Abstract
The eutF locus of Salmonella typhimurium LT2 was identified as a locus necessary for the utilization of ethanolamine as a sole carbon source. Initial models suggested that EutF was involved in either ethanolamine transport or was a transcriptional regulator of an ethanolamine transporter. Phenotypic characterization of eutF mutants suggested EutF was somehow involved in 1,2-propanediol, propionate, and succinate utilization. Here we provide evidence that two alleles defining the eutF locus, Δ903 and eutF1115, are partial-loss-of-function tonB alleles. Both mutations were complemented by plasmids containing a wild-type allele of the Escherichia coli tonB gene. Immunoblot analysis using TonB monoclonal antibodies detected a TonB fusion protein in strains carrying eutF alleles. Molecular analysis of the Δ903 allele identified a deletion that resulted in the fusion of the 3′ end of tonB with the 3′ end of trpA. In-frame translation of the tonB-trpA fusion resulted in the final 9 amino acids of TonB being replaced by a 45-amino-acid addition. We isolated a derivative of a strain carrying allele Δ903 that regained the ability to grow on ethanolamine as a carbon and energy source. The molecular characterization of the mutation that corrected the Eut− phenotype caused by allele Δ903 showed that the new mutation was a deletion of two nucleotides at the tonB-trpA fusion site. This deletion resulted in a frameshift that replaced the 45-amino-acid addition with a 5-amino-acid addition. This change resulted in a TonB protein with sufficient activity to restore growth on ethanolamine and eut operon expression to nearly wild-type levels. It was concluded that the observed EutF phenotypes were due to the partial loss of TonB function, which is proposed to result in reduced cobalamin and ferric siderophore transport in an aerobic environment; thus, the eutF locus does not exist.
Salmonella typhimurium and Escherichia coli can use the nonfermentable amino alcohol ethanolamine as the sole carbon and/or nitrogen source (8, 21). The initial step in the catabolism of ethanolamine involves the cleavage of ethanolamine into acetaldehyde and ammonia by the adenosylcobalamin (AdoCbl)-dependent enzyme ethanolamine ammonia-lyase (5, 7, 8). In addition to the requirement of AdoCbl for the enzymatic degradation of ethanolamine, work with S. typhimurium has shown that AdoCbl is also required for the induction of the genetically defined eut operon (35, 36, 45). This operon encodes proteins involved in ethanolamine catabolism in this bacterium and E. coli (5, 6, 37, 48). The requirement of AdoCbl for both ethanolamine catabolism and eut operon expression presents a challenge to these organisms growing aerobically, since S. typhimurium can synthesize AdoCbl de novo only under anaerobic conditions and E. coli is unable to synthesize the complete coenzyme de novo (20, 24). Both organisms meet this challenge by using transport systems to acquire exogenous complete and incomplete corrinoids under aerobic conditions.
Transport of exogenous cobalamin (Cbl) and other corrinoids from the environment into the cytoplasm of S. typhimurium or E. coli requires two independently functioning transport systems; the first actively transports Cbl across the outer membrane, while the second transports Cbl across the cytoplasmic membrane (10). Transport across the outer membrane involves BtuB, a high-affinity outer membrane receptor for Cbl, and the TonB-dependent energy-transducing complex consisting of the cytoplasmic membrane proteins TonB, ExbB, ExbD, and other, yet to be identified proteins (4, 18, 32, 46). TonB is anchored in the cytoplasmic membrane and spans the periplasm to interact directly with a number of outer membrane receptors involved in Cbl or ferric siderophore transport (32). The TonB-dependent energy-transducing complex couples electrochemical potential from the cytoplasmic membrane to the active transport of Cbl and ferric siderophores across the outer membrane. In the absence of a functional transport system, aerobically growing cells become starved for iron and respond by hypersecreting siderophores in a futile attempt to access iron. More relevant to ethanolamine utilization, these cells cannot access exogenous Cbl unless Cbl is present in a concentration high enough to overcome the transport defect (4, 34). Transport across the cytoplasmic membrane is carried out by the ABC transport system of BtuB, BtuC, and BtuD and functions independently of the TonB-dependent system (10).
eutF mutants were originally identified by the inability to grow on ethanolamine as a sole source of carbon, and EutF was proposed to play a role in ethanolamine transport or regulation of an ethanolamine transporter (28). Since then, we have also observed other phenotypes associated with eutF mutations which included the inability to grow on 1,2-propanediol as a sole carbon source and reduced growth rates on the nonfermentable carbon sources propionate and succinate (30). Here we present evidence that these phenotypes are the result of partial-loss-of-function tonB alleles and are not due to a new gene locus.
MATERIALS AND METHODS
Bacteria, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Carbon source utilization was tested on no-carbon medium E (NCE) (9, 49) supplemented with 1 mM MgSO4, 0.3 mM methionine, 0.1 mM tryptophan, 15 nM cyanocobalamin (CNCbl), and, as a carbon source, 30 mM ethanolamine, 1,2-propanediol, propionate, succinate, or glycerol. Concentrations were the same regardless of whether growth was tested on solid or liquid medium. The final concentrations of antibiotics in complex medium were 20 (tetracycline), 50 (kanamycin), 20 (chloramphenicol), and 100 (ampicillin) μg/ml. In NCE medium, the final concentrations used were 10, 100, 10, and 50 μg/ml, respectively. Growth of cultures at 37°C was monitored with a Spectronic 20D spectrophotometer (Milton Roy Co., Rochester, N.Y.) at 650 nm. CAS medium was a gift from Michelle R. Rondon. All experiments were done under aerobic conditions.
