P.S., Flores-Gonzalez M., Shippy T., et al. 2019, Chromosomal length reference assembly for Diaphorina citri using single-molecule sequencing and Hi-C proximity ligation with manually curated genes in developmental, structural and immune pathways, bioRxiv. 10.1101/869685. [DOI] [Google Scholar]
- 23. Lang D., Zhang S., Ren P., et al. 2020, Comparison of the two up-to-date sequencing technologies for genome assembly: HiFi reads of Pacific Biosciences Sequel II system and ultralong reads of Oxford Nanopore, GigaScience, 9, giaa123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rao S.S.P., Huntley M.H., Durand N.C., et al. 2014, A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping, Cell, 159, 1665–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schneider C., Woehle C., Greve C., et al. 2021, Two high-quality de novo genomes from single ethanol-preserved specimens of tiny metazoans (Collembola). GigaScience, 10, giab035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Labina E.S., Maryańska-Nadachowska A., Kuznetsova V.G.. 2007, Meiotic karyotypes in males of nineteen species of Psylloidea (Hemiptera) in the families Psyllidae and Triozidae, Folia Biol. (Krakow), 55, 27–34. [DOI] [PubMed] [Google Scholar]
- 27. Galdeano D. M., Breton M.C., Lopes J.R.S., Falk B.W., Machado M.A.. 2017, Oral delivery of double-stranded RNAs induces mortality in nymphs and adults of the Asian citrus psyllid, Diaphorina citri,. PLoS One, 12, e0171847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bolger A.M., Lohse M., Usadel B.. 2014, Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 30, 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H. 2018, Minimap2: pairwise alignment for nucleotide sequences, Bioinformatics, 34, 3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li H., Handsaker B., Wysoker A., et al. ; 1000 Genome Project Data Processing Subgroup. 2009, The sequence alignment/map format and SAMtools, Bioinformatics, 25, 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng H., Concepcion G.T., Feng X., Zhang H., Li H.. 2021, Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm, Nat. Methods, 18, 170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li H., Durbin R.. 2009, Fast and accurate short read alignment with Burrows-Wheeler transform, Bioinforma, 25, 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Durand N.C., Shamim M.S., Machol I., et al. 2016, Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments, Cell Syst., 3, 95–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dudchenko O., Batra S.S., Omer A.D., et al. 2017, De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds, Science, 356, 92–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Robinson J.T., Turner D., Durand N.C., Thorvaldsdóttir H., Mesirov J.P., Aiden E.L.. 2018, Juicebox.js provides a cloud-based visualization system for Hi-C data, Cell Syst., 6, 256–8.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gurevich A., Saveliev V., Vyahhi N., Tesler G.. 2013, QUAST: quality assessment tool for genome assemblies, Bioinformatics, 29, 1072–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M.. 2015, BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs, Bioinformatics, 31, 3210–2. [DOI] [PubMed] [Google Scholar]
- 38. Yu Y., Ouyang Y., Yao W.. 2018, shinyCircos: an R/Shiny application for interactive creation of Circos plot, Bioinformatics, 34, 1229–31. [DOI] [PubMed] [Google Scholar]
- 39. Johnston J.S., Bernardini A., Hjelmen C.E.. 2019, Genome size estimation and quantitative cytogenetics in insects. In: Brown S. J., Pfrender M. E., (eds.), Insect Genomics. Springer: New York, NY, pp. 15–26. [DOI] [PubMed] [Google Scholar]
- 40. Flynn J.M., Hubley R., Goubert C., et al. 2020, RepeatModeler2 for automated genomic discovery of transposable element families, Proc. Natl. Acad. Sci. USA, 117, 9451–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen N. 2004, Using RepeatMasker to identify repetitive elements in genomic sequences, Curr. Protoc. Bioinforma, 5, 4–10. [DOI] [PubMed] [Google Scholar]