TABLE 1.
S. typhimurium LT2 strains and plasmids used
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| S. typhimurium strains | ||
| TR6583a | metE205 ara-9 | K. Sanderson via J. R. Roth |
| Derivatives of TR6583 | ||
| JE1291 | trpC3484::Tn10d(Tc)b | 28 |
| JE1418 | Δ903 | 28 |
| JE1684 | eutE18::Mud1-8 | Lab collection |
| JE1685 | eutE18::Mud1-8 Δ903 | 28 |
| JE1690 | tonB251 derivative of JE1291 | 28 |
| JE2123 | Δ1235 zde-6396::Tn10d(Tc) Δ903 | |
| JE4163 | trpC3480::Tn10d (Cmr) Δ903 | |
| JE4386 | Δ1235 | |
| JE4387 | eutE18::Mud1-8 Δ1235 | |
| Plasmids | ||
| pRZ526 | tonB+ (E. coli tonB+) (Kmr) | 33 |
| pRZ531 | tonB+ (E. coli tonB+) (Kmr) | 33 |
| pKP292 | tetp/o-tonB-phoA (E. coli tonB+) (Apr) | 38 |
| pBST324 | Tet repressor for regulation of tonB from pKP292 (Kmr) | 38 |
| pTONB1 | tonB+ (E. coli tonB+) (Cmr) | |
| pYCIBC | tonB (E. coli yciB+ yciC+) (Cmr) |
Formerly strain SA2979.
Abbreviation for Tn10Δ16Δ17.
Genetic techniques. (i) Transductions.
All transductional crosses were performed with mutant phage P22 HT105 int-201 (42, 43), and all phage manipulations were performed as previously described (9).
(ii) Isolation of allele Δ1235.
The derivative of strain JE1418 capable of growing on ethanolamine as a carbon and energy source was isolated by plating JE1418 on NCE minimal medium supplemented with ethanolamine as a carbon source. Portions of 10 independent cultures (∼109 cells) were plated individually. After incubation for 3 days, we recovered a single colony with a reversion rate estimated to be 10−9. The mutant strain JE4386 was reconstructed by growing phage P22 on the revertant, using the phage lysate as the donor to transduce strain JE1418 to Eut+, and then further characterized. A Tn10Δ 16Δ17 element (50) [referred to as Tn10d(Tc)] located near Δ1235 was isolated by genetic means as described elsewhere (12). This transposon [zde-6396::Tn10d(Tc)] was ca. 30% cotransducible with Δ1235 by phage P22.
Recombinant DNA techniques. (i) Plasmid constructions.
To make pTONB1, pRZ526 was digested to completion with HpaI and the ∼4.9-kb fragment containing the original insert was gel purified by the QIAquick gel extraction protocol as instructed by the manufacturer (Qiagen Inc., Valencia, Calif.). This fragment was digested to completion with BglII; the ca. 3,100-bp fragment containing tonB, yciC, yciB, and yciA was gel purified by QIAquick gel extraction and cloned into the BamHI/HincII site of vector pSU19, resulting in plasmid pTONB1.
To make pYCIBC, pTONB1 was digested to completion with SnaB1 and SmaI. Digested DNA was religated, and resulting plasmids were screened for the loss of the SnaB1/SmaI fragment containing tonB and yciA. The resulting plasmid, pYCIBC, contained yciB and yciC cloned in the direction of the lac promoter of pSU19.
(ii) PCR amplification of tonB from TR6583 and JE1291.
tonB was PCR amplified from chromosomal DNA of boiled whole cells of TR6583 and JE1291. An 860-bp amplified product was generated by using the following primers in a standard PCR mixture: 5′tonB (5′ TTCAGCTCTGGTTTTTCA 3′, corresponding to bases 82 to 99 of the published tonB sequence [16]) and 3′tonB (5′ TCCGACGGTAAACCTCGC 3′; corresponding to bases 941 to 924 of the published tonB sequence [16]). The amplification profile was as follows: 94°C for 5 min; 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min; 72°C for 10 min; 4°C for 12 h. All primers used in this study were obtained from Integrated DNA Technology, Inc. (Coralville, Iowa).
(iii) PCR amplification of tonB from strain JE1418 (Δ903).
tonB was amplified from strain JE1418 chromosomal DNA by using boiled whole cells as the template. The universal Tn10 primer (2) was used in conjunction with the 5′tonB primer described above to amplify the DNA between trpC3480::Tn10d(Cmr) to the 5′ end of tonB. The amplification profile was as follows: 94°C for 5 min; 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 3 min; 72°C for 10 min; 4°C for 12 h.
(iv) PCR amplification of tonB from strain JE2123 (Δ1235).
tonB was amplified from JE2123 chromosomal DNA by using boiled whole cells as the template. The trpA primer 5′ AGCTTAAAGAGTACCATGCC 3′ (corresponding to bp 665 to 685 of the trpA sequence [26]) was used in amplifications with the 5′tonB primer described above to amplify from the trpA coding region, across the deletion, to the 5′ end of tonB. The amplification protocol was the same as for the universal Tn10 and 5′tonB primers discussed above.
(v) DNA sequencing of PCR products.
All PCR products used for sequencing were purified by using a QIAquick gel extraction kit as instructed by the manufacturer. All sequencing was done by nonradioactive sequencing at the Nucleic Acid and Protein Facility at the University of Wisconsin Biotechnology Center. Primers used for sequencing include those described above in addition to the primer tonB DEL (5′ GCATCGGCGACCAGCAAG 3′, corresponding to bases 538 to 555 of the S. typhimurium tonB sequence [16]).
Biochemical procedures. (i) β-Galactosidase assays.
β-Galactosidase activity assays were performed by a modification of the method of Miller (25) as described elsewhere (14).
(ii) Immunoblot analysis of TonB.
Immunoblot analysis of TonB was done as previously described (23), with minor modifications. Briefly, cells were grown in NCE medium supplemented with glycerol (30 mM), MgSO4 (1 mM), methionine (0.3 mM), and tryptophan 0.1 mM, in the presence or absence of FeSO4 (45 μM). Cells were grown to A650 of 0.5 in 5 ml of NCE medium. A 1-ml sample of culture was removed, diluted with 0.5 ml of 15% trichloroacetic acid, and incubated on ice for 30 min. The acid-precipitated material was pelleted, washed once with 1.0 M Tris-Cl (pH 8.0) at 25°C, resuspended in 50 μl of 2× sample buffer, and boiled for 5 min. Samples were then resolved by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis (SDS-PAGE) (22) and transferred to an Immobilon-P membrane (Millipore, Bedford, Mass.) by means of a Mini Trans-Blot apparatus (Bio-Rad, Richmond, Calif.) at 100 V for 1 h according to the manufacturer’s specifications. Immunoblot analyses were performed according to the Phototope-horseradish peroxidase Western blot detection kit (New England Biolabs, Inc., Beverly, Mass.) protocol, with minor modifications. Membranes were blocked at 25°C overnight, and 1× Tris-buffered saline–0.1% Tween 20–5% dry milk was used for all incubations. Mouse monoclonal antibody 4H4 (a gift from Kathleen Postle) was used at a 1:5,000 dilution. The secondary antibody was horseradish peroxidase-conjugated donkey anti-mouse immunoglobulin G (a gift from Heidi Goodrich-Blair) diluted 1:20,000. X-ray film (XAR-50; Kodak) was used to detect signal.
RESULTS
eutF mutants were identified by the inability to grow on ethanolamine as source of carbon (28). Two of the mutants, one putative point mutant (strain JE1690) and a deletion mutant (strain JE1418), are characterized in this study.
The strain carrying allele Δ903 (JE1418) was reported to be able to use ethanolamine as a sole nitrogen source but not as a sole carbon source, suggesting the eutF locus was only partially affected by this mutation. Characterization of strain JE1418 revealed that the eutF locus mapped between the trp operon and tonB. Strain JE1418 was found to be a tryptophan auxotroph, thus defining one end of the deletion somewhere within the trp operon. The other end of the deletion was determined based on the following tests for TonB protein function. First, strain JE1418 was sensitive to infection by bacteriophage ES18, which requires a functional TonB protein for infection (47). Second, strain JE1418 could use CNCbl or AdoCbl for the synthesis of methionine at a concentration of 15 nM. tonB mutants require micromolar concentrations of Cbl to overcome the transport defect (4). The putative point mutant showed TonB function as defined by these two criteria (data not shown). Further phenotypic analysis of the eutF locus determined that eutF mutants were unable to grow on 1,2-propanediol as the sole carbon source and had reduced growth rates on the nonfermentable carbon sources propionate and succinate (30).
Complementation of eutF mutants.
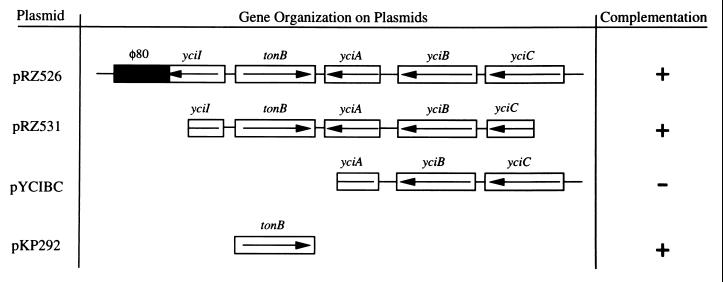
Attempts to isolate a complementing clone from an S. typhimurium library were unsuccessful. However, complementation of the EutF phenotypes was achieved with a plasmid carrying an ∼4,900-bp fragment containing the E. coli tonB locus and surrounding loci. This plasmid, pRZ526, also contained a fragment of bacteriophage φ80 DNA (33). Plasmid pRZ526 complemented strains JE1418 and JE1690 for growth on ethanolamine, suggesting that pRZ526 contained the eutF locus (Fig. 1). This finding suggested that the eutF locus was complemented by one (or more) of five possible E. coli genes or possibly by a φ80 gene. To narrow the possibilities, complementation was tested with plasmid pRZ531 (Fig. 1). pRZ531 complemented both JE1418 and JE1690, suggesting eutF was not yciA, yciC, or φ80 DNA. Previous minicell analysis of protein expression from pRZ531 determined that only two proteins were synthesized from this cloned region of E. coli DNA (33). The apparent molecular weights of the products, based on SDS-PAGE analysis, suggested these proteins were TonB and YciB. To address whether yciB was eutF, yciB and yciC were subcloned from pRZ526 resulting in plasmid pYCIBC (Fig. 1). Plasmid pYCIBC failed to complement strain JE1418 or JE1690, suggesting that yciB was not eutF. These results supported the previous finding that yciC was not involved in the complementation of EutF phenotypes.