- 42. Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L.. 2019, Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype, Nat. Biotechnol., 37, 907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kriventseva E.V., Kuznetsov D., Tegenfeldt F., et al. 2019, OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs, Nucleic Acids Res., 47, D807–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barnett D.W., Garrison E.K., Quinlan A.R., Stromberg M.P., Marth G.T.. 2011, BamTools: a C++ API and toolkit for analyzing and managing BAM files, Bioinformatics, 27, 1691–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brůna T., Lomsadze A., Borodovsky M.. 2020, GeneMark-EP+: eukaryotic gene prediction with self-training in the space of genes and proteins, NAR Genomics Bioinform., 2, lqaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brůna T., Hoff K.J., Lomsadze A., Stanke M., Borodovsky M.. 2021, BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database, NAR Genomics Bioinform., 3, lqaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Buchfink B., Xie C., Huson D.H.. 2015, Fast and sensitive protein alignment using DIAMOND, Nat. Methods., 12, 59–60. [DOI] [PubMed] [Google Scholar]
- 48. Gotoh O., Morita M., Nelson D.R.. 2014, Assessment and refinement of eukaryotic gene structure prediction with gene-structure-aware multiple protein sequence alignment, BMC Bioinformatics, 15, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hoff K.J., Lange S., Lomsadze A., Borodovsky M., Stanke M.. 2016, BRAKER1: unsupervised RNA-seq-based genome annotation with GeneMark-ET and AUGUSTUS: Table 1, Bioinformatics, 32, 767–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hoff K.J., Lomsadze A., Borodovsky M., Stanke M.. 2019, Whole-genome annotation with BRAKER. In: Kollmar M., (ed.), Gene Prediction. Springer: New York, NY, pp. 65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Iwata H., Gotoh O.. 2012, Benchmarking spliced alignment programs including Spaln2, an extended version of Spaln that incorporates additional species-specific features, Nucleic Acids Res., 40, e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lomsadze A., Ter-Hovhannisyan V., Chernoff Y.O., Borodovsky M.. 2005, Gene identification in novel eukaryotic genomes by self-training algorithm, Nucleic Acids Res., 33, 6494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stanke M., Schöffmann O., Morgenstern B., Waack S.. 2006, Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources, BMC Bioinformatics, 7, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stanke M., Diekhans M., Baertsch R., Haussler D.. 2008, Using native and syntenically mapped cDNA alignments to improve de novo gene finding, Bioinformatics, 24, 637–44. [DOI] [PubMed] [Google Scholar]
- 55. Lagesen K., Hallin P., Rødland E.A., Staerfeldt H.-H., Rognes T., Ussery D.W.. 2007, RNAmmer: consistent and rapid annotation of ribosomal RNA genes, Nucleic Acids Res., 35, 3100–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lowe T.M., Eddy S.R.. 1997, tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence, Nucleic Acids Res., 25, 955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kanehisa M., Goto S.. 2000, KEGG: Kyoto Encyclopedia of Genes and Genomes, Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hart A.J., Ginzburg S., Xu M.S., et al. 2020, EnTAP: bringing faster and smarter functional annotation to non-model eukaryotic transcriptomes, Mol. Ecol. Resour., 20, 591–604. [DOI] [PubMed] [Google Scholar]
- 59. Kanehisa M., Sato Y., Kawashima M.. 2022, KEGG mapping tools for uncovering hidden features in biological data, Protein Sci., 31, 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kanehisa M., Sato Y.. 2020, KEGG mapper for inferring cellular functions from protein sequences, Protein Sci., 29, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xu L., Dong Z., Fang L., et al. 2019, OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species, Nucleic Acids Res., 47, W52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Heller D., Vingron M.. 2021, SVIM-asm: structural variant detection from haploid and diploid genome assemblies, Bioinformatics, 36, 5519–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Quinlan A.R., Hall I.M.. 2010, BEDTools: a flexible suite of utilities for comparing genomic features, Bioinformatics, 26, 841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pedersen B.S., Quinlan A.R.. 2018, Mosdepth: quick coverage calculation for genomes and exomes, Bioinformatics, 34, 867–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nigg J.C., Kuo Y.