FIG. 1.
Gene organization of plasmids tested for complementation. All represented reading frames are based on the E. coli sequence (6). Complementation is scored as growth (+) or no growth (−) on NCE plates supplemented with ethanolamine (30 mM), Met (0.3 mM), Trp (0.1 mM), MgSO4 (1 mM), and CNCbl (15 nM).
These results led to the hypothesis that complementation of the eutF phenotype by pRZ526 and pRZ531 was due to tonB. To test this hypothesis, a plasmid containing only tonB (pKP292) was introduced in strains JE1418 and JE1690. Plasmid pKP292 complemented all eutF phenotypes described above (Fig. 1). Complementation of the growth defects on ethanolamine, 1,2-propanediol, propionate, and succinate could be due either to complementation of tonB mutations in strains JE1418 and JE1690 or to multicopy suppression of a mutation that affects TonB function.
eutF alleles result in a TonB phenotype.
To show that strains JE1418 and JE1690 contained lesions that affected tonB function, we tested both strains for the hypersecretion of siderophores. Disruption of TonB function causes a hypersecretion of siderophores due to iron starvation, and the presence of the secreted siderophores can be indirectly detected by the use of CAS medium (44). Both strains (JE1418 and JE1690) grown on CAS medium resulted in large orange-yellow zones around the colonies, diagnostic of siderophore hypersecretion. tonB+ control strains TR6583 and JE1291 showed no significant zones (data not shown).
Taken together, the complementation of EutF phenotypes by wild-type tonB+ and siderophore hypersecretion by strains JE1418 and JE1690 strongly suggested eutF and tonB were allelic.
Altered TonB proteins in eutF mutants.
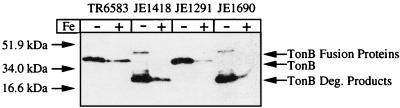
To analyze directly whether TonB was altered in eutF mutants, immunoblotting using an anti-TonB monoclonal antibody was performed to detect chromosomally expressed TonB protein in wild-type and mutant strains (Fig. 2).
FIG. 2.
Immunoblot analysis of TonB protein in strains TR6583 (tonB+), JE1418 (Δ903), JE1291 (tonB+), and JE1690 (eutF1115). The amount of material loaded in each lane was the equivalent of 0.1 A650 of whole cells. Proteins were separated by SDS-PAGE. Samples were grown in the presence or absence of 45 μM FeSO4. Deg., degradation.
The monoclonal antibody used in these studies, 4H4, was specific for the region near amino acids 60 to 103 of the E. coli TonB (23). This antibody cross-reacts with the S. typhimurium TonB, and it is assumed the same epitope of TonB is recognized. In the two wild-type tonB+ strains TR6583 and JE1291, an approximately 40-kDa protein, corresponding to the electrophoretic behavior of TonB in an SDS-PAGE system (16, 23), was detected. The two mutant strains tested, however, also contained an aberrant TonB protein of approximately 45 kDa in addition to a series of bands with molecular masses of ca. 25 kDa (Fig. 2). These bands were not present in the tonB+ control lanes, which suggested that strains JE1418 and JE1690 synthesized a TonB fusion protein that was unstable, with the 25-kDa protein being a stable degradation product of the 45-kDa fusion protein.
These results suggested that the deletion present in strain JE1418 generated a TonB fusion protein. Interestingly, strain JE1690, which was isolated during hydroxylamine mutagenesis experiments aimed at isolating point mutations in eutF, also contained a TonB fusion protein. The molecular characterization of both mutants is discussed below.
Iron-dependent regulation of tonB expression in eutF mutants.
TonB protein levels in S. typhimurium and E. coli are reduced in the presence of high concentrations of iron due to Fur regulation of tonB expression (31, 51). To ensure that iron regulation of tonB was still functional, we grew strains JE1418 and JE1690, along with the wild-type controls, in the presence or absence of 45 μM FeSO4 and detected TonB by immunoblotting as discussed above. Since we routinely use NCE medium without iron supplementation, all strains were expected to express TonB at some increased level to scavenge residual iron in the medium, as seen in studies of tonB regulation in E. coli (51). Upon addition of FeSO4 to the medium, and assuming that the promoter region of tonB is intact, there should be a reduction in the level of TonB. All strains showed iron regulation of TonB levels (Fig. 2), consistent with the promoter region of tonB being unaffected in both eutF mutants.
The analysis of our data was initially complicated by the confusion in the literature regarding the orientation of the tonB gene in S. typhimurium. The S. typhimurium genetic maps published after 1988 all show tonB expression in the same direction as the trp operon (39, 40). However, the original publication discussing the orientation of tonB (19) and the S. typhimurium genetic map published in 1988 (41) showed tonB and the trp operon as convergently transcribed. The results shown here are consistent with the proposal for convergent transcription (discussed further below). Therefore, the TonB fusion protein detected in JE1418 was concluded to be the result of a fusion to the carboxy terminus of TonB.
Physical characterization of tonB in strains JE1418 and JE1690.
To show that tonB was eutF, we attempted amplification and sequencing of chromosomal tonB alleles in strains JE1418 and 1690. Primers flanking tonB allowed amplification and sequencing of the complete coding region of tonB. For both of the tonB+ strains TR6583 and JE1291, an 860-bp product was amplified as expected from the published sequence (16) (data not shown). The product from TR6583 was sequenced, and the results were consistent with the amplified product being tonB (data not shown). In a control experiment, the cobU gene of S. typhimurium was amplified in a parallel reaction and always resulted in successful amplification from both wild-type and mutant DNAs (data not shown).
(i) DNA sequence analysis of the tonB allele in strain JE1418.
Reactions using DNA from strain JE1418 failed to produce an amplified tonB product. These results supported the hypothesis that tonB was affected by the Δ903 allele in this strain. To characterize the Δ903 allele in strain JE1418, the transposition-deficient element trpC3480::Tn10d(Cm) (>90% cotransducible with allele Δ903) was recombined into the chromosome of JE1418, with retention of the Δ903 mutation (strain JE4163). The DNA between the insertion and the 5′ end of tonB was amplified by using a primer specific for the end of the insertion and the 5′tonB primer. The amplification product (ca. 3,000 bp) was sequenced by using the transposon primer. DNA sequence information determined the trpC sequence on one side of the PCR product, while the 5′tonB primer determined the tonB sequence as expected (data not shown). Primer walking from the tonB side of the PCR product identified the deletion site of strain JE4163 (Fig. 3A). Sequence data demonstrated that the fusion of tonB with trpA resulted in the removal of the last nine codons of tonB. In-frame translation across the tonB-trpA junction predicted the removal of the final 9 amino acids of TonB and the addition of 45 amino acids after amino acid 231 of wild-type TonB (Fig. 3B). These data confirmed that tonB was disrupted in strains carrying the Δ903 allele and that the orientation of tonB transcription was toward the trp operon as originally proposed (19).
FIG. 3.
Partial nucleotide and predicted amino acid sequences of tonB genes from TR6583, JE4163, and JE2123. (A) Nucleotide sequence flanking the tonB-trpA fusion site. The TR6583 sequence corresponds to bases 790 to 837 of wild-type tonB (16). Boldface bases correspond to bases from trpA; boldface underlined bases correspond to bases deleted in a strain carrying allele Δ1235. (B) Predicted amino acid sequence of TonB. The starting proline residue corresponds to amino acid 221 of the published sequence (16). Boldface residues are predicted amino acids from in-frame translation of tonB from the Δ903 allele; boldface underlined residues are predicted amino acids from in-frame translation of the Δ1235 allele. DEL903 and DEL1235 are referred to as Δ903 and Δ1235, respectively, throughout the text.
Interestingly, Anton and Heller previously constructed C-terminally altered TonB proteins from E. coli in which the final 8 amino acids of TonB were replaced with either 19 or 2 amino acids (3). In that study, the TonB fusion proteins were also rapidly degraded to an approximately 29-kDa degradation product likely similar to that seen in this study (Fig. 2). Surprisingly, the TonB fusions in the previous study did not affect TonB function, based on φ80 infectivity or growth response to ferrichrome or vitamin B12 (3). Therefore, we believe that the phenotype of JE1418 is not due to the rapid degradation of the TonB fusion protein but instead is due to the addition of 45 amino acids to the C-terminus causing a partial-loss-of-function TonB fusion protein.
(ii) DNA sequence analysis of the tonB allele in strain JE1690.
Surprisingly, tonB could not be amplified from the putative point mutant, strain JE1690, which suggested that the inability to amplify tonB from JE1690 was probably due to the deletion of one of the tonB primer sites. We hypothesized that the 3′ end of tonB in this strain was affected since the 5′tonB primer was designed to hybridize immediately downstream of the −10 region, and hence a deletion that removed this site would abolish tonB expression.
Results from Western blot analysis discussed above confirmed that tonB expression was regulated by iron (Fig. 2). The inability to amplify tonB from strain JE1690 was an unexpected result since strain JE1690 was isolated during an experiment aimed at isolating hydroxylamine-generated point mutants (28). Although unusual, it is not unprecedented to generate deletions with this procedure (27). Regardless of how this deletion arose, PCR amplification and immunoblot analysis clearly showed that strain JE1690 carried a deletion that affected the tonB gene. It is also clear that the lesion in strain JE1690 is not the same as the Δ903 allele in JE1418, since the two strains are phenotypically distinct (28).
Isolation and characterization of a mutation that restores growth of strain JE1418 on ethanolamine.
Before determining that tonB was disrupted in strain JE1418, we isolated a derivative of it capable of growing on ethanolamine as a sole carbon source. This revertant strain displayed eut operon expression to ca. 60% of the level measured in a wild-type strain in the presence of ethanolamine and Cbl (Table 2). A Tn10d(Tc) insertion linked to the mutation causing this reversion was isolated and found to be ca. 30% cotransducible with the Δ903 allele by phage P22. This result raised the possibility that the mutation causing the reversion was in tonB.
TABLE 2.
Effects of allele Δ1235 on eut operon expression and growth on ethanolamine
| Relevant genotype | Addition | Sp acta (U/A650) | Growth on EAb |
|---|---|---|---|
| tonB+ | None | 4 | NTc |
| EA/Cbld | 189 | + | |
| Δ903 | None | 3 | NT |
| EA/Cbl | 16 | − | |
| Δ1235 | None | 6 | NT |
| EA/Cbl | 113 | + |
Specific activity of β-galactosidase from the lacZ gene fused to the eut operon.
Growth on ethanolamine (EA) as a sole carbon source in a derivative of the strain listed with a wild-type eut operon. The medium used was NCE supplemented with tryptophan and MgCl2.
NT, not tested.
EA/Cbl, ethanolamine (20 mM) plus Cbl (15 nM).