-W., Falk B.W.. 2020, Endogenous viral element-derived piwi-interacting RNAs (piRNAs) are not required for production of ping-pong-dependent piRNAs from Diaphorina citri densovirus, mBio, 11, e02209–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rosenkranz D., Zischler H.. 2012, proTRAC - a software for probabilistic piRNA cluster detection, visualization and analysis, BMC Bioinformatics, 13, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. ter Horst A.M., Nigg J.C., Dekker F.M., Falk B.W.. 2019, Endogenous viral elements are widespread in arthropod genomes and commonly give rise to piwi-interacting RNAs, J. Virol., 93, e02124–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Edgar R.C. 2010, Search and clustering orders of magnitude faster than BLAST, Bioinformatics, 26, 2460–1. [DOI] [PubMed] [Google Scholar]
- 69. Li D., Luo R., Liu C.-M., et al. 2016, MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices, Methods, 102, 3–11. [DOI] [PubMed] [Google Scholar]
- 70. Allio R., Schomaker‐Bastos A., Romiguier J., Prosdocimi F., Nabholz B., Delsuc F.. 2020, MitoFinder: efficient automated large‐scale extraction of mitogenomic data in target enrichment phylogenomics, Mol. Ecol. Resour., 20, 892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wu F., Kumagai L., Cen Y., et al. 2017, Analyses of mitogenome sequences revealed that Asian citrus psyllids (Diaphorina citri) from California were related to those from Florida, Sci. Rep., 7, 10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jühling F., Pütz J., Bernt M., et al. 2012, Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements, Nucleic Acids Res., 40, 2833–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Thompson J.D., Higgins D.G., Gibson T.J.. 1994, CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice, Nucleic Acids Res., 22, 4673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kumar S., Stecher G., Li M., Knyaz C., Tamura K.. 2018, MEGA X: molecular evolutionary genetics analysis across computing platforms, Mol. Biol. Evol., 35, 1547–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J.C., et al. 2017, DnaSP 6: DNA sequence polymorphism analysis of large data sets, Mol. Biol. Evol., 34, 3299–302. [DOI] [PubMed] [Google Scholar]
- 76. Nei M. 1987, Molecular Evolutionary Genetics. Columbia University Press: New York, NY. [Google Scholar]
- 77. Strobeck C. 1987, Average number of nucleotide differences in a sample from a single subpopulation: a test for population subdivision, Genetics, 117, 149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fu Y.X., Li W.H.. 1993, Statistical tests of neutrality of mutations, Genetics, 133, 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Leigh J.W., Bryant D.. 2015, popart: full-feature software for haplotype network construction, Methods Ecol. Evol., 6, 1110–6. [Google Scholar]
- 80. Clement M., Posada D., Crandall K.A.. 2000, TCS: a computer program to estimate gene genealogies, Mol. Ecol., 9, 1657–9. [DOI] [PubMed] [Google Scholar]
- 81. Li Y., Zhang B., Moran N.A.. 2020, The aphid X chromosome is a dangerous place for functionally important genes: diverse evolution of hemipteran genomes based on chromosome-level assemblies, Mol. Biol. Evol., 37, 2357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ferrarezi R.S., Vincent C.I., Urbaneja A., Machado M.A.. 2020, Editorial: unravelling citrus huanglongbing disease, Front. Plant Sci., 11, 609655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Guan D., McCarthy S.A., Wood J., Howe K., Wang Y., Durbin R.. 2020, Identifying and removing haplotypic duplication in primary genome assemblies, Bioinformatics, 36, 2896–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pryszcz L.P., Gabaldón T.. 2016, Redundans: an assembly pipeline for highly heterozygous