We used PCR amplification and sequencing to characterize the reversion mutation. A trpA primer was designed and used in combination with the 5′tonB primer to amplify tonB from strain JE2123 (Δ1235). Analysis of DNA sequence data determined that the revertant strain carried a deletion of two bases at the junction site of tonB and trpA (Fig. 3A). This deletion (Δ1235) resulted in a frameshift in the coding region of the tonB allele generated by Δ903. The net result was a replacement of the 45-amino-acid addition seen in strain with the Δ903 allele by 5 amino acids in strain JE2123 (Fig. 3B). The restored ability of strain JE2123 to grow on ethanolamine, and the near-wild-type eut operon expression observed in this strain clearly showed that the observed EutF phenotypes were due to mutations in tonB and not to the deletion of an unidentified gene.
DISCUSSION
Data presented herein show that the mutations initially defining the eutF locus are partial-loss-of-function tonB alleles. This conclusion is based on the following findings: (i) mutations in the eutF locus can be complemented by a plasmid containing only the wild-type allele of the tonB gene from E. coli; (ii) eutF mutant strains hypersecrete siderophores, consistent with lesions in tonB; (iii) immunoblot analyses of TonB from both mutant strains suggest the synthesis of an unstable TonB fusion protein; (iv) sequencing of tonB from a eutF mutant, JE1418 (Δ903), showed that the mutation resulted in the replacement of the final 9 amino acids of TonB with 45 amino acids from a tonB-trpA gene fusion; and (v) a 2-bp deletion (Δ1235) in the coding region of the tonB-trpA gene fusion of strain JE1418 removed the bulky addition to the carboxy terminus of TonB, resulting in a partially functional TonB protein.
Why were eutF mutants not identified as mutants partially defective in tonB function?
The initial characterization of eutF mutants originally eliminated tonB from consideration for two reasons. First, eutF strains were still sensitive to infection by bacteriophage ES18, which requires a functional tonB locus for infection (47). Second, both strains were able to grow in the presence of CNCbl or AdoCbl at a concentration of 15 nM. tonB mutants require micromolar concentrations of Cbl to overcome the TonB transport mutation (4). We now believe that the original phenotypic screen for eutF alleles led to the isolation of partial-loss-of-function tonB alleles. Strain JE1418 was isolated in a screen for deletions of cobA, a gene encoding the adenosyltransferase enzyme involved in AdoCbl biosynthesis. cobA mutants do not grow on ethanolamine as a sole carbon source because (i) ethanolamine ammonia-lyase requires AdoCbl as a coenzyme and (ii) cobA mutants cannot synthesize AdoCbl (15). Strain JE1418 did not grow on ethanolamine but was found to be wild type for cobA, which suggested that an alternative locus was responsible for the phenotype; this locus was named eutF (28).
The strain used in our laboratory to study cobalamin biosynthesis is TR6583, which contains a metE205 mutation that disrupts the Cbl-independent methionine synthase gene. Strain TR6583, therefore, is dependent on exogenous methionine or must use the Cbl-dependent methionine synthase, which requires the addition of exogenous Cbl in an aerobic environment. All strains in this and the original eutF study are TR6583 derivatives, and exogenous Cbl was added in all media for complementation of the metE205 allele. In the phenotypic screen that isolated strain JE1418, a positive control was used to screen out unwanted auxotrophs. This positive control was growth on minimal medium containing glucose as a carbon source, plus 15 nM Cbl to complement the metE205 mutation. Therefore, the screen required a functional Cbl transport system for growth on the positive control plate. This growth requirement would therefore eliminate all tonB mutations that completely abolished TonB function. Unexpectedly, however, this would allow partial-loss-of-function tonB mutants to grow as long as enough Cbl was transported for the methionine requirements. The amount of Cbl needed to meet the methionine requirement is approximately 25-fold less than the amount required for growth on ethanolamine (13). Therefore, it was possible to isolate tonB mutants that meet the Cbl requirement for methionine synthesis but do not meet the increased demand for Cbl for growth on ethanolamine. The sensitivity to ES18 infection would still occur because of the much higher sensitivity of phage infection as a TonB function assay (1). This would explain the isolation of strain JE1418, and we believe that a similar type of selection was the reason for the isolation of strain JE1690.
Explaining EutF phenotypes in terms of partially functional TonB proteins.
Based on the conclusion that eutF mutants were actually partial-loss-of-function tonB alleles, the various carbon source utilization phenotypes can be explained in the following way. As described above, ethanolamine utilization requires Cbl for breakdown of ethanolamine and the induction of the genetically defined eut operon (35, 36, 45). Recent molecular characterization of a portion of the putative eut operon identified a proposed ethanolamine permease (48). Based on the findings presented here, the expression of this permease is likely to be dependent on the genetically defined regulator eutR (35). Therefore, a strain unable to access exogenous Cbl would not fully express the ethanolamine permease and would exhibit decreased ethanolamine transport. The combination of reduced Cbl acquisition and decreased ethanolamine transport would result in decreased expression of eutR-controlled genes and reduced growth rates on ethanolamine. This phenotype is exactly what was observed in eutF mutants (28).
The first enzyme involved in 1,2-propanediol degradation, diol dehydratase, also requires AdoCbl as a coenzyme. Aerobically growing S. typhimurium, therefore, requires exogenous Cbl for growth on 1,2-propanediol. Partial-loss-of-function tonB alleles would be expected to have growth defects on 1,2-propanediol, again as seen for the eutF alleles.
The final two phenotypes, reduced growth on the nonfermentable carbon sources succinate and propionate, are more difficult to explain. Growth on succinate or propionate does not require Cbl; however, mutations in fur have been shown to affect growth on succinate as a sole carbon source (17). Fur is a regulatory protein involved in controlling expression of a wide number of genes involved in iron acquisition (11). It is assumed that tonB mutations will decrease iron levels in the cell and lead to phenotypes similar to that of a fur mutant. The observed phenotypes may be caused by disruption of the Fur regulon, or, alternatively, growth on nonfermentable carbon sources may be more sensitive to iron levels in the cell due to the requirement of iron for oxidative phosphorylation.
ACKNOWLEDGMENTS
This work was supported by NIH grant RO1-GM40313 to J.C.E.-S. and by the College of Agricultural and Life Sciences of the University of Wisconsin—Madison. We thank K. Postle for anti-TonB monoclonal antibodies and plasmids, H. Goodrich-Blair for the secondary antibody, and M. R. Rondon for CAS medium. We thank the anonymous reviewers for their constructive and insightful criticism of this work.
REFERENCES
- 1.Ahmer B M, Thomas M G, Larson R A, Postle K. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J Bacteriol. 1995;177:4742–4747. doi: 10.1128/jb.177.16.4742-4747.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman E, Roth J R, Hessel A, Sanderson K E. Transposons currently in use in genetic analysis of Salmonella species. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: American Society for Microbiology; 1996. pp. 2613–2626. [Google Scholar]
- 3.Anton M, Heller K J. Functional analysis of a C-terminally altered TonB protein of Escherichia coli. Gene. 1991;105:23–29. doi: 10.1016/0378-1119(91)90509-a. [DOI] [PubMed] [Google Scholar]
- 4.Bassford P J, Jr, Kadner R J. Genetic analysis of components involved in vitamin B12 uptake in Escherichia coli. J Bacteriol. 1977;132:796–805. doi: 10.1128/jb.132.3.796-805.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackwell C M, Turner J M. Microbial metabolism of amino alcohols: formation of coenzyme B12-dependent ethanolamine ammonia-lyase and its concerted induction in Escherichia coli. Biochem J. 1978;176:751–757. doi: 10.1042/bj1760751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.Bradbeer C. The clostridial fermentation of choline and ethanolamine. II. Requirement for a cobamide coenzyme by an ethanolamine deaminase. J Biol Chem. 1965;240:4675–4681. [PubMed] [Google Scholar]
- 8.Chang G W, Chang J T. Evidence for the B12-dependent enzyme ethanolamine deaminase in Salmonella. Nature. 1975;254:150–151. doi: 10.1038/254150a0. [DOI] [PubMed] [Google Scholar]
- 9.Davis R W, Botstein D, Roth J R. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 10.DeVeaux L C, Clevenson D S, Bradbeer C, Kadner R J. Identification of the BtuCED polypeptides and evidence for their role in vitamin B12 transport in Escherichia coli. J Bacteriol. 1986;167:920–927. doi: 10.1128/jb.167.3.920-927.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Earhart C F. Uptake and metabolism of iron and molybdenum. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 1075–1090. [Google Scholar]
- 12.Elliott T, Roth J R. Characterization of Tn10d-Cam: a transposition-defective Tn10 specifying chloramphenicol resistance. Mol Gen Genet. 1988;213:332–338. doi: 10.1007/BF00339599. [DOI] [PubMed] [Google Scholar]
- 13.Escalante-Semerena, J. C. Unpublished results.
- 14.Escalante-Semerena J C, Roth J R. Regulation of cobalamin biosynthetic operons in Salmonella typhimurium. J Bacteriol. 1987;169:2251–2258. doi: 10.1128/jb.169.5.2251-2258.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escalante-Semerena J C, Suh S-J, Roth J R. cobA function is required for both de novo cobalamin biosynthesis and assimilation of exogenous corrinoids in Salmonella typhimurium. J Bacteriol. 1990;172:273–280. doi: 10.1128/jb.172.1.273-280.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hannavy K G, Barr C, Dorman C J, Adamson J, Mazengera L R, Gallagher M P, Evans J S, Levine B A, Trayer I P, Higgins C F. TonB protein of Salmonella typhimurium: a model for signal transduction between membranes. J Mol Biol. 1990;216:987–910. doi: 10.1016/S0022-2836(99)80009-6. [DOI] [PubMed] [Google Scholar]
- 17.Hantke K. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 18.Heller K, Kadner R J. Nucleotide sequence of the gene for the vitamin B12 receptor protein in the outer membrane of Escherichia coli. J Bacteriol. 1985;161:904–908. doi: 10.1128/jb.161.3.904-908.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiles I D, Powell L M, Higgins C F. Peptide transport in Salmonella typhimurium: molecular cloning and characterization of the oligopeptide permease genes. Mol Gen Genet. 1987;206:101–109. doi: 10.1007/BF00326543. [DOI] [PubMed] [Google Scholar]
- 20.Jeter R M, Olivera B M, Roth J R. Salmonella typhimurium synthesizes cobalamin (vitamin B12) de novo under anaerobic growth conditions. J Bacteriol. 1984;159:206–213. doi: 10.1128/jb.159.1.206-213.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones P W, Turner J M. A model for the common control of enzymes of ethanolamine catabolism in Escherichia coli. J Gen Microbiol. 1984;130:849–860. doi: 10.1099/00221287-130-4-849. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage and structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Larsen R A, Myers P S, Skare J T, Seachord C L, Darveau R P, Postle K. Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J Bacteriol. 1996;178:1363–1373. doi: 10.1128/jb.178.5.1363-1373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence J, Roth J R. The cobalamin (coenzyme B12) biosynthetic genes of Escherichia coli. J Bacteriol. 1995;177:6371–6380. doi: 10.1128/jb.177.22.6371-6380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 26.Nichols B P, Yanofsky C. Nucleotide sequences of trpA of Salmonella typhimurium and Escherichia coli: an evolutionary comparison. Proc Natl Acad Sci USA. 1979;76:5244–5248. doi: 10.1073/pnas.76.10.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Toole G A, Escalante-Semerena J C. Ph.D. thesis. Madison, Wis: University of Wisconsin—Madison; 1994. [Google Scholar]
- 28.O’Toole G A, Escalante-Semerena J C. Identification and initial characterization of the eutF locus of Salmonella typhimurium. J Bacteriol. 1991;173:5168–5172. doi: 10.1128/jb.173.16.5168-5172.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Toole G A, Rondon M R, Trzebiatowski J R, Suh S-J, Escalante-Semerena J C. Biosynthesis and utilization of adenosyl-cobalamin (coenzyme B12) In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 710–720. [Google Scholar]
- 30.O’Toole, G. A., M. G. Thomas, and J. C. Escalante-Semerena. 1996. Unpublished results.
- 31.Postle K. Aerobic regulation of the Escherichia coli tonB gene by changes in iron availability and the fur locus. J Bacteriol. 1990;172:2287–2293. doi: 10.1128/jb.172.5.2287-2293.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Postle K. TonB protein and energy transduction between membranes. J Bioenerg Biomembr. 1993;25:591–601. doi: 10.1007/BF00770246. [DOI] [PubMed] [Google Scholar]
- 33.Postle K, Reznikoff W. Identification of the Escherichia coli tonB gene product in minicells containing tonB hybrid plasmids. J Mol Biol. 1979;131:619–636. doi: 10.1016/0022-2836(79)90011-1. [DOI] [PubMed] [Google Scholar]
- 34.Rioux C R, Friedrich M J, Kadner R J. Genes of the 90-kilobase plasmid of Salmonella typhimurium confer low-affinity cobalamin transport: relationship to fimbria biosynthesis genes. J Bacteriol. 1990;172:6217–6222. doi: 10.1128/jb.172.11.6217-6222.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roof D M, Roth J R. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J Bacteriol. 1992;174:6634–6643. doi: 10.1128/jb.174.20.6634-6643.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roof D M, Roth J R. Ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1988;170:3855–3863. doi: 10.1128/jb.170.9.3855-3863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roof D M, Roth J R. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1989;171:3316–3323. doi: 10.1128/jb.171.6.3316-3323.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roof S K, Allard J D, Bertrand K P, Postle K. Analysis of Escherichia coli TonB membrane topology by use of PhoA fusions. J Bacteriol. 1991;173:5554–5557. doi: 10.1128/jb.173.17.5554-5557.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanderson K E, Hessel A, Liu S-H, Rudd K E. The genetic map of Salmonella typhimurium, edition VIII. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: American Society for Microbiology; 1996. pp. 1903–1999. [Google Scholar]
- 40.Sanderson K E, Hessel A, Rudd K E. Genetic map of Salmonella typhimurium, edition VIII. Microbiol Rev. 1995;59:241–303. doi: 10.1128/mr.59.2.241-303.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanderson K E, Roth J R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988;52:485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmieger H. A method for detection of phage mutants with altered transduction ability. Mol Gen Genet. 1971;100:378–381. doi: 10.1007/BF00438281. [DOI] [PubMed] [Google Scholar]
- 43.Schmieger H, Bakhaus H. The origin of DNA in transducing particles of P22 mutants with increased transduction frequencies (HT-mutants) Mol Gen Genet. 1973;120:181–190. doi: 10.1007/BF00267246. [DOI] [PubMed] [Google Scholar]
- 44.Schwyn B, Neilands J B. Universal assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 45.Sheppard D E, Roth J R. A rationale for autoinduction of a transcriptional activator: ethanolamine ammonia-lyase (EutBC) and the operon activator (EutR) compete for adenosyl-cobalamin in Salmonella typhimurium. J Bacteriol. 1994;176:1287–1296. doi: 10.1128/jb.176.5.1287-1296.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skare J T, Ahmer B M M, Seachord C L, Darveau R P, Postle K. Energy transduction between membranes. TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J Biol Chem. 1993;268:16302–16308. [PubMed] [Google Scholar]
- 47.Stocker B A D, Nurminen M, Makela P H. Mutants defective in the 33K outer membrane protein of Salmonella typhimurium. J Bacteriol. 1979;139:376–383. doi: 10.1128/jb.139.2.376-383.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stojiljkovic I, Bäumler A J, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutj eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vogel H J, Bonner D M. Acetylornithase of Escherichia coli: partial purification, and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 50.Way J C, Davis M A, Morisato D, Roberts D E, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- 51.Young G M, Postle K. Repression of tonB transcription during anaerobic growth requires Fur binding at the promoter and a second factor binding upstream. Mol Microbiol. 1994;11:943–954. doi: 10.1111/j.1365-2958.1994.tb00373.x. [DOI] [PubMed] [Google Scholar]