genomes, Nucleic Acids Res., 44, e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Driscoll T.P., Verhoeve V.I., Gillespie J.J., et al. 2020, A chromosome-level assembly of the cat flea genome uncovers rampant gene duplication and genome size plasticity, BMC Biol., 18, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nouri S., Salem N., Nigg J.C., Falk B.W.. 2015, Diverse array of new viral sequences identified in worldwide populations of the Asian citrus psyllid (Diaphorina citri) using viral metagenomics, J. Virol., 90, 2434–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Matsumura E.E., Nerva L., Nigg J.C., Falk B.W., Nouri S.. 2016, Complete genome sequence of the largest known flavi-like virus, Diaphorina citri flavi-like virus, a novel virus of the Asian citrus psyllid, Diaphorina citri, Genome Announc., 4, e00946-16. https://doi.org/10.1128/genomeA.00946-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Brennecke J., Aravin A.A., Stark A., et al. 2007, Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila, Cell, 128, 1089–103. [DOI] [PubMed] [Google Scholar]
- 89. Rahman R., Chirn G., Kanodia A., et al. 2015, Unique transposon landscapes are pervasive across Drosophila melanogaster genomes, Nucleic Acids Res., 43, 10655–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Song J., Liu J., Schnakenberg S.L., Ha H., Xing J., Chen K.C.. 2014, Variation in piRNA and transposable element content in strains of Drosophila melanogaster, Genome Biol. Evol., 6, 2786–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wierzbicki F., Kofler R., Signor S.. 2021, Evolutionary dynamics of piRNA clusters in Drosophila, Mol. Ecol., 1–17. 10.1111/mec.16311 [DOI] [PubMed] [Google Scholar]
- 92. Boykin L.M., Barro P.D., Hall D.G., et al. 2012, Overview of worldwide diversity of Diaphorina citri Kuwayama mitochondrial cytochrome oxidase 1 haplotypes: two Old World lineages and a New World invasion, Bull. Entomol. Res., 102, 573–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lashkari M., Manzari S., Sahragard A., Malagnini V., Boykin L.M., Hosseini R.. 2014, Global genetic variation in the Asian citrus psyllid, Diaphorina citri (Hemiptera: Liviidae) and the endosymbiont Wolbachia: links between Iran and the USA detected, Pest Manag. Sci., 70, 1033–40. [DOI] [PubMed] [Google Scholar]
- 94. de León J.H., Sétamou M., Gastaminza G.A., et al. 2011, Two separate introductions of Asian Citrus Psyllid Populations Found in the American Continents, Ann. Entomol. Soc. Am., 104, 1392–8. [Google Scholar]
- 95. Luo Y., Agnarsson I.. 2018, Global mtDNA genetic structure and hypothesized invasion history of a major pest of citrus, Diaphorina citri (Hemiptera: Liviidae), Ecol. Evol., 8, 257–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nigg J.C., Falk B.W.. 2020, Diaphorina citri densovirus is a persistently infecting virus with a hybrid genome organization and unique transcription strategy, J. Gen. Virol., 101, 226–39. [DOI] [PubMed] [Google Scholar]
- 97. Britt K., Gebben S., Levy A., Al Rwahnih M., Batuman O.. 2020, The detection and surveillance of Asian citrus psyllid (Diaphorina citri)—associated viruses in Florida citrus groves, Front. Plant Sci., 10, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Grant W., Bowen B.. 1998, Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation, J. Hered., 89, 415–26. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing datasets have been deposited in the NCBI Sequence Read Archive (BioProject PRJNA800468). Genome assemblies and protein-coding genes have been deposited in DDBJ/ENA/GenBank under BioProject PRJNA800468, with accession numbers JAKMOW000000000, JAKMOV000000000, and JAKMOU000000000. The versions described in this article are versions JAKMOW010000000, JAKMOV010000000, and JAKMOU010000000. Mitogenomes have been deposited in GenBank, accession numbers OM181945-OM181947. All genome annotation files and the custom viral protein database have been deposited in Dryad (https://datadryad.org/, doi:10.25338/B8MW7P). The custom EVE Python scripts produced for this study are available on GitHub (https://github.com/Mrbland/Dcitri_genome_EVE_scripts_